Significance
The canonical targeting signal for degrading proteins by the ubiquitin (Ub) system—a chain composed of multiple Ub moieties—has remained a mystery. The structure of the proteasome, the enzyme that recognizes the signal and degrades the target substrate cannot explain why such a long chain is needed. To better understand this problem, we synthesized α-globin to which chains with different number of Ubs were attached. In long adducts, the proximal Ub remains on the substrate, likely securing its attachment to the proteasome, and is degraded with it. The distal Ub protects the proximal from removal by deubiquitinating enzymes and is then removed and recycled. In short adducts, the Ub moieties are rapidly removed, and the substrate remains stable.
Keywords: ubiquitination, deubiquitination, 26S proteasome, protein degradation, chemical synthesis
Abstract
One of the enigmas in the ubiquitin (Ub) field is the requirement for a poly-Ub chain as a proteasomal targeting signal. The canonical chain appears to be longer than the distance between the two Ub-binding proteasomal receptors. Furthermore, genetic manipulation has shown that one receptor subunit is sufficient, which suggests that a single Ub can serve as a degradation signal. To shed light on this mystery, we chemically synthesized tetra-Ub, di-Ub (K48-based), and mono-Ub adducts of HA-α-globin, where the distal or proximal Ub moieties were tagged differentially with either Myc or Flag. When incubated in a crude cell extract, the distal Ub moiety in the tetra-Ub adduct was mostly removed by deubiquitinating enzymes (DUBs) and reconjugated to other substrates in the extract. In contrast, the proximal moiety was most likely degraded with the substrate. The efficacy of degradation was proportionate to the chain length; while tetra-Ub globin was an efficient substrate, with mono-Ub globin, we observed rapid removal of the Ub moiety with almost no degradation of the free globin. Taken together, these findings suggest that the proximal moieties are necessary for securing the association of the substrate with the proteasome along the proteolytic process, whereas the distal moieties are important in protecting the proximal moieties from premature deubiquitination. Interestingly, when the same experiment was carried out using purified 26S proteasome, mono- and tetra-Ub globin were similarly degraded, highlighting the roles of the entire repertoire of cellular DUBs in regulating the degradation of proteasomal substrates.
Recent advances in the synthesis and semisynthesis of ubiquitin (Ub)-based conjugates, such as ubiquitinated proteins and activity-based probes, paved the road for various challenging studies in the Ub research field (1). Of particular interest is the role of the Ub chain and its structure in the different functions of the Ub system (2, 3). One unsolved mystery is the requirement for a poly-Ub chain for degradation of many proteasomal substrates, which cannot be explained based on the number and distance of the Ub-binding receptors in the 19S regulatory complex of the 26S proteasome (4–7). True, some substrates are degraded following monoubiquitination (8, 9), and a chain length of ∼7 Ub moieties has been reported for yeast proteins (10); however, the canonical chain is still considered to be composed of multiple Ub moieties. We have shown that some of these Ub moieties are degraded along with the substrate (11), whereas others are recycled (12). We also have shown that the length of the Ub chain is proportionate to the size of the target substrate, and that a long chain is probably required to stabilize a large substrate on the 26S proteasome and secure its processive destruction (13).
To shed light on the enigma of the proteolytic signal, we have developed different synthetic strategies for selectively modifying expressed proteins with Ub chains via disulfide, thioether, and oxime linkages (14–16). For example, we reported the semisynthesis of poly-Ub substrates such as poly-Ub α-synuclein, which enabled us to comprehend the role of ubiquitination in regulating α-synuclein stability, aggregation, phosphorylation, and clearance (13, 14). In a different study, we used synthetic ubiquitinated α-globin, in which the linkage between the Ub moiety and the substrate was noncleavable. We demonstrated that the activity of Rpn11 (an integral 26S proteasome DUB) and removal of the chain are not prerequisites for proteasomal degradation (16).
To study the possibility that the different Ub moieties in the chain have different fates and roles, we decided to synthesize a tetra-Ub α-globin, a protein known to be degraded via the Ub-proteasome system (UPS) (17, 18). In the synthetic adduct, we differentially tagged the proximal Ub and the distal Ub, allowing us to study the fate of each independently. The globin moiety was tagged with a third tag. We showed that while the proximal moiety is degraded along with the substrate, the distal moiety is recycled. However, the distal Ub moiety is required to protect the proximal moiety from removal by DUBs, because in contrast to tetra-Ub globin, which is degraded, in mono-Ub globin the Ub moiety is rapidly removed, and the protein remains stable. An important point is that the chemically synthesized tetra-Ub α-globin, composed of 472 residues, is an example of a large synthetic protein (19, 20), representing a significant advance in the field of chemical synthesis (21).
Results
Chemical Synthesis and Characterization of Free and Differentially Tagged Poly-Ubiquitinated α-Globin.
Free, mono-, di-, and tetra-Ub HA-α-globin, in which the distal and proximal Ub moieties are differentially tagged, were synthesized and characterized as described in Materials and Methods, Figs. 1 and 2, and SI Appendix.
