Significance
Our understanding of the factors that affected the diversification of passerines, the most diverse and widespread bird order (Passeriformes), is limited. Here, we reconstruct passerine evolutionary history and produce the most comprehensive time-calibrated phylogenetic hypothesis of the group using extensive sampling of the genome, complete sampling of all passerine families, and a number of vetted fossil calibration points. Our phylogenetic results refine our knowledge of passerine diversity and yield divergence dates that are consistent with the fossil record, and our macroevolutionary analyses suggest that singular events in Earth history, such as increases in Cenozoic global temperature or the colonization of new continents, were not the primary forces driving passerine diversification.
Keywords: Passeriformes, diversification, macroevolution, climate, biogeography
Abstract
Avian diversification has been influenced by global climate change, plate tectonic movements, and mass extinction events. However, the impact of these factors on the diversification of the hyperdiverse perching birds (passerines) is unclear because family level relationships are unresolved and the timing of splitting events among lineages is uncertain. We analyzed DNA data from 4,060 nuclear loci and 137 passerine families using concatenation and coalescent approaches to infer a comprehensive phylogenetic hypothesis that clarifies relationships among all passerine families. Then, we calibrated this phylogeny using 13 fossils to examine the effects of different events in Earth history on the timing and rate of passerine diversification. Our analyses reconcile passerine diversification with the fossil and geological records; suggest that passerines originated on the Australian landmass ∼47 Ma; and show that subsequent dispersal and diversification of passerines was affected by a number of climatological and geological events, such as Oligocene glaciation and inundation of the New Zealand landmass. Although passerine diversification rates fluctuated throughout the Cenozoic, we find no link between the rate of passerine diversification and Cenozoic global temperature, and our analyses show that the increases in passerine diversification rate we observe are disconnected from the colonization of new continents. Taken together, these results suggest more complex mechanisms than temperature change or ecological opportunity have controlled macroscale patterns of passerine speciation.
The diversification of modern birds (Neornithes) was shaped by numerous factors, including a mass extinction event (1–3), shifts in connectivity between continents (4, 5), and changes in global climate (4, 6). Specific ecological, geological, and climatological events proposed to be associated with the diversification and global distribution of Neornithes include opening of ecological niches following the Cretaceous-Paleogene (K-Pg) mass extinction event (2, 3, 7, 8), establishment of dispersal corridors linking the geographic origin of modern birds to other landmasses during the Paleogene (4), rapid continental drift and island formation in Wallacea allowing the dispersal of songbirds out of Australia (5, 9), fragmentation of tropical habitats during cooling events of the Late Cretaceous and Cenozoic (4, 10), and expansion of temperate habitats and retreat of glaciers during the Miocene (6). Although we are beginning to understand how Earth history affected the early diversification of modern birds, we understand much less about the macroevolutionary factors that affected diversification within specific major avian subclades, including the hyperdiverse passerines (order Passeriformes).
Passerines comprise more than 6,000 extant species, representing more than 60% of extant avian diversity. During the past two decades, new data have elucidated the diversity of (11, 12) and relationships among (5, 9, 13–17) a number of major passerine lineages (summarized in ref. 18), resulting in the expansion of the number of recognized passerine families from 46 to 137 (11, 12). However, our understanding of the relationships among these major subclades remains incomplete. Numerous apparently rapid divergence events within passerines have hindered reconstruction of their evolutionary interrelationships when sampling few loci (<10) (4, 9, 14, 15, 19) or few taxa (<70% of passerine families) (5, 17, 20).
Knowledge of the time frame of passerine evolution has changed substantially in recent years as well. Historically, because of a dearth of reliably vetted fossils, the age of crown passerines was calibrated based on the geological separation of New Zealand from the rest of Gondwana ∼82 Ma (9, 14, 15). This Late Cretaceous calibration for crown passerines not only predates the earliest fossils that may belong to stem passerines (21) by 27 Ma but also exceeds the age of the earliest known crown bird fossil (22) by 15 Ma. Subsequent studies frequently used this age estimate, or associated secondary calibrations, entrenching major discrepancies between the inferred timing of evolutionary events and the fossil record (23–25). More recently, studies that used fossil calibration-based divergence time analyses (2–4, 19, 26) or combined fossils with biogeographic calibrations (27) estimated a much younger Eocene origin for crown passerines, bringing age estimates closer in line with the fossil record. However, the timing of diversification across passerine families has remained unclear because of sparse sampling (<25% of passerine families) (2, 3) or because analyses have been performed using phylogenetic hypotheses with considerable uncertainty about the placement of passerine lineages (4, 19, 26, 27). Moreover, the number of fossil calibrations within passerines has often been small (fewer than five) (2–4) or the choice of calibration points has been questionable (19, 26, 27), because many of the fossils used had ambiguous phylogenetic affinities or incompletely vetted phylogenetic placement, stratigraphic position, or priors in light of best practices for calibration justification (28).
Uncertainty regarding the relationships among passerine families and the timing of passerine evolution has prevented robust tests of hypotheses explaining the roles of geology, climate, and ecology in diversification of this clade. For example, precise age estimates are critical for determining the extent to which passerine diversification was affected by major climatological events during the Paleogene (29–32) or more gradual global climate shifts throughout the Cenozoic (4). Similarly, robust and taxonomically well-sampled phylogenetic hypotheses are critical for accurately estimating shifts in diversification rates (33). Finally, a combination of better age estimates and more accurate phylogeny is important for examining various historical biogeographic hypotheses in light of geological events (34–36).
Here, we analyze genome-scale DNA sequence data from thousands of nuclear loci to infer a phylogenetic hypothesis for all 137 currently recognized passerine families (12). Based on this phylogenetic hypothesis and the largest set of vetted (28) passerine fossil calibrations used to date, we estimate a time frame for passerine diversification using a variety of analytical approaches. We then use our time-calibrated phylogeny and fossil data to investigate passerine biogeography and diversification rates, as well as the influence of major events in Earth history on the evolution of passerines.
Results and Discussion
Data Characteristics.
By collecting new sequence data and integrating them with data from other studies, we assembled a set of 4,060 ultraconserved element (UCE) loci from 221 individuals representing all passerine families and closely related outgroup families of parrots, falcons, and seriemas (Dataset S1). Our sampling included the extinct Hawaiian honeyeaters (Mohoidae), as well as the recently established monotypic family of the spotted elachura (Elachuridae) (37). The concatenated alignment consisted of 2,464,926 nucleotide sites, 811,688 of which were parsimony-informative, and the data matrix contained 7.3% missing characters. Individual alignments of trimmed UCE loci averaged 607 bp in length (range: 155–1,410 bp) and contained an average of 200 parsimony-informative sites (range: 2–631 sites).
Phylogeny.
Concatenation and coalescent analyses, supplemented by analyses of subsets of the data matrix (Materials and Methods), produced highly resolved (bootstrap support >70% for 95% of 220 nodes) and largely congruent estimates of phylogenetic relationships among passerine families, with only 3–4% of nodes exhibiting conflict between the coalescent and concatenation results (Figs. 1 and 2, SI Appendix, Figs. S1–S5, and Dataset S2). Our analyses resolved three well-established main clades of passerines (9, 13–15) (Figs. 1 and 2): the New Zealand wrens (Acanthisittidae), the suboscines (Tyranni), and the oscines (Passeri). Our results also confirmed the main subdivisions among suboscines (38, 39): the Old World suboscines (Eurylaimides), the ovenbirds and allies (Furnariida), and the tyrant flycatchers and allies (Tyrannida). Our improved sampling allowed us to delineate membership of the oscine infraorders Corvides and Passerides more robustly than in earlier work (5). At a finer scale, our results resolved relationships in a number of clades that have been difficult to place and support the recognition of one family of enigmatic Afrotropical species and five other families recognized in some taxonomic checklists (40, 41). A more complete description of these and other phylogenetic results is provided in SI Appendix.
