Significance
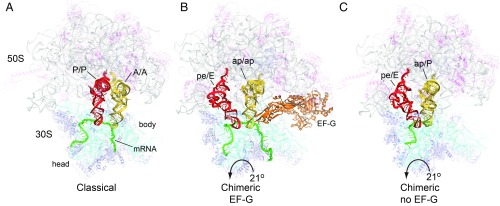
The crystal structure of a pretranslocation complex containing mRNA and two tRNAs shows spontaneous movement of the mRNA and tRNAs from the classical state to a chimeric hybrid state in the absence of elongation factor G (EF-G) or GTP. This finding suggests that many steps of translocation, including rotation of the 30S subunit head domain and movement of the A-site tRNA into the position of the classical-state P-site tRNA, are supported by the ribosome itself. However, movement of mRNA and tRNA has become uncoupled, resulting in negative slippage of the translational reading frame. These findings have implications for our current view of the role of EF-G, suggesting that its main function may be to stabilize codon–anticodon pairing during translocation.
Keywords: ribosomes, translocation, EF-G, frame shifting, translation
Abstract
The elongation factor G (EF-G)–catalyzed translocation of mRNA and tRNA through the ribosome is essential for vacating the ribosomal A site for the next incoming aminoacyl-tRNA, while precisely maintaining the translational reading frame. Here, the 3.2-Å crystal structure of a ribosome translocation intermediate complex containing mRNA and two tRNAs, formed in the absence of EF-G or GTP, provides insight into the respective roles of EF-G and the ribosome in translocation. Unexpectedly, the head domain of the 30S subunit is rotated by 21°, creating a ribosomal conformation closely resembling the two-tRNA chimeric hybrid state that was previously observed only in the presence of bound EF-G. The two tRNAs have moved spontaneously from their A/A and P/P binding states into ap/P and pe/E states, in which their anticodon loops are bound between the 30S body domain and its rotated head domain, while their acceptor ends have moved fully into the 50S P and E sites, respectively. Remarkably, the A-site tRNA translocates fully into the classical P-site position. Although the mRNA also undergoes movement, codon–anticodon interaction is disrupted in the absence of EF-G, resulting in slippage of the translational reading frame. We conclude that, although movement of both tRNAs and mRNA (along with rotation of the 30S head domain) can occur in the absence of EF-G and GTP, EF-G is essential for enforcing coupled movement of the tRNAs and their mRNA codons to maintain the reading frame.
During protein synthesis, the mRNA and tRNAs must be moved synchronously through the ribosome, to vacate the A site for the next incoming aminoacyl-tRNA while preserving the translational reading frame. This process, called translocation, is catalyzed by the GTPase elongation factor G (EF-G) (1, 2). Following peptide bond formation, the deacylated acceptor end of the P-site tRNA moves into the 50S subunit E site, forming the P/E binding state, and the peptide-bearing acceptor stem of the A-site tRNA moves into the 50S P site, creating the A/P state (3–5). The P/E and A/P states are called hybrid binding states, because the tRNAs are bound to different sites with respect to the 30S and 50S subunits. These movements can occur in the absence of EF-G or GTP (3, 6–9) and are accompanied by an ∼7° rotation of the 30S subunit relative to the 50S subunit (10–13). There is no translocation of the mRNA or the anticodon ends of the tRNAs on the 30S subunit during this step.
Unlike hybrid-state formation, completion of translocation requires the action of EF-G (3, 9, 14–16). This phase of translocation involves movement of the mRNA, which must be tightly coordinated with movement of the associated tRNA anticodon ends through the 30S subunit to preserve the translational reading frame. The discovery that EF-G can catalyze translocation of tRNAs in the absence of mRNA suggests that the mechanism of translocation acts primarily on the tRNAs, and that the mRNA moves passively, via its base-paired interactions with the tRNA anticodons (17, 18). Accordingly, preservation of codon–anticodon pairing during movement is crucial.
