Significance
Biologists now recognize that animal microbiomes have strong impacts not only on the organs with which they associate, but also anatomically remote tissues; however, the precise triggers underlying these impacts remain unknown. Here, using the squid–vibrio light-organ association, which affords unparalleled resolution of a natural binary partnership, we report both near-field (light organ) and far-field (eye and gill) symbiont-driven effects on host gene expression. Colonization by the symbiont results in unique transcriptional signatures for each organ. Furthermore, distinct organ-specific patterns arise over the day/night cycle, and across the host’s developmental trajectory. Most strikingly, the loss of a single genetic locus in the symbiont, that encoding bioluminescence, triggers a dominant and biologically relevant change of gene expression across the host’s body.
Keywords: squid–vibrio, symbiosis, development, daily rhythm, bioluminescence
Abstract
The colonization of an animal’s tissues by its microbial partners creates networks of communication across the host’s body. We used the natural binary light-organ symbiosis between the squid Euprymna scolopes and its luminous bacterial partner, Vibrio fischeri, to define the impact of colonization on transcriptomic networks in the host. A night-active predator, E. scolopes coordinates the bioluminescence of its symbiont with visual cues from the environment to camouflage against moon and starlight. Like mammals, this symbiosis has a complex developmental program and a strong day/night rhythm. We determined how symbiont colonization impacted gene expression in the light organ itself, as well as in two anatomically remote organs: the eye and gill. While the overall transcriptional signature of light organ and gill were more alike, the impact of symbiosis was most pronounced and similar in light organ and eye, both in juvenile and adult animals. Furthermore, the presence of a symbiosis drove daily rhythms of transcription within all three organs. Finally, a single mutation in V. fischeri—specifically, deletion of the lux operon, which abrogates symbiont luminescence—reduced the symbiosis-dependent transcriptome of the light organ by two-thirds. In addition, while the gills responded similarly to light-organ colonization by either the wild-type or mutant, luminescence was required for all of the colonization-associated transcriptional responses in the juvenile eye. This study defines not only the impact of symbiont colonization on the coordination of animal transcriptomes, but also provides insight into how such changes might impact the behavior and ecology of the host.
Recent studies of animal and plant microbiomes have demonstrated that they can have far reaching effects, influencing both the internal and external environments of the host (1, 2). For example, the human microbiota impacts both tissues with which it directly interacts and more remote tissues of the body (3), as well as the built and natural environment in which the human host resides (4, 5). These complex microbial networks profoundly influence host development, from embryogenesis through senescence, while maintaining physiological homeostasis along this trajectory (2).
The best-studied nexus of these complex interactions is the mammalian gut microbiota (6), which affects not only the gut tissues themselves, but also the immune system (7), brain (8), liver (9), heart (10, 11), kidney (12), lung (13, 14), and eye (15–17). The microbiota also helps coordinate the activities of these tissues and organs: for example, the strong association of the gut microbiota with the control of host circadian rhythms (18, 19). Furthermore, axes of influence between the gut and other organs have revealed that dysbiosis of the microbiota is a critical driver of seemingly unrelated diseases (3).
Thus far, the mechanisms underlying these wide-ranging effects remain poorly studied. The integration of the gut microbiota into host biology is reflected in the transcriptomic regulation of genes in tissues both in direct contact with (20–22) and distant from (23, 24) the microbial assemblage. Available data suggest that the metabolomes of the blood, sweat, and urine carry products of the gut microbiota, such as short-chain fatty acids and microbe-associated molecular patterns (25, 26), to which these remote tissues respond. The complexity of the mammalian gut microbiota, however, renders it difficult to investigate the impact of a particular microbe on host biology under natural conditions, because the responses of adjacent and remote host tissues are the result of the cumulative effects of microbe–microbe and host–microbe interactions with hundreds to thousands of microbial phylotypes. In contrast, here we use the binary light-organ symbiosis between the Hawaiian bobtail squid, Euprymna scolopes, and its luminous bacterial partner, Vibrio fischeri (27, 28), to define the impact of a single symbiotic partner on the transcriptomic responses of host tissues, both those housing the symbiont population and those remote from the symbionts.
The squid host acquires its symbiont each generation from the surrounding environment and, similar to the mammalian gut microbiota, the bacteria reside extracellularly along the apical surfaces of epithelium-lined crypts (29). Also analogous to the mammalian gut microbiota, the squid–vibrio symbiosis undergoes significant development and maturation. Within hours following initial colonization of the juvenile animal, the symbionts trigger the regression of superficial ciliated fields of cells that promote light-organ inoculation (28). The symbionts also induce development of the crypt cells with which they directly associate throughout the animal’s life, notably an increase in microvillar density and a swelling of the cells lining the crypts (Fig. 1A). A dark mutant derivative of V. fischeri (Δlux), defective in light production, the principal “currency” of the symbiosis, is also defective in the induction of this latter hallmark event of early light-organ development (30–32).
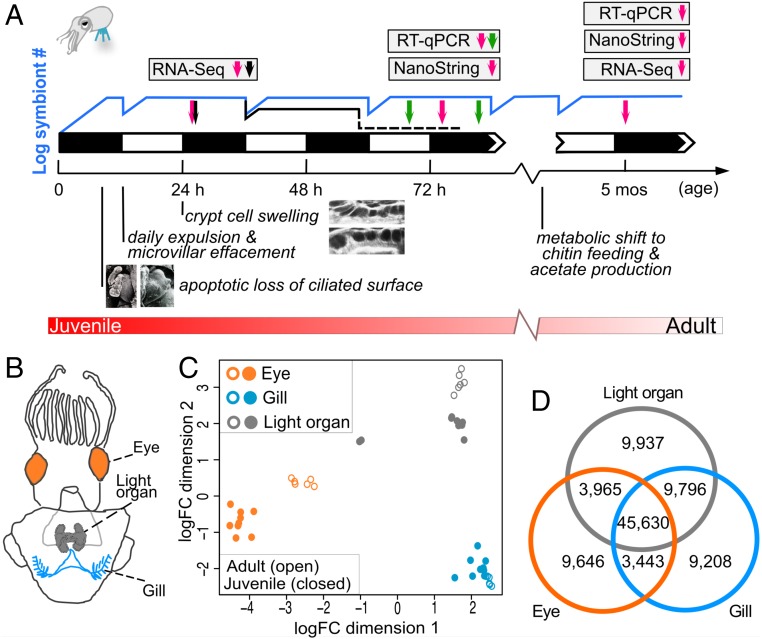
Fig. 1.