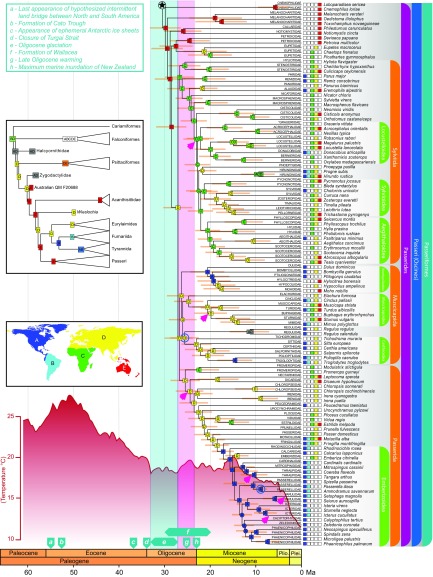
Fig. 1.
Family-level phylogenetic relationships in passerines reconciled from concatenation and coalescent analyses (connects to top of Fig. 2 at the circled star). Maximum likelihood bootstrap support (BS) values are indicated by boxes (BS > 70) or circles (BS < 70) at nodes. Node ages were estimated using 13 fossil calibrations in BEAST on nodes indicated by empty blue circles; 95% credible intervals are shown with orange bars. Ancestral areas were estimated under the DEC + j model using BioGeoBEARS from the distribution of clades represented by each tip [shown by boxes at tips coded according to the map (Inset)]. Light blue and pink bars indicate Oligocene glaciation and warming events, respectively. The estimate of Cenozoic global surface temperatures (red curve) was taken from ref. 99. Branches with the strongest support for diversification rate shifts are indicated by pink arrows in their descendant node for internal branches or at the base of the branch for terminal branches. Geological and climatic events are indicated above the timeline with descriptions provided in the key (Inset). Plei., Pleistocene; Plio., Pliocene.
Fig. 2.
Family-level phylogenetic relationships in passerines reconciled from concatenation and coalescent analyses (connects to bottom of Fig. 1 at the circled star). Biogeographic reconstruction including fossil taxa (Inset, tree) yields identical ancestral areas for crown lineages of passerines, suboscines, and oscines (also SI Appendix, Fig. S8). Plei., Pleistocene; Plio., Pliocene.
Time Frame of Passerine Diversification.
Our comprehensive taxonomic sampling allowed us to estimate the timing of origin for all passerine families. The dating analyses (Figs. 1 and 2) suggest that crown passerines began to diversify during the Middle Eocene (∼47 Ma) and suboscines and oscines diverged ∼44 Ma. We inferred Late Eocene origins of crown suboscines (∼39 Ma) and oscines (∼38 Ma). These dates are approximately half the age of previously published estimates based on the separation of New Zealand from the rest of Gondwana (14, 15, 42) but are much closer to more recent estimates that used fossil calibrations or fossil-derived secondary calibrations (2–5, 19, 20, 26) (Fig. 3 and SI Appendix, Fig. S6). Although the extent of taxon sampling has varied among previous studies, the credible intervals of our date estimates within passerines generally had broader overlap with the earlier estimates of Prum et al. (3) and Selvatti et al. (19) than with those of Claramunt and Cracraft (4) (5–10 Ma older) or Moyle et al. (5) (2–7 Ma younger).
Fig. 3.
Comparison of date estimates of key nodes in passerine phylogeny across three fossil calibration schemes (sets A–C; a detailed description is provided in Materials and Methods) using log-normally distributed priors. Bars represent 95% credible intervals of joint priors [Markov chain Monte Carlo (MCMC) without data] and posteriors of random samples of 25 loci. Color boxes in the heading for each node indicate the set(s) of calibrations in which the node was used as a calibration point. Date estimates from previous studies (3–5, 19) for the same node are also shown, if available.
Our divergence time estimates are generally consistent (overlapping 95% credible intervals) across randomly sampled datasets and are robust to the choice of prior distribution (log-normal or uniform), maximum age constraint, loci sampled (random versus most clock-like), and statistical methodology (BEAST versus MCMCTree; Fig. 3 and SI Appendix, Figs. S6 and S7). Posterior distributions of age estimates for passerine nodes were distinguishable from joint prior distributions (Fig. 3 and SI Appendix, Figs. S6 and S7), suggesting that our age estimates were driven by data rather than priors alone, and the mean of the coefficient of variation of clock rates among lineages ranged from 0.28 to 0.44 across analyses, with 95% credible interval limits as low as 0.24 and as high as 0.48, suggesting that our choice of a relaxed clock model was appropriate. This level of rate heterogeneity across the tree is lower than those reported in recent avian phylogenomic studies that sampled more broadly across extant birds (4, 43). Removal of some or most fossil calibrations did not significantly change divergence time estimates at uncalibrated nodes but had mixed effects on the estimated ages of calibrated nodes (Fig. 3 and SI Appendix, Fig. S6). For example, removing fossil calibrations for Falconidae and Acanthisittidae resulted in younger estimates for each calibrated node, while the removal of the fossil calibrations for Menuridae-Atrichornithidae and Strigopidae resulted in slightly older estimates.
Biogeography.
Diversification history of passerines.
Biogeographic reconstructions under both the dispersal-extinction-cladogenesis (DEC) and DEC + j models suggest that the ancestral area of crown passerines was the Australo-Pacific region (Fig. 1). These results are robust to the signal from passerine and outgroup fossils from Europe and North America (7): In iterative analyses exploring six possible topologies created by grafting four fossil taxa onto our tree, the DEC + j model continued to yield an Australo-Pacific origin of crown passerines (SI Appendix, Fig. S8). Reconstructions using the DEC model were more sensitive to the phylogenetic position of fossil taxa (SI Appendix, Fig. S9). These biogeographic results are consistent with previous hypotheses suggesting that passerines originated in the Southern Hemisphere (9, 15, 19, 44, 45), as well as with the discovery of the oldest putative passerine fossils from the early Eocene of Australia (21). Our results contrast with one analysis (4) suggesting that crown passerines originated in South America ∼53 Ma. The difference among studies could be attributed to methodological differences [parsimony versus maximum likelihood (ML)], the included fossil and outgroup taxa, or alternative codings of the geographic regions. Choice of method seems to have played a large role, because the alternative ML analysis of Claramunt and Cracraft (figure S3A of ref. 4) also showed the highest probability for an Australian origin of crown passerines. Another interpretation of the differences between the two studies is simply that these results reflect the difficulty of pinpointing where passerines originated, because South America, Antarctica, and Australia were connected during the Paleocene and Eocene, although exact details of these connections are debated (46, 47). It is also important to consider that biogeographic reconstructions can be biased by model choice (48), better sampling of fossils in the Northern Hemisphere compared with the Southern Hemisphere (49), and extinction (50). Thus, we suggest that biogeographic reconstructions at the deeper nodes of passerine phylogeny be interpreted cautiously and updated as new information from the fossil record comes to light.
Diversification history of crown suboscines.
