Significance
Biomolecular condensates comprise multiple protein and RNA molecules that are typically organized into complex, multilayered structures. Although recent studies have shown that multilayered condensates can arise spontaneously, the interactions that drive the spontaneous transitions remain unclear. Further, can molecules within the same layer have different dynamical properties or do such complex features require inputs of energy? Here, we report results from in vitro studies, which show that coexisting liquid- and solid-like material properties and multilayered architectures can result from spontaneous, sequence-encoded driving forces. Our studies, which are directed at the simplest biologically relevant protein and RNA sequences, suggest that spontaneous processes make key contributions to the formation of condensates with complex morphologies and diverse material properties.
Keywords: phase transitions, biomolecular condensates, complex coacervation, RNA, intrinsically disordered proteins
Abstract
Phase separation of multivalent protein and RNA molecules underlies the biogenesis of biomolecular condensates such as membraneless organelles. In vivo, these condensates encompass hundreds of distinct types of molecules that typically organize into multilayered structures supporting the differential partitioning of molecules into distinct regions with distinct material properties. The interplay between driven (active) versus spontaneous (passive) processes that are required for enabling the formation of condensates with coexisting layers of distinct material properties remains unclear. Here, we deploy systematic experiments and simulations based on coarse-grained models to show that the collective interactions among the simplest, biologically relevant proteins and archetypal RNA molecules are sufficient for driving the spontaneous emergence of multilayered condensates with distinct material properties. These studies yield a set of rules regarding homotypic and heterotypic interactions that are likely to be relevant for understanding the interplay between active and passive processes that control the formation of functional biomolecular condensates.
Phase transitions of protein and RNA molecules are implicated as drivers of spatial and temporal organization of cellular matter (1–3). The relevant phase transitions include first-order transitions such as phase separation and higher-order transitions such as sol−gel transitions (4, 5). These phase transitions are thought to be drivers of the biogenesis of membraneless organelles and other nonstoichiometric assemblies that are collectively referred to as biomolecular condensates (4, 6, 7). Key protein and RNA molecules, referred to as scaffolds, are found to be necessary and sufficient to drive phase separation in vitro (1–3). However, transferring insights from in vitro studies to an understanding of in vivo condensates requires an understanding of the interplay between driven (active) and spontaneous (passive) processes (2, 4, 8). Several in vivo observations regarding condensates are thought to involve active processes that incur the expenditure of energy while maintaining condensates in metastable states (2). Relevant observations include distinct material properties, inhibition of aging and fusion of condensates, the selective partitioning or extraction of molecular components, the dissolution of condensates, and the maintenance of spatially organized structures within condensates. For example, stress granules are cytoplasmic RNA−protein granules that form when cells are under stress and dissolve when the stresses abate (9, 10). Their functions/dysfunctions have been implicated in viral infections, cancers, and neurodegenerative disorders (11–16). Data from mammalian cells suggest that stress granules consist of dense solid-like cores of RNA and proteins surrounded by a labile liquid shell (17, 18). Similarly, the nucleolus consists of three distinct layers, and each layer is characterized by distinct inward and outward fluxes of protein and RNA molecules. Recent work suggests that the structure of the nucleolus could arise through differences in surface tensions associated with the distinct liquid phases (19). Indeed, it appears that many spatially organized structures can arise through spontaneous processes alone (20, 21). In contrast, in vivo regulation and dissolution of these architectures, as opposed to their formation, might require the intervention of active processes (22). For example, the layered topology of nucleoli is reversed during transcriptional inhibition, suggesting that additional molecular interactions might be involved in nucleolar regulation (23).
The majority of known biomolecular condensates contain multiple protein and RNA molecules. Even condensates that are not directly involved in RNA metabolism or harbor canonical RNA binding proteins seem to contain or even require RNA for their formation. For example, G bodies are reversible assemblies of glycolytic enzymes that form under hypoxia (24). Not only do these condensates stain positively for specific RNA molecules, but the treatment of cells with nucleases results in the dissociation of G bodies, thus suggesting a role for RNA molecules as scaffolds that drive their formation (24). Recent studies suggest that RNA molecules undergo conformational transitions such as chain compaction in the context of stress granules. This is to be contrasted with the expanded conformations formed by the same RNA molecules in the cytosol (25). Indeed, roughly 103 RNA species are preferentially associated with stress granules, and these RNAs are enriched for specific sequence and structural features (26, 27). RNA molecules also have the ability to undergo phase separation even in the absence of protein partners (28). RNA phase separation appears to underlie RNA foci formation in repeat expansion disorders (29). Additionally, RNA also controls the dynamic properties of RNA transport granules (30) and nucleoli (31, 32), implying that RNA molecules also regulate the material properties of condensed phases. To what extent do spontaneous processes governed by sequence-intrinsic properties of protein and RNA sequences explain in vivo observations? In this work, we seek answers to this question by investigating the impact of spontaneous processes governed by the interactions among the simplest, biologically relevant proteins and archetypal RNA molecules.
We deployed in vitro model systems for phase separation that were designed to uncover the rules governing RNA- and protein-driven interactions. For the protein system, we used biologically relevant arginine-rich and lysine-rich sequences that are intrinsically disordered (33). We investigated the interplay between these protein systems and a series of homopolymeric RNA sequences. Our results show that purine versus pyrimidine contents of RNA molecules directly influence condensate morphology and dynamics. We trace these effects to the details of nucleobase composition of RNA molecules and the affinities of protein−RNA interactions. Strikingly, the differences in molecule-specific protein−protein, protein−RNA, and RNA−RNA interactions lead to the spontaneous generation of spatially organized multilayered condensates with subcompartments. These findings provide insights regarding spontaneous, sequence-encoded driving forces for condensates with complex architectures and dynamics.
Results
Polyanions Determine Morphological and Dynamic Properties of Arginine-Based Protein Phase Separation.
Arginine-rich, intrinsically disordered, low-complexity domains (LCDs) have been established as key regions that drive protein phase separation in vitro and in vivo (33–41). Previous studies quantified the phase transitions of two types of toxic arginine-rich dipeptide repeats that are generated by aberrant translation of a hexanucleotide repeat expansion in the C9orf72 gene, which is the major genetic cause underlying amyotrophic lateral sclerosis (ALS) (42–44). Protein products of these repeats include poly(proline−arginine) and poly(glycine−arginine), which we refer to hereafter as PR and GR, respectively. These proteins target and perturb various condensates, including the nucleolus and stress granules (33–35). Mechanistic studies of PR phase separation in vitro revealed that phase separation of arginine-rich systems is driven by a combination of electrostatic interactions, and interactions involving pi systems (33, 45).
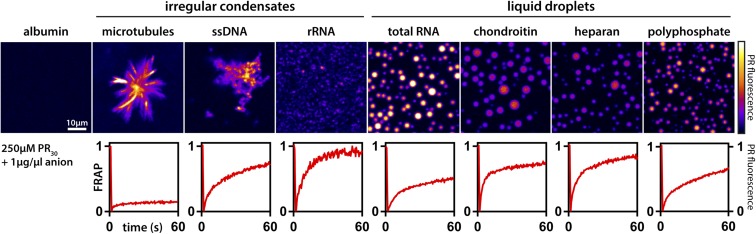
Phase separation driven by interactions among complexing polyions is known as complex coacervation (46). We examined PR-mediated phase separation via complex coacervation using a wide array of biological polyanions. Whereas albumin, a negatively charged protein, could not induce phase separation, long polyanions or negatively charged protein assemblies robustly drive either condensate formation or PR recruitment (Fig. 1). The morphologies of the resulting condensates were dependent on the specific type of polyanion and their assembly state: Microtubule asters or filaments provided a solid scaffold for PR recruitment, while flexible polyanions such as total yeast RNA or polyphosphate lead to the formation of spherical droplets, indicative of a minimization of the surface energy that is consistent with liquid−liquid phase separation driven by complex coacervation (47). Additionally, although the same PR polycation containing 30 PR repeats was used in all cases, we observed substantially different PR dynamics within condensates that depends on the polyanion in question, as assayed by fluorescence recovery after photobleaching (FRAP) (Fig. 1). These results indicate that chemical details of the complexing polyanions are important determinants of the material properties associated with protein−polyanion assemblies, and that the details of complex coacervation are influenced by the chemical details of the constituent cations and anions within polycations and polyanions, respectively. It would appear that a combination of generic and specific driving forces could be involved in the formation of condensates via complex coacervation. Our observations in our polyanion−protein model system are in line with observations for phase separation being driven by the combination of protein−protein, RNA−protein, and RNA−RNA interactions. Indeed, a molecular grammar for protein−protein phase separation highlights the importance of arginine residues (40, 48, 49). Phase separation of RNA in the absence of protein−RNA interactions appears to be driven mainly by base pairing interactions (28, 29). However, the details underlying protein−RNA interactions that give rise to protein−RNA condensates, especially with disordered proteins, remain unclear.
