Significance
In chloroplasts, five enzymes of the Calvin–Benson (CB) cycle are regulated by thioredoxins (TRXs). These enzymes have all been structurally characterized, with the notable exception of phosphoribulokinase (PRK). Here, we determined the crystal structure of chloroplast PRK from two model photosynthetic organisms. Regulatory cysteines appear distant from each other and are linked by a long loop that is present only in plant-type PRKs and allows disulfide-bond formation and subsequent conformational rearrangements. Structural comparisons with ancient PRKs indicate that the presence of flexible regions close to regulatory cysteines is a unique feature that is shared by TRX-dependent CB cycle enzymes, suggesting that the evolution of the PRK structure has resulted in a global increase in protein flexibility for photosynthetic eukaryotes.
Keywords: Calvin–Benson cycle, phosphoribulokinase, 3D structure, redox regulation, thioredoxin
Abstract
In land plants and algae, the Calvin–Benson (CB) cycle takes place in the chloroplast, a specialized organelle in which photosynthesis occurs. Thioredoxins (TRXs) are small ubiquitous proteins, known to harmonize the two stages of photosynthesis through a thiol-based mechanism. Among the 11 enzymes of the CB cycle, the TRX target phosphoribulokinase (PRK) has yet to be characterized at the atomic scale. To accomplish this goal, we determined the crystal structures of PRK from two model species: the green alga Chlamydomonas reinhardtii (CrPRK) and the land plant Arabidopsis thaliana (AtPRK). PRK is an elongated homodimer characterized by a large central β-sheet of 18 strands, extending between two catalytic sites positioned at its edges. The electrostatic surface potential of the catalytic cavity has both a positive region suitable for binding the phosphate groups of substrates and an exposed negative region to attract positively charged TRX-f. In the catalytic cavity, the regulatory cysteines are 13 Å apart and connected by a flexible region exclusive to photosynthetic eukaryotes—the clamp loop—which is believed to be essential for oxidation-induced structural rearrangements. Structural comparisons with prokaryotic and evolutionarily older PRKs revealed that both AtPRK and CrPRK have a strongly reduced dimer interface and an increased number of random-coiled regions, suggesting that a general loss in structural rigidity correlates with gains in TRX sensitivity during the molecular evolution of PRKs in eukaryotes.
Photosynthetic CO2 fixation supports life on Earth and is a fundamental source of food, fuel, and chemicals for human society. In the vast majority of photosynthetic organisms, carbon fixation occurs via the Calvin–Benson (CB) cycle (SI Appendix, Fig. S1), a pathway that has been extensively studied at the physiological, biochemical, and structural levels. The CB cycle consists of 13 distinct reactions catalyzed by 11 enzymes (1) that are differentially regulated to coordinate the two stages of photosynthesis—electron transport and carbon fixation—and to couple them with the continuous changes in environmental light.
Thioredoxins (TRXs) are small thiol oxidoreductase enzymes that are widely distributed among the kingdoms of life. They reduce disulfide bonds in target proteins, controlling the redox state and modulating the activity of target enzymes (2). Compared with animals that usually possess two or three different TRXs, the plant thioredoxin system is much more complex. Looking only at the chloroplast, five different classes of TRXs are known (i.e., TRX-f, -m, -x, -y, and -z), with several isoforms composing many of these classes (2). Despite this diversity, TRXs are ancient and strongly evolutionarily conserved, consisting of a central core of a five-stranded β-sheet surrounded by four α-helices and an active site that protrudes from the protein’s surface (3).
In TRX-dependent regulation, a small fraction of photosynthetic-reducing equivalents are transferred from photosystem I to several chloroplast TRXs, which in turn reduce key enzymes of the CB cycle, regulating their activity (1). Among the five classes of chloroplast TRXs, class f displays a high specificity toward CB cycle enzymes. In both plants and algae, the activities of fructose-1,6-bisphosphatase (FBPase) (4, 5), sedoheptulose-1,7-bisphosphatase (SBPase) (5), phosphoribulokinase (PRK) (6), and the AB isoform of NADP-glyceraldehyde 3-phosphate dehydrogenase (AB-GAPDH) (7) in plants and of phosphoglycerate kinase (PGK) (8) in Chlamydomonas are directly linked to light by a thiol-based mechanism driven by TRX (SI Appendix, Fig. S1). In addition, PRK forms a regulatory complex with the redox-sensitive CP12 and the photosynthetic GAPDH (A4-GAPDH and AB-GAPDH) (9–15). The formation of this ternary complex occurs in both plants and algae, and causes an almost complete inhibition of PRK activity (11, 16).
In contrast to the canonical WCGPC active site of TRXs, no thioredoxin-recognition signature is found in these enzymes. A structural comparison of SBPase and FBPase highlights how evolution has adopted different strategies for modeling protein structure to achieve the same TRX-dependent redox control (5). In the three available crystal structures of enzymes regulated by TRX, the pair of redox-sensitive cysteines was found to be located in different regions. In AB-GAPDH, these cysteines are located in a C-terminal extension (CTE) (7); in SBPase, they are present at the dimer interface (5); and in FBPase, they are solvent-exposed and distant from the sugar-binding site (4, 5).
To complete the structural redox proteome of the CB cycle enzymes and identify the common feature behind the acquisition of TRX-mediated redox control, the crystal structures of PRK from two model species—the green alga Chlamydomonas reinhardtii and the land plant Arabidopsis thaliana—were determined and compared with those of other TRX-controlled enzymes. Our results strongly suggest that the acquisition of a TRX-targeted motif positively correlates with the acquisition of flexibility in the target proteins, consistent with the observation that the entropic contribution obtained from the reduction of the oxidized target enzyme is the main driving force of the redox control mediated by TRXs (17).