Structural studies of trapped EF-G–bound translocation intermediates (19–22), stopped-flow ensemble FRET experiments (23–25), and single-molecule FRET studies (9) reveal that this movement is coupled to large-scale (∼21°) rotation of the head domain of the 30S subunit (Fig. 1). During head rotation, the anticodon stem loop (ASL) of the P-tRNA remains bound to P-site elements of the 30S head domain, moving into a position that is nearly juxtaposed with E-site elements of the 30S body, while its acceptor end moves fully into the 50S E site; accordingly, this has been called the pe/E chimeric hybrid state (19, 20). P-tRNA movement is exactly coupled to head rotation, but trapped complexes containing two tRNAs and EF-G show that the A-site tRNA moves further toward the P site than would be predicted simply from head rotation (21, 22). The anticodon end of the transiting A-tRNA is positioned roughly between the A site of the 30S head and P site of the body, while its CCA end is bound in the 50S P site; this state is called the ap/P chimeric hybrid state (21). In another trapped intermediate, termed the ap/ap state, the acceptor end of the EFG-bound tRNA made simultaneous contacts with the A and P loops of the 50S subunit, facilitated by rearrangements in the conformations of the tRNA binding sites themselves (22).
Fig. 1.
Spontaneous movement of the translocation complex into a chimeric hybrid state. (A) Classical-state ribosome complex (32). (B and C) Chimeric state formed (B) in the presence of EF-G (22) or (C) spontaneously (this work). The spontaneous chimeric hybrid-state translocation intermediate (C) has the same overall conformation as that obtained in the presence of bound EF-G, including a 21° rotation of the head domain of the 30S subunit. Components are 23S rRNA (white), 16S rRNA (cyan), 5S rRNA (light blue), 50S proteins (magenta), 30S proteins (blue), P/P-tRNA and pe/E intermediate (red), A/A-tRNA and ap/ap or ap/P intermediates (yellow), and mRNA (green). Arrow indicates direction of rotation of the 30S subunit head domain.
Previously, the presence of two tRNAs in the chimeric hybrid state has only been observed in the presence of bound EF-G. Here, we present the 3.2-Å crystal structure of a translocation intermediate prepared in the absence of EF-G, GTP, or antibiotics. The complex, containing a defined mRNA and two tRNAs bound initially to the A and P sites of the 70S ribosome, was crystallized under conditions expected to stabilize the hybrid state. Unexpectedly, the structure reveals that the tRNAs have moved spontaneously into ap/P and pe/E chimeric hybrid states, along with an ∼21° rotation of the head domain of the 30S ribosomal subunit, closely resembling, but not identical to, previously determined structures of trapped chimeric hybrid-state intermediates containing bound EF-G (21, 22). Most interestingly, translocation of the tRNAs has become uncoupled from that of their mRNA codons in the absence of EF-G. These findings have strong implications for the respective roles of EF-G and the ribosome in translocation.
Results
Complexes were constructed from Thermus thermophilus 70S ribosomes, using a 40-nucleotide defined mRNA designed to position a Val GUA codon in the P site and a Tyr UAC codon in the A site. Deacylated tRNAVal and tRNATyr were bound and incubated at a magnesium-ion concentration (5 mM) that was expected to stabilize hybrid-state binding (3, 26). Under these conditions, the complex was fully occupied with tRNA and mRNA and capable of EF-G–dependent translocation (SI Appendix, Fig. S1). The complex crystallized in the P212121 space group and was solved by molecular replacement at 3.2-Å resolution (Methods and SI Appendix, and Table S1). The asymmetric unit contains two ribosome complexes (A and B); since no electron density is present for tRNATyr in the A complex, we focus here on our findings for the B complex.
Unexpectedly, the 30S subunit head domain has undergone spontaneous large-scale (21°) rotation (Fig. 1), virtually identical to that previously seen in trapped translocation intermediates containing two tRNAs and EF-G (21, 22); this is accompanied by a modest (2°) rotation of the 30S body, relative to the 50S subunit. The L1 stalk of the 50S subunit moves inward, creating a stacking contact between the noncanonical G2112–A2169 purine–purine base pair of 23S rRNA and the tertiary Watson–Crick G19–C56 pair in the elbow of the pe/E tRNA, similar to that seen in EF-G–containing hybrid-state (13, 27–29) and chimeric hybrid-state translocation intermediates (21, 22, 30) (SI Appendix, Fig. S2).