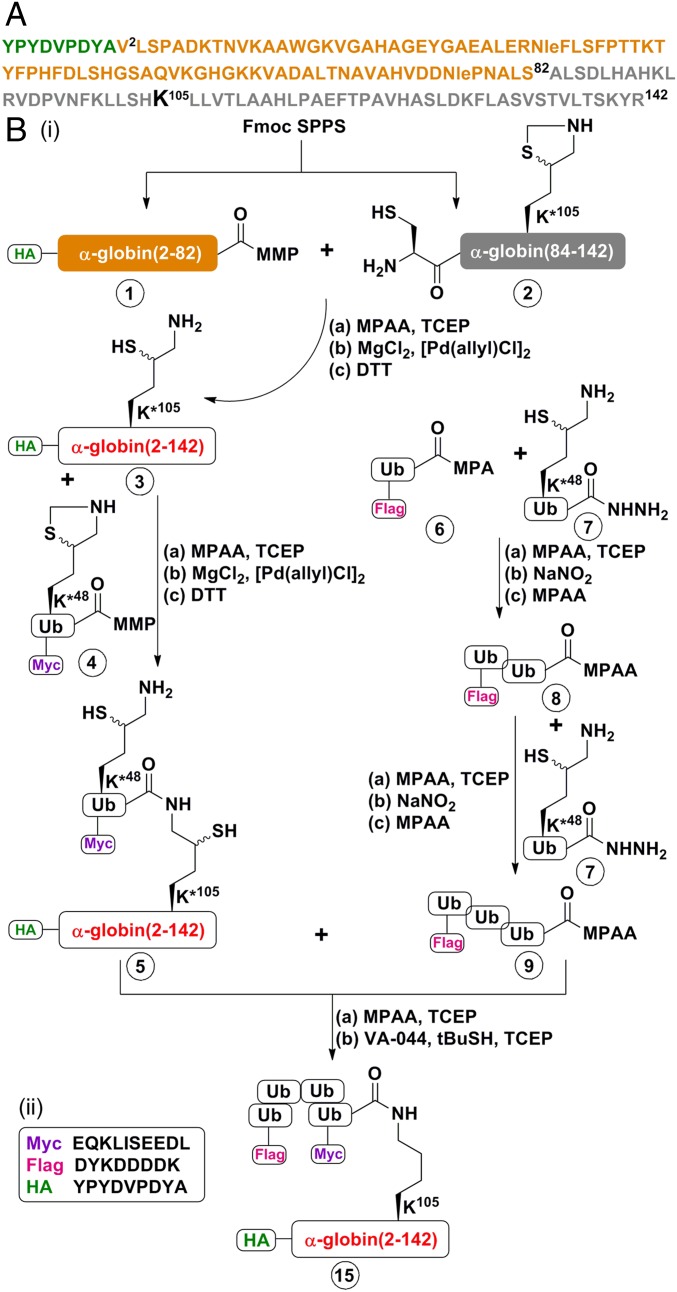
Fig. 1.
Representative scheme of the strategy used for chemical synthesis of differentially tagged tetra-Ub HA-α-globin. A detailed description of the complete synthesis of the building blocks and intermediates for the generation of HA-α-globin constructs is provided in SI Appendix, Figs. S1–S23. (A) Sequence of human HA-α-globin. The HA tag sequence is in green. The fragment 1 sequence (residues 2–82) is in light brown, and the fragment 2 sequence (residues Cys83-84–142) is in gray. Note that Ala83 in the native α-globin was substituted with Cys to enable chemical ligation and later desulfurized back to Ala. (B, i) Chemical synthesis steps of tetra-Ub HA-α-globin (construct 15). HA-α-globin K*105 (intermediate 3; K*105 denotes the presence of δ-mercaptolysine at this position) was generated from the ligation of fragments 1 and 2 of α-globin. Note that native α-globin contains a Cys residue at position 105. The ubiquitination sites of globins have not been identified; a previous study reported successful ubiquitination at position 105, generating a thiolester bond on the native Cys residue (15). In the present study, to generate an isopeptide bond, the native Cys residue was replaced with δ-mercaptolysine. Intermediate 3 was next ligated with (intermediate 4Myc-Ub(K*48)-MMP; K*48 denotes the presence of δ-mercaptolysine at this position) to obtain intermediate 5, which contains the whole HA-α-globin to which one Myc-Ub moiety is bound. In parallel, Flag-tri-Ub-MPAA (intermediate 9) was assembled from Ub monomers. Intermediates 5 and 9 were then ligated and desulfurized to obtain the final Flag-Ub-Ub-Ub-Myc-Ub-(tetra-Ub)-HA-α-globin (construct 15). (a), (b), and (c) denote the reagents used for each reaction. Details of the materials used and the synthetic steps are provided in SI Appendix, Materials and Methods). (B, ii) Sequences of Myc, Flag, and HA tags.
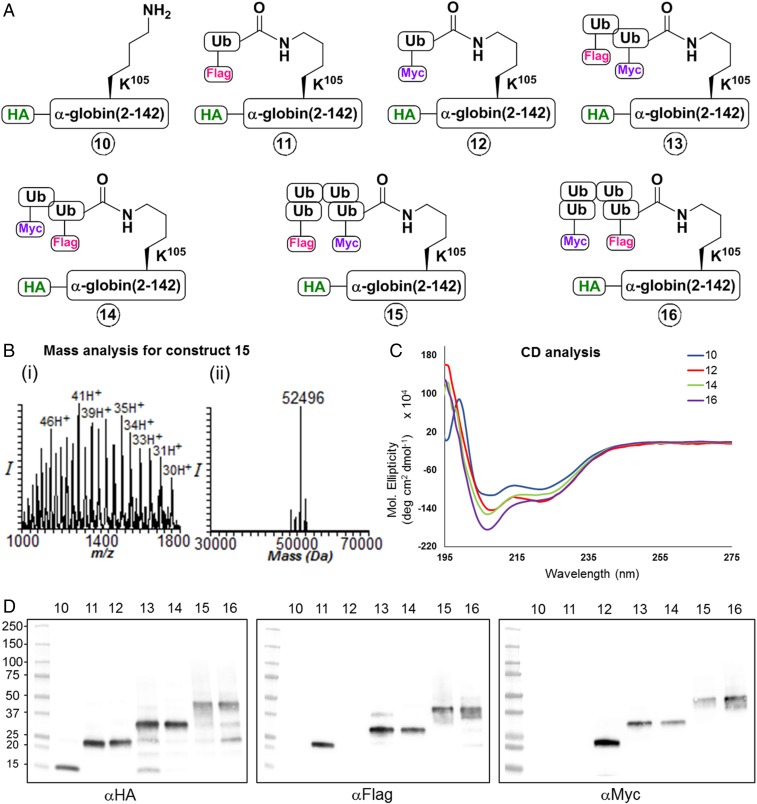
Fig. 2.
