Significance
Control of multiple life-critical physiological and fundamental behavioral functions requires the hypothalamus. Here, we provide a comprehensive description and analysis of mammalian intrahypothalamic network organization at the level of gray matter regions (macroscale). Network analysis revealed deep top-level network integration, but regional organization of two second-level subsystems (or subnetworks) indicated partial separation between control of physiological functions and behavioral functions; furthermore, inclusion of dominant network nodes in the subnetwork associated prominently with physiological functions suggests network primacy for physiological control. Comparing network organization to inhibitory (GAD65) and excitatory (VGLUT2) neurotransmission-associated gene markers revealed a significant positive correlation between network centrality (dominance) and the inhibitory marker. We introduce a novel intrahypothalamic network model to guide future hypothesis-driven investigations into hypothalamic structure and function.
Keywords: hypothalamus, mammal, neuronal connections, neurome, neuroinformatics
Abstract
Control of multiple life-critical physiological and behavioral functions requires the hypothalamus. Here, we provide a comprehensive description and rigorous analysis of mammalian intrahypothalamic network architecture. To achieve this at the gray matter region (macroscale) level, macroscale connection (macroconnection) data for the rat hypothalamus were extracted from the primary literature. The dataset indicated the existence of 7,982 (of 16,770 possible) intrahypothalamic macroconnections. Network analysis revealed that the intrahypothalamic macroconnection network (its macroscale subconnectome) is divided into two identical top-level subsystems (or subnetworks), each composed of two nested second-level subsystems. At the top-level, this suggests a deeply integrated network; however, regional grouping of the two second-level subsystems suggested a partial separation between control of physiological functions and behavioral functions. Furthermore, inclusion of four candidate hubs (dominant network nodes) in the second-level subsystem that is associated prominently with physiological control suggests network primacy with respect to this function. In addition, comparison of network analysis with expression of gene markers associated with inhibitory (GAD65) and excitatory (VGLUT2) neurotransmission revealed a significant positive correlation between measures of network centrality (dominance) and the inhibitory marker. We discuss these results in relation to previous understandings of hypothalamic organization and provide, and selectively interrogate, an updated hypothalamus structure–function network model to encourage future hypothesis-driven investigations of identified hypothalamic subsystems.
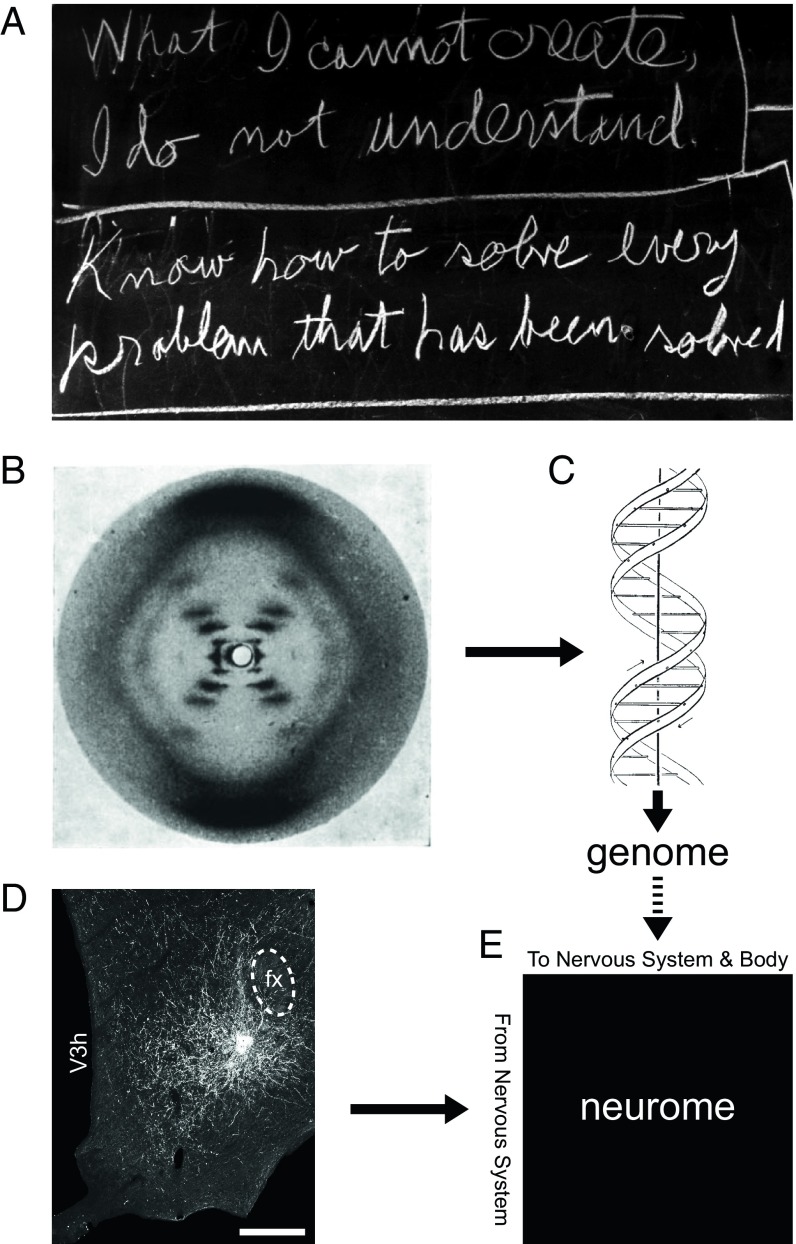
The renowned theoretical physicist Richard P. Feynman, whose birth centenary was in 2018, also explored far afield (1) and valued modeling problems to gain understanding—a philosophy alluded to in two statements he wrote on his office chalkboard that were present there on his last day of life (Fig. 1A): “What I cannot create, I do not understand,” and “Know how to solve every problem that has been solved.” The first statement supports using models to gain understanding; the second recognizes the value to future progress in understanding how solutions (leading to models) are arrived at. In biology, these tenets are exemplified by the discovery of the structure of DNA (2), with empirical evidence from X-ray crystallography experiments (3) (Fig. 1B) leading to a structural model (Fig. 1C). Evidence-based models are also a mainstay of systems neuroscience, in which the system being modeled—the nervous system—is generally considered to be the most complex biological system (4). A central goal, supported by models, is to understand the connectional organization of the nervous system both intrinsically (its connectome) (5, 6) and in relation to the body (the neurome; Fig. 1E) (6, 7) at different scales of granularity. These scales range from gray matter region (macroscale), to neuron type (mesoscale), to single neuron (microscale), to synaptic (nanoscale) (6). Macroscale neuronal connection data are typically obtained from pathway-tracing experiments (Fig. 1D).
Fig. 1.
Empirical models advance understanding. (A) Interrelated statements by Richard Feynman conveying how knowledge-based modeling can advance scientific understanding. (B) The X-ray diffraction pattern of DNA obtained by Rosalind Franklin (B) was instrumental for determining the double helical structure of DNA by James Watson and Francis Crick (C), enabling genome determination. (D) Visualized injection site of the anterograde neuronal pathway tracer Phaseolus vulgaris-leucoagglutinin in the rat hypothalamus. fx, fornix; V3h, third ventricle, hypothalamic part. (Scale bar: 250 µm.) (E) Data obtained from pathway-tracing experiments can be used to construct a network model for the complete nervous system that describes connections between all parts of the nervous system and between the nervous system and the rest of the body—a neurome (7); genome structure is a fundamental determinant of neurome structure (dashed arrow). (A) Image courtesy of the Archives, California Institute of Technology. (B) Reproduced by permission from ref. 3, Springer Nature: Nature, copyright (1953). (C) Reproduced by permission from ref. 2, Springer Nature: Nature, copyright (1953). (D) Reproduced from ref. 18.