Results and Discussion
Three-Dimensional Structures of CrPRK and AtPRK in Solution and in Crystal Form.
To gain insight into the molecular mechanism of regeneration of the five-carbon sugar ribulose-1,5-bisphosphate (SI Appendix, Fig. S1) and of its redox control by dithiol–disulfide exchange, the crystal structures of CrPRK and AtPRK were determined at 2.6 and 2.5 Å, respectively, and compared with those obtained in solution by size-exclusion chromatography–small-angle X-ray scattering (SEC-SAXS).
Both enzymes are dimers of two identical monomers connected by a twofold noncrystallographic axis (Fig. 1). They are structurally similar, and their superimposition results in a root-main-square deviation of 0.61 Å (on 330 aligned residues) for CrPRK and 1.03 Å (on 328 aligned residues) for AtPRK.
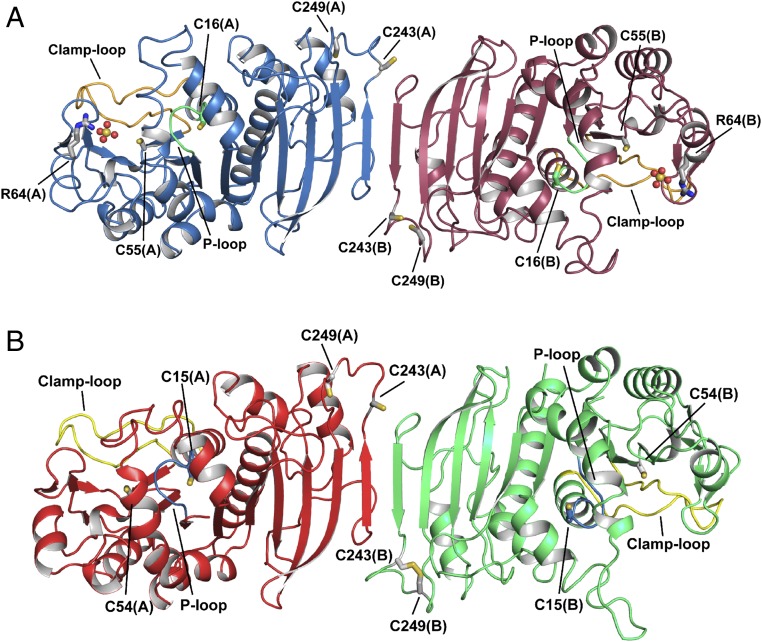
Fig. 1.
Crystal structure of photosynthetic PRK. Representation of the overall crystal structure of reduced phosphoribulokinase from (A) C. reinhardtii and (B) A. thaliana. The P loop is highlighted in green and light blue, and the clamp loop in orange and yellow, respectively. Each monomer contains two pairs of cysteines, one in the active site (Cys16 and Cys55 for CrPRK, and Cys15 and Cys54 for AtPRK) and one in the dimer interface close to the C-terminal end of the protein chain (Cys243 and Cys249). Cysteine residues are indicated and represented as sticks. In AtPRK, a pair of C-terminal cysteines forms a disulfide bond. CrPRK binds two sulfate ions (one for each monomer), represented as spheres, from the crystallization solution. Arg64, represented as sticks, is one of the residues stabilizing the anions.
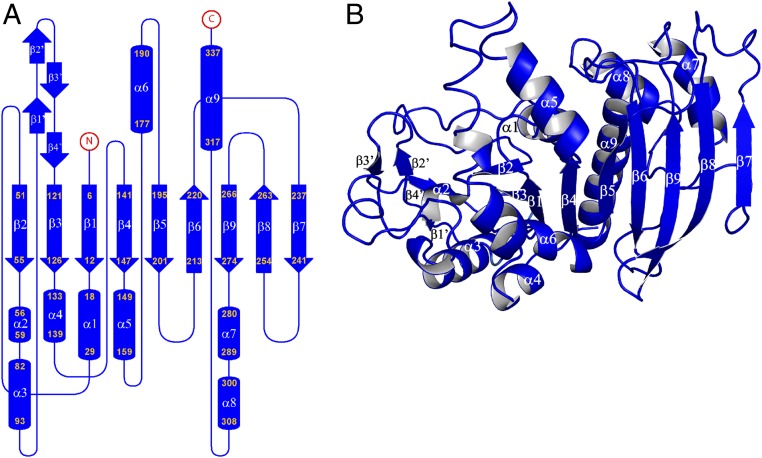
Each monomer contains a central mixed β-sheet of nine strands (β1 to β9) surrounded by eight α-helices (α1, α3 to α9), four additional β-strands (β1′ to β4′), and one additional α-helix (α2) (Fig. 2 and SI Appendix, Fig. S2).
Fig. 2.
Crystal structure of the CrPRK monomer. (A) Topology diagram: The monomer is composed of a mixed β-sheet of nine strands, nine α-helices, and four additional small β-strands indicated by β′. (B) Representation of the structure of the monomer: The central β-sheet is sandwiched between helices α3, α4, and α6 and helices α1, α7, α8, and α9. Strand β7 is located in the dimer interface, whereas the four additional β-strands (β1′ to β4′) form the edge of the dimer.