Characterization of the chemically synthesized HA-α-globin and its ubiquitinated derivatives. (A) Sketches of the seven different synthetic HA-α-globin constructs used in the study (constructs 10–16): 10, HA-α-globin; 11, Flag-Ub-HA-α-globin; 12, Myc-Ub-HA-α-globin; 13, Flag-Ub-Myc-Ub-HA-α-globin; 14, Myc-Ub-Flag-Ub-HA-α-globin; 15, Flag-Ub-Ub-Ub-Myc-Ub-HA-α-globin; 16, Myc-Ub-Ub-Ub-Flag-Ub-HA-α-globin. The synthesis of all intermediates and final constructs is described in SI Appendix, Figs. S1–S23). (B, i and ii) Representative mass analysis of tetra-Ub HA-α-globin (construct 15). (C) Circular dichroism analysis of HA-α-globin constructs 10, 12, 14, and 16. (D) Western blot analysis for HA-α-globin constructs 10–16 with αHA, αFlag, and αMyc antibodies.
Different Fates of the Distal and Proximal Ub Moieties from Tetra- and di-Ub HA-α-Globin in Crude Cell Extract.
To study the fate of the distal and proximal Ub moieties in chains of different lengths, we synthesized HA-tagged α-globin to which we attached a K48-based tetra- or di-Ub chain. In each chain, the Ub moiety adjacent to the substrate (proximal) and the one most remote from it (distal) were tagged with either Flag or Myc fused to the N-terminal residue of Ub. Thus, we synthesized two constructs for each adduct, one in which the proximal moiety was tagged with Myc and the distal one was tagged with Flag, and another in which the tagging was reversed (Figs. 2A, 3 A and B1 i and ii, and 4 A, i and ii). We incubated the two tetra-Ub adducts in the presence of rabbit fraction II, a crude cell extract containing all the UPS components required for conjugation and degradation, including E1, most of the E2s and E3s, the proteasome, and a broad array of DUBs, but does not contain Ub and certain specific E2s (12, 22, 23), and followed the fate of each tag. As shown in Fig. 3 A, 2, Flag-tagged high-molecular-mass conjugates are clearly observed when the Flag is distal but much less so when it is proximal (see the anti-Flag panel). The same was true for Myc; high-molecular-mass Myc-tagged conjugates are observed when the Myc-tagged Ub is distal, but not when it is proximal (see the anti-Myc panel). Importantly, none of the conjugates contained HA-tagged globin, which shows unequivocally that they are all derived from free Ub released by DUBs present in crude fraction II and then reconjugated to endogenous protein substrates contained in this fraction (Fig. 3A, 2, anti-HA). For each conjugate, and regardless of whether the Myc or Flag-tagged Ub was distal or proximal along the chain, the distal moiety was conjugated ∼10-fold more efficiently to the exogenous substrates compared with its proximal counterpart (as quantified in Fig. 3A, 2).
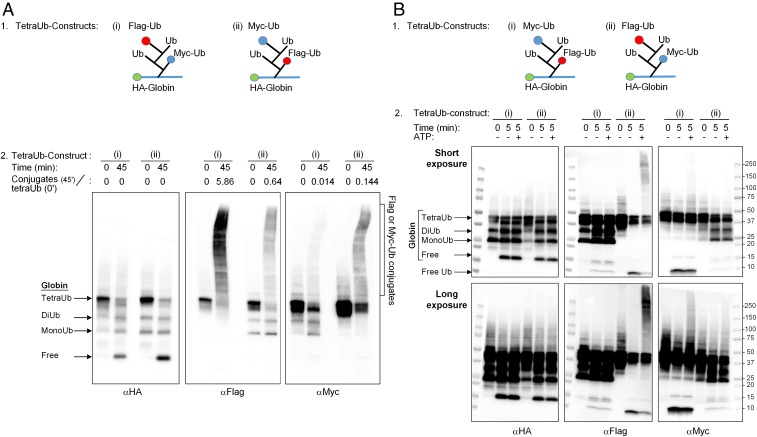
Fig. 3.
Fates of proximal and distal Ub moieties in a tetra-Ub chain. (A, 1) Sketches of differentially tagged tetra-Ub HA-α-globin: (i) tetra-Ub HA-α-globin with distal Flag-Ub and proximal Myc-Ub and (ii) tetra-Ub HA-α-globin with distal Myc-Ub and proximal Flag-Ub (also see Fig. 2A, constructs 15 and 16, respectively). (A, 2) Differentially tagged tetra-Ub HA-α-globin constructs were incubated for the indicated times in the presence of crude fraction II and ATP. Following SDS/PAGE, reactions were analyzed by Western blot analysis with αHA (Left), αFlag (Middle), and αMyc (Right). Quantification of the Flag-Ub- and Myc-Ub conjugates is presented as the ratio of the Flag or Myc signal in the generated conjugates at 45 min (minus the signal at time 0) relative to the Flag or Myc signal in the tetra-Ub HA-α-globin at time 0. (B, 1) Sketches of differentially tagged tetra-Ub HA-α-globins: (i) tetra-Ub HA-α-globin with distal Myc-Ub and proximal Flag-Ub and (ii) tetra-Ub HA-α-globin with distal Flag-Ub and proximal Myc-Ub (also see Fig. 2A, constructs 16 and 15, respectively). (B, 2) Differentially tagged tetra-Ub HA-α-globin constructs were incubated for the indicated times in the presence of fraction II with and without ATP. Following SDS/PAGE, reactions were analyzed by Western blot analysis as described in A, 2. The same image is shown with short (Upper) and long (Lower) exposures.
