Significance
Efficient nonviral delivery of macromolecules including mRNA, DNA plasmids, Cas9 ribonucleoproteins, and functional protein into both adherent cells and suspension cells with high cell viability is crucial for cellular manipulation, cellular imaging, and medical applications. However, the conventional delivery methods are limited to a certain range of molecules and cell types and often reduce cell viability. Here, we demonstrate effective delivery of macromolecules by nanopore electroporation (NanoEP) using a water-filter nanoporous membrane. As compared with conventional electroporation that porates the entire cell, NanoEP induces localized electroporation on a nanosized area of the cell membrane, which preserves cell viability. Additionally, our delivery method does not require specialized equipment, which allows for easy access across laboratories and medical facilities.
Keywords: intracellular delivery, electroporation, nanotechnology, nanopore, genome engineering
Abstract
We present a simple nanopore-electroporation (NanoEP) platform for delivery of nucleic acids, functional protein, and Cas9 single-guide RNA ribonucleoproteins into both adherent and suspension cells with up to 80% delivery efficiency and >95% cell viability. Low-voltage electric pulses permeabilize a small area of cell membrane as a cell comes into close contact with the nanopores. The biomolecule cargo is then electrophoretically drawn into the cells through the nanopores. In addition to high-performance delivery with low cell toxicity, the NanoEP system does not require specialized buffers, expensive materials, complicated fabrication processes, or cell manipulation; it simply consists of a generic nanopore-embedded water-filter membrane and a low-voltage square-wave generator. Ultimately, the NanoEP platform offers an effective and flexible method for universal intracellular delivery.
Delivery of biomacromolecules, such as mRNA, DNA, and proteins, into living cells is crucial for cellular manipulation (1, 2), genome engineering (3–5), cellular imaging (6), and medical applications (7, 8). While viral-mediated approaches efficiently transfect DNA into various cell types, their application remains a considerable safety concern (8, 9). Chemical-mediated delivery methods, including lipofectamine (LFN) and positively charged polymers, allow intracellular delivery of biomolecules. However, these methods are often toxic to cells and they are limited to particular molecules and cell types (8). Bulk electroporation (BEP) has been used for effective DNA transfection in suspension cells (10) and it has shown promise for genome engineering of T cells (5). However, to permeabilize individual cell membranes, the bulk cell solution is subjected to a strong electric field that leads to the destruction of a large population of cells due to excessive pore formation on the cell membrane (8, 11).
An alternate approach employs the diminutive scale of nanostructures to induce localized electroporation, which allows for the effective use of a low-voltage electric field. Therefore, cell damage may be reduced (12, 13). Several groups have demonstrated nanomaterials-meditated electroporation to transfect adherent cells while maintaining high cell viability (12, 14–20). “Three-dimensional nanochannel” electroporation delivers small molecules and transfects large DNA plasmids into mouse embryonic fibroblasts with >90% cell viability (15); “nanostraw” electroporation enables 80% plasmid transfection with cell viability of >95% (18, 20, 21). Although these promising results indicate that the cell toxicity of electroporation can be minimized by careful control of the electric field distribution, the equipment and sophisticated fabrication needed for realizing these devices are complicated and costly. This may hamper the accessibility of the nanostructured delivery technology and constrain its adoption by medical and research laboratories. Therefore, an easily employable nano-based electroporation delivery system is needed. Appropriately, Kang et al. (16) established on-chip localized electroporation based on a nanoporous membrane. While the device retains >90% of cell viability and can be easily fabricated, its delivery efficiency is only up to 50% for DNA plasmid. Moreover, the technology is limited to adherent cells, which prevents it from gaining a broad application in basic research and clinics.
Here, we demonstrate a simple nanopore-electroporation (NanoEP) method building on a nanopore-embedded water filter for the universal delivery of nucleic acids, functional proteins, and Cas9 ribonucleoproteins (RNPs) (Fig. 1). The local electric field is enhanced through the nanopores, and therefore close contact between the cell membrane and nanopores is key for highly efficient localized electroporation (16, 19, 20). To improve delivery, we tested different surface coatings to facilitate the formation of tight cell membrane/nanopore contact. With an optimal surface coating, NanoEP is able to delivery macrobiomolecules to both adherent and suspension cells with up to 80% transfection efficiency and >95% cell viability.
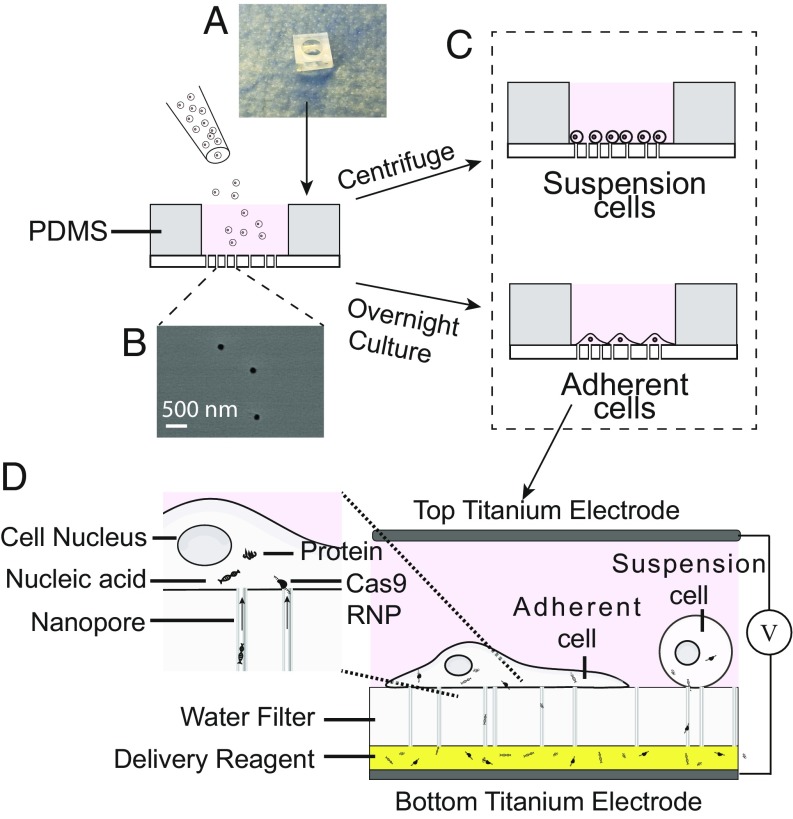
Fig. 1.