Fig. 1.
Morphologies and dynamics of condensates formed via complex coacervation of PR30 are governed by the chemistry of polyanions. (Top) Each panel shows PR30 (PR) fluorescence in the context of condensates formed by complex coacervation. All measurements were performed in the presence of 250 µM PR30 and 1 µg/µL of polyanion. Under these conditions, albumin, a generic protein macroanion, does not promote phase separation. However, all other polyanions in the current study promote phase separation, although the morphologies of the condensates are specific to the type of polyanion used to drive phase separation. (Bottom) Normalized kinetic traces from FRAP experiments. In each of the experiments, the fluorescently labeled PR30 molecules are photobleached, and the recovery of fluorescence is monitored as a function of time. FRAP dynamics of PR30 molecules exhibit a clear dependence on polyanions. Scale bar applies for all images.
RNA Structure Determines Coacervate Morphology.
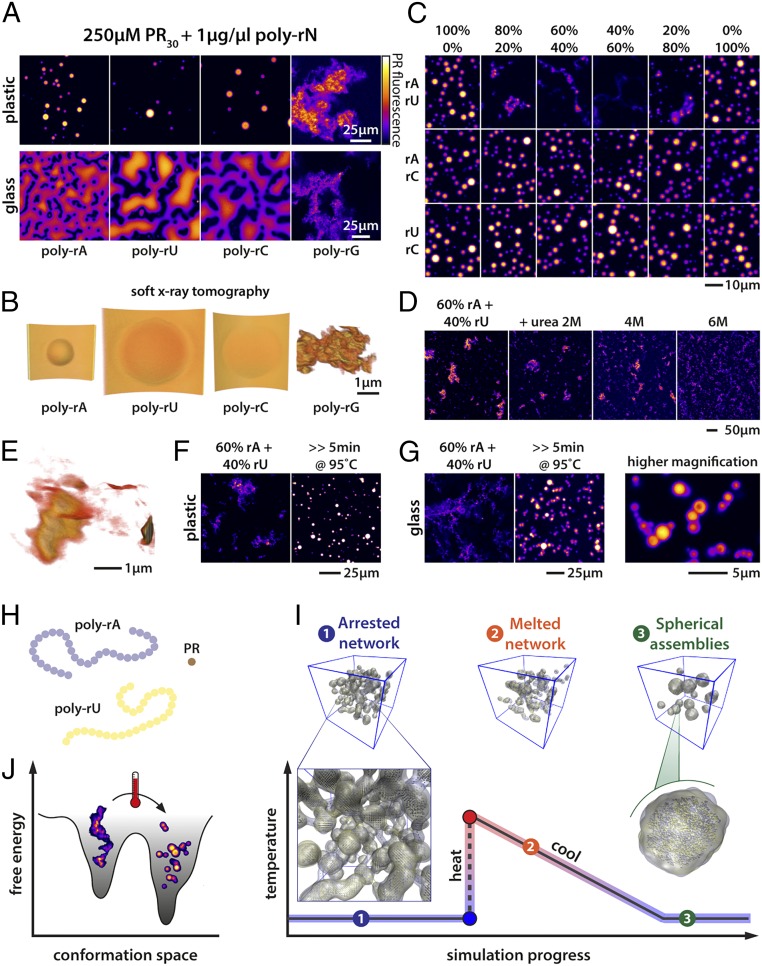
To uncover the physicochemical determinants of generic versus specific driving forces in complex coacervation involving protein and RNA molecules, we focused our attention on uncovering the rules associated with complex coacervation of PR as influenced by different types of homopolymeric RNA molecules. Mixing PR30 with homopolymeric RNAs (Mw = 600 kDa to 1,000 kDa) (37) leads to the spontaneous formation of condensates at room temperature. As can be seen in Fig. 2 A and B (and Movie S1), mixing of PR with homopolymeric RNAs (except poly-rG) results in the formation of liquid-like condensates. We make this assessment based on the spherical shapes of the assemblies, the wetting of glass surfaces, and the fusion of condensates. Poly-rA, poly-rC, and poly-rU are unlikely to form persistent higher-order (tertiary) structures, although there is evidence that intramolecular base stacking can drive the formation of short helical regions interspersed by random-coil−like segments (50). In contrast, poly-rG is known to form G quadruplexes, and these structural elements are highly stable (51). Interestingly, unlike the spherical coacervates formed by complexation of each of poly-rA, poly-rC, and poly-rU with PR, the assemblies formed by poly-rG and PR peptides are characterized by an open, fractal-like network structure that is resistant to thermal denaturation at high salt concentrations (SI Appendix, Fig. S1E). A deep quench into the two-phase regime can give rise to continuous transitions such as sol−gel transitions, which can be a manifestation of kinetically arrested phase separation (52). The morphologies we observe for the assemblies of PR and poly-rG are concordant with kinetically arrested phase separation that can lead to the formation of gels. Interestingly, however, the PR molecules within open networked structures remain dynamic, indicating that the noncovalent physical cross-links among PR−poly-rG molecules are labile on the timescale of the experiments (SI Appendix, Figs. S1A and S2D). The addition of RNA molecules such as poly-rC, which makes complementary base pairing interactions with poly-rG, reduces the formation of these open networks. This reduction occurs in a dose-dependent manner and points to the competitive formation of structures that displace the hyper-stable poly-rG−poly-rG interactions (SI Appendix, Fig. S1 A–D and Movie S2).
Fig. 2.
RNA structure determines condensate morphology. (A) All RNA homopolymers except poly-rG induce the formation of spherical liquid droplets. The liquid droplets can fuse and wet a glass surface. In contrast, poly-rG−PR complex coacervation yields distinct morphologies that are akin to large, open, fractal-like networks. (B) SXT images of PR-homopolymeric RNA coacervates. Poly-rA, poly-rC, and poly-rU droplets wet the surface of the capillary. In contrast, poly-rG−PR systems form open 3D networks. (C) Mixing homopolymeric RNA molecules that can make complementary base pairs indicates that base pairing can induce morphological transitions in PR−RNA coacervates, and this is tuned by the stoichiometry of the interacting RNAs. In contrast, morphological transitions were not observed in the presence of homopolymeric RNA molecules that cannot make complementary base pairs. (D) Urea interferes with poly-rA:rU base pairing and disrupts the formation of large networks. (E) SXT of PR−poly-rA:rU (60:40) coacervates illustrates formation of aspherical granules linked by sheet-like low-density connections. (F) Boiling PR−poly-rA:rU coacervates melts A:U base pairing and results in spherical assemblies. (G) Boiled PR−poly-rA:rU coacervates fail to fuse and wet the glass surface. (H) In our minimal polymer model, poly-rA and poly-rU are represented as polymers of multivalent beads, while PR is a single bead, reflecting the relative differences in molecular weight and contour length. (I) Schematic showing the formation of open but arrested networks when mixtures of polymers that engage in strong intermolecular interactions (as is the case for base pairing RNAs) are simulated together. These arrested networks resolve to spherical assemblies through a heating and cooling process, as observed in simulations here and experimentally in F and G. PR is not shown, for clarity. (J) Schematic illustrating a putative energy landscape associated with complementary homopolymeric RNAs in the presence of PR. The open networks observed in C–G are metastable states corresponding to minima on the left, while lower energy spherical assemblies are shown on the right. Heating and cooling facilitates network rearrangement into spherical assemblies. Scale bars apply for all images in the panel.