Using data-driven approaches, we recently investigated the macroscale network of the cerebral hemispheres (8) and their principle parts: the cerebral cortex (9) and cerebral nuclei (10). This led to novel network models for two of the four major divisions of the forebrain that play an essential role in the cognitive control of behavior. However, cerebral hemisphere function also requires ancillary neuronal networks that enable cognitively directed motor actions to occur in concert with sensory cues and behavioral state. Prominent supporting networks include those in the thalamus and hypothalamus (the two other main divisions of the forebrain). The thalamus plays a major role in supporting cognition by processing sensory information en route to the cerebral cortex, whereas the hypothalamus is vital for subcognitive control of fundamental physiological processes and survival behaviors (11).
Experimental evidence acquired over more than a century has established the necessity of the hypothalamus for the control of those behavioral and physiological functions of the body that are essential for survival and reproduction (for review, see refs. 12–14). Hypothalamic involvement in this control is diverse and includes all three nervous system divisions for motor output: neuroendocrine, autonomic, and somatomotor. This is illustrated by (i) hypothalamic neuroendocrine control of the pituitary gland, (ii) autonomic control of the cardiovascular system and abdominal viscera via preautonomic (sympathetic and parasympathetic preganglionic) connections, and (iii) (via other polysynaptic connections) hypothalamic somatomotor control of motivated behaviors that are fundamental to survival (defensive and aggressive, ingestive, reproductive, and exploratory behaviors) (11, 12).
Hypothalamic functional diversity reflects its structural and connectional complexity, from embryonic development to adult. A systematic study of mammalian brain development in the mid-1990s concluded that embryological differentiation of the rat hypothalamus was “an unusually complex, little understood process” (15). More recent investigations have advanced our understanding of the underlying genetics, but (as noted in a recent review), “the development of the hypothalamus remains poorly understood, with large and obvious gaps in the literature at every developmental stage” (16). Hypothalamic cytoarchitecture is highly differentiated, generally more so than other central nervous system (CNS) divisions (17). Illustrative of hypothalamic connectional complexity, one of its major subdivisions—the lateral hypothalamic area (LHA)—contains the most highly connected CNS regions identified to date [in terms of macroscale connections (macroconnections)] (18–20).
Challenges notwithstanding, some general organizing principles have emerged. A classical cytoarchitecturally based structural description of the hypothalamus divides it into three longitudinal zones: periventricular, medial, and lateral (21); and four transverse rostral-to-caudal levels: preoptic, supraoptic (latterly referred to as anterior), tuberal, and mammillary (22) (for review, see refs. 12 and 13). Developmental analysis to some extent supports an outside-in sequence with respect to the differentiation of the three longitudinal zones (lateral to periventricular), and differentiation of the four rostral-to-caudal levels is understood primarily in relation to adjacent structures (15).
Synthesis of structural and functional data has engendered various models of hypothalamic participation in the control of different fundamental behaviors, including ingestive (23), defensive (24), reproductive (25), and exploratory (26). However, despite persistent efforts, these models remain quite rudimentary, partly due to a lack of basic data and partly due to fragmentary synthesis of the available data. Nevertheless, it is pertinent to mention a general model for hypothalamic involvement in patterned motor actions, which places it at the top of a motor control hierarchy that includes successive motor pattern initiators, generators, and finally motoneuron pools—with “control” being used in the sense of “a certain level of endogenous activity, (perhaps some form of ‘set-point’)” (see figure 9 and text of ref. 11). The model is supported by loss/preservation-of-function experiments and is exemplified by control of locomotion. However, general applicability is suggested, providing a basis for understanding hypothalamic organization in relation to the three categories of motor output (neuroendocrine, autonomic, and somatic) that enable and sustain diverse behavioral expression (11, 27).
Given the diverse and complex nature of the hypothalamus, a holistic model of its intrinsic connection network would provide a foundation for future hypothesis-driven investigations into how hypothalamic circuits relate not only to specific sensory, cognitive, behavioral state, and motor functions, but also to the overall function of the hypothalamus as it relates to the interdependent motivations of individual and species survival. Accordingly, here, we investigated the organizing principles of the mammalian intrahypothalamic network based on rat macroconnection data that were obtained from pathway-tracing experiments and published in the primary literature. This is complemented with comparative analysis of gene markers associated with inhibitory [glutamic acid decarboxylase 65 (GAD65)] and excitatory [vesicular glutamate transporter 2 (VGLUT2)] neurotransmission. Network analysis is based on a weighted and directed connection matrix (for all 130 hypothalamic gray matter regions; 65 on each side), which is a macroscale intrahypothalamic subconnectome, and follows a strategy we have employed previously (8–10).
Results
Analysis Framework.
The analysis is based on macroconnections reported in the primary literature between all regions of the adult rat hypothalamus, including connections originating and terminating on the same side (ipsilateral) or opposite sides (contralateral) of the hypothalamus. It is independent of possible left/right or male/female differences because, with one exception, none of the reports investigated such differences [one report included a semiquantitative male/female analysis (28) that indicated minor differences, at most]. A recently revised rat brain atlas (17) was used to identify hypothalamic gray matter regions that were considered as individual nodes for subsequent network analysis. Anatomical nomenclature follows the reference atlas, which was chosen because it is the only available standard, hierarchically organized, annotated parcellation and nomenclature for the rat brain; in addition, it has a nomenclature designed to have panmammalian applicability.
A total of 65 hypothalamic gray matter regions were identified on each side of the brain, resulting in a total of 16,770 possible macroconnections: 8,320 (652 × 2 − 65) within each side, and 8,450 (652 × 2) between each side. A dataset of 9,164 macroconnection reports was extracted and collated by J.D.H. from 66 publications in the primary literature spanning 40 y (from 1975) for the 8,385 possible intrahypothalamic ipsilateral and contralateral macroconnections originating in one side (2 × 652 − 65) (Dataset S1). In the absence of reports of statistically significant right/left differences, these numbers are doubled to give 18,328 macroconnection reports for the 16,770 possible macroconnections.
Data were obtained from experiments using monosynaptic anterograde and retrograde axonal pathway-tracing methods (15 in total), recorded for each macroconnection report (Dataset S1). Approximately 33 laboratories generated the data (56% of it from the L.W.S. laboratory—5,025 reports for macroconnections originating on one side), which were published in 11 different journal and book sources (51% in the Journal of Comparative Neurology).
Macroconnection Numbers and Data Validity.
For the hypothalamus on one side (numbers double for both sides), analysis of the collated data indicated 2,303 intrinsic hypothalamic ipsilateral macroconnections as present, and 1,375 as absent, between the 65 regions of the hypothalamus; this yields an ipsilateral macroconnection density of 62.6% [present ÷ (present + absent)]. In contrast, 1,194 intrinsic hypothalamic contralateral (between-sides) macroconnections were identified as present, and 2,478 as absent, equating to a contralateral macroconnection density of 32.5%. For the entire hypothalamus, 3,497 macroconnections (2,303 + 1,194) were present, and 3,853 were absent (1,375 + 2,478), yielding an overall macroconnection density of 47.6%. For network analysis, as in previous work (8–10), reports categorized as “no data” and “unclear” were assigned to and binned with reports in the “absent” category; reports categorized as “axons-of-passage” were assigned the nominal weight of “weak” and binned with other reports similarly categorized (Dataset S2). The resultant macroconnection densities were 55.3% (2,303 ÷ 4,160) for ipsilateral macroconnections, 28.3% (1,194 ÷ 4,225) for contralateral macroconnections, and 41.7% when combined.