Schematic of NanoEP device and delivery process. (A) Nanopore-embeded PC water-filter membrane sealed at the base of PDMS holder. (B) Randomly distributed 100-nm nanopores on the commercial PC membrane are shown in the scanning electronic microscopic image. (C) Cells of interest are either overnight-cultured (adherent cells) or are directly centrifuged (suspension cells) in the NanoEP device before electroporation. (D) Delivery reagent is first loaded onto the bottom electrode plate before the NanoEP device is placed onto it. Square-wave electric pulses are then applied to the cells by sandwiching the device between two titanium electrodes.
Results and Discussion
NanoEP Delivery Process.
The NanoEP device consists of two flat titanium electrodes and a polydimethylsiloxane (PDMS) holder with a track-etched polycarbonate (PC) water-filter membrane embedded with nanopores 100 nm (±10 nm) in diameter (Methods and Fig. 1 A and B). The PC membrane is commercially available, which allows for easy fabrication of the device. Cells of interest are either cultured overnight (adherent cells) or centrifuged (suspension cells) in the NanoEP device that is precoated with poly-l-lysine (PL) or fibronectin (FN) of 1 to 100 ng/mL (Fig. 1C). To perform delivery, we first load 2 to 5 µL of the delivery reagent at serial concentrations on the bottom titanium electrode and immediately place the NanoEP device on top of the delivery reagent. The second titanium electrode is placed on top of the NanoEP device and put in contact with the cell culture media. Finally, DC square-waved electric pulses of 20 Hz, 200-µs pulse durations, and voltage between 15 and 90 V are applied to the cells via the two electrodes for 20 s (Methods and Fig. 1D). As supported by our numerical simulation, the porous membrane structure enhances the local electric field in and around the nanopores (10, 11). Cell membranes that form tight contact to the nanopores experience adequate electric field (>3 kV/cm) for poration under >15-V dc pulses (SI Appendix, Fig. S1). Since the electric field diminishes quickly away from the nanopores, there will only be a weak to no electric field that is incapable of breaking down the cell membrane of cells not in contact with the nanopores. In comparison with BEP, which disrupts the integrity of the whole cell membrane, the NanoEP with a membrane of 2 × 107 pores per cm2 pore density porates only 0.05% (±0.025%) of the whole cell membrane, which significantly reduces cell damage. Conveniently, native cell culture media is used as the electroporation buffer. The transfected cells can be either directly cultured in the NanoEP device or transferred to routine cell-culture dishes for further analysis.
Highly Efficient Transfection of Nucleic Acids into Adherent and Suspension Cells.
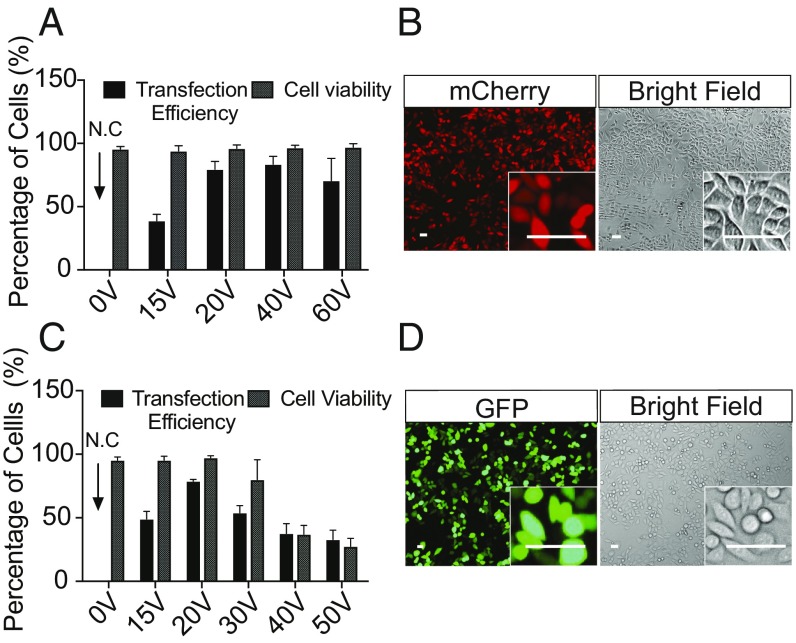
To evaluate if the NanoEP could efficiently deliver macromolecules into living cells, we first transfected a HeLa cell line with 100 ng/mL mCherry mRNA (Fig. 2 A and B) and 200 ng/mL GFP-expressing plasmid (Fig. 2 C and D). We tested a range of voltage intensities from 15 to 60 V to find the optimal field strength. After delivery, cells were incubated for 6 h to allow protein expression (Methods). Cell viability was analyzed by trypan blue exclusion method before transfection efficiency analysis (Methods). The mRNA and DNA expression was analyzed by fluorescence imaging analysis and flow cytometry assay. The results show up to 80% transfection efficiency at 20 V for both mRNA and DNA plasmids with >95% cell viability (Fig. 2 and SI Appendix, Fig. S2). However, 15 V is insufficient for effective transfection, suggesting that there might be a gap weakening the field strength between the cell membrane and the nanopore opening (SI Appendix, Fig. S1). This gap could be caused by surface proteins and molecules that protrude from the cell membrane (22). Increasing the voltage to 60 V maintains the mRNA transfection efficiency and cell viability at 80% and >95%, respectively, indicating that NanoEP allows for effective mRNA delivery and limits the cellular damage even at high voltage intensities. However, as voltage increases from 20 to 60 V, both transfection efficiency and cell viability for DNA plasmid delivery decrease significantly (Fig. 2C and SI Appendix, Fig. S2). This drop could be due to the high toxicity of DNA plasmid at high dosages. As voltage increases, more DNA plasmid is electrophoretically drawn into cells, which causes cell death (10).
Fig. 2.
