Significance
The three endocrine fibroblast growth factors (FGFs)—FGF19, FGF21, and FGF23—are circulating hormones that regulate critical metabolic processes in liver, adipocytes, hypothalamus, and kidney. Endocrine FGFs mediate their cellular responses by binding to and stimulating complexes composed of an FGF receptor bound to either an α-Klotho or β-Klotho receptor. This paper describes the three-dimensional structure of FGF19CT bound to sKLB and provides mechanistic insights into the mode of action of endocrine FGFs. Detailed understanding of the molecular mechanism of FGF19 provides valuable information and a roadmap for development of agonists or antagonists for treatment of patients with metabolic diseases or cancers that may benefit from either stimulation (FGF21) or inhibition (FGF19, FGF23) of endocrine FGF cell signaling.
Keywords: structural biology, cell signaling, endocrine FGF, phosphorylation, metabolism
Abstract
The three members of the endocrine fibroblast growth factor (FGF) family designated FGF19, FGF21, and FGF23 mediate their pleiotropic cellular effects by binding to and activating binary complexes composed of an FGF receptor (FGFR) bound to either α-Klotho or β-Klotho receptors. Structural analyses of ligand-occupied Klotho extracellular domains have provided important insights concerning mechanisms underlying the binding specificities of FGF21 and FGF23 to β-Klotho or α-Klotho, respectively. They have also demonstrated that Klotho proteins function as primary high-affinity receptors while FGFRs function as the catalytic subunits that mediate intracellular signaling. Here we describe the crystal structure the C-terminal tail of FGF19 (FGF19CT) bound to sKLB and demonstrate that FGF19CT and FGF21CT bind to the same binding site on sKLB, via a multiturn D-P motif to site 1 and via a S-P-S motif to the pseudoglycoside hydrolase region (site 2). Binding affinities to sKLB and cellular stimulatory activities of FGF19CT, FGF21CT, and a variety of chimeric mutants to cells expressing β-Klotho together with FGFR1c or FGFR4 were also analyzed. These experiments as well as detailed comparison of the structures of free and ligand-occupied sKLB to the structure of ligand-occupied sKLA reveal a general mechanism for recognition of endocrine FGFs by Klotho proteins and regulatory interactions with FGFRs that control their pleiotropic cellular responses.
The three members of the endocrine fibroblast growth factor (FGF) family designated FGF19, FGF21, and FGF23 are circulating hormones that regulate a variety of essential metabolic processes in specific tissues (1–6). Unlike canonical FGFs, which require additional interactions with heparin or heparan sulfate proteoglycan (HSPG) for FGF receptor (FGFR) activation and cell stimulation (7–9), cell stimulation by endocrine FGFs requires binding to Klotho receptors for FGFR activation (10–15). A variety of genetic and biochemical experiments have demonstrated that binding of FGF23 to α-Klotho is essential for FGFR activation in target tissues (10, 11) and that binding of FGF19 or FGF21 to β-Klotho is essential for activation of FGFR in their target tissues (12–15). It was proposed that Klotho proteins function as coreceptors of endocrine FGF in a similar way to the role played by heparin or HSPG in mediating the action of canonical FGFs (9–14). Overall, high-affinity binding of endocrine FGF to Klotho receptors defines the specific tissues in which a distinct set of critical metabolic processes are regulated by FGF19, FGF21, or FGF23 stimulation.
Klotho proteins are type-I membrane proteins composed of a signal sequence, a large extracellular ligand-binding region, a single transmembrane domain, and a small cytoplasmic region (16). The extracellular ligand-binding regions of both α-Klotho and β-Klotho are composed of tandem repeats, designated KL1 and KL2, with amino acid sequences similar to glycoside hydrolase family 1 (GH1) enzymes. GH1 enzymes contain two highly conserved glutamic acid residues shown to be essential for enzymatic activity via a double-replacement mechanism in which one glutamic acid functions as nucleophilic residue and a second glutamic acid enables acid/base catalysis (17, 18). As each of the KL1 or KL2 domains of α-Klotho or β-Klotho contain only a single glutamic acid, both KL1 or KL2 domains of Klotho proteins do not possess enzymatic activities and can be described as pseudoglycoside hydrolases. Indeed, the crystal structure of β-Klotho extracellular domain (sKLB) in complex with the C-terminal tail of FGF21 (FGF21CT) revealed strong similarities to the structure of β-glucosidase bound to an oligosaccharide substrate, providing molecular insights into how a glycoside hydrolase has evolved to become a hormone receptor that regulates a variety of metabolic processes (19).
In this report, we describe the crystal structure of sKLB in complex with the C-terminal tail of FGF19 (FGF19CT) and compare it to the structure of sKLB in complex with FGF21CT. Detailed comparison of sKLB structures occupied by two different ligands and quantitative analyses of the binding properties of a variety of FGF19 and FGF21 mutants together with analyses of their cellular properties provide insights into the molecular mechanism underlying distinct and common factors underlying the action of FGF19 and FGF21. Moreover, on the basis of these results, together with structural analyses of the ligand-binding regions of α-Klotho, we propose a general mechanism of how endocrine FGFs recognize and activate their cognate Klotho/FGFR complexes.
