Significance
The fundamental biological functions of a living cell are stored within the DNA sequence of its genome. Classical genetic approaches dissect the functioning of biological systems by analyzing individual genes, yet uncovering the essential gene set of an organism has remained very challenging. It is argued that the rewriting of entire genomes through the process of chemical synthesis provides a powerful and complementary research concept to understand how essential functions are programed into genomes.
Keywords: Caulobacter crescentus, chemical genome synthesis, genome rewriting, synonymous recoding, de novo DNA synthesis
Abstract
Understanding how to program biological functions into artificial DNA sequences remains a key challenge in synthetic genomics. Here, we report the chemical synthesis and testing of Caulobacter ethensis-2.0 (C. eth-2.0), a rewritten bacterial genome composed of the most fundamental functions of a bacterial cell. We rebuilt the essential genome of Caulobacter crescentus through the process of chemical synthesis rewriting and studied the genetic information content at the level of its essential genes. Within the 785,701-bp genome, we used sequence rewriting to reduce the number of encoded genetic features from 6,290 to 799. Overall, we introduced 133,313 base substitutions, resulting in the rewriting of 123,562 codons. We tested the biological functionality of the genome design in C. crescentus by transposon mutagenesis. Our analysis revealed that 432 essential genes of C. eth-2.0, corresponding to 81.5% of the design, are equal in functionality to natural genes. These findings suggest that neither changing mRNA structure nor changing the codon context have significant influence on biological functionality of synthetic genomes. Discovery of 98 genes that lost their function identified essential genes with incorrect annotation, including a limited set of 27 genes where we uncovered noncoding control features embedded within protein-coding sequences. In sum, our results highlight the promise of chemical synthesis rewriting to decode fundamental genome functions and its utility toward the design of improved organisms for industrial purposes and health benefits.
In the early 2000s, the template-independent chemical synthesis of the 7.4-kb polio virus (1) and 5.4-kb bacteriophage phiX174 genomes (2) using oligonucleotides has ushered in the field of synthetic genomics. The initial progress on moderately sized viral genomes has spurred whole-genome synthesis of more complex organisms. In 2008 and 2010, the Craig Venter Institute reported the chemical synthesis of genome replicas from Mycoplasma genitalium (583 kb) and Mycoplasma mycoides (1.1 Mb) (3, 4), respectively. These efforts expanded the chemical synthesis scale to megabases and improved in vitro DNA assembly strategies and genome transplantation methods. However, the work also highlighted the challenges of whole-genome synthesis, as a single missense mutation within the dnaA gene initially prevented boot up. To gain insights into a minimal gene set for cellular life, the teams of Craig Venter built a 473-gene reduced version of the M. mycoides genome (5).
Along with these accomplishments, the concept of whole-genome synthesis and genome minimization has been expanded toward the rebuilding of all 16 chromosomes of Saccharomyces cerevisiae driven by an international consortium composed of 21 institutions. In 2014, the consortium reported synthesis of the artificial yeast chromosome synIII (273 kb) (6). Subsequently, five additional chromosomes (7–11) were generated, and as of 2018, roughly 40% of the entire yeast genome has been covered. The redesigned chromosomes removed repetitive sequences (tRNA genes, introns, and transposons) to increase targeting fidelity during stepwise homologous replacement as well as included the seeding of loxP sites to permit iterative genome reduction on completion of yeast chromosomes. In the beginning of the yeast 2.0 synthesis project, CRISPR had not yet entered the stage, but today, it offers an alternative approach for progressive genome reduction.
The redundancy of the genetic code defining the same amino acid by multiple synonymous codons offers the possibility to erase and reassign codons throughout an entire genome. Such rewriting efforts are used to engineer organisms with altered genetic codes and free up codons for incorporation of artificial amino acids, which do not occur within natural organisms. To date, genome-wide rewriting efforts have been primarily reported for viral genomes (12–14), and a few are focused on the rewriting of microbial genomes of Escherichia coli, Salmonella, and S. cerevisiae. Using oligo-mediated recombineering (15), all 321 instances of the TAG stop codon in E. coli were altered to TAA, demonstrating the dispensability of a stop codon within the genetic code (16). In an extension of this approach, rewriting of 13 sense codons across a set of ribosomal genes (17) and genome-wide rewriting of 123 instances of the arginine rare codons AGA and AGG (18) were accomplished in E. coli. These studies unearthed unexpected recalcitrant synonymous rewriting events that occurred primarily in the vicinity of 5′ and 3′ termini of protein-coding sequences (18, 19). Recently, to investigate the impact of more complex rewriting schemes, de novo DNA synthesis methods have been used for the rewriting of gene cassettes in conjunction with genomic replacement strategies (15, 20). Ongoing de novo synthesis toward a 57-codon E. coli genome was reported (21), with the complete genome synthesis underway.
Despite this progress, the underlying rewriting design principles have remained ill defined, and debugging has remained challenging (17, 19). It has been speculated that presence of embedded transcriptional and translational control signals at the termini of coding sequences (CDSs) as well as imprecise genome annotations are the underlying cause. We hypothesized that massive synonymous rewriting in conjunction with a systematic investigation of error causes will shed light onto the general sequence design principles of how biological functions are programed into genomes. However, while some progress has been made to study recoding schemes using individual genes and gene clusters (21), the field currently lacks a broadly applicable high-throughput error diagnosis approach to probe the rewriting of entire genomes.
Here, we report the chemical synthesis of Caulobacter ethensis-2.0 (C. eth-2.0), a bacterial minimized genome composed of the most fundamental functions of a bacterial cell. We present a broadly applicable design–build–test approach to program the most fundamental functions of a cell into a customized genome sequence. By rebuilding the essential genome of Caulobacter crescentus (Caulobacter hereafter) through the process of chemical synthesis writing, we studied the genetic information content at the level of its essential genes.
Results
Essential Part List to Build C. eth-1.0.