Biogeographic analyses under the DEC + j model suggest a Eurasian origin of crown suboscines (Tyranni; Fig. 1) and a North/Central American origin of crown New World suboscines (Tyrannides; Fig. 1), regardless of the placement of fossil taxa (SI Appendix, Fig. S8). Reconstructions using the DEC model were equivocal (SI Appendix, Fig. S9). The DEC + j results are consistent with a suboscine diversification hypothesis based on paleontological data (7): As the Turgai Strait closed between Asia and Europe (event d, Figs. 1 and 2), nonoscine passerine fossils abruptly appear in Europe during the earliest Oligocene (51), which suggests that their ancestors may have dispersed from Australia to Europe through Asia (7, 52). The Beringian land bridge and the warm temperatures of the Late Eocene (53) could have provided a plausible route for dispersal to North America from Asia. In addition, the passerine fossil record in the Americas is more consistent with suboscine dispersal from North America to South America (7). However, the hypothesis of a Eurasian origin of crown suboscines hinges on two critical assumptions: (i) transoceanic dispersal of suboscine ancestors from Australia to Asia and from North America to South America during the Eocene (notably, only a few extant lineages of suboscines are known to disperse over large oceanic expanses) and (ii) a failure of these ancestors to cross the Turgai Strait from Asia to Europe during the same epoch. This hypothesis is an alternative to the idea that crown suboscines originated in South America after trans-Antarctic separation from oscines in Australia (4, 19). Although unsupported by our biogeographic reconstructions, the South American origin hypothesis is compatible with our dating results. Our estimate of the split between oscines and suboscines ∼44 Ma occurs well before the appearance of ephemeral ice sheets in Antarctica during the Late Eocene (54) (event c, Fig. 1), which would have made the trans-Antarctic route implausible. Given the differences among these competing hypotheses, and until additional Paleogene fossils of nonoscine passerines are collected from Asia and South America, arguments regarding the biogeographic origins of crown suboscines remain unresolved.
Origin of Old World suboscines.
The phylogenetic placement of the monotypic Neotropical family Sapayoidae within the Old World suboscines (Eurylaimides) is critical to understanding the origins of Eurylaimides and has been the subject of active research (16, 23, 38, 39, 55, 56). Our phylogenetic analyses place Sapayoidae firmly as sister to Old World pittas (Pittidae), well embedded within the Eurylaimides (Fig. 1 and Dataset S2). Our DEC + j results unequivocally suggest crown Eurylaimides originated in Eurasia (SI Appendix, Fig. S8), whereas our DEC results are again sensitive to the placement of fossil taxa (SI Appendix, Fig. S9). Variance around the point estimates of divergence times suggests the rapid splitting of Eurylaimides lineages among Africa, Eurasia, and the New World potentially occurred during a period of Late Oligocene warming (event g, Fig. 1), as ancestral species took advantage of warmer temperatures and the resulting habitats that connected these continents. Continental connectivity during this period is supported by fossil data that show the movement of mammals from Africa into Eurasia during the Late Oligocene (57, 58) and from Asia into North America during the Oligocene (59). Our phylogenetic results do not support the Atlantogea hypothesis (56), which invokes a Paleogene island chain between Africa and South America and relies on a sister relationship between Sapayoidae and the rest of Eurylaimides. Our estimated divergence date between Eurylaimides and the New World suboscines also conflicts with the use of intermittent land bridges between North and South America by ancestors of Eurylaimides during the Early Paleogene (4) (event a, Fig. 1). Finally, our results are compatible with the hypothesis of Laurasian diversification within Eurylaimides (4, 23), although our time frame differs by as much as 25 Ma (23).
Oligocene glaciation and passerine diversification.
Our passerine chronogram exhibits potential signatures of the Oligocene glaciation (event e, Figs. 1 and 2) on passerine diversification. This extended period of reduced temperature, which started at the Eocene-Oligocene boundary and lasted until the Late Oligocene warming, led to the formation of the first permanent Antarctic ice sheets and a global decline of broadleaf forests (54). The climatic changes that occurred at the beginning of the Oligocene glaciation caused extensive turnover of mammal species in Europe and Asia, known as the “Grande Coupure” and “Mongolian Remodelling” (29, 31), yet the effect of the Oligocene glaciation on Cenozoic avifaunas is poorly known (60). We infer long branches subtending the suboscine clades Eurylaimides, Furnariida, and Tyrannida spanning 38–28 Ma, followed by rapid diversification of these crown clades during a period in which the credible intervals of our estimates overlap the Late Oligocene warming. These observations are consistent with the hypothesis that extinctions during the Oligocene glaciation drastically reduced diversity in each of these mostly tropical bird groups, followed by rapid diversification of each group as temperatures rose again. Another explanation for the long branches leading to crown Eurylaimides, Furnariida, and Tyrannida could be low diversification rates early in the evolutionary history of suboscines, yet the fossil record of suboscines during the Early and Late Oligocene of Europe (7, 61) shows that suboscines were more diverse during the Oligocene than the number of surviving lineages from that period suggests. Although the immigration of mammal predators from Asia to Europe during the Grande Coupure is one alternative to explain the extinction of some avian lineages in Europe during the Paleogene (60), this hypothesis cannot explain apparent suboscine extinctions in the New World. A long branch also leads to crown oscines, but this clade comprised threefold as many surviving lineages compared with suboscines by the Middle Oligocene. The higher survival rate of oscine lineages during the Oligocene glaciation could be tied to a combination of factors, including differences in food preference, habitat availability, and geographic location. The presence of species that occur in temperate and montane habitats among deeply diverging oscine lineages, such as the Menuridae and Ptilonorhynchidae, could also indicate their greater viability in colder temperatures during the Oligocene glaciation.
Oscine diversification out of Australia.
Our biogeographic analyses suggest that the ancestors of two distantly related oscine lineages, the family of rockfowl, rockjumpers, and the rail-babbler (Eupetidae) and the clade formed by Sylviida, Muscicapida, and Passerida, were the first oscines (and the only lineages in Passerides) to reach Eurasia from Australia (Figs. 1 and 2). Our estimate for the arrival of oscines in Eurasia (∼27 Ma) is consistent with the fossil record: No crown passerine fossils have been reported from the Eocene of Europe despite extensive collecting of small bird fossils, whereas oscines appear in Europe ∼24 Ma and are well represented from the Late Oligocene onward (61, 62). Our age estimate for the arrival of oscines in Asia is slightly older than the 23 Ma estimated in a recent oscine study (5). However, our results remain consistent with the idea that uplift in Wallacea occurring 20–30 Ma (63) (event f, Figs. 1 and 2) produced island chains that served as a dispersal corridor for oscines from Australia to Asia (5), and they contrast with the hypothesis of direct dispersal from Australia to Africa during the Eocene (64, 65). We inferred three other independent dispersal events of oscine lineages out of Australia in the Corvides clade, all involving dispersal to Eurasia during the Early Miocene (Fig. 1). These invasions were made by oscines in the family Vireonidae and the superfamilies Corvoidea and Malaconotoidea, with members of the first two clades eventually reaching the New World. Finally, our results suggest two instances of back-colonization to Australia from Eurasia in the superfamily Corvoidea and several reinvasions of Australia from Passerides lineages, but denser sampling of taxa is required to clarify the details of these events.
New Zealand biogeography.
Our age estimates provide insight into how the inundation of the New Zealand landmass during the Oligocene (30, 32) (event h, Figs. 1 and 2) might have affected the endemic parrot and passerine families of New Zealand. Assuming Strigopidae, Acanthisittidae, and the oscine lineages Callaeidae and Notiomystidae are autochthonous, our Oligocene estimate of time of origin for each of these New Zealand endemic families (Figs. 1 and 2) is consistent with the idea that these birds survived the Oligocene drowning event (66, 67). Our estimate of the age of crown passerines ∼47 Ma also suggests that the divergence of the Acanthisittidae from all other passerines was not driven by the formation of the Cato Trough (event b, Fig. 1) between New Zealand and Australia (19). In contrast to these more general results for New Zealand passerine families, our age estimates suggest that the New Zealand-endemic mohouas (Mohouidae) in Corvides diverged from the sittellas (Neosittidae) of Australasia during the Early Miocene ∼20 Ma, a substantially younger age for mohouas than previously estimated (20). This divergence estimate implies that mohoua ancestors dispersed to New Zealand following the Oligocene drowning event. It is also notable that the split of Callaeidae and Notiomystidae from other oscines in Passerides has a credible interval that overlaps with the divergence of Strigopidae from all other parrots during the Oligocene 29–31 Ma (Figs. 1 and 2). The coincidence of these splits is likely random, arising from events like over-water dispersal, because no geological evidence supports the existence of a land bridge connecting Australia and New Zealand 29–31 Ma.