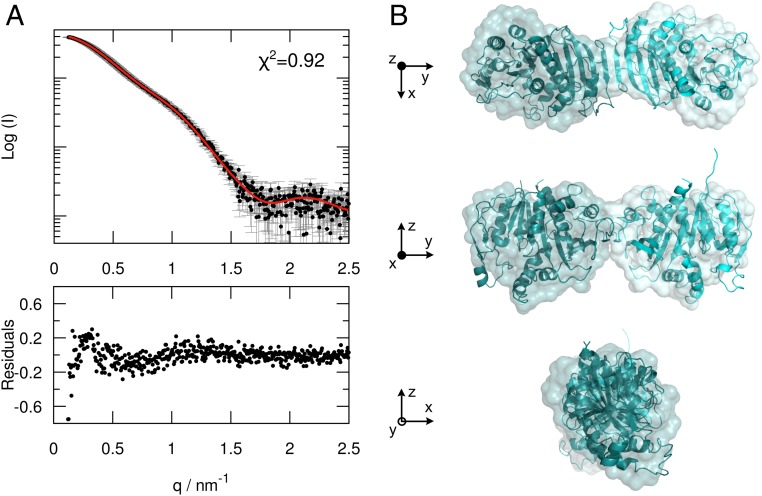
The AtPRK crystal structure confirms the elongated shape of the oxidized protein previously observed by SAXS analysis (12) but appears less bent and twisted. Structural information regarding CrPRK in solution provides a single recognizable species with an Rg value of 35 ± 1 Å and an estimated molecular mass around 70 kDa (SI Appendix, Fig. S3 and Table S1). The SAXS profile can be completely superimposed on the theoretical profile calculated from the CrPRK crystal structure (Fig. 3A), indicating that the solved crystal structure closely matches the structure of the protein in solution (Fig. 3B).
Fig. 3.
SAXS analysis of CrPRK. (A, Upper) Superimposition of the experimental SAXS profile (black) and the theoretical SAXS curve (red), calculated from the CrPRK crystal structure. (A, Lower) The residuals obtained by the difference between the experimental and calculated intensities. (B) Comparison between the ab initio model (surface representation) and the crystal structure (ribbon representation) of CrPRK, showing an excellent agreement between the two models.
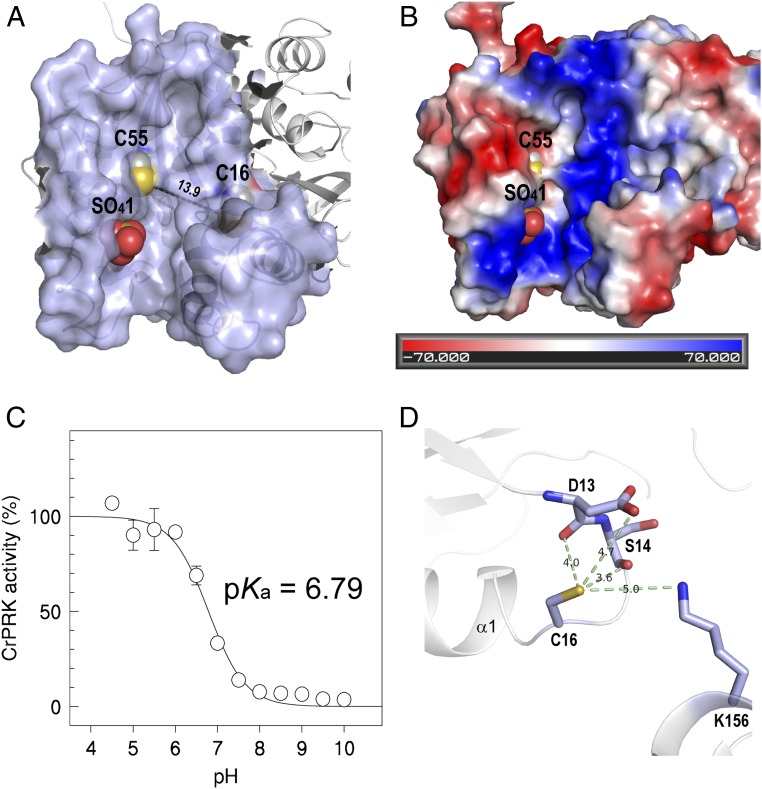
The active sites (one per monomer) are located in an elongated cavity at the edges of the dimer (Fig. 4A and SI Appendix, Fig. S4A). In the CrPRK structure, the active-site cavity is marked by a sulfate ion derived from the crystallization medium that highlights the position occupied by the phosphate group of the substrate ribulose 5-phosphate (Ru5P) (Fig. 4 A and B). The anion is stabilized by the side chains of Arg64, Arg67, and Tyr103, and by the main-chain carbonyl group of His105 (SI Appendix, Fig. S5). Except for Arg67, these residues are strictly conserved in PRKs (SI Appendix, Fig. S6A). The catalytic role of the invariant Arg64 was also confirmed in C. reinhardtii (18). In addition, Arg64 was found to play a major role in the interaction of PRK with A4-GAPDH during the formation of the regulatory ternary complex (18). Consistent with this finding, Arg64 faces out at the far end of the dimer (Fig. 1 and SI Appendix, Table S2).
Fig. 4.
Active-site and TRX-dependent regulation of CrPRK. (A) The catalytic cavity is shown. The distance between the regulatory cysteines is greater than 13 Å. (B) The electrostatic surface potential of the catalytic cavity is shown. The bottom of the catalytic cavity is marked by a positive potential. The negative potential observed in the left side of the cavity is thought to be involved in the correct positioning of TRX close to the regulatory cysteines. (C) The pKa of Cys16 was determined by measuring the IAM-mediated inactivation as a function of pH. (D) The molecular environment of Cys16, considering a sphere of 5 Å centered on its thiol group. The hydrogen bonds between the thiol group and the neighboring residues and the corresponding distances are shown.