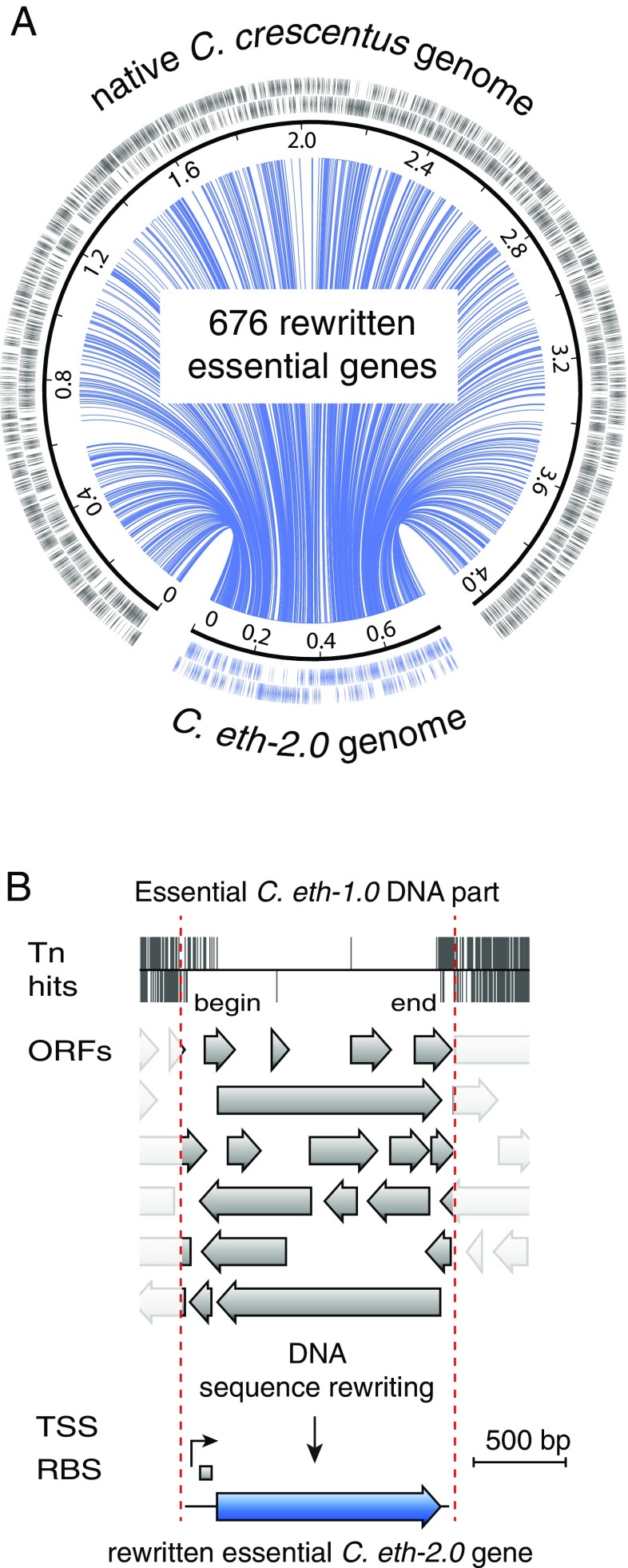
We conceived a bacterial genome design encoding the entire set of essential DNA sequences from the freshwater bacterium Caulobacter (Fig. 1A). Caulobacter is recognized as an exquisite cell cycle model organism (22–25) for which multidimensional omics (26) and transcriptome- (27) and ribosome-profiling measurements have been integrated into a well-annotated genome model (28, 29). We computationally generated the entire list of essential DNA parts for building a bacterial genome from a previously published high-resolution transposon sequencing dataset (30) that identified with base pair resolution the precise coordinates of essential genes, including endogenous promoter sequences. DNA parts were extracted from the native Caulobacter NA1000 genome sequence [National Center for Biotechnology Information (NCBI) accession no. NC_011916.1] according to predefined design rules and concatenated into a digital genome design preserving gene organization and orientation (Fig. 1A and SI Appendix) (31). The resulting 785,701-bp genome design termed Caulobacter ethensis-1.0 (C. eth-1.0) encodes for the most fundamental functions of a bacterial cell. Cumulatively, C. eth-1.0 consists of 1,761 DNA parts, including 676 protein-coding, 54 noncoding, and 1,015 intergenic sequences. To select for faithful assembly and permit stable maintenance in S. cerevisiae, auxotrophic marker genes (TRP1, HIS3, MET14, LEU2, ADE2) and a set of 10 autonomous replicating sequences (ARSs) were seeded across the genome design (Table 1 and SI Appendix). Furthermore, the pMR10Y (31) shuttle vector sequence, permitting stringent low copy replication in S. cerevisiae, E. coli, and Caulobacter, was inserted at the native location of the Caulobacter origin of replication.
Fig. 1.
Part design, compilation, and chemical synthesis rewriting of the C. eth-1.0 genome. (A) Schematic representation of the digital design process; 1,745 DNA parts were extracted from the native Caulobacter NA1000 genome (gray) and reorganized into a rewritten genome design (blue) comprising the entire list of essential genes required to run the basic operating system of a bacterial cell. Lines (blue) connect positions of DNA parts between native and rewritten genomes. (B) Workflow of the part identification and chemical synthesis rewriting process. Transposon sequencing was used to identify the entire set of essential DNA parts of Caulobacter at a resolution of a few base pairs. Absence of transposon insertions [transposon (Tn) hits are plotted as gray lines] pinpoints the nondisruptable DNA regions within the native Caulobacter genome. Such essential DNA parts may encode for putative alternative ORFs, TSSs, or ribosome binding sites (RBSs) that are not required for functionality of the essential DNA part itself. Computational sequence rewriting (Materials and Methods) was used to erase putative sequence features that have not been assigned to a specific biologic function. The resulting rewritten DNA parts are fully defined and only encode for their desired function.
Table 1.
Part list used to build the C. eth-1.0 genome design
| DNA part category | Quantity | Size (bp) | Fraction (%) |
| Protein-coding sequences | 676 | 660,789 | 83.9 |
| Essential | 462 | 471,072 | 59.8 |
| Semiessential | 113 | 114,270 | 14.5 |
| Redundant | 15 | 14,970 | 1.9 |
| Nonessential | 86 | 60,477 | 7.7 |
| Noncoding sequences | 54 | 9,726 | 1.2 |
| tRNA | 44 | 3,455 | 0.4 |
| rRNA | 3 | 4,387 | 0.6 |
| ncRNA | 7 | 1,884 | 0.2 |
| Intergenic sequences | 1,015 | 96,043 | 12.2 |
| Genome replication and assembly | 16 | 19,143 | 2.5 |
| Click markers* | 5 | 6,352 | 0.8 |
| ARS | 10 | 2,121 | 0.3 |
| pMR10Y† | 1 | 10,670 | 1.4 |
| Total no. of DNA parts | 1,761 | 785,701 |
Auxotrophic selection markers that were used to direct the assembly and maintenance of the genome design in yeast. ncRNA, noncoding RNA.
The pMR10Y shuttle vector contains a broad host-range RK2-based low copy replicon (GenBank accession no. AJ606312.1), a kanamycin selection marker, and oriT function for conjugational transfer from E. coli to Caulobacter as well as URA3 marker, ARS, and centromere (CEN) elements for selection and replication in yeast.
Sequence Rewriting of C. eth-1.0 to Enable de Novo Genome Synthesis.
We were unable to obtain 3- to 4-kb DNA building blocks of C. eth-1.0 from commercial DNA suppliers due to a multitude of synthesis constraints. Synthesis constraints are a common problem of natural genome sequences, which have evolved to maintain biological information rather than facilitate chemical synthesis. Recent bioinformatics work (31) showed that more than three-quarters of all deposited bacterial genome sequences are not amenable for low-cost synthesis. We hypothesized that computational synonymous rewriting into an easy to synthesize sequence would facilitate chemical synthesis of the 785-kb genome while maintaining the encoded biological functions (SI Appendix, Fig. S1A). We used our previously reported computational DNA design algorithms (31, 32) and generated a synthesis-optimized genome design termed C. eth-2.0. Cumulatively, we introduced 10,172 base substitutions and removed 5,668 synthesis constraints (Table 2). These are composed of 1,233 repeats, 93 homopolymeric stretches, and 4,342 regions of high guanine-cytosine (GC) content (Table 2), known to hinder chemical DNA synthesis. Moreover, we erased additional 1,045 endonuclease restriction sites to facilitate standardized assembly of the DNA building blocks into the 785-kb chromosome of C. eth-2.0.
Table 2.