Rates of Passerine Diversification.
Diversification rate and global temperature.
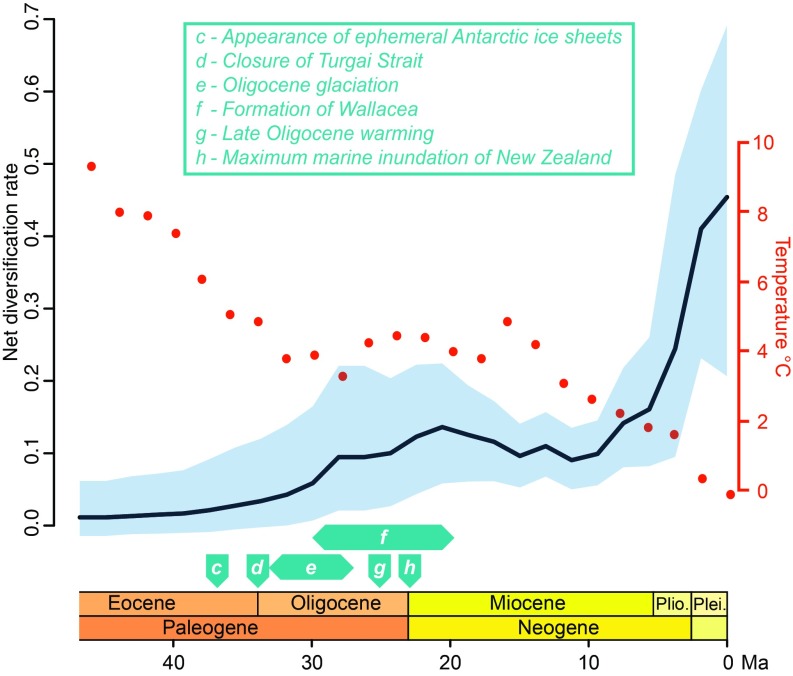
Previous research suggested an inverse relationship between global temperature during the Cenozoic and the diversification rate of modern birds (4). To determine the strength of this relationship in passerines, we estimated episodic diversification rates [i.e., one overall rate for each 2-million-y period of the passerine tree]. Passerine net diversification rates increased slightly during the Middle Oligocene to the Early Miocene; decreased during the Middle Miocene; and rose sharply during the Late Miocene, Pliocene, and Pleistocene (Fig. 4). The sharp rise in diversification rate from the Late Miocene onward should be interpreted with caution: Although our method accounted for missing taxa, rate estimates during this period are based on few estimated splits along our backbone tree. Despite these rate fluctuations, we did not find strong support for an inverse relationship between passerine diversification rate and global temperature. The probability of a negative correlation between the logarithm of speciation rate and global temperature change was only 0.56, and support for the correlation was negligible (68) [Bayes factor (BF) = 1.46]. Support for this correlation remained negligible (BF = 1.00) when we considered slightly longer episodes of 5 million y, as in the estimates of Claramunt and Cracraft (4), or weak (BF = 3.54) when we used a tree dated with uniform priors (in which some nodes were 1–6 Ma older than those estimated using log-normally distributed priors). Taken together, these results suggest that the inverse relationship between global temperature and diversification rate may be strong across Neornithes generally (4) but is not evident among passerines.
Fig. 4.
Estimate of episodic diversification rate of passerines (solid line) and 95% credible interval (light blue band) based on RevBayes analysis of our chronogram (Figs. 1 and 2) across time periods of 2 million y. Global deep ocean temperature data used in the analysis (dotted red line) were taken from ref. 99. Geological and climatic events from Figs. 1 and 2 (excluding events a and b) are indicated above the time line with descriptions provided in the key (Inset). Plei., Pleistocene; Plio., Pliocene.
Diversification rate shifts.
Global correlations of diversification with climate assume a uniform response among lineages, an assumption likely to be violated in an ecologically heterogeneous and rapidly diversifying lineage such as passerines. To test for diversification rate shifts among passerine lineages, we conducted branch-specific diversification rate analyses using our well-resolved and taxonomically well-sampled phylogeny. These analyses yielded strong evidence of multiple higher level rate shifts across the passerine tree (Figs. 1 and 2 and SI Appendix, Fig. S10 and Table S1). The posterior distribution of the number of rate shifts had a mode of 19, ranging from a minimum of 14 to a maximum of 25. The majority of rate shifts were rate increases in specific lineages relative to the passerine background. The three clades with the highest support for a rate shift belong to Muscicapoidea, Sylviida, and Passerida (SI Appendix, Fig. S10 and Table S1), roughly corresponding to oscine radiations originally recognized by DNA/DNA hybridization data (13). Additional clades with strong evidence of rate increases include the New World oscine families Thraupidae and Parulidae, the Neotropical suboscine family Thamnophilidae, and the globally distributed Corvidae. The largest shift in diversification rate occurred in the Parulidae, with a more than sixfold net increase relative to the passerine background. The next two largest rate shifts (4.6- and 4.2-fold the background net diversification rate) were in the same higher clade (the Emberizoidea, a subclade of the Passerida), whereas the remaining shifts ranged from approximately two- to fourfold the background rate and were spread across the passerine tree. Nine of the 14 clades with the highest support for rate shifts were represented by at least two tips on our phylogeny, and all were well supported by both concatenation and coalescent approaches to topology estimation. None of the 14 clades exhibiting rate shifts appear to coincide with range shifts estimated from our biogeographic analyses (Figs. 1 and 2), suggesting that the rate increases we observed were not associated with colonization of new landmasses. Three rate shifts appear to have occurred almost simultaneously during the Oligocene-Miocene transition in three different oscine clades on three different continents (Figs. 1 and 2): near the base of Corvides, within Sylviida, and near the base of Passerida. Together, these clades comprise 3,396 extant species, roughly 56% of extant passerine diversity. The coincidence of these rate shifts suggests a relationship between the climatic and ecological transition of the Oligocene-Miocene boundary and oscine diversification. The expansion of open areas, especially grasslands, during the Miocene (69) would have created a suitable habitat for ancestors of these groups while simultaneously fragmenting forest areas and creating new edge-habitats around forest fragments. Both novel habitats and fragmentation of old habitats could have stimulated speciation events.
A large macroevolutionary study (6) reconstructed diversification rates across a synthetic tree of avian relationships and found at least some evidence for 14 rate shifts within the passerine portion of the phylogeny. We corroborated only five of these 14 rate shifts, including two that were strongly supported (shift detected in >50% of their sampled trees) and three that were weakly supported (shift detected in 25–50% of their sampled trees) in their analyses (SI Appendix, Table S1). Importantly, nine of our 14 (65%) best-supported rate shifts were not detected in previous analyses. The two rate shifts we corroborated that had strong support in previous analyses occurred (i) at the branch leading to the Furnariidae and Dendrocolaptidae (node N in figure 2 of ref. 6) and (ii) at the base of the Passerida (node X in figure 2 of ref. 6). Our analysis differed slightly regarding the location of the rate shift within Passerida, placing the shift closer to the initial divergence in crown Passerida than in the analyses of Jetz et al. (6) (SI Appendix, Fig. S10). It is likely that both analyses are detecting the same event, although our results suggest the rate shift occurred several million years earlier. Given the biology of the included taxa, this rate shift could be associated with adaptation to more carbohydrate-rich diets. The only other rate shift within passerines strongly supported by Jetz et al. (6), at the base of the Timaliidae (inclusive of the zosteropids; node T in figure 2 of ref. 6), was not supported by our analyses (SI Appendix, Fig. S10). It is likely that we were unable to recover the rate shift identified by Jetz et al. (6) in Zosteropidae (node T in figure 2 of ref. 6) because we sampled only one representative of the family, but we found no evidence for the remaining eight passerine rate shifts suggested in that study. Although our tree contains fewer sequenced taxa than the tree of Jetz et al. (6), our extensive sampling of loci and sites likely allowed us to infer relationships more consistently across analyses with higher support and to more accurately estimate branch lengths. Consequently, our analyses offer an updated perspective on lineage-specific patterns of diversification rate in passerines, although some of the differences between the two studies can be attributed to methodological approach (SI Appendix, Table S2). We expect that comprehensive reanalyses of avian relationships and diversification using similar approaches will yield additional insights into the evolution of all birds, although we recognize that future work should examine how diversification rate shifts could be confounded by correlated shifts in diversification and molecular clock rates (70) or extinction dynamics (50).