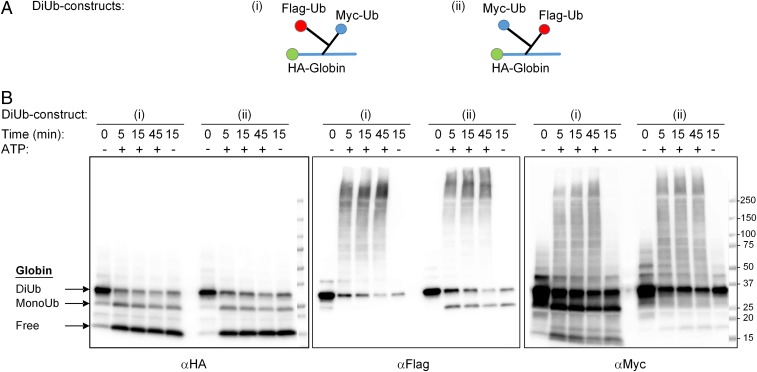
Fig. 4.
Similar fates of distal and proximal Ub moieties from di-Ub HA-α-globin. (A) Sketches of differentially tagged di-Ub HA-α-globins: (i) di-Ub HA-α-globin with distal Flag-Ub and proximal Myc-Ub and (ii) di-Ub HA-α-globin with distal Myc-Ub and proximal Flag-Ub (also seem Fig. 2A, constructs 13 and 14, respectively). (B) Differentially tagged di-Ub HA-α-globin constructs were incubated for the indicated times in the presence of crude fraction II with or without ATP. Following SDS/PAGE, reactions were analyzed by Western blot analysis with αHA (Left), αFlag (Middle), and αMyc (Right).
To demonstrate the free released Ub reconjugated to the endogenous substrates, and to show that the distal moiety is preferably released by DUBs, we incubated a larger amount of the same tetra-Ub HA-α-globin adducts for a shorter time (5 min vs. 45 min; Fig. 3B, 2 vs. Fig. 3A, 2). The free released Ub could be clearly seen when both adducts were incubated in the presence of the crude fraction. Importantly, its appearance strongly correlated with the position of the tagged Ub along the chain. Thus, a strong signal of free Ub almost always appears only when the Flag-tagged moiety or the Myc-tagged moiety is in the distal position of the adduct, not in the proximal one (Fig. 3B, 2, the anti-Flag and anti-Myc panels). It should be noted that here too, similar to the experiment described in Fig. 3A, it is mostly the distal Ub moiety that is reconjugated, as observed after only a short (5 min) incubation time.
We next carried out a similar experiment using the di-Ub adducts. As shown in Fig. 4B, both the distal and the proximal Ub moieties were reconjugated to the same extent following their deubiquitination; similar conjugate formation was observed regardless of the position of either the Flag-tagged or Myc-tagged Ub in the chain. This result strongly suggests that the proximal Ub moieties are relatively less protected from the activity of DUBs in short chains than in longer chains.
Substrates with Tetra-Ub Chains Are Degraded More Efficiently Than Substrates with Shorter Chains in Crude Cell Extract but Not by the 26S Proteasome.
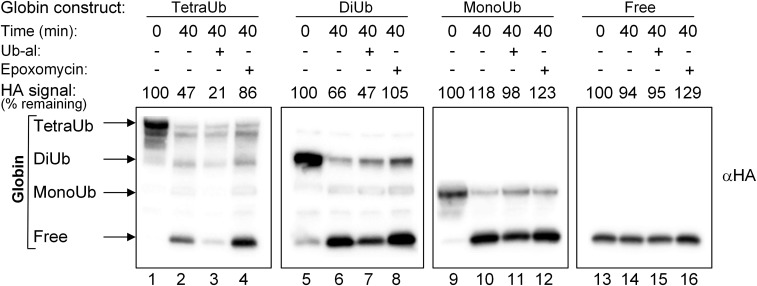
The observation that the proximal Ub moiety is more resistant to the activity of DUBs in long chain Ub adducts prompted us to test whether these substrates are degraded more efficiently. As shown in Fig. 5, the globin moiety in tetra-Ub globin is efficiently degraded, with almost no release of free globin (lane 2). The di-Ub adduct is also degraded, but less efficiently, with a significant fraction of the globin released by DUBs (lane 6). The globin moiety in the mono-Ub adduct is completely released and not degraded (lane 10). Not surprisingly, free globin is resistant to degradation (lane 14; it is not degraded in crude fraction II, which lacks Ub). The degradation is mediated by the 26S proteasome; addition of the proteasome inhibitor epoxomicin repressed degradation and, importantly, shifted the balance toward the activity of the DUBs as evidenced by the increased release of free globin (lanes 4, 8, 12, and 16).
Fig. 5.
Ub-al inhibits the release of free α-globin from Ub-globin adducts and accelerates the degradation of tetra-Ub and di-Ub HA-α-globin. Tetra-Ub HA-α-globin with distal Flag-Ub and proximal Myc-Ub, di-Ub HA-α-globin with distal Flag-Ub and proximal Myc-Ub, mono-Ub HA-α-globin with Flag-Ub, and free HA-α-globin (Fig. 2A, constructs 15, 13, 11, and 10, respectively) were incubated for the indicated times in the presence of crude fraction II and ATP. Ub-al (1 μM) and epoxomicin (40 μM) were added to the reaction mixtures as indicated. Following SDS/PAGE, reactions were analyzed by Western blot analysis using αHA. Quantification of degradation was calculated as the HA signal along the lane at 40 min minus the background signal at time 0 and expressed as percentage of the total HA signal in the HA-α-globin construct at time 0.