Sequence rewriting of C. eth-1.0 into C. eth-2.0 leads to massive reduction of genetic features
| Type | C. eth-1.0 | C. eth-2.0 | Fraction (%) |
| Sequence rewriting | |||
| Base substitutions | None | 133,313 | 17.0 |
| Rewritten codons* | None | 123,562 | 56.1 |
| Codons | |||
| TTG | 1,154 | 0 | 100 |
| TTA | 46 | 0 | 100 |
| TAG | 173 | 10 | 94.2 |
| Alternative genetic features | |||
| ORFs† | 3,229 | 407 | 87.4 |
| TSS‡ | 1,730 | 82 | 95.3 |
| RBS§ | 1,331 | 310 | 76.7 |
| Remaining genetic features¶ | 6,290 | 799 | |
| DNA synthesis constraints | |||
| High GC regions# | 4,342 | 0 | 100 |
| Direct repeats 8 bp | 880 | 113 | 87.2 |
| Hairpins 8 bp | 606 | 140 | 76.9 |
| Homopolymers | 139 | 46 | 66.9 |
| Restriction sites‖ | 1,047 | 2 | 99.8 |
| Synthesis constraints** | 7,014 | 301 |
Number of synonymous codon substitutions introduced on sequence rewriting.
Number of alternative ORFs residing within the 676 CDSs of C. eth-1.0 and C. eth-2.0.
Number of TSSs internal to CDSs.
Number of ribosome binding sites (RBSs) internal to CDSs.
Number of remaining genetic features within CDS of C. eth-1.0 and C. eth-2.0.
Regions of high GC content > 0.8 within a 100-bp window.
Total number of type IIS restriction sites that were removed (AarI, BsaI, BspQI, PacI, PmeI, I-CeuI, I-SceI). Note that the two unique PmeI and PacI sites remained within the pMR10Y backbone to facilitate linearization of the final assembled chromosome for subsequent analysis by pulsed field gel electrophoresis.
Number of DNA synthesis constraints of C. eth-1.0 and C. eth-2.0.
Sequence Rewriting to Minimize the Number of Genetic Features.
We reasoned that chemical synthesis rewriting offers a powerful experimental approach to probe the accuracy of existing genome annotations and study where additional layers of information exist beyond the primary amino acid code. Furthermore, fundamental functions encoded within the essential genomes can be identified. In addition to the base substitutions introduced for synthesis streamlining, we used computational sequence design algorithms (31, 32) to deliberately add 123,141 base substitutions within protein-coding sequences to yield the rewritten C. eth-2.0 design (Table 2) (33). In C. eth-2.0, we replaced 56.1% of all codons by synonymous versions. While the amino acid sequence of the 676 annotated genes was maintained, rewriting enabled us to minimize the number of hypothetical genetic elements present within protein-coding sequences of C. eth-2.0. These elements include alternative ORFs, predicted gene internal transcriptional start sites (TSSs), and sequence motifs (predicted or cryptic) that may fine tune translation rates (Fig. 1B and Materials and Methods). Overall, we removed 87.4% of all putative ORFs (2,822 of 3,229) (SI Appendix, Fig. S2C), 95.3% of all internal TSSs (1,648 of 1,730), and 76.7% of all predicted ribosome stalling motifs (1,021 of 1,331) (Table 2). Testing whether rewritten genes remain functional will identify genes in which additional information beyond the amino acid code is necessary for proper functioning. Achieving functional C. eth-2.0 genes, however, will provide fully defined artificial genes composed of a minimized number of genetic elements. The precise knowledge of which genes remain functional and the subsequent repair of nonfunctional genes will ultimately lead to a fully defined artificial cell.
Chemical Synthesis of C. eth-2.0.
We computationally devised a four-tier DNA assembly strategy starting from 3- to 4-kb assembly blocks to build the complete C. eth-2.0 chromosome in yeast (32) (Fig. 2A). Demonstrating the ease of genome-scale synthesis on sequence rewriting, 235 of 236 blocks were successfully manufactured (SI Appendix), and only a single DNA block required custom synthesis. We progressively assembled these initial 236 DNA blocks into 37 chromosome segments (19–22 kb in size) and further into 16 megasegments (38–65 kb in size) (Fig. 2A and SI Appendix) using yeast transformation. To select for the complete chromosome assembly, we applied a click marker strategy by introducing five auxotrophic yeast genes (TRP1, HIS3, MET14, LEU2, and ADE2) split between adjacent megasegments. On correct chromosome assembly in an engineered yeast strain lacking all auxotrophic marker genes (YJV04), click markers will form functional genes and reconstitute prototrophy (Fig. 2B). Initial attempts to assemble the C. eth-2.0 chromosome from 16 megasegments were not successful. Sequencing of yeast clones with partial C. eth-2.0 assemblies identified two defective ARS elements (ARS416 and ARS1213), which prevented replication of the full-length chromosome. We corrected these design errors and added five additional ARS sequences to promote efficient replication of the GC-rich C. eth-2.0 chromosome in yeast.
Fig. 2.
Assembly of C. eth-2.0 in S. cerevisiae. (A) Schematic representation of the circular 785,701-bp C. eth-2.0 chromosome with six auxotrophic selection markers (red), 11 ARSs (black), and the restriction sites for PmeI and PacI (blue); 236 DNA blocks (green boxes) were assembled into 37 genome segments (blue boxes) and 16 megasegments (orange boxes) and further assembled into the complete C. eth-2.0 genome (outermost gray track). (B) The complete C. eth-2.0 chromosome was assembled in a single reaction from 16 megasegments by yeast spheroplast transformation and subsequent growth selection for auxotrophic TRP1 and LEU2 markers. (C) Growth selection on medium lacking Ura, Trp, His, Met, Leu, and Ade identified yeast clone 2 (C. eth-2.0) positive for all auxotrophic markers, while the parental strain (YJV04) fails to grow and requires synthetic defined (SD) medium. (D) Size validation of the 785-kb C. eth-2.0 chromosome by pulsed field gel electrophoresis. Digestion with PmeI and PacI releases a 771-kb portion of the C. eth-2.0 chromosome (arrow) from the shuttle vector pMR10Y. Undigested (marker) and PmeI- and PacI-digested yeast chromosomes (YJV04 digest) serve as controls. (E) DNA sequencing coverage at segment level (Top) and megasegment level (Middle) and the complete chromosome assembly (Bottom) are shown.
One-step transformation of the 16 corrected megasegments into yeast spheroplasts yielded two clones, one of which restored prototrophy for all six auxotrophic click markers, indicating complete assembly of C. eth-2.0 (Fig. 2C). We subsequently confirmed the presence of C. eth-2.0 as a single circular chromosome by pulsed field gel electrophoresis (Fig. 2D), diagnostic PCR (SI Appendix, Fig. S3), and whole-genome sequencing (Fig. 2E). C. eth-2.0 has a high GC content exceeding 57%, while previous chemically synthesized chromosomes (5, 8, 34) exhibit low GC contents closely matching the native yeast genome. So far, attempts to clone high GC sequences in yeast have proven to be difficult (35).
To assess whether C. eth-2.0 is stably maintained in yeast, we performed whole-genome sequencing on prolonged cultivation. After propagation for over 60 generations, we found no occurrences of adaptive mutations or chromosomal rearrangements within C. eth-2.0, indicating stable maintenance in YJV04 (SI Appendix, Fig. S4A). In agreement with this observation, electron micrographs showed normal yeast cell morphologies for parental cells and YJV04 bearing the C. eth-2.0 chromosome (SI Appendix, Fig. S4B).