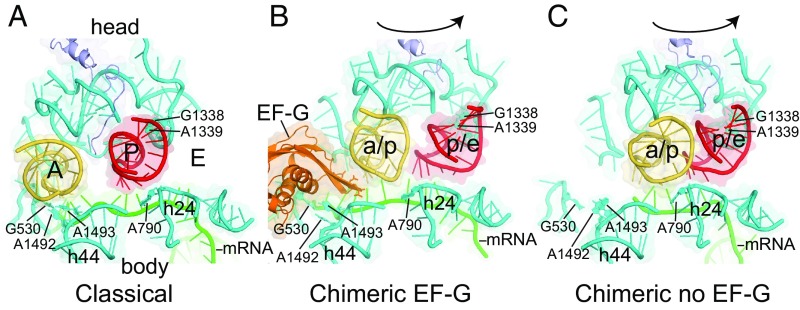
Both tRNAs undergo spontaneous large-scale translocation into ap/P and pe/E chimeric hybrid states. As observed for previous EF-G–containing complexes, the P- tRNAVal moves precisely with rotation of the 30S head domain, due to its tight binding to 16S rRNA residues G1338 and A1339 and other P-site elements of the head domain (Fig. 2). Its position is identical to the pe/E state seen in the EF-G complexes (20, 22) (Fig. 3B).
Fig. 2.
Movement of tRNA ASLs in chimeric hybrid-state complexes. Closeup views of the positions of the tRNA ASLs in the 30S subunit, viewed from the subunit interface. (A) Classical-state complex (32). (B) EF-G–containing chimeric hybrid state (22). (C) Spontaneous chimeric hybrid state (this work). In both chimeric hybrid structures, the P-tRNA ASL moves precisely with rotation of the 30S head domain, but in the no–EF-G complex (C), the A-tRNA (a/p) moves further than in the EF-G complex (B). Structures were aligned relative to the 30S subunit body domain.
Fig. 3.
Positions of tRNAs in chimeric hybrid-state complexes. Positions of the ap/P (orange) and pe/E (red) spontaneous chimeric hybrid-state tRNAs compared with (A) classical-state P/P (cyan) and E/E (blue) tRNAs (32) and (B) EF-G–containing ap/ap (cyan) and pe/E (blue) chimeric hybrid-state tRNAs (22). (A) In the absence of EF-G, the A-site tRNATyr (orange) moves into the ap/P chimeric hybrid state, resulting in direct contact (circle) with the pe/E-tRNAVal. Note that its position is identical to that of the fully translocated classical P/P-tRNA (32). (B) In the absence of EF-G, the P-site tRNAVal moves into the same position as the pe/E-tRNA observed in the presence of EF-G (22). Structures were aligned relative to the 50S subunit.
The anticodon end of the A-site tRNATyr translocates even further toward the P site than in the ap/ap–tRNA–EF-G–containing chimeric-hybrid complex (22), moving into contact with the pe/E tRNAVal (Figs. 2–4). Remarkably, this movement results in complete translocation, moving the A-site tRNATyr into a position that is virtually indistinguishable from that of the classical-state P-site tRNA (31, 32), using the 50S subunit as the frame of reference (Fig. 3A), and is similar to that of the ap/P-tRNA seen in the presence of EF-G (21).
Fig. 4.
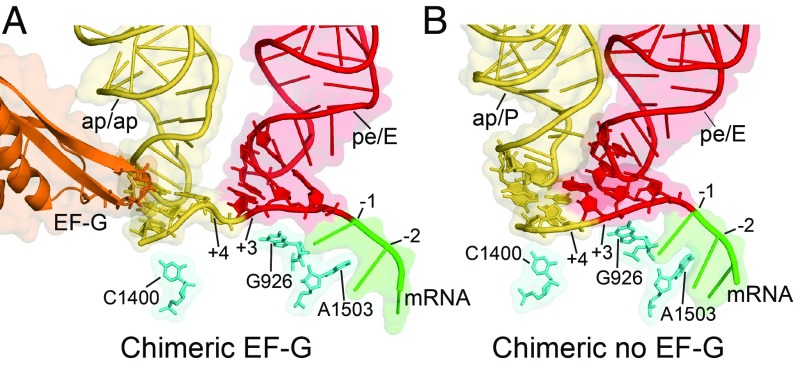
Extended movement of the A-tRNA ASL in the absence of EF-G. Interface views of tRNA ASLs and mRNA in (A) EF-G-bound (22) and (B) spontaneous chimeric hybrid states. In the absence of EF-G, the A-ASL moves further than in its presence, suggesting that EF-G restrains the movement of the A-tRNA, rather than pushing it. The 16S rRNA bases A1503, C1400, and G926 are shown as reference points. Structures were aligned on the 30S subunit body domain.
