Significance
Despite great success of deep learning a question remains to what extent the computational properties of deep neural networks are similar to those of the human brain. The particularly nonbiological aspect of deep learning is the supervised training process with the backpropagation algorithm, which requires massive amounts of labeled data, and a nonlocal learning rule for changing the synapse strengths. This paper describes a learning algorithm that does not suffer from these two problems. It learns the weights of the lower layer of neural networks in a completely unsupervised fashion. The entire algorithm utilizes local learning rules which have conceptual biological plausibility.
Keywords: biological deep learning, backpropagation, Hebbian-like plasticity
Abstract
It is widely believed that end-to-end training with the backpropagation algorithm is essential for learning good feature detectors in early layers of artificial neural networks, so that these detectors are useful for the task performed by the higher layers of that neural network. At the same time, the traditional form of backpropagation is biologically implausible. In the present paper we propose an unusual learning rule, which has a degree of biological plausibility and which is motivated by Hebb’s idea that change of the synapse strength should be local—i.e., should depend only on the activities of the pre- and postsynaptic neurons. We design a learning algorithm that utilizes global inhibition in the hidden layer and is capable of learning early feature detectors in a completely unsupervised way. These learned lower-layer feature detectors can be used to train higher-layer weights in a usual supervised way so that the performance of the full network is comparable to the performance of standard feedforward networks trained end-to-end with a backpropagation algorithm on simple tasks.
Supervised learning with backpropagation at its core works extremely well on an immense diversity of complicated tasks (1). Using conventional techniques, the earliest layers of neurons in deep neural networks learn connections that yield the neuron’s receptive fields qualitatively described as feature detectors. In visual tasks, the resemblance of some of the features found by backpropagation in convolutional neural networks to the simple observed selectivities of the response of neurons in early visual processing areas in the brains of higher animals is intriguing (2). For simplicity, we always describe the task to be performed as a visual task, but none of our methods have any explicit limitation to vision. All of the methods discussed below are pixel and color permutation invariant.
In concept, the learning rule that shapes the early artificial neural network responses through supervised end-to-end training using backpropagation and the learning rules which describe the development of the early front-end neural processing in neurobiology are unrelated. There are two conceptual reasons for this. First, in real biological neural networks, the neuron responses are tuned by a synapse-change procedure that is physically local and thus describable by local mathematics. Consider the synaptic connection between an input neuron and an output neuron . In the backpropagation training the alteration of depends not only on the activities of neurons and , but also on the labels and the activities of the neurons in higher layers of the neural network, which are not directly knowable from the activities of neurons and . Thus, the learning rule is nonlocal, i.e., requires information about the states of other specific neurons in the network in addition to the two neurons that are connected by the given synapse. In biology, the synapse update depends on the activities of the presynaptic cell and the postsynaptic cell and perhaps on some global variables such as how well the task was carried out (state of animal attention, arousal, fear, etc.), but not on the activities other specific neurons.
Second, higher animals require extensive sensory experience to tune the early (in the processing hierarchy) visual system into an adult system. This experience is believed to be predominantly observational, with few or no labels, so that there is no explicit task. The learning is said to be unsupervised. By contrast, supervised training of a deep artificial neural network (ANN) with backpropagation requires a huge amount of labeled data.
The central question that we investigate in this paper is the following: Can useful early layer representations be learned in an ANN context without supervision and using only a local “biological” synaptic plasticity rule? We propose a family of learning rules that have conceptual biological plausibility and make it possible to learn early representations that are as good as those found by end-to-end supervised training with backpropagation on Modified National Institute of Standards and Technology (MNIST) dataset. On Canadian Institute for Advanced Research 10 dataset (CIFAR-10) the performance of our algorithm is slightly poorer than end-to-end training, but it still demonstrates very good results in line with some of the previously published benchmarks on biological learning. We demonstrate these points by training the early layer representations using our algorithm and then freezing the weights in that layer and adding another layer on top of it, which is trained with labels using conventional methods of stochastic gradient decent (SGD), to perform the classification task.
End-to-end training with backpropagation can also be used to generate useful early representations in an unsupervised way. An autoencoder can be trained to carry out a self-mapping task on input patterns (3). This is very different from our system which requires no backpropagation of signals, receives all its information directly from forward-propagating signals, and has a local rule of synapse update.
A large body of research is dedicated to constructing algorithms that approximate backpropagation training, without grossly violating the “rules” of neurobiology. These diverse ideas include the feedback alignment algorithms (4, 5), target propagation algorithms (6), equilibrium propagation (7), predictive coding network algorithms (8), alternating minimization techniques (9), etc. There are also online videos summarizing some of these approaches (10, 11). Some of these algorithms use local learning rules (6–8, 12). The main difference between our approach and the aforementioned ideas is that the algorithms of refs. 4–8 and 10–12 use top–down propagation of information in the neural network to adjust the weights in the lower layers of that network (the task can be supervised or unsupervised, depending on the specific paper cited above). In contrast, in our algorithm the synapse learning procedure uses only the bottom–up information about activities of the neurons.
Another set of ideas, more closely related to the current proposal, is studied in a series of papers (13–16). Similarly to our approach this work takes as fundamental two ideas: that the learning be unsupervised and that the synaptic plasticity rule be local. From that common starting point the two approaches diverge. Studies (13–16) postulate in addition that the desired hidden unit synapses should minimize a particular mathematical measure of similarity of data. Their equations describing synapse change, although motivated by biology, are tailored to implement minimization of this postulated measure. Our algorithm for synapse development does not assume such a restriction. Instead the goal of our approach is to engineer a plasticity rule that leads to a good generalization performance of the neural network. Because of the common starting point and the focus of interest, for particular network architectures, plasticity rules, learning protocols, etc., there might be special cases in which the conclusions of the two approaches are similar. But this will not generally be the case.