Transfection of mCherry mRNA and GFP-expressing plasmid into HeLa cells. (A) The average efficiency of mRNA transfection to HeLa cells at and above 20 V is >80% (error bars indicate SDs of experimental replicates, n = 2 to 3). Transfection at 15 V is significantly lower than other three conditions (P < 0.05, ANOVA test). The transfection efficiencies at and above 20 V are independent to the delivery voltages (P > 0.05, ANOVA test). The cell viabilities for the tested delivery conditions are >95%. N.C, negative control (B) Fluorescent and bright-field microscopic cell images for HeLa mCherry expression were taken from the transfection at 20 V. (C) The best efficiency of plasmid transfection is >80% at 20 V; 15 V is insufficient to porate cell membrane, resulting in less than 50% transfection. Voltage greater that 20 V is less efficient. The cell viabilities drop to <30% as the voltage increase to 60 V (error bars indicate SDs of experimental replicates, n = 2 to 3). (D) Fluorescent and bright-field microscopic cell images for GFP expression were taken from the transfection at 20 V. (Scale bars, 50 μm.)
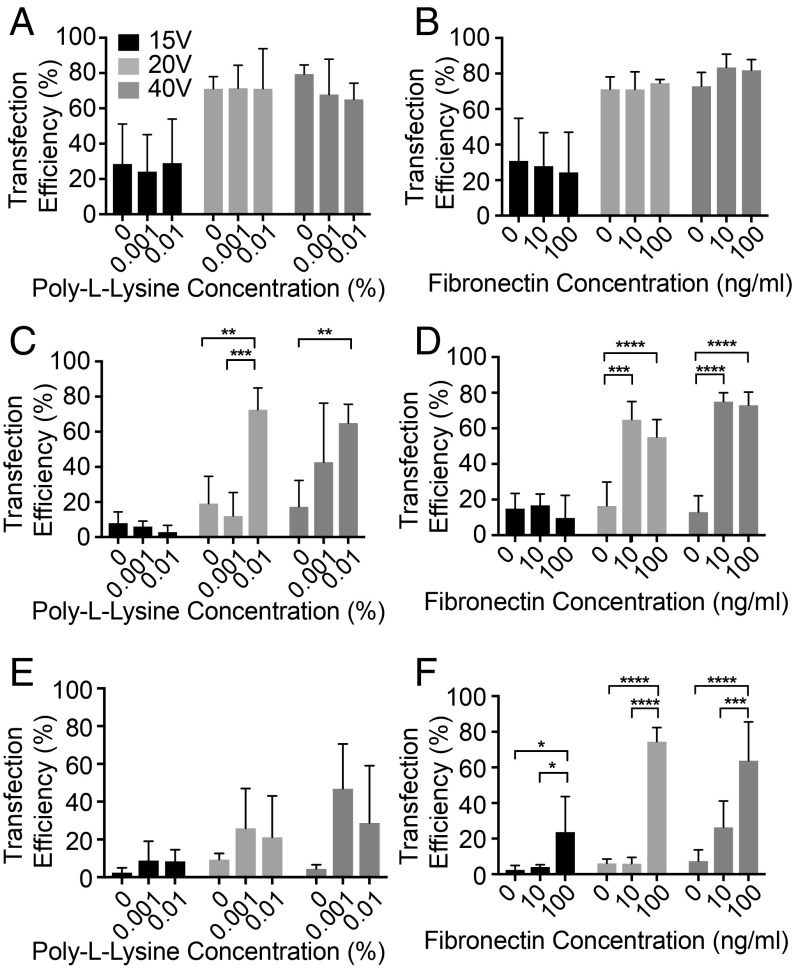
The electric field strength diminishes quickly away from the nanopore (SI Appendix, Fig. S1). A tight cell/nanopore interface is crucial for effective electroporation. Next, we examined if PL (Fig. 3 A, C, and E) and FN (Fig. 3 B, D, and F) coating could enhance cell/nanopore contact and improve NanoEP-mediated delivery efficiency (Methods). Besides HeLa cells, we selected human embryonic kidney (HEK 293) cells and a mouse embryo fibroblast (3t3) cell line that are less adhesive than HeLa cells to test the efficacy of coating (23, 24). The NanoEP culture chamber was first coated with 0 to 0.1% PL or 0 to 100 ng/mL FN for 4 h. mCherry mRNA of 100 ng/mL was delivered into the cells by applying 15-, 20-, or 40-V electric pulses for 20 s. The results show that the surface coated with 100 ng/mL FN allows >75% transfection in all tested cell types at both 20 V and 40 V (Fig. 3 B, D, and F). Transfection efficiency of HeLa remains ∼80% for PL and FN coating and noncoating surface at 20 V and 40 V, suggesting that HeLa cells are able to form tight contact to the nanopore without additional binding support (Fig. 3 A and B). With the 0.1% FN surface coating, the mRNA transfection efficiency in HEK and 3T3 cells is 80% and 75%, respectively. Although 0.01% and 0.001% PL and 10 ng/mL FN coating also improved the transfection efficiency of HEK 293 cells to up to 80% (Fig. 3 C and D), no significant transfection improvement for 3T3 cells was observed (Fig. 3 E and F). Note that even with 100 ng/mL FN surface coating, a 15-V pulse is insufficient for high-rate mRNA transfection of all tested cell types. The efficiency of plasmid DNA transfection for the two cell types is more than 65%, and 40%, respectively (SI Appendix, Fig. S3 A and B), with cell viability of >90% for both cell types (SI Appendix, Fig. S3C).
Fig. 3.
Analysis of PL and FN surface coating on the NanoEP device. (A–F) mCherry mRNA transfection efficiency of HeLa (A and B), HEK (C and D), and 3T3 cells (E and F) on PL- (A, C, and E) and FN- (B, D, and F) coated surfaces at 15-, 20-, and 40-V electroporation voltages (error bars indicate SDs. of experimental replicates, **P < 0.01; ***P < 0.001, ****P < 0.0001, post hoc Tukey test, n = 3).