To further test the impact of stable RNA structures as determinants of condensate morphology, we combined base-pairing and non−base-pairing RNA species before induction of PR phase separation (Fig. 2C). Combining poly-rA and poly-rU (poly-rA:rU), but not poly-rA−poly-rC or poly-rU−poly-rC, resulted in the formation of large granular and filament/sheet-like assemblies. A detailed 3D view of the interconnected network formed by the base-pairing RNAs can be seen in Fig. 2E (Movie S3), as visualized using soft X-ray tomography (SXT). Interestingly, when preformed PR−poly-rA and PR−poly-rU condensates/coacervates were mixed together, we did not observe the formation of filamentous structures. Instead, small spherical assemblies are observed (SI Appendix, Fig. S1G). This suggests that RNA base pairing arrests complex coacervation of RNA and PR molecules by sequestering the RNA molecules into stable base-paired structures, leading to networks that incorporate PR molecules via physical cross-links such as hydrogen-bonding to the RNA phosphate backbones (53). Therefore, it appears plausible that at least one of the two phases observed for mixtures of poly-rA + poly-rU + PR is a metastable phase. Given that base pairing appears to drive extended network formation, we reasoned that interfering with base pairing should disrupt the formation of networked structures and promote phase separation. Urea is known to destabilize protein and RNA structure (54). Addition of urea to open network assemblies helped prevent the formation of networked structures, but this did not result in droplet formation (Fig. 2D). However, thermal denaturation of the poly-rA:rU-PR assemblies followed by an annealing reaction helped convert preformed open networks into spherical assemblies. This is suggestive of a disruption of metastable, networked structures and an annealing reaction that yields the thermodynamic ground state, which is the spherical coacervate (Fig. 2F and SI Appendix, Fig. S1F). Despite their spherical nature, under ambient temperature, these annealed assemblies are solid-like, as evidenced by their inability to fuse or wet a glass surface (Fig. 2G and SI Appendix, Fig. S1F).
Next, we asked whether simulations based on phenomenological models could reproduce results from experiments and thereby provide a coherent explanation for the findings. To this end, we performed simulations using a coarse-grained, four-component model consisting of simple representations of PR, poly-rA, poly-rU, and solvent. Long-range electrostatic interactions are effectively screened in the simulations (see Materials and Methods for details). In these simulations, poly-rA, poly-rU, and PR form homogeneous dispersed phases in binary mixtures of these molecules with solvent. However, in simulations of ternary systems that include poly-rA, PR, and solvent or poly-rU, PR, and solvent, we observed the formation of spherical condensates (SI Appendix, Fig. S3 A–C). Simulations suggest that spherical condensates readily deform and the molecules within them undergo rapid rearrangements (Movie S4). In contrast, in simulations of four-component systems that include poly-rA, poly-rU, PR, and solvent, we observed the formation of open, percolated networks that are characterized by complementary poly-rA:rU interactions. While PR partitions into these networks, the resulting assemblies relax slowly, and this behavior is indicative of a kinetically trapped phase. However, upon heating, this open network melts and undergoes a rearrangement to form spherical droplets (Fig. 2I and Movie S5). The effective mean-field energy associated with the spherical droplet phase is significantly lower than the energies associated with the open network (SI Appendix, Fig. S4). These results are consistent with the hypothesis that metastable mesophases that drive a percolated network form due to the networking of base-paired RNA molecules. Additionally, simulations with a reduced poly-rA−poly-rU interaction strength directly lead to spherical assemblies, bypassing the formation of open networks (SI Appendix, Fig. S3E). Taken together, our simulations show that higher-order RNA assemblies can provide a stable scaffold for the recruitment of a dynamic protein phase, but also that the resulting protein−RNA assemblies can exist in a metastable phase, determined by the competition between RNA base pairing and RNA−peptide interactions. These observations are concordant with physical theories that describe gelation in colloidal systems wherein the formation of open, percolated networks is viewed as kinetically arrested phase separation (55).
RNA−Peptide and RNA−RNA Interactions Determine Condensate Viscosity and Protein Dynamics.
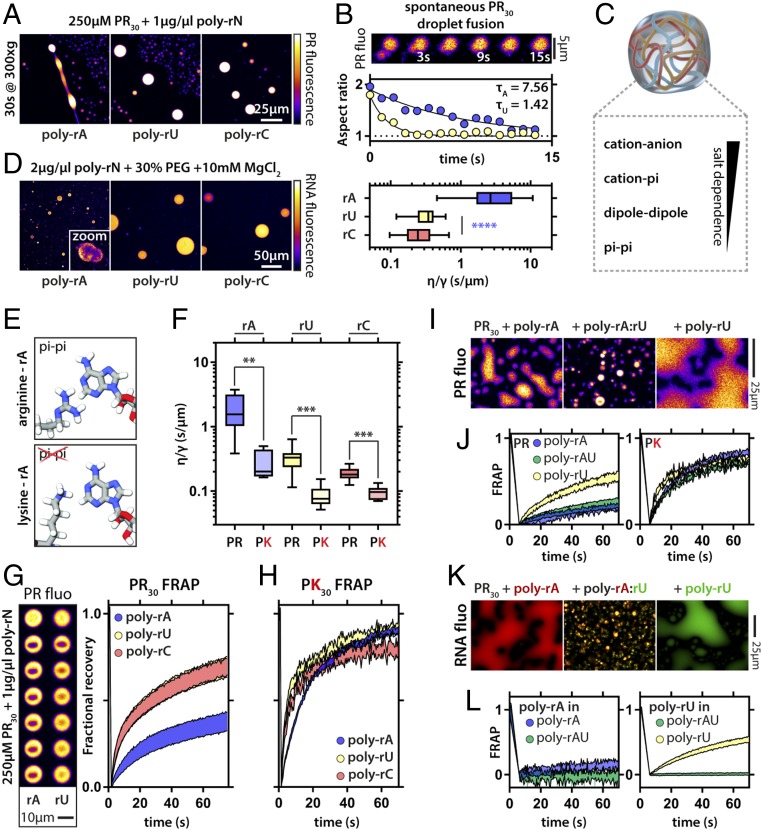
Poly-rA, poly-rU, and poly-rC molecules can drive the formation of condensates via complex coacervation with PR molecules. The resultant condensates have similar micrometer-scale morphologies. Dynamical properties within condensates are governed by molecular densities, the extent of physical cross-linking, and the timescales associated with these cross-links (56). Accordingly, we asked whether condensates formed by different RNA molecules and similar proteins have intracondensate dynamics that vary depending on the type of RNA. Condensates formed with poly-rA were more viscous than those formed by poly-rC and poly-rU, as assessed by induced and spontaneous fusion of condensates (Fig. 3 A and B). Next, we investigated the set of interactions that were the main contributors to the observed differences in condensate dynamics (Fig. 3C). The differences in viscosity associated with distinct polyribonucleotide−PR condensates may be caused by stronger polyribonucleotide−PR interactions, by stronger intrinsic interactions among polyribonucleotides, or by some combination of the two. To disentangle these effects, we asked whether different RNA homopolymers had different intrinsic interaction strengths with themselves. We induced RNA phase separation in the absence of PR by adding a crowding agent [30% polyethylene glycol (PEG)] and magnesium chloride (Fig. 3D). Under these conditions, poly-rA, poly-rC, and poly-rU form spherical, liquid-like droplets. However, poly-rA droplets fuse more slowly compared with poly-rU and poly-rC RNA droplets, implying a difference in the strengths of intradroplet interactions and the interfacial tension between the droplet and dispersed phases.