What is meant by “biological plausibility” for synapse change algorithms in ANNs? We take as fundamental the idea coming from psychologists that the change of the efficacy of synapses is central to learning and that the most important aspect of biological learning is that the coordinated activity of a presynaptic cell and a postsynaptic cell will produce changes in the synaptic efficacy of the synapse between them (17). In the years since Hebb’s original proposal, a huge amount of neurobiology research has fleshed out this idea. We now know that neurons make either excitatory or inhibitory outgoing synapses, but not both (18) (this constraint is not respected by the proposed algorithm); that for excitatory synapses the change in efficacy can either increase or decrease, depending on whether the coordinated activity of the postsynaptic cell is strong or weak [long-term potentiation (LTP) and long-term depression (LTD)] (19); that homeostatic processes regulate overall synaptic strength; that prepost timings of neuronal action potentials can be very important (STDP) (20); that changes in synaptic efficacy may be quantized (21); that there are somewhat different rules for changing the strength of excitatory and inhibitory synapses (22), etc. For the ANN purposes, we subsume most of such details with four ideas:
-
i)
The change of synapse strength during the learning process is proportional to the activity of the presynaptic cell and to a function of the activity of the postsynaptic cell. Both LTP (Hebbian-like plasticity) and LTD (anti–Hebbian-like plasticity) are important. The synapse update is locally determined.
-
ii)
Lateral inhibition between neurons within a layer, which makes the network not strictly feedforward during training, is responsible for developing a diversity of pattern selectivity across many cells within a layer.
-
iii)
The effect of limitations of the strength of a synapse and homeostatic processes will bound possible synaptic connection patterns. The dynamics of our weight-change algorithm express such a bound, so that eventually the input weight vector to any given neuron converges to lie on the surface of a (unit) sphere.
-
iv)
The normalization condition could emphasize large weights more than small ones. To achieve this flexibility we construct (local) dynamics of the synapse development during learning so that the fixed points of these dynamics can be chosen to lie on a Lebesgue sphere with norm, for .
Throughout the paper, biological algorithm is used as a metaphorical name, meaning an algorithm obeying the most important constraints of biological neural networks. We recognize that our proposed algorithm omits many known aspects of neurobiology.
Mathematical Framework
A standard feedforward architecture is considered with a layer of visible neurons , a layer of hidden neurons , and a layer of output neurons . In this case the forward pass is specified by the equations (summation over repeated indexes and is assumed)
| [1] |
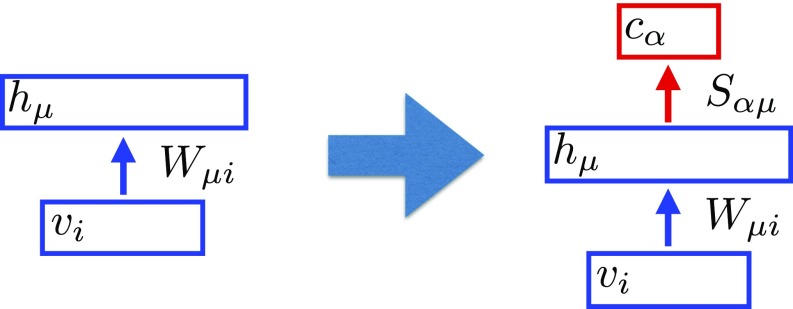
The power of the activation function is a hyperparameter of the model (23, 24), corresponds to rectified linear unit (ReLU), and is a numerical constant. The receptive fields of the hidden layer are learned using our local unsupervised algorithm, described below, without any information about the labels. Once this unsupervised part of the training is complete, the second set of weights is learned using conventional SGD techniques. This is the only part of the training algorithm where labels are used. This logic is illustrated in Fig. 1.
Fig. 1.
Lower layers of the neural network (weights ) are trained using the proposed biological learning algorithm. Once this phase is complete, the weights are plugged into a fully connected perceptron. The weights of the top layer are then learned using SGD in a supervised way.
It has been known since the work of Hubel and Wiesel that many neurons in the visual cortex are tuned to detect certain elementary patterns of activity in the visual stimulus. There is a body of literature dedicated to the study of biologically plausible mechanisms for development of orientation selectivity in the visual cortex, for example refs. 25–29. For our purposes the work of Bienenstock, Cooper, and Munro (BCM) (26) is particularly important. The idea of BCM theory is that for a random sequence of input patterns a synapse is learning to differentiate between those stimuli that excite the postsynaptic neuron strongly and those stimuli that excite that neuron weakly. Learned BCM feature detectors cannot, however, be simply used as the lowest layer of a feedforward network so that the entire network is competitive to a network of the same size trained with backpropagation algorithm end-to-end. The main reason for this is that by using BCM alone, without inhibition between the hidden neurons, synapses of many hidden units can converge to the same pattern. In other words, BCM is a theory of the development of the pattern selectivity of a single cell. Highly specific pattern responses in BCM come about because there is a temporal competition between patterns seeking to drive a single neuron. This competition is controlled by the dynamics of an adjustable learning threshold parameter. In our system, the neurons compete with each other for patterns, and there is no adjustable threshold. The competition is between neurons, not between patterns. When one neuron becomes tuned to some pattern of inputs, the within-layer lateral inhibition keeps other neurons from becoming selective to that same pattern. Thus, a layer of diverse early feature detectors can be learned in a completely unsupervised way without any labels.