We sequence verified C. eth-2.0 at each assembly level to assess the performance of the genome synthesis process (Fig. 2E). Across the 785-kb genome design, we detected a total of 21 nonsynonymous mutations (SI Appendix, Table S3). Thereof, 17 emanated from nonsequence perfect DNA blocks that were provided by one of two commercial suppliers. Only four additional missense mutations within the genes argS (arginyl-tRNA synthetase) and fabI (acyl-carrier protein) and the ribosomal genes S7P and L12P were introduced during segment and megasegment assembly in yeast and E. coli, respectively. No additional mutations occurred in the final assembly of the C. eth-2.0 chromosome, indicating a high sequence fidelity in the genome build process.
Mapping of Toxic Genes.
It was previously reported in clone-based genome sequencing studies that natural microbial genomes contain genes encoding for toxic and dosage-sensitive expression products (36, 37). We speculated that toxic genes residing on C. eth-2.0 would prevent chromosomal maintenance in Caulobacter. Therefore, we tested the design in the form of the 37 individual C. eth-2.0 chromosome segments for the presence of toxic genes. Quantification of conjugational transfer from E. coli to Caulobacter in conjunction with sequencing demonstrated that toxic genes were absent in 25 of 37 chromosome segments (SI Appendix, Fig. S5 and Table S4). However, we observed a drastic reduction in transfer efficiency for 12 segments, suggesting presence of toxic genes that collectively cover 18.9 ± 3.6 kb in sequence (SI Appendix, Table S4). We carried out genetic suppressor analysis and identified evolved strains that tolerated formerly toxic genome segments (Materials and Methods). Whole-genome sequencing of suppressor strains led to the identification of 14 toxic genetic loci (SI Appendix, Table S5) that bear mutations alleviating toxicity. An additional three chromosome segments acquired small deletions on selection for fast growth (SI Appendix, Table S6). Among the toxic genes, we found three chromosome replication genes (dnaQ, dnaB, rarA), six genes involved in LPS and fatty acid biosynthesis (fabB, lptD, lpxD, accC, murU, waaF), two genes encoding interacting RNA polymerase components (rpoC, topA), the S10-spc-alpha ribosomal protein gene cluster (CETH_01304-01323), and the sodium-proton antiporter nhaA. Multiple of the identified toxic genes encode for interacting proteins that form complexes, suggesting an imbalance in subunit dosage as a likely cause for toxicity. Overall, the observed fraction of 1.9% (14 of 730) toxic genes found in C. eth-2.0 is well in agreement with the previously reported average of 2.15 ± 0.8% of toxic genes identified among seven E. coli strains (36). We concluded that computational sequence rewriting as part of the chemical synthesis rewriting process does not induce additional gene toxicity. In agreement with this hypothesis, 6 genes among the identified 14 toxic rewritten genes have been previously identified as “unclonable genes” in E. coli (37). Furthermore, misbalanced expression of rpoC, rarA, dnaB, and accC has previously been reported to elicit toxicity due to imbalance in protein complex subunit stoichiometry (38–41). Given the precedence of toxicity for wild-type genes, we argue that the toxicity of these genes when ectopically expressed is likely a general property and is not attributed to the rewriting process that maintains identical proteins.
Genome-Wide Functionality Assessment of C. eth-2.0.
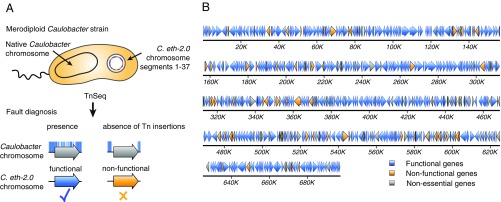
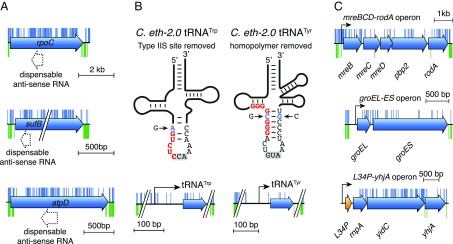
While throughout the build process, the C. eth-2.0 genome was maintained in heterologous hosts, we next investigated whether rewritten genes resume their anticipated function on introduction into Caulobacter. Functionality assessment and error diagnosis of large-scale DNA constructs are major challenges for bioengineering of synthetic genomes. To permit parallel functionality assessment of rewritten C. eth-2.0 genes, we developed a transposon-based testing approach. This approach assesses the functionality of rewritten genes in merodiploid test strains, which harbor episomal copies of C. eth-2.0 chromosome segments in addition to the native chromosome. The testing approach measures the functional equivalence between native and rewritten C. eth-2.0 genes through genetic complementation. In the presence of functional C. eth-2.0 genes, previously essential native genes become dispensable and acquire disruptive transposon insertions (Fig. 3A). In contrast, native genes remain essential and do not tolerate disruptive transposon insertions in the presence of nonfunctional rewritten genes (Fig. 3A). In the case of functional C. eth-2.0 genes, such an analysis will prove that rewritten gene variants are functionally equivalent to essential native Caulobacter genes. Failure in complementation will identify specific genes where sequence rewriting in C. eth-2.0 erases additional genetic control elements that are important for proper gene functioning. We asked whether rewritten genes are functionally equivalent to native genes despite the massive level of sequence modification introduced. We subjected 37 merodiploid test strains bearing C. eth-2.0 chromosome segments as episomal copies along the native chromosome to transposon mutagenesis to test gene functionality. We compared transposon insertion patterns obtained between complementing and noncomplementing conditions and assessed the functionality of C. eth-2.0 (Materials and Methods and Dataset S1). Nucleotide substitutions introduced on rewriting and sequence optimization of the C. eth-2.0 genome allowed us to unambiguously assign transposon insertions to the native Caulobacter genome and C. eth-2.0 chromosome segments. Cumulatively, we found 81.5% (432 of 530) of all essential and semiessential C. eth-2.0 genes to be functional (Fig. 3B and Table 3). Functional rewritten C. eth-2.0 genes encompass a drastic reduction in the number of genetic features (annotated, cryptic, or predicted) compared with the wild-type Caulobacter genome annotation. Maintenance of biologic functionality within rewritten genes suggests dispensability of these genetic features, and hence, it will lead to refinement of the current genome annotation. During the design process of C. eth-2.0, we have reduced the number of genetic features within CDS from 6,290 to 799 (Table 2). The high functionality level of 81.5% observed within the rewritten C. eth-2.0 suggests that the large majority of the 6,290 previously annotated and predicted genetic features do not adopt essential function. Among the genetic features found to be dispensable were the three formerly assigned antisense transcripts (sRNAs) CCNA_R0109, R0151, and R0194 internal to rpoC, sufB, and atpD that acquired 16, 17, and 62 base substitutions during the rewriting process, respectively (Fig. 4A). Dispensability of the formerly assigned antisense transcripts suggests that the majority of chromosomally encoded sRNAs identified by transcriptome analysis (42, 43) do not elicit an essential function.
Fig. 3.
Fault diagnosis and error isolation across the C. eth-2.0 chromosome. (A) Functionality assessment of the C. eth-2.0 chromosome. Merodiploid strains bearing episomal C. eth-2.0 chromosome segments (orange and blue circle) are subjected to transposon sequencing (TnSeq). Presence of transposon insertions (blue marks) in a previously essential chromosomal gene (gray arrows) indicates functionality of the homologous C. eth-2.0 gene (blue arrow), while absence of insertions indicates a nonfunctional C. eth-2.0 gene (orange arrow). (B) Functionality map of the C. eth-2.0 chromosome with functional genes (blue arrows), nonfunctional genes (orange arrows), and nonessential control genes (gray arrows).
Table 3.