Fig. 3.
RNA−peptide and RNA−RNA interactions determine viscosity and protein dynamics within condensates. (A) PR−poly-rA condensates are more viscous than poly-rU and poly-rC condensates upon induced fusion via mild centrifugation. (B) PR−poly-rA condensates are more viscous as assayed by spontaneous fusion events. The exponential decay of the aspect ratio of two droplets during a fusion event can be characterized by the time constant τ. Example relaxation curves for two fusion events are shown. This value allows for the calculation of the (viscosity/surface tension) ratio (see Materials and Methods); n = 10 fusion events per condition, one-way ANOVA. Boxplots; whiskers indicate range. (C) Potential molecular interactions involved in PR−RNA coacervation. (D) Pure RNA droplets display droplets arrested in fusion for poly-rA. (E) Scheme indicating that arginine but not lysine can engage in pi-like interactions with the adenine base. (F) PK−polynucleotide droplets are uniformly less viscous than PR−polynucleotide droplets as assayed by spontaneous fusion events; n = 8, one-way ANOVA. Boxplots; whiskers indicate range. (G) PR is less dynamic in PR−poly-rA droplets compared with PR in PR−poly-rU and in PR−poly-rC droplets, as assayed by FRAP. Normalized traces also show the SDs. Two examples of bleach experiments for PR−poly-rA and PR−poly-rU droplets are shown. (H) PK dynamics assayed using FRAP experiments are largely similar for different homopolymeric RNAs. Traces indicate SEM. (I) Examples of PR fluorescence in PR−poly-rA and PR−poly-rU droplets and PR−poly-rA:rU gels. (J) FRAP dynamics for PR and PK in PR/PK−poly-rA and PR/PK−poly-rU droplets and PR/PK−poly-rA:rU gels. Traces indicate SEM. (K) Examples of poly-rA (red) and poly-rU (green) fluorescence in PR−poly-rA and PR−poly-rU droplets and PR−poly-rA:rU solid-like gels. (L) FRAP dynamics for poly-rA and poly-rU in PR−poly-rA and PR−poly-rU droplets and PR−poly-rA:rU solid-like gels. Traces indicate SEM. **P < 0.01, ***P < 0.001. Scale bars apply for all images in the panel.
Previous work showed that PR proteins engage in specific interactions with aromatic amino acids (33). Purine-containing nucleobases (adenine and guanine) contain a double-ring system while pyrimidines (cytosine and uracil) contain a single ring (SI Appendix, Fig. S5). Therefore, we hypothesized that, in addition to the electrostatic interactions among PR and RNA molecules, PR may also engage in short-range, directional, so-called cation−pi interactions with the aromatic RNA bases. In proteins, tryptophan has been proposed to mediate stronger cation−pi interactions compared with the single-ring systems of phenylalanine and tyrosine (45). It is also worth emphasizing that the strength and nature of cation−pi interactions depend not only on the aromatic system but also on the identity of the cation and properties of the solvent. The guanidinium groups of Arg sidechains are Y-shaped planar moieties that are best described as cross-conjugated pi systems, implying that six pi electrons are likely to be delocalized across the functional group. From a classical perspective, moieties with pi-like structures will have high intrinsic quadrupole moments (57). Sidechains such as Tyr and Trp are also endowed with high intrinsic dipole moments, whereas Arg will have monopole, dipole, and quadrupole moments. In contrast, the tertiary amine of Lys, which is also a cation, will have a monopole moment that is identical to that of Arg, but its dipole and quadrupole moments are negligible (57). These features enable a hierarchy of protein−protein and protein−RNA interactions (48, 49). Specifically, it has been shown that arginine drives stronger interactions with tyrosine and phenylalanine than does lysine (40, 45, 48, 49). These findings, in conjunction with our initial observations regarding the intrinsic interactions among poly-rA molecules, lead to two testable hypotheses. First, we propose that PR should interact more strongly with poly-rA than with either poly-rU or poly-rC. Second, RNA should interact more strongly with PR than an equivalent length proline−lysine (PK) dipeptide repeat. We used biolayer interferometry to quantify the apparent strengths of PR interactions with different RNA molecules. These data suggest that the PR−poly-rA interaction is the strongest, while PR−poly-rU and PR−poly-rC interactions are almost identical to one another (SI Appendix, Fig. S2). Interestingly, the apparent dissociation constant (Kd) was highest (lowest apparent affinity) for poly-rG. We interpret these results to mean that the effects of rG-rich RNA sequences on the PR phase behavior derives from the inability of PR to outcompete base stacking interactions associated with G-quadruplex formation.
Next, we compared the phase behaviors of PR with a peptide that contains 30 PK repeats (PK). Lysine has a similar positive charge to arginine, and also engages in cation−pi interactions. However, it lacks the apparent Y aromaticity of the guanidinium group and cannot encode the hierarchy of interactions attributable to the higher-order multipole moments of Arg (48) (Fig. 3E). We find that condensates formed using the PK peptide displayed reduced viscosity irrespective of the RNA sequence. Interestingly, we observe the largest decrease in viscosity for PK−poly-rA droplets, implying a reduction in the strength of interaction between PK and RNA compared with PR (Fig. 3F). These findings suggest that networks of interactions among arginine residues and the nucleobases are involved in tuning droplet dynamics. When we examined PR dynamics, rather than droplet dynamics, we obtained similar results (Fig. 3G). PR was less mobile in poly-rA droplets compared with the others, in agreement with inferences based on biolayer interferometry (SI Appendix, Fig. S2). As for dynamics, PK was more mobile than PR, and the differences between the RNAs were largely reduced (Fig. 3H). Taken together, these findings illustrate that hierarchies of molecular interactions among the protein and RNA components influence both nanoscale (intradroplet diffusion) and micrometer-scale dynamics (droplet fusion). These results are analogous to the recent findings of Wang et al. (48), who showed that a hierarchy of interactions combined with a so-called “stickers” and “spacers” model explains their data for the phase behavior of FUS/FET family proteins.
RNA Concentration Determines Protein Dynamics.
Previous studies showed that PR undergoes phase separation in the absence of RNA, providing that a suitable macromolecular crowder was added (33). Under these conditions, the positive charges of the arginine residues appear to be neutralized by anions present in the buffer (e.g., phosphate groups). By driving PR-only droplet formation with PEG as a molecular crowder, additional RNA (poly-rU) molecules should partition preferentially into the condensate by enabling the release of neutralizing mobile anions. This could provide a route to increase the intradroplet concentration of RNA, as opposed to adding different concentrations of RNA to soluble PR, which would just result in more or less PR−RNA droplets, as we have observed before (33). As can be seen in SI Appendix, Fig. S6, the stepwise increase in RNA concentration reduces the FRAP recovery and dynamics of PR, but only in the PEG-induced droplets (SI Appendix, Fig. S6). Indeed, the half-life for FRAP correlates with the total concentration of added RNA. This observation indicates that the number density of interaction sites provided by protein and RNA can determine coacervate dynamics in a manner that is analogous to the dependence of molecular dynamics on interaction strengths.
RNA Gelation Induces Coexistence of Material States.