No (or no adequate) data were found for 11.6% of possible ipsilateral macroconnections (482/4,160), resulting in a matrix coverage (fill ratio) of 88.4%. Fill ratio for contralateral macroconnections was slightly lower at 86.9% (553 no-data reports for 4,225 possible macroconnections). The complete matrix fill ratio was 87.7%. Taking the collated data as a representative sample of the 65-region matrix, the complete intrinsic macroconnection dataset for one side of the hypothalamus would contain ∼2,604 ipsilateral macroconnections (4,160 × macroconnection density ratio of 0.626) and ∼1,373 contralateral macroconnections (4,225 × macroconnection density ratio of 0.325). Combining the ipsilateral and contralateral macroconnection data, it follows that each side of the hypothalamus generates ∼3,991 intrinsic macroconnections (8,385 × macroconnection density ratio of 0.476) and, thus, ∼7,982 intrinsic macroconnections (of 16,770 possible) are generated in the complete hypothalamus. We also applied a metric for the validity of pathway-tracing methods (as described previously) (8, 9). The average validity of the pathway-tracing methods was 6.6 [on a scale of 1 (lowest) to 7 (highest)] for macroconnection reports of present intrahypothalamic macroconnections selected for network analysis, and the average validity was 6.2 for selected reports of macroconnections that do not exist (absent) (SI Appendix, Fig. S1 and Dataset S1).
Contralateral Heterotopic and Homotopic Macroconnections.
Contralateral hypothalamic macroconnections are generally weak, and contralateral heterotopic macroconnections (connecting different regions) are slightly weaker on average than homotopic macroconnections (connecting the same regions). On a 1 to 7 ordinal weight scale [1 (very weak) to 7 (very strong); absence of signal denoted by 0], average contralateral hetero- vs. homotopic macroconnection weights are 1.5 vs. 2.1; this slight difference is also reflected in average individual-region contralateral macroconnection weights that range from 0 to 3.4 for contralateral heterotopic macroconnections and from 0 to 4 for homotopic macroconnections. However, exceptions above the average are more pronounced for contralateral heterotopic macroconnections, and most notable are four with a weight of 7 (very strong): ventrolateral preoptic nucleus (VLP) to tuberomammillary nucleus (TM), LHA juxtadorsomedial region (LHAjd) to dorsal premammillary nucleus (PMd), LHA subfornical region anterior zone to PMd, and retina to suprachiasmatic nucleus (SCH). In contrast, the strongest hypothalamic homotopic macroconnections each have a weight of 4 (weak to moderate) and are formed by the following four regions: parastrial nucleus, SCH, arcuate hypothalamic nucleus (ARH), and ventral premammillary nucleus (PMv).
About half (47.7%, 31/65) of hypothalamic regions (on each side) form a homotopic connection, whereas all but two hypothalamic regions [subthalamic nucleus (STN) and preparasubthalamic nucleus (PSTN)] form at least one heterotopic contralateral connection (96.9%, 63/65; a total of 1,163 heterotopic macroconnections). This gives an average heterotopic contralateral in/out-degree (combined input and output connection number) of 18 [i.e., each hypothalamic region on one side connects with an average of 18 (1,163 ÷ 65) different hypothalamic regions on the opposite side]. Furthermore, of the 1,163 heterotopic contralateral macroconnections, 1,155 have an ipsilateral counterpart (>99%). In addition, in terms of macroconnection weights, all but 17 of the 1,163 heterotopic contralateral macroconnections have weights that are equal to or lower than their ipsilateral counterparts (i.e., about 98.5% of heterotopic macroconnections are stronger within a side than between sides). Lastly, the stronger an ipsilateral macroconnection, the more likely it is to also have a matching contralateral counterpart: If an ipsilateral macroconnection has a weight of 7 (very strong), the likelihood of there being a contralateral twin is >90%.
Multiresolution Consensus Clustering Analysis.
For network analysis, we employed a recently developed algorithmic community detection method called multiresolution consensus clustering (MRCC) (29). This method analyses directed weighted connections between network nodes (gray matter regions, in this case) across multiple network resolutions, to generate a coclassification matrix that is also a hierarchical nested network solution.
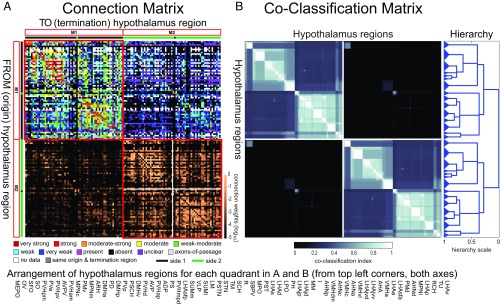
Application of MRCC to the complete 130 × 130 region matrix of connections within and between the right and left sides of the hypothalamus (HY2) yielded a two-subsystem (or two-module) top-level solution, with each bilateral top-level module composed of two second-level subsystems (Fig. 2A), whereas MRCC applied to the 65 regions on either side (HY1) yielded a top-level three-module solution for either side (for a total of six modules, with identical sets of three on each side) (SI Appendix, Fig. S2A). The term “module” is used here to refer to top-level subsystems (or subnetworks) as determined by the network analysis. The complete coclassification matrix for HY2 comprises a 21-level hierarchy of nested partitions with 30 bottom-level subsystems (15 on each side of the brain) (Fig. 2B). For HY1, the coclassification matrix comprises a 12-level hierarchy of nested partitions with 17 bottom-level subsystems (SI Appendix, Fig. S2B).
Fig. 2.
(A and B, Top) Connection and coclassification network matrices for the bilateral intrahypothalamic subconnectome (HY2). (Bottom) Arrangement of hypothalamus regions for each quadrant in A and B. (A) Directed and weighted monosynaptic macroconnection matrix for the rat hypothalamus, with gray matter region sequence in a subsystem arrangement derived from MRCC analysis (shown in B). Connection weights are represented by descriptive values (Upper half, color key shown Below), and on a log10 scale (Lower half, scale shown at right edge); sides 1 and 2 are indicated by green and black bars, respectively. Two bilateral top-level subsystems (M1 and M2) are outlined in red; two second-level subsystems are delineated by a white cross (shown only for M2 for clarity but applies to both M1 and M2). (B) Complete coclassification matrix obtained from MRCC (as in A) for the 130 regions (65 per side) of the hypothalamus. A linearly scaled coclassification index (shown Below) gives a range between 0 (no coclassification at any resolution) and 1 (perfect coclassification across all resolutions). Ordering and hierarchical arrangement are determined after building a hierarchy (Right) of nested solutions that recursively partition each cluster (i.e., subsystem), starting with the two top-level subsystems. The 30 subsystems obtained for the finest partition are indicated on the left edge of the dendrogram, while the two identical top-level subsystems (corresponding to M1 and M2) appear at the root of the tree (far right edge). A total of 21 distinct hierarchical levels are present, as determined by the sum of vertical cuts through each unique set of branches. The length of each distinct set of branches represents a distance between adjacent solutions in the hierarchical tree that may be interpreted as its persistence along the entire spectrum; dominant solutions extend longer branches, while fleeting or unstable solutions extend shorter branches. All solutions plotted in the tree survive the statistical significance level of α = 0.05. Abbreviations are defined in Dataset S2.