The active site also contains a pair of regulatory cysteines (Fig. 4 A and B and SI Appendix, Fig. S4 A and B); Cys15 in AtPRK and Cys16 in CrPRK are located within the P loop (Walker A) (19) and belong to the ATP-binding site (20). Additionally, Cys54 in AtPRK and Cys55 in CrPRK are located at the C-terminal end of strand β2 and compose the intermolecular mixed disulfide with TRX-f (6) (Fig. 2 and SI Appendix, Fig. S2).
Redox Properties of CrPRK and AtPRK.
Despite the role that PRK plays in photosynthesis, putative PRK sequences have also been found in heterotrophic prokaryotes. Phylogenetic analyses performed on 69 PRK sequences, including 31 from nonphotosynthetic prokaryotes, clearly show a first evolutionary separation between Bacteria and Archaea. Cyanobacterial and eukaryotic PRKs emerged as the third clade from Archaea (SI Appendix, Fig. S6B). Single PRK structures from the anoxygenic photosynthetic bacterium Rhodobacter sphaeroides [RsPRK; Protein Data Bank (PDB) ID code 1A7J] (21) and the nonphotosynthetic Archaea Methanospirillum hungatei (MhPRK; PDB ID code 5B3F) (22) have been reported. Both RsPRK and MhPRK sequences are shorter than plant PRKs, with a low sequence identity (SI Appendix, Fig. S7). All PRKs from bacteria (type I PRKs) (23), including phototrophic organisms, are octamers (∼32-kDa subunits) that are allosterically activated by NADH and inhibited by AMP (24). Archaeal, cyanobacterial, and eukaryotic (i.e., plant-type) PRKs are dimers (∼40-kDa subunits; type II PRKs) (23) with no allosteric regulation (15, 25). While little is known about the regulation of archaeal PRKs, in oxygenic phototrophs from ancient cyanobacteria to modern flowering plants it is well-established that PRK forms a complex with A4-GAPDH and the regulatory protein CP12 at low NADP(H)/NAD(H) ratios (26, 27). Moreover, cyanobacterial PRKs are inhibited by AMP via the cyanobacteria-specific fusion protein CBS-CP12 (28), whereas plant-type PRKs are directly regulated by the TRX-dependent interconversion of specific dithiol–disulfide bridges (6). To date, no experimental evidence of the direct regulation of cyanobacterial PRKs by TRXs has been reported.
Regulatory cysteines Cys15/16 and Cys54/55 in AtPRK and CrPRK, respectively, protrude from the catalytic cavity (Fig. 4A and SI Appendix, Fig. S4A), and are thus available to interact with TRX (SI Appendix, Table S2). In addition, both PRKs share similar redox properties with comparable midpoint redox potentials (−312 ± 3 mV for CrPRK and −330 mV for AtPRK) (27) and pKa values (6.79 ± 0.01 for CrPRK and 6.95 ± 0.13 for AtPRK) (Fig. 4C and SI Appendix, Fig. S4C). These slightly acidic values are supported by the molecular environment around Cys15/16, as the thiol groups form hydrogen bonds with the main-chain atoms of two nearby residues (i.e., Asp13 and Ser14 in CrPRK and Asp12 and Ser13 in AtPRK) and with the amino and carboxyl groups of Lys156 and Asp13 in CrPRK or Lys151 in AtPRK (Fig. 4D and SI Appendix, Fig. S4D). All of these residues are strictly conserved in type II PRKs (SI Appendix, Fig. S6A). Furthermore, the proximity of Cys15/16 to the positive N-terminal end of helix α1 contributes to a low pKa value (Fig. 4D and SI Appendix, Fig. S4D).
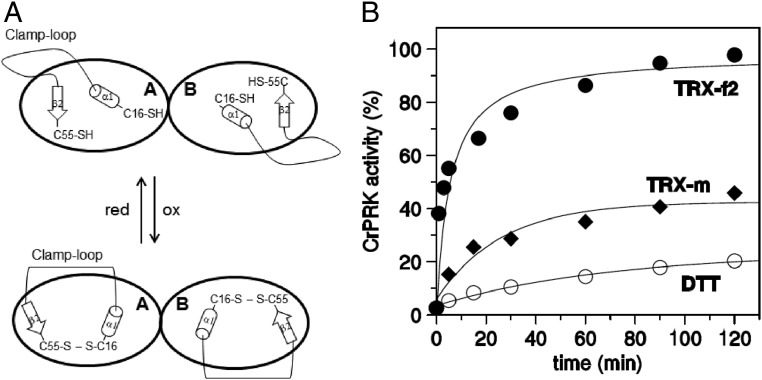
The pH and temperature dependence of AtPRK and CrPRK are also comparable, although the algal enzyme has a wider range of activity at higher temperatures and lower pH values (SI Appendix, Fig. S8). The pair of cysteines involved in TRX regulation is also present in the TRX-insensitive PRK in cyanobacteria (SI Appendix, Fig. S6A). However, cyanobacterial PRKs lack an unstructured amino acid stretch (29)—here named the clamp loop—which connects the regulatory cysteines whose thiol groups are separated by more than 13 Å, as found in plant-type PRKs (Fig. 4A and SI Appendix, Fig. S4A). The clamp loop connects α1 and β2 (Fig. 1 and SI Appendix, Fig. S7), and possibly confers the flexibility necessary to permit the conformational changes required for disulfide formation. This bond would thus allow the narrowing of the catalytic cavity and the inhibition of enzymatic activity (Fig. 5A). Similarly, a comparison of the chloroplast and redox-sensitive isoforms of FBPase (5) and AB-GAPDH (7) with their cytosolic and redox-insensitive counterparts reveals that the acquisition of TRX-mediated regulation is accompanied by the appearance of loops presumably required for the approach of the two cysteines involved in the formation of the regulatory disulfide bond.