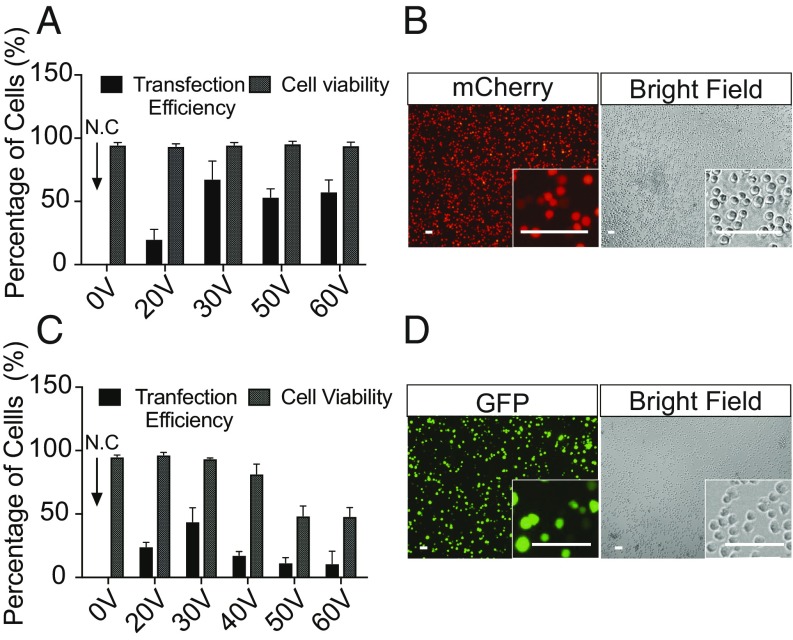
Intracellular delivery of biomacromolecules into T cells such as chimeric antigen receptor T cells is a limiting step for implementing immunotherapy. Therefore, we evaluated whether the NanoEP platform could be used for the effective transfection of nonadherent Jurkat cells, an immortal human T cell lymphoma cell line used to model patient-derived T cells. We transfected Jurkat cells with either mCherry mRNA or GFP plasmid DNA (Fig. 4). To enable the cells to form tight contact with the nanoporous PC membrane, they were centrifuged in the NanoEP culture chamber at 150 × g for 5 min (Methods). We then transfected the Jurkat cells under different voltages (ranging from 20 V to 60 V). Cell viability was analyzed before transfection efficiency analysis 24 h after delivery by trypan blue exclusion methods. The results show the transfection efficiency of the mRNA and the DNA plasmid into Jurkat cells is as high as 75% and 50%, respectively, with cell viabilities of >95% (Fig. 4 and SI Appendix, Fig. S4). We found the most effective delivery at 30 V (Fig. 4 B and D). The higher voltage required for delivery in Jurkat than in adherent cells suggests that the gap between the cell membrane and nanopore is larger. The DNA plasmid transfection efficiency and cell viability drops as the voltage increases (Fig. 4C), which is consistent with the previous demonstration in HeLa cells (Fig. 2C).
Fig. 4.
Transfection of mCherry mRNA and GFP-expressing plasmid into Jurkat cells. (A–D) The best transfection efficiency for mRNA and DNA plasmid are 75% (A) and 52% (C) at 30 V with >95% of cell viability. The efficiency and cell viability of mCherry mRNA to Jurkat cells is independent of voltages at and above 30 V (A, P > 0.05, ANOVA test; error bars indicate SDs of experimental replicates, n = 2 to 3), while the transfection and cell viability of plasmid DNA is voltage-dependent (C, P < 0.05, ANOVA test; error bars indicate SDs of experimental replicates, n = 2 to 3). N.C, negative control. Fluorescent and bright-field cell images for mCherry (B) and GFP expression (D) were taken from the transfection at 30 V. (Scale bars, 50 μm.)
Effective Delivery of Functional Proteins and Cas9 RNPs.
To test if the NanoEP device could be used for protein delivery, we delivered mCherry-tagged cytosolic fragment of Stromal Interaction Molecule 1 (STIM 1, 98 kDa; Methods) into overnight-cultured HEK cells at different voltages ranging from 30 to 90 V for 20 s. After delivery, cells were transferred into a 96-well plate for further cell imaging analyses (Fig. 5). The imaging results show that 30-V NanoEP of the STIM1 protein allows up to 80% delivery efficiency (Fig. 5A and SI Appendix, Fig. S5). Increasing the voltage intensity does not improve the delivery efficiency of STIM1 (SI Appendix, Fig. S5).
Fig. 5.
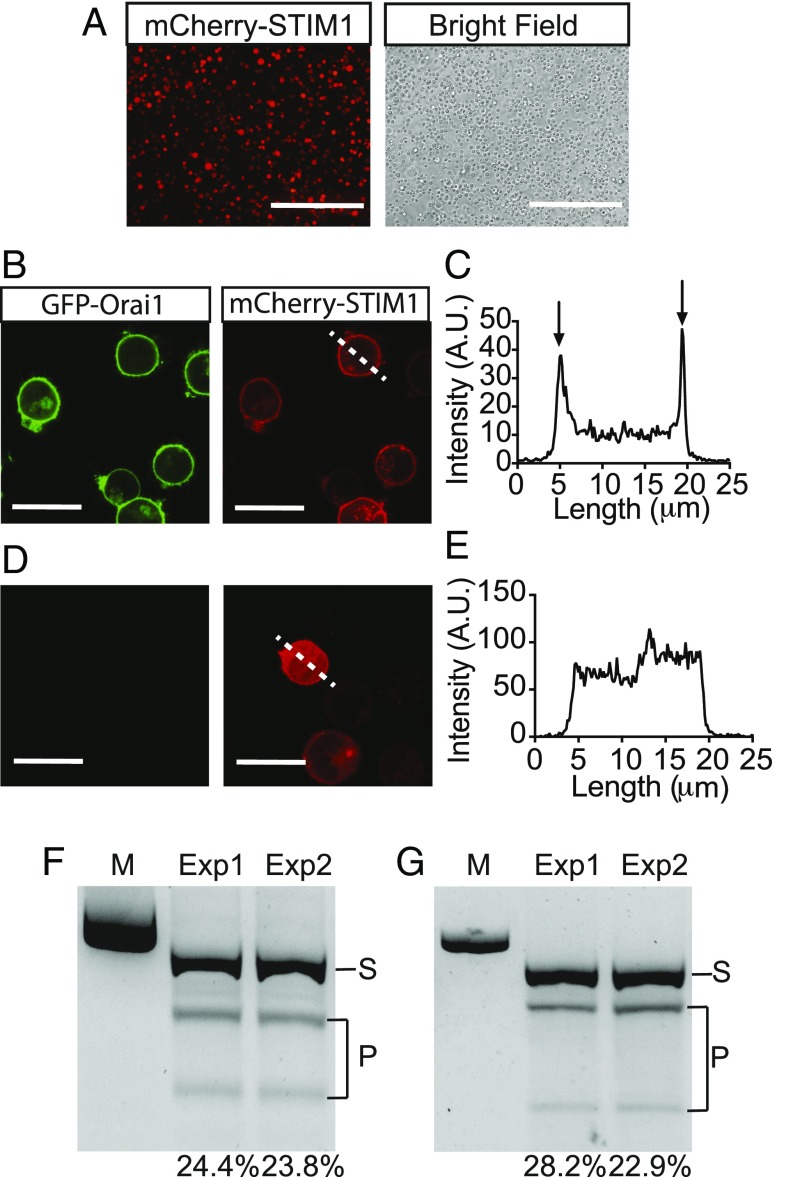
Intracellular delivery of functional mCherry-tagged STIM1 protein and Cas9-sgRNA RNPs. (A) Fluorescent cell images show mCherry-tagged STIM1 protein is effectively delivered into HEK cells. (Scale bars, 400 μm.) (B) Colocalization of transfected mCherry-tagged STIM1 protein with the GFP-Orai1 protein on the cell membrane indicates that functional STIM1 interacts with the membrane protein GFP-Orai1. (Scale bars, 20 μm.) (C) The two peaks at the edges (arrows) of the cell across the dashed trace line indicate mCherry-tagged STIM1 protein binds to the GFP-Orai1 protein on the cell membrane. (D) In the control HEK cells, the STIM1 protein is uniformly distributed throughout the cytosol. (Scale bars, 20 μm.) (E) No shape peak is observed across the dashed trace, indicating no mCherry signal accumulates on the edges of the control HEK cell. (F and G) SpyCas9-sgRNA RNPs targeting the PPIB locus were delivered into HEK (F) and Jurkat cells (G) via the NanoEP device. Each experiment was preformed twice (Exp1 and Exp2). T7E1 cleavage assay suggests the averaged gene editing efficiencies of HeLa and Jurkat cells were 24.1 ± 0.3% and 25.6 ± 2.65%, respectively. S indicates original PCR products (505 bp); P indicates T7E1 cleavage products of edited DNA. M indicates 639-bp size marker.