Synaptic Plasticity Rule.
Consider the case of only one hidden unit. Then the matrix becomes a vector , which has components . We use similar notations for other variables; for example, is a vector with components . It is convenient to define a metric and an inner product ( is Kronecker delta)
| [2] |
where is the parameter defining the Lebesgue norm. In the case of this inner product is the conventional dot product between two vectors, and if the contribution of the weights in the metric becomes important. The plasticity rule that we study can then be written as
| [3] |
The constant defines the time scale of the learning dynamics. It should be larger than the time scale of presentation of an individual training example, as well as the time scale of evolution of individual neurons, which is defined below. The function is a nonlinear learning activation function that is discussed below (Eq. 9) and plotted in Fig. 2, are visible neurons or training examples, and is a constant that is discussed below.
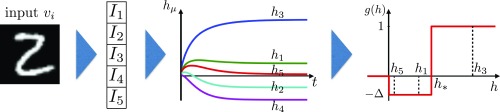
Fig. 2.
The pipeline of the training algorithm. Inputs are converted to a set of input currents . These currents define the dynamics [8] that lead to the steady-state activations of the hidden units. These activations are used to update the synapses using the learning rule [3]. The learning activation function changes the sign at , which separates the Hebbian and anti-Hebbian learning regimes. The second term in the plasticity rule [3], which is the product of the input current and the weight , corresponds to another path from the data to the synapse update. This path is not shown here and does not go through Eq. 8.
The plasticity rule [3] is an extension of the famous Oja rule (30). It implements three of the four ideas outlined in the Introduction. The first term of the plasticity rule is the product of the activity of the presynaptic neuron and a function of the postsynaptic activity . Thus, it is an example of Hebbian-like plasticity. As we discuss below this function has both positive and negative values, resulting in both Hebbian and anti-Hebbian learning. The first term in Eq. 3 implements the first idea from the Introduction. The second term in [3] ensures that the weights of a hidden neuron connected to all of the visible units converge to vectors of length . This is an implementation of the third and the fourth ideas outlined in the Introduction. Initially the weights are initialized from a standard normal distribution. Then a sequence of training examples is presented one at a time and the weights are updated according to [3]. It can be shown that as the weights converge to a sphere of radius , defined using the norm:
| [4] |
To see this consider the time derivative of the norm of the weight vector
| [5] |
where in the second line the time derivative of the weight vector is substituted from [3]. Provided that , if the length of vector is less than , its length increases on the dynamics, and if it is greater than , its length decreases. Thus, although the training procedure starts with random values of the synaptic weights, eventually these weights converge to a sphere. The positivity bound is satisfied for a broad class of activation functions. In practical applications, we use this learning rule even in situations when the positivity constraint is violated. It turns out that if the violation is weak, which is justified by the small parameter (Fig. 2), the weights still converge to a sphere of radius .
In situations when the network has more than one hidden unit, which are enumerated by index , each vector will have an external index, as will the inner product:
| [6] |
In this case the fixed points of the plasticity dynamics are such that synapses of each hidden unit converge to their own unit vector on a sphere.
It can also be shown that plasticity rule [3] has a Lyapunov function (Appendix A)
| [7] |
that monotonically increases on the dynamical trajectory of synapses (the sum over data runs over all of the training examples). Finally, if one works on the ordinary Euclidean sphere with and the function is linear, our learning rule reduces to the famous Oja rule (30). In the following the radius of the sphere is set to .
To summarize, the learning rule [3] implements the Hebbian-like plasticity (idea i from the Introduction) subject to homeostatic constraints on the length of synaptic weights (ideas iii and iv). The intuition behind different choices of the Lebesgue parameter is the following. For the weights are normalized to lie on the ordinary Euclidean sphere. If is increased, only large values of the weights contribute to the normalization condition [4], while small values become suppressed, since the power function grows very rapidly. This results in weights that lie almost on the surface of a hypercube with slightly rounded corners. This allows the learning algorithm to find a good set of weights that are smaller than the maximal weight without too many restrictions from the homeostatic constraint [4]. If those small weights increase and approach the maximal weight, however, they begin to contribute strongly to the normalization condition [4].
A Biologically Inspired Learning Algorithm.
A good set of weights in the neural network should be such that different hidden units detect different features of the data. Neither the BCM algorithm nor the plasticity rule [3] addresses this issue of differential selectivity. To do this, we use a neural network with global inhibition between the hidden neurons and dynamical equations
| [8] |
In Eq. 8 the activities of the hidden neurons are denoted by , are the corresponding firing rates (we use a ReLU), is the input current from the visible layer, is a parameter that defines the strength of the global inhibition, and constant defines the dynamical time scale of individual neurons.
The pipeline of the unsupervised part of the algorithm is shown in Fig. 2. Using the weights a raw input is converted into a set of input currents . These currents drive the dynamics of hidden units, as is shown in Fig. 2. The strength parameter of the global inhibition is set so that in the final state only a small fraction of hidden units have positive activity (in this case , , and ). The values of these steady-state activities are then used as arguments in the nonlinear learning activation function in [3]. This function implements temporal competition between the patterns, so that it is positive for the activities exceeding a threshold and negative for the activities in the range . Activities that are below zero do not contribute to training:
| [9] |
The intuitive idea behind this choice of the activation function is that the synapses of hidden units that are strongly driven are pushed toward the patterns that drive them, while the synapses of those hidden units that are driven slightly less are pushed away from these patterns. Given a random temporal sequence of the input stimuli, this creates a dynamic competition between the hidden units and results in the synaptic weights that are different for each hidden unit and specific to features of the data. The idea of having an activation function that is positive for activations above the threshold and negative below the threshold is inspired by the BCM theory (26) and the existence of LTP and LTD (19).
