Abstract
Repetitive satellite DNA (satDNA) sequences are abundant in eukaryote genomes, with a structural and functional role in centromeric function. We analyzed the nucleotide sequence and chromosomal location of the five known cattle (Bos taurus) satDNA families in seven species from the tribe Tragelaphini (Bovinae subfamily). One of the families (SAT1.723) was present at the chromosomes’ centromeres of the Tragelaphini species, as well in two more distantly related bovid species, Ovis aries and Capra hircus. Analysis of the interaction of SAT1.723 with centromeric proteins revealed that this satDNA sequence is involved in the centromeric activity in all the species analyzed and that it is preserved for at least 15–20 Myr across Bovidae species. The satDNA sequence similarity among the analyzed species reflected different stages of homogeneity/heterogeneity, revealing the evolutionary history of each satDNA family. The SAT1.723 monomer-flanking regions showed the presence of transposable elements, explaining the extensive shuffling of this satDNA between different genomic regions.
Keywords: satellite DNA, centromeric function, SAT1.723, Bovinae, Caprinae, Bovidae genomes
Introduction
Tandemly repeated or satellite DNA (satDNA) represents a major fraction of most eukaryotic genomes, as one of the classes of repetitive sequence (Charlesworth et al. 1994; Slamovits and Rossi 2002; Biscotti, Canapa, et al. 2015; Biscotti, Olmo, et al. 2015). Consisting of tandemly repeated DNA motifs, typically arranged in large arrays of hundreds or thousands of copies, satDNA is often (although not exclusively) located in blocks at the heterochromatic regions of chromosomes, at centromeres, near telomeres, or in interspersed locations (Vourc’h and Biamonti 2011; Plohl et al. 2012; Garrido-Ramos 2015).
Most eukaryotic species include multiple, unrelated, families of satDNA that differ in sequence, nucleotide composition, monomer unit length, abundance, and chromosomal location (Garrido-Ramos 2015; Rojo et al. 2015). Each satDNA family consists in a library of monomer variants that can be shared by related species. Expansions and/or elimination of different variants from this library may result in rapid changes in satDNA distribution and abundance profiles, even among closely related species (Kuhn et al. 2008; Plohl et al. 2012; Rojo et al. 2015). Consequent on the dynamic changes in satDNA during an evolutionary period, these sequences can be species or genus specific (Garrido-Ramos 2015). Nevertheless, some satDNA sequences seem to have been preserved over long evolutionary periods in some genomes, being considered as “frozen” satDNAs (Mravinac et al. 2002, 2005; Kuhn and Heslop-Harrison 2011; Biscotti, Canapa, et al. 2015; Petraccioli et al. 2015; Chaves et al. 2017). This long-term conservation of ancestral repeats can be explained by the influence of selective constraints imposed on functional motifs, or on structural features of satellites monomers possibly involved in any of the putative roles of satDNAs (Plohl et al. 2012; Rojo et al. 2015; Chaves et al. 2017), including heterochromatin formation and maintenance, chromosome pairing and segregation, chromatin elimination, chromosome rearrangements’ promoters, cell cycle control, or gene expression regulation (Slamovits and Rossi 2002; Plohl et al. 2008; Adega et al. 2009; Ugarkovic 2009; Pezer et al. 2012; Enukashvily and Ponomartsev 2013; Paço et al. 2013; Ferreira et al. 2015; Louzada et al. 2015). Evolutionary changes in satDNA can drive population and species divergence (Ugarković and Plohl 2002; Adega et al. 2009; López-Flores and Garrido-Ramos 2012; Paço et al. 2013; Feliciello et al. 2015; Vieira-da-Silva et al. 2015). We found that centromeric satDNA was involved in the mechanics of chromosomal fusion in Bovidae (Chaves et al. 2000, 2003, 2005; Adega et al. 2006, 2009; Di Meo et al. 2006; Kopecna et al. 2012, 2014; Nieddu et al. 2015). SatDNA sequences are also useful phylogenetically where changes in composition, organization, and/or physical location can allow inference of evolutionary relationships (Chaves et al. 2004, 2005; Adega et al. 2006, 2009; Kopecna et al. 2012, 2014).
The Bovidae is one of the most important mammalian families in the Cetartiodactyla order, comprising ∼140 species, whose evolutionary relationships are often obscure (MacEachern et al. 2009; Groves and Grubb 2011). Chromosome evolution studies use the domestic cattle (Bos taurus, 2n = 60) karyotype as a reference (Gallagher and Womack 1992). In the cattle genome, eight abundant centromeric satDNA families were initially described (Macaya et al. 1978), representing nearly 25% of all the DNA, with interrelated evolutionary histories (Macaya et al. 1978; Taparowsky and Gerbi 1982a, 1982b; Modi et al. 1996; Chaves et al. 2000, 2005). Some families are ancient and shared by descent in other bovid species (Modi et al. 1993, 1996, 2004; Chaves et al. 2000, 2004, 2005; Adega et al. 2006; Kopecna et al. 2012, 2014); the bovine SATI is present in all Pecoran genomes (Modi et al. 1993, 1996; Chaves et al. 2000, 2005), unlike families named SAT1.723, SATIV, SAT1.711a, and SAT1.711b that are not in all Pecora, although results are somewhat equivocal (Escudeiro et al. 2019).
Study of the structure and function of centromeric satDNAs (Schmidt and Heslop-Harrison 1998; Plohl et al. 2008; Adega et al. 2009; Giannuzzi et al. 2012; Cerutti et al. 2016) at the primary constriction (centromere) of mammalian chromosomes shows the sequences interacting with the centromere-specific histone H3 variant (CENP-A) (Aldrup-Macdonald and Sullivan 2014). The ability to bind CENP-A is considered a marker of active centromeres (Zhang et al. 2013; Plohl et al. 2014; Henikoff et al. 2015; Purgato et al. 2015; Steiner and Henikoff 2015; Cerutti et al. 2016; Talbert et al. 2018).
To understand the nature, conservation, evolution and functional role of cattle satDNA, we selected the most abundant families (SATI, SATIV, SAT1.723, SAT1.711a, and SAT1.711b) and studied these in seven species representative of the genera Tragelaphus and Taurotragus; all were medium- to large-bodied taxa distributed through the savannah and forested regions of sub-Saharan Africa (Groves and Grubb 2011). The orthologous sequences and the differences in sequence similarity, chromosome location, and distribution provide important information on the evolutionary history of these species since their divergence from a common ancestor. We also investigated the association with the CENP-A histone protein to test the involvement of specific sequences in centromeric function.
Materials and Methods
Cell Culture, Chromosome Preparation, and Genomic DNA Isolation
Our investigation included representatives of three bovid tribes (Hassanin and Douzery 1999): cattle (B.taurus, BTA, tribe Bovini), the spiral horned antelope species (tribe Tragelaphini), Tragelaphus angasii (TAN, Nyala), Tragelaphus imberbis (TIM, Lesser kudu), Tragelaphus scriptus (TSC, Bushbuck), Tragelaphus spekii (TSP, Sitatunga), Tragelaphus strepsiceros (TST, Greater kudu), Taurotragus derbianus (TDE, Derby Eland), and Taurotragus oryx (TOR, Common Eland). The third tribe, the Caprini, was represented by the sheep Ovis aries (OAR) and goat Capra hircus (CHI). Cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 13% AmnioMax C-100 Basal Medium, 2% AmnioMax C-100 supplement, 15% Fetal Bovine Serum (FBS), 100 U/ml and 100 μg/ml of penicillin/streptomycin antibiotic mixture, and 200-mM l-glutamine (all from Gibco, Thermo Fisher Scientific). Chromosome harvesting and metaphase preparations followed routine procedures. Genomic DNA isolation was performed using Quick-Gene DNA Tissue Kit S (Fujifilm Life Science) according to the manufacturer’s instructions.