Retaining protein functionality is critical for effective protein delivery (6, 25). We therefore investigated if STIM1 remained functional in cells after transfection via our NanoEP device by delivering mCherry-tagged STIM1 into GFP-tagged Orai1-expressing cells. Wild-type HEK293 cells were used as a control. Since the cytosolic domain of STIM1 has strong binding affinity to the membrane protein Orai1 calcium channel (26), we expected that GFP and mCherry would colocalize on the cell membrane after delivery of mCherry-STIM1 into the Orai1-GFP–expressing cells if STIM1 maintains functionality. Indeed, the mCherry-STIM1–delivered cells show the mCherry signal accumulates on the cell membrane (Fig. 5 B and C), suggesting that functional STIM1 protein interacts with the membrane-expressed GFP-Orai1 protein. In the control cells, as expected, the mCherry signal is uniformly distributed in the cytoplasm (Fig. 5 D and E), indicating STIM1 does not bind to cell membrane in the absence of Orai1 protein.
CRISPR technology is a powerful tool for genome editing (27, 28). Here, we show that NanoEP also allows for the delivery of Cas9 RNPs to both adherent and suspension cells. We designed and constructed the Cas9 RNPs to knock out PPIB, a housekeeping gene, and delivered the Cas9 RNPs into HeLa and Jurkat cells with the same protocol as was previously used for nucleic acids and proteins (Methods). The gene-editing efficiency was measured via T7 endonuclease (T7E1) cleavage assay 48 h after transfection. The bands of 330 and 175 bp displayed are the products from the edited portion of 505-bp PCR products, indicating a single-site mutation occurred in the PPIB gene after Cas9 RNPs genome editing. The estimated editing efficiency in HeLa (Fig. 5F) and Jurkat (Fig. 5G) cells is ∼24.1% and 25.6%, respectively. The editing efficiencies of PPIB targeting Cas9 RNPs in HeLa and Jurkat cells without NanoEP were treated as negative control. No bands of 330 and 175 bp are found in the DNA gel after T7E1 treatment (SI Appendix, Fig. S6).
Cell-Viability Analysis of NanoEP.
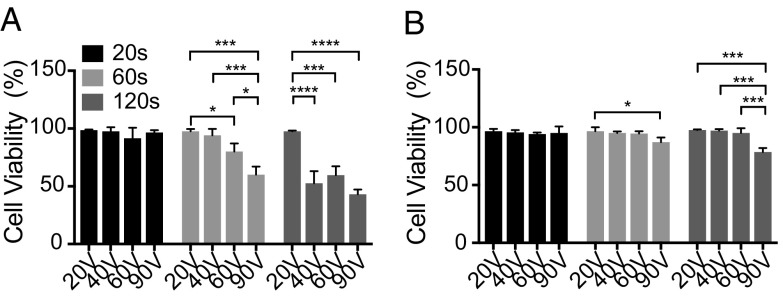
Although general pulse conditions (20 to 60 V, for 20 s) allow for >95% cell viability, cell-viability dependence on voltage intensity and delivery duration have not been examined. To systematically study the relationship between NanoEP delivery conditions and cell viability, we evaluated the cell viability in both HeLa and Jurkat after NanoEP with different combinations of voltage intensity (from 20 to 90 V) and delivery duration (from 20 to 120 s). Since the cargos (mRNA, plasmid, and proteins) may disrupt cellular homeostasis and impact cell viability, no cargo is delivered in this cell-viability analysis. After NanoEP of different conditions, both HeLa and Jurkat were resuspended in normal cell culture dish and incubated under 5% CO2, 37 °C overnight before viability testing. Short delivery duration (20 s) preserves >95% cell viability for both cell types at all voltage intensities (Fig. 6); 20 V and 40 V continue to allow for >95% cell viability for both cell types when the delivery duration increases to 60 s. However, HeLa cell viability starts to decrease to 80% and 60% at 60 V and 90 V, respectively (Fig. 6A). The viability of Jurkat also drops to 85% at 90 V and 60-s delivery duration (Fig. 6B). The HeLa cell viability decreases to 50% at 40 V, 60 V, and 90 V as the delivery duration increases to 120 s. The high delivery duration does not impact Jurkat viability at 20 to 60 V, but it drops to 75% at 90 V. Notably, the cell viability of Jurkat is less sensitive to voltage and duration than HeLa’s, which could be due to the looser contact between Jurkat and the nanopore membrane.