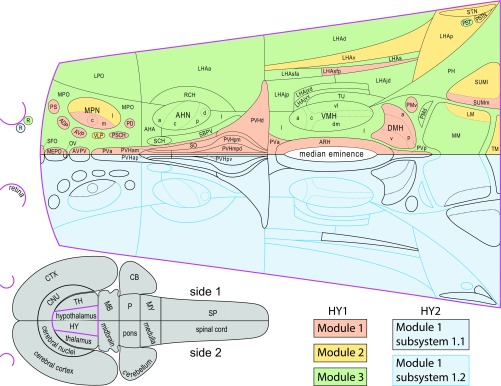
A flatmap representation (Fig. 3) facilitates comparison of the top-level modules of HY1 and HY2 (and the latter’s two second-level subsystems). A first point to note is that the bilateral structure of each top-level HY2 module is the result of the predominantly crossed connections of a single outlying region—the retina. Second, it is apparent when comparing HY1 to HY2 that regions in HY1 module (M)1 or M3 assign respectively and exclusively to HY2 M1/2 second-level subsystems 1.1 (HY1 M1) or 1.2 (HY1 M3), whereas regions in HY1 M2 assign to one or the other of these HY2 subsystems (Fig. 3). Considering region number/module for HY1, M1 and M3 are the largest and each contains a similar number of regions (27 for HY1 M1 and 29 for HY1 M3) compared with nine regions for HY1 M2. Given that regions in HY1 M1 and HY1 M3 assign exclusively to either subsystem 1.1 or 1.2 of HY2 M1/2, while those in HY1 M2 split between these HY2 subsystems, the number of regions in each is also similar (33 for HY2 M1/2 subsystem 1.1, and 32 for HY2 M1/2 subsystem 1.2). Third, a medial–lateral topological division is apparent: regions in HY1 M1 and HY2 M1/2 subsystem 1.1 are mostly medial, whereas regions in HY1 M3 and HY2 M1/2 subsystem 1.2 are mostly lateral; by comparison, longitudinal axis region subsystem assignment is evenly distributed (Fig. 3). However, the nine regions of HY1 M2 are an exception to this general pattern (they are mostly caudal and include both medial and lateral components).
Fig. 3.
Comparison of bilateral (HY2) and unilateral (HY1) intrahypothalamic subsystems. (Top) Organization of within and between-sides (bilateral) top-level and second-level subsystems (HY2, Lower half of flatmap), and within-side (unilateral) top-level subsystems (HY1, Upper half of flatmap), for the hypothalamus macroscale subconnectome. To facilitate comparison, each bilaterally mirrored dataset is represented on one side of a flatmap of the rat hypothalamus (17)—the hypothalamus (magenta delineated) and its spatial relation to the CNS is represented on the gray flatmap at Lower Left. Top-level subsystems (modules) (for HY1 and HY2) and second-level subsystems (for HY2) are color coded (key at Lower Right). For HY1, there are six modules (three per side, as shown); for HY2, there are two bilateral modules (one shown). Each HY2 module (pastel blue) has two second-level subsystems: mostly medial HY2 subsystem 1.1 (black delineated) includes all regions in HY1 module 1 (pastel pink); mostly lateral HY2 subsystem 1.2 (blue delineated) includes all regions in HY1 module 3 (pastel green). HY1 module 2 (pastel yellow) includes fewer regions than the other HY1 modules, and these separate into one or the other HY2 second-level subsystem. Abbreviations are defined in Dataset S2.
Centrality Metrics and Additional Network Attributes.
In addition to MRCC analysis, we investigated hypothalamic network properties using four common network centrality metrics (measures that indicate the dominance/“importance” of each node in the network): degree, strength, betweenness, and closeness (8–10). The centrality metric of degree measures the number of input (in-degree) or output (out-degree) connections for each network node (here, each gray matter region); strength represents the total weight of each node’s macroconnections; and the related centrality measures of betweenness and closeness take account of the shortest path between nodes and are considered to provide an indication of node centrality with respect to information flow.
With respect to degree, considering macroconnections reported as present yielded an average per-region single-side in-degree or out-degree value of 35 (2,303 ÷ 65) for ipsilateral macroconnections, 18 (1,194 ÷ 65) for contralateral macroconnections, and 54 (3,497 ÷ 65) for both. However, individual-region in-degree values ranged from 0 to 56 for ipsilateral macroconnections, from 0 to 40 for contralateral macroconnections, and from 0 to 94 for both; out-degree values for individual regions ranged from 1 to 58 for ipsilateral macroconnections, from 0 to 51 for contralateral macroconnections, and from 1 to 108 for both.
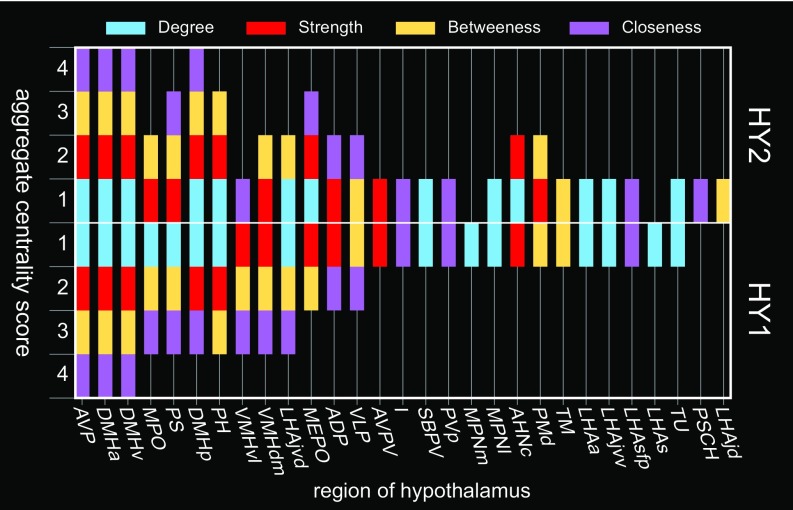
To identify regions with the highest overall centrality, an aggregate score was calculated for regions scoring in the top 20th percentile for each centrality metric (Fig. 4 and SI Appendix, Fig. S3). Regions scoring in the top 20th percentile for all four measures of centrality were considered candidate hubs. For HY2, four regions met this criterion: Anteroventral periventricular nucleus (AVP) and dorsomedial hypothalamic nucleus anterior- (DMHa), posterior- (DMHp), and ventral (DMHp) parts (Fig. 4). The top 20th percentile aggregate centrality scores for HY1 and HY2 were mostly similar, but there were also notable differences, indicative of a substantial contribution of contralateral connections to the whole network (Fig. 4).
Fig. 4.
Central nodes of the intrahypothalamic network. Identification of candidate hub regions (and others with high network centrality) for the bilateral (HY2) and unilateral (HY1) hypothalamic subconnectomes. Regions are assigned a score of 0 to 4 according to the number of times they fall within the top 20th percentile for each of four measures of centrality (degree, strength, betweenness, and closeness) and are arranged from left to right by HY1 descending aggregate centrality and topographically (17). Regions with a centrality score of 4 are considered candidate hubs. For individual-region centrality values for each measure of centrality (for HY2), see SI Appendix, Fig. S3. Note that aggregate centrality scores are modulated between HY1 and HY2, indicative of the relevance of HY2 contralateral connections to the overall structure of the network. Abbreviations are defined in Dataset S2.
We investigated two additional network properties: the attributes of “small-world” and “rich-club.” Previously, we investigated these for macroscale cerebral hemisphere subconnectomes (8–10). The small-world attribute is characteristic of networks with clustered nodes connected via short paths, whereas rich-club, in network theory parlance, refers to a group (subgraph) of well-connected network nodes that are also densely connected with one another. For both HY1 and HY2, there was only weak indication of small-world and rich-club organization. With respect to small-world attributes, we computed weighted clustering coefficients (HY1, 0.0155; HY2, 0.0080) and path lengths (HY1, 259.4; HY2, 260.6) and compared these values to a population of 1,000 randomly rewired networks preserving degree sequence (values are mean ± SD) [clustering: 0.0127 ± 3 × 10−4 and 0.0049 ± 1 × 10−4; path length: 240.0 ± 2.9 and 241.4 ± 5.1]. These metrics are only weakly indicative of small-world organization, mainly due to only modest levels of clustering relative to the null model. With respect to rich-club attributes, neither HY1 nor HY2 was found to contain a densely connected subgraph of hubs. Analysis of the HY1 network revealed a 48-node subgraph with significantly greater density compared with 1,000 randomized networks; however, the excess in density (over the null model) was less than 7% and the subgraph included almost three-fourths of the entire network, thus excluding it from consideration as a rich-club (no rich-club was found in HY2).
Comparison of Network Centrality with Markers of Excitatory and Inhibitory Neurotransmission.