Fig. 5.
Clamp-loop and TRX-dependent regulation of CrPRK. (A) A schematic representation of the proposed role for the clamp loop and conformational changes occurring in the chloroplast PRK upon TRX binding. (B) Activation kinetics by plastidial C. reinhardtii TRX-f2 and -m and DTT.
The electrostatic surface potential of the catalytic cavity reveals an elongated positive region on the bottom and a negative region on the side (Fig. 4B and SI Appendix, Fig. S4B), the former suitable for binding the phosphate groups of substrates (i.e., ATP and Ru5P) and the latter relevant for the recognition and positioning of TRX, as proposed for FBPase (17, 30). Compared with TRX-m, TRX-f2 is more efficient in the reductive activation of both AtPRK (31) and CrPRK (Fig. 5B). Accordingly, the structural model of AtTRX-f1 and the crystal structure of CrTRX-f2 (32) show that the catalytic cysteines are surrounded by a large positive region, less extended in TRX-m (SI Appendix, Fig. S9). This finding suggests that the early stages of pairing between TRX and PRK are mainly governed by electrostatic interactions between exposed regions of the two proteins (30).
Moreover, it has been reported that the oxidized state of target proteins is an essential feature for TRX target recognition, since disulfide formation does not confer any specific conformational change in TRX structure (17). Indeed, the disulfide bond between the two distant regulatory cysteines introduces a significant conformational constraint that decreases the overall entropy of PRK. Therefore, a favorable entropic contribution is proposed to be the main driving force of the reduction of PRK by TRXs.
Dimer Interface and Flexible Regions.
Dimer interfaces consist of large contact areas in octameric RsPRK (1,667 Å2) (21) and dimeric MhPRK (1,695 Å2) (SI Appendix, Fig. S10) (22), and a small area in eukaryotic PRK. The dimer interfaces in CrPRK (545.6 Å2) and AtPRK (560.3 Å2) are exclusively formed by β7, which is located in an antiparallel position to its partner (Figs. 1 and 2 and SI Appendix, Fig. S2), suggesting a less rigid structure. Since cyanobacterial and eukaryotic PRKs are found together with CP12, the greater flexibility of PRK could enable the formation of regulatory GAPDH–CP12–PRK complexes (9–15). Moreover, AtPRK and CrPRK are characterized by an increased number of exposed random-coiled regions (Fig. 1 and SI Appendix, Table S3), some of which show a poor or absent electron density, indicative of highly flexible and disordered regions (SI Appendix, Fig. S11). Two such regions are common to algal and plant enzymes, whereas the presence of a third region is exclusive to AtPRK.
One of these common regions corresponds to the long loop between helices α5 and α6 (SI Appendix, Fig. S11) that contains the highly conserved catalytic Asp160 or Arg159 and Arg164 residues involved in Ru5P binding (33, 34). A second common flexible region, corresponding to the loop between β7 and β8, contains Cys243 and Cys249 (Fig. 1 and SI Appendix, Fig. S11). In both PRK structures, this portion is so flexible that the two thiol groups are far from each other in one monomer and closer to one another in the other; however, in contrast to AtPRK, they are not oxidized in CrPRK (Fig. 1 and SI Appendix, Fig. S12). This result is in agreement with the observation that in CrPRK, the disulfide bond between Cys243 and Cys249 only forms via an interaction with CP12 (35). This interaction is an essential step for the assembly of PRK and A4-GAPDH, leading to the regulatory complex GAPDH–CP12–PRK (36, 37).
Concluding Remarks
TRXs are widespread regulatory proteins whose structure is strongly conserved throughout evolution. Fascinating experiments in paleobiochemistry conducted on seven “resurrected” TRXs dating back to the Precambrian Era showed that only small differences in the length of helix α1 were found to occur over 4 Gy of evolution (3). How evolution affected the TRX dependence of enzymes in the CB cycle is still a mystery. After studying the five CB cycle enzymes whose 3D structures have been solved or modeled (SI Appendix, Fig. S1), several strategies were adopted to establish their dependency on the same regulator. In fact, each of these five proteins is regulated by TRX in a different way. In FBPase and SBPase, the TRX-binding site is far from the conserved sugar-binding domain (5). In FBPase, upon reduction, the substrate-binding site adopts a catalytically competent conformation due to the loosening of secondary structures (4, 5). In AB-GAPDH, the redox-active cysteines are found in the highly flexible CTE, which is characteristic of B subunits and, when oxidized, the last residue in the CTE blocks the NADPH-binding site (7). Similarly, a change in protein flexibility is believed to enable the regulation of Chlamydomonas PGK, in which the formation of the regulatory disulfide appears to lower catalytic efficiency by decreasing the flexibility required to approach the substrates (8). Finally, we report that the regulatory pair of cysteines in PRK is embedded in the substrate-binding site and that oxidation can be achieved by the presence of the flexible clamp loop.
We conclude that a common strategy has allowed the acquisition of TRX dependence of the CB cycle enzymes during evolution—the introduction of flexible regions. This strategy is in sharp contrast to the evolutionary rigidity of the TRX structure.
Materials and Methods
Protein Expression and Purification.