The phylogenetic hypothesis of passerine relationships and the time line we estimate for passerine diversification reconcile the evolutionary history of this group with paleontological, geological, and climatic data. We find that passerine diversification is driven by dynamics that are more complex than simply Cenozoic temperature change or ecological opportunity associated with the colonization of new landmasses. Still, the drivers of diversification within this group remain incompletely understood. Denser sampling of passerine lineages, combined with improved sampling of the passerine fossil record (particularly outside of Europe and North America), is needed to refine our estimates of lineage-splitting events and tease apart those macro- and microevolutionary factors that were responsible for the diversification of this extraordinary group.
Materials and Methods
Sampling Design.
For this study, we used phylogenetic analysis of 4,060 UCEs to infer a time-calibrated phylogenetic hypothesis of 209 passerine lineages and 12 outgroup lineages. Specifically, we used targeted enrichment of UCEs (71) to collect new sequence data from 113 passerines (Dataset S1), and we combined these with existing UCE sequence data collected from 104 passerine lineages as part of earlier studies (5, 72, 73). We also included UCE loci harvested, in silico, from four publicly available genomes (74). To identify and collect data in silico, we followed an established protocol (71, 75) to align the bait set of 5,060 UCE loci to each genome and extract matching loci plus or minus 500 bp of flanking sequence. We integrated previously collected and in silico data into the analysis pipeline during the data assembly steps. Full details of laboratory methods, data assembly steps, topology estimation procedures, divergence time estimation approaches, biogeographic methods, and diversification rate analyses are provided in SI Appendix, Extended Materials and Methods.
Library Preparation and Sequencing.
We extracted DNA from tissues of 113 vouchered museum specimens (Dataset S1) using the Qiagen DNeasy Blood and Tissue Kit, and we prepared sequencing libraries for the Illumina platform using a commercial kit (Kapa Biosystems, Inc.), 1/2 reactions, and dual indexes (76). Before sequencing, we enriched pools of sequencing libraries for a set of UCE loci using commercially synthesized baits targeting 5,060 loci (Mycroarray MYbaits Kit for Tetrapods UCE 5K, version 1). After enrichment, we used 18 cycles of PCR to recover enriched loci, we measured fragment size of libraries using an Agilent 2100 Bioanalyzer, and we quantified final libraries using an Illumina Eco qPCR System with a commercial quantification kit (Kapa Biosystems, Inc.). We sequenced enriched libraries using a paired-end run of 300 cycles (150 bp in each read direction) on an Illumina HiSeq 3000 System at the Oklahoma Medical Research Facility.
Data Assembly.
We trimmed the raw fastq data from each library using illumiprocessor, version 2 (https://github.com/faircloth-lab/illumiprocessor). We assembled reads from tissues using Trinity, version trinityrnaseq-r2013-02-25 (77), and we assembled reads from toepads using Spades, version 3.9.0 (78). After assembly, we grouped new assemblies with those from previous work and the in silico data collected from genomes (Dataset S1), and we performed the remaining data preparation steps using PHYLUCE (79) following a standard protocol (80). These steps produced trimmed alignments of UCE loci where all loci included data from at least 80% of all 221 taxa.
Topology Estimation.
We used concatenation and coalescent approaches to estimate phylogenetic relationships among passerines. First, we concatenated the trimmed alignments and performed ML inference using ExaML, version 3.0.15 (81), assuming a general time-reversible model of rate substitution and gamma-distributed rates among sites. We evaluated node support using 100 bootstrap replicates, and we tested for convergence of bootstrap replicates a posteriori using the “autoMRE” option in RAxML, version 8.2.8 (82). Second, we used four coalescent programs to estimate species trees: (i) SVDquartets (83, 84); (ii) ASTRID, version 1.3 (85); (iii) ASTRAL-II, version 4.10.11 (86); and (iv) STEAC (phybase version 1.4) (87). We evaluated nodal support for ASTRID, ASTRAL, and STEAC by generating 100 multilocus bootstrap replicates (88), and we estimated nodal support for SVDquartets trees using the same 100 bootstrap replicates we generated for ML analysis.
We observed conflicts among species trees produced by the two analytical paradigms (concatenation and coalescent) for the placement of 27 taxa (SI Appendix, Figs. S1–S5 and Dataset S2). To investigate this issue, we performed additional ASTRID, ASTRAL, and STEAC analyses of subsets of the data matrix (Datasets S2 and S3). These analyses eliminated most of the highly supported inconsistencies across methods; only five taxa had highly supported conflicting placements following these analyses (SI Appendix, Fig. S1). For these remaining inconsistencies, we adopted the ML topology, unless coalescent approaches provided a consistent contradicting topology, which was observed only in the placement of Peltops and Calyptophilus (SI Appendix, Figs. S2–S5). We used this “reconciled” topology for subsequent divergence time, biogeographic, and diversification analyses.
Divergence Time Estimation.
Fossil calibrations.
We followed best practices for justifying fossil calibrations (28) to select and assign nine passerine and four nonpasserine fossils to specific nodes of our reconciled topology (SI Appendix). To examine the sensitivity of our date estimates to the inclusion or exclusion of particular calibration points, we performed analyses with three sets of fossil calibrations: set A, which included all 13 fossils; set B, which included a calibration at the root of the phylogeny, one at the split of passerines with parrots, and five others at key passerine nodes; and set C, which included a single calibration at the root (Fig. 3 and SI Appendix). We also investigated the effects of prior distribution choice on our date estimates by performing dating analyses using two prior settings for each set of calibration points (an uninformative, uniform prior and a log-normal prior), and we examined the robustness of age estimates for deeper nodes by performing one analysis using an excessively old maximum age (80 Ma) with the log-normal prior. We used dating results from analyses of log-normally distributed priors with a maximum age constraint of 56 Ma in subsequent biogeographic and diversification analyses because, based on theoretical and empirical considerations, the uncertainty regarding clade age is represented better by a continuously decreasing probability function rather than discrete upper bounds (4, 89).
Approach.
After selecting calibration points and defining priors, we took two analytical approaches to divergence time estimation: (i) We analyzed small, random subsamples of loci using BEAST, version 1.8.4 (90), and a more complex model of sequence evolution, and (ii) we analyzed the concatenated dataset using MCMCTree (PAML, version 4.8 package) (91) and a simpler model of sequence evolution. For both approaches, we fixed the tree topology to the reconciled topology between ML and coalescent analyses described above. Additionally, to ensure that among-lineage rate variation was not affecting our date estimates, we analyzed a subsample of the most clock-like UCE loci identified using SortaDate (92) and input to BEAST. We checked each run for convergence of parameter values and age estimates by inspecting traces and effective sample sizes in Tracer, version 1.6.0 (93), and we compared divergence time estimates between the two approaches by comparing them with each other and with other studies (3–5, 19), using R, version 3.3.2, and the R package ggplot2, version 2.2.1. We also generated joint prior distributions for each analytical approach and each set of fossil calibrations by running the analyses with no data (94).
Biogeographic Analysis.