SatDNA Isolation, Cloning, and Sequencing from Bovidae Species
We choose the most abundant cattle satDNA sequences to study in other Bovinae species. Five satDNA families (SATI, SATIV, SAT1.723, SAT1.711a, and SAT1.711b) of the domestic cattle genome and the orthologous satDNAs of seven species from the Tragelaphini were isolated in the current work by polymerase chain reaction (PCR) amplification of genomic DNA obtained from cryopreserved cells of these species using specific primers (supplementary table S1, Supplementary Material online) designed following Nijman and Lenstra (2001). Between 100 and 300 ng of genomic DNA from each species was used as template with an initial denaturing step at 94 °C for 10 min, followed by 30 cycles of 94 °C for 1 min (denaturation), 55/57/59 °C for 45 s (annealing), and 72 °C for 45 s (extension). The reaction terminates with a final extension at 72 °C for 10 min. The annealing temperature was optimized for each set of primers used: 55 °C for the SATI and SATIV primers, 57 °C for the SAT1.723 and SAT1.711b primers, and 59 °C for the SAT1.711a primers. PCR products were cloned and 20 clones of each SatDNA from each species were sequenced and submitted to GenBank with the references MK499473 to MK499615.
SatDNAs Clones Sequences Analysis
Sequence analysis was performed using BLAST in the NCBI database. Multiple alignments were obtained with the CLUSTALW cost matrix in Geneious R9 version 9.1.2 (Biomatters); parameters were set to default values. For the in silico analysis, we used the B. taurus satellite sequences with NCBI accession numbers: SATI, AJ293510.1; SATIV, AF446392.1; SAT1.711a, AF162491.1; SAT1.711b, AF162499.1; and SAT1.723, M36668.1. The Guanine-Cytosine (GC) content and distribution of the satDNAs monomers was calculated using the Biologicscorp facility (https://www.biologicscorp.com/tools/GCContent/; last accessed January 12, 2019). Bendability/curvature–propensity plots were determined using the bend.it server, applying the DNase I-based bendability parameters (Brukner et al. 1995) and the consensus bendability scale (Gabrielian et al. 1996). Monomer-flanking regions of the satDNAs sequences clones from each Tragelaphini species were screened for the presence of repetitive elements in the Eukaryota Repbase using the Censor software, and the repetitive elements found were mapped in the satDNAs clones sequences using Geneious program. The presence of the mammalian CENP-B box motif (17 bp, YTTCGTTGGAAACGGGA) in these satDNA sequences was also investigated using Geneious tools.
Fluorescent In Situ Hybridization with SatDNA Clones
Physical mapping of Bovidae SatDNA sequences (SATI, SATIV, SAT1.723, SAT1.711a, and SAT1.711b) onto the chromosomes used standard in situ hybridization methods (Schwarzacher and Heslop-Harrison 2000). Metaphases from BTA and the Tragelaphini species were hybridized with cloned sequences isolated from the same species except OAR and CHI were hybridized in situ with SAT1.723 clone isolated from BTA. The sequences corresponding to the SatDNAs analyzed were labeled with digoxigenin-11-dUTP or biotin-16-dUTP (both from Roche Biochemical reagents, Sigma-Aldrich) by PCR. The most stringent posthybridization wash was 50% formamide/2× saline sodium citrate (SSC) at 42 °C. Digoxigenin-labeled probes were detected with antidigoxigenin-5′-TAMRA (Roche Biochemical reagents, Sigma-Aldrich) and biotin-labeled probes were detected with Fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories). Preparations were mounted with Vectashield containing 4′-6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) to counterstain chromosomes.
CENP-A Immunolocalization and FISH
Immunostaining on metaphase chromosomes from BTA, the Tragelaphini species, CHI, and OAR was performed as described by Piras et al. (2010) with slight modifications. Cells were incubated overnight with 40 ng/ml Colcemid (Gibco, Thermo Fisher Scientific). The cells were harvested, washed once with phosphate-buffered saline, and resuspended in 0.075-M KCl for 20 min at 37 °C following which 200 µl of cell suspension was cyto-spun (Hettich rotofix 32A Benchtop) onto slides at 1,400 rpm for 10 min. Slides were incubated in KCM (120-mM KCl, 20-mM NaCl, 10-mM Tris–HCl, 0.5-mM Disodium ethylenediaminetetraacetate dihydrate (Na2EDTA), 0.1% [v/v] Triton X-100) for 10 min at room temperature. A crosslinking treatment was performed with Ultraviolet radiation (UVs) radiant exposure of 150 mJ/cm3. The primary antibody, mouse anti-human centromere protein A (CENP-A) monoclonal antibody (ab13939, Abcam) was added at a concentration of 1:100 and the slides were incubated at 37 °C for 1 h. Slides were then washed in KCM for 10 min at room temperature. The secondary antibody, anti-mouse monoclonal FITC (81-6511, Zymed) was added at a concentration of 1:200 and the slides incubated for a further hour at 37 °C. Following another wash in KCM, slides were fixed in 4% formaldehyde for 10 min at room temperature and washed again in KCM. Chromosomes were mounted with coverslips and counterstained with DAPI (Vector Laboratories). For combined immunofluorescence (IF)/FISH, the slides were washed in 4× SSC, 0.05% Tween 20 at room temperature for 4 h with agitation and equilibrated in 50% formamide/2× SSC for 7 days at 4 °C. Colocalization analysis was performed with AutoQuant X3 software (Media Cybernetics) using Pearson’s Correlation and Manders’ Overlap Coefficients.
Image Capture and Processing
FISH images were observed using a Zeiss ImagerZ microscope coupled to an Axiocam digital camera using AxioVision software (version Rel. 4.5, Zeiss). Digitized photos were prepared for printing in Adobe Photoshop (version 7.0).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed for all the species using the Pierce Agarose ChIP Kit (Thermo Scientific), following the manufacturer’s recommendations. Immunoprecipitation was carried out using 2 µg of mouse anti-human CENP-A monoclonal antibody (ab13939, Abcam) and normal rabbit Immunoglobulin G (IgG) to control nonspecific binding. One-tenth of starting material was reserved as input DNA control. DNA immunoprecipitated (IP) and input samples were analyzed by a PCR amplification with specific primers for the several satDNA sequences (supplementary table S1, Supplementary Material online). The input/IP ratio was quantified using Image J software.
Statistical Analysis
GraphPad Prism 6 (version 6.01) was used in the statistical analysis. The Pearson’s correlation and Manders’ overlap test was performed to determine the presence of colocalization between CENP-A antibody signals and the studied satDNA sequences in each species’ centromeres. As the samples did not present a Gaussian distribution, the values were transformed with the log function in order to normalize the values distribution.