The CCA acceptor ends of both tRNAs are fully translocated into the P and E sites of the 50S subunit, respectively. C74 and C75 of the ap/P-tRNATyr are base paired with G2252 and G2251, respectively, of the P loop of 23S rRNA, as in the classical P/P state (SI Appendix, Fig. S3). The 2′-hydroxyl of ribose 76 of the pe/E tRNAVal H bonds to the N3 and O2 positions of C2394 of 23S rRNA, and its adenosine 76 is stacked between bases G2421 and A2422 of 23S rRNA (SI Appendix, Fig. S4). The N-terminal β-hairpin of protein L28 contacts the backbone of the CCA tail of the tRNAVal around position 74.
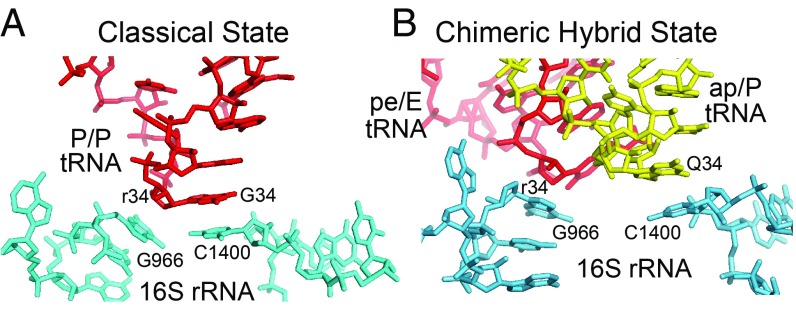
Contacts between P-site elements of 16S rRNA and the wobble nucleotide (position 34) at the apex of the anticodon loops of the two tRNAs are rearranged from those seen previously for either the classical or chimeric hybrid states. In the classical state, nucleotide 34 of the P-site tRNA is held in position by stacking of 16S rRNA C1400 on the wobble base and packing of G966 against its ribose moiety (33, 34). In the EF-G–containing chimeric hybrid intermediate (22), these contacts are rearranged, so that both G966 and C1400 contact the anticodon loop of the ap/ap chimeric hybrid tRNA. Here, in the absence of EF-G, we observe a third configuration, in which C1400 stacks on wobble base 34 of the ap/P-tRNA, as in the EF-G chimeric-state structure, but G966 remains packed against ribose 34 of the pe/E-tRNA (Fig. 5). G966 in the 30S head domain has moved with the P-site tRNA with rotation of the head, whereas C1400, in the body, makes contact with the translocating A-tRNA as it moves into the P site of the 30S body domain.
Fig. 5.
Rearranged stacking of C1400 on the tRNA anticodon. In the spontaneous chimeric hybrid-state complex, C1400 shifts from its (A) classical-state contact with base 34 of the P-site tRNA (32) to stacking on (B) base 34 of the ap/P-tRNA. G966 remains packed against ribose 34 of the pe/E-tRNA, instead of moving to the ap/P-tRNA, as in the EF-G–containing complex (22).
The conformations of the 16S rRNA bases A1492 and A1493, which contact the minor groove of the A-tRNA codon–anticodon duplex in the 30S decoding site, also differ from those seen previously (SI Appendix, Fig. S5). In this structure, both bases are flipped out from their stacked positions in helix 44 seen in ribosomes with a vacant A site, but do not reach to make contact with G530, as in ribosomes containing a bound cognate A-tRNA. Furthermore, 23S rRNA base A1913 assumes the same tucked-in conformation as seen in ribosomes with a vacant A site (SI Appendix, Fig. S5). The positions of A1492 and A1493 more closely resemble those of an EF-G–containing chimeric hybrid-state complex (22), which, however, were likely stabilized by the presence of neomycin. Thus, in the absence of antibiotics, A1492, A1493, and A1913 assume a conformation intermediate to those seen in vacant or A-site-bound classical complexes.