Functionality of C. eth-2.0 genes according to cellular processes
| Category | Functional C. eth-2.0 genes* | P value† |
| Translation‡ | 73.6% (81/110) | 5.49E-03 |
| Ribosomal proteins‡ | 60.6% (20/33) | 2.01E-03 |
| tRNA synthetases | 81.8% (18/22) | 6.14E-01 |
| tRNAs | 67.8% (19/28) | 4.73E-02 |
| Translation factors | 88.9% (24/27) | 2.22E-01 |
| Transcription | 86.7% (13/15) | 4.51E-01 |
| DNA replication | 83.9% (26/31) | 4.69E-01 |
| Cellular processes | 87.2% (123/141) | 1.12E-02 |
| Cell cycle | 87.5% (28/32) | 2.52E-01 |
| Cell envelope | 86.9% (73/84) | 8.48E-02 |
| Protein turnover | 88.0% (22/25) | 2.81E-01 |
| Energy production | 73.9% (34/46) | 1.03E-01 |
| Metabolism§ | 90.3% (121/134) | 2.60E-04 |
| Hypothetical proteins‡ | 64.2% (34/53) | 7.45E-04 |
| Total | 81.5% (432/530) |
Fraction of functional C. eth-2.0 genes as assessed by transposon sequencing. Numbers of functional genes vs. total gene numbers per class are shown in parentheses.
P values for functionality enrichment and deenrichment of different gene Categories.
Categories of genes that display a significant decrease in functionality.
Categories of genes that display a significant increase in functionality.
Fig. 4.
Sequence design flexibility within rewritten C. eth-2.0 genes. (A) Dispensability of antisense RNAs. Schematic depicting dispensable antisense transcripts embedded with CDSs of genes rpoC, sufB, and atpD (blue arrows). On synonymous rewriting, antisense transcripts CCNA_R0109, CCNA_R0151, and CCNA_R0194 (doted arrows) internal to rpoC, sufB, and atpD acquired 16, 17, and 62 base substitutions, respectively. Essential chromosomal genes rpoC, sufB, and atpD carry disruptive transposon insertion (blue marks) in the presence of complementing C. eth-2.0 chromosome segments (blue marks) compared with the transposon insertion pattern of the wild-type control strain (green marks), indicating that antisense transcripts are nonessential. (B) Schematic depiction of the secondary structure of the rewritten tRNATrp and tRNATyr. Type IIS restriction sites (red letters; Left) and homopolymeric sequences (red letters; Right) hindering chemical synthesis of tRNA genes were erased by introducing base substitutions (blue) in the anticodon arms while maintaining the anticodons (gray box). Transposon testing reveals functionality of C. eth-2.0 tRNA genes. (C) Functionality testing of C. eth-2.0 operons. On complementation with C. eth-2.0 operons, chromosomal genes tolerate disruptive transposon insertions (blue marks) throughout the native operon, leading to simultaneous inactivation of multiple native genes.
Sequence Design Flexibility Beyond Protein-Coding Sequences.
The large majority of the 133,313 base substitutions were introduced within protein-coding sequences of C. eth-2.0. However, a significant number of nonsynonymous substitutions were inserted within intergenic and noncoding regions, such as tRNA genes, to facilitate the de novo DNA synthesis process. We found that base substitutions within noncoding sequences were frequently tolerated and did not impair gene functionality. For example, the two tRNA genes tRNATrp and tRNATyr remained functional despite base changes that were introduced within the anticodon arm to erase DNA synthesis constraints present within the wild-type sequences (Fig. 4B). In the case of tRNATrp, we removed a Type IIS restriction site, and in tRNATyr, a homopolymeric sequence pattern hindering DNA synthesis was removed. Both rewritten tRNA genes retained their function as revealed by our transposon-based complementation measurements (Fig. 4B). These findings suggest that, even apart from protein-coding sequences, a high level of sequence design flexibility exists to imprint biological functions into DNA.
Level of Gene Functionality Among Cellular Processes.
We reasoned that the analysis of gene functionality among different cellular processes would permit identification of gene classes harboring high levels of transcriptional and translational control elements within CDS. Assignment of gene functionality among cellular processes revealed that metabolic genes were enriched with over 90.3% functionality (P value of 2.60E-4) (Table 3). This supports the idea that metabolic genes contain a low level of transcriptional and translational control elements embedded within their CDS. This finding correlates with the observation that regulation of bacterial metabolism mainly occurs at the enzymatic level (44, 45). However, hypothetical and ribosomal genes were underrepresented, with 64.2 and 60.6% functional C. eth-2.0 genes, respectively (P values of 5.45E-4 and 2.01E-3, respectively) (Table 3). Based on these findings, we estimated that one-third of the hypothetical essential genes encode for genetic features other than the annotated protein-coding sequence. Likewise, close to 40% of all ribosomal genes likely contain additional regulatory elements embedded within their protein-coding sequence. From a gene regulatory perspective, this is not surprising, as protein synthesis is the major consumer of cellular energy in bacteria (46). Furthermore, the biogenesis of functional ribosome complexes depends on the concerted transcriptional control of many ribosomal operon genes (47). In sum, our analysis suggests that a low level of additional essential regulatory elements is embedded within the protein-coding sequences of metabolic genes. However, a high number of regulatory elements are embedded within coexpressed ribosomal genes and other multigene core modules of the bacterial cell.
Rewritten C. eth-2.0 Operons Encompass Fully Functional Biological Modules.
Although a significant amount of chemical synthesis rewritten C. eth-2.0 genes are functional on an individual basis, we hypothesized that additive fitness effects might arise when multiple synthetic genes were combined. We thus searched for chromosomal transposon insertions within essential Caulobacter operons leading to simultaneous inactivation of multiple gene products due to truncation of a polycistronic mRNA transcript. We observed such transposon insertions within 41 formerly essential Caulobacter operon genes (Fig. 4C and Dataset S3), suggesting that the chemical synthesis rewritten C. eth-2.0 genes may indeed fully encompass functional biological modules complementing the function of their native counterparts. One example includes the mreBCD-rodA operon, which is involved in the coordination of cell wall peptidoglycan biosynthesis machinery. This complex is critical for the generation and maintenance of bacterial cell shape (48). We found transposon insertions disrupting the polycistronic mRNA, suggesting that the function of the chromosomal mreBCD-rodA operon is complemented by the C. eth-2.0 counterparts (Fig. 4C). Similar patterns of transposon insertions were obtained for the groEL-ES operon (49) and the membrane protein chaperone operon yidC-yidA (50). Both tolerated disruptive transposon insertions throughout the native sequence, leading to simultaneous inactivation of multiple genes. These findings support the idea that additive fitness effects do not likely arise when multiple synthetic genes are combined into functional modules, which will ultimately simplify the build process of artificial chromosomes by using chemical synthesis rewritten DNA.
Discovery of Essential Genetic Features Within CDS.
We reasoned that the process of chemical synthesis rewriting offers a powerful experimental approach to map hitherto unknown genetic regulatory elements encoded within the protein-coding sequence and validate the annotation accuracy of an organism’s genome. Discovery of genes that lost their function on rewriting suggested the presence of additional essential genetic features, which have evaded previous genome annotation efforts. Error cause classification of the 98 nonfunctional C. eth-2.0 genes (Materials and Methods) pinpointed to 52 instances of imprecise annotation of the ancestral Caulobacter genome including misannotated promoter regions and incorrect TSSs predictions. This implies that a significant number of protein-coding genes remain misannotated within curated genomes. We found evidence for 27 transcriptional and translational control signals embedded within protein-coding sequences that were erased due to sequence rewriting. This finding suggests that internal transcriptional and translational control elements do not often occur within CDS of the Caulobacter genome. In only 13 instances, we detected nonfunctional genes due to base substitutions introduced outside of protein-coding sequences to optimize synthesis. Furthermore, six genes acquired deleterious mutations during the build and boot-up process (Dataset S2). These findings suggest that inaccurate annotation of protein-coding sequences is the main cause for losing functionality on synonymous rewriting.