The idea of using lateral inhibition combined with Hebbian-like plasticity goes back to a series of papers (27–29). In the machine-learning literature it is known as competitive learning or “winner-takes-all” learning (31, 32). Formally our approach reduces to those methods if anti-Hebbian learning is switched off ( limit) and the second term in the plasticity rule [3], which describes homeostatic constraints on synaptic strengths and leads to normalized weights, is removed. The global inhibition motif has also been used in a number of unsupervised learning algorithms (13, 25, 33–35).
A Fast Implementation
We view the algorithm presented in the previous section as a conceptual idea of how it might be possible to learn good early layer representations given the biological constraints of the sensory cortex. A computational advantage of the presented algorithm, compared with the standard supervised training with backpropagation, is that it is unsupervised. The question then arises whether this algorithm might be valuable from the artificial intelligence (AI) perspective even if we forget about its biological motivation.
From the AI point of view the main drawback of the presented algorithm is that it is slow. There are two reasons for this. First, it is an online algorithm so that training examples are presented one at a time, unlike SGD where training examples can be presented in minibatches. Second, for any training example one has to wait until the set of hidden units reaches a steady state. This requires numerically solving Eq. 8, which is time consuming.
An approximation of this computational algorithm has been found which circumvents these two drawbacks and works extremely well in practice. First, instead of solving dynamical equations we use the currents as a proxy for ranking of the final activities. Given that ranking, the unit that responds the most to a given training example is pushed toward that example with activation . The unit that is second (or more generally th) in ranking is pushed away from the training example with activation . The rest of the units remain unchanged (here is the total number of hidden units, corresponds to the least-activated hidden unit, and is the strongest-driven hidden unit):
| [10] |
This heuristic significantly speeds up the algorithm. Second, training examples can then be organized in minibatches, so that the ranking is done for the entire minibatch and the weight updates that result from the learning rule [3] are averaged over all examples in the minibatch.
In this “fast” implementation the sorting of the input currents has a computational complexity , where is the number of hidden units, and is the ranking parameter.
Testing the Model
The presented algorithm was tested on the MNIST and CIFAR-10 datasets. All of the tasks that are discussed below are pixel and (in the case of CIFAR-10) color permutation invariant. The hyperparameters are reported in Appendix B. The benchmarks discussed below use the fast implementation of the proposed algorithm, described in A Fast Implementation. The code can be found in the GitHub repository (36).
MNIST.
A standard training set of 60,000 examples was randomly split into a 50,000-examples training set and a 10,000-examples validation set that was used for tuning the hyperparameters. After the hyperparameters were fixed, the training and the validation sets were combined to train the final model on 60,000 examples. The final performance of the models was evaluated on the standard held-out test set of 10,000 examples.
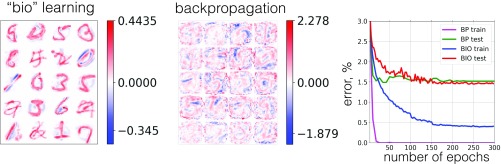
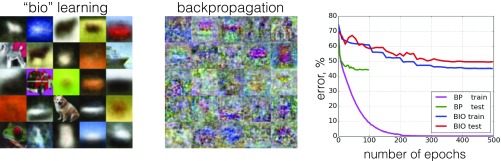
In the first set of experiments a network of 2,000 hidden units was trained using the proposed biological algorithm to find the weights . The initial values for weights were drawn from the standard normal distribution. As training progresses the weights eventually converge to the surface of a unit sphere. Final learned weights, connected to 20 randomly chosen hidden units, are shown in Fig. 3, Left. After the unsupervised phase of the training was complete, the learned weights were frozen and used in the network [1]. The second set of weights was learned using a standard SGD procedure described in Appendix B. The decrease of the errors on the training and the test sets during this second (supervised) phase of the training is shown in Fig. 3, Right (blue and red curves). The performance of this biologically trained network was compared with the performance of the feedforward network of the same size () trained end-to-end using the Adam optimizer starting from random weights (training and test errors are shown in Fig. 3, Right, magenta and green curves). Twenty randomly chosen feature detectors learned by the standard network trained end-to-end are shown in Fig. 3, Center.
Fig. 3.
(Left) The weights learned by the biological network using the unsupervised learning algorithm. Twenty randomly chosen feature detectors of 2,000 are shown. (Center) The weights learned by the network trained end-to-end with the backpropagation algorithm. Twenty randomly chosen feature detectors of 2,000 are shown. (Right) Error rate on the training and test sets as training progresses for the proposed biological algorithm and for the standard network trained end-to-end.
The network trained with the backpropagation end-to-end reaches the well-known benchmarks: training error , test error . Training error of the biological network is . The most surprising aspect of this plot is that the test error of the biological algorithm, which is , is as low as the test error of the network trained end-to-end. This result can be compared with the previously published benchmarks on other biologically inspired algorithms, for example in ref. 7, in ref. 12, in ref. 6, in ref. 37, and in ref. 38. There is also a recent paper (39), where multiple biologically inspired algorithms are assessed on this task with results ranging from to . When comparing these numbers, it should be noted that the algorithms of refs. 6, 7, 12, 37, and 39 are using the information about the labels all of the way during the training. In contrast, our algorithm learns the first-layer representations in a completely unsupervised way. Thus, it demonstrates a comparable or better performance despite solving a more challenging task. Finally, we note that the performance of networks trained end-to-end with backpropagation can be significantly enhanced using dropout, injecting noise, augmenting the dataset, etc. See for example ref. 40, which reports error, or ref. 41, which reports error. Additional work is required to see whether similar (or different) approaches can affect the performance of the proposed biological training algorithm.