To study the balance between degradation and deubiquitination, two competing activities that affect the fate of the Ub adduct in the cell, we incubated the different Ub adducts in the absence and presence of Ub aldehyde (Ub-al), a known inhibitor of DUBs (24). Not surprisingly, Ub-al inhibited the release of free globin from the adducts and, consequently, increased the efficiency of degradation (Fig. 5). This increase was slightly higher for the tetra-Ub globin compared with the di-Ub adduct, likely reflecting more efficient proteasomal degradation of substrates with longer chains. In addition, the inhibitor appears to be more efficient in protecting longer chains from DUB-mediated disassembly, as evidenced by the lack of release of free globin from the tetra-Ub adduct and its marked release from the di-UB and especially the mono-Ub adducts (lanes 3, 7, 11, and 15).
We next tested the importance of Ub chain length on the efficiency of degradation in the presence of purified 26S proteasome. The crude extract contains a broad array of DUBs that play different roles in different modes of modification by Ub and in protein degradation (12, 25, 26). Therefore, we hypothesized that a long chain is more critical in the intact cellular environment than in the purified system. It should be noted that one of the subunits of the 26S proteasome is a DUB (Rpn11 in yeast and PSMD14 in mammalian cells). The proteasome also contains associated DUBs (e.g., Ubp6 in yeast, USP14 and Uch37 in mammalian cells) (4, 27, 28). These DUBs appear to have defined specific roles, and thus the deubiquitinating activity of the 26S complex is much more limited compared with that in the crude extract containing ∼100 DUBs.
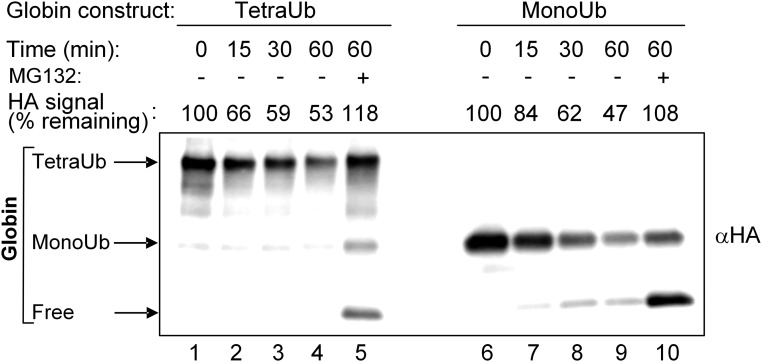
As shown in Fig. 6, the degradation of tetra-Ub and mono-Ub α-globins was similar in a cell free system containing purified 26S proteasome complex, unlike the behavior of the two adducts in the crude system (Fig. 5). Not surprisingly, the degradation was sensitive to proteasomal inhibition and shifted the balance toward the activity of DUBs, as evidenced by the increased release of free globin. Importantly, DUB activity was significantly lower in the presence of the purified proteasome than in the crude extract and could be detected only when proteasome activity was inhibited (compare Fig. 6, lanes 2–4 and 7–9 and Fig. 5, lanes 2, 6, and 10). This is again due to the multitude DUBs present in the crude extract, compared with their paucity in the proteasome.
Fig. 6.
Similar degradation of tetra-Ub HA-α-globin and mono-Ub HA-α-globin by purified 26S proteasome. Tetra-Ub HA-α-globin with distal Flag-Ub and proximal Myc-Ub and mono-Ub HA-α-globin with Flag-Ub (Fig. 2A, constructs 15 and 11, respectively) were incubated for the indicated times in the presence of purified human 26S proteasome (1.5 μg). MG132 (100 μM) was added as indicated. Following SDS/PAGE, reactions were analyzed by Western blot analysis using αHA. Quantification of degradation at the different time points was calculated as described in Fig. 5.
Discussion
The underlying mechanism and evolution of a long Ub chain as a signal for proteasomal degradation has remained elusive. Certain proteins can be recognized by a single Ub-binding subunit in the 19S regulatory complex of the 26S proteasome. Even for substrates that may require the two binding receptors (Rpn 10 and 13), the chain appears to be longer than the distance between the receptors. One can argue that the chain may be anchored to a Lys residue buried deep underneath the surface of the protein (although this is unlikely for hydrophilic Lys residues, which typically reside on the surface of proteins), but even that cannot explain the canonical long chains that constitute the proteasomal recognition signals. To approach this problem from a different angle, we used the power of synthetic organic chemistry and generated tetra-, di-, and mono-Ub derivatives of α-globin in which the distal and proximal Ub moieties in the relevant adducts were differentially tagged. We could not synthesize longer chains, owing to technical limitations. Nonetheless, synthesis of a 472-residue protein posed a formidable technical challenge. Our hope was that even a short oligo-Ub chain would be able to shed light on this mystery. The tetra-Ub adducts were incubated in fraction II, a crude extract containing all the enzymes necessary for UPS-mediated conjugation and degradation of protein substrates except for several involved in targeting specific proteins. We found that the distal moiety is released from the adduct in a reaction catalyzed by a DUB, followed by reconjugation to endogenous substrates in the extract. In striking contrast, the proximal moiety is released to a significantly lesser extent, and thus undergoes much less reconjugation. It stays on, likely to ensure processive translocation of the substrate into the proteolytic chamber of the 20S complex and its subsequent degradation. The identity of the tag—distal or proximal—had no effect on the results (Figs. 3 and 4). Importantly, when we used di-Ub adducts, the difference in behavior between the distal and proximal Ub moieties disappeared, and they were released and reconjugated to the same extent. This finding strongly suggests that proximal moieties are less protected from the activity of DUBs in short chains than in longer chains (Fig. 4). Taken together, these results indicate that in the tetra-Ub chain, the proximal moiety is more protected than the distal moiety and thus is less sensitive to the activity of DUBs. This property may make the longer chain substrates more efficient proteasomal targets. In contrast, in shorter Ub chain substrates (e.g., di-Ub), rapid removal of the proximal, relatively unprotected, Ub moieties can affect the efficient destruction of such adducts, resulting in stabilization of the substrate moiety.