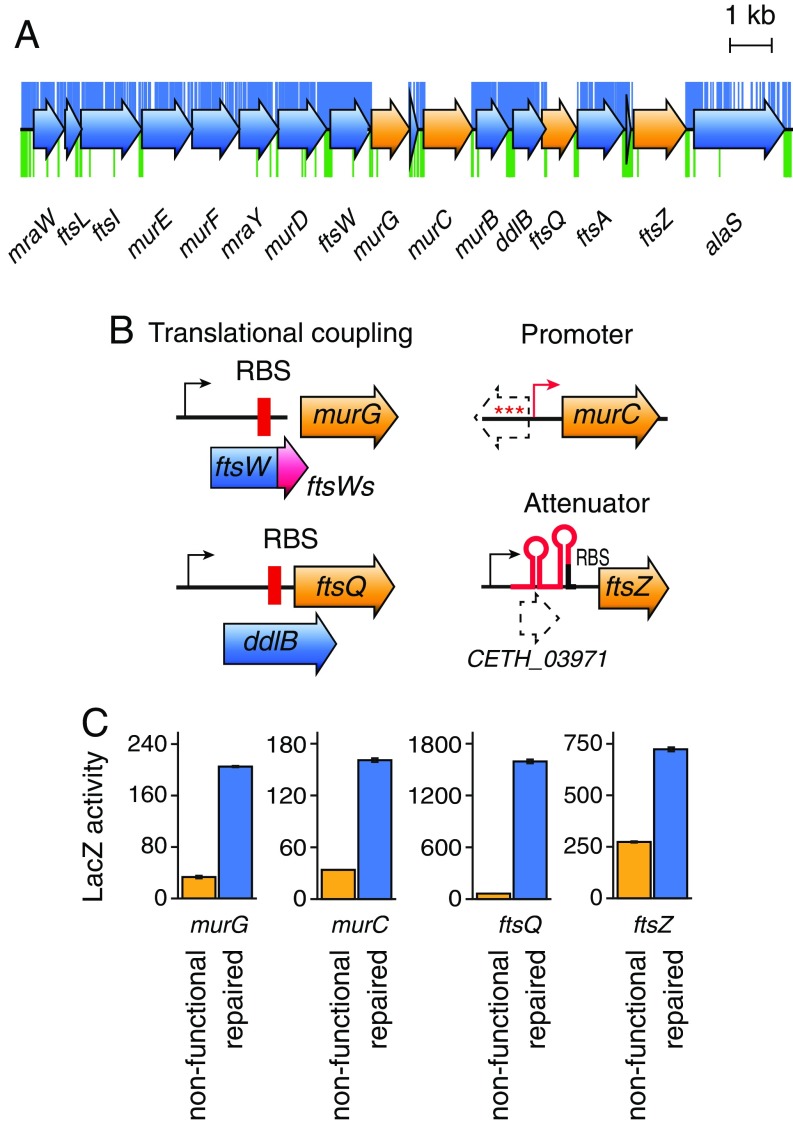
Genetic Control Features Within the Cell Division Genes.
We next investigated the presence of additional genetic features within the cell division genes murG, murC, ftsQ, and ftsZ, in which genetic complementation failed with the corresponding C. eth-2.0 counterparts (Fig. 5A). We hypothesized that computational sequence rewriting likely erased critical control elements needed for proper gene expression. Indeed, we found that rewriting of an overlapping CDS upstream of murC corrupted the associated promoter region. Similarly, sequence rewriting of ddlB erased an internal ribosome binding site necessary for translation of the downstream gene ftsQ (Fig. 5B). The rewriting of ftsW erased an embedded short transcript (ftsWs) necessary for murG translation (29) (Fig. 5B). Finally, we found a short annotated CDS (29) upstream of the nonfunctional ftsZ gene. However, sequence analysis revealed that the wild-type sequence contains a hairpin secondary structure, which resembles a transcriptional attenuation element. This may control ftsZ expression depending on the metabolic conditions (Fig. 5B). While additional studies are needed to unravel the exact molecular functions of these genetic control elements within the cell division gene cluster, we found that repair of the sequence upstream of the ftsZ gene restored the ftsZ expression levels (Fig. 5C). Similarly, insertion of the wild-type sequence elements into the C. eth-2.0 genes murC, ftsQ, and murG also restored gene expression (Fig. 5C). This suggests that, after missing essential genetic elements are identified, error causes can rapidly be deduced to allow for rational repair of genome designs. Furthermore, identification and error diagnosis of noncomplementing genes will provide a formidable opportunity to uncover DNA design principles that will further improve our capabilities in programing biological functions into synthetic chromosomes.
Fig. 5.
Fault diagnosis and repair across the C. eth-2.0 chromosome. (A) Fault diagnosis across the C. eth-2.0 cell division gene cluster. Transposon insertions in the wild-type control (green marks) and on complementation with C. eth-2.0 cell division genes (blue marks) are shown. With the exception of the four nonfunctional genes murG, murC, ftsQ, and ftsZ (orange arrows), the large majority of rewritten genes are functional (blue arrows). (B) Chemical synthesis rewriting reveals genetic control elements present within the cell division gene cluster, including translational coupling signals (murG), internal ribosome binding sites (RBSs; ftsQ), extended promoter regions (murC), and attenuator sequences upstream of ftsZ. (C) Insertion of the wild-type sequence elements upstream of nonfunctional cell division genes restores gene expression as measured by -galactosidase assays using lacZ reporter gen fusions.
Discussion
C. crescentus has emerged as an important model organism for understanding the regulation of the bacterial cell cycle (25, 51, 52). A notable feature of Caulobacter is that the regulatory events that control polar differentiation and cell cycle progression are highly integrated and occur in a temporally restricted order (53). The advent of genomic technologies has enabled global analyses that have revolutionized our understanding of Caulobacter genetic core networks that control the lifecycle (26–29). In recent years, many components of the regulatory circuit have been identified, and simulation of the circuitry has been reported (25, 54). More recent experimental work using transposon sequencing has shown that 12% of the Caulobacter genome is essential for survival under laboratory conditions (30). The identified set of essential sequences included not only protein-coding sequences but also, regulatory regions and noncoding elements that collectively store the genetic information necessary to run a living cell. Of the individual DNA regions identified as essential, 91 were noncoding regions of unknown function, and 49 were genes presumably coding for hypothetical proteins with function that is unknown.
Although classical genetic approaches dissect the functioning of biological systems by analyzing individual native genes, uncovering the function of essential genes has remained very challenging. Herein, we show that the rewriting of entire genomes through the process of chemical synthesis provides a powerful and complementary research concept to understand how essential functions are programed into genomes. Contemporary synthetic genome projects (3, 5, 8) have largely maintained natural genome sequences, implementing only modest design changes to increase the likelihood of functionality. However, conservative genome design misses a key opportunity of chemical DNA synthesis: the rewriting of DNA to advance our understanding of how fundamental biological functions are encoded within genomes. Indeed, synthetic autonomous bacteria, such as M. mycoides strain JCVI-syn3.0 made up of 473 genes within a 531-kb genome (5), resulted in the creation of a replicative cell. However, it also encompasses 149 genes with unknown functions (84 labeled as “generic,” and 65 labeled as “unknowns”) (55). This corresponds to over one-third of its gene set. While these studies were highly valuable to experimentally determine the core set of genes for an independently replicating cell, they did not probe the genetic information content of its essential genes.