Results
Bos taurus satDNA Families and Their Chromosomal Locations in the Bovidae
In the present study, five previously characterized satDNA families, SATI, SATIV, SAT1.723, SAT1.711a, and SAT1.711b (Macaya et al. 1978), were isolated by PCR (fig. 1a; supplementary table S1, Supplementary Material online), cloned, sequenced, and physically mapped by in situ hybridization. The five satDNA sequences showed that all bovine satDNAs analyzed display a pericentromeric to centromeric location in BTA cattle autosomes (fig. 1b). The orthologous SATI, SATIV, SAT1.723, SAT1.711a, and SAT1.711b sequences were amplified from seven species of the Tragelaphini using the same PCR primers (fig. 1a and supplementary table S1, Supplementary Material online). The amplicons were subsequently cloned and sequenced. All showed high sequence similarity to the corresponding BTA satellites. The satellite clones were mapped by in situ hybridization to the chromosomes of the seven Tragelaphini species (fig. 1c–i). All species shared the (peri)centromeric location of SATI and SAT1.723 families with BTA chromosomes. SATI was found in all the chromosomes from T. angasii (2n = 55/56) (fig. 1c), T. strepsiceros (2n = 31/32) (fig. 1g), T.derbianus (2n = 31/32) (fig. 1h), and T. oryx (2n = 31/32) (fig. 1i), and it was also found in some of the chromosomes from T. spekii (2n = 30) (fig. 1f), a species with a large block of SATI in a submetacentric chromosome (fig. 1f). SAT1.723 was found at the (peri)centromeric regions of all the chromosomes in T. angasii, Tragelaphusimberbis (2n = 38) (fig. 1d), T. spekii, T. strepsiceros, T.derbianus, and T. oryx. In Tragelaphusscriptus (2n = 33/34) (fig. 1e), the SAT1.723 was present only in about half of the chromosomes. In contrast to the cattle genome, where no satellite signals were detected on either sex chromosomes, the other seven Tragelaphini species carried SATI and SAT1.723 sequences on the X chromosome. Two species of Taurotragus similarly presented SATIV at the (peri)centromeric regions (fig. 1h and i). In T. derbianus and T. oryx, SATIV orthologous sequences were on more than half of the chromosomes. After finding SAT1.723 in all seven Tragelaphini species, we tested its presence in Capra hircus (CHI) and Ovis aries (OAR) (fig. 1j and l). SAT1.723 present in these distant species, exhibiting also a (peri)centromeric location.
Fig. 1.
—SatDNA isolation and mapping onto Bos taurus (BTA), Tragelaphini, Capra hircus (CHI), and Ovis aries (OAR) chromosomes. (a) satDNAs amplicons obtained by PCR from the genomic DNA of the species analyzed. PCR amplicons were SATI, 400 bp; SATIV, 604 bp; SAT1.723, 680 bp; SAT1.711a, 400 bp; and SAT1.711b, 975 bp. SATI amplicons from the Tragelaphini genomes are 500 bp long. SAT1.723 amplicons from TAN, TIM, TDE, and TOR are ∼600 bp in length, and those of the SAT1.723 amplicons from TSC, TSP, and TST are ∼750 bp. TOR and TDE SATIV sequences revealed an amplicon size of 700 bp. (b–l) Physical mapping of the satDNAs present at pericentromeric and centromeric regions by in situ hybridization (red or green) in the respective species chromosomes (blue, DAPI). The name and color of each probe were indicated within each metaphase. Scale bar represents 10 μm.
SAT Variation across Species
An analysis of the intrinsic features of the nucleotide sequences of the satDNA families isolated in each species was performed. The GC distribution in SAT1.723, SATI, and SATIV monomers showed substantial differences (fig. 2a–c): SAT1.723 from 57% to 67%; SATI from 51% to 54%, and SatIV from 40% to 45%. In fact, the GC distribution is significantly higher and more constant across SAT1.723 monomer length (fig. 2a), in comparison with the other two satDNAs (fig. 2b and c). Moreover, the GC periodicity across the SAT1.723 monomer seems to be ∼10 bp, which is in agreement with the nucleosomal organization (Kogan and Trifonov 2005; Kaplan et al. 2009; Zhang et al. 2013).
Fig. 2.
—Intrinsic features of satDNAs monomers. GC content distribution across SAT1.723 (a), SATI (b), and SATIV (c) monomers. Curvature/bendability propensity plots of SAT1.723 (d), SATI (e), and SATIV (f) monomers. The sequences used in these analyses were M36668, AJ293510, and AF446392.
Differences were also detected in the curvature–propensity and bendability of SAT1.723, SATI, and SATIV across the monomers (fig. 2d–f). SAT1.723 monomer presents the higher values of bendability (fig. 2d), whereas those of SATIV were lower (fig. 2f). Examination of the curvature–propensity plot calculated with DNase I-based trinucleotide parameters, reveals only one peak of a potential curvature around the 120-bp position in SAT1723 monomer (fig. 2d). In SATI (fig. 2e) and SATIV (fig. 2f) monomers, at least two peaks with similar curvature–propensity were detected.
A pairwise alignment of the cloned satDNA sequences isolated from each Tragelaphini species was performed with the related BTA sequences deposited in GenBank (fig. 3a–c and supplementary figs. S1–S3, Supplementary Material online). SATI is conserved among the species analyzed, revealing low intra- and inter-sequence variability overall (fig. 3a; light and medium blue coloring showing sequence similarity >70%; supplementary table S2, Supplementary Material online). SAT1.723 sequences show higher discrepancies in similarity values when comparing the clones from all the species (fig. 3b; dark blue, dark green and yellow showing a sequence similarity range of 100–40%; supplementary table S2, Supplementary Material online). This color palette shows that this sequence is much more conserved in some of the species. SATIV monomers (fig. 3c) revealed to be highly different between TDE and TOR (39–20% similarity; supplementary table S2, Supplementary Material online). Finally, SATIV from BTA is more similar to the monomer from TDE than from the one of TOR.
Fig. 3.
—Orthologous bovine satDNA sequence identity. Distance matrix of pairwise alignments of SATI (a), SAT1.723 (b), and SATIV (c) clones from BTA and the Tragelaphini species analyzed in the present study. Cells showing nucleotide identities of 90–100% are in dark blue; 80–89%, medium blue; 70–79%, light blue; 60–69%, dark green; 50–59%, light green; 40–49%, yellow; 30–39%, orange; and <30% in red. The multiple alignment of all the clones is shown in supplementary figures S1–S3, Supplementary Material online.
SAT1.723 Is Associated with CENP-A Protein across the Bovidae Family
We used IF with an anti-CENP-A antibody combined with in situ hybridization using probes for each isolated satDNA family to characterize the association of sequences and CENP-A in metaphase chromosomes (fig. 4a). SAT1.723 colocalized more closely with CENP-A antibody signals compared with the hybridization signals of the other satDNA families (supplementary fig. S4, Supplementary Material online). These results are similar both for BTA and Tragelaphini species’ genomes analyzed. The phylogenetically more distant species CHI and OAR genomes were also analyzed and revealed analogous results. Pearson’s correlation coefficient (fig. 4b) and Manders’ overlap coefficient (supplementary fig. S5, Supplementary Material online) confirmed the existence of a strong colocalization between SAT1.723 and CENP-A in the combined IF-FISH experiment.
Fig. 4.
—The centromeric function of SAT1.723 in BTA, Tragelaphini, CHI, and OAR genomes. (a) Representative images of IF with CENP-A antibody (green; DNA DAPI blue) followed by DNA-FISH with SAT1.723 (red) in BTA, Tragelaphini, CHI, and OAR species. A colocalization spot was amplified 300% (top, right). Scale bar represents 10 μm. (b) Graphic validation of the colocalization of the CENP-A antibody signals with the satDNA sequence signals in BTA, the Tragelaphini species, CHI, and OAR. Each colocalization spot in each cell was analyzed by Pearson’s correlation coefficient. A minimum of 15 spots per cell in at least 10 images of each species and satDNA FISH experiment were analyzed (a minimum of 150 spots per variable). As the samples did not present a Gaussian distribution, the values were transformed with the log function in order to normalize the values distribution. The correlogram was made with GraphPad Prism 6 (version 6.01). All values are expressed as mean ± SD (standard deviation). (c) Relative quantification of band intensity from ChIP sample analysis by PCR with specific primers for the satDNA sequences isolated in each species’ (peri)centromeric regions. This analysis was performed using the software Image J. The area of each band was determined, and the value of each IP sample was compared with the value of Input band. (d) In silico search for the CENP-B boxlike motif in the SAT1.723 monomer from BTA and the seven Tragelaphini species.