Recombinant AtPRK and CrPRK were expressed in Escherichia coli BL21(DE3) strain (New England Biolabs) harboring the pET-28 expression vector (Novagen) containing the coding sequence of the mature form of both enzymes (SI Appendix, Fig. S7). AtPRK was expressed and purified as previously described (37). The sequence coding for CrPRK was cloned in-frame with a 6×His tag and a thrombin cleavage site at the 5′ end of the nucleotide sequence. CrPRK was purified by a Chelating Sepharose Fast Flow (GE Healthcare) column, following the manufacturer’s instructions. Immediately after elution, CrPRK was desalted in 30 mM Tris⋅HCl, 1 mM EDTA (pH 7.9) by PD-10 desalting columns (GE Healthcare). The 6×His tag was removed by overnight digestion with thrombin protease. The removal of the 6×His tag was controlled by MALDI-TOF mass spectrometry analysis. Sample concentration was evaluated by absorbance at 280 nm (NanoDrop; Thermo Scientific) using the extinction coefficients εAtPRK 31,985 M−1⋅cm−1 and εCrPRK 35,995 M−1⋅cm−1 and molecular masses AtPRK 38784.2 g⋅mol−1 and CrPRK 40790.7 and 38908.7 g⋅mol−1, respectively, before and after removal of the 6×His tag. Sample purity was checked by 12.5% SDS/PAGE, and proteins were stored at −80 °C.
When required, oxidized and reduced AtPRK and CrPRK were prepared by incubation at 25 °C for 2 to 3 h in the presence of 40 mM trans-4,5-dihydroxy-1,2-dithiane (oxidized DTT) or 1,4-DTT (reduced DTT). Following incubation, samples were desalted in 30 mM Tris⋅HCl, 1 mM EDTA (pH 7.9) through NAP-5 columns (GE Healthcare) and brought to the desired concentration either by dilution or by concentration through an Amicon Ultra device (cutoff 10 kDa; Millipore).
Activity Assay and Determination of pH and Temperature Dependence.
Phosphoribulokinase activity was measured spectrophotometrically (Cary 60 UV-Vis; Agilent), coupling the formation of ADP to NADH oxidation via pyruvate kinase and lactate dehydrogenase (38). All of the details for pH and temperature dependence experiments are reported in SI Appendix, Materials and Methods.
Thioredoxin Specificity and Redox Titration.
Thioredoxin specificity for activation of oxidized CrPRK (2 µM) was measured in the presence of 0.2 mM reduced DTT and C. reinhardtii recombinant TRX-f2 and TRX-m (5 µM each), as previously described (31). At different incubation times, PRK activity was measured. To determine the midpoint redox potential of CrPRK, three independent redox titrations were performed. Briefly, a mixture containing pure recombinant CrPRK (12 µM), commercial E. coli thioredoxin (1 mg⋅ml−1), and 20 mM DTT, at different reduced-to-oxidized ratios, was incubated for 3 h at 25 °C before measuring PRK activity. The obtained curves were fitted by nonlinear regression (CoStat; Cohort Software) to the Nernst equation.
Determination of the pKa Values of the Catalytic Cysteines.
The determination of pKa values for both CrPRK and AtPRK was performed as previously described (39). Briefly, PRK activity was measured on reduced PRK samples (2 µM) following 20 min of incubation at 25 °C in the presence or absence of 0.2 mM iodoacetamide (IAM). Incubations were performed in different buffers within a 4.5 to 10 pH range. Aliquots of 10 to 20 µL were used to measure PRK activity in 1 mL of assay mixture. The residual activity was expressed as the percentage of inhibition between IAM-treated and untreated samples, and expressed as a function of pH. The obtained curves were fitted by nonlinear regression (CoStat; Cohort Software) to a modified Henderson–Hasselbalch equation.
Crystallization and Structure Determination.
Reduced CrPRK was concentrated to 10 mg⋅mL−1 in 30 mM Tris⋅HCl, 1 mM EDTA (pH 7.9). Reduced AtPRK was concentrated to 10 mg⋅mL−1 in 25 mM potassium phosphate buffer solution (pH 7.5). They were crystallized by the vapor-diffusion method at 293 K with sitting drop in 22% (wt/vol) PEG MME 5K, 0.1 M Mes (pH 6.5), 0.2 M ammonium sulfate for CrPRK, and hanging drop in 1.5 M sodium malonate (pH 5.0) for AtPRK. The crystals obtained were analyzed by X-ray diffraction, and the structures were solved by molecular replacement. The refinement was performed with REFMAC 5.8.0135 (40), selecting 5% of reflections for Rfree (for details, see SI Appendix, Materials and Methods).
Small-Angle X-Ray Scattering Data Collection and Analysis.
Small-angle X-ray scattering data were collected at the BioSAXS beamline BM29 (41) at the European Synchrotron Radiation Facility (ESRF, Grenoble), and the data-collection parameters are reported in SI Appendix, Table S4. A size-exclusion chromatography–SAXS experiment was performed using an HPLC system (Shimadzu) directly connected to the measurement capillary. All of the details about experimental procedures and data analysis are listed in SI Appendix, Materials and Methods.
Modeling from SAXS Data.
The experimental SAXS profile of the CrPRK dimer was fitted with the theoretical profile calculated from the atomic coordinates using CRYSOL3 (42) with default parameters. In addition, low-resolution models of the CrPRK dimer based on the SAXS profile were built using the ab initio program GASBOR (43). All details are reported in SI Appendix, Materials and Methods.
Data Availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under PDB ID codes 6H7G (44) and 6H7H (45) for CrPRK and AtPRK, respectively. The SEC-SAXS data of CrPRK have been deposited in the Small Angle Scattering Biological Data Bank with ID code SASDDH9 (46).