It is instructive to examine the weights (Fig. 3, Left) learned by the unsupervised phase of the proposed biological algorithm. The color coding uses the white color to represent zero weights, the red color to represent positive weights, and the blue color to represent negative weights. Training examples are normalized so that they are always positive . The weights associated with each hidden unit live on a unit sphere, i.e., satisfy the constraint , which is the fixed point of the learning rule [3]. Although some of the learned feature detectors resemble certain training examples, they are not simply copies of the individual training data points. The easiest way to see this is to note that the largest (in absolute value) negative weight among the 20 shown feature detectors, , is almost as big as the largest positive weight 0.444, while the training examples are always positive. Thus, the learned feature detectors encode both where the ink is in the data and where the ink is not. Mathematically, these negative weights arise because of the anti-Hebbian piece, proportional to , in the learning rule [3] (Fig. 2 and Eq. 10).
Another important aspect is that the proposed biological network is not doing a simple template matching to classify the data. For example, feature detector 9 (counting is left to right, top to bottom) encodes subdigit features—a slanted line with a gap in the middle. Feature detector 3 “votes” for class 5 and against class 0. Feature detectors 6 and 13 are activated by classes 4, 7 and 2, 7, respectively. When a network is presented with an input, many hidden units are activated. Activities of these hidden units are then used in the next layer to arrive at the classification decision. Thus, the network learns a distributed representation of the data over multiple hidden units. This representation, however, is very different from the representation learned by the network trained end-to-end, as is clear from comparison of Fig. 3, Left and Center.
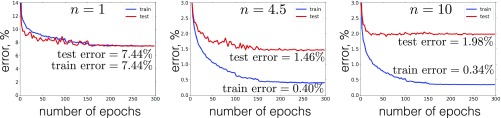
It is instructive to learn the first-layer weights for the optimal values of the hyperparameters , , and (on the validation set), then freeze those weights, and then learn the second-layer weights for different values of power in [1]. The remaining hyperparameters associated with the second supervised phase of the training, from Eq. 12 and from Eq. 1, are optimized for each value of power individually. The results are shown in Fig. 4. There is an optimal value of power for which the validation error and the test error are the smallest. The errors increase for both smaller and larger values of power . This indicates that given the weights, which are learned by the unsupervised phase of the algorithm, the architecture of higher layers should be tuned so that the network could take most advantage of those unsupervised weights.
Fig. 4.
Error on the training set (blue) and the test set (red) for three networks with different powers of the activation function . All networks were trained for 300 epochs with the schedule of the learning-rate annealing described in Appendix B. The hyperparameters of the unsupervised phase of the training are , , .
For completeness we tested our algorithm in the limit , when it reduces to familiar competitive learning with additional dynamical convergence of weight vectors to a unit sphere. The weights in this limit are shown in Fig. 5. They are all positive and, unlike the weights shown in Fig. 3, represent prototypes of hand-written digits. The performance of the algorithm in this limit is error rate, which is significantly worse than the performance of our algorithm with nonzero , which is error rate. The two networks that are compared have the same capacity (2,000 hidden units).
Fig. 5.
Weights of the first layer in the limit . All synaptic connections are positive and represent prototypes of digits.
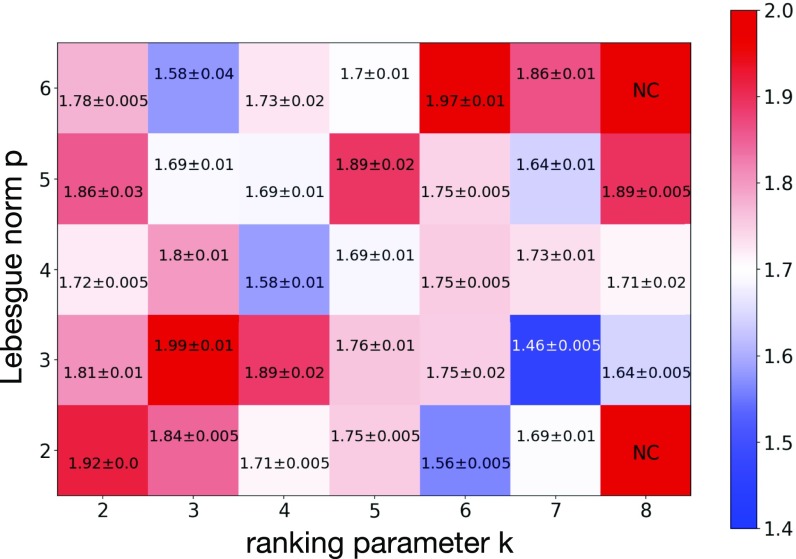
We systematically investigated the performance of the “fast AI” implementation for different values of Lebesgue norm and the ranking parameter . To do this, for each value of and (with increment 1 in both parameters) the proposed biological algorithm was used to find a set of weights . For each and the top-layer weights were trained for different values of the hyperparameters from Eq. 1, from Eq. 12, and from Eq. 1. For each combination of the parameters the network was trained three times for different random initializations of the weights, different sampling of minibatches, and different validation sets (10,000 examples sampled from the standard training set of 60,000 examples). Altogether this results in 360 training runs per each combination of and and 12,600 runs in total. For each combination of and a combination of , , and that worked best on the validation set was selected. For those “optimal” hyperparameters the training was repeated on the complete training set of 60,000 examples three times to determine the performance on the held-out test set. The results are shown in Fig. 6, where for each combination of and the mean performance together with the SD for the three training runs is reported. As is clear from Fig. 6, the algorithm demonstrates competitive results for close to all values of and . If one further optimizes the performance (on the validation set) over the parameters , , and , the best combination ends up being , , . This gives the final performance of our proposed biological algorithm, .