To further confirm the association of SAT1.723 with centromeric function, chromatin from all the species analyzed was immunoprecipitated with the anti-CENP-A antibody. The ChIP assay showed that the satDNA sequence was able to form DNA–protein complexes with CENP-A in living cells (Piras et al. 2010; Hayden and Willard 2012; Melters et al. 2013; Zhang et al. 2013; Henikoff et al. 2015; Cerutti et al. 2016; Khademi 2017; Talbert et al. 2018). The input DNA and the immunoprecipitated sample (IP CENP-A) from each species was analyzed by PCR using specific primers for each satDNA family in our study (supplementary fig. S6, Supplementary Material online). As shown in figure 4c, the ratio between IP CENP-A and input values for SAT1.723 ranged between 5.0 and 16.6, confirming that this satDNA is enriched in CENP-A bound chromatin for all taxa. On the contrary, no enrichment of the other satDNA families was observed, as reflected by the IP CENP-A/Input ratio values which ranged from 0.1 to 0.4.
In order to identify features of centromeric activity, an in silico search for the CENP-B boxlike motifs (a conserved short sequence acting as the binding site for CENP-B, which directly interacts with CENP-A to maintain kinetochore nucleation) (Dumont and Fachinetti 2017; Schalch and Steiner 2017) was also performed in SAT1.723 sequence monomers from BTA and the seven Tragelaphini species. In fact, this analysis revealed the presence of a CENP-B boxlike motif in the monomers of this satDNA family in all species examined (fig. 4d). The in silico search was performed allowing the occurrence of a maximum of four mismatches within the 17-bp CENP-B box motif used.
SAT1.723 Monomer-Flanking Regions Are Enriched in Transposable Elements
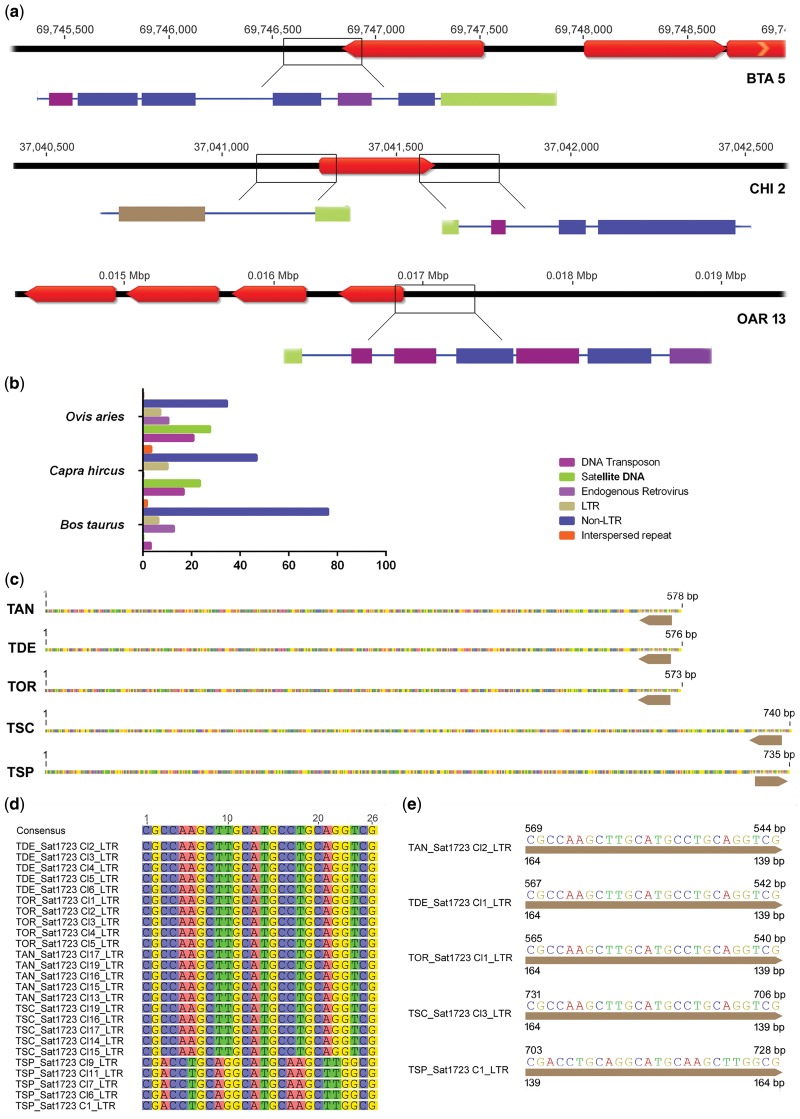
The conservation of SAT1.723 in the Bovidae family suggests an essential function, so a detailed analysis of the genomic context of ends of satellite sequences was made. An in silico analysis of SAT1.723 was performed on the assembled genomes of BTA (Btau_5.0.1 assembly, GenBank assembly accession GCA_000003205.6), CHI (ARS1 assembly, GenBank assembly accession: GCA_001704415.1), and OAR (Oar_V4.0, GenBank assembly accession: GCA_000298735.2) as there are no Tragelaphini genome sequence assemblies currently available. This analysis showed an interspersed presence of SAT1.723 in the three species’ genomes (∼300, 30, and 50 BLAST hits distributed on all the autosomes and on the X chromosome from BTA, CHI, and OAR, respectively) (supplementary tables S3–S5, Supplementary Material online). The SAT1.723 neighbor sequences revealed that the monomers from this satDNA family present in the genome assemblies are flanked by transposable elements, both in the isolated and clustered SAT1.723 BLAST hits (fig. 5a). The global annotation revealed that non-LTR sequences are the most represented TEs in all the SAT1.723 BLAST hits-flanking regions of all the chromosomes of the three species (fig. 5b). An additional BlastN search for SATI, SATIV, SAT1.711a, and SAT1.711b was also performed using BTA, CHI, and OAR sequencing data, and this revealed that these satDNAs present an interspersed distribution pattern in these genomes (supplementary tables S3–S5, Supplementary Material online), being similarly flanked by TEs (supplementary fig. S7, Supplementary Material online).
Fig. 5.
—In silico analysis of the flanking regions of SAT1.723 hits mapped in BTA, CHI, and OAR genomes and the Tragelaphini clones. (a) Representative image of a BTA, CHI, and OAR chromosome showing the annotation of SAT1.723 BLAST hits (in red) on the top line and the annotation of the repetitive sequences found in the flanking sequences of SAT1.723 on the bottom bar (Repbase). (b) Quantification of the different classes of repetitive sequences flanking the SAT1.723 monomers along the BTA, CHI, and OAR genomes. The flanking regions of all the BLAST hits of SAT1.723 mapped onto BTA, CHI, and OAR chromosomes were screened for the presence of repetitive elements in the Eukaryota Repbase using the Censor software. (c) Representative SAT1.723 clones from TAN, TDE, TOR, TSC, and TSP with the TCR1_LTR motif mapped in the monomer sequence. The SAT1.723 clones’ sequences from each species were screened for the presence of repetitive elements in the Eukaryota Repbase using the Censor software, and the TCR1_LTR elements found were mapped in clone sequences. (d) Multiple alignment of the TCR1_LTR motifs found in the SAT1.723 clones analyzed. (e) Location of the TCR1_LTR motif in the representative SAT1.723 monomers from TAN, TDE, TOR, TSC, and TSP.
The SAT1.723 monomer-flanking regions were also analyzed in the sequenced clones of the seven Tragelaphini species. Only the flanking regions of the SAT 1.723 repeats with centromeric location were analyzed, as these were the ones isolated by PCR due to their higher abundance. SAT1.723 clone sequences from each species were screened for the presence of repetitive elements in the Eukaryota Repbase using Censor software. This revealed that a sequence of 26 bp from a specific LTR, the TCR1_LTR was present in almost all SAT1.723 clones from TAN, TSC, TSP, TDE, and TOR (fig. 5c and d). The LTR sequence was found in the terminal region of SAT1.723 monomers and the 26 bp of the LTR corresponded to the last 26 bp of the complete sequence of TCR1_LTR (164 bp) (fig. 5e). No significant sequence similarity was found between the primers used for isolating SAT1.723 sequences with the sequence of TCR1_LTR.