In assemblies formed from complementary RNAs and PR, RNA base pairing induces the formation of solid-like gels that can be either metastable open networks or globally stable spherical solid-like assemblies (Fig. 2F). Accordingly, we asked whether the dynamics of either PR or PK would be influenced by the material states of RNA. To investigate this, we first examined the dynamics of PK and PR in solid-like spherical assemblies formed by base pairing RNA molecules (poly-rA:rU) after heating and those in liquid-like droplets with poly-rA or poly-rU. Surprisingly, we found the dynamics of the PR molecules to be completely independent of RNA dynamics (Fig. 3 I and J). In addition, PK was significantly more labile in poly-rA and poly-rA:rU droplets than was PR. These results illustrate that an assembly with apparent solid-phase behavior, as gleaned from the dynamics of RNA molecules, can still support a fluid phase. This points to the coexistence of liquid-like properties for the proteins and solid-like properties for the cRNA molecules.
Having examined protein dynamics within protein−RNA coacervates, we next examined the dynamics associated with distinct types of RNAs in different condensates (Fig. 3K). Comparative assessments of the timescales for FRAP show that poly-rA molecules are less dynamic than poly-rU molecules in their respective condensates (Fig. 3L). Specifically, within a 60-s window, we observed limited recovery of poly-rA fluorescence in poly-rA condensates and a recovery of ca. 70% of the original fluorescence for poly-rU in poly-rU condensates. This result is in agreement with the finding that, for both pure RNA condensates and PR/PK−RNA condensates, those containing poly-rA are always more viscous than those containing poly-rU (Fig. 2 D–F). Further, in the poly-rA:rU RNA gels, both RNA species were completely immobile. This is in stark contrast to the PR and PK peptides, which remain fully mobile. This finding, which is in agreement with results in Fig. 1, demonstrates that multicomponent condensates can support overlapping spatial distributions and distinct, essentially decoupled dynamics of the molecular components. Therefore, even in unitary condensates, that is, condensates where the protein and RNA molecules are apparently well mixed, the dynamical behaviors of protein and RNA molecules can be different from one another. This is achieved purely through the nature of the underlying molecular interactions and does not require active processes. The behavior we observe for differential dynamics within an apparently well-mixed condensate is reminiscent of the postulates of viscoelastic phase separation, which is governed by the differential mobilities of constituent molecules (58).
Mixing RNA Species Generates Complex Multilayered Condensates.
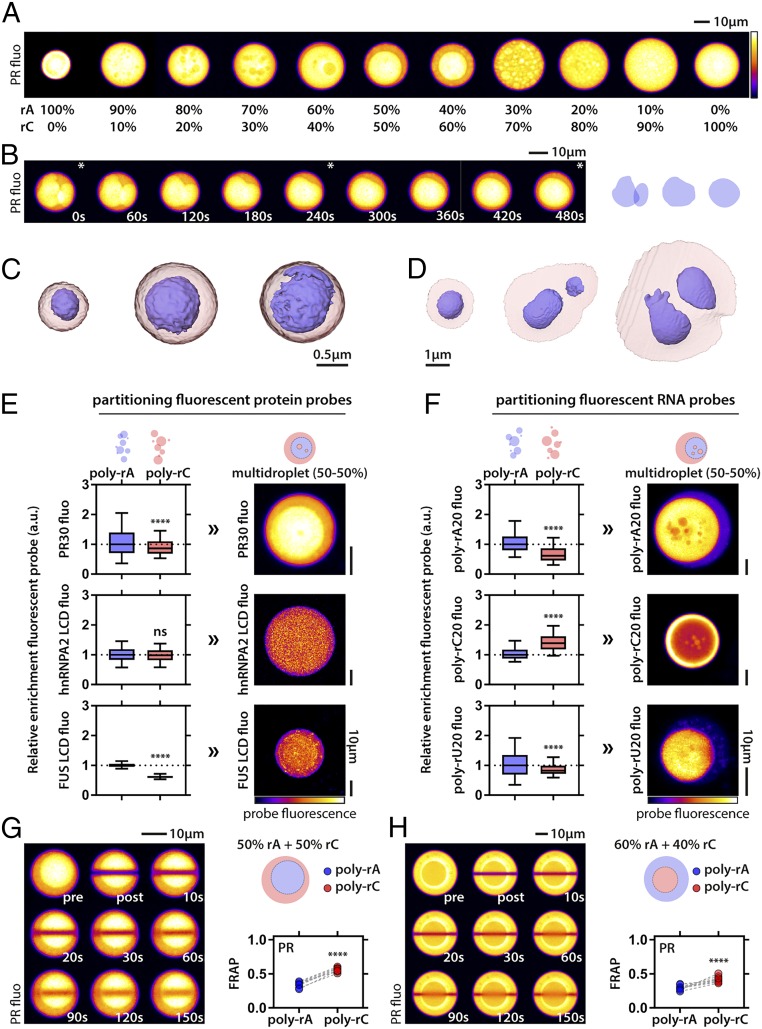
We have shown that RNA concentration, sequence, and structure can alter the driving forces for phase separation and the dynamics of the resultant condensates. Mixing non−base-pairing RNAs does not alter condensate formation. However, when we induce the fusion of these condensates to enable the formation of larger-scale droplets through gentle centrifugation, we observe the formation of multilayered topologies (Fig. 4 A and B and SI Appendix, Fig. S7). Interestingly, we also observed different topologies when varying the stoichiometry of poly-rA and poly-rC. On both ends of the stoichiometry spectrum, we obtain uniform condensates (as observed by PR fluorescence), yet, gradually, they evolve to a core/shell topology observed around a 50:50 mix of the RNAs (Fig. 4A). To get a better 3D view of these multilayered assemblies, we resorted to SXT. SXT has emerged as a 3D imaging technique to visualize internal structure of single cells (59, 60). In the soft X-ray regime, water is relatively transparent, and the contrast arises from X-ray absorption of biomolecules containing carbon and nitrogen without any need for fixing or staining of the samples (61, 62). With spatial resolution of about a few tens of nanometers, SXT bridges the resolution gap between traditional imaging methods, such as light and electron microscopy (63).
Fig. 4.
PR molecules combined with RNA mixtures generate multilayered condensates with differential partitioning of biomolecules. (A) Condensates show PR fluorescence. Mixing poly-rA and poly-rC results in subcompartmentalization, as seen via differences in PR intensity. The subcompartmentalization is tuned by the stoichiometry of poly-rA to poly-rC. Exposure settings are different between condensates to enhance visibility of subcompartments. (B) Internal droplets behave as liquids as probed by their fusion dynamics (Left). Stars indicate frames corresponding to scheme on Right. Data are shown for 50:50 ratio of poly-rA−poly-rC. (C and D) The 3D models of multilayered condensates (50:50) as imaged by SXT. (C) Free multilayered condensates. (D) Multilayered condensates that wet the capillary surface. (E) Differential protein partitioning in multilayered condensates is predicted by partitioning in single RNA−PR droplets. Fluorescence of the indicated probe molecule is shown. (F) Differential partitioning of RNA also follows partitioning preference of single RNA−PR condensates. In E and F, multilayered condensates are generated by 50:50 poly-rA−poly-rC mixtures. Boxplots; whiskers indicate range. Statistical analysis was performed using the Student t test. (G) Internal poly-rA droplets are less dynamic than their corresponding poly-rC shells. Data are shown for 50:50 poly-rA−poly-rC; n = 7. (H) Internal poly-rC droplets are more dynamic than their corresponding poly-rA shells. Data are shown for 60:40 poly-rA−poly-rC; n = 8. In G and H, paired values for poly-rA and poly-rC compartment are shown for each multidroplet. Statistical analysis was performed using the Student t test. ns, nonsignificant; ****P < 0.0001.
The high resolution and intrinsic contrast to biomolecules makes SXT a unique technique for direct 3D visualization of partitioning in phase-separated droplets. Using SXT, we observe multilayered topologies in condensates with dimensions that are reminiscent of membraneless organelles in vivo (Fig. 4 C and D and Movie S6). Our data clearly show that multilayered assemblies can arise from spontaneous fusion and Ostwald ripening (64). It is worth emphasizing that these events are not an artifact of centrifugation. Importantly, we identify subcompartments based on differences in X-ray absorbance, suggesting that the multilayered architectures have defined and observable differences in molecular density.