Fig. 6.
Cell viability of HeLa and Jurkat at different voltage intensity and delivery duration. (A) The viability of HeLa cells keeps at >95% after 20-s NanoEP delivery with 20 to 90 V. As delivery duration increases to 120 s, only 20-V pulse intensity preserved high-level cell viability. (B) The Jurkat cell viability is less sensitive to voltage and delivery duration. Jurkat viability drops significantly from 95% only at the condition of 90 V and 60- to 120-s delivery duration (error bars indicate SDs of experimental replicates, *P < 0.05, ***P < 0.001, ****P < 0.0001, post hoc Tukey test, n = 2).
Conventional methods (BEP and LFN) often cause higher rates of cell death or cellular damage after transfection (29, 30). We evaluated the cell viability by trypan blue exclusion in both HeLa and Jurkat cells after GFP plasmid transfection via 20-V, 20-s NanoEP and compared the results to those from the cells transfected with LFN and BEP. To make a fair comparison, we optimized LFN and BEP transfection according to the manufacturer’s instructions. The delivery conditions that gave the best transfection efficiency for each of the three methods were selected for cell viability analyses. After delivery, both HeLa and Jurkat cells were incubated under 5% CO2, 37 °C overnight before analysis (Methods). The results show that NanoEP preserved more than 95% of cell viability for both HeLa and Jurkat cells (SI Appendix, Fig. S7). Although BEP with its specialized electroporation buffer also reaches 90 to 95% cell viability in both cell lines, these values drop significantly to 50 to 55% viability when either PBS or cell culture media is used as the electroporation buffer (SI Appendix, Fig. S7). Cell toxicity in LFN transfection into HeLa cells was the highest among these three approaches.
The degree of membrane leakage of lactate dehydrogenase (LDH) in the culture media and the gene expression of inducible transcript 3 gene (DDIT3) are indicators of the damage induced by transfection. We therefore measured the activity of LDH in the culture media and studied the expression profile of DDIT3 gene in transfected HeLa cells with qPCR (Methods). The LDH activity assay demonstrates that the LDH leakage from the cells transfected with NanoEP is significantly lower than from the cells transfected with LFN (P < 0.05, SI Appendix, Fig. S8). Similarly, qPCR shows that the level of expression of DDIT3 gene in the cells transfected with NanoEP is significantly lower than in the cells transfected with LFN (P < 0.01, SI Appendix, Fig. S8).
Conclusion
We demonstrated that NanoEP enables effective delivery of various biomolecules, such as mRNA, DNA, proteins, and a large RNP complex of Cas9 RNPs, into both adhesive and suspension cells, including difficult-to-transfect fibroblast cells 3T3 and T lymphocyte Jurkat cells. Unlike other conventional delivery methods that often require specialized delivery buffer, expensive materials, a complicated fabrication process, and multiple cell manipulations, the NanoEP is based on a nanpore-embedded water-filter membrane platform and a square-wave function generator which are easily accessible to general laboratories. With its high delivery performance and great simplicity, NanoEP offers an effective method for universal intracellular delivery.
Methods
Assembly of the NanoEP Device.
The NanoEP device consists of two titanium-plate electrodes, a 20-µm-thick track-etched PC water-filter membrane with nanopore density of 2 × 107 nanopores per cm2 and a PDMS (Dow Corning) square (1.5 × 1.5 × 0.2 cm) with a hole 0.5 cm in diameter in the middle. We chose the 100-nm nanopore PC membrane for its strength and its adequate pore size for the passage of our target biomolecules. In addition, this membrane is inexpensive and accessible. We used uncured PDMS [ratio of of 10 (base): 1(curing agent)] to glue the PC membrane onto one end of the PDMS hole to make a well and cured the construct at 100 °C for 5 min.
Standard NanoEP Intracellular Delivery Protocol.
The cells used in this study are HEK293, HeLa, 3T3, and Jurkat cells. All of the cells are cultured via standard cell culture protocols (18, 20) in corresponding media (SI Appendix, Table S1) with addition of 10% FBS (Thermo Fisher Scientific) and 1% penicillin–streptomycin (Thermo Fisher Scientific).
To perform intracellular delivery, the NanoEP device is first coated with 20 μL of 1 to 100 ng/mL FN and incubated at 37 °C for 3 to 4 h. After two washes with deionized water and one wash with cell culture medium, the device is ready to use, and 5,000 to 15,000 cells of interest are pipetted into the device well in 50 μL of the corresponding culture medium (SI Appendix, Table S1). To form a tight cell–nanopore interface, adherent cells are grown on the PC surface overnight under 5% CO2, 37 °C. For suspension cells, cells are centrifuged at 150 × g for 5 min to establish tight cell contact with the nanopores before delivery. The device is then placed on a titanium electrode plate (2 × 2 cm) which is preloaded with 3 to 5 μL of the delivery sample of a desired concentration. For instance, 500 ng/mL GFP-expressing plasmid DNA was used in the plasmid transfection experiments for various cell types and 10 μM Cas9 RNPs was used for gene editing of HeLa and Jurkat cells. The second titanium electrode plate (1.5 × 2 cm) is then placed on the top of the device filled with cell culture media. For delivery, square-wave dc pulses of 20 Hz, 200 μs and a range of voltage intensities are generated by a square-pulse stimulator (Grass Instruments) and applied between the two titanium electrodes for 20 to 120 s. The square frequency and pulse duration are selected based on previous work (18, 20). Electrophoresis is considered the dominant mechanism to transport biomolecules across the nanopore membrane. Therefore, electric field polarity is primarily determined by the charges on delivery molecules. In the mRNA and DNA and Cas9 RNPs delivery, a positive electrode is placed on the top of the device. In the mCherry-STIM1 protein delivery, a negative electrode is placed on the top of the device. After delivery, the delivered cells are either directly placed into a 24-well plate for further incubation or suspended in cell culture media for analysis.
Flow Cytometry Analysis.