The presence of the TCR1_LTR in the monomer-flanking regions of SAT 1.723 clone sequences in almost all Tragelaphini species (except for TIM and TST), may be due to the biased character of the PCR technique which may not have allowed the isolation of terminal monomers in these two genomes. No transposable elements were found in the monomers-flanking regions of the other satDNAs sequence clones (SATI and SATIV).
Discussion
SatDNA Families and Their Chromosomal Location
We present an analysis of the five most abundant satDNAs families in bovid species not restricted to Bos. Orthologous bovine satDNA sequences were isolated from seven Tragelaphini genomes, molecularly characterized and mapped (fig. 1a–i). SAT1.723 has not previously been analyzed outside BTA; the presence of bovine SATI (Kopecna et al. 2012; 2014) and SATIV (Adega et al. 2006) in species from the Tragelaphini has previously been reported. Nevertheless the SATI isolated from cattle and used for FISH analysis by Kopecna et al. (2012) produced only a weak hybridization signal on the Tragelaphini chromosomes, and later the same authors reported that SATI sequences isolated specifically from Tragelaphini genomes were present in all the acrocentric chromosomes of those species (Kopecna et al. 2014), but not the biarmed chromosomes in T. spekii and T. strepsiceros. This contrasts with the results obtained from the newly isolated sequences from these species (fig. 1f and g). Although of the same satDNA family, different sequence variants were probably isolated in each study, suggesting that SATI family is composed of subfamilies/variants of sequences that are not necessarily identical to each other: studies on the Bovinae initially suggested that SATIV was specific for Bovini genomes (Modi et al. 2004) but Adega et al. (2006) reported the presence of this sequence in Tragelaphini indicating that the ancestral sequence most likely predates the divergence of the Bovini and Tragelaphini. In addition to the expected presence of SATI (considered the oldest bovine satDNA and present in all pecorans), and the confirmation of a SATIV presence in Taurotragus, we found SAT1.723 sequences at the (peri)centromeric region in all the genomes exhibiting typical satDNA sequence features, that is, characterized by a tandemly repetitive pattern at the constitutive heterochromatin regions (fig. 1). This suggests that this satDNA family was likely preserved in other subfamilies of Bovidae (including Caprini, subfamily Caprinae, as well as Bovini and Tragelaphini, subfamily Bovinae). Modi et al. (1996) dated the origin of SATI family to 20–40 Ma, whereas Adega et al. (2006) reported the origin of SATIV at 10 Ma. We now propose that SAT1.723 family predated the separation of the Bovidae subfamilies Bovinae and Caprinae (Chaves et al. 2005) by at least 15–20 Ma.
SatDNA Evolution on Bovidae Reflects the Different Stages of the Library Model
The sequence similarity among the satDNAs isolated sequences from BTA (Bovini) and the Tragelaphini revealed significant differences in the homogeneity/heterogeneity of each satDNA family, probably reflecting different stages of in their evolution (fig. 6). The high sequence conservation among SATI clones (fig. 3a) suggested that this sequence is the oldest bovine satDNA (fig. 5) and is moving into the “homogenization” stage. The multiple alignment of SAT1.723 clone sequences (fig. 3b) showed that this family has not yet reached the homogenization stage, being much more similar between some species than in others. The high discrepancy in the similarity values when comparing the clones from all the species strongly suggests that this satDNA family is in a “degeneration” stage of the satDNA nucleotide sequence evolutionary process (fig. 6).
Fig. 6.
—Schematic representation of the different stages of satDNA evolutionary process, Library model. SATI, SAT1.723, and SATIV are represented in the scheme as satDNAs families in different stages of the evolutionary process. The origin of each family was inferred considering the presence/absence in Bovinae and Caprinae analyzed species.
The nucleotide sequence variability of SATIV monomers (fig. 3c), even between phylogenetically related species, reflects its dynamic evolution and mutation rate which generally characterizes the initial, “amplification,” stage of satDNA evolution (López-Flores and Garrido-Ramos 2012). Bovine SATIV is considered the evolutionarily youngest satDNA family in the Bovinae (Jobse et al. 1995; Modi et al. 1996, 2004; Adega et al. 2006), and being in the amplification/contraction stage has probably undergone independent amplification events from the other bovine satDNA sequences (Lenstra et al. 1993) (fig. 6).
Despite the overall level of homogeneity, we found some intra- and inter-specific variability in the clones of the three satDNAs (supplementary table S2, Supplementary Material online), a finding that is consistent with the existence of different satDNA sequence variants as allowed for in the library model (Fry and Salser 1977; Mestrović et al. 1998; Ugarković and Plohl 2002). Each satDNA family consists in a library of monomer variants shared by related species, and each species presents a specific repeats’ profile shaped by expansions and/or elimination of different variants from the library (Fry and Salser 1977; Mestrović et al. 1998; Ugarković and Plohl 2002). In agreement with this model, the three satDNA families analyzed presented distinct subfamilies differing by sequence length and composition (supplementary figs. S1–S3, Supplementary Material online). Moreover, different turnover rates of each satellite repeat, even among closely related species, can result in profound differences in overall sequence homogeneity. Genomic constraints such as karyotype architecture as well as the evolutionary age of a satDNA family may influence the turnover rates of satDNA sequences (Plohl et al. 2010; Paço et al. 2013; Louzada et al. 2015).
SAT1.723 Has a Centromeric Function in the Bovidae Family
The localization of SAT1.723 at the centromeres in all the species analyzed suggested an involvement in centromeric function. It is accepted that centromeres are defined by epigenetic factors and through interactions between centromeric satDNA sequences and proteins (Rocchi and Archidiacono 2006; Heslop-Harrison and Schwarzacher 2013; Plohl et al. 2014; Purgato et al. 2015). In the majority of eukaryotes, the centromere identity is defined epigenetically by the presence of the histone H3 variant centromere protein A, CENP-A in the centromeric nucleosomes (Plohl et al. 2014; Steiner and Henikoff 2015; McKinley and Cheeseman 2016; Talbert et al. 2018), and genetically by the presence of satDNA sequences containing CENP-B box motifs (Dumont and Fachinetti 2017). Our analysis of the interaction of SAT1.723 sequences with CENP-A (fig. 4a–c) and CENP-B (fig. 4d) shows centromeric activity of this satDNA in BTA (Bovini), the seven Tragelaphini species and the two Caprini species suggesting that this satDNA may have been retained (conserved) due to functional constrains in bovid evolution. The CENP-B boxes found in the SAT1.723 monomers were demonstrated to be functional as these monomers interact with CENP-A (see IF-FISH and ChIP experiments). Despite the existence of different satDNA families at the centromeric and pericentromeric regions of these species’ chromosomes, only SAT1.723 seems to be involved with centromeric function. Although the finding that only one specific satDNA family is capable of binding CENP-A has been described in other species (humans, Plohl et al. [2014] and horses, Cerutti et al. [2016]), ours is the first report of this functional satDNA sequence in the centromeres of bovids. Additionally, because SAT1.723 seems to be associated with centromere function, this satDNA is most probably located at the centromeric region (at least in some of the monomers). Thus, only a fraction of SAT1.723 monomers may be associated with CENP-A, similar to the alpha-satellite in the human genome, where only a few sequences are associated to CENP-A (Sullivan et al. 2011).
The mechanism responsible for the activity of the SAT1.723 centromeric sequence is unknown. In humans, alpha-satellite is the preferred component of the active centromeres (Aldrup-MacDonald et al. 2016). Recent studies on the architecture of centromeres have reported the presence of specific secondary structures such as DNA loops suggesting that active centromeric sequences were selected for their ability to form secondary structures, rather than for the nucleotide sequence itself (Aze et al. 2016; Kasinathan and Henikoff 2017). In fact, a bioinformatic analysis on the prediction of secondary structures showed that SAT1.723 can indeed form DNA loops and G-quadruplexes (data not shown; see Kejnovsky et al. [2015]). Moreover, the high GC content of SAT1.723 is in agreement with recent reports that GC richness is compatible with the centromeric function (Melters et al. 2013; Cerutti et al. 2016).