The mRNA can be traced from its 5′ end (position −15) to the decoding site, after which it becomes abruptly disordered, but is very well ordered from positions −11 to +5) (SI Appendix, Fig. S6). Shine–Dalgarno pairing is formed between the mRNA GGAGG sequence (residues −10 to −6) and the 16S rRNA sequence CCUCC (residues 1535–1539). Base A1503 of 16S rRNA is intercalated between adjacent mRNA bases at positions −1 and −2 (SI Appendix, Fig. S7) as was previously observed in EF-G–containing chimeric hybrid-state structures (20, 22). As in the previous EF-G–containing complexes (20, 22), the bulged G926 contacts phosphate +3 (residue 18) of the mRNA (SI Appendix, Fig. S8) instead of phosphate +1 as in the classical-state complex (32, 33) indicating a net movement of the mRNA by two nucleotides (relative to the 30S body domain) as the P-site tRNA nears completion of its translocation into the 30S E site.
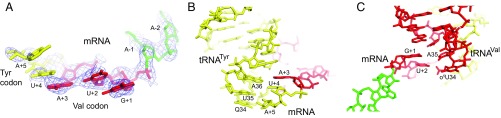
Although the backbone positions of the first two nucleotides of the GUA Val codon and the pe/E-tRNAVal anticodon are roughly juxtaposed for pairing, the codon and anticodon bases are pulled apart and are no longer precisely aligned, resulting in slippage of the P-codon register by about one-half position (Fig. 6). A kink in the mRNA backbone following U+2 orients the wobble base A+3 of the Val codon to diverge from the path of its 5′-GU to stack on U+4 of the following Tyr UAC codon (Fig. 6A). Thus, G+1 of the Val codon is aligned between C36 and A35 of the tRNAVal anticodon, and U+2 between A35 and cmU34 (Fig. 6C and SI Appendix, Fig. S9B). This leads to a nonpairing coplanar juxtaposition of the tRNATyr anticodon with the resulting AUA triplet (nucleotides 18–20), creating a −1 slippage of the reading frame (Fig. 6B and SI Appendix, Fig. S9A). These codon–anticodon slippages are summarized schematically in Fig. 7. The result is consistent with the overall difference in movement of mRNA and tRNA relative to the 30S body domain. As described above, G926 of 16S rRNA is juxtaposed with phosphate +3 of the mRNA (SI Appendix, Fig. S8), instead of phosphate +4 (which would correspond to translocation by one full codon), whereas the A-site tRNA has moved fully into the position of classical P-site tRNA (Fig. 3A). Thus, movement of the A-site tRNA (and to a lesser extent the P-site tRNA) in the absence of EF-G appears to be uncoupled from movement of the mRNA, in contrast to that seen in the EF-G–containing chimeric hybrid-state complex (Fig. 7 and SI Appendix, Fig. S10) (22).
Fig. 6.
Disruption of the translational reading frame. (A) Stacking of A+3 of the GUA Val codon on U+4 of the UAC Tyr codon. The Val (GUA) codon is shown in red and the Tyr (UAC) codon in yellow. Base C+6 of the Tyr codon is disordered. (B) Rearrangement of A+3 brings it nearly into alignment with A36 of the Tyr anticodon, creating a −1 frameshift for the Tyr codon. (C) G+1 and U+2 of the Val codon have slipped into juxtaposition with A35 and o5U of the Leu anticodon, respectively, creating a frameshift of about −1/2. Note that many of the codon and anticodon bases are pulled out of H-bonding distance of each other. (See also SI Appendix, Fig. S7.)
Fig. 7.