Results and Discussion
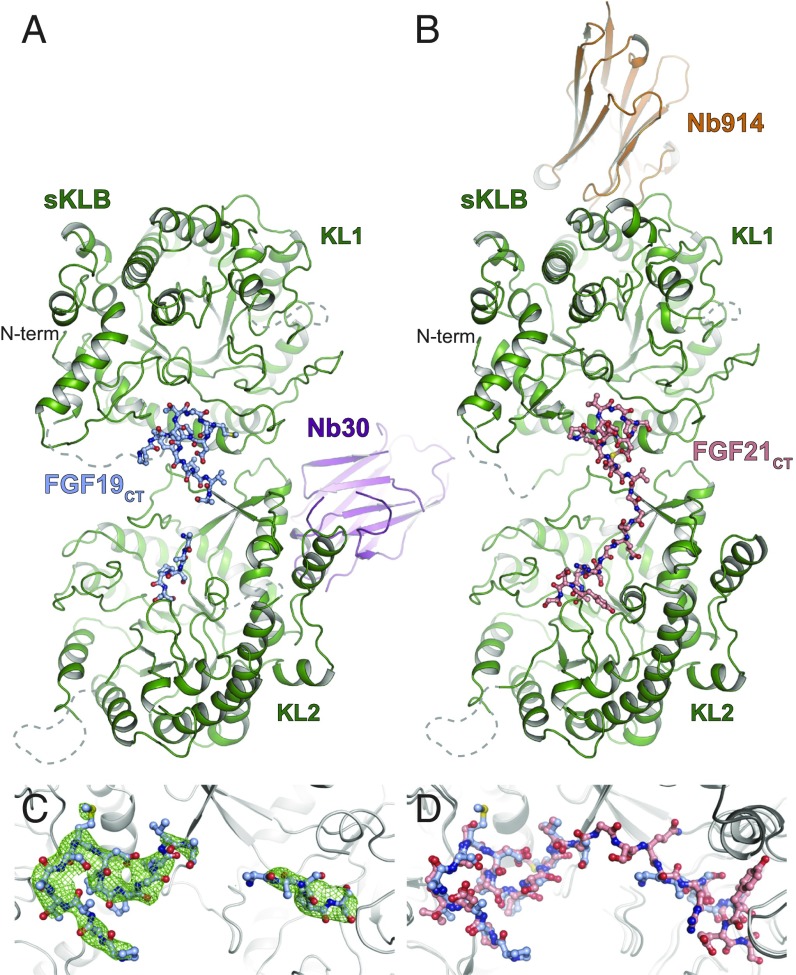
While stimulation of FGFRs by both FGF19 and FGF21 depends upon interactions with β-Klotho, the two endocrine FGFs stimulate different cellular responses even in the same cells or tissues, raising the possibility that FGF19 and FGF21 may interact differentially with β-Klotho and/or FGFRs. Indeed, previously described binding experiments using surface plasmon resonance (SPR) analysis proposed that FGF19 exhibits higher binding affinity than FGF21 toward β-Klotho (20). To provide molecular details about the nature of the interactions between β-Klotho and FGF19, we embarked on determination of the crystal structure of sKLB in complex with FGF19CT. We previously determined the crystal structure of sKLB in complex with FGF21CT by soaking preformed crystals of sKLB:Nb914 nanobody complexes with FGF21CT (19). However, extensive attempts to soak crystals of the sKLB:Nb914 complex using various preparations of the C-terminal tails of FGF19 did not yield visible electron densities of bound FGF19CT molecules. Also, crystallization trials with the sKLB:Nb914 complex in the presence of FGF19CT did not yield any diffraction-quality crystals, as seen previously when FGF21CT was used in similar crystallization trials. To overcome this problem, we searched for alternative nanobodies that may serve as crystallization chaperones of sKLB in the presence of FGF19CT (or FGF21CT). Indeed, nanobody Nb30, together with sKLB and FGF19CT, produced crystals that diffract to a resolution of 3.2 Å (SI Appendix, Table S1). Molecular replacement using separate KL1 or KL2 domains as search models of sKLB followed by a round of rigid body refinement enabled identification of the Nb30-binding region on sKLB, which is distinct from the epitope region recognized by Nb914 (Fig. 1 A and B). After several rounds of refinement, electron densities that correspond to FGF19CT (Fig. 1C) could be confidently modeled. A final model composed of a complex of two molecules of sKLB:Nb30:FGF19CT where we were able to place amino acid residues P191-T204 and V209-S213 of FGF19 is presented in Fig. 1A. One of the two complex molecules in the asymmetric unit, owing to its more extensive packing interfaces, is more ordered and is presented in Figs. 1 and 2.
Fig. 1.
Crystal structures of sKLB in complex with FGF19CT. Crystal structures of sKLB (green) in complex with (A) FGF19CT (light blue) and nanobody Nb30 (purple), and with (B) FGF21CT (salmon) and nanobody Nb914 (orange) (PDB ID: 5VAQ). sKLB and nanobodies are shown as ribbon representations, and FGF19CT and FGF21CT are shown as ball-and-stick representations. Regions that do not exhibit significant electron densities are shown as gray dashed lines. (C) FGF19CT binding site showing |Fo|-|Fc| omit map (in green mesh) contoured at 3.0 σ for FGF19CT. (D) FGF19CT is superimposed with FGF21CT with an overall Cα rmsd of 1.02 Å. sKLB is shown as a gray ribbon in C and D.
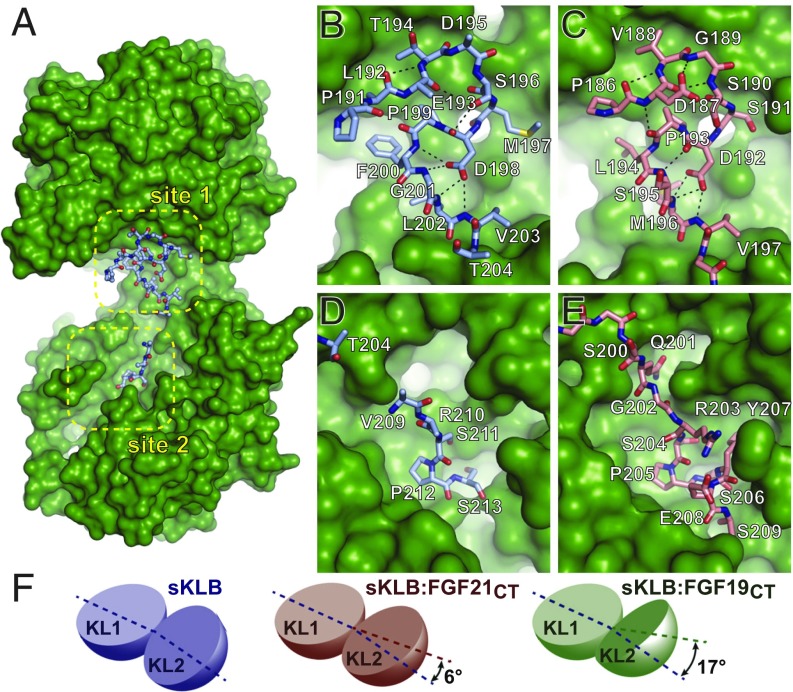
Fig. 2.