Multilayered Condensates Support Differential Partitioning of Biomolecules.
Our initial observation was that, depending on the RNA stoichiometry, certain compartments had a higher PR intensity. Given that PR has a stronger binding affinity for poly-rA than for poly-rC (SI Appendix, Fig. S2), and a higher partitioning ratio for poly-rA than for poly-rC droplets (Fig. 4E), we reasoned that, for a 50:50 stoichiometry, we were observing a poly-rA droplet (core) surrounded by a poly-rC shell. We subsequently tested the effects of LCDs derived from two well-studied RNA-binding proteins and RNAs for similar preferential partitioning into one compartment over another. These experiments allowed us to test how RNA might drive differential partitioning in multilayered topologies. The FUS and hnRNPA2 LCDs undergo phase separation independently (39, 65), and showed modest partitioning into PR−RNA droplets. We observed that the hnRNPA2 LCD did not have a preference for poly-rA over poly-rC condensates, whereas, for the FUS LCD, we observed a clear preference for poly-rA over poly-rC (Fig. 4E). Accordingly, we predicted that, in a multilayered condensate comprising poly-rA and poly-rC, the hnRNPA2 LCD would not show differential partitioning, while FUS LCD should display a preference for the poly-rA core. As can be seen in Fig. 4E, this was indeed the case. These preferences are interpretable in terms of sequence-encoded interactions of LCDs. The LCD of FUS lacks charged residues, but it contains a large number of tyrosine residues. These tyrosine residues appear to drive preferential interactions with poly-rA via a hierarchy of interactions. This proposal is consistent with results indicating that poly-rA is able to engage in stronger interactions due to its double-ringed nucleobase (SI Appendix, Fig. S8). In contrast, the hnRNPA2 LCD contains fewer tyrosine residues and also includes cationic arginine residues. These features appear to engender equivalent interactions for both poly-rA and poly-rC (SI Appendix, Fig. S8).
We made similar observations for short RNA probes. While oligo-rA and oligo-rU preferred condensates formed by poly-rA as opposed to poly-rC, oligo-rC showed a preference for condensates formed by poly-rC (Fig. 4F). Hence, multilayered poly-rA−poly-rC condensates are predicted to enrich poly-rA and poly-rU in the core versus poly-rC in the shell. These predicted preferences from the single-condensate observations were congruent with observations in the multilayered condensate system (Fig. 4F). In contrast, rather than a preference for poly-rC, we propose that poly-rC oligomers are unable to outcompete stronger poly-rA−poly-rA interactions and are preferentially excluded from poly-rA condensates. This gives rise to an effective preferential partitioning into poly-rC condensates where this competition is no longer relevant. These findings indicate that partitioning into complex topological settings might follow predictable trends for the respective phases in simpler single-phase condensates, an observation that is reminiscent of the findings from the work of Feric et al. (19).
Since poly-rA and poly-rC layers seem to preserve their affinity for certain biomolecules, we also asked whether the characteristic dynamics of these layers would be retained in multilayered condensates. We tested the dynamics of FRAP within the core versus the shell in different multilayered droplets. For droplets that had a core of poly-rA and a shell of poly-rC (50:50 poly-rA:poly-rC), PR was less dynamic in the core compared with the shell, as can be seen in the example droplet and the quantification (Fig. 4G). Performing the same assay, but on multilayered droplets with poly-rC cores surrounded by poly-rA shells (often observed in 60:40 poly-rA:poly-rC mixtures), we found that PR was still less dynamic in the poly-rA phase compared with the poly-rC phase (Fig. 4H), even though their relative positions in the multilayered assembly were inverted. Hence, we conclude that PR is always less dynamic in poly-rA than in poly-rC, irrespective of their relative topology. This indicates that single-compartment behaviors are predictive of the compartmental dynamics associated with more-complex condensate topologies.
Equivalent Multilayered Topologies Can Be Achieved Through Multiple Independent Routes.
Given the observed multilayered topologies and the range of distinct interactions, we next asked whether we could unambiguously determine the molecular origins of the multilayered topologies. Specifically, given a ternary mixture of PR, poly-rA, poly-rC, and solvent, what types of interactions could give rise to the observed core/shell architecture? To answer this question, we again turned to coarse-grained simulations (see Materials and Methods for additional details). Equivalent core/shell topologies could be achieved through a number of entirely independent interaction mechanisms. For example, if poly-rA preferentially binds PR over poly-rC, then a dense PR−poly-rA core forms and a PR−poly-rC shell surrounds the core. The formation of this topology depends both on the relative mismatch in PR binding and on the absolute concentration of free PR. If poly-rA−PR binding is too strong and there is insufficient PR, then poly-rA will effectively sequester the majority of the PR, preventing poly-rC from coalescing or only allowing a thin shell of poly-rC to form over the poly-rA condensate periphery (SI Appendix, Fig. S9 A and B). An alternative route to the observed core−shell topology is realized if poly-rA interacts with itself more strongly than with poly-rC. Importantly, for this to give rise to a single multilayered condensate, poly-rA−poly-rC interactions must be more favorable than poly-rC−poly-rC interactions. If this is not the case, then two distinct droplets will form (SI Appendix, Fig. S9C). Finally, if poly-rA experiences less favorable interactions with the solvent compared with poly-rC, then a poly-rA core will form to reduce the solvent−poly-rA interactions, while an enveloping poly-rC shell will engulf the core. Examples of these three distinct scenarios are shown in Fig. 5, along with the associated interaction tables that give rise to the resulting topologies. In each of the three cases, a single set of interactions is modulated while the others remain fixed. Our simulation results suggest that the spatial organization of condensates can be tuned in distinct, synergistic, and antagonistic ways, thus providing routes for condensate assembly and disassembly.
Fig. 5.
Core−shell multilayered condensates can be formed via several distinct mechanisms. (A) Schematic of the molecular entities included in simulations based on the coarse-grained model that includes poly-rA, poly-rC, and PR. (B and C) Simulations are run with 70:30 split of poly-rC−poly-rA and give rise to droplets that are significantly larger than individual polymers. (D–G) A suitable interaction table defines the attractive and repulsive interactions between individual beads. These effective pairwise interactions represent the cumulative interplay among all bead−bead and bead−solvent interactions. Yellow boxes highlight differential values compared with D. When poly-rA and poly-rC are equivalent to one another, a single well-mixed droplet is formed (in D). (E) If poly-rA−PR interaction is stronger than poly-rC−PR interaction, (F) if poly-rA−poly-rA interaction is stronger than poly-rC−poly-rA interaction which, in turn, is stronger than poly-rC−poly-rC interaction, or (G) if poly-rA−solvent is more repulsive than poly-rC−solvent, then topologically equivalent multilayered condensates are formed. Experimental data suggest that all three mechanisms may occur, to varying degrees.