A critical determinant of neuronal network function is whether it is inhibitory or excitatory. To investigate this property, we mapped gene markers associated with inhibitory (GAD65) (30), and excitatory (VGLUT2) (31) synaptic neurotransmission. The levels of mRNA for GAD65 and VGLUT2 were analyzed for 64 of 65 hypothalamic regions—the retina was not included, but it was previously reported that its output neurons express VGLUT2 (32).
The results indicate that GAD65 and VGLUT2 are both highly and heterogeneously expressed in the hypothalamus, consistent with and confirmatory to previous work (33, 34). GAD65 and/or VGLUT2 mRNA was detected in all regions, and most regions (81.5%, 53/65) express both markers (Dataset S3). However, one region expressed GAD65, but not VGLUT2 (i.e., anterior hypothalamic nucleus dorsal region), and six regions (seven if including the retina) expressed VGLUT2, but not GAD65 [i.e., supraoptic nucleus, PMd, medial mammillary nucleus, lateral mammillary nucleus (LM), PSTN, and STN]. Moreover, in regions that expressed both GAD65 and VGLUT2, there was considerable inter- and intraregion variation in expression levels, and equal expression levels of both markers in the same region was rare (nine regions, 13.8%). Nevertheless, aggregate regional expression levels for the hypothalamus were approximately equal [expression levels of GAD65 were just 2% higher than those of VGLUT2 (Dataset S3)—a slim margin further reduced by inclusion of the retina].
To relate expression of GAD65 and VGLUT2 to the results of the network analysis, their regional expression levels were compared with the aggregate centrality scores for each hypothalamic region in HY2 (Fig. 5). This comparative analysis revealed a significant positive correlation between GAD65 and all four measures of centrality (degree: ρ = 0.256, P = 0.04; strength: ρ = 0.317, P = 0.01; betweenness: ρ = 0.343, P = 0.005; and closeness: ρ = 0.400, P = 0.001; Spearman’s rank order correlation). In contrast, only closeness presented a significant (negative) correlation with VGLUT2 (ρ = −0.276, P = 0.02). (SI Appendix, Fig. S4).
Fig. 5.
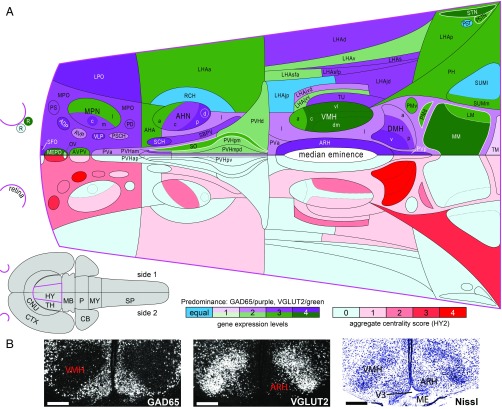
Comparison of intrahypothalamic network centrality with GAD65 and VGLUT2 mRNA expression. (A, Lower half of flatmap) Regional distribution of aggregate centrality measures for the bilateral hypothalamus macroscale subconnectome (HY2) (color key shown Below Right). (A, Upper half of flatmap) mRNA expression levels of GAD65 and VGLUT2 (color key shown Below Middle). For comparison, each bilateral dataset is represented on one side of a rat hypothalamus flatmap. The hypothalamus (magenta) and its spatial relation to the CNS is represented on the gray flatmap at Lower Left. A five-point index is used for both centrality and mRNA expression levels: for centrality, regions are assigned a score of 0 to 4 according to the number of times they fall within the top 20th percentile for each of four measures of centrality (degree, strength, betweenness, and closeness); for gene expression, regions are assigned a score of 1 to 4 according to binned data for their average expression levels ranging from 0 to 7 (1, 0 to 1.75; 2, >1.75 to 3.5; 3, >3.5 to 5.25; and 4, >5.25 to 7). Flatmap coloration for aggregate centrality follows the corresponding color scale (candidate hubs have a score of 4 and are bright red). Flatmap coloration for mRNA expression indicates region predominance (GAD65/purple, VGLUT2/green) and expression level (1 to 4); regions with equal expression levels of both genes are blue (these regions had a binned score of 2); regions outlined in white express either GAD65 or VGLUT2. (B) Representative darkfield photomicrographs of mRNA expression for GAD65 (Left) and VGLUT2 (Middle) at a midrostrocaudal level of the VMH nucleus (shown in corresponding Nissl-stained section image, Right). Note the inverse relationship between GAD65 and VGLUT in the VMH and ARH. (Scale bars: 500 µm.) ME, median eminence; V3, third ventricle. Abbreviations for the hypothalamus flatmap are defined in Dataset S2 and those for the gray flatmap are defined in Fig. 3. Maps adapted from ref. 17.
Discussion
Analysis of macroconnection data extracted from the primary literature for the rat intrahypothalamic subconnectome (HY2) indicated the existence of 7,982 of 16,770 possible connections (from 87.7% data coverage), a connection density of 47.6%. Comparable analysis recently applied to the endbrain (EB) and its principle divisions—cerebral cortex (CTX) and cerebral nuclei (CNU)—indicated connection densities of 17.9% (EB), 22.8% (CNU), and 24.5% (CTX) (8). Evidently, HY2 is markedly more connection dense than the intrinsic networks (subconnectomes) for these other divisions of the forebrain. Connection density differences for the component ipsilateral and contralateral subconnectomes are even greater. For example, the connection density of the ipsilateral intra-CTX subconnectome is 37.7% compared with 62.6% for the hypothalamus (a 66% increase); for the contralateral subconnectome, it is 10.2% for the CTX compared with 32.5% for the hypothalamus (a 219% increase) (8). The comparatively high hypothalamic connection density accounts for the marginal expression of the small-world network attribute (characterized by simultaneous high clustering and short path length) compared with relatively robust small-worldness exhibited by EB2, CTX2, and CNU2 (SI Appendix, Fig. S5). This organization may reflect reduced wiring cost afforded by the greater spatial compactness of hypothalamic regions compared with CNU and CTX regions (17). Similar considerations may underlie the absence of intrahypothalamic rich-club expression (a feature of networks whose most highly connected nodes are highly interconnected).
Comparing the current MRCC analysis to an earlier-alluded-to hypothalamic structure–function model (11, 35) facilitates exploration of possible functional interpretations. To elaborate, the earlier model identifies subgroupings based on structural and functional properties, giving five divisions: (i) a neuroendocrine motor zone associated with pituitary gland control; (ii) medial zone nuclei forming part of a putative behavior control column; (iii) a highly interconnected group of five rostrally located regions and the three regions of the DMH that together are considered to form a theoretical “visceromotor network” (35) for generating patterned autonomic and neuroendocrine motor output; (iv) a periventricular region related to (and possibly expanding) the visceromotor network, defined essentially by what remains of the periventricular and medial zones after subtracting the neuroendocrine motor zone, medial zone nuclei, and visceromotor network; and (v) a lateral zone associated prominently with the behavior control column, which is supported by more recent data (18–20, 36).
The arrangement of regions in the two second-level HY2 subsystems largely follows the five divisions of the earlier model, with HY2 M1/2 subsystem 1.1 including (and mostly composed of) all regions of the neuroendocrine motor zone and the visceromotor network, and with HY2 M1/2 subsystem 1.2 mostly composed of medial zone nuclei (behavior control column), the periventricular region, and the lateral zone (Fig. 6). This grouping of regions supports an updated structure–function model for the hypothalamus comprising two longitudinal divisions: one that is mostly periventricular and associated prominently with physiological control, and the other mostly medial–lateral and associated prominently with behavioral control (Fig. 7). The suggested divisions are supported further by inclusion in HY2 M1/2 subsystem 1.1 of neuroendocrine-related hypothalamic circumventricular organs (organum vasculosum of the lamina terminalis and subfornical organ) as well as four candidate hub regions that all are members of the visceromotor network: AVP, DMHa, DMHp, and DMHv. However, the existence of only two bilateral top-level modules for HY2 underscores integration of the complete intrahypothalamic network—congruent with the functional understanding that behavioral and physiological homeostatic processes are mutually supportive and that both support the overarching function of the hypothalamus to support survival and reproduction (11) (Fig. 7).