Comparison of ligand-binding sites in crystal structures of sKLB-ligand complexes. (A) Surface representation of sKLB (green) highlighting two binding sites, site 1 and site 2, of FGF19CT (light blue, ball-and-stick). Nb30 is omitted for clarity. (B–E) Regions of (B and D) FGF19CT (light blue, stick) and (C and E) FGF21CT (salmon, stick) that interact with (B and C) site 1 of sKLB and (D and E) site 2 of sKLB. Site 1-interacting regions in both (B) FGF19CT and (C) FGF21CT form consecutive turns through intramolecular hydrogen bonds indicated as black dashed lines. S-P-S motifs in site 2-interacting regions in both (D) FGF19CT and (E) FGF21CT interact with a pseudosubstrate-binding pocket in KL2 of sKLB. (F) Diagrams depicting change interdomain angles across various crystal structures of sKLB. Angles are shown as a difference in comparison with the structure of sKLB:Nb914 (PDB ID: 5VAN). Changes in sKLB interdomain angles upon binding FGF19 or FGF21 vary not only in absolute angle values (16.9° and 5.9°, respectively), but also in the direction of movement of KL2 in respect to KL1, which are calculated as different contributions of twist and closure components to the resulting movement. Twist is movement around the axis parallel to the line joining the centers of mass of domains, and closure is movement around the axis perpendicular to this line (21). The contributions from twist and closure components are 41 and 59% for sKLB:FGF19CT and 23 and 77% for sKLB:FGF21CT, respectively.
The overall structure of sKLB in complex with FGF19CT (Fig. 1A) is similar to the structure of sKLB in complex with FGF21CT (Fig. 1B); both ligand-occupied sKLB complexes exhibit the glycoside hydrolase-like domains KL1 and KL2, which are similarly occupied by FGF19CT or FGF21CT (Figs. 1 and 2 A–E). However, comparison of the crystal structures shows that the distances and angles between KL1 and KL2 vary among those seen in the structures of free sKLB, sKLB bound to FGF21CT, and sKLB bound to FGF19CT (Fig. 2F and Movie S1). A change of 17° in the angle between the KL1 and KL2 domains was detected in the structure of the sKLB:Nb30:FGF19CT complex in comparison with the structure of the sKLB:Nb914 complex, and a change of 6° was detected in the angle between KL1 and KL2 domains in the structure of the sKLB:Nb914:FGF21CT compared with the structure of the sKLB:Nb914 complex (21). These results demonstrate that the linker connecting KL1 to KL2 is flexible, enabling interdomain conformational changes that likely affect FGF19 and FGF21 binding to KL1 and KL2 and complex formation between sKLB and FGFRs (Fig. 2F and Movie S1). It is noteworthy that the changes in interdomain angles and distances seen in different crystal structures may also be influenced by the binding of the nanobodies because of their impact on crystal packing interfaces. In any event, the existence of intrinsic flexibilities in sKLB and the dynamic nature of interdomain interaction suggest that conformational changes upon ligand binding may contribute to how sKLB interacts with FGFR and how signaling occurs by endocrine FGF stimulation.
FGF19 and FGF21 Occupy the Same Binding Site on β-Klotho.
Comparison of the crystal structure of the sKLB:Nb30:FGF19CT complex to the previously described crystal structure of the sKLB:Nb914:FGF21CT complex shows that FGF19 binds to the same sites occupied by FGF21 (Fig. 2 B–E). Because of the difference in the angles and distances between the KL1 and KL2 domains of the two structures, overlaying of the full-length sKLB structures (excluding nanobodies) yielded an overall Cα rmsd of 1.80 Å. By contrast, overlaying individual domains yielded an overall Cα rmsd of 0.438 Å and 0.516 Å for KL1 and KL2, respectively, revealing that FGF19CT and FGF21CT indeed bind to the same site on sKLB. While the two structures utilize different crystal packing interfaces, the two ligands occupy the same site in sKLB of the two structures, providing further validation for the authenticity of the binding site of FGF19 and FGF21 to β-Klotho.
Similarly to FGF21CT, FGF19CT binds to an elongated shallow binding site along both KL1 and KL2 and the linker connecting the two domains on sKLB, named site 1 and site 2, respectively (Fig. 2A). Site 1 and site 2 defined in the structure of sKLB:Nb30:FGF19CT correspond to the same site 1 and site 2 in the sKLB:Nb914:FGF21CT structure. Site 1 is composed of a large hydrophobic surface that is occupied by a multiturn element created by P191-V203 in FGF19 (Fig. 2B), exhibiting an almost identical binding mode of P186-V197 in FGF21 with site 1 of sKLB (Fig. 2C). Site 2, on the other hand, exhibits limited contacts with FGF19CT, while FGF21CT showed extensive interactions with sKLB (Fig. 2 D and E). Because of the lack of electron densities in this site, we were able to model only V209-S213 of FGF19. Possible explanations for the lack of molecular contacts seen in site 2 of the sKLB:Nb30:FGF19CT structure include differences in binding affinities between FGF19 and FGF21 toward sKLB (see below), flexibilities of the site 2-interacting region (SIR2) in FGF19, and the resolution limit of our current crystal form. Nonetheless, the sKLB:Nb30:FGF19CT structure also shows that the pseudosubstrate binding region in site 2 interacts with a sugar-mimicking motif, S211-P212-S213, in FGF19, in a manner similar to the interaction with S204-P205-S206 in FGF21 (Fig. 2 D and E).
Amino acid sequence alignment of the C-terminal regions of all endocrine FGFs (SI Appendix, Fig. S1) reveals two signature motifs in FGF19 and FGF21: (i) the D-P motif, which is crucial for maintaining intramolecular hydrogen bonds in the site 1-interacting region (SIR1), and (ii) the S-P-S motif, which is important for recognizing the pseudosubstrate region in site 2. Additionally, the alignment shows that the SIR1 in FGF23 also contains the D-P motif. Since these two signature sequences are conserved in both FGF19 and FGF21 across many species (SI Appendix, Fig. S1), it is perhaps not surprising that the molecular interactions seen in site 1 or site 2 in these crystal structures are similar. The sequence alignment also reveals that the amino acid sequences of the C-terminal regions of FGF19 and FGF15 (mouse ortholog of FGF19) are variable except for the two signature motifs, while FGF21 sequences are well-conserved across species. This observation, combined with the high conservation of β-Klotho’s site 1 and site 2 between human and mouse (SI Appendix, Fig. S2A), may suggest that FGF21CT is more “evolutionarily optimized” for binding to β-Klotho, compared with FGF19CT, a hypothesis consistent with the binding affinity measurements of FGF19 and FGF21 to β-Klotho (see below).
The interactions between FGF19CT and sKLB seen in the crystal structure were validated by comparing the ability of FGF19 to stimulate cells harboring wild-type (WT) or various β-Klotho mutants (SI Appendix, Fig. S3). In these experiments, amino acids in β-Klotho that make contacts with FGF19 in the structure were changed to corresponding amino acids in α-Klotho. L6 cells stably expressing matched levels of FGFR1c and various β-Klotho mutants were generated and stimulated with FGF19. Lysates from unstimulated or ligand-stimulated cells were subjected to immunoprecipitation with anti-FGFR antibody, followed by SDS/PAGE and immunoblotting with either antiphosphotyrosine antibody or an anti-FGFR antibody. Results shown in SI Appendix, Fig. S3, support the molecular contacts in the crystal structure of sKLB:Nb30:FGF19CT and are consistent with our previous results using L6 cell lines harboring the same set of β-Klotho mutants stimulated with FGF21 (19), further demonstrating that FGF19 and FGF21 bind to the same binding sites on β-Klotho.
Reduced Binding Affinity of FGF19 Versus FGF21 Binding to β-Klotho.
Binding kinetics of FGF19 and FGF21 to sKLB were measured using bio-layer interferometry (BLI) (Fig. 3 A and B and SI Appendix, Table S2). BLI biosensors coated with anti-His tag antibodies were used to capture FGF19 or FGF21, the amino termini of which were engineered to contain a hexa-histidine tag. The sensors were then transferred into solutions containing various concentrations of sKLB. The experiment presented in Fig. 3B depicts BLI traces that fit a 1:1 ligand:receptor stoichiometry with dissociation constants (KD) of 210 ± 13 and 23.9 ± 0.7 nM for binding of sKLB to FGF19 and FGF21, respectively. The nearly 10-fold increase in FGF21 binding affinity is primarily driven by a slower dissociation rate (koff) of 6.7 × 10−3 versus 6.1 × 10−2 s−1 for FGF21 versus FGF19 binding, respectively (Fig. 3B and SI Appendix, Table S2). Similar differences in dissociation constants were also determined using microscale thermophoresis (MST) measurements of fluorescently labeled FGF19 or FGF21 to sKLB (SI Appendix, Fig. S4). The kinetic parameters and dissociation constants of GST fusion proteins containing the C-terminal tails (CTs) of FGF21 or FGF19 toward sKLB were also measured using BLI biosensors (SI Appendix, Fig. S5), which provided binding results consistent with the measurements carried on using the full-length ligands and confirmed that binding of FGF19 or FGF21 to β-Klotho is entirely mediated by interactions between the CT regions of FGF19 or FGF21 and β-Klotho.
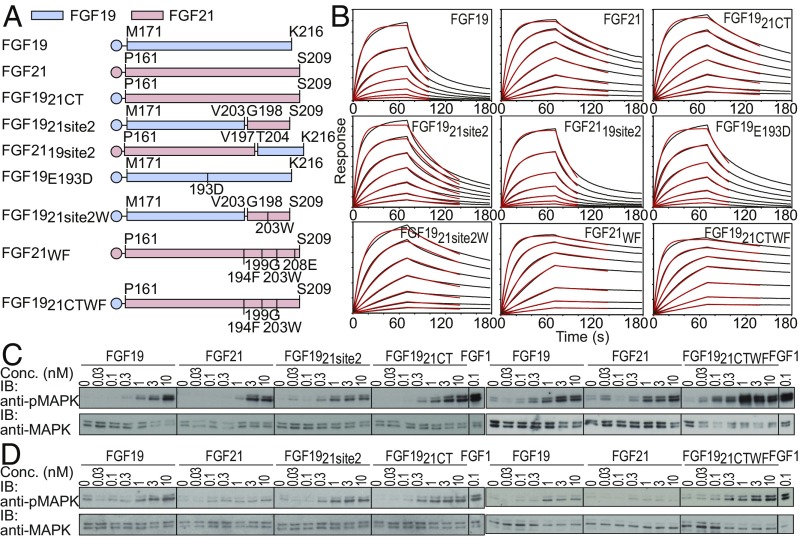
Fig. 3.
Binding parameters and cellular activities of FGF19, FGF21, and their chimeric variants. (A) Schematic diagram of FGF19, FGF21, and their variants tested for their binding affinity, kinetic parameters, and abilities to stimulate MAPK responses. Sequences derived from FGF19 and FGF21 are shown in light blue and light red, respectively. (B) Sensorgrams from BLI measurements. Biosensors coated with anti-histidine antibody were used to capture hexa-histidine–tagged FGF19 or FGF21 variants and dipped into solutions containing a series of concentrations of sKLB (400, 200, 100, 50, 25, 12.5, and 6.25 nM). Sensorgrams were fitted with a 1:1 ligand:receptor–binding model (red lines) to calculate kinetic parameters. (C and D) Comparison of MAPK responses of FGF19, FGF21, and their chimeric ligands. Stably transfected L6 cells expressing either (C) FGFR1c or (D) FGFR4 together with β-Klotho were incubated with various concentrations of FGF19, FGF21, or their chimeric proteins for 10 min and analyzed for MAPK activation.
The binding experiments presented in Fig. 3B confirm previous studies demonstrating that the C-terminal tails of endocrine FGFs are responsible for complex formation with Klotho receptors (22–24). However, the nearly 10-fold increase in the binding affinity of FGF21 over FGF19 to sKLB disagrees with the binding affinity obtained using earlier SPR-based inhibition measurements reporting a higher binding affinity of FGF19 versus FGF21 to sKLB (20). Overall, SPR-binding experiments of FGF19 or FGF21 to sKLB suffer from high background signals, raising concerns about the validity of these measurements. BLI measurements, on the other hand, utilize sensors that exhibit minimal background signals for binding experiments of FGF19, FGF21, or other FGFs to Klotho proteins, allowing determination of accurate binding kinetics and dissociation constant measurements with great reproducibility. Therefore, all of the kinetic parameter and the dissociation constants of FGF19, FGF21, or their variants to sKLB were derived from data using BLI or MST measurements.
Cooperative Interaction of FGF19CT with Two Binding Sites on β-Klotho.
To investigate the roles played by each of the binding regions of FGF19 and FGF21 as well as to identify regions responsible for mediating their distinct binding characteristics to sKLB, we have generated a series of chimeric molecules composed of a different combination of SIR1 and SIR2 of FGF19 and FGF21 (Fig. 3A) and measured their binding kinetics and dissociation constants to sKLB (Fig. 3B and SI Appendix, Table S2). The results show that a chimeric molecule composed of the “FGF core” of FGF19 linked to FGF21CT, FGF1921CT, exhibits binding characteristics similar to FGF21 binding to sKLB (Fig. 3B and SI Appendix, Table S2). Moreover, when SIR2 in FGF19 is replaced by the corresponding region of FGF21, the resulting FGF19 variant (FGF1921site2) exhibits nearly fivefold enhanced binding affinity compared with FGF19 binding, i.e., 210 and 43 nM for FGF19 and FGF1921site2, respectively, whereas an FGF21 variant that contains SIR2 in FGF19 (FGF2119site2) exhibits a binding affinity of 165 nM, similar to FGF19 binding. The BLI sensorgrams presented in Fig. 3B show that the differences in overall binding affinities of these proteins are primarily derived from their dissociation rates, koff to sKLB, of 6.1 × 10−2, 1.4 × 10−2, and 4.6 × 10−2 s−1 for FGF19, FGF1921site2, and FGF2119site2, respectively. The lower binding affinity of FGF2119site2 compared with the binding affinity of FGF1921site2 to sKLB is probably caused by different amino acids flanking the conserved R-S-P-S motifs in the two endocrine FGFs. This might also explain the lack of electron densities in SIR2 of the sKLB:Nb30:FGF19CT structure (Fig. 1C).
We also examined SIR1 for structural features that might explain the differences in the binding affinities to sKLB. Since the binding of SIR1 depends greatly on intramolecular interactions, we introduced a single amino acid substitution that might stabilize the multiturn element in SIR1 of FGF19. Indeed, an E193D mutant of FGF19 exhibits an approximately three- to fourfold slower koff compared with the koff of WT FGF19 (Fig. 3 A and B and SI Appendix, Table S2). It is noteworthy that carboxyl groups of either D187 in FGF21 or E193 in FGF19 do not make any contact with sKLB (Fig. 2 B and C). E193D substitution in FGF19 would most likely affect an intramolecular interaction, possibly creating hydrogen bonds between backbone nitrogen and the side-chain carboxyl group, resulting in formation of a more rigid β-turn, potentially facilitating an entropically favorable binding event. Thus, for both FGF19 and FGF21, SIR1 and SIR2 act in a cooperative manner to stabilize FGF19 and FGF21 binding to β-Klotho.
We have previously described an FGF21 variant with improved binding affinity to sKLB (19), FGF21WF, in which two structure-guided amino acid substitutions were introduced in the CT region. When SIR2 of FGF19 was replaced by the corresponding region of FGF21WF, the resulting FGF1921site2 mutant (FGF1921site2W) exhibited a 10-fold slower koff to sKLB binding compared with WT FGF19 binding, resulting in a dissociation constant similar to WT FGF21 (Fig. 3B and SI Appendix, Table S2). However, because of the cooperative contribution from the SIR1 and SIR2 regions to overall binding affinities, replacing the whole CT region in FGF19 with FGF21WF (FGF1921CTWF) reduced koff even further, rendering its KD to sKLB more than 30-fold lower compared with WT FGF19 binding (Fig. 3B and SI Appendix, Table S2).
We next compared the cellular activities of WT FGF19, FGF21, and several FGF19 chimeric molecules to stimulate MAPK response in L6 cells that express β-Klotho together with either FGFR1c or FGFR4. The cells were stimulated with increasing concentrations of FGF19, FGF21, FGF1921CT, FGF1921site2, FGF1921CTWF, or FGF1 as a control (Fig. 3 C and D). Lysates of unstimulated or ligand-stimulated cells (matched for expression levels of β-Klotho, FGFR1c, or FGFR4) were subjected to immunoblotting with anti-pMAPK antibodies or anti-MAPK antibodies to monitor MAPK stimulation and MAPK expression, respectively. The results presented in Fig. 3C show only modest increases in MAPK activation in cells coexpressing β-Klotho and FGFR1c in response to stimulation with the chimeric FGF19 ligands, which exhibit approximately up to 30-fold enhanced binding affinity toward β-Klotho. It is noteworthy that FGF19 binds with an ∼10-fold reduced binding affinity from FGF21 to β-Klotho, yet similar MAPK activation was measured for cells expressing FGFR1c stimulated with FGF19 or FGF21. In addition, despite exhibiting a 10-fold reduced binding affinity (compared with FGF21) to β-Klotho, FGF19 stimulated a robust MAPK response while FGF21 stimulated a negligible MAPK response in L6 cells coexpressing β-Klotho and FGFR4 (Fig. 3D). Moreover, approximately fivefold enhanced MAPK activation (compared with WT FGF19) was measured in cells stimulated by the chimeric FGF1921site2 or FGF1921CTWF ligands, which exhibit up to a 30-fold enhanced binding affinity to β-Klotho (Fig. 3 C and D). While it is well established that FGF receptor and Klotho protein expressions are indispensable for FGFR activation and cellular responses stimulated by endocrine FGFs, detailed molecular mechanisms underlying cell selectivity and signal robustness induced by endocrine FGFs remain to be elucidated.
Structural Insights Reveal a Conserved Mode of Endocrine FGF Recognition by Klotho Family Proteins.
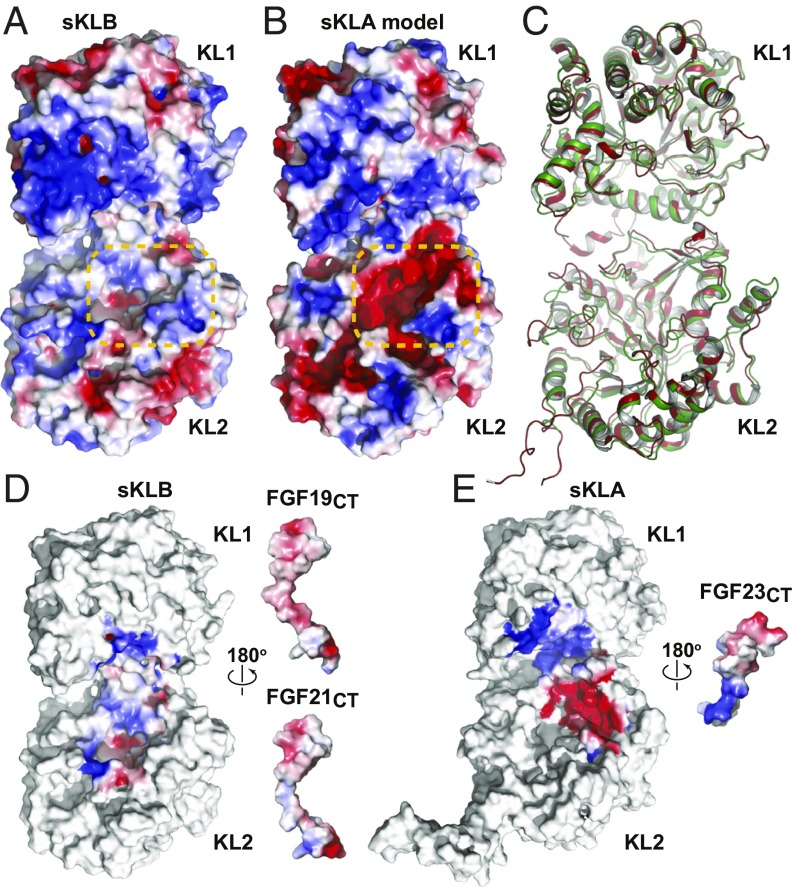
Sequence alignment of sKLB and the extracellular domain of α-Klotho (sKLA) shows a high similarity (SI Appendix, Fig. S2B, 48.9% identity), suggesting that a homology model of sKLA structure can be built based on the crystal structure of sKLB with reasonable accuracy. To visualize potential interactions between FGF23 and α-Klotho, we used the crystal structure of sKLB as a modeling template to generate a homology model of sKLA (Fig. 4 A and B). Comparison of the residues in sKLB that interact with FGF21CT or FGF19CT with the corresponding residues in sKLA reveals two important features: (i) residues in site 1 that are critical for hydrophobic interactions with the ligand are well conserved in β-Klotho and α-Klotho, and (ii) phenylalanine residues (F826 and F931) in site 2 of sKLB that are critical for maintaining hydrophobic interaction with the sugar-mimicking motif, S-P-S, are substituted by tyrosine residues in α-Klotho (Y809 and Y915). These two features of the sKLA model are consistent with the amino acid sequences of the CT regions of ligands: the multiturn element in FGF21CT or FGF19CT may be supported by a hydrophobic surface created by F377, W417, and F418 in sKLA; substitution of phenylalanine residues in sKLB (F826 and F931 that supports S-P-S motif in FGF21CT or FGF19CT) by tyrosine residues in sKLA (Y809 and Y915) may create a surface with negative electrostatic potential in site 2, which might prevent hydroxyl groups in the S-P-S motif from approaching the polar pocket in sKLA.
Fig. 4.
Homology model of sKLA built based on the crystal structure of the sKLB:FGF21CT complex. Electrostatic potentials of (A) sKLB (PDB ID: 5VAQ, chain A) and (B) homology model of sKLA are color-coded on the surface representation. Site 2 areas that exhibit a major difference in electrostatic potentials are highlighted with orange dashed lines. (C) Comparison of crystal structures of sKLB and sKLA. Coordinates of the sKLB molecule from the sKLB:FGF19CT:Nb30 structure (this work) are overlaid with the coordinates of the sKLA molecule from the sKLA:FGF23:FGFR1cD2D3 structure (PDB ID: 5W21) with an overall Cα rmsd of 1.17 Å. Only sKLB (green) and sKLA (red) are shown for clarity. (D and E) Electrostatic complementarity of ligand–Klotho interactions in site 2 may determine ligand specificities. Open-book representations of (D) sKLB:FGF21CT (PDB ID: 5VAQ) and sKLB:FGF19CT (this work) and (E) sKLA:FGF23CT (PDB ID: 5W21) showing surface electrostatic potentials of regions in which ligand–Klotho interactions occur. Note that coordinates of missing FGF19 residues (G205-A208) in the sKLB:FGF19CT structure were built using the crystal structure of sKLB:FGF21CT (PDB ID: 5VAQ) to examine electrostatic complementarity.
The CT region of FGF23 contains 72 amino acids compared with the shorter CT regions of FGF21 and FGF19, which contain 38 and 39 amino acids, respectively (SI Appendix, Fig. S1C). It was previously reported that residues S180–S205 of the C-terminal region of FGF23 are critical for mediating interaction between FGF23 with α-Klotho (23). As shown in SI Appendix, Fig. S1C, the sequence alignment of the CT regions of FGF21, FGF19, and FGF23 shows that FGF23CT exhibits weak sequence similarities to FGF21CT or FGF19CT; however, residues critical for maintaining the multiturn element in SIR1, D-P, are conserved in FGF23, suggesting that FGF23 may also contain a multiturn element in FGF23CT that binds to site 1 of α-Klotho. By contrast, the sugar-mimicking motif, S-P-S in FGF21 or FGF19, is absent in FGF23, suggesting that different interactions may take place between amino acid residues of FGF23CT that bind to site 2 of α-Klotho from those seen in the crystal structure of FGF21CT or FGF19CT bound to site 2 of β-Klotho. As such, we surmise that amino acids residues that form site 1 may function as a common hydrophobic surface for ligand binding, while amino acids located in site 2 may represent a pocket playing a critical role in determining ligand-binding selectivity. It is noteworthy that the structural model of sKLA described in this report was prepared before publication of the crystal structure of sKLA:FGF23:FGFR1cD2D3 (25). The homology model of sKLA and the sKLA:FGF23:FGFR1cD2D3 crystal structure [Protein Data Bank (PDB) ID code 5W21] overlaid well (overall Cα rmsd of 0.73 and 0.78 Å when overlaid with respect to KL1 and KL2, respectively) and displayed a similar surface electrostatic potential distribution (SI Appendix, Fig. S6), validating the quality of the homology model and increasing our confidence in our analysis. The interactions between FGF23CT and sKLA in the crystal structure are restricted to site 1 and some part of site 2; however, only a minimal region of FGF23CT (which is only half of the C-terminal region; SI Appendix, Fig. S1C) has been included in the sKLA:FGF23:FGFR1 crystal structure, raising a question of whether other regions of sKLA could be occupied by the rest of FGF23CT.
Analysis on the surface electrostatic potential of sKLA and sKLB (Fig. 4 A and B) suggests that differences in the electrostatic potential of site 2 are important for determining ligand-binding specificity. Remarkably, the CT regions of FGF19, FGF21, and FGF23 have electrostatic potential complementary to that of their corresponding receptor (Fig. 4 D and E). FGF23CT has a strong positive potential centered on R196 and R198 that matches the strong negative electrostatic potential of the site 2 surface on sKLA (Fig. 4E), while both FGF19CT and FGF21CT have a weak negative electrostatic potential preceding the S-P-S motif that complements the weak positive electrostatic potential of the site 2 surface on sKLB (Fig. 4D). FGF19CT exhibits slightly stronger negative electrostatic potential than FGF21CT due to the difference in the residue preceding the S-P-S motif (i.e., E207 in FGF19 and S200 in FGF21). These results agree with the specificity of the FGFs to the corresponding Klothos, where both FGF19 and FGF21 bind specifically to β-Klotho, while their subtle differences in electrostatics in SIR2 and rigidities in SIR1 may explain the 10-fold difference in their binding affinity to β-Klotho (SI Appendix, Table S2).
Conclusion.
Experiments presented in this paper demonstrate that FGF19CT and FGF21CT bind to the same binding site on sKLB. Both FGF21CT and FGF19CT bind to site 1 primarily via a multiturn D-P motif, which is conserved in complex formation between FGF23CT and sKLA, and to site 2 via an S-P-S motif that binds to the pseudoglycoside hydrolase region in KL2 that is not conserved in the interaction between FGF23CT and sKLA. While the overall structures of sKLB and sKLA are highly similar, site 2 of the two Klotho proteins exhibits a different electrostatic potential that is complementary to those of their corresponding ligands. The structural analyses of sKLB in complex with either FGF21CT and FGF19CT provide a clear and consistent picture about the molecular interactions that govern the binding and specificities of FGF21CT and FGF19CT to sKLB. In contrast, the molecular interactions that govern the binding and specificity of FGF23CT to sKLA are not fully understood; only site 1 was shown to be occupied in the crystal structure of FGF23CT in complex with sKLA. Moreover, although FGF23CT is nearly twice as long as FGF21CT and FGF19CT, a truncated form of FGF23CT was used in the structural analysis of the sKLA:FGF23:FGFR1 complex, raising the possibility that additional interactions may still exist between sKLA and FGF23CT.
Our results show that, while FGF19CT binds to sKLB with a nearly 10-fold lower binding affinity, a robust MAP kinase response was measured for cells coexpressing FGFR4 with β-Klotho in response to FGF19 stimulation, and a weak MAP kinase response was detected when the same cells were stimulated with the more potent engineered FGF21 mutant. Moreover, a similar MAP kinase response was measured for cells coexpressing FGFR1c with β-Klotho in response to FGF19 or FGF21 stimulation. Similar profiles of MAP kinase stimulation were detected in cells coexpressing FGFR4 with β-Klotho or in cells coexpressing FGFR1c with β-Klotho when stimulated with more potent engineered FGF19 or FGF21 variants. The scheme presented in SI Appendix, Fig. S7, shows that the dissociation constants of FGF19 and FGF21 to sKLB are ∼220 and 24 nM, respectively, and dissociation constants of FGFR1c and FGFR4 to sKLB are ∼4 and 2 µM, respectively. It is noteworthy that the binding affinities of FGF19 and FGF21 to FGFR4 or FGFR1c are too low to be reliably determined. However, FGF19 induces a robust mitogenic response and FGF21 induces a negligible mitogenic response when stimulating the same cells. While Klotho proteins serve as primary high-affinity binding sites to endocrine FGFs, these experiments demonstrate that the cellular activities of FGF19 and FGF21 depend not only on their binding affinities to Klotho proteins but also on additional interactions between endocrine FGFs with FGFRs, interactions between FGFRs with Klotho proteins, as well as the tyrosine kinase activities of activated FGFRs.
Materials and Methods
sKLB, expressed and purified as previously described (19), was mixed with Nb30, concentrated, and injected into an Superdex 200 Increase 10/300 GL (GE Healthcare) column pre-equilibrated with 20 mM Hepes and 150 mM NaCl, pH 7.5. Fractions containing the complex were pooled, concentrated to 6 mg/mL, mixed with 3× molar excess of FGF19CT peptide (amino acids 167–216) and screened for crystallization using Mosquito Crystal liquid handler (TTP Labtech). The 96-well plates were incubated and imaged at 20 °C using Rock Imager 1000 (Formulatrix). The sKLB:Nb30:FGF19CT complex produced plate-shaped crystals when mixed with an equal volume of well solution containing 21% PEG1500, 0.1 M ammonium sulfate, 0.2 M NDSB-201, 0.1 M Tris⋅HCl, pH 7.5, and equilibrated for 8–10 d using the hanging drop vapor diffusion method. The crystals were cryopreserved by gradually transferring crystals to the mother liquor containing 35% PEG1500 before being flash-frozen in liquid nitrogen. The diffraction dataset was collected at beamline 24-ID-E (Advanced Photon Source) and processed using XDS (26). The initial phase was calculated by molecular replacement with PHASER (27) using the coordinates of sKLB:Nb914 (PDB ID: 5VAQ). Iterative rounds of refinement were performed using PHENIX (28) with manual inspection using COOT (29). All of the figures containing the structures were generated using the PyMOL Molecular Graphics System, Version 1.8 (Schrödinger).
Full details about procedures for cloning, expression and purification of recombinant proteins used in the study, biophysical measurements, cellular activities, and homology modeling are provided in SI Appendix.
Supplementary Material
Acknowledgments
This work is based upon research utilizing NE-CAT beamlines (Grant GM124165), a Pilatus detector (Grant RR029205), and an Eiger detector (Grant OD021527) at the Advanced Photon Source (Grant DE-AC02-06CH11357). We thank Instruct-ERIC, part of the European Strategy Forum on Research Infrastructures (ESFRI), and the Fund for Scientific Research Flanders for their support to the nanobody discovery.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6NFJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822055116/-/DCSupplemental.
References
- 1.Kir S, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kir S, Kliewer SA, Mangelsdorf DJ. Roles of FGF19 in liver metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:139–144. doi: 10.1101/sqb.2011.76.010710. [DOI] [PubMed] [Google Scholar]
- 3.Owen BM, Mangelsdorf DJ, Kliewer SA. Tissue-specific actions of the metabolic hormones FGF15/19 and FGF21. Trends Endocrinol Metab. 2015;26:22–29. doi: 10.1016/j.tem.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher FM, Maratos-Flier E. Understanding the physiology of FGF21. Annu Rev Physiol. 2016;78:223–241. doi: 10.1146/annurev-physiol-021115-105339. [DOI] [PubMed] [Google Scholar]
- 5.Kuro-O M, Moe OW. FGF23-αKlotho as a paradigm for a kidney-bone network. Bone. 2017;100:4–18. doi: 10.1016/j.bone.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Razzaque MS. The FGF23-Klotho axis: Endocrine regulation of phosphate homeostasis. Nat Rev Endocrinol. 2009;5:611–619. doi: 10.1038/nrendo.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spivak-Kroizman T, et al. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell. 1994;79:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 8.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb Perspect Biol. 2013;5:a015958. doi: 10.1101/cshperspect.a015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urakawa I, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 11.Kurosu H, et al. Regulation of fibroblast growth factor-23 signaling by Klotho. J Biol Chem. 2006;281:6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogawa Y, et al. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proc Natl Acad Sci USA. 2007;104:7432–7437. doi: 10.1073/pnas.0701600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of FGF19 require klotho beta. J Biol Chem. 2007;282:27277–27284. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- 14.Kharitonenkov A, et al. FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. J Cell Physiol. 2008;215:1–7. doi: 10.1002/jcp.21357. [DOI] [PubMed] [Google Scholar]
- 15.Adams AC, et al. Fundamentals of FGF19 & FGF21 action in vitro and in vivo. PLoS One. 2012;7:e38438. doi: 10.1371/journal.pone.0038438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuro-o M. Klotho and βKlotho. Adv Exp Med Biol. 2012;728:25–40. doi: 10.1007/978-1-4614-0887-1_2. [DOI] [PubMed] [Google Scholar]
- 17.D.E. K Stereochemistry and the mechanism of enzymatic reactions. Biol Rev Camb Philos Soc. 1953;28:416–436. [Google Scholar]
- 18.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, et al. Structures of β-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature. 2018;553:501–505. doi: 10.1038/nature25010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goetz R, et al. Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands. Mol Cell Biol. 2012;32:1944–1954. doi: 10.1128/MCB.06603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee RA, Razaz M, Hayward S. The DynDom database of protein domain motions. Bioinformatics. 2003;19:1290–1291. doi: 10.1093/bioinformatics/btg137. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, et al. C-terminal tail of FGF19 determines its specificity toward klotho co-receptors. J Biol Chem. 2008;283:33304–33309. doi: 10.1074/jbc.M803319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetz R, et al. Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23-FGFR-klotho complex formation. Proc Natl Acad Sci USA. 2010;107:407–412. doi: 10.1073/pnas.0902006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Micanovic R, et al. Different roles of N- and C- termini in the functional activity of FGF21. J Cell Physiol. 2009;219:227–234. doi: 10.1002/jcp.21675. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, et al. α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature. 2018;553:461–466. doi: 10.1038/nature25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams PD, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.