We examined broad patterns of avian dispersal across the following major landmasses: North and Central America and the Caribbean (A, referred to as North/Central America); South America (B); Africa and Madagascar (C, referred to as Africa); the Palearctic and Indomalaya east to Wallace’s line (D, referred to as Eurasia); and Wallacea, Australia, New Guinea, New Zealand, and the Pacific (E, referred to as the Australo-Pacific). Using the R package BioGeoBEARS, version 0.2.1 (95), we compared ancestral area estimates using likelihood versions of the DEC model (96, 97) and the DEC + j model (98) under two scenarios (SI Appendix, Table S3): one that allowed dispersal between all areas and another that limited dispersal to movement between adjacent areas. We also performed a series of biogeographic analyses that included four fossil taxa to examine the effects of including closely related fossil taxa on ancestral area estimates of major passerine lineages (SI Appendix, Figs. S8 and S9).
Diversification Rate Analysis.
Because we were interested in understanding whether passerine diversification was correlated with Cenozoic global temperature (99), we estimated episodic diversification rates (100, 101) as implemented in RevBayes, version 1.0.3 (102), using an empirical taxon sampling strategy (103). We employed a reverse-jump Markov chain Monte Carlo analysis between a model in which changes in the logarithm of speciation (and extinction) rates were proportional to changes in global temperature (103) and a model that did not include global temperature as a factor. We assessed model support using BFs (103).
To examine diversification rate shift configurations within passerines, we generated 100 trees with diversities matching the number of extant passerines (6,054 species) using code in R, version 3.3.2 (104), by replacing each tip in our reconciled dated tree with a randomly generated phylogeny of taxon size equal to the species diversity of the sampled tip. We estimated lineage-specific diversification rates on each replicate tree using BAMM, version 2.5.0 (105). To assess where shifts were best supported, we estimated branch-specific marginal odds ratios, eliminated branches not found on the original tree, and then calculated the median ratio for each remaining branch across replicates (SI Appendix, Fig. S10).
Data Availability.
Raw sequencing reads are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (NCBI BioProjects PRJNA304409 and PRJNA480834), and UCE nucleotide sequences are accessioned in the NCBI Genbank (Dataset S1). The PHYLUCE computer code used in this study is available from https://github.com/faircloth-lab/phyluce. Other custom computer code, DNA alignments, analysis inputs, and analysis outputs are available from the Dryad Digital Repository (doi:10.5061/dryad.2vd01gr) (106).
Supplementary Material
Acknowledgments
We thank the curators, staff, and field collectors at the institutions listed in Dataset S1 for tissue samples used in this project; without their hard work, this study would not have been possible. We also thank Van Remsen for comments on earlier drafts, and we thank our three reviewers and the editor for their comments, which improved this manuscript. This study was supported by setup funds from Louisiana State University (to B.C.F.) and by funds from the National Science Foundation: Grant DEB-1655624 (to B.C.F. and R.T.B.), Grant DEB-1655736 (to B.T.S., D.T.K., and R.T.C.), Grants DEB-1655559 and DEB-1541312 (to F.K.B.), Grant DEB-1655683 (to R.T.K. and E.L.B.), Grants DEB-1241181 and DEB-1557053 (to R.G.M.), Grant DEB-1146265 (to R.T.B., A.A., R.T.C., and F.H.S.), Grant DEB-1241066 (to J.C.), and Grant DEB-1146423 (to E.P.D.). M.J.B., N.D.W., T.C.G., R.T.B., E.L.B., and B.C.F. were supported by grants from the Smithsonian Grand Challenges Consortia. G.A.B. and L.F.S. were supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (Grants 2012-23852-0 and 56378-0). L.F.S. and A.A. were supported by the Conselho Nacional de Pesquisas (Grants 302291/2015-6 and 306843/2016-1). P.A. was supported by the Swedish Research Foundation (Grant 2015-04402) and Jornvall Foundation. Portions of this research were conducted with high-performance computing resources provided by Louisiana State University (www.hpc.lsu.edu). Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.E.A. is a guest editor invited by the Editorial Board.
Data deposition: Raw sequencing reads and ultraconserved element (UCE) nucleotide sequences are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive and Genbank as part of BioProjects PRJNA304409 and PRJNA480834. NCBI BioSample accession numbers are available in Dataset S1. The PHYLUCE computer code used in this study is available from https://github.com/faircloth-lab/phyluce. Other custom computer code, DNA alignments, analysis inputs, and analysis outputs are available from the Dryad Digital Repository database, datadryad.org/ (doi: 10.5061/dryad.2vd01gr).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813206116/-/DCSupplemental.
References
- 1.Feduccia A. Explosive evolution in tertiary birds and mammals. Science. 1995;267:637–638. doi: 10.1126/science.267.5198.637. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prum RO, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526:569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 4.Claramunt S, Cracraft J. A new time tree reveals Earth history’s imprint on the evolution of modern birds. Sci Adv. 2015;1:e1501005. doi: 10.1126/sciadv.1501005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyle RG, et al. Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat Commun. 2016;7:12709. doi: 10.1038/ncomms12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 7.Mayr G. Avian Evolution. Wiley; Chichester, UK: 2017. [Google Scholar]
- 8.Field DJ, et al. Early evolution of modern birds structured by global forest collapse at the end-Cretaceous mass extinction. Curr Biol. 2018;28:1825–1831.e2. doi: 10.1016/j.cub.2018.04.062. [DOI] [PubMed] [Google Scholar]
- 9.Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci USA. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field DJ, Hsiang AY. A North American stem turaco, and the complex biogeographic history of modern birds. BMC Evol Biol. 2018;18:102. doi: 10.1186/s12862-018-1212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibley CG, Monroe B., Jr . Distribution and Taxonomy of the Birds of the World. Yale Univ Press; New Haven, CT: 1990. [Google Scholar]
- 12.Dickinson E, Christidis L. In: The Howard & Moore Complete Checklist of the Birds of the World. 4th Ed. Dickinson E, Christidis L, editors. Vol 2 Aves Press; Eastbourne, UK: 2014. [Google Scholar]
- 13.Sibley CG, Ahlquist J. Phylogeny and Classification of the Birds of the World. Yale Univ Press; New Haven, CT: 1990. [Google Scholar]
- 14.Barker FK, Barrowclough GF, Groth JG. A phylogenetic hypothesis for passerine birds: Taxonomic and biogeographic implications of an analysis of nuclear DNA sequence data. Proc Biol Sci. 2002;269:295–308. doi: 10.1098/rspb.2001.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericson PGP, et al. A Gondwanan origin of passerine birds supported by DNA sequences of the endemic New Zealand wrens. Proc Biol Sci. 2002;269:235–241. doi: 10.1098/rspb.2001.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 17.Barker FK. Mitogenomic data resolve basal relationships among passeriform and passeridan birds. Mol Phylogenet Evol. 2014;79:313–324. doi: 10.1016/j.ympev.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Cracraft J. Avian higher-level relationships and classification: Passeriforms. In: Dickinson E, Christidis L, editors. The Howard & Moore Complete Checklist of the Birds of the World. 4th Ed. Vol 2. Aves Press; Eastbourne, UK: 2014. pp. xvii–xlv. [Google Scholar]