Fig. 6.
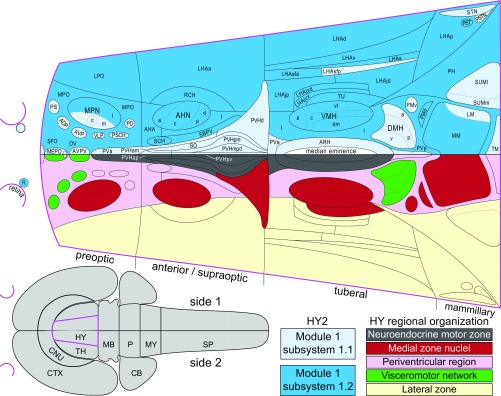
Comparison of current (network) and previous (cell group-based) models of hypothalamic organization. An established cell group-based model of hypothalamic organization (Lower half of flatmap) divides it into four transverse levels (preoptic, anterior/supraoptic, tuberal, mammillary) and three longitudinal zones (periventricular, medial, lateral). Refinements to this model include the identification of a neuroendocrine motor zone (gray), medial zone nuclei (red) considered to form the rostral end of a behavior control column, and a periventricular region (pink) containing a putative visceromotor network (green), leaving the remaining lateral zone (yellow) (11, 35). The Upper half of the flatmap shows the two top-level partitions of the bilateral hypothalamus macroscale subconnectome (HY2). For comparison, each organization schema is presented on a single side. Most HY2 subsystem 1.1 regions (lighter blue) are in the periventricular zone and include all regions of the neuroendocrine motor zone and a putative visceromotor network, whereas most HY2 subsystem 1.2 regions (darker blue) are in the lateral and medial zones. The hypothalamus (magenta) and its spatial relation to the CNS is represented on the gray flatmap at Lower Left.
Fig. 7.
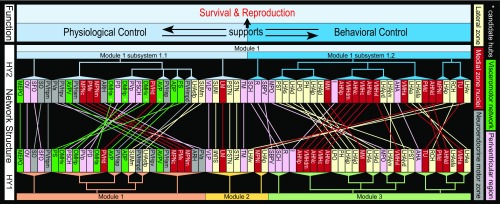
Hypothalamus structure–function network model. Network structures emerging from MRCC analysis of the bilateral (HY2) and unilateral (HY1) hypothalamus macroscale subconnectomes (colored dendrograms) are compared with one another and with an earlier model of hypothalamic cell group and network organization (11, 35). Colored lines connecting regions for HY1 and HY2 show correspondence between subsystem assignment determined by MRCC. Two-thirds of HY2 M1/2 subsystem 1.1 regions are in the periventricular zone, which includes the periventricular region (pink); included in this partition are all regions of a putative hypothalamic visceromotor pattern generator network (green), all regions of the neuroendocrine motor zone (gray), and both hypothalamic circumventricular organs [organum vasculosum of the lamina terminalis (OV) and subfornical organ (SFO)]; in contrast, four-fifths of HY2 M1/2 subsystem 1.2 regions are in the medial (dark red) and lateral (light yellow) hypothalamic zones. Overall, this organization suggests a prominent functional association for HY2 M1/2 subsystem 1.1 with physiological control (especially neuroendocrine signaling), and for subsystem 1.2 with behavioral control (especially somatomotor signaling). Moreover, primacy of physiological control is suggested by inclusion of all four candidate hubs in HY2 M1/2 subsystem 1.1 (white asterisks). However, both HY2 top-level subsystems 1.1 and 1.2 include regions involved in autonomic and behavioral-state control, and the existence of only two bilateral HY2 top-level modules underscores deep intrahypothalamic integration. Communication between HY2 M1/2 subsystems 1.1 and 1.2 may be considered mutually supportive, and both support the prime function of the hypothalamus to support survival and sexual reproduction. Abbreviations are defined in Dataset S2.
Whichever level of network resolution is interrogated, the general comparative (and hypothesis-generating) approach is to consider how a novel network structure compares to existing models. We have followed this approach at a high level (above discussion) and, in the remaining discussion, consider selected examples at lower nested levels of the network hierarchy.
In the earlier model (see figure 12 of ref. 11), regions of the behavior control column division are subdivided by structural connectivity and by association with different categories of goal-directed behavior. For example, reproductive behavior involves the medial preoptic nucleus (MPN) lateral part, the ventromedial hypothalamic nucleus (VMH) ventrolateral part, and the PMv; defensive behavior involves the anterior hypothalamic nucleus, the VMH dorsomedial part, and the PMd. More recent evidence indicates defensive behavior may also involve the VMH central part (24) and the LHAjd (37). With one exception (PMv), all these regions are in HY2 M1/2 subsystem 1.2, consistent with an updated model (Fig. 7).
Additional comparison with lower-level HY2 subsystems reveals novel associations. For example, the PMv previously appeared to be an outlier, but is grouped in a third-level subsystem that includes several other regions associated with reproductive function, including the two other MPN divisions (central and lateral) and especially the anteroventral periventricular nucleus and ARH involved in critical neuroendocrine reproductive control (12). In fact, while the PMv is associated strongly with both neuroendocrine and somatomotor control (12, 24, 27), the current model emphasizes the former. This illustrates further the highly integrated intrahypothalamic network structure and also suggests that theoretical functional interpretation of an MRCC nested network hierarchy is informed by its consensus basis, analogous to a multilayered functional heatmap. With respect to the third-level PMv-included HY2 subsystem, it also suggests that other regions in this subsystem not clearly associated with reproductive function merit reevaluation. This strategy could also be applied to other regions and subsystems, such as the DMH candidate hub that is indicated to integrate behavioral and behavioral-state control (Fig. 7) (38).
As a second example, note that HY1 has six top-level modules (i.e., M1 to M3, each side) and that one of these modules, HY1 M2, splits between HY2 M1/2 subsystems 1.1 and 1.2 (unlike HY1 M1 and HY1 M3 that transpose directly to HY2 M1/2 subsystems 1.1 and 1.2, respectively) (Figs. 3 and 7). This distinguishes HY1 M2 from HY1 M1 and HY1 M3, and HY1 M2 is further distinguished by having only nine regions compared with 27 and 29 for HY1 M1 and HY1 M3, respectively. At the second hierarchical level, HY1 M2 has two subsystems of seven and two regions. All but one region in the seven-region subsystem are in HY2 M1/2 subsystem 1.1, with the remaining region and the two-region subsystem in HY2 M1/2 subsystem 1.2 (Fig. 7). At face value, this indicates that HY1 M2 is less robust than either HY1 M1 or HY1 M3. Given also the suggested general differentiation between physiological and behavioral control for HY2 M1/2 subsystems 1.1 and 1.2, it is salient to consider functional associations for the HY1 M2 regions that assign differently to these HY2 subsystems. We consider here the six regions of HY1 M2 that are in HY2 M1/2 subsystem 1.1: VLP, supramammillary nucleus (SUM) lateral part (SUMl), LM, TM, PSTN, and STN. In addition, in HY2, these six regions are in a partition that includes a seventh region: the SUM medial part (SUMm) (Fig. 7).