Models for predicting nucleosomes have been developed using DNA sequence properties, such as dinucleotide periodicity (Segal et al. 2006; Kaplan et al. 2009; Ioshikhes et al. 2011; Zhang et al. 2013) and curvature pattern (Liu et al. 2008; 2011). The observation of a 10-bp periodicity of GC dinucleotides (in agreement to the DNA helical repeat ∼10.4 bp) across the SAT1.723 monomer (fig. 2a) could be considered as a facilitator of DNA bendability and nucleosome formation (Kogan and Trifonov 2005; Kaplan et al. 2009; Ioshikhes et al. 2011). Moreover, the predicted curvature for the SAT1.723 monomer (fig. 2d) resembles the “curvature pattern” for the nucleosomal DNA helix proposed by Liu et al. (2008). In this satDNA monomer, the two ends have a large curvature, whereas the middle has a small curvature which provides powerful evidence for a periodicity characteristic of core DNA (Liu et al. 2008; 2011). The less variable and higher bendability values detected across the SAT1.723 monomer (fig. 2d), in contrast with the other two satDNAs (fig. 2e and f), suggest this monomer to comprise the more bendable and flexible sequence, a factor which could facilitate CENP-A nucleosomal organization (Heslop-Harrison and Schwarzacher 2013; Zhang et al. 2013; Steiner and Henikoff 2015). The differences in curvature/bendability patterns of the three satDNA sequences could potentially reflect different modes of nucleosomes packaging, suggesting a more relaxed conformation of the SAT1.723 monomer. We propose that the SAT1.723 sequence intrinsically favors the translational and rotational phasing of the CENP-A nucleosomes in bovids, similar to that proposed for CentO on rice by Zhang et al. (2013).
SAT1.723 Monomer-Flanking Regions
Current whole genome assemblies collapse most copies of the satellite sequences and do not show the long arrays present at the centromeres of chromosomes. However, assembly algorithms would be expected to assemble regions flanking satellite monomer fragments correctly, regardless of the ability to assemble either satellite arrays, or small contigs including satellite fragments into larger scaffolds. Thus, the analysis of satDNA monomer-flanking regions can provide important insights on the organization and mode of evolution of these sequences (Satović et al. 2016; Chaves et al. 2017). An analysis of the flanking regions of the SAT1.723 monomers identified the frequent presence of transposable elements with both non-LTR and LTR sequences (fig. 5). Moreover, the in silico mapping of the bovine satDNAs (supplementary tables S3–S5, Supplementary Material online) showed that these sequences are present not only at the (peri)centromeric regions, but they also occur in an interspersed fashion on the chromosomes of BTA, CHI and OAR (although in too low copy numbers and density to be detected by in situ hybridization). These findings point to an intense intragenomic reshuffling of satDNAs mediated by the TEs found in the satDNAs monomer flanks. Several studies have reported a physical association between satDNAs and TEs (Heslop-Harrison and Schwarzacher 2011; Louzada et al. 2015; Petraccioli et al. 2015; Chaves et al. 2017), suggesting a role for these elements in satDNA evolution probably by promoting their intragenomic movement and expansion in the genomes (López-Flores et al. 2004; Biscotti et al. 2008; Kuhn et al. 2008; Macas et al. 2009; Šatović and Plohl 2013; Scalvenzi and Pollet 2014; Petraccioli et al. 2015; Satović et al. 2016; Chaves et al. 2017). Our analysis reveals that the TE association seems to be the rule for all the bovine satDNAs analyzed, as all are embedded in TEs, with a particular emphasis for non-LTR elements (supplementary fig. S7, Supplementary Material online). The presence of the same class of TEs in the monomer-flanking regions of all the five satDNAs suggests that their movement may occur by the same transposition mechanism.
The presence of LTR sequences flanking the SAT1.723 monomers (fig. 5c) reinforces the centromeric activity of this sequence. It has been hypothesized that retrotransposons, particularly LTRs, may accumulate at active centromeres because of their favored integration into an epigenetically modified centromeric “environment” or, alternatively, due to the preferred association with CENP-A nucleosomes in both animals and plants (Wolfgruber et al. 2009; Plohl et al. 2014). Similarly, as reported for maize and wheat (both of which present species-specific centromeric retrotransposons), the TCR1_LTR sequence found in the SAT1.723 monomer-flanking regions could be a specific centromeric retrotransposon in several of the Tragelaphini species (fig. 5d and e). However, additional work is needed to disclose if there are any specific centromeric retrotransposon.
It is important to highlight that the different classes of TEs associated with the dispersed SAT1.723 hits mapped in the BTA, OAR, and CHI genomes (fig. 5a and b), and at the flanking regions of the SAT1.723 centromeric monomers in Tragelaphini (fig. 5c), could reflect their different chromosome locations. Alternatively, these differences could be due to the limited length of flanking sequences present in the SAT1.723 cloned monomers. Our data for transcriptional activity of SAT1.723 agree with reports of transcription of other centromeric repetitive sequences (Carone et al. 2009; Gent and Dawe 2012; Hall et al. 2012; Quénet and Dalal 2014), potentially having a role in kinetochore assembly and maintenance. Centromeric transcripts have been shown to be required for CENP-A loading in humans, as depletion of these transcripts leads to mitotic defects (Quénet and Dalal 2014).
Bovids are ecologically, economically, and biologically important animals. Whole genome sequencing generally gives information on low copy sequence evolution, but the data here show the value of understanding the evolution of repetitive DNA copy number, sequence motif, and chromosomal location, on both autosomes and sex chromosomes from different Bovidae species.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by PhD grant SFRH/BD/98122/2013 from the Science and Technology Foundation (FCT) from Portugal. This work was funded by the BioISI project UID/Multi/04046/2019.
This project data have been deposited at GenBank (NCBI) under the acession numbers MK499473 to MK499615.