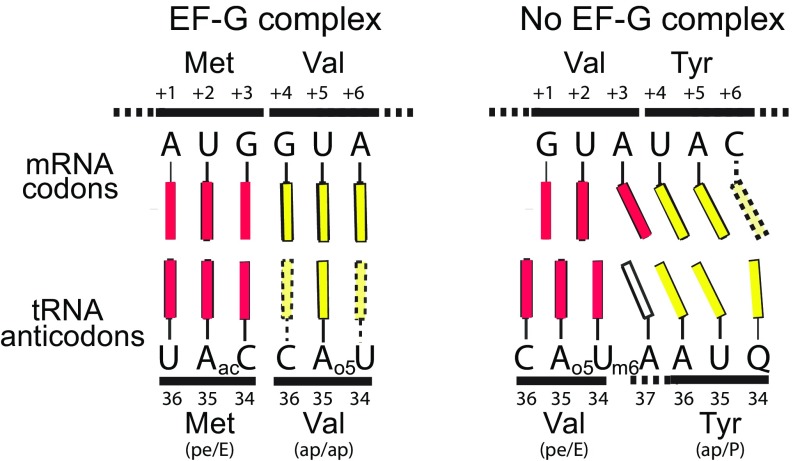
Schematic representation of codon–anticodon alignments in chimeric hybrid-state complexes. mRNA codons are shown at the Top and tRNA anticodons at the Bottom. Bases are represented by colored rectangles; dotted rectangles indicate disordered bases. (Left) In the EF-G–containing chimeric hybrid-state complex (22), codon–anticodon base pairing is normal, except that C36 and o5U of the Val anticodon are disordered. (Right) In the chimeric hybrid-state complex formed in the absence of EF-G, codons and anticodons have shifted out of alignment, creating slippage of the reading frame by about −1 for tRNATyr and −1/2 for tRNAVal. C+6 of the Tyr codon is disordered. Several of the bases are outside of base-pairing distance and misaligned, with the tRNAs moving further than their mRNA codons in the absence of EF-G. G+1 of the Val GUA codon is aligned between C36 and A35, and U+2 between A35 and o5U34 of the RNAVal anticodon. Note that A+3 is displaced from the other two bases (G+1 and U+2) of the Val codon to stack on U+4 of the Tyr codon, and aligns with A36 of the Tyr anticodon. U+4 of the Tyr codon aligns with U35 of the tRNATyr anticodon, creating a −1 frameshift.
Discussion
Previous studies have shown that the ribosome is capable of complete translocation in the absence of EF-G or GTP, albeit at greatly reduced rates (35–37) or if stimulated by antibiotics (38, 39), or in reverse (40). Here, a pretranslocation complex assembled under conditions intended to favor hybrid-state formation, appears to have moved spontaneously into a chimeric hybrid state (Fig. 1). Since this occurred during the several-day period of crystallization, the rate of this translocation event is unknown. This fortuitous trapping of the complex in the chimeric intermediate state may be due to the relatively low (∼5 mM) Mg2+ concentration conditions, compared with the higher (∼10–15 mM) concentrations used in many studies where complete spontaneous translocation was observed. In any case, toeprinting analysis shows that in the initial complex, the tRNAs were bound to the A and P sites and were competent for EF-G–catalyzed translocation (SI Appendix, Fig. S1).
The structure of a ribosome complex containing two tRNAs bound in chimeric hybrid states in the absence of EF-G or GTP provides an unexpected window into the role of EF-G in the mechanism of translocation. If we ask which steps of chimeric hybrid-state formation do not require EF-G, we can begin to narrow down the list of essential functions of EF-G, and so distinguish them from those that are defined by the ribosome itself.
It has been known for some time that movement of tRNA into the A/P and P/E hybrid states, coupled to intersubunit rotation, can occur independently of EF-G and GTP (3, 6, 7). We can now add the following major structural changes that can also take place spontaneously in the absence of EF-G: (i) release of the codon–anticodon duplex from the decoding site; (ii) large-scale (21°) rotational movement of the 30S subunit head domain; (iii) complete translocation of the A-site tRNA into the position of the classical P-site tRNA, relative to the 50S subunit; (iv) translocation of the P-site tRNA into the pe/E chimeric hybrid state; and (v) intercalation of the universally conserved A1503 of 16S rRNA between mRNA bases −1 and −2. In light of this newly expanded list of EF-G–independent functions, we can now revisit the question, Which steps of translocation actually require EF-G?
Extensive evidence shows that domain IV of EF-G is structurally mobile (21, 22, 27, 28, 41–45) and required for translocation (16, 46). One model is that domain IV acts early in translocation to release the A-tRNA from the decoding site by disrupting codon–anticodon interaction with 16S rRNA bases 1492 and 1493 (19, 45, 47–50), freeing the 30S subunit head to rotate. Our structure shows that EF-G is not required to release the A-tRNA, although it may catalyze the event.