Discussion
Biomolecular condensates harbor a complex mixture of hundreds of different RNA and protein species in vivo (17, 26, 27, 66, 67). This makes it challenging to uncover specific molecular functions and/or quantify interactions of specific RNA or protein molecules within condensates. These molecular functions are likely to be important for mesoscale functions of condensates. Here, we resorted to a simple test tube model of RNA−protein condensates that are based on biologically relevant PR proteins. These experiments allow us to dissect the underlying physicochemical forces that drive specific behaviors. We show that biological polyanions drive phase separation of Arg-rich proteins through complex coacervation, albeit with significant chemical specificity. The morphologies and material states of the resulting phases vary dramatically with polyanion type. To understand the role of RNA sequence and structure, we focused on simple homopolymeric RNA molecules. We found that stable RNA secondary structure formation leads to rigid open networks. In contrast, RNA molecules lacking stable secondary structure promote the formation of liquid-like condensates. Based on denaturing−annealing reactions and computer simulations, we show that the open networks represent metastable phases that arise from barriers governed by a competition between RNA base pairing and RNA−protein interactions. When released from their metastable phases, the resulting molecular rearrangements lead to an annealing toward spherical condensates. However, solid-like behavior of RNA molecules can persist within spherical condensates, and we propose, based on extant data, that this derives from the formation of condensate-spanning (percolated) networks via physical cross-links that determine rheological properties. Despite the immobility of RNA within the assemblies, the condensates support a protein phase that exhibits rapid dynamics that are indistinguishable compared with liquid droplets. Our findings indicate that RNA base pairing can decouple RNA and protein dynamics and lead to a coexistence of different molecular level dynamics within the same condensate. This observation has major implications for the description of cellular condensates: Rapid dynamics of one (protein or RNA) component does not necessarily mean the condensate as a whole behaves as a liquid.
Besides RNA base pairing, RNA sequence and the purine/pyrimidine contents are also key regulators of the material properties of condensed phases. Nanoscale and micrometer-scale dynamics of protein−RNA condensates are tuned by the interplay among RNA−RNA, RNA−protein, RNA−solvent, and protein−solvent interactions. We showed that, by mixing different homopolymeric RNAs, we can induce spontaneous subcompartmentalization within condensates, and that this can be tuned by varying RNA stoichiometries. These multilayered condensates enable differential partitioning of other biomolecules while also displaying differential protein dynamics. Our findings provide a touchstone for understanding the in vivo phase behavior of heterogeneous mixtures of protein and RNA molecules. Additionally, the coarse-grained simulations indicate that core−shell architecture can arise readily from the differential effects of RNA−protein, RNA−RNA, RNA−solvent, and protein−solvent interactions.
Multilayered condensates have previously been reported for mixtures of nucleolar protein and RNA molecules (19) and synthetic multivalent proteins (21, 68). As was shown previously for nucleolar proteins (19), we also found that differential surface energies determine the preferential partitioning of molecules into the cores versus shells in multilayered condensates. These findings are borne out in coarse-grained simulations. However, simulations do not show the formation of multilayered condensates in which the denser poly-rA phase exists as a shell around a less dense poly-rC core, as observed in Fig. 4 A and H. The formation of this “inverted” topology would be at odds with a simple equilibrium thermodynamic model. However, for multilayered condensates in which the difference in surface energies between distinct phases is low and those phases are relatively viscous, these inverted topologies could form in response to some initial mixing or other energy input. Moreover, once formed, there may be a large energetic barrier associated with topological reorganization. Since we did not observe these apparently metastable states in smaller multilayered condensates (as imaged with SXT), but did find them in larger multilayered condensates formed via mild centrifugation, it is possible that the inverted multilayered condensates derive from the energy that is supplied during the centrifugation process. It is worth noting that similar inverted topologies have been observed for some condensates in cells. Overexpression of a TDP-43−derived fusion protein results in the formation of large nuclear droplets that contain “vacuoles” of nucleoplasm (69). Vacuolization has also been observed when nucleoli fuse together due to destabilization of the nuclear actin cytoskeleton (19). This suggests that fusion events, especially of larger more viscous droplets, may provide enough energy to drive multilayered condensates into metastable states, both in the test tube and in cells.
Our findings, combined with recent studies of Maharana et al. (70) and Langdon et al. (71), show that RNA structure, RNA-to-protein ratios, and the interplay of RNA−RNA, protein−protein, and protein−RNA interactions contribute to the complex topologies of protein−RNA condensates as well as their material properties. It appears that many of the features observed in vivo for protein−RNA condensates can be recapitulated in vitro and that these features are the result of spontaneous (passive), sequence-encoded driving forces rather than driven (active) processes. The rules we have gleaned help refine the questions we get to ask about the interplay between spontaneous (passive) versus driven (active) processes in vivo. A proposal that emerges from our work is that driven processes mainly modulate or maintain the sequence-encoded properties of condensates that form spontaneously. Additionally, despite the fact that we used model RNA molecules, it is worth emphasizing that poly-rA tails are an integral part of mRNA molecules. It is therefore tempting to speculate that some of the observations in our test tube model may apply to the behavior of poly-rA tails in cells. For example, stress granules have been shown to contain dense cores that stain positively for poly-rA (17). Our results also highlight key design challenges and opportunities that could inspire new nanomaterials and drug delivery strategies. This is relevant given the rising popularity of nucleic acid-based nanotechnology (72–75) and therapies (76, 77), including arginine-rich, peptide-nucleic acid coacervates (78–81).
Materials and Methods
Full details of all procedures, including specific compounds used, specific details surrounding phase separation assays, biolayer interferometry, FRAP assays, droplet fusion experiments, partitioning assays, SXT, and coarse-grained simulations are provided in SI Appendix.
For phase separation assays, polyanions were diluted to 1 µg/µL in 100 mM K2HPO4/KH2PO4 buffer at pH 7. Peptides were added at 250-µM concentrations. For partitioning experiments, probe molecules were spiked in at 100 nM. For the generation of pure RNA liquid droplets, homopolymeric RNAs were diluted to 2 µg/µL in 1x PBS buffer with 30% PEG and 10 mM MgCl2 at pH 7. For intradroplet FRAP, a circular area of 1-μM radius was bleached in droplets with a radius between 5 μM and 10 μM. For SXT 3D reconstructions, 92 projection images, with 200-ms exposure time each, were acquired sequentially around a rotation axis with 2° increment angles. For droplet fusion assays, confocal time-lapse images were taken of droplets settling on the coverslip and fusing together. Coarse-grained simulations are lattice-based and utilize a simple neighbor−neighbor interaction potential. For all other details, please see SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Broder Schmidt, Dr. Frederic Rousseau, Dr. Joost Schymkowitz, Dr. Sean Friedowitz, Dr. Jian Qin, Dr. Jeong-Mo Choi, and Dr. Andrea Soranno for helpful discussions. We thank Dr. Nicolas L. Fawzi for his kind gift of the FUS LCD and hnRNPA2 LCD recombinant proteins, and Dr. Andrew Olson [Stanford Neuroscience Microscopy Service, supported by National Institutes of Health (NIH) Grant NS069375]. L.V.D.B. is supported by KU Leuven (“Opening the future” and C1), the Fund for Scientific Research Flanders [Fonds Wetenschappelijk Onderzoek (FWO)-Vlaanderen], the Agency for Innovation by Science and Technology (IWT) in Flanders (IWT-Vlaanderen), and the ALS Liga. S.B. acknowledges a long-term fellowship from European Molecular Biology Organization. P.S.T. is supported by Odysseus Grant G.0029.12 from FWO-Vlaanderen. V.W. is supported by German Research Foundation Research Fellowship WE 6221/1-1. C.L. is supported by NIH−National Institute on Drug Abuse Grant U01 DA040582. The National Center for X-ray Tomography is supported by NIH Grant P41GM103445 and Department of Energy’s Office of Biological and Environmental Research DE-AC02-5CH11231. A.D.G. is supported by the National Institutes of Health (Grant R35NS097263). The contributions of A.S.H. and R.V.P. were supported by Human Frontier Science Program Grant RGP0034/2017, US National Science Foundation Grant MCB-1614766, and the St. Jude Children’s Research Hospital sponsored collaborative on membraneless organelles.
Footnotes
Conflict of interest statement: R.V.P. is a member of the scientific advisory board of Dewpoint Therapeutics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821038116/-/DCSupplemental.