Literature Cited
- Adega F, Chaves R, Guedes-Pinto H, Heslop-Harrison J.. 2006. Physical organization of the 1.709 satellite IV DNA family in Bovini and Tragelaphini tribes of the Bovidae: sequence and chromosomal evolution. Cytogenet Genome Res. 114(2):140–146. [DOI] [PubMed] [Google Scholar]
- Adega F, Guedes-Pinto H, Chaves R.. 2009. Satellite DNA in the karyotype evolution of domestic animals–clinical considerations. Cytogenet Genome Res. 126(1-2):12–20. [DOI] [PubMed] [Google Scholar]
- Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA.. 2016. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26(10):1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrup-Macdonald ME, Sullivan BA.. 2014. The past, present, and future of human centromere genomics. Genes 5(1):33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V.. 2016. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat Cell Biol. 18(6):684.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscotti MA, Canapa A, Forconi M, Olmo E, Barucca M.. 2015. Transcription of tandemly repetitive DNA: functional roles. Chromosome Res. 23(3):463–477. [DOI] [PubMed] [Google Scholar]
- Biscotti MA, Olmo E, Heslop-Harrison JP.. 2015. Repetitive DNA in eukaryotic genomes. Chromosome Res. 23(3):415–420. [DOI] [PubMed] [Google Scholar]
- Biscotti MA, et al. 2008. Molecular and cytogenetic characterization of repetitive DNA in the Antarctic polyplacophoran Nuttallochiton mirandus. Chromosome Res. 16(6):907–916. [DOI] [PubMed] [Google Scholar]
- Brukner I, Sanchez R, Suck D, Pongor S.. 1995. Sequence‐dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 14(8):1812–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone DM, et al. 2009. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118(1):113–125. [DOI] [PubMed] [Google Scholar]
- Cerutti F, et al. 2016. The major horse satellite DNA family is associated with centromere competence. Mol Cytogenet. 9:35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W.. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371(6494):215–220. [DOI] [PubMed] [Google Scholar]
- Chaves R, Adega F, Heslop-Harrison J, Guedes-Pinto H, Wienberg J.. 2003. Complex satellite DNA reshuffling in the polymorphic t (1; 29) Robertsonian translocation and evolutionarily derived chromosomes in cattle. Chromosome Res. 11(7):641–648. [DOI] [PubMed] [Google Scholar]
- Chaves R, Ferreira D, Mendes-da-Silva A, Meles S, Adega F.. 2017. FA-SAT is an old satellite DNA frozen in several Bilateria genomes. Genome Biol Evol. 9(11):3073–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves R, Guedes-Pinto H, Heslop-Harrison J, Schwarzacher T.. 2000. The species and chromosomal distribution of the centromeric α-satellite I sequence from sheep in the tribe Caprini and other Bovidae. Cytogenet Genome Res. 91(1-4):62–66. [DOI] [PubMed] [Google Scholar]
- Chaves R, Guedes-Pinto H, Heslop-Harrison JS.. 2005. Phylogenetic relationships and the primitive X chromosome inferred from chromosomal and satellite DNA analysis in Bovidae. Proc R Soc Lond B Biol Sci. 272(1576):2009–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves R, Santos S, Guedes-Pinto H.. 2004. Comparative analysis (Hippotragini versus Caprini, Bovidae) of X-chromosome’s constitutive heterochromatin by in situ restriction endonuclease digestion: X-chromosome constitutive heterochromatin evolution. Genetica 121(3):315–325. [DOI] [PubMed] [Google Scholar]
- Di Meo G, et al. 2006. Cattle rob (1; 29) originating from complex chromosome rearrangements as revealed by both banding and FISH-mapping techniques. Chromosome Res. 14(6):649–655. [DOI] [PubMed] [Google Scholar]
- Dumont M, Fachinetti D.. 2017. DNA Sequences in centromere formation and function In: Centromeres and kinetochores. Philadelphia:Springer; p. 305–336. [DOI] [PubMed] [Google Scholar]
- Enukashvily NI, Ponomartsev NV.. 2013. Mammalian satellite DNA: a speaking dumb. Adv Protein Chem Struct Biol. 90:31–65. [DOI] [PubMed] [Google Scholar]
- Escudeiro A, et al. 2019. Bovine satellite DNAs—a history of the evolution of complexity and its impact in the Bovidae family. Euro Zool J. 86(1):20–37. [Google Scholar]
- Feliciello I, Akrap I, Ugarković Đ.. 2015. Satellite DNA modulates gene expression in the beetle Tribolium castaneum after heat stress. PLoS Genet. 11(8):e1005466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D, et al. 2015. Satellite non-coding RNAs: the emerging players in cells, cellular pathways and cancer. Chromosome Res. 23(3):479–493. [DOI] [PubMed] [Google Scholar]
- Fry K, Salser W.. 1977. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 12(4):1069–1084. [DOI] [PubMed] [Google Scholar]
- Gabrielian A, Simoncsits A, Pongor S.. 1996. Distribution of bending propensity in DNA sequences. FEBS Lett. 393(1):124–130. [DOI] [PubMed] [Google Scholar]
- Gallagher D, Womack J.. 1992. Chromosome conservation in the Bovidae. J Hered. 83(4):287–298. [DOI] [PubMed] [Google Scholar]
- Garrido-Ramos MA. 2015. Satellite DNA in plants: more than just rubbish. Cytogenet Genome Res. 146(2):153–170. [DOI] [PubMed] [Google Scholar]
- Gent JI, Dawe RK.. 2012. RNA as a structural and regulatory component of the centromere. Annu Rev Genet. 46:443–453. [DOI] [PubMed] [Google Scholar]
- Giannuzzi G, Catacchio CR, Ventura M.. 2012. Chapter 5. Centromere evolution: digging into mammalian primary constriction. In: Current frontiers and perspectives in cell biology. INTECH Open Access Publisher. Available: http://www.intechopen.com/books/current-frontiers-andperspectivesin-cell-biology/centromere-evolution-digging-into-mammalianprimary-constriction. last accessed September 4, 2018
- Groves C, Grubb P.. 2011. Ungulate taxonomy: Baltimore, Maryland:JHU Press. [Google Scholar]
- Hall LE, Mitchell SE, O’Neill RJ.. 2012. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res. 20(5):535–546. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Douzery EJ.. 1999. The tribal radiation of the family Bovidae (Artiodactyla) and the evolution of the mitochondrial cytochrome b gene. Mol Phylogenet Evol. 13(2):227–243. [DOI] [PubMed] [Google Scholar]
- Hayden KE, Willard HF.. 2012. Composition and organization of active centromere sequences in complex genomes. BMC Genomics. 13:324.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Thakur J, Kasinathan S, Henikoff S.. 2015. A unique chromatin complex occupies young α-satellite arrays of human centromeres. Sci Adv. 1(1):e1400234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J, Schwarzacher T.. 2011. Organisation of the plant genome in chromosomes. Plant J. 66(1):18–33. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JP, Schwarzacher T.. 2013. Nucleosomes and centromeric DNA packaging. Proc Natl Acad Sci U S A. 110(50):19974–19975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes I, Hosid S, Pugh F.. 2011. Variety of genomic DNA patterns for nucleosome positioning. Genome Res. 21(11):1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobse C, et al. 1995. Evolution and recombination of bovine DNA repeats. J Mol Evol. 41(3):277–283. [PubMed] [Google Scholar]
- Kaplan N, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458(7236):362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan S, Henikoff S.. 2017. Non-B-form DNA structures mark centromeres. Mol Biol Evol. 35(4):949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E, Tokan V, Lexa M.. 2015. Transposable elements and G-quadruplexes. Chromosome Res. 23(3):615–623. [DOI] [PubMed] [Google Scholar]
- Khademi TG. 2017. A re-evaluation of phylogenetic relationships within the tribe Tragelaphini (Bovinae: Bovidae), based on complete mitochondrial genomes. Flora 33:34. [Google Scholar]
- Kogan S, Trifonov EN.. 2005. Gene splice sites correlate with nucleosome positions. Gene 352:57–62. [DOI] [PubMed] [Google Scholar]
- Kopecna O, et al. 2012. Isolation and comparison of tribe-specific centromeric repeats within Bovidae. J Appl Genet. 53(2):193–202. [DOI] [PubMed] [Google Scholar]
- Kopecna O, et al. 2014. Tribe-specific satellite DNA in non-domestic Bovidae. Chromosome Res. 22(3):277–291. [DOI] [PubMed] [Google Scholar]
- Kuhn G, Heslop-Harrison J.. 2011. Characterization and genomic organization of PERI, a repetitive DNA in the Drosophila buzzatii cluster related to DINE-1 transposable elements and highly abundant in the sex chromosomes. Cytogenet Genome Res. 132(1-2):79–88. [DOI] [PubMed] [Google Scholar]