Fig. 6.
For each pair of hyperparameters and of the proposed unsupervised algorithm the hyperparameters of the top layer (, , ) were optimized on the validation set. For these optimal (, , ), the mean error together with the SD of the individual runs on the held-out test set is shown for each pair of Lebesgue norm and the ranking parameter . In these experiments the hyperparameter was set to the optimal value determined on the validation set. The unsupervised algorithm did not converge for (, ) and (, ), indicating that a smaller value of is required for those hyperparameters.
CIFAR-10.
A standard training set of 50,000 examples was randomly split into a 45,000-examples training set and a 5,000-examples validation set. After tuning the hyperparameters on the validation set, the final models were trained on the entire 50,000 data points and evaluated on the held-out test set of 10,000 examples. No preprocessing of the data was used, except that each input image was normalized to be a unit vector in the -dimensional space.
In analogy with MNIST the performance of two networks was compared. One trained using the proposed biological algorithm in two phases: unsupervised training of the network, followed by the supervised training of the top layer, . The other one, , trained with Adam end-to-end. The results are shown in Fig. 7. The optimal duration of the backpropagation training for the conventional network (see Appendix B for details) was 100 epochs, which gave the error on the test set of . At that point the error on the training set is still nonzero. To demonstrate that the capacity of the standard network is sufficient to fit all of the training examples, we kept training it until the training error reached . The proposed biological network achieves training error and test error . Unfortunately, previously published benchmarks for biologically inspired training algorithms are very scarce on CIFAR. The study (6) reports . A recent paper (39) reports error rates in the range . It also reports a benchmark of for the networks trained end-to-end, which is better than the result that we achieved in our end-to-end trained network. This is perhaps explained by the fact that a deeper architecture was used in ref. 39 for this task. It is worth emphasizing again that the algorithms of refs. 6 and 39 are supervised all of the way throughout training and for this reason solve a simpler task compared with our algorithm that learns the first set of weights in an entirely unsupervised way. In Fig. 7 the weights of 25 randomly selected hidden units learned by the unsupervised phase of the algorithm are shown. They represent a diverse collection of features and prototypes of the training images. These weights have large negative elements and for this reason are not just copies of the training examples, which are all positive. The classification decision of the full biological network is done in a distributed way involving votes of multiple hidden units. This distributed representation is very different, however, from the end-to-end feature detectors shown in Fig. 7, Center. Finally, although the network is dealing with the pixel-color permutation invariant problem, the unsupervised algorithm discovers the continuity of color in the data. This is different from the end-to-end trained feature detectors, which look speckled.
Fig. 7.
(Left) The weights learned by the network using the unsupervised learning algorithm. Twenty-five randomly chosen feature detectors of 2,000 are shown. (Center) The weights learned by the network trained end-to-end with the backpropagation algorithm. Twenty-five randomly chosen feature detectors of 2,000 are shown. (Right) Error rate on the training and test sets as training progresses for the proposed biological algorithm and for the standard network trained end-to-end.
Discussion and Conclusions
Historically, neurobiology has inspired much research on using various plasticity rules to learn useful representations from the data. This line of research chiefly disappeared after 2014 because of the success of deep neural networks trained with backpropagation on complicated tasks like ImageNet. This has led to the opinion that neurobiology-inspired plasticity rules are computationally inferior to networks trained end-to-end and that supervision is crucial for learning useful early layer representations from the data. By consequence, the amount of attention given to exploring the diversity of possible biologically inspired learning rules, in the present era of large datasets and fast computers, has been rather limited. Our paper challenges this opinion by describing an unsupervised learning algorithm that demonstrates a very good performance on MNIST and CIFAR-10. The core of the algorithm is a local learning rule that incorporates both LTP and LTD types of plasticity and a network motif with global inhibition in the hidden layer.
The SGD training of the top layer was used in this paper to assess the quality of the representations learned by the unsupervised phase of our algorithm. This does not invalidate the biological plausibility of the entire algorithm, since SGD in one layer can be written as a local synaptic plasticity rule involving only pre- and postsynaptic cell activities. Thus, it complies with the locality requirement that we took as fundamental.
In the present paper all of the experiments were done on a network with one hidden layer. The proposed unsupervised algorithm, however, is iterative in nature. This means that after a one-layer representation is learned, it can be used to generate the codes for the input images. These codes can be used to train another layer of weights using exactly the same unsupervised algorithm. There are many possibilities of how one could organize those additional layers, since they do not have to be fully connected. This line of research requires further investigation. At this point it is unclear whether or not the proposed algorithm can lead to improvements on the single hidden-layer network, if applied iteratively in deeper architectures.
Another limitation of the present work is that all of the experiments were done on MNIST and CIFAR-10 and only in the fully connected setting. Many biologically plausible approaches to deep learning fail on more complicated datasets, like ImageNet, even if they work well on MNIST and CIFAR-10 (39).
Finally, this paper does not claim that end-to-end training is incompatible with biological plausibility. There is a large body of literature dedicated to designing biologically plausible variants of end-to-end training approximating backpropagation (4–8, 10–12). The main difference between those approaches and the present proposal is that the algorithms of refs. 4–8 and 10–12 require the top–down propagation of information and craft the synaptic weights in the early layers of neural networks to solve some specific task (supervised or unsupervised). In our algorithm there is no top–down propagation of information, the synaptic weights are learned using only bottom–up signals, and the algorithm is agnostic about the task that the network will have to solve eventually in the top layer. Despite this lack of knowledge about the task, the algorithm finds a useful set of weights that leads to a good generalization performance on the standard classification task, at least on simple datasets like MNIST and CIFAR-10.