The VLP and TM are implicated (opposingly) in behavioral-state control (VLP in sleep state; TM in awake state) (39). The LM has an established role in processing directional heading information during locomotion (40). The SUMm and SUMl are also associated with locomotion via their generative role for the hippocampal theta rhythm (41) present during locomotion (42); further SUM association is with REM sleep, which is also positively correlated to the hippocampal theta rhythm (43). The PSTN is indicated to play a broad role that includes coordination of parasympathetic responses associated with cardiovascular function and ingestive behavior, and may also influence central relay of sensory information relating to the latter (44, 45). Lastly, the STN is implicated in somatomotor control, notably of orofacial and limb movements (46). With respect to limb movements, STN lesion is associated with the rare movement-disorder ballism, characterized by uncontrollable “throwing” of the limbs (47). The STN has also received attention as a therapeutic target site for other movement disorders, including Parkinson’s disease (47, 48). In sum, these regions are associated with goal-directed behavior, locomotion necessary to obtain a goal, and behavioral state that determines when these are active (broad functions germane to multiple specific behaviors). Given indicated STN involvement in several diseases affecting locomotor control and its use as a therapeutic target, corelative investigation of other regions in the same HY2 third-level subsystem appears relevant.
Our analysis of the macroscale intrinsic network architecture of the hypothalamus reveals structure–function relations that tend to increase in specificity with increasing network resolution, but the whole network is richly integrated. More generally, we have demonstrated how data-driven network modeling approaches can be employed as hypothesis-generating tools, with selected examples provided by interrogation of an updated model of the intrahypothalamic network, and we hope this encourages further investigation of the multiple intrahypothalamic subnetworks described here. Thus far, we have investigated macroscale subconnectomes for the cerebral hemispheres (8–10) and for the hypothalamus (this study). Future investigations will be aided by a more comprehensive understanding of the network architecture of the nervous system.
Materials and Methods
Network Analysis.
All network analysis methods used here follow those described previously (8–10), including a recently introduced method for MRCC analysis (8, 29). All macroconnection data obtained from the primary literature were interpreted in relation to the current version of the only available standard, hierarchically organized, annotated parcellation and nomenclature for the rat brain (17). Within- and between-sides connection reports were assigned ranked qualitative connection weights (reported values) according to their description; an ordinal scale [1 (very weak) to 7 (very strong)] was used. Connection report data and annotations are provided in a Microsoft Excel spreadsheet (Dataset S1), as are the data extracted from these reports to construct connection matrices (Dataset S2). To facilitate access to, and use of, the connection-report data, they are freely available as a searchable resource at The Neurome Project (http://www.neuromeproject.org). For weighted network analysis, an exponential scale was applied to the ordinal weight categories. As in previous work (8–10), the scale spanned 4 orders of magnitude and is consistent with quantitative pathway-tracing data in rats (7). Network analyses were carried out on the directed and log-weighted rat intrahypothalamic macroconnection matrix (Dataset S2, worksheet “HY topographic bins”) using tools collected in the Brain Connectivity Toolbox (www.brain-connectivity-toolbox.net/).
In Situ Hybridization for GAD65 and VGLUT2.
Methods and controls for isotopic in situ hybridization for detection of GAD65 and VGLUT2 mRNA were fully described previously (49). Tissue sections were obtained from an adult male Sprague-Dawley rat (all procedures were approved by the Institutional Animal Care and Use Committee at the University of Southern California). cDNA probes used to generate 35S-UTP-labeled cRNA probes for in situ hybridization were obtained from the following sources: GAD65 from M. G. Erlander, University of California, Los Angeles, CA (50), and VGLUT2 from D. R. Ziegler, University of Pikeville, Pikeville, KY (34). For analysis of VGLUT2 and GAD65 mRNA expression, an ordinal value ranging from 1 (very weak) to 7 (very strong) was recorded that qualitatively reflected signal strength (silver grains visible under darkfield microscopic illumination) relative to the overall range of signal observed independently for each gene marker (absence of signal was denoted by a 0). Data acquisition was aided with the use of a specialized Microsoft Excel template (Axiome C, created by J.D.H.) that facilitated data entry across multiple levels for each hypothalamic gray matter region as described in a rat brain reference atlas (17). Accordingly (excepting the retina), data were acquired for all hypothalamic regions across 22 transverse levels of the hypothalamus, with individual values recorded for each region at atlas-level resolution (average of four atlas levels per region) (Dataset S3). To negate the effects of interanimal variability, series of sections from the same rat brain were analyzed for each mRNA sequence and matched with an adjacent series of Nissl-stained sections to enable data transposition to the reference atlas. Analysis encompassed expression of markers on either side of the brain (no appreciable difference in signal between sides was observed for any region).
Supplementary Material
Acknowledgments
We thank Graciela Sanchez-Watts for her preparation of material that was used for analysis of GAD65 and VGLUT2 mRNA expression. This work was supported, in part, by grant funding from the Kavli Foundation (L.W.S. and J.D.H.) and by the National Institutes of Health Grant R01 NS029728 (to A.G.W.). The manuscript is dedicated by J.D.H. to Herbert C. Hahn, whose life’s work illumined for many a path to understanding their motivations and emotions.
Footnotes
Conflict of interest statement: L.W.S. and C.B.S. were coauthors of a 2016 Commentary.
Data deposition: An implementation of the network analysis method of multiresolution consensus clustering that was used in the present work is available at https://github.com/LJeub/HierarchicalConsensus. All connection reports used for this study are provided as supporting information (SI) in spreadsheet format (Microsoft Excel) in SI Appendix. The connection reports (including associated methodological information) are also deposited as a searchable resource at The Neurome Project (http://www.neuromeproject.org).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819448116/-/DCSupplemental.
References
- 1.Feynman RP, Phillips R, Gates B, Leighton R, Hutchings E. “Surely You’re Joking, Mr. Feynman!”: Adventures of a Curious Character. Reissue Ed W. W. Norton & Company; New York: 2018. [Google Scholar]
- 2.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 4.Bota M, Dong HW, Swanson LW. From gene networks to brain networks. Nat Neurosci. 2003;6:795–799. doi: 10.1038/nn1096. [DOI] [PubMed] [Google Scholar]
- 5.Sporns O, Tononi G, Kötter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swanson LW, Lichtman JW. From cajal to connectome and beyond. Annu Rev Neurosci. 2016;39:197–216. doi: 10.1146/annurev-neuro-071714-033954. [DOI] [PubMed] [Google Scholar]
- 7.Bota M, Sporns O, Swanson LW. Architecture of the cerebral cortical association connectome underlying cognition. Proc Natl Acad Sci USA. 2015;112:E2093–E2101. doi: 10.1073/pnas.1504394112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson LW, Hahn JD, Jeub LGS, Fortunato S, Sporns O. Subsystem organization of axonal connections within and between the right and left cerebral cortex and cerebral nuclei (endbrain) Proc Natl Acad Sci USA. 2018;115:E6910–E6919. doi: 10.1073/pnas.1807255115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson LW, Hahn JD, Sporns O. Organizing principles for the cerebral cortex network of commissural and association connections. Proc Natl Acad Sci USA. 2017;114:E9692–E9701. doi: 10.1073/pnas.1712928114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanson LW, Sporns O, Hahn JD. Network architecture of the cerebral nuclei (basal ganglia) association and commissural connectome. Proc Natl Acad Sci USA. 2016;113:E5972–E5981. doi: 10.1073/pnas.1613184113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886:113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- 12.Simerly RB. Organization of the hypothalamus. In: Paxinos G, editor. The Rat Nervous System. 4th Ed. Elsevier; Amsterdam: 2015. pp. 267–294. [Google Scholar]
- 13.Swanson LW. 1987. The hypothalamus. Handbook of Chemical Neuroanatomy, Integrated Systems of the CNS, Part 1, eds Hökfelt T, Björklund A, Swanson LW (Elsevier, Amsterdam), Vol 5, pp 1–124.