- Kuhn GC, Sene FM, Moreira-Filho O, Schwarzacher T, Heslop-Harrison JS.. 2008. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Res. 16(2):307–324. [DOI] [PubMed] [Google Scholar]
- Lenstra J, Boxtel J, Zwaagstra K, Schwerin M.. 1993. Short interspersed nuclear element (SINE) sequences of the Bovidae. Anim Genet. 24(1):33–39. [DOI] [PubMed] [Google Scholar]
- Liu H, Duan X, Yu S, Sun X.. 2011. Analysis of nucleosome positioning determined by DNA helix curvature in the human genome. BMC Genomics. 12:72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. 2008. Characteristics of nucleosome core DNA and their applications in predicting nucleosome positions. Biophys J. 94(12):4597–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Flores I, Garrido-Ramos M.. 2012. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 7:1–28. [DOI] [PubMed] [Google Scholar]
- López-Flores I, et al. 2004. The molecular phylogeny of oysters based on a satellite DNA related to transposons. Gene 339:181–188. [DOI] [PubMed] [Google Scholar]
- Louzada S, et al. 2015. A novel satellite DNA sequence in the Peromyscus genome (PMSat): evolution via copy number fluctuation. Mol Phylogenet Evol. 92:193–203. [DOI] [PubMed] [Google Scholar]
- Macas J, Koblížková A, Navrátilová A, Neumann P.. 2009. Hypervariable 3′ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene 448(2):198–206. [DOI] [PubMed] [Google Scholar]
- Macaya G, Cortadas J, Bernardi G.. 1978. An analysis of the bovine genome by density‐gradient centrifugation. Eur J Biochem. 84(1):179–188. [DOI] [PubMed] [Google Scholar]
- MacEachern S, McEwan J, Goddard M.. 2009. Phylogenetic reconstruction and the identification of ancient polymorphism in the Bovini tribe (Bovidae, Bovinae). BMC Genomics. 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM.. 2016. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 17(1):16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestrović N, Plohl M, Mravinac B, Ugarković D.. 1998. Evolution of satellite DNAs from the genus Palorus—experimental evidence for the “library” hypothesis. Mol Biol Evol. 15(8):1062–1068. [DOI] [PubMed] [Google Scholar]
- Modi WS, Gallagher DS, Womack JE.. 1993. Molecular organization and chromosomal localization of six highly repeated DNA families in the bovine genome. Anim Biotechnol. 4(2):143–161. [Google Scholar]
- Modi WS, Gallagher DS, Womack JE.. 1996. Evolutionary histories of highly repeated DNA families among the Artiodactyla (Mammalia). J Mol Evol. 42(3):337–349. [DOI] [PubMed] [Google Scholar]
- Modi WS, Ivanov S, Gallagher DS.. 2004. Concerted evolution and higher-order repeat structure of the 1.709 (satellite IV) family in bovids. J Mol Evol. 58(4):460–465. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Mestrović N, Ugarković Đ.. 2002. Sequence of PRAT satellite DNA “frozen” in some Coleopteran species. J Mol Evol. 54(6):774–783. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Ugarković Ð.. 2005. Preservation and high sequence conservation of satellite DNAs suggest functional constraints. J Mol Evol. 61(4):542–550. [DOI] [PubMed] [Google Scholar]
- Nieddu M, et al. 2015. Evolution of satellite DNA sequences in two tribes of Bovidae: a cautionary tale. Genet Mol Biol. 38(4):513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman IJ, Lenstra JA. 2001. Mutation and recombination in cattle satellite DNA: a feedback model for the evolution of satellite DNA repeats. J Mol Evol. 52:361–371. [DOI] [PubMed] [Google Scholar]
- Paço A, Chaves R, Vieira-da-Silva A, Adega F.. 2013. The involvement of repetitive sequences in the remodelling of karyotypes: the Phodopus genomes (Rodentia, Cricetidae). Micron 46:27–34. [DOI] [PubMed] [Google Scholar]
- Petraccioli A, et al. 2015. A novel satellite DNA isolated in Pecten jacobaeus shows high sequence similarity among molluscs. Mol Genet Genomics. 290(5):1717–1725. [DOI] [PubMed] [Google Scholar]
- Pezer Z, Brajkovic J, Feliciello I, Ugarkovc D.. 2012. Satellite DNA-mediated effects on genome regulation. Genome Dyn. 7:153–169. [DOI] [PubMed] [Google Scholar]
- Piras FM, et al. 2010. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 6(2):e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero) chromatin. Gene 409(1-2):72–82. [DOI] [PubMed] [Google Scholar]
- Plohl M, Mestrovic N, Mravinac B.. 2012. Satellite DNA evolution. Genome Dyn. 7:126–152. [DOI] [PubMed] [Google Scholar]
- Plohl M, Mestrovic N, Mravinac B.. 2014. Centromere identity from the DNA point of view. Chromosoma 123(4):313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, et al. 2010. Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 104(6):543–551. [DOI] [PubMed] [Google Scholar]
- Purgato S, et al. 2015. Centromere sliding on a mammalian chromosome. Chromosoma 124(2):277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quénet D, Dalal Y.. 2014. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 3: e03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N.. 2006. Genome plasticity in evolution In: Genomic disorders. Humana Press. New Jersey:Springer; p. 153–165. [Google Scholar]
- Rojo V, et al. 2015. Evolutionary dynamics of two satellite DNA families in rock lizards of the genus Iberolacerta (Squamata, Lacertidae): different histories but common traits. Chromosome Res. 23(3):441–461. [DOI] [PubMed] [Google Scholar]
- Šatović E, Plohl M.. 2013. Tandem repeat-containing MITEs in the clam Donax trunculus. Genome Biol Evol. 5(12):2549–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satović E, Zeljko TV, Luchetti A, Mantovani B, Plohl M.. 2016. Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements. BMC Genomics. 17:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalvenzi T, Pollet N.. 2014. Insights on genome size evolution from a miniature inverted repeat transposon driving a satellite DNA. Mol Phylogenet Evol. 81:1–9. [DOI] [PubMed] [Google Scholar]
- Schalch T, Steiner FA.. 2017. Structure of centromere chromatin: from nucleosome to chromosomal architecture. Chromosoma 126(4):443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Heslop-Harrison J.. 1998. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends Plant Sci. 3(5):195–199. [Google Scholar]
- Schwarzacher T, Heslop-Harrison P. 2000. Practical in situ hybridization: BIOS Scientific Publishers Ltd. [Google Scholar]
- Segal E, et al. 2006. A genomic code for nucleosome positioning. Nature 442(7104):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits CH, Rossi MS.. 2002. Satellite DNA: agent of chromosomal evolution in mammals. A review. Mastozool Neotrop. 9:297–308. [Google Scholar]
- Steiner FA, Henikoff S.. 2015. Diversity in the organization of centromeric chromatin. Curr Opin Genet Dev. 31:28–35. [DOI] [PubMed] [Google Scholar]
- Sullivan LL, et al. 2011. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 19:457.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P, Kasinathan S, Henikoff S.. 2018. Simple and complex centromeric satellites in Drosophila sibling species. Genetics 208(3):977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky EJ, Gerbi SA.. 1982a. Sequence analysis of bovine satellite I DNA (1.715 gm/cm3). Nucleic Acids Res. 10(4):1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taparowsky EJ, Gerbi SA.. 1982b. Structure of 1.711 b gm/cm3 bovine satellite DNA: evolutionary relationship to satellite I. Nucleic Acids Res. 10(18):5503–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarković Ð, Plohl M.. 2002. Variation in satellite DNA profiles—causes and effects. EMBO J. 21(22):5955–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic DI. 2009. Centromere-competent DNA: structure and evolution. Prog Mol Subcell Biol. 48:53–76. [DOI] [PubMed] [Google Scholar]
- Vieira-da-Silva A, Louzada S, Adega F, Chaves R.. 2015. A high-resolution comparative chromosome map of Cricetus cricetus and Peromyscus eremicus reveals the involvement of constitutive heterochromatin in breakpoint regions. Cytogenet Genome Res. 145(1):59–67. [DOI] [PubMed] [Google Scholar]
- Vourc’h C, Biamonti G.. 2011. Transcription of satellite DNAs in mammals In: Long non-coding RNAs. Berlin, Heidelberg:Springer; p. 95–118. [DOI] [PubMed] [Google Scholar]
- Wolfgruber TK, et al. 2009. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 5(11):e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, et al. 2013. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc Nat Acad Sci U S A. 110(50):E4875–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.