Appendix A
Below we prove that [7] monotonically increases on the dynamics [3]. The temporal derivative of [7] is given by Eq. 11:
| [11] |
The last expression in [11] is positive due to the Cauchy–Schwarz inequality. Thus, [7] monotonically increases on the dynamics. Also, the Lyapunov function [7] is upper bounded by the Cauchy–Schwarz inequality.
Appendix B
The unsupervised part of the training for MNIST was done for the following parameters: Lebesgue norm , ranking parameter , and anti-Hebbian learning parameter . Training was done using minibatches of size 100 examples for 1,000 epochs. Learning rate linearly decreased from the maximal value 0.04 at the first epoch to 0 at the last epoch. The learning rate here is defined as the numerical coefficient that is used to multiply the right-hand side of [3] to obtain the increment in weights.
The supervised part of the training was done using the loss function (labels are one-hot–encoded vectors of units of )
| [12] |
The Adam optimizer was used to minimize the loss function during 300 epochs with the following schedule of learning rate change: 0.001 for the first 100 epochs and after that the learning rate decreased every 50 epochs as 0.0005, 0.0001, 0.00005, 0.00001. Minibatch size was 100.
For the unsupervised part of CIFAR-10 experiments, a similar setting was used with , , and . Training was done for 1,000 epochs with minibatches of size 1,000. Learning rate linearly decreased from 0.02 to 0.
For the supervised part of the biological network in Fig. 7 the Adam optimizer was used with minibatch size 10, , for 500 epochs. The learning-rate schedule: 0.004 for 100 epochs and then each 50 epochs the learning rate changed as 0.002, 0.001, 0.0005, 0.0002, 0.0001, 0.00005, 0.00002, 0.00001. For the supervised part of the conventional end-to-end trained network the Adam optimizer was used with minibatch size 100 and , for 100 epochs. Learning rate: 0.004 for 50 epochs and then 0.001 for another 50 epochs.
The code used in the biological unsupervised phase of the algorithm is available on GitHub (36).
Acknowledgments
We thank Yoshua Bengio, Shiyu Chang, David Cox, Kristjan Greenwald, Leopold Grinberg, Guy Gur-Ari, Arnold Levine, Ralph Linsker, Sijia Liu, Falk Pollok, and Irina Rish for helpful discussions. The work of D.K. at the Institute for Advanced Study was partly supported by the Starr Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The code used in this paper has been deposited in the GitHub repository, https://github.com/DimaKrotov/Biological_Learning.
References
- 1.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 2.Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. European Conference on Computer Vision. Springer; Cham, Switzerland: 2014. pp. 818–833. [Google Scholar]
- 3.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 4.Lillicrap TP, Cownden D, Tweed DB, Akerman CJ. 2014. Random feedback weights support learning in deep neural networks. arXiv:1411.0247. Preprint, posted November 2, 2014. [DOI] [PMC free article] [PubMed]
- 5.Lillicrap TP, Cownden D, Tweed DB, Akerman CJ. Random synaptic feedback weights support error backpropagation for deep learning. Nat Commun. 2016;7:13276. doi: 10.1038/ncomms13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DH, Zhang S, Fischer A, Bengio Y. 2015. Difference target propagation. Joint European Conference on Machine Learning and Knowledge Discovery in Databases, eds Appice A., et al. (Springer, Cham, Switzerland), pp 498–515.
- 7.Scellier B, Bengio Y. Equilibrium propagation: Bridging the gap between energy-based models and backpropagation. Front Comput Neurosci. 2017;11:24. doi: 10.3389/fncom.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whittington JC, Bogacz R. An approximation of the error backpropagation algorithm in a predictive coding network with local Hebbian synaptic plasticity. Neural Comput. 2017;29:1229–1262. doi: 10.1162/NECO_a_00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choromanska A, et al. 2018. Beyond backprop: Alternating minimization with co-activation memory. arXiv:1806.09077. Preprint, posted February 1, 2019.
- 10.Bengio Y. 2017 Deep learning and backprop in the brain (online video). Available at https://www.youtube.com/watch?v=FhRW77rZUS8. Accessed October 13, 2017.
- 11.Hinton G. 2016 Can sensory cortex do backpropagation? (online video). Available at https://www.youtube.com/watch?v=cBLk5baHbZ8. Accessed April 16, 2016.
- 12.Sacramento J, Costa RP, Bengio Y, Senn W. 2017. Dendritic error backpropagation in deep cortical microcircuits. arXiv:1801.00062. Preprint, posted December 30, 2017.
- 13.Pehlevan C, Hu T, Chklovskii DB. A Hebbian/anti-Hebbian neural network for linear subspace learning: A derivation from multidimensional scaling of streaming data. Neural Comput. 2015;27:1461–1495. doi: 10.1162/NECO_a_00745. [DOI] [PubMed] [Google Scholar]
- 14.Pehlevan C, Sengupta AM, Chklovskii DB. Why do similarity matching objectives lead to Hebbian/anti-Hebbian networks? Neural Comput. 2018;30:84–124. doi: 10.1162/neco_a_01018. [DOI] [PubMed] [Google Scholar]
- 15.Pehlevan C, Chklovskii DB. A Hebbian/anti-Hebbian network derived from online non-negative matrix factorization can cluster and discover sparse features. In: Matthews MB, editor. 2014 48th Asilomar Conference on Signals, Systems and Computers. IEEE; New York: 2014. pp. 769–775. [Google Scholar]
- 16.Sengupta A, Pehlevan C, Tepper M, Genkin A, Chklovskii D. 2018. Manifold-tiling localized receptive fields are optimal in similarity-preserving neural networks. Advances in Neural Information Processing Systems, eds Bengio S, et al. (Neural Information Processing Systems Foundation, Inc., La Jolla, CA), pp 7077–7087.