The transfected cells are incubated under 5% CO2, 37 °C overnight. The adherent cells are treated with trypsin-EDTA 0.05% (Thermo Fisher Scientific) followed by three washes with 1× PBS via centrifugation at 150 × g for 5 min. Suspension cells are also washed three times with 1× PBS via centrifugation. After final wash, the cells are resuspended in 1× PBS. GFP and mCherry cells are analyzed by an LSR II analyzer (BD Biosciences).
NanoEP System Simulation.
To quantitatively study the electric field distribution through nanopores, we simulated the local electric field intensity using the AC/DC Module (steady state) of the COMSOL Multiphysics finite-element-analysis software (COMSOL Inc). We assumed each nanopore is an independent system and identical to the others, and therefore we studied the electric distribution of individual nanopores. In the 3D simulations, the nanopore’s geometry (20-μm height and 150 nm in diameter; see SI Appendix, Fig. S1) is consistent with the experimental setup. The cell culture reservoir (on the top of the nanopore) and the delivery sample (under the nanopore) are both simplified into cubic media (10 × 10 × 3 μm). The voltage (ranging from 5 V to 80 V) is applied between the top of the cell culture and the bottom of the delivery sample. The property of the media (defined by 1× PBS solution) is considered homogeneous. The simulated electric field intensity is plotted as a function of the distance from top of the nanopore (the gap between the cytoplasm and the nanopore). The electric field strength increases with applied voltage and diminishes quickly along the gap. Considering the roughness of the cell surface (22), we assume that a 50-nm gap is still present when the cells adhere well to the PC membrane. As a result, applied voltage of 20 V is sufficient to generate a ∼3 kV/cm electric field [typical electric strength for permeabilizing cell membranes (31)] at the cytoplasm.
Expression of His6-mCherry-STIM1 Protein.
His6-mCherry-STIM1 (residues 342–469) is expressed in Hi5 insect cells with the Bac-to-Bac Baculovirus Expression System (Thermo Fisher Scientific). Cells are lysed by sonication in a buffer containing 20 mM Hepes, 300 mM NaCl, pH 7.5, and 10 mM imidazole (buffer A), and the supernatant is collected after centrifugation at 12,000 × g for 45 min. The supernatant is incubated with nickel-NTA beads (Qiagen) for 1 h. The beads are washed with buffer A mixed with 50 mM imidazole. The mCherry-STIM1 protein is eluted with buffer A supplemented with 300 mM imidazole. The elution is then desalted with a column packed with Sephadex G-50 beads (Sigma-Aldrich) to remove imidazole.
Expression and Purification of SpyCas9.
The experimental protocol was described previously (32). Briefly, DNA sequence encoding SpyCas9 proteins was cloned into a custom pET-based expression vector containing an N-terminal 10× His-tag, maltose-binding protein, and tobacco etch virus (TEV) protease cleavage site. The proteins were purified as described (1), with some modifications. The SpyCas9 protein was expressed in Escherichia coli BL21(DE3) cells. Five hundred milliliters of culture (Terrific broth, containing 100 mg/L ampicillin) was inoculated with 5 mL of overnight culture grown in Luria broth. The culture was induced by addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to final concentration of 0.5 mM at an OD of 0.5 to 0.6. This induced culture was transferred to a 16 °C incubator and further incubated overnight at 16 °C before harvest. E. coli cells were harvested and resuspended in bacterial lysis buffer [LB: 20 mM Tris⋅HCl, pH 7.5, 500 mM NaCl, 5% (vol/vol) glycerol, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), and two tablets of Roche protease inhibitor mixture per 100 mL of LB]. The cells were disrupted by sonication, and SpyCas9 proteins were purified using Ni-NTA resin. After overnight TEV cleavage at 4 °C, the SpyCas9 proteins were purified over an ortho-HisTrap HP column. The HisTrap column flow-through was further purified via a HiTrap Heparin HP column for cation exchange chromatography. The final gel filtration step (Superdex 200) was carried out in filtration buffer containing 20 mM Tris⋅HCl, pH 7.5, 200 mM NaCl, 5% (vol/vol) glycerol, and 1 mM TCEP.
In Vitro Transcription of PPIB Single-Guide RNA (sgRNA) and Assembly of SpyCas9-PPIB-sgRNA RNP Complex.
PPIB-sgRNA DNA templates are PCR-amplified from overlapping primers containing a T7 promoter, 20-nt target sequence (GUGUAUUUUGACCUACGAAU), and an sgRNA scaffold. The amplified PCR products serve as the DNA templates for in vitro transcription reactions, which are performed as described (32). In vitro-transcribed sgRNAs are separated in 12% urea-PAGE and eluted overnight at 4 °C with diethylpyrocarbonate (DEPC) water. After several changes with DEPC water, the RNA is concentrated before use.
To assemble the SpyCas9-PPIB-sgRNA RNP complex, purified SpyCas9 protein is slowly added to PPIB-sgRNA which is buffered with the SpyCas9 size-exclusion buffer (the molar ratio of Cas9 to sgRNA = 1 to 1.2), and the mixture is further incubated for 10 min at 37 °C to form active RNPs as previously described (31, 32).
T7E1 Assay.
T7E1 assays are performed as previously described with slight modification (32). Briefly, cells are pelleted and resuspended in 100 μL QuickExtract buffer (Lucigen) and DNA is extracted according to the manufacturer’s protocol. The 200 to 300 ng of genomic DNA is directly used for PCR amplification with the pair of primers for the PPIB locus. For qPCR, the forward and reverse primers are GAACTTAGGCTCCGCTCCTT and CTCTGCAGGTCAGTTTGCTG, respectively. Approximately 200 nanograms of PCR product is denatured, annealed, and digested with T7E1 (NEB). The digested DNA samples are separated in a 2% agarose gel stained with SybrGold (Thermo Fisher Scientific). Cleaved products are quantified by ChemiDoc MP Imaging System (Bio-Rad).
Cell Membrane Leakage Analysis.
Cellular membrane leakage in transfected cells was studied by measuring LDH activity in the cell media. In detail, the media were collected after 12 h of transfection and LDH activity was measured. Specifically, the LDH assay was carried out according to the manufacturer’s instructions provided by the Lactate Dehydrogenase Assay kit (Abcam). The color readings were measured with Cytation 5 Imaging Reader (BioTek) at OD 450 nm in a kinetic mode, every 2 min, for 60 min at 37 °C. The LDH activity was calculated based on these color readings against a standard curve.