By rebuilding the essential genome of Caulobacter through the process of chemical synthesis rewriting, we assessed the essential genetic information content of a bacterial cell on the level of its protein-coding sequences. Within the 785,701-bp genome of C. eth-2.0, we used sequence rewriting to reduce the number of genetic features present within protein-coding sequences from 6,290 to 799. Overall, we introduced 133,313 base substitutions, resulting in the synonymous rewriting of 123,562 codons. We speculated that synonymous rewriting of protein-coding sequences maintains the encoded amino acid sequences but likely erases additional genetic information layers. These include alternative reading frames as well as hidden control elements embedded within protein-coding sequences of essential genes.
Rewriting of 56% of all codons resulted in complete rewriting of the essential Caulobacter transcriptome. Despite incorporating such drastic changes at the level of mRNA, our functionality analysis revealed that over 432 of the transcribed essential genes of C. eth-2.0 corresponding to 81.5% of all rewritten essential genes are equal in functionality to natural counterparts to support viability. This result suggests that, in most essential genes, the primary mRNA sequence, the secondary structure, or the codon context has no significant influence on biological functionality. This finding is surprising given the fact that previous studies on individual genes reported that codon translation in vivo is controlled by many factors, including codon context (56). Furthermore, our findings suggest that the vast majority of the probed ORFs encode exclusively for proteins and that other layers of genetic control do not seem to play a significant role. Among the 134 enzyme-encoding genes that make up the metabolic core network of C. eth-2.0, the level of functional genes is even over 90%, suggesting that rewritten biosynthetic pathways retain their functionality in most cases. A possible explanation for the high proportion of functional metabolic genes might be the fact that regulation of essential metabolic functions occurs rather by allosteric interactions at the level of enzymes than at the level of gene expression.
In addition to 432 functional rewritten genes, our study precisely mapped 98 genes that lost functionality on synonymous rewriting as detected by our transposon-based functionality assessment. Since retaining solely the protein-coding sequences of these genes is not sufficient for their functionality, it is reasonable to conclude that these genes are misannotated or contain hitherto unknown essential genetic elements embedded within their CDS. Alternatively, it is also possible that a subset of these genes encode for RNA rather than protein-coding functions. Taken together, our genome rewriting approach can be used to experimentally validate the annotation fidelity of entire genomes.
Altogether, the identified set of 98 nonfunctional genes corresponds to less than 20% of the essential genome of C. eth-2.0 and precisely revealed where we currently have gaps in our knowledge that persisted despite previous omics-informed genome reannotation efforts. In the future, it will be interesting to unravel why rewriting renders particular genes nonfunctional. These studies will shed light onto hitherto unknown transcriptional and translational control layers embedded within protein-coding sequences that are of fundamental importance for proper gene functioning. Targeted repair of identified nonfunctional C. eth-2.0 genes, as exemplified within the subset of the four faulty cell division genes murG, murC, ftsQ, and ftsZ, will lead to the discovery of genetic features, such as the essential attenuator element identified upstream of the ftsZ gene, the function of which is currently unknown. We acknowledge that the 98 identified nonfunctional genes are still poorly understood, yet our findings on C. eth-2.0 serve as an excellent starting point to close current knowledge gaps in essential genome functions toward rational construction of a synthetic organism with a fully defined genetic blueprint.
On the level of de novo DNA synthesis, we herein demonstrate how chemical synthesis rewriting facilitates the genome synthesis process. To simplify the entire genome build process, we used sequence design algorithms (31, 32) and collectively introduce 10,172 base substitutions to remove 5,668 DNA synthesis constraints, including 1,233 repeats, 93 homopolymeric stretches, and 4,342 regions of high GC content. Successful low-cost synthesis and subsequent higher-order assembly of C. eth-2.0 into the complete chromosome exemplify the utility of our approach to rapidly produce designer genomes.
Our results highlight the promise of chemical synthesis rewriting of entire genomes to understand how the most fundamental functions of a cell are programed into DNA. On the systems engineering level, our design–build–test approach enables us to harness massive design flexibility to produce rewritten genomes that are customized in sequence while maintaining their biological functionality. On the level of genome synthesis, our findings also highlight how chemical synthesis facilitates rewriting of biological information into DNA sequences that can be physically manufactured in a highly reliable manner, thereby reducing costs and increasing effectiveness of the genome build process. In sum, our results highlight the promise of chemical synthesis rewriting to decode fundamental genome functions and its utility toward design of improved organisms for industrial purposes and health benefits.
Materials and Methods
Detailed materials and methods are in SI Appendix. The sequence of the C. eth-2.0 genome has been deposited in the NCBI database (GenBank accession no. CP035535).
Design of C. eth-1.0 Genome and Sequence Rewriting into C. eth-2.0.
To streamline the C. eth-1.0 design (30) for DNA synthesis, the previously reported Genome Caligrapher algorithm and sequence design pipeline (31) were applied at a codon recoding probability of 0.56. The streamlined C. eth-2.0 design contains a low amount of both synthesis constraints and unnecessary genetic features. To enable the retrosynthetic assembly route, C. eth-2.0 was partitioned into 3- to 4-kb DNA blocks using the previously published Genome Partitioner algorithm (32).
Synthesis and Hierarchical Assembly of the C. eth-2.0 Genome.
The partitioned 3- to 4-kb DNA blocks for the hierarchical assembly of C. eth-2.0 were ordered from two commercial suppliers of low-cost de novo DNA synthesis. The blocks were assembled into 20-kb segments and subsequently, into 40- to 60-kb megasegments using yeast homologous gap repair. To verify the assemblies, a junction-amplifying PCR was conducted.
To assemble the megasegments into the 785-kb C. eth-2.0 genome, homologous gap repair was done by the newly generated S. cerevisiae strain YJV04. To transform the segments into the yeast cells, a spheroplast procedure was applied. The assembly was verified by a junction-amplifying PCR. The correct size of the construct was verified using pulse field agarose gel electrophoresis by lysing the yeast cells inside an agarose plug.
The sequence of the C. eth-2.0 construct was verified using the Illumina NextSeq and iSeq systems.
Construction of Merosynthetic Caulobacter Test Strains.
Sequence-confirmed C. eth-2.0 segments were conjugated from E. coli S17-1 into Caulobacter NA1000 to generate a panel of 37 merosynthetic test strains. The occurrence of toxic C. eth-2.0 genes was measured by the conjugation frequency of the different segments. To pinpoint the toxic genes, the C. eth-2.0 segments were sequenced on an Illumina system after the boot up in Caulobacter. Using the sequencing data, the mutations within the evolved C. eth-2.0 segments were analyzed, yielding the precise coordinates of toxic genes.
Fault Diagnosis of C. eth-2.0 by Transposon Sequencing.
To benchmark the functionality of the C. eth-2.0 genes, transposon sequencing was applied (30). The analysis was conducted using hypersaturated transposon libraries and an Illumina system. The sequencing data were mapped onto the original Caulobacter genome, resulting in a set of all functional C. eth-2.0 genes. After analyzing the nonfunctional genes, a repair of the sequence was done using standard cloning techniques. To test the repaired C. eth-2.0 genes, a on-galactosidase reporter assay was conducted.