References
- 1.Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. [Google Scholar]
- 2.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 3.Boeynaems S, et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 5.Harmon TS, Holehouse AS, Rosen MK, Pappu RV. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife. 2017;6:e30294. doi: 10.7554/eLife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 7.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry J, Brangwynne CP, Haataja M. Physical principles of intracellular organization via active and passive phase transitions. Rep Prog Phys. 2018;81:046601. doi: 10.1088/1361-6633/aaa61e. [DOI] [PubMed] [Google Scholar]
- 9.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boeynaems S, Bogaert E, Van Damme P, Van Den Bosch L. Inside out: The role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol. 2016;132:159–173. doi: 10.1007/s00401-016-1586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeynaems S, Tompa P, Van Den Bosch L. Phasing in on the cell cycle. Cell Div. 2018;13:1. doi: 10.1186/s13008-018-0034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain S, et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. eLife. 2016;5:e18413. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feric M, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei J, et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci. 2017;130:4180–4192. doi: 10.1242/jcs.206854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmon TS, Holehouse AS, Pappu RV. Differential solvation of intrinsically disordered linkers drives the formation of spatially organized droplets in ternary systems of linear multivalent proteins. New J Phys. 2018;20:045002. [Google Scholar]
- 22.Holehouse AS, Pappu RV. Functional implications of intracellular phase transitions. Biochemistry. 2018;57:2415–2423. doi: 10.1021/acs.biochem.7b01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin M, et al. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep. 2017;20:895–908. doi: 10.1016/j.celrep.2017.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adivarahan S, Sr, Zenklusen D. 2017. Spatial organization of single mRNPs at different stages of the gene expression pathway. bioRxiv:10.1016/j.molcel.2018.10.010. Preprint, posted July 9, 2018.
- 26.Khong A, et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell. 2017;68:808–820.e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namkoong S, Ho A, Woo YM, Kwak H, Lee JH. Systematic characterization of stress-induced RNA granulation. Mol Cell. 2018;70:175–187.e8. doi: 10.1016/j.molcel.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Treeck B, et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci USA. 2018;115:2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, et al. RNA controls PolyQ protein phase transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci USA. 2015;112:E5237–E5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caudron-Herger M, et al. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 2015;34:2758–2774. doi: 10.15252/embj.201591458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boeynaems S, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–1055.e5. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee KH, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788.e17. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, et al. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802.e12. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitrea DM, et al. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife. 2016;5:e13571. doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aumiller WM, Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- 38.Elbaum-Garfinkle S, et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan VH, et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell. 2018;69:465–479.e7. doi: 10.1016/j.molcel.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nott TJ, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogaert E, et al. Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 2018;24:529–537.e4. doi: 10.1016/j.celrep.2018.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 43.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pak CW, et al. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aumiller WM, Jr, Pir Cakmak F, Davis BW, Keating CD. RNA-based coacervates as a model for membraneless organelles: Formation, properties, and interfacial liposome assembly. Langmuir. 2016;32:10042–10053. doi: 10.1021/acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brady JP, et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci USA. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys Rev Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 51.Petrovic AG, Polavarapu PL. The quadruplex-duplex structural transition of polyriboguanylic acid. J Phys Chem B. 2008;112:2245–2254. doi: 10.1021/jp0758723. [DOI] [PubMed] [Google Scholar]
- 52.Sciortino F, Bansil R, Stanley HE, Alstrom P. Interference of phase separation and gelation: A zeroth-order kinetic model. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1993;47:4615–4618. doi: 10.1103/physreve.47.4615. [DOI] [PubMed] [Google Scholar]
- 53.Mascotti DP, Lohman TM. Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry. 1997;36:7272–7279. doi: 10.1021/bi970272n. [DOI] [PubMed] [Google Scholar]
- 54.Priyakumar UD, Hyeon C, Thirumalai D, Mackerell AD., Jr Urea destabilizes RNA by forming stacking interactions and multiple hydrogen bonds with nucleic acid bases. J Am Chem Soc. 2009;131:17759–17761. doi: 10.1021/ja905795v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaccarelli E, Lu PJ, Ciulla F, Weitz DA, Sciortino F. Gelation as arrested phase separation in short-ranged attractive colloid-polymer mixtures. J Phys Condens Matter. 2008;20:494242. [Google Scholar]
- 56.Wei MT, et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat Chem. 2017;9:1118–1125. doi: 10.1038/nchem.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burley SK, Petsko GA. Weakly polar interactions in proteins. Adv Protein Chem. 1988;39:125–189. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka H, Araki T, Koyama T, Nishikawa Y. Universality of viscoelastic phase separation in soft matter. J Phys Condens Matter. 2005;17:S3195. [Google Scholar]
- 59.McDermott G, Le Gros MA, Knoechel CG, Uchida M, Larabell CA. Soft X-ray tomography and cryogenic light microscopy: The cool combination in cellular imaging. Trends Cell Biol. 2009;19:587–595. doi: 10.1016/j.tcb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harkiolaki M, et al. Cryo-soft X-ray tomography: Using soft X-rays to explore the ultrastructure of whole cells. Emerging Top Life Sci. 2018;2:81–92. doi: 10.1042/ETLS20170086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDermott G, et al. Visualizing and quantifying cell phenotype using soft X-ray tomography. BioEssays. 2012;34:320–327. doi: 10.1002/bies.201100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Gros MA, et al. Soft X-ray tomography reveals gradual chromatin compaction and reorganization during neurogenesis In vivo. Cell Reports. 2016;17:2125–2136. doi: 10.1016/j.celrep.2016.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider G, et al. Three-dimensional cellular ultrastructure resolved by X-ray microscopy. Nat Methods. 2010;7:985–987. doi: 10.1038/nmeth.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lifshitz IM, Slyozov VV. The kinetics of precipitation from supersaturated solid solutions. J Phys Chem Solids. 1961;19:35–50. [Google Scholar]
- 65.Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hubstenberger A, et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Simon JR, Carroll NJ, Rubinstein M, Chilkoti A, López GP. Programming molecular self-assembly of intrinsically disordered proteins containing sequences of low complexity. Nat Chem. 2017;9:509–515. doi: 10.1038/nchem.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt HB, Rohatgi R. In vivo formation of vacuolated multi-phase compartments lacking membranes. Cell Rep. 2016;16:1228–1236. doi: 10.1016/j.celrep.2016.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maharana S, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langdon EM, et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seeman NC, Sleiman HF. DNA nanotechnology. Nat Rev Mater. 2017;3:17068. [Google Scholar]
- 73.Han D, et al. Single-stranded DNA and RNA origami. Science. 2017;358:eaao2648. doi: 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Geary C, Rothemund PW, Andersen ES. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014;345:799–804. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- 75.Liu K, et al. Solvent-free liquid crystals and liquids from DNA. Chemistry. 2015;21:4898–4903. doi: 10.1002/chem.201500159. [DOI] [PubMed] [Google Scholar]
- 76.Lundin KE, Gissberg O, Smith CI. Oligonucleotide therapies: The past and the present. Hum Gene Ther. 2015;26:475–485. doi: 10.1089/hum.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein CA, Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol Ther. 2017;25:1069–1075. doi: 10.1016/j.ymthe.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu SW, et al. Arginine-rich intracellular delivery peptides synchronously deliver covalently and noncovalently linked proteins into plant cells. J Agric Food Chem. 2010;58:2288–2294. doi: 10.1021/jf903039j. [DOI] [PubMed] [Google Scholar]
- 79.Liu BR, Lin MD, Chiang HJ, Lee HJ. Arginine-rich cell-penetrating peptides deliver gene into living human cells. Gene. 2012;505:37–45. doi: 10.1016/j.gene.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 80.Schmidt N, Mishra A, Lai GH, Wong GCL. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 81.Cordier C, et al. Delivery of antisense peptide nucleic acids to cells by conjugation with small arginine-rich cell-penetrating peptide (R/W)9. PLoS One. 2014;9:e104999. doi: 10.1371/journal.pone.0104999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.