Supplementary Material
Acknowledgments
We thank Elettra and the European Synchrotron Radiation Facility for allocation of X-ray diffraction and SAXS beam time (BAG Proposals MX686 and MX1750) and the staff of beamlines XRD1 (Elettra) and ID14-4 and BM29 (ESRF) for technical support. S.F. thanks the Consorzio Interuniversitario di Ricerca in Chimica dei Metalli nei Sistemi Biologici (CIRCMSB). This work was supported by Italian Ministry of Education, Research and University (MIUR) Grant FIRB2003 (to P.T.); University of Bologna Grant FARB2012 (to F.S., S.F., and M.Z.); CNRS, Sorbonne Université, Agence Nationale de la Recherche Grant 17-CE05-0001; and CalvinDesign and LABEX DYNAMO ANR-LABX-011 (to J.H., S.D.L., and P.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6H7G and 6H7H for CrPRK and AtPRK, respectively). The SEC-SAXS data of CrPRK reported in this paper have been deposited in the Small Angle Scattering Biological Data Bank, https://www.sasbdb.org (ID code SASDDH9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820639116/-/DCSupplemental.
References
- 1.Michelet L, et al. Redox regulation of the Calvin-Benson cycle: Something old, something new. Front Plant Sci. 2013;4:470. doi: 10.3389/fpls.2013.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geigenberger P, Thormählen I, Daloso DM, Fernie AR. The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci. 2017;22:249–262. doi: 10.1016/j.tplants.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Ingles-Prieto A, et al. Conservation of protein structure over four billion years. Structure. 2013;21:1690–1697. doi: 10.1016/j.str.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiadmi M, Navaza A, Miginiac-Maslow M, Jacquot JP, Cherfils J. Redox signalling in the chloroplast: Structure of oxidized pea fructose-1,6-bisphosphate phosphatase. EMBO J. 1999;18:6809–6815. doi: 10.1093/emboj/18.23.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gütle DD, et al. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc Natl Acad Sci USA. 2016;113:6779–6784. doi: 10.1073/pnas.1606241113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandes HK, Larimer FW, Hartman FC. The molecular pathway for the regulation of phosphoribulokinase by thioredoxin f. J Biol Chem. 1996;271:3333–3335. doi: 10.1074/jbc.271.7.3333. [DOI] [PubMed] [Google Scholar]
- 7.Fermani S, et al. Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 2007;104:11109–11114. doi: 10.1073/pnas.0611636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morisse S, et al. Thioredoxin-dependent redox regulation of chloroplastic phosphoglycerate kinase from Chlamydomonas reinhardtii. J Biol Chem. 2014;289:30012–30024. doi: 10.1074/jbc.M114.597997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouche F, Gontero B, Callebaut I, Mornon J-P, Boisset N. Striking conformational change suspected within the phosphoribulokinase dimer induced by interaction with GAPDH. J Biol Chem. 2002;277:6743–6749. doi: 10.1074/jbc.M106401200. [DOI] [PubMed] [Google Scholar]
- 10.Howard TP, Metodiev M, Lloyd JC, Raines CA. Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc Natl Acad Sci USA. 2008;105:4056–4061. doi: 10.1073/pnas.0710518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard TP, Lloyd JC, Raines CA. Inter-species variation in the oligomeric states of the higher plant Calvin cycle enzymes glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase. J Exp Bot. 2011;62:3799–3805. doi: 10.1093/jxb/err057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Giudice A, et al. Unravelling the shape and structural assembly of the photosynthetic GAPDH-CP12-PRK complex from Arabidopsis thaliana by small-angle X-ray scattering analysis. Acta Crystallogr D Biol Crystallogr. 2015;71:2372–2385. doi: 10.1107/S1399004715018520. [DOI] [PubMed] [Google Scholar]
- 13.Moparthi SB, et al. FRET analysis of CP12 structural interplay by GAPDH and PRK. Biochem Biophys Res Commun. 2015;458:488–493. doi: 10.1016/j.bbrc.2015.01.135. [DOI] [PubMed] [Google Scholar]
- 14.López-Calcagno PE, Abuzaid AO, Lawson T, Raines CA. Arabidopsis CP12 mutants have reduced levels of phosphoribulokinase and impaired function of the Calvin-Benson cycle. J Exp Bot. 2017;68:2285–2298. doi: 10.1093/jxb/erx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mcfarlane C, et al. 2018. Structural basis of light-induced redox regulation in the Calvin cycle. bioRxiv:10.1101/414334. Preprint, posted September 11, 2018.