The finding that the proximal moiety is more protected in longer chains than in shorter chains prompted us to test whether substrates with longer chains are also degraded more efficiently. Indeed, as shown in Fig. 5, globin moiety was degraded more efficiently in the tetra-Ub adduct than in the di-Ub adduct. Unsurprisingly, in the mono-Ub derivative, α-globin was completely released by the activity of DUBs and consequently was stable. Performing a similar experiment in the presence of purified 26S proteasome rather than a crude cell extract abrogated the effect of the long chain on the efficiency of degradation; the tetra-Ub and the mono-Ub derivatives were degraded to the same extent.
Taken together, the foregoing results define distinct roles for the proximal and distal Ub moieties in the chain. The proximal moieties are required to ensure efficient anchoring of the substrate to the proteasome and its processive translocation and degradation. The role of the distal moieties is to protect the substrate from premature removal of the proximal ones (mediated by the broad repertoire of cytosolic DUBs). Once anchoring, translocation, and initiation of degradation are secured, the distal Ub moieties can be released and recycled.
An interesting point is the balance between proteasomal degradation and deubiquitination, the two “competing” processes that affect the extent and pace of degradation of ubiquitinated proteins in the cell. The addition of a proteasome inhibitor allowed the DUBs more time to remove the Ub moieties from the substrate. Conversely, the addition of a DUB inhibitor enhanced proteasomal degradation (Figs. 5 and 6). This finding also has been reported by others in a different cell-based experiment (29).
A last point related to our findings involves the final fate of the Ub moieties. In previous work, we noted that at least part of the conjugated Ub is degraded along with the substrate, but could not identify the specific location of the moieties along the chain (11). In the present study, we have shown unequivocally that the proximal moiety is neither released from the substrate nor recycled, in striking contrast to the distal moiety. The proximal moiety appears to be degraded with the substrate, whereas the distal moiety is also degraded but much more slowly, after several cycles of deconjugation and reconjugation. It will be interesting to show that the catabolism of Ub is mediated mostly via its random incorporation into proximal positions in poly-Ub chains. It was difficult to directly demonstrate fast degradation of the proximal moiety and a slower degradation of the distal one, because the signal of either Flag or Myc was much stronger in conjugates of endogenous substrates than in the tetra-Ub synthetic adduct, which made quantification of disappearance of the signal impossible. (This is illustrated in, e.g., Fig. 3A, 2, the anti-Flag panel). This may be due to better exposure of the tag to the antibody in the longer Ub chains generated and/or the variety of conjugated endogenous fraction II substrates that differ from the single globin and well-defined adduct.
An interesting question is related to the complexity and specificity of deubiquitination compared with Ub conjugation. Clearly, ubiquitination is more complex, requiring an entire cohort of substrate-specific recognition elements—the ubiquitin ligases (E3s). Of equal importance is the chain elongation process that requires peptide bond synthesis in a continuously increasing distance from the active center of the E2-E3 complex. Therefore, it is not surprising that accessory factors have been identified (e.g., Cue1) that assist in aligning the elongating Ub chain with the Ubc7 E2 to accelerate the elongation pace in ERAD substrates (30, 31). Other authors also have suggested that the binding of Cue to the short Ub chain can protect it from premature deubiquitination. In contrast, deubiquitination is analogous to endoproteolytic activity typically mediated by a single enzyme (e.g., trypsin, chymotrypsin). Therefore, despite the myriad substrates degraded by the proteasome, only a handful of DUBs are known to be an integral part of or associated with the 26S proteasome (28). Even the close to 100 nonproteasomal cytosolic DUBs cannot account for the high substrate specificity. The specificity of the DUBs is likely related to the type of chains attached to the protein target. The chains can be of different internal linkages, mixed (i.e., different linkages within one chain), branched (i.e., two or more Ub moieties attached to a single moiety), a single moiety, or a mixture of all or some of these. It is possible that soluble DUBs catalyze the initial trimming of the substrate, which may be already bound to the proteasome, whereas the proteasomal DUBs complete the process. We admit that this process is difficult to understand given our short tetra-ubiquitinated substrate in which structural data are also missing.
The issue of deSUMOylation is different. Typically, SUMOylated substrates are not characterized by long chains and are not substrates for proteasomal degradation. However, unlike Ub, SUMO molecules take three different forms. In addition, SUMO proteases are known to be organelle-specific, with many localized to the nucleus. Therefore, it is difficult to compare deSUMOylation and deubiquitination (32, 33).
Many questions concerning deubiquitination remain open. One question is related to the energy cost of the process. Conjugation of each Ub “costs” two ATP molecules, and degradation of each Ub molecule is much costlier. Therefore, the energy cost and ascertainment of efficient and specific proteasomal degradation must be balanced. The degradation of the most proximal Ub moieties along with the substrate appears to be essential for removal; however, sparing of the more distal Ub moieties—which is necessary initially to protect the proximal ones from deubiquitination—is important for saving energy. The fine-tuning between the two processes, along with the energy cost of the proteasomal activity, contribute to the overall cost of energy necessary for protein degradation.
Experimental Procedures
Materials.