- 17.Hebb DO. The Organization of Behavior: A Neurophysiological Approach. Wiley; New York: 1949. p. 62. [Google Scholar]
- 18.Eccles J. From electrical to chemical transmission in the central nervous system. Notes Rec R Soc Lond. 1976;30:219–230. doi: 10.1098/rsnr.1976.0015. [DOI] [PubMed] [Google Scholar]
- 19.Koch C. Biophysics of Computation: Information Processing in Single Neurons. Oxford Univ Press; New York: 2004. [Google Scholar]
- 20.Gerstner W, Kistler WM, Naud R, Paninski L. Neuronal Dynamics: From Single Neurons to Networks and Models of Cognition. Cambridge Univ Press; Cambridge, UK: 2014. [Google Scholar]
- 21.Petersen CC, Malenka RC, Nicoll RA, Hopfield JJ. All-or-none potentiation at CA3-CA1 synapses. Proc Natl Acad Sci USA. 1998;95:4732–4737. doi: 10.1073/pnas.95.8.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo L. Principles of Neurobiology. Garland Science, Taylor & Francis Group, LLC; New York: 2015. [Google Scholar]
- 23.Krotov D, Hopfield JJ. Dense associative memory for pattern recognition. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R, editors. Advances in Neural Information Processing Systems. Neural Information Processing Systems Foundation, Inc.; La Jolla, CA: 2016. pp. 1172–1180. [Google Scholar]
- 24.Krotov D, Hopfield J. Dense associative memory is robust to adversarial inputs. Neural Comput. 2018;30:3151–3167. doi: 10.1162/neco_a_01143. [DOI] [PubMed] [Google Scholar]
- 25.Von der Malsburg C. Self-organization of orientation sensitive cells in the striate cortex. Kybernetik. 1973;14:85–100. doi: 10.1007/BF00288907. [DOI] [PubMed] [Google Scholar]
- 26.Bienenstock EL, Cooper LN, Munro PW. Theory for the development of neuron selectivity: Orientation specificity and binocular interaction in visual cortex. J Neurosci. 1982;2:32–48. doi: 10.1523/JNEUROSCI.02-01-00032.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linsker R. From basic network principles to neural architecture: Emergence of spatial-opponent cells. Proc Natl Acad Sci USA. 1986;83:7508–7512. doi: 10.1073/pnas.83.19.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linsker R. From basic network principles to neural architecture: Emergence of orientation-selective cells. Proc Natl Acad Sci USA. 1986;83:8390–8394. doi: 10.1073/pnas.83.21.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linsker R. From basic network principles to neural architecture: Emergence of orientation columns. Proc Natl Acad Sci USA. 1986;83:8779–8783. doi: 10.1073/pnas.83.22.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oja E. Simplified neuron model as a principal component analyzer. J Math Biol. 1982;15:267–273. doi: 10.1007/BF00275687. [DOI] [PubMed] [Google Scholar]
- 31.Rumelhart DE, Zipser D. Feature discovery by competitive learning. Cogn Sci. 1985;9:75–112. [Google Scholar]
- 32.Carpenter GA, Grossberg S. A massively parallel architecture for a self-organizing neural pattern recognition machine. Comput Vision Graphics Image Process. 1987;37:54–115. [Google Scholar]
- 33.Kohonen T. The self-organizing map. Proc IEEE. 1990;78:1464–1480. [Google Scholar]
- 34.Földiak P. Forming sparse representations by local anti-Hebbian learning. Biol Cybernetics. 1990;64:165–170. doi: 10.1007/BF02331346. [DOI] [PubMed] [Google Scholar]
- 35.Seung HS, Zung J. 2017. A correlation game for unsupervised learning yields computational interpretations of Hebbian excitation, anti-Hebbian inhibition, and synapse elimination. arXiv:1704.00646. Preprint, posted April 3, 2017.
- 36.Krotov D, Hopfield JJ. 2019 Data from “Biological Learning.” GitHub. Available at https://github.com/DimaKrotov/Biological_Learning. Deposited January 28, 2019.
- 37.Nøkland A. Direct feedback alignment provides learning in deep neural networks. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R, editors. Advances in Neural Information Processing Systems. Neural Information Processing Systems Foundation, Inc.; La Jolla, CA: 2016. pp. 1037–1045. [Google Scholar]
- 38.Diehl PU, Cook M. Unsupervised learning of digit recognition using spike-timing-dependent plasticity. Front Comput Neurosci. 2015;9:99. doi: 10.3389/fncom.2015.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartunov S, Santoro A, Richards BA, Hinton GE, Lillicrap T. 2018. Assessing the scalability of biologically-motivated deep learning algorithms and architectures. arXiv:1807.04587. Preprint, posted November 20, 2018.
- 40.Tang Y. 2013. Deep learning using linear support vector machines. arXiv:1306.0239. Preprint, posted February 21, 2015.
- 41.Rasmus A, Berglund M, Honkala M, Valpola H, Raiko T. 2015. Semi-supervised learning with ladder networks. Advances in Neural Information Processing Systems, pp 3546–3554.