- 14.Swanson LW. Organization of the mammalian neuroendocrine system. In: Bloom FE, editor. Handbook of Physiology, the Nervous System. Vol 4. Waverly Press; Baltimore: 1986. pp. 317–363. [Google Scholar]
- 15.Alvarez-Bolado G, Swanson LW. Developmental Brain Maps: Structure of the Embryonic Rat Brain. Elsevier; New York: 1996. [Google Scholar]
- 16.Bedont JL, Newman EA, Blackshaw S. Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip Rev Dev Biol. 2015;4:445–468. doi: 10.1002/wdev.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanson LW. Brain maps 4.0-Structure of the rat brain: An open access atlas with global nervous system nomenclature ontology and flatmaps. J Comp Neurol. 2018;526:935–943. doi: 10.1002/cne.24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn JD, Swanson LW. Connections of the juxtaventromedial region of the lateral hypothalamic area in the male rat. Front Syst Neurosci. 2015;9:66. doi: 10.3389/fnsys.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn JD, Swanson LW. Connections of the lateral hypothalamic area juxtadorsomedial region in the male rat. J Comp Neurol. 2012;520:1831–1890. doi: 10.1002/cne.23064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn JD, Swanson LW. Distinct patterns of neuronal inputs and outputs of the juxtaparaventricular and suprafornical regions of the lateral hypothalamic area in the male rat. Brain Res Brain Res Rev. 2010;64:14–103. doi: 10.1016/j.brainresrev.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crosby EC, Woodburne RT. The comparative anatomy of the preoptic area and the hypothalamus. Proc Assoc Res Nervous Mental Dis. 1940;20:52–169. [Google Scholar]
- 22.Le Gros Clark WE. Morphological aspects of the hypothalamus. In: Le Gros Clark WE, Beattie J, Riddoch G, Dott NM, editors. The Hypothalamus: Morphological, Functional, Clinical and Surgical Aspects. Oliver and Boyd; Edinburgh: 1938. pp. 1–68. [Google Scholar]
- 23.Sternson SM, Eiselt AK. Three pillars for the neural control of appetite. Annu Rev Physiol. 2017;79:401–423. doi: 10.1146/annurev-physiol-021115-104948. [DOI] [PubMed] [Google Scholar]
- 24.Canteras NS. Hypothalamic survival circuits related to social and predatory defenses and their interactions with metabolic control, reproductive behaviors and memory systems. Curr Opin Behav Sci. 2018;24:7–13. [Google Scholar]
- 25.Simerly RB. Wired for reproduction: Organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu Rev Neurosci. 2002;25:507–536. doi: 10.1146/annurev.neuro.25.112701.142745. [DOI] [PubMed] [Google Scholar]
- 26.Stuber GD, Britt JP, Bonci A. Optogenetic modulation of neural circuits that underlie reward seeking. Biol Psychiatry. 2012;71:1061–1067. doi: 10.1016/j.biopsych.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risold PY, Thompson RH, Swanson LW. The structural organization of connections between hypothalamus and cerebral cortex. Brain Res Brain Res Rev. 1997;24:197–254. doi: 10.1016/s0165-0173(97)00007-6. [DOI] [PubMed] [Google Scholar]
- 28.Cavalcante JC, Bittencourt JC, Elias CF. Distribution of the neuronal inputs to the ventral premammillary nucleus of male and female rats. Brain Res. 2014;1582:77–90. doi: 10.1016/j.brainres.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeub LGS, Sporns O, Fortunato S. 2018. Multiresolution consensus clustering in networks. Sci Rep 8:3259, arXiv:1710.02249. [DOI] [PMC free article] [PubMed]
- 30.Soghomonian JJ, Martin DL. Two isoforms of glutamate decarboxylase: Why? Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- 31.Takamori S, Rhee JS, Rosenmund C, Jahn R. Identification of differentiation-associated brain-specific phosphate transporter as a second vesicular glutamate transporter (VGLUT2) J Neurosci. 2001;21:RC182. doi: 10.1523/JNEUROSCI.21-22-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Land PW, Kyonka E, Shamalla-Hannah L. Vesicular glutamate transporters in the lateral geniculate nucleus: Expression of VGLUT2 by retinal terminals. Brain Res. 2004;996:251–254. doi: 10.1016/j.brainres.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 33.Bowers G, Cullinan WE, Herman JP. Region-specific regulation of glutamic acid decarboxylase (GAD) mRNA expression in central stress circuits. J Neurosci. 1998;18:5938–5947. doi: 10.1523/JNEUROSCI.18-15-05938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziegler DR, Cullinan WE, Herman JP. Distribution of vesicular glutamate transporter mRNA in rat hypothalamus. J Comp Neurol. 2002;448:217–229. doi: 10.1002/cne.10257. [DOI] [PubMed] [Google Scholar]
- 35.Thompson RH, Swanson LW. Structural characterization of a hypothalamic visceromotor pattern generator network. Brain Res Brain Res Rev. 2003;41:153–202. doi: 10.1016/s0165-0173(02)00232-1. [DOI] [PubMed] [Google Scholar]
- 36.Goto M, Canteras NS, Burns G, Swanson LW. Projections from the subfornical region of the lateral hypothalamic area. J Comp Neurol. 2005;493:412–438. doi: 10.1002/cne.20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rangel MJ, Jr, Baldo MV, Canteras NS, Hahn JD. Evidence of a role for the lateral hypothalamic area juxtadorsomedial region (LHAjd) in defensive behaviors associated with social defeat. Front Syst Neurosci. 2016;10:92. doi: 10.3389/fnsys.2016.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou TC, et al. Critical role of dorsomedial hypothalamic nucleus in a wide range of behavioral circadian rhythms. J Neurosci. 2003;23:10691–10702. doi: 10.1523/JNEUROSCI.23-33-10691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 40.Blair HT, Cho J, Sharp PE. Role of the lateral mammillary nucleus in the rat head direction circuit: A combined single unit recording and lesion study. Neuron. 1998;21:1387–1397. doi: 10.1016/s0896-6273(00)80657-1. [DOI] [PubMed] [Google Scholar]
- 41.Vertes RP. Major diencephalic inputs to the hippocampus: Supramammillary nucleus and nucleus reuniens. Circuitry and function. Prog Brain Res. 2015;219:121–144. doi: 10.1016/bs.pbr.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buzsáki G, Leung LW, Vanderwolf CH. Cellular bases of hippocampal EEG in the behaving rat. Brain Res. 1983;287:139–171. doi: 10.1016/0165-0173(83)90037-1. [DOI] [PubMed] [Google Scholar]
- 43.Montgomery SM, Sirota A, Buzsáki G. Theta and gamma coordination of hippocampal networks during waking and rapid eye movement sleep. J Neurosci. 2008;28:6731–6741. doi: 10.1523/JNEUROSCI.1227-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto M, Swanson LW. Axonal projections from the parasubthalamic nucleus. J Comp Neurol. 2004;469:581–607. doi: 10.1002/cne.11036. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, van den Pol AN. Rapid binge-like eating and body weight gain driven by zona incerta GABA neuron activation. Science. 2017;356:853–859. doi: 10.1126/science.aam7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mora F, Mogenson GJ, Rolls ET. Activity of neurons in the region of the substantia nigra during feeding in the monkey. Brain Res. 1977;133:267–276. doi: 10.1016/0006-8993(77)90763-6. [DOI] [PubMed] [Google Scholar]
- 47.Hamani C, Saint-Cyr JA, Fraser J, Kaplitt M, Lozano AM. The subthalamic nucleus in the context of movement disorders. Brain. 2004;127:4–20. doi: 10.1093/brain/awh029. [DOI] [PubMed] [Google Scholar]
- 48.Kalia SK, Sankar T, Lozano AM. Deep brain stimulation for Parkinson’s disease and other movement disorders. Curr Opin Neurol. 2013;26:374–380. doi: 10.1097/WCO.0b013e3283632d08. [DOI] [PubMed] [Google Scholar]
- 49.Watts AG, Sanchez-Watts G. Physiological regulation of peptide messenger RNA colocalization in rat hypothalamic paraventricular medial parvicellular neurons. J Comp Neurol. 1995;352:501–514. doi: 10.1002/cne.903520403. [DOI] [PubMed] [Google Scholar]
- 50.Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.