Another possibility is that movement of domain IV of EF-G pushes the A-site tRNA toward the P site, possibly powered by GTP hydrolysis (16, 22, 25, 51–53). However, our structure shows that in the absence of EF-G the A-tRNA actually moves further than in the EF-G–containing complex, to the point of physically contacting the P-tRNA, in apparent contradiction of this model (Figs. 2 and 4). This finding suggests the possibility that domain IV actually restricts movement of the A-tRNA, to avoid potential slippage of the reading frame.
Previously determined structures of trapped EF-G–containing complexes showed that the tip of domain IV of EF-G maintains contact with the minor groove of the A-site codon–anticodon duplex as it moves from the hybrid state (27) into the chimeric hybrid ap/P or ap/ap states (21, 22) and into the posttranslocation P/P state (45). These and other observations have led to a proposal for yet another crucial role for domain IV: preserving codon–anticodon pairing (and thus the translational reading frame) during the vulnerable transition between the 30S A and P sites, when the weak triplet helix cannot rely on its interactions with the ribosome for stabilization (21, 22, 45).
Our findings provide direct evidence for a role for EF-G in maintaining the reading frame during translocation by stabilization of pairing between the A-tRNA anticodon and its mRNA codon. In the absence of EF-G, movement of the A-site tRNA is uncoupled from that of its mRNA codon, resulting in a −1 slippage of the reading frame (Figs. 6 and 7), presumably due to the absence of stabilization by domain IV of EF-G. Codon–anticodon interactions are also disrupted for the P-tRNA, although slipping by less than one full position.
The structure presented here, unlike previously determined EF-G–containing chimeric hybrid-state structures, was trapped in the absence of antibiotics or nonhydrolyzable GTP analogs, so must represent a stable state. One further role for EF-G must then be to move the ribosome out of the chimeric hybrid state toward completion of translocation. In particular, the mechanism of release of the E-site tRNA from the ribosome, which is likely coupled to completion of translocation, remains poorly understood. One possibility is suggested by the so far unexplained intercalation of 16S rRNA base A1503 between bases −2 and −1 of the mRNA (SI Appendix, Fig. S6) (20, 22), which correspond to bases 2 and 3 of the E-site codon. This intercalation, which has so far only been observed in chimeric hybrid-state complexes, would likely destabilize codon–anticodon pairing and thus favor release of the E-site tRNA.
A recent ribosome structure by Dunham and coworkers (54) shows that binding of a frameshift-inducing tRNA containing an eight-nucleotide anticodon loop can also lead to a spontaneous large-scale rotation of the 30S subunit head. In this structure, containing a single tRNA bound in the e*/E state, A1503 is not intercalated within the mRNA, suggesting a possible role for this base in reading-frame maintenance.
Although the P-tRNA moves into the pe/E state, it remains a few ångstroms short of its full classical E-site position. For the A-tRNA, which now occupies the position of the classical P site in our structure (Fig. 3A), translocation is essentially complete except for reverse rotation of the 30S head domain. We still have no clear explanation for how the 30S head domain undergoes reverse rotation without reversing the movement of tRNA and mRNA. This event, which represents the final step of translocation, must embody a true (yet unexplained) ratchet-like behavior.
As our understanding of the basic molecular mechanisms of protein synthesis evolves, it becomes ever clearer that, despite the involvement of numerous essential factors, translation is fundamentally based on the properties of the ribosome itself. Moreover, as we examine the basis for these ribosome-specific mechanisms, we discover that they appear to be, virtually exclusively, functions of ribosomal RNA, in keeping with the idea that the ribosome emerged from an RNA world.
Materials and Methods
Complexes were constructed from T. thermophilus 70S ribosomes, a 40-nucleotide mRNA, tRNAVal and tRNATyr, and crystallized as described in SI Appendix, in the P212121 space group. Diffraction data were integrated and scaled, and the structure was solved by molecular replacement and refined at a resolution of 3.2 Å. Small subunit head and body rotation were calculated as described by Mohan et al. (11).
All other details are as described in SI Appendix.
Supplementary Material
Acknowledgments
We thank the beamline staffs at the Advanced Light Source (Lawrence Berkeley National Laboratory), the Stanford Synchrotron Radiation Laboratory (SLAC National Accelerator Laboratory), and the Advanced Photon Source (Argonne National Laboratory) for their expert support during screening and data collection. This work was supported by Grant R35-GM118156 from the NIH and funds from the Robert L. Sinsheimer Chair.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, www.rcsb.org (RCSB PDB ID code 6N1D).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901310116/-/DCSupplemental.