Transcriptome profiling of E. scolopes organs. (A) Transcriptome sampling scheme during symbiotic development. On the night (black squares) that they hatch, juvenile squid become inoculated by V. fischeri cells, which proliferate, (blue line), filling the light-organ crypts and producing bioluminescence. Colonization triggers developmental events in the light organ’s tissues, including apoptosis of the surface epithelium (Left Inset, APO; Right Inset, SYM), and edematous swelling of the crypt epithelial cells (Upper Inset, APO; Lower Inset, SYM). Each dawn, the nocturnally active host effaces the crypt-cell microvilli and expels most of its symbiont population, which grows back up by noon. A dark mutant (Δlux) colonizes normally, but is unable to persist in the organ (dotted black line), and doesn’t induce normal crypt cell swelling. After 1 mo, the host begins providing chitin to the symbionts, which ferment it to acetate. Transcriptomes of light organ, eye, and gill were constructed by RNA-seq or NanoString from organs sampled at 2000 hours (magenta arrows) in APO and SYM hosts of juvenile and adult animals. SYM-dark hosts, colonized by a dark-mutant strain, were also sampled (black arrow). For day/night comparisons, APO and SYM organs were sampled at 1600 and 0400 hours (green arrows) as well. Levels of transcripts of interest were confirmed by NanoString and qRT-PCR. (B) Schematic drawing of E. scolopes indicating tissue types collected from both juvenile and adult squid. (C) Multidimensional scaling plot of gene expression for the E. scolopes reference transcriptome. (D) Venn diagram of the number of shared and specific genes, expressed by tissue type. A gene is considered expressed when FPKM (fragments per kilobase million) > 0.5 in at least two samples per tissue.
Development of the light organ also involves the onset of diel cycles. Beginning during the first day of colonization and thereafter, ∼90% of the symbiont population is vented each day at dawn into the surrounding environment (33). Furthermore, in response to luminous (but not Δlux) symbionts, the organ’s cryptochrome-encoding clock gene, escry1, begins a day/night cycling, and the host concomitantly imposes a cycling of the symbiont’s luminescence levels, which peak in the hours of the early evening (1900–2000 hours), when the nocturnal squid host begins to forage. Then, after 3–4 wk of colonization, the symbiosis undergoes a final maturation, with the onset of a strong daily rhythm of metabolic processes (34, 35), not unlike the circadian rhythms of metabolism described in the mammalian gut symbioses (18, 36). Specifically, the animal becomes fully nocturnal, and the symbiont metabolism begins a day/night fluctuation between respiration and fermentation in response to a change in nutrients provided by the host.
Here we compared the transcriptomes of three highly vascularized organs of the squid host: the light organ itself and the eye and gill, manipulating both their symbiotic state and the genetics of the bacterial partner, in both juvenile and adult animals, and over the day/night cycle (Fig. 1A). The eye was chosen because, like the symbiotic organ, it is a light-sensing organ, and shows convergence in morphology, biochemistry (37), molecular biology (38, 39), and developmental pathways (40). As an immune organ, the gill, like the light organ, responds to bacterial colonization (41). Here, we present evidence that both light-organ colonization and luminescence influence gene regulation of not only symbiotic tissue, but also host organs remote from the symbionts.
Results
De Novo Transcriptome Assembly, Annotation, and Validation Provide the Resources for Analyses of Symbiosis Effects on Host Gene Expression.
To determine the extent to which symbiotic colonization impacts host gene expression, we sequenced transcripts isolated from the squid light organ, eye, and gill (Fig. 1 A and B). Samples were collected for RNA-sequencing (RNA-seq) analysis from both juvenile (24-h posthatch) and mature adult (5-mo-old) animals, under two colonization states: symbiont-free (i.e., aposymbiotic, or APO) or colonized by the wild-type V. fischeri light-organ isolate ES114 (i.e., symbiotic, or SYM). An additional condition that was analyzed in juvenile animals was colonization by an isogenic, nonluminous (Δlux) mutant of ES114 (i.e., SYM-dark) (42) (see SI Appendix, SI Results for details). The 2.2 billion paired-end reads obtained by Illumina sequencing, were de novo assembled to create a reference transcriptome (SI Appendix, Fig. S1 and Dataset S1). The large number of assembled transcripts is a common trait found in other de novo assembled transcriptomes of E. scolopes (43, 44). Cephalopods are known to expand certain gene families (45, 46). This feature, together with the high levels of heterozygosity and transcript editing (47) that are known to challenge assembly software (48), contributed to the high number of observed expressed transcripts. Transcriptomic profiles clustered by tissue type and, within each tissue type, by developmental stage, with a higher degree of variation among juvenile replicates (Fig. 1C and SI Appendix, Fig. S2). Irrespective of developmental stage, eye-derived samples showed the most divergent transcriptional profile. The light organ and gill displayed a more highly correlated expression pattern (Fig. 1C and SI Appendix, Fig. S2A), and shared more total expