Significance
Diseases that infect domestic and wild species cause severe human health burdens and exacerbate declines of endangered species. However, we currently lack theory for predicting the mortality of multihost parasites in different hosts: Some of these diseases rarely harm their hosts, while others are nearly always fatal. Using global case-fatality data for multiple diseases of domestic mammals, we show that the evolutionary relationship among infected and susceptible hosts is a strong predictor of disease-induced mortality. We find that parasites infecting domestic species that are more evolutionarily distant from their other known hosts have a higher probability of resulting in lethal infections.
Keywords: host specificity, domestication, virulence, phylogeny, infectious disease
Abstract
Infectious diseases of domesticated animals impact human well-being via food insecurity, loss of livelihoods, and human infections. While much research has focused on parasites that infect single host species, most parasites of domesticated mammals infect multiple species. The impact of multihost parasites varies across hosts; some rarely result in death, whereas others are nearly always fatal. Despite their high ecological and societal costs, we currently lack theory for predicting the lethality of multihost parasites. Here, using a global dataset of >4,000 case-fatality rates for 65 infectious diseases (caused by microparasites and macroparasites) and 12 domesticated host species, we show that the average evolutionary distance from an infected host to other mammal host species is a strong predictor of disease-induced mortality. We find that as parasites infect species outside of their documented phylogenetic host range, they are more likely to result in lethal infections, with the odds of death doubling for each additional 10 million years of evolutionary distance. Our results for domesticated animal diseases reveal patterns in the evolution of highly lethal parasites that are difficult to observe in the wild and further suggest that the severity of infectious diseases may be predicted from evolutionary relationships among hosts.
Infectious diseases that cross species barriers are responsible for severe human health burdens (1) and act as direct and synergistic drivers of species extinctions (2). Many of these diseases infect domesticated animals and impact human well-being via loss of food security, labor and livelihoods, costs of prevention and control programs, and increased human infection (3). However, the severity of disease can vary dramatically among parasites. Canine rabies alone results in 59,000 human deaths and 8.6 billion US dollars in economic losses annually (4). By contrast, other diseases rarely result in death. For example, bovine brucellosis largely impacts cattle by causing abortion, infertility, and reduced growth, but disease-induced mortality in adult cows is uncommon (5).
Well-established theory on single-host parasites predicts that the reduction in host fitness due to infection (termed “virulence”) should evolve to an optimal level determined by a trade-off with transmission (6). For multihost parasites, optimal virulence may be subject to additional trade-offs, with selection for high or low virulence depending on the ecologies and evolutionary histories of each susceptible host species (7–9). In the absence of trade-offs, a wider host breadth should provide a larger pool of susceptible individuals, increasing opportunities for transmission and the evolution of higher virulence (10). However, adaptation to novel hosts may reduce a parasite’s ability to use resources of their coevolved hosts (11, 12), resulting in limited replication and decreased virulence (13). This trade-off is supported by comparative studies of plant RNA viruses and avian malaria parasites, in which specialist parasites tend to be more virulent than generalists (14, 15). Yet generalist parasites remain highly virulent in some host species (16).
Our ability to predict the outcome of a given host–parasite interaction is currently limited because the full suite of traits underlying virulence is either poorly estimated or unknown for the vast majority of host–parasite interactions. However, our understanding of evolutionary relationships is often much better, and host phylogeny can be used as a proxy for latent traits and evolutionary histories that have shaped contemporary host–parasite associations (17). For example, closely related hosts suffer similar impacts for some parasites of Drosophila (18, 19), consistent with the prediction that parasite virulence should covary with host phylogeny. However, there have been few studies that have developed and tested theories of how host evolutionary relationships influence disease outcomes across multiple host–parasite combinations.
As parasites adapt to infect novel host species increasingly distant from their coevolved hosts, they are expected to experience increased fitness costs (13), leading to the prediction of lowered virulence following greater phylogenetic jumps. This pattern, termed “nonhost resistance” (13), may act in opposition to resistance evolved by hosts in response to infection, which is expected to decrease with evolutionary distance from a parasite’s coevolved hosts and lead to phylogenetically distant hosts experiencing more intense disease (13). The relative strengths of these opposing relationships will likely influence the virulence of a given host–parasite interaction.
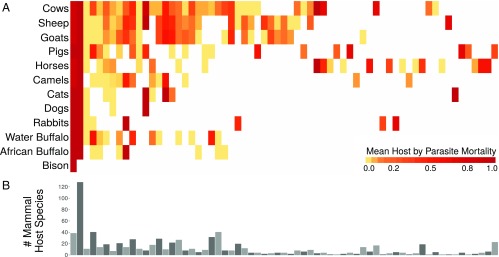
Infectious diseases of domestic species, many of which have severe economic impacts (3), present a unique opportunity to explore the links between virulence, host specificity, and the evolutionary relationships among hosts. While virulence can take many forms, mortality is most widely reported. The World Organisation for Animal Health (OIE) publishes yearly reports documenting the numbers of cases and deaths caused by diseases of importance for international trade (20), providing a remarkable dataset of disease-induced mortality for multiple parasites across different host species. We examined data from 4,157 reports (in which no host culling was recorded) from 155 countries across 7 years, representing 202 unique host–parasite combinations with large variation in average mortality (Fig. 1A).
Fig. 1.
(A) Heatmap of mean host by parasite mortality derived from the OIE World Animal Health yearly reports from 2005 to 2011 (data from refs. 26–32; full heatmap with disease common names is included in SI Appendix, Fig. S1). (B) Bar plot of the number of documented mammal host species per parasite derived from the Global Mammal Parasite Database 2.0 (21) and the Enhanced Infectious Disease Database (22). The order of parasites matches the column order in A.
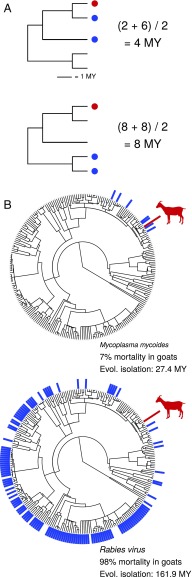
For each parasite, we identified the set of documented mammal host species from two recently published global host–parasite databases (21, 22), returning 788 unique host–parasite interactions (Fig. 1B). For each host–parasite combination, we then calculated the mean phylogenetic distance from all documented host species to the infected species, which we refer to as “host evolutionary isolation” (Fig. 2). This metric is analogous to measures of mean phylogenetic relatedness to a focal species, which have been used to analyze species invasions (23) and predict disease pressure in plant communities (24). Since parasites typically infect closely related species (13, 17), we assume that the phylogenetic centroid of susceptible species indicated the most likely position of hosts to which a parasite is best adapted, and that the distance from a host to this centroid may provide a reasonable proxy for the relative extent of coadaptation between parasite and host. However, for the vast majority of these diseases, there is limited knowledge of reservoir species or ancestral hosts. We modeled the probability of death as a function of host evolutionary isolation and number of documented host species (host species richness) using a hierarchical Bayesian approach (25) that allowed us to adjust for additional factors, including the number of cases per report and the effects of parasite, host, country, and year.
Fig. 2.
(A) Example of how host evolutionary isolation is calculated. Red circles indicate the infected host; blue circles indicate documented hosts. Host evolutionary isolation is calculated as the mean phylogenetic distance from the infected host to all documented host species. (B) Examples with Mycoplasma mycoides and rabies virus. Documented hosts are indicated by blue bars on the host phylogeny, with host evolutionary isolation (Evol. isolation) and average mortality calculated for goats (Capra hircus; shown in red).
Results
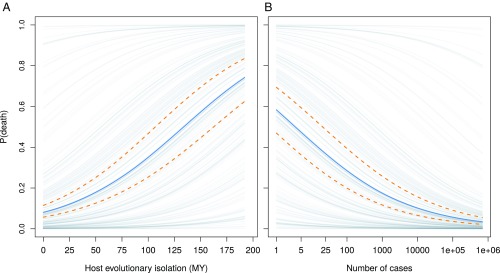
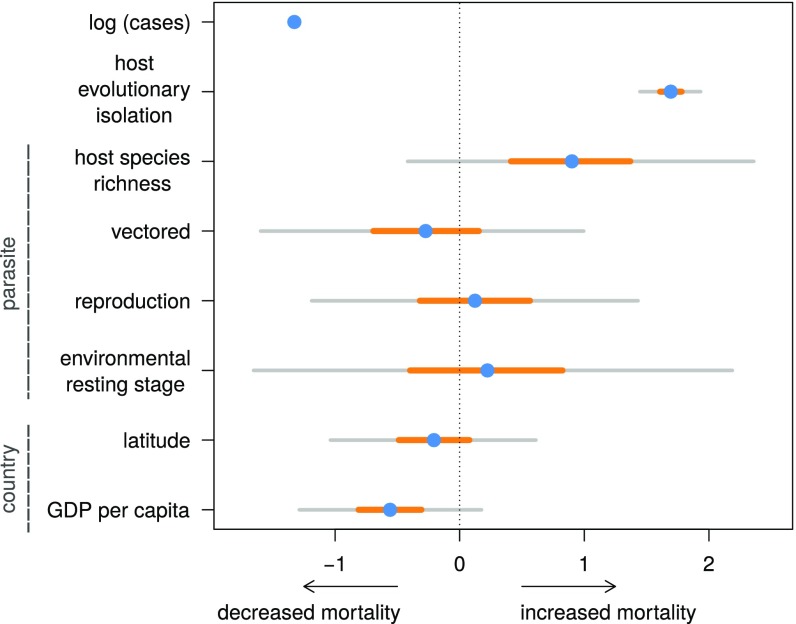
We found that disease-induced mortality was highest when infected hosts were evolutionarily distant from other documented hosts (Figs. 3A and 4 and SI Appendix, Table S3), with an increase of 10 million years of evolutionary isolation resulting in a doubling in the odds of host death (odds ratio 50% credible interval: 1.99–2.15). This predicts that a parasite infecting an Artiodactyl, which otherwise infects only Primate hosts, would have 4.8 times higher odds of host death than a parasite that otherwise infects hosts in the order Carnivora. This effect size is comparable in magnitude only to the number of cases and much greater than the effect of all ecological and socioeconomic predictors in our model. The effect of host evolutionary isolation became stronger when single-host parasites were excluded (SI Appendix, section 2.2.1), indicating that the results are not driven simply by differences between single-host and multihost parasites.
Fig. 3.
Posterior predictions of the probability of death as a function of host evolutionary isolation (in millions of years) (A) and the number of cases (B). Solid blue lines represent the mean logistic curve; dashed yellow lines represent the upper and lower bounds of the 50% credible interval. Gray lines depict equivalent mean curves offset by the posterior mean effects for each country.
Fig. 4.
Estimated regression coefficients for continuous predictors. Blue circles represent posterior means; yellow horizontal lines represent 50% credible intervals; gray horizontal lines represent 95% credible intervals. Predictors at the parasite and country level are indicated.
We found some support for a positive relationship between mortality and host species richness (50% credible interval does not overlap zero), opposite to what would be predicted if there was a trade-off between parasite generalism and virulence (parasites with larger host richness causing lower mortality). However, there was large variability in the strength of this relationship, as was the case for all parasite-level predictors. Reports with high host mortality were associated with fewer infected individuals (Figs. 3B and 4), consistent with the upper extreme of a virulence–transmission trade-off. High mortality may naturally limit transmission; however, human interventions to limit spread may also be strongest for deadlier outbreaks in domesticated animals.
Our model also revealed large variation in mortality among countries (Fig. 3 and SI Appendix, Fig. S2), indicating that effective disease-management practices from one nation could be identified and introduced to other nations. Countries with large positive effects—higher mortality than otherwise predicted—may have lower capacities for detection and prevention of outbreaks. For example, top-ranked Sri Lanka and Kyrgyzstan have struggled to develop legislation and infrastructure for addressing veterinary public health issues (33) and have deteriorated veterinary and sanitation systems (34). In contrast, nations with large negative country effects (e.g., the former Yugoslav Republic of Macedonia, China, and Iran) suffer considerable infectious-disease burdens, but have made great improvements in surveillance, control, and eradication programs (35–37). In addition, we found support for a negative relationship between mortality and gross domestic product (GDP) per capita (Fig. 4), indicating that wealthier countries may allocate greater resources toward animal-health and disease-control efforts.
Discussion
We found that as parasites infect domesticated species outside of their typical evolutionary host range, they have a higher probability of resulting in lethal infections. However, high mortality was also associated with fewer infected individuals. Our findings suggest that disease spillover into evolutionary isolated hosts is marked by increased virulence, but potentially at the cost of decreased transmission. The high mortality observed in our data likely occurs through multiple pathways, including the maladaptation of both host and parasite, and the decoupling of transmission from virulence. While it is difficult to isolate the different mechanisms leading to high mortality, we suggest that the evolutionary distances among infected and susceptible hosts can, to some extent, capture these multiple dimensions.
For some host–parasite combinations, elevated mortality may be explained by a decoupling of virulence from transmission. Consistent with this hypothesis, many vertebrate arboviruses commonly use birds as reservoir hosts, but fail to transmit after spillover into mammal hosts, such as humans and horses, where they are regularly fatal (38). To investigate this, we ran an additional model identifying parasites associated with avian reservoirs, but found no strong evidence that these parasites cause higher mortality (SI Appendix, section 2.2.3 and Table S6). It is possible that the positive relationship between virulence and evolutionary isolation breaks down at these larger phylogenetic distances. For example, nonhost resistance may be more common following large phylogenetic jumps (39). Expanding our framework to include nonmammal hosts may provide additional insight into trade-offs faced by parasites exhibiting extreme phylogenetic generalism.
In evolutionarily isolated hosts, mortality may result from a combination of direct damage caused by parasites and damage caused by the host’s immune response to infection, which may impose different selective pressures on the evolution of virulence (40). Hosts that contribute little to transmission provide one pathway by which transmission can become decoupled from virulence, resulting in parasites experiencing little or no selection to reduce hypervirulence (7, 13). This can occur when the majority of transmission is facilitated by a reservoir host, as has been suggested for foot and mouth disease in southern Africa, which uses asymptomatic African buffalo as a reservoir, but causes severe outbreaks after spillover in domestic cattle (41). In the example of rinderpest in Africa, cattle facilitated the sustained transmission of the virus, which caused widespread mortality following spillover into wild ungulates (42). Many of the domesticated-animal diseases we analyzed here may represent spillover of infections from wildlife reservoirs; however, identifying reservoir species can be challenging, and, for many parasites, the reservoir species are unknown.
Virulence may also become decoupled from transmission if parasites infect tissues unrelated to transmission, such as bacterial meningitis infection of the CNS (43) or when parasite stages can persist for long periods of time in the environment (6). While there was no clear relationship between transmission mode and host mortality in our model (Fig. 4), parasite identity had an important effect (SI Appendix, Table S3 and Figs. S2 and S3), suggesting that other parasite traits modify virulence.
The animal diseases for which we have multiple case-fatality estimates are weighted toward those that have large impacts on international trade. Our analysis is also focused on host–parasite interactions that have measurable effects (i.e., we do not consider, for example, cases where a pathogen failed to infect a host or where infections were largely asymptomatic). Gathering additional data on asymptomatic infections would be challenging, but would provide further insights into limits of parasite host range. While the OIE-listed diseases may more often display high mortality, we suggest that they provide a window into the evolution of virulence that would otherwise be hard to observe. In natural systems, spillovers of highly virulent diseases often display stuttering chains of transmission before burning out (12), and thus instances of deadly disease in wildlife may frequently go undocumented (16). High host densities allow parasites to maintain transmission, despite causing high mortality (44), and artificially high densities of domesticated animals may facilitate the maintenance of more deadly diseases, allowing us to better observe their behavior.
Predicting the outcomes of novel host–parasite interactions presents a major challenge in disease ecology. There is a pressing need to address this challenge, given rapid rates of ecosystem transformation, which can generate communities never before seen in evolutionary history and promote disease emergence in novel hosts. Proactive approaches to document wildlife hosts (45) may help predict mortality of emerging diseases, and disease burdens may be reduced by implementing effective disease-management practices. As a step toward this, we have shown host evolutionary isolation to be a strong predictor of infection-induced mortality in domesticated mammals and quantified the potential for country-level initiatives to reduce animal death.
Materials and Methods
With a global database of infection-induced mortality rates, we used a Bayesian hierarchical modeling framework to examine the relationship between host specificity and mortality for diseases of domesticated mammals. To separate the importance of our two aspects of host specificity (host evolutionary isolation and host species richness) from other factors that might also influence host mortality, we included copredictors and hierarchical terms in our model. At the parasite level, these included traits for major modes of transmission, plus hierarchical effects of parasite type to account for parasite traits not measured directly. We also included hierarchical effects for host, host taxonomic order, country, and year of reporting. Environmental conditions, which include socio-economic factors such as the ability of local peoples to maintain animal health, effects of ambient temperature on parasite growth rate, or coinfection with other parasites, may also influence host mortality. To adjust for these additional country-level effects, we included per capita GDP and latitude per country, in addition to modeling variation among countries. The virulence-transmission trade-off suggests that outbreaks resulting in large numbers of infected individuals are unlikely to be associated with high mortality, as premature host death restricts transmission rate, ultimately resulting in lower case numbers for more lethal diseases (46). We therefore also included the number of cases per report as an offset variable. We estimated the effect sizes of these predictors on host mortality with a Bayesian hierarchical binomial-logit model. For additional information on materials and methods, see SI Appendix, section 1.
To assess the sensitivity of our model to exclusion of subsets of the data, and inclusion of additional or substitute predictors, we constructed five alternative models described in SI Appendix, section 2.2. These comprise models that exclude single-host parasites, exchange host species richness for host taxonomic diversity, include whether parasites are known to have avian reservoirs, exchange parasite type for parasite taxonomic family, and include the number of citation counts per parasite as a measure of study effort. We show that, for each of our alternative models, the effect sizes of our main model predictors remain qualitatively unchanged.
Data and Code
All data and code necessary to reproduce the results can be found at doi.org/10.6084/m9.figshare.7497137 (47).
This archive includes underlying data and R scripts used to download and process additional covariate data, Stan model code for the main model and four alternative models, and R scripts used to run models, generate simulated data to validate the main model, and generate model plots and tables.
Supplementary Material
Acknowledgments
We thank Vanessa Ezenwa, Charlie Nunn, Elizabeth Wolkovich, Ria Ghai, Jan Gogarten, and Carl Boodman for helpful feedback on the manuscript; Elizabeth Wolkovich, Margaret Kosmala, the Harvard Stanleyi group, Will Pearse, and Bob Carpenter for feedback on the model; and Debarun Gupta and Madeleine McGreer for help with data entry. M.J.F. was supported by a National Sciences and Engineering Research Council of Canada Vanier Canada Graduate Scholarship, the Canadian Institutes of Health Research Systems Biology Training Program, the Quebec Centre for Biodiversity Science, and the McGill Biology Department.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data and code necessary to reproduce our analyses have been deposited in Figshare (doi:10.6084/m9.figshare.7497137).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817323116/-/DCSupplemental.
References
- 1.Hotez PJ, et al. The global burden of disease study 2010: Interpretation and implications for the neglected tropical diseases. PLOS Negl Trop Dis. 2014;8:1–9. doi: 10.1371/journal.pntd.0002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heard MJ, et al. The threat of disease increases as species move toward extinction. Conservation Biol. 2013;27:1378–1388. doi: 10.1111/cobi.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dehove A, Commault J, Petitclerc M, Teissier M, Macé J. Economic analysis and costing of animal health: A literature review of methods and importance. Revue Sci Tech (International Off Epizootics) 2012;31:605–617. doi: 10.20506/rst.31.2.2146. [DOI] [PubMed] [Google Scholar]
- 4.Hampson K, et al. 2015. Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis 9:e0003709, and erratum (2015) 9:0003786.
- 5.McDermott JJ, Grace D, Zinsstag J. Economics of brucellosis impact and control in low-income countries. Sci Tech Rev Off Int des Epizooties (Paris) 2013;32:249–261. doi: 10.20506/rst.32.1.2197. [DOI] [PubMed] [Google Scholar]
- 6.Cressler CE, McLeod DV, Rozins C, Van den Hoogen J, Day T. The adaptive evolution of virulence: A review of theoretical predictions and empirical tests. Parasitology. 2016;143:915–930. doi: 10.1017/S003118201500092X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 8.Gandon S. Evolution of multihost parasites. Evolution. 2004;58:455–469. [PubMed] [Google Scholar]
- 9.Rigaud T, Perrot-Minnot M-J, Brown MJF. Parasite and host assemblages: Embracing the reality will improve our knowledge of parasite transmission and virulence. Proc R Soc Biol Sci. 2010;277:3693–3702. doi: 10.1098/rspb.2010.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett LG, Kniskern JM, Bodenhausen N, Zhang W, Bergelson J. Continua of specificity and virulence in plant host-pathogen interactions: Causes and consequences. New Phytol. 2009;183:513–529. doi: 10.1111/j.1469-8137.2009.02927.x. [DOI] [PubMed] [Google Scholar]
- 11.Ebert D. Experimental evolution of parasites. Science. 1998;282:1432–1436. doi: 10.1126/science.282.5393.1432. [DOI] [PubMed] [Google Scholar]
- 12.Longdon B, Brockhurst M, Russell C, Welch JJ, Jiggins FM. The evolution and genetics of virus host shifts. PLoS Pathog. 2014;10:e1004395. doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonovics J, et al. The origin of specificity by means of natural selection: Evolved and nonhost resistance in host-pathogen interactions. Evolution. 2013;67:1–9. doi: 10.1111/j.1558-5646.2012.01793.x. [DOI] [PubMed] [Google Scholar]
- 14.Zsolt Garamszegi L. The evolution of virulence and host specialization in malaria parasites of primates. Ecol Lett. 2006;9:933–940. doi: 10.1111/j.1461-0248.2006.00936.x. [DOI] [PubMed] [Google Scholar]
- 15.Agudelo-Romero P, Elena SF. The degree of plant resilience to infection correlates with virus virulence and host-range. Spanish J Agric Res. 2008;6:160–169. [Google Scholar]
- 16.Leggett HC, Buckling A, Long GH, Boots M. Generalism and the evolution of parasite virulence. Trends Ecol Evol. 2013;28:592–596. doi: 10.1016/j.tree.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc Biol Sci-R Soc. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longdon B, et al. The causes and consequences of changes in virulence following pathogen host shifts. PLoS Pathog. 2015;11:e1004728. doi: 10.1371/journal.ppat.1004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perlman SJ, Jaenike J. Infection success in novel hosts: An experimental and phylogenetic study of Drosophila-parasitic nematodes. Evolution. 2003;57:544–557. doi: 10.1111/j.0014-3820.2003.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 20.World Organisation for Animal Health (OIE) 2016. Terrestrial animal health code: general provisions (World Organization for Animal Health, Paris), Technical Report.
- 21.Stephens PR, et al. Global mammal parasite database version 2.0. Ecology. 2017;98:42–49. doi: 10.1002/ecy.1799. [DOI] [PubMed] [Google Scholar]
- 22.Wardeh M, Risley C, McIntyre MK, Setzkorn C, Baylis M. Database of host-pathogen and related species interactions, and their global distribution. Sci Data. 2015;2:150049. doi: 10.1038/sdata.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauss SY, Webb CO, Salamin N. Exotic taxa less related to native species are more invasive. Proc Natl Acad Sci USA. 2006;103:5841–5845. doi: 10.1073/pnas.0508073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker IM, et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature. 2015;520:542–544. doi: 10.1038/nature14372. [DOI] [PubMed] [Google Scholar]
- 25.The Stan Development Team 2017 The Stan core library, Version 2.17.0, Available at mc-stan.org.
- 26.OIE . World Animal Health in 2005. World Organisation for Animal Health; Paris: 2007. [Google Scholar]
- 27.OIE . World Animal Health in 2006. World Organisation for Animal Health; Paris: 2008. [Google Scholar]
- 28.OIE . World Animal Health in 2007. World Organisation for Animal Health; Paris: 2008. [Google Scholar]
- 29.OIE . World Animal Health in 2008. Vol 1–2 World Organisation for Animal Health; Paris: 2009. [Google Scholar]
- 30.OIE . World Animal Health in 2009. Vol 1–2 World Organisation for Animal Health; Paris: 2010. [Google Scholar]
- 31.OIE . World Animal Health in 2010. Vol 1–2 World Organisation for Animal Health; Paris: 2011. [Google Scholar]
- 32.OIE . World Animal Health in 2011. Vol 1–2 World Organisation for Animal Health; Paris: 2012. [Google Scholar]
- 33.Dissanayake DMRB, Stephen C, Daniel S, Abeynayake P. Gap assessment of animal health legislation in Sri Lanka for emerging infectious disease preparedness. Outlook Agric. 2012;41:203–208. [Google Scholar]
- 34.Counotte MJ, Minbaeva G, Usubalieva J, Abdykerimov K, Torgerson PR. The burden of zoonoses in Kyrgyzstan: A systematic review. PLoS Negl Trop Dis. 2016;10:e0004831. doi: 10.1371/journal.pntd.0004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stojmanovski Z, Zdravkovska M, Taleski V, Jovevska S, Markovski V. Human brucellosis in the republic of Macedonia by regions depending on vaccination procedures in sheep and goats. Macedonian J Med Sci. 2014;7:135–140. [Google Scholar]
- 36.Hotez PJ, Savioli L, Fenwick A. Neglected tropical diseases of the Middle East and North Africa: Review of their prevalence, distribution, and opportunities for control. PLoS Negl Trop Dis. 2012;6:e1475. doi: 10.1371/journal.pntd.0001475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, et al. Emergence and control of infectious diseases in China. Lancet. 2008;372:1598–1605. doi: 10.1016/S0140-6736(08)61365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver SC, Barrett ADT. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Baarlen P, van Belkum A, Summerbell RC, Crous PW, Thomma BPHJ. Molecular mechanisms of pathogenicity: How do pathogenic microorganisms develop cross-kingdom host jumps? FEMS Microbiol Rev. 2007;31:239–277. doi: 10.1111/j.1574-6976.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- 40.Graham AL, Allen JE, Read AF. Evolutionary causes and consequences of immunopathology. Annu Rev Ecol Evol Syst. 2005;36:373–397. [Google Scholar]
- 41.Michel AL, Bengis RG. The African buffalo: A villain for inter-species spread of infectious diseases in Southern Africa. Onderstepoort J Vet Res. 2012;79:453. doi: 10.4102/ojvr.v79i2.453. [DOI] [PubMed] [Google Scholar]
- 42.Barrett T, Rossiter PB. Rinderpest: The disease and its impact on humans and animals. Adv Virus Res. 1999;53:89–110. doi: 10.1016/s0065-3527(08)60344-9. [DOI] [PubMed] [Google Scholar]
- 43.Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 44.Mennerat A, Nilsen F, Ebert D, Skorping A. Intensive farming: Evolutionary implications for parasites and pathogens. Evol Biol. 2010;37:59–67. doi: 10.1007/s11692-010-9089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrell MJ, Berrang-Ford L, Davies TJ. The study of parasite sharing for surveillance of zoonotic diseases. Environ Res Lett. 2013;8:015036. doi: 10.1088/1748-9326/8/1/015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: History, current state of affairs and the future. J Evol Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 47.Farrell M, Davies TJ. 2019 Data & code: “Disease mortality in domesticated animals is predicted by host evolutionary relationships.” Available at https://figshare.com/articles/Data_Code_Disease_mortality_in_domesticated_animals_is_predicted_by_host_evolutionary_relationships_/7497137. Deposited March 15, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.