The materials used for chemical protein synthesis are described in SI Appendix, Materials. The reagents used for biochemical analyses are described in Methods. All other reagents were of analytical grade.
Methods.
Chemical synthesis and characterization of differentially tagged tetra-Ub HA-α-globin.
The detailed synthesis of tetra-Ub HAα-globin is described in Fig. 1 and SI Appendix, Figs. S1–S23. To generate HA-α-globin with a tetra-Ub chain attached to it via a native isopeptide bond and in which the distal and proximal Ub moieties are differentially tagged, we began with the chemical synthesis of α-globin. We first divided α-globin, a 142-aa protein, into two fragments: fragment 1 (Fig. 1 and SI Appendix, Fig. S1), with HA tag (YPYDVPDYA) connected N-terminally to residues 2–82 of the protein (we omitted the initiator Met), and fragment 2 (Fig. 1 and SI Appendix, Fig. S2), containing protein residues 83–142. Both fragments were prepared using standard 9-fluorenylmethoxycarbonyl–based-solid phase peptide synthesis. Specifically, fragment 1 was prepared using an N-acyl-benzimidazolinone approach, which enabled the introduction of thioester functionality in fragment 1. In parallel, fragment 2 was synthesized similarly, with Ala83 substituted with Cys for native chemical ligation (NCL), with the aim of desulfurizing it back to the native Ala at the completion of synthesis (SI Appendix, Fig. S2). In fragment 2, Cys105 was replaced with δ-mercaptolysine (K*) to enable ubiquitination of α-globin via a native isopeptide bond (15). The purified fragments were ligated to one another using standard NCL. This was followed by thiazolidine deprotection of the δ-mercaptolysine (K*105) residue using Pd(allyl)Cl2 in one-pot fashion (34) to yield synthetic intermediate 3 (Fig. 1 and SI Appendix, Fig. S3). Myc-Ub(K*48) thioester intermediate 4 (Fig. 1 and SI Appendix, Fig. S6) was synthesized as reported previously for unmodified Ub thioesters (15). Following its synthesis and purification, it was ligated to intermediate 3, and K*48 of Ub was deprotected in one pot to obtain intermediate 5 (Fig. 1 and SI Appendix, Fig. S12).
To prepare the tetra-Ub derivative, we synthesized in parallel Flag-tagged tri-Ub thioester (intermediate 9; Fig. 1 and SI Appendix, Fig. S19). For this, we ligated Flag-Ub thioester (intermediate 6; Fig. 1 and SI Appendix, Fig. S5) with Ub-NHNH2 (intermediate 7; Fig. 1 and SI Appendix, Fig. S4) bearing K*48, followed by one-pot oxidative switching of the hydrazide to 4-mercaptophenylacetic acid (MPAA) thioester derivative (intermediate 8; Fig. 1 and SI Appendix, Fig. S18). Next, Flag-di-Ub-MPAA intermediate 8, was ligated with intermediate 7, followed by hydrazide switching to yield Flag-tri-Ub-MPAA (intermediate 9). Finally, intermediate 9 was ligated with intermediate 5, followed by one-pot desulfurization to yield the differentially tagged tetra-Ub HA-α-globin (construct 15; Figs. 1 and 2 and SI Appendix, Fig. S20). Using the same strategy, we were able to synthesize different analogs of ubiquitinated α-globin, including free globin (Fig. 2A, constructs 10–16 and SI Appendix; synthesis of constructs 10, 11, 12, 13, 14, 15, and 16, is described under Figs. S3, S11, S12, S15, S17, S20, and S23, respectively).
The molecular mass of the tetra-Ub globin was confirmed by mass spectrometry (Fig. 2B, i and ii). The constructs were folded in a buffer containing 50 mM Tris and 150 mM NaCl pH 7.4, and the proper folding was confirmed using circular dichroism (Fig. 2C). The samples were first dissolved in 6 M Gn·HCl (samples 5% of the total volume) and then diluted in the folding buffer. The tagging was confirmed following SDS/PAGE and Western blot analysis using the appropriate antibodies (Fig. 2D).
Degradation of chemically synthesized α-globin constructs in a cell-free system.
α-globin constructs 10–16 (Fig. 2A; ∼100 ng) were incubated in the presence of rabbit reticulocyte crude fraction II (25 μg), prepared as described previously (22, 35), or purified human 26S proteasome (1.5 μg; Enzo Life Sciences), as indicated. Reactions were carried out in a volume of 15 μL containing 50 mM Tris⋅HCl pH 7.6, 5 mM MgCl2, 2 mM DTT, and 5 mM ATP (as indicated). Reactions carried out in the absence of ATP were incubated in the presence of fraction II that had been pretreated with 4.0 μg hexokinase (Roche) and 10 mM 2-deoxy-d-glucose for 5 min at 37 °C. Ub-al (1 μM; Enzo Life Sciences), MG132 (100 μM; EMD Millipore), or epoxomicin (40 μM; Sigma-Aldrich) was added as indicated. Mixtures were incubated at 37 °C, with incubation terminated at the indicated times by the addition of fourfold concentrated sample buffer. They were then resolved via SDS/PAGE (4–20% gradient; Bio-Rad), followed by Western blot analysis on 0.2- μm PVDF using αHA (BioLegend), αFlag (M2; Sigma-Aldrich), and αMyc (9E10; Santa Cruz Biotechnology) mouse monoclonal antibodies. Reactions detected with more than one antibody were resolved on separate gels, and each was detected independently. Enhanced chemiluminescence imaging was performed using the Fusion FX Western blot and chemiluminescence imaging system (Vilber). Quantification of the bands was performed using ImageQuant TL image analysis software (GE Healthcare).