References
- 1.Noller HF, Lancaster L, Zhou J, Mohan S. The ribosome moves: RNA mechanics and translocation. Nat Struct Mol Biol. 2017;24:1021–1027. doi: 10.1038/nsmb.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtkamp W, Wintermeyer W, Rodnina MV. Synchronous tRNA movements during translocation on the ribosome are orchestrated by elongation factor G and GTP hydrolysis. BioEssays. 2014;36:908–918. doi: 10.1002/bies.201400076. [DOI] [PubMed] [Google Scholar]
- 3.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 4.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma H, et al. Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Rep. 2016;16:2187–2196. doi: 10.1016/j.celrep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 9.Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 11.Mohan S, Donohue JP, Noller HF. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci USA. 2014;111:13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belardinelli R, et al. Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol. 2016;23:342–348. doi: 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- 15.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 17.Tnalina GZh, Belitsina NV, Spirin AS. [Template-free polypeptide synthesis from aminoacyl-tRNA in Escherichia coli ribosomes] Dokl Akad Nauk SSSR. 1982;266:741–745. [PubMed] [Google Scholar]
- 18.Yusupova GZ, Belitsina NV, Spirin AS. Template-free ribosomal synthesis of polypeptides from aminoacyl-tRNA. Polyphenylalanine synthesis from phenylalanyl-tRNALys. FEBS Lett. 1986;206:142–146. doi: 10.1016/0014-5793(86)81356-4. [DOI] [PubMed] [Google Scholar]
- 19.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramrath DJ, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belardinelli R, Sharma H, Peske F, Wintermeyer W, Rodnina MV. Translocation as continuous movement through the ribosome. RNA Biol. 2016;13:1197–1203. doi: 10.1080/15476286.2016.1240140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtkamp W, et al. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci USA. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol. 2013;20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 30.Mohan S, Noller HF. Recurring RNA structural motifs underlie the mechanics of L1 stalk movement. Nat Commun. 2017;8:14285. doi: 10.1038/ncomms14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 32.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 34.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 35.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 36.Southworth DR, Brunelle JL, Green R. EFG-independent translocation of the mRNA:tRNA complex is promoted by modification of the ribosome with thiol-specific reagents. J Mol Biol. 2002;324:611–623. doi: 10.1016/s0022-2836(02)01196-8. [DOI] [PubMed] [Google Scholar]
- 37.Pestka S. Assay for nonenzymatic and enzymatic translocation with Escherichia coli ribosomes. Methods Enzymol. 1974;30:462–470. doi: 10.1016/0076-6879(74)30046-8. [DOI] [PubMed] [Google Scholar]
- 38.Ermolenko DN, Cornish PV, Ha T, Noller HF. Antibiotics that bind to the A site of the large ribosomal subunit can induce mRNA translocation. RNA. 2013;19:158–166. doi: 10.1261/rna.035964.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 40.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 41.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160:219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salsi E, Farah E, Netter Z, Dann J, Ermolenko DN. Movement of elongation factor G between compact and extended conformations. J Mol Biol. 2015;427:454–467. doi: 10.1016/j.jmb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc Natl Acad Sci USA. 2014;111:15060–15065. doi: 10.1073/pnas.1410873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valle M, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 45.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- 47.Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol. 2011;18:1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu G, et al. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon-anticodon duplex. Nat Struct Mol Biol. 2014;21:817–824. doi: 10.1038/nsmb.2869. [DOI] [PubMed] [Google Scholar]
- 49.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 50.Lancaster LE, Savelsbergh A, Kleanthous C, Wintermeyer W, Rodnina MV. Colicin E3 cleavage of 16S rRNA impairs decoding and accelerates tRNA translocation on Escherichia coli ribosomes. Mol Microbiol. 2008;69:390–401. doi: 10.1111/j.1365-2958.2008.06283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor DJ, et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, et al. Elongation factor G initiates translocation through a power stroke. Proc Natl Acad Sci USA. 2016;113:7515–7520. doi: 10.1073/pnas.1602668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong S, et al. Mechanism of tRNA-mediated +1 ribosomal frameshifting. Proc Natl Acad Sci USA. 2018;115:11226–11231. doi: 10.1073/pnas.1809319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.