Supplementary Material
Acknowledgments
We thank R. Schlapbach and L. Poveda from Zürich Functional Genomics Center (ZFGC) for sequencing support; B. Maier and members from ScopeM for electron microscopy support; S. Nath from the Joint Genome Institute (JGI) for DNA synthesis and sequencing support; F. Rudolf for assistance with yeast marker design; H. Christen for conception of computational algorithms; and Samuel I. Miller, Markus Aebi, and Uwe Sauer for critical comments. This work received institutional support from Community Science Program (CSP) DNA Synthesis Award Grants JGI CSP-1593 (to M.C. and B.C.) and CSP-2840 (to M.C. and B.C.) from the US Department of Energy Joint Genome Institute, Swiss Federal Institute of Technology (ETH) Zürich ETH Research Grant ETH-08 16-1 (to B.C.), and Swiss National Science Foundation Grant 31003A_166476 (to B.C.). The work conducted by the US Department of Energy Joint Genome Institute, a Department of Energy Office of Science User Facility, is supported by Office of Science of the US Department of Energy Contract DE-AC02-05CH11231.
Footnotes
Conflict of interest statement: Eidgenössische Technische Hochschule holds a patent application (WO2017085249A1) with M.C. and B.C. as inventors that covers functional testing of synthetic genomes. M.C. and B.C. hold shares from Gigabases Switzerland AG.
This article is a PNAS Direct Submission.
Data deposition: The sequence of the C. eth-2.0 genome reported in this paper has been deposited in the National Center for Biotechnology Information database (GenBank accession no. CP035535).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818259116/-/DCSupplemental.
References
- 1.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cdna: Generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 2.Smith HO, Hutchison CA, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 4.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 5.Hutchison CA, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 6.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell LA, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: Synvi and beyond. Science. 2017;355:eaaf4831. doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 8.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y, et al. Deep functional analysis of synii, a 770-kilobase synthetic yeast chromosome. Science. 2017;355:eaaf4791. doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017;355:eaaf4706. doi: 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie ZX, et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017;355:eaaf4704. doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 12.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez MA, Jordan-Paiz A, Franco S, Nevot M. Synonymous virus genome recoding as a tool to impact viral fitness. Trends Microbiol. 2016;24:134–147. doi: 10.1016/j.tim.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28:723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lajoie M, et al. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 18.Napolitano MG, et al. Emergent rules for codon choice elucidated by editing rare arginine codons in escherichia coli. Proc Natl Acad Sci USA. 2016;113:E5588–E5597. doi: 10.1073/pnas.1605856113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau YH, et al. Large-scale recoding of a bacterial genome by iterative recombineering of synthetic dna. Nucleic Acids Res. 2017;45:6971–6980. doi: 10.1093/nar/gkx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 22.Holtzendorff J, et al. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–987. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- 23.McGrath PT, et al. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- 24.Christen M, et al. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X, et al. Architecture and inherent robustness of a bacterial cell-cycle control system. Proc Natl Acad Sci USA. 2008;105:11340–11345. doi: 10.1073/pnas.0805258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrader JM, et al. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 2014;10:e1004463. doi: 10.1371/journal.pgen.1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, et al. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 2015;11:e1004831. doi: 10.1371/journal.pgen.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nierman WC, et al. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrader JM, et al. Dynamic translation regulation in Caulobacter cell cycle control. Proc Natl Acad Sci USA. 2016;113:E6859–E6867. doi: 10.1073/pnas.1614795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christen B, et al. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528–528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christen M, Deutsch S, Christen B. Genome calligrapher: A web tool for refactoring bacterial genome sequences for de Novo DNA synthesis. ACS Synth Biol. 2015;4:927–934. doi: 10.1021/acssynbio.5b00087. [DOI] [PubMed] [Google Scholar]
- 32.Christen M, Del Medico L, Christen H, Christen B. Genome partitioner: A web tool for multi-level partitioning of large-scale DNA constructs for synthetic biology applications. PLoS One. 2017;12:e0177234. doi: 10.1371/journal.pone.0177234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christen M, Christen B. 2019 Caulobacter ethensis CETH2.0 genome sequence. GenBank. Available at https://www.ncbi.nlm.nih.gov/search/all/?term=CP035535. Deposited January 31, 2019.
- 34.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 35.Noskov VN, et al. Assembly of large, high G+C bacterial DNA fragments in yeast. ACS Synth Biol. 2012;1:267–273. doi: 10.1021/sb3000194. [DOI] [PubMed] [Google Scholar]
- 36.Kimelman A, et al. A vast collection of microbial genes that are toxic to bacteria. Genome Res. 2012;22:802–809. doi: 10.1101/gr.133850.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorek R, et al. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–1452. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 38.Izard J, et al. A synthetic growth switch based on controlled expression of rna polymerase. Mol Syst Biol. 2015;11:840. doi: 10.15252/msb.20156382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata T, et al. Functional overlap between reca and mgsa (rara) in the rescue of stalled replication forks in escherichia coli. Genes Cells. 2005;10:181–191. doi: 10.1111/j.1365-2443.2005.00831.x. [DOI] [PubMed] [Google Scholar]
- 40.Allen G, Kornberg A. Fine balance in the regulation of dnab helicase by dnac protein in replication in escherichia coli. J Biol Chem. 1991;266:22096–22101. [PubMed] [Google Scholar]
- 41.Choi-Rhee E, Cronan JE. The biotin carboxylase-biotin carboxyl carrier protein complex of escherichia coli acetyl-coa carboxylase. J Biol Chem. 2003;278:30806–30812. doi: 10.1074/jbc.M302507200. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harbor Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz G, Vogel J, Wassarman KM. Regulation by small rnas in bacteria: Expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 45.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 46.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul BJ, Ross W, Gaal T, Gourse RL. Rrna transcription in escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 48.Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z. Two independent spiral structures control cell shape in caulobacter. Proc Natl Acad Sci USA. 2005;102:18608–18613. doi: 10.1073/pnas.0507708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Susin MF, Baldini RL, Gueiros-Filho F, Gomes SL. Groes/groel and dnak/dnaj have distinct roles in stress responses and during cell cycle progression in caulobacter crescentus. J Bacteriol. 2006;188:8044–8053. doi: 10.1128/JB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiefer D, Kuhn A. Yidc as an essential and multifunctional component in membrane protein assembly. Int Rev Cytol. 2007;259:113–138. doi: 10.1016/S0074-7696(06)59003-5. [DOI] [PubMed] [Google Scholar]
- 51.McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- 52.McAdams HH, Shapiro L. System-level design of bacterial cell cycle control. FEBS Lett. 2009;583:3991. doi: 10.1016/j.febslet.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lasker K, Mann TH, Shapiro L. An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr Opin Microbiol. 2016;33:131–139. doi: 10.1016/j.mib.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skerker JM, Laub MT. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat Rev Microbiol. 2004;2:325–337. doi: 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- 55.Danchin A, Fang G. Unknown unknowns: Essential genes in quest for function. Microb Biotechnol. 2016;9:530–540. doi: 10.1111/1751-7915.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chevance FFV, Hughes KT. Case for the genetic code as a triplet of triplets. Proc Natl Acad Sci USA. 2017;114:4745–4750. doi: 10.1073/pnas.1614896114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.