- 19.Selvatti AP, Gonzaga LP, Russo CADM. A Paleogene origin for crown passerines and the diversification of the Oscines in the New World. Mol Phylogenet Evol. 2015;88:1–15. doi: 10.1016/j.ympev.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Gibb GC, et al. New Zealand passerines help clarify the diversification of major songbird lineages during the Oligocene. Genome Biol Evol. 2015;7:2983–2995. doi: 10.1093/gbe/evv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boles WE. The world’s oldest songbird. Nature. 1995;374:21–22. [Google Scholar]
- 22.Clarke JA, Tambussi CP, Noriega JI, Erickson GM, Ketcham RA. Definitive fossil evidence for the extant avian radiation in the Cretaceous. Nature. 2005;433:305–308. doi: 10.1038/nature03150. [DOI] [PubMed] [Google Scholar]
- 23.Moyle RG, Chesser RT, Prum RO, Schikler P, Cracraft J. Phylogeny and evolutionary history of Old World suboscine birds (Aves: Eurylaimides) Am Mus Novit. 2006;3544:1–22. [Google Scholar]
- 24.Jønsson KA, Fabre P-H, Ricklefs RE, Fjeldså J. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc Natl Acad Sci USA. 2011;108:2328–2333. doi: 10.1073/pnas.1018956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohlson JI, Irestedt M, Ericson PG, Fjeldså J. Phylogeny and classification of the New World suboscines (Aves, Passeriformes) Zootaxa. 2013;3613:1–35. doi: 10.11646/zootaxa.3613.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Hooper DM, Price TD. Chromosomal inversion differences correlate with range overlap in passerine birds. Nat Ecol Evol. 2017;1:1526–1534. doi: 10.1038/s41559-017-0284-6. [DOI] [PubMed] [Google Scholar]
- 27.Price TD, et al. Niche filling slows the diversification of Himalayan songbirds. Nature. 2014;509:222–225. doi: 10.1038/nature13272. [DOI] [PubMed] [Google Scholar]
- 28.Parham JF, et al. Best practices for justifying fossil calibrations. Syst Biol. 2012;61:346–359. doi: 10.1093/sysbio/syr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stehlin H. Remarques sur les faunules de mammifères des couches éocènes et oligocènes du Bassin de Paris. Bulletin de la Société Géologique de France. 1909;9:488–520. French. [Google Scholar]
- 30.Cooper A, Cooper RA. The Oligocene bottleneck and New Zealand biota: Genetic record of a past environmental crisis. Proc Biol Sci. 1995;261:293–302. doi: 10.1098/rspb.1995.0150. [DOI] [PubMed] [Google Scholar]
- 31.Meng J, McKenna MC. Faunal turnovers of Palaeogene mammals from the Mongolian Plateau. Nature. 1998;394:364–367. [Google Scholar]
- 32.Landis CA, et al. The Waipounamu erosion surface: Questioning the antiquity of the New Zealand land surface and terrestrial fauna and flora. Geol Mag. 2008;145:173–197. [Google Scholar]
- 33.Title PO, Rabosky DL. Do macrophylogenies yield stable macroevolutionary inferences? An example from squamate reptiles. Syst Biol. 2017;66:843–856. doi: 10.1093/sysbio/syw102. [DOI] [PubMed] [Google Scholar]
- 34.Hoorn C, et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science. 2010;330:927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- 35.Smith BT, et al. The drivers of tropical speciation. Nature. 2014;515:406–409. doi: 10.1038/nature13687. [DOI] [PubMed] [Google Scholar]
- 36.Feng Y-J, et al. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Paleogene boundary. Proc Natl Acad Sci USA. 2017;114:E5864–E5870. doi: 10.1073/pnas.1704632114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alström P, et al. Discovery of a relict lineage and monotypic family of passerine birds. Biol Lett. 2014;10:20131067. doi: 10.1098/rsbl.2013.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fjeldså J, Zuccon D, Irestedt M, Johansson US, Ericson PGP. Sapayoa aenigma: A New World representative of ‘Old World suboscines’. Proc Biol Sci. 2003;270(Suppl 2):S238–S241. doi: 10.1098/rsbl.2003.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesser RT. Molecular systematics of New World suboscine birds. Mol Phylogenet Evol. 2004;32:11–24. doi: 10.1016/j.ympev.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 40.del Hoyo J, Collar N. HBW and BirdLife International Illustrated Checklist of the Birds of the World. Passerines. Vol 2 Lynx Edicions; Barcelona: 2016. [Google Scholar]
- 41.Gill FB, Donsker D. 2018 doi: 10.14344/IOC.ML.8.1. IOC World Bird List (v. 8.1). Available at . . Accessed February 27, 2018. [DOI]
- 42.Ericson PGP, Klopfstein S, Irestedt M, Nguyen JMT, Nylander JAA. Dating the diversification of the major lineages of Passeriformes (Aves) BMC Evol Biol. 2014;14:8. doi: 10.1186/1471-2148-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berv JS, Field DJ. Genomic signature of an avian Lilliput effect across the K-Pg extinction. Syst Biol. 2018;67:1–13. doi: 10.1093/sysbio/syx064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cracraft J. Continental drift, paleoclimatology, and the evolution and biogeography of birds. J Zool. 1973;169:455–545. [Google Scholar]
- 45.Olson SL. Oligocene fossils bearing on the origins of the Todidae and the Momotidae (Aves: Coraciiformes) Smithson Contrib to Paleobiol. 1976;27:111–119. [Google Scholar]
- 46.Li ZX, Powell CM. An outline of the palaeogeographic evolution of the Australasian region since the beginning of the Neoproterozoic. Earth Sci Rev. 2001;53:237–277. [Google Scholar]
- 47.Scher HD, Martin EE. Timing and climatic consequences of the opening of Drake Passage. Science. 2006;312:428–430. doi: 10.1126/science.1120044. [DOI] [PubMed] [Google Scholar]
- 48.Ree RH, Sanmartín I. Conceptual and statistical problems with the DEC+J model of founder-event speciation and its comparison with DEC via model selection. J Biogeogr. 2018;45:741–749. [Google Scholar]
- 49.Cracraft J, Claramunt S. Conceptual and analytical worldviews shape differences about global avian biogeography. J Biogeogr. 2017;44:958–960. [Google Scholar]
- 50.Marshall CR. Five palaeobiological laws needed to understand the evolution of the living biota. Nat Ecol Evol. 2017;1:0165. doi: 10.1038/s41559-017-0165. [DOI] [PubMed] [Google Scholar]
- 51.Mayr G, Manegold A. New specimens of the earliest European passeriform bird. Acta Palaeontol Pol. 2006;51:315–323. [Google Scholar]
- 52.Mayr G. Paleogene Fossil Birds. Springer; Berlin: 2009. [Google Scholar]
- 53.Sanmartín I, Enghoff H, Ronquist F. Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol J Linn Soc Lond. 2001;73:345–390. [Google Scholar]
- 54.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 55.Irestedt M, Ohlson JI, Zuccon D, Källersjö M, Ericson PGP. Nuclear DNA from old collections of avian study skins reveals the evolutionary history of the Old World suboscines (Aves, Passeriformes) Zool Scr. 2006;35:567–580. [Google Scholar]
- 56.Selvatti AP, Galvão A, Pereira AG, Pedreira Gonzaga L, Russo CA. An African origin of the Eurylaimides (Passeriformes) and the successful diversification of the ground-foraging pittas (Pittidae) Mol Biol Evol. 2017;34:483–499. doi: 10.1093/molbev/msw250. [DOI] [PubMed] [Google Scholar]
- 57.Antoine P-O, et al. First record of Paleogene Elephantoidea (Mammalia, Proboscidea) from the Bugti Hills of Pakistan. J Vertebr Paleontol. 2003;23:977–980. [Google Scholar]
- 58.Kappelman J, et al. Oligocene mammals from Ethiopia and faunal exchange between Afro-Arabia and Eurasia. Nature. 2003;426:549–552. doi: 10.1038/nature02102. [DOI] [PubMed] [Google Scholar]
- 59.Samuels JX, Albright LB, Fremd TJ. The last fossil primate in North America, new material of the enigmatic Ekgmowechashala from the Arikareean of Oregon. Am J Phys Anthropol. 2015;158:43–54. doi: 10.1002/ajpa.22769. [DOI] [PubMed] [Google Scholar]
- 60.Mayr G. Two-phase extinction of “Southern Hemispheric” birds in the Cenozoic of Europe and the origin of the Neotropic avifauna. Palaeobiodivers Palaeoenviron. 2011;91:325–333. [Google Scholar]
- 61.Manegold A. Passerine diversity in the late Oligocene of Germany: Earliest evidence for the sympatric coexistence of suboscines and oscines. Ibis. 2008;150:377–387. [Google Scholar]
- 62.Mourer-Chauvire C, Hugueney M, Jonet P. Discovery of passeriformes in the Upper Oligocene of France. C R Acad Sci II. 1989;309:843–849. [Google Scholar]