- 16.Marri L, et al. CP12-mediated protection of Calvin-Benson cycle enzymes from oxidative stress. Biochimie. 2014;97:228–237. doi: 10.1016/j.biochi.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Palde PB, Carroll KS. A universal entropy-driven mechanism for thioredoxin-target recognition. Proc Natl Acad Sci USA. 2015;112:7960–7965. doi: 10.1073/pnas.1504376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avilan L, Gontero B, Lebreton S, Ricard J. Information transfer in multienzyme complexes. 2. The role of Arg64 of Chlamydomonas reinhardtii phosphoribulokinase in the information transfer between glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase. Eur J Biochem. 1997;250:296–302. doi: 10.1111/j.1432-1033.1997.0296a.x. [DOI] [PubMed] [Google Scholar]
- 19.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandes HK, Hartman FC, Lu TY, Larimer FW. Efficient expression of the gene for spinach phosphoribulokinase in Pichia pastoris and utilization of the recombinant enzyme to explore the role of regulatory cysteinyl residues by site-directed mutagenesis. J Biol Chem. 1996;271:6490–6496. doi: 10.1074/jbc.271.11.6490. [DOI] [PubMed] [Google Scholar]
- 21.Harrison DH, Runquist JA, Holub A, Miziorko HM. The crystal structure of phosphoribulokinase from Rhodobacter sphaeroides reveals a fold similar to that of adenylate kinase. Biochemistry. 1998;37:5074–5085. doi: 10.1021/bi972805y. [DOI] [PubMed] [Google Scholar]
- 22.Kono T, et al. A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat Commun. 2017;8:14007. doi: 10.1038/ncomms14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin W, Schnarrenberger C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: A case study of functional redundancy in ancient pathways through endosymbiosis. Curr Genet. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
- 24.Kung G, Runquist JA, Miziorko HM, Harrison DH. Identification of the allosteric regulatory site in bacterial phosphoribulokinase. Biochemistry. 1999;38:15157–15165. doi: 10.1021/bi991033y. [DOI] [PubMed] [Google Scholar]
- 25.Porter MA, Hartman FC. Exploration of the function of a regulatory sulfhydryl of phosphoribulokinase from spinach. Arch Biochem Biophys. 1990;281:330–334. doi: 10.1016/0003-9861(90)90452-5. [DOI] [PubMed] [Google Scholar]
- 26.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42:504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 27.Marri L, Trost P, Pupillo P, Sparla F. Reconstitution and properties of the recombinant glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase supramolecular complex of Arabidopsis. Plant Physiol. 2005;139:1433–1443. doi: 10.1104/pp.105.068445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackenberg C, et al. Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria. Proc Natl Acad Sci USA. 2018;115:7141–7146. doi: 10.1073/pnas.1806668115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi D, Tamoi M, Iwaki T, Shigeoka S, Wadano A. Molecular characterization and redox regulation of phosphoribulokinase from the cyanobacterium Synechococcus sp. PCC 7942. Plant Cell Physiol. 2003;44:269–276. doi: 10.1093/pcp/pcg048. [DOI] [PubMed] [Google Scholar]
- 30.Mora-García S, Rodríguez-Suárez R, Wolosiuk RA. Role of electrostatic interactions on the affinity of thioredoxin for target proteins. Recognition of chloroplast fructose-1, 6-bisphosphatase by mutant Escherichia coli thioredoxins. J Biol Chem. 1998;273:16273–16280. doi: 10.1074/jbc.273.26.16273. [DOI] [PubMed] [Google Scholar]
- 31.Marri L, et al. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- 32.Lemaire SD, et al. Crystal structure of chloroplastic thioredoxin f2 from Chlamydomonas reinhardtii reveals distinct surface properties. Antioxidants. 2018;7:E171. doi: 10.3390/antiox7120171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlier HA, Jr, Runquist JA, Miziorko HM. Evidence supporting catalytic roles for aspartate residues in phosphoribulokinase. Biochemistry. 1994;33:9343–9350. doi: 10.1021/bi00197a039. [DOI] [PubMed] [Google Scholar]
- 34.Runquist JA, Harrison DH, Miziorko HM. Rhodobacter sphaeroides phosphoribulokinase: Identification of lysine-165 as a catalytic residue and evaluation of the contributions of invariant basic amino acids to ribulose 5-phosphate binding. Biochemistry. 1999;38:13999–14005. doi: 10.1021/bi9910326. [DOI] [PubMed] [Google Scholar]
- 35.Thieulin-Pardo G, Remy T, Lignon S, Lebrun R, Gontero B. Phosphoribulokinase from Chlamydomonas reinhardtii: A Benson-Calvin cycle enzyme enslaved to its cysteine residues. Mol Biosyst. 2015;11:1134–1145. doi: 10.1039/c5mb00035a. [DOI] [PubMed] [Google Scholar]
- 36.Graciet E, et al. The small protein CP12: A protein linker for supramolecular complex assembly. Biochemistry. 2003;42:8163–8170. doi: 10.1021/bi034474x. [DOI] [PubMed] [Google Scholar]
- 37.Marri L, et al. Spontaneous assembly of photosynthetic supramolecular complexes as mediated by the intrinsically unstructured protein CP12. J Biol Chem. 2008;283:1831–1838. doi: 10.1074/jbc.M705650200. [DOI] [PubMed] [Google Scholar]
- 38.Roesler KR, Ogren WL. Chlamydomonas reinhardtii phosphoribulokinase: Sequence, purification, and kinetics. Plant Physiol. 1990;93:188–193. doi: 10.1104/pp.93.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bedhomme M, et al. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem J. 2012;445:337–347. doi: 10.1042/BJ20120505. [DOI] [PubMed] [Google Scholar]
- 40.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 41.Brennich ME, et al. Online data analysis at the ESRF bioSAXS beamline, BM29. J Appl Crystallogr. 2016;49:203–212. [Google Scholar]
- 42.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franke D, et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fermani S, et al. 2018 Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii. Protein Data Bank. Available at https://www.rcsb.org/structure/6H7G. Deposited July 31, 2018.
- 45.Fermani S, Sparla F, Gurrieri L, Falini G, Trost P. 2018 Crystal structure of redox-sensitive phosphoribulokinase (PRK) from Arabidopsis thaliana. Protein Data Bank. Available at www.rcsb.org/structure/6H7H. Deposited July 31, 2018.
- 46.Del Giudice A. 2018 Photosynthetic CO2 assimilation: The last gap in the structural proteome is closed. Small Angle Scattering Biological Data Bank. Available at https://www.sasbdb.org/data/SASDDH9/. Deposited July 5, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under PDB ID codes 6H7G (44) and 6H7H (45) for CrPRK and AtPRK, respectively. The SEC-SAXS data of CrPRK have been deposited in the Small Angle Scattering Biological Data Bank with ID code SASDDH9 (46).