Supplementary Material
Acknowledgments
Research in the A.C. laboratory is supported by grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Israel Science Foundation (ISF), the German Israeli Foundation for Research and Development (GIF), the University of Michigan and Technion Collaborative Program, and a special fund for research at Technion established by Mr. Albert Sweet. A.C. is an Israel Cancer Research Fund USA Professor. A.B. is supported by a grant from the ISF-National Science Foundation of China Joint Program and is an incumbent of the Jordan and Irene Tark Academic Chair.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 7614.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822148116/-/DCSupplemental.
References
- 1.Mali SM, Singh SK, Eid E, Brik A. Ubiquitin signaling: Chemistry comes to the rescue. J Am Chem Soc. 2017;139:4971–4986. doi: 10.1021/jacs.7b00089. [DOI] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Kulathu Y, Komander D. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 4.Finley D, Chen X, Walters KJ. Gates, channels, and switches: Elements of the proteasome machine. Trends Biochem Sci. 2016;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budenholzer L, Cheng CL, Li Y, Hochstrasser M. Proteasome structure and assembly. J Mol Biol. 2017;429:3500–3524. doi: 10.1016/j.jmb.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saeki Y. Ubiquitin recognition by the proteasome. J Biochem. 2017;161:113–124. doi: 10.1093/jb/mvw091. [DOI] [PubMed] [Google Scholar]
- 8.Braten O, et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci USA. 2016;113:E4639–E4647. doi: 10.1073/pnas.1608644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livneh I, Kravtsova-Ivantsiv Y, Braten O, Kwon YT, Ciechanover A. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. BioEssays. 2017;39:1700027. doi: 10.1002/bies.201700027. [DOI] [PubMed] [Google Scholar]
- 10.Tsuchiya H, et al. Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat Commun. 2018;9:524. doi: 10.1038/s41467-018-02869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shabek N, Herman-Bachinsky Y, Ciechanover A. Ubiquitin degradation with its substrate, or as a monomer in a ubiquitination-independent mode, provides clues to proteasome regulation. Proc Natl Acad Sci USA. 2009;106:11907–11912. doi: 10.1073/pnas.0905746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci USA. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Haj-Yahya M, et al. Synthetic polyubiquitinated α-synuclein reveals important insights into the roles of the ubiquitin chain in regulating its pathophysiology. Proc Natl Acad Sci USA. 2013;110:17726–17731. doi: 10.1073/pnas.1315654110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemantha HP, et al. Nonenzymatic polyubiquitination of expressed proteins. J Am Chem Soc. 2014;136:2665–2673. doi: 10.1021/ja412594d. [DOI] [PubMed] [Google Scholar]
- 16.Singh SK, et al. Synthetic uncleavable ubiquitinated proteins dissect proteasome deubiquitination and degradation, and highlight distinctive fate of tetraubiquitin. J Am Chem Soc. 2016;138:16004–16015. doi: 10.1021/jacs.6b09611. [DOI] [PubMed] [Google Scholar]
- 17.Bank A, O’Donnell JV. Intracellular loss of free alpha chains in beta thalassemia. Nature. 1969;222:295–296. doi: 10.1038/222295a0. [DOI] [PubMed] [Google Scholar]
- 18.Shaeffer JR. Turnover of excess hemoglobin alpha chains in beta-thalassemic cells is ATP-dependent. J Biol Chem. 1983;258:13172–13177. [PubMed] [Google Scholar]
- 19.Tang S, et al. Practical chemical synthesis of atypical ubiquitin chains by using an isopeptide-linked Ub isomer. Angew Chem Int Ed Engl. 2017;56:13333–13337. doi: 10.1002/anie.201708067. [DOI] [PubMed] [Google Scholar]
- 20.Xu W, et al. Total chemical synthesis of a thermostable enzyme capable of polymerase chain reaction. Cell Discov. 2017;3:17008. doi: 10.1038/celldisc.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondalapati S, Jbara M, Brik A. Expanding the chemical toolbox for the synthesis of large and uniquely modified proteins. Nat Chem. 2016;8:407–418. doi: 10.1038/nchem.2476. [DOI] [PubMed] [Google Scholar]
- 22.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 23.Blumenfeld N, et al. Purification and characterization of a novel species of ubiquitin-carrier protein, E2, that is involved in degradation of non-“N-end rule” protein substrates. J Biol Chem. 1994;269:9574–9581. [PubMed] [Google Scholar]
- 24.Hershko A, Rose IA. Ubiquitin-aldehyde: A general inhibitor of ubiquitin-recycling processes. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 26.Fraile JM, Quesada V, Rodríguez D, Freije JMP, López-Otín C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 27.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Poot SAH, Tian G, Finley D. Meddling with fate: The proteasomal deubiquitinating enzymes. J Mol Biol. 2017;429:3525–3545. doi: 10.1016/j.jmb.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee B-H, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Delbrück M, et al. The CUE domain of Cue1 aligns growing ubiquitin chains with Ubc7 for rapid elongation. Mol Cell. 2016;62:918–928. doi: 10.1016/j.molcel.2016.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Bagola K, et al. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol Cell. 2013;50:528–539. doi: 10.1016/j.molcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ovaa H, Vertegaal ACO. Probing ubiquitin and SUMO conjugation and deconjugation. Biochem Soc Trans. 2018;46:423–436. doi: 10.1042/BST20170086. [DOI] [PubMed] [Google Scholar]
- 34.Maity SK, Jbara M, Mann G, Kamnesky G, Brik A. Total chemical synthesis of histones and their analogs, assisted by native chemical ligation and palladium complexes. Nat Protoc. 2017;12:2293–2322. doi: 10.1038/nprot.2017.049. [DOI] [PubMed] [Google Scholar]
- 35.Hershko A, Heller H, Elias S, Ciechanover A. Components of the ubiquitin-protein ligase system: Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.