- 63.Hall R. Southeast Asia’s changing palaeogeography. Blumea J Plant Taxon Plant Geogr. 2009;54:148–161. [Google Scholar]
- 64.Fuchs J, Fjeldså J, Bowie RCK, Voelker G, Pasquet E. The African warbler genus Hyliota as a lost lineage in the Oscine songbird tree: Molecular support for an African origin of the Passerida. Mol Phylogenet Evol. 2006;39:186–197. doi: 10.1016/j.ympev.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 65.Jønsson KA, Fjeldså J. Determining biogeographical patterns of dispersal and diversification in oscine passerine birds in Australia, Southeast Asia and Africa. J Biogeogr. 2006;33:1155–1165. [Google Scholar]
- 66.Mitchell KJ, et al. Ancient mitochondrial genomes clarify the evolutionary history of New Zealand’s enigmatic acanthisittid wrens. Mol Phylogenet Evol. 2016;102:295–304. doi: 10.1016/j.ympev.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 67.Worthy TH, Tennyson AJD, Scofield RP. An early Miocene diversity of parrots (Aves, Strigopidae, Nestorinae) from New Zealand. J Vertebr Paleontol. 2011;31:1102–1116. [Google Scholar]
- 68.Kass B, Raftery A. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 69.Strömberg CAE. Evolution of grasses and grassland ecosystems. Annu Rev Earth Planet Sci. 2011;39:517–544. [Google Scholar]
- 70.Duchêne DA, Hua X, Bromham L. Phylogenetic estimates of diversification rate are affected by molecular rate variation. J Evol Biol. 2017;30:1884–1897. doi: 10.1111/jeb.13148. [DOI] [PubMed] [Google Scholar]
- 71.McCormack JE, et al. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Res. 2012;22:746–754. doi: 10.1101/gr.125864.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manthey JD, Campillo LC, Burns KJ, Moyle RG. Comparison of target-capture and restriction-site associated DNA sequencing for phylogenomics: A test in cardinalid tanagers (Aves, genus: Piranga) Syst Biol. 2016;65:640–650. doi: 10.1093/sysbio/syw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campillo LC, Oliveros CH, Sheldon FH, Moyle RG. Genomic data resolve gene tree discordance in spiderhunters (Nectariniidae, Arachnothera) Mol Phylogenet Evol. 2018;120:151–157. doi: 10.1016/j.ympev.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 74.Zhang G, et al. Avian Genome Consortium Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faircloth BC. 2018 Tutorial III: Harvesting UCE Loci from Genomes. Available at https://phyluce.readthedocs.io/en/latest/tutorial-three.html. Accessed August 6, 2018.
- 76.Glenn TC, et al. 2016. Adapterama I: Universal stubs and primers for thousands of dual-indexed Illumina libraries (iTru & iNext). bioRxiv:10.1101/049114.
- 77.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nurk S, et al. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. Research in Computational Molecular Biology. RECOMB 2013, Lecture Notes in Computer Science, eds Deng M, Jiang R, Sun F, Zhang X (Springer, Berlin), Vol 7821, pp 158–170.
- 79.Faircloth BC. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics. 2016;32:786–788. doi: 10.1093/bioinformatics/btv646. [DOI] [PubMed] [Google Scholar]
- 80.Faircloth BC. 2018 Tutorial I: UCE Phylogenomics. Available at https://phyluce.readthedocs.io/en/latest/tutorial-one.html. Accessed August 6, 2018.
- 81.Kozlov AM, Aberer AJ, Stamatakis A. ExaML version 3: A tool for phylogenomic analyses on supercomputers. Bioinformatics. 2015;31:2577–2579. doi: 10.1093/bioinformatics/btv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chifman J, Kubatko L. Quartet inference from SNP data under the coalescent model. Bioinformatics. 2014;30:3317–3324. doi: 10.1093/bioinformatics/btu530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chifman J, Kubatko L. Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites. J Theor Biol. 2015;374:35–47. doi: 10.1016/j.jtbi.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 85.Vachaspati P, Warnow T. ASTRID: Accurate species TRees from internode distances. BMC Genomics. 2015;16:S3. doi: 10.1186/1471-2164-16-S10-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mirarab S, Warnow T. ASTRAL-II: Coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics. 2015;31:i44–i52. doi: 10.1093/bioinformatics/btv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu L, Yu L, Pearl DK, Edwards SV. Estimating species phylogenies using coalescence times among sequences. Syst Biol. 2009;58:468–477. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- 88.Seo TK. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol Biol Evol. 2008;25:960–971. doi: 10.1093/molbev/msn043. [DOI] [PubMed] [Google Scholar]
- 89.Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- 90.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.dos Reis M, Yang Z. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 2011;28:2161–2172. doi: 10.1093/molbev/msr045. [DOI] [PubMed] [Google Scholar]
- 92.Smith SA, Brown JW, Walker JF. So many genes, so little time: A practical approach to divergence-time estimation in the genomic era. PLoS One. 2018;13:e0197433. doi: 10.1371/journal.pone.0197433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rambaut A, Drummond AJ. 2014 Tracer v.1.6. Available at tree.bio.ed.ac.uk/software/tracer/. Accessed April 25, 2017.
- 94.Brown JW, Smith SA. The past sure is tense: On interpreting phylogenetic divergence time estimates. Syst Biol. 2018;67:340–353. doi: 10.1093/sysbio/syx074. [DOI] [PubMed] [Google Scholar]
- 95.Matzke NJ. 2013. BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R scripts. PhD dissertation (University of California, Berkeley, CA)
- 96.Clark JR, et al. A comparative study in ancestral range reconstruction methods: Retracing the uncertain histories of insular lineages. Syst Biol. 2008;57:693–707. doi: 10.1080/10635150802426473. [DOI] [PubMed] [Google Scholar]
- 97.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 98.Matzke NJ. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst Biol. 2014;63:951–970. doi: 10.1093/sysbio/syu056. [DOI] [PubMed] [Google Scholar]
- 99.Hansen J, Sato M, Russell G, Kharecha P. Climate sensitivity, sea level and atmospheric carbon dioxide. Philos Trans A Math Phys Eng Sci. 2013;371:20120294. doi: 10.1098/rsta.2012.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stadler T. Mammalian phylogeny reveals recent diversification rate shifts. Proc Natl Acad Sci USA. 2011;108:6187–6192. doi: 10.1073/pnas.1016876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Höhna S. The time-dependent reconstructed evolutionary process with a key-role for mass-extinction events. J Theor Biol. 2015;380:321–331. doi: 10.1016/j.jtbi.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 102.Höhna S, et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst Biol. 2016;65:726–736. doi: 10.1093/sysbio/syw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Höhna S, et al. 2017 Statistical phylogenetic inference using RevBayes. Available at https://raw.githubusercontent.com/revbayes/revbayes_tutorial/master/tutorial_TeX/RB_Manual.pdf. Accessed January 16, 2017.
- 104.R Development Core Team 2008 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Version 3.3.2. Available at www.r-project.org. Accessed February 1, 2017.
- 105.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One. 2014;9:e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oliveros CH, et al. 2019 doi: 10.5061/dryad.2vd01gr. Data from: “Earth history and the passerine superradiation.” Dryad Digital Repository. . Deposited March 7, 2019. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (NCBI BioProjects PRJNA304409 and PRJNA480834), and UCE nucleotide sequences are accessioned in the NCBI Genbank (Dataset S1). The PHYLUCE computer code used in this study is available from https://github.com/faircloth-lab/phyluce. Other custom computer code, DNA alignments, analysis inputs, and analysis outputs are available from the Dryad Digital Repository (doi:10.5061/dryad.2vd01gr) (106).