Cell Viability Test.
The trypan blue exclusion method is used to determine the cell viability. After overnight incubation under 5% CO2 and 37 °C, cells are resuspended in fresh cell culture media. The cell density is first determined by using a hemacytometer. The cells area then diluted to desirable density. A 0.4% stock solution of trypan blue (Thermo Fisher Scientific) in 1× PBS buffer, pH 7.2 (ThermoFisher Scientific), is prepared and added to the cells solution with 1:1 volume ratio. A hemacytometer is loaded with 10 μL of the mixture. The blue-stained cells and total number of cells are counted. The percent cell viability is determined according to the following formula:
Supplementary Material
Acknowledgments
We thank Dr. Meredith Triplet and Dr. Jennifer Hamilton for their critical editing of the manuscript and the Cell Culture Facility of the University of California, Berkeley for the cell lines used in this work. This work was supported by Keck Foundation Grant 89208-31150-44-X-IQJED.
Footnotes
Conflict of interest statement: Y.C., E.M., J.A.D., and P.Y. are inventors on patent applications (filed by the University of California, Berkeley) related to the nanopore electroporation systems and uses thereof. J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences and is a member of the board of directors of Driver and Johnson & Johnson and a member of the scientific advisory boards for Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Metagenomi and Inari. Y.C., S.C.-B., P.Y., and Bozhi Tian are coauthors on a 2018 review article.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818553116/-/DCSupplemental.
References
- 1.DiGiandomenico A, Wylezinski LS, Hawiger J. Intracellular delivery of a cell-penetrating SOCS1 that targets IFN-gamma signaling. Sci Signal. 2009;2:ra37. doi: 10.1126/scisignal.1162191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Roth TL, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 5.Reinshagen C, et al. CRISPR-enhanced engineering of therapy-sensitive cancer cells for self-targeting of primary and metastatic tumors. Sci Transl Med. 2018;10:eaao3240. doi: 10.1126/scitranslmed.aao3240. [DOI] [PubMed] [Google Scholar]
- 6.Erazo-Oliveras A, et al. Protein delivery into live cells by incubation with an endosomolytic agent. Nat Methods. 2014;11:861–867. doi: 10.1038/nmeth.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Zienkiewicz J, DiGiandomenico A, Hawiger J. Suppression of acute lung inflammation by intracellular peptide delivery of a nuclear import inhibitor. Mol Ther. 2009;17:796–802. doi: 10.1038/mt.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart MP, et al. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 9.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 10.Chu G, Hayakawa H, Berg P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987;15:1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canatella PJ, Karr JF, Petros JA, Prausnitz MR. Quantitative study of electroporation-mediated molecular uptake and cell viability. Biophys J. 2001;80:755–764. doi: 10.1016/S0006-3495(01)76055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim W, Ng JK, Kunitake ME, Conklin BR, Yang P. Interfacing silicon nanowires with mammalian cells. J Am Chem Soc. 2007;129:7228–7229. doi: 10.1021/ja071456k. [DOI] [PubMed] [Google Scholar]
- 13.Shalek AK, et al. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc Natl Acad Sci USA. 2010;107:1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boukany PE, et al. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nat Nanotechnol. 2011;6:747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 15.Chang L, et al. 3D nanochannel electroporation for high-throughput cell transfection with high uniformity and dosage control. Nanoscale. 2016;8:243–252. doi: 10.1039/c5nr03187g. [DOI] [PubMed] [Google Scholar]
- 16.Kang W, et al. Microfluidic device for stem cell differentiation and localized electroporation of postmitotic neurons. Lab Chip. 2014;14:4486–4495. doi: 10.1039/c4lc00721b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding X, et al. High-throughput nuclear delivery and rapid expression of DNA via mechanical and electrical cell-membrane disruption. Nat Biomed Eng. 2017;1:0039. doi: 10.1038/s41551-017-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X, et al. Nanostraw-electroporation system for highly efficient intracellular delivery and transfection. ACS Nano. 2013;7:4351–4358. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- 19.Fan R, et al. DNA translocation in inorganic nanotubes. Nano Lett. 2005;5:1633–1637. doi: 10.1021/nl0509677. [DOI] [PubMed] [Google Scholar]
- 20.Cao Y, et al. Universal intracellular biomolecule delivery with precise dosage control. Sci Adv. 2018;4:eaat8131. doi: 10.1126/sciadv.aat8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y, et al. Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc Natl Acad Sci USA. 2017;114:E1866–E1874. doi: 10.1073/pnas.1615375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S, Jung GB, Kim KS, Lee GJ, Park HK. Medical applications of atomic force microscopy and Raman spectroscopy. J Nanosci Nanotechnol. 2014;14:71–97. doi: 10.1166/jnn.2014.9112. [DOI] [PubMed] [Google Scholar]
- 23.Potthoff E, et al. Rapid and serial quantification of adhesion forces of yeast and mammalian cells. PLoS One. 2012;7:e52712. doi: 10.1371/journal.pone.0052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalili AA, Ahmad MR. A review of cell adhesion studies for biomedical and biological applications. Int J Mol Sci. 2015;16:18149–18184. doi: 10.3390/ijms160818149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Astolfo DS, et al. Efficient intracellular delivery of native proteins. Cell. 2015;161:674–690. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Deng X, Gill DL. Calcium signaling by STIM and Orai: Intimate coupling details revealed. Sci Signal. 2010;3:pe42. doi: 10.1126/scisignal.3148pe42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 28.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan PH, et al. Immunolipoplexes: An efficient, nonviral alternative for transfection of human dendritic cells with potential for clinical vaccination. Mol Ther. 2005;11:790–800. doi: 10.1016/j.ymthe.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Lesueur LL, Mir LM, André FM. Overcoming the specific toxicity of large plasmids electrotransfer in primary cells in vitro. Mol Ther Nucleic Acids. 2016;5:e291. doi: 10.1038/mtna.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weaver JC, Smith KC, Esser AT, Son RS, Gowrishankar TR. A brief overview of electroporation pulse strength-duration space: A region where additional intracellular effects are expected. Bioelectrochemistry. 2012;87:236–243. doi: 10.1016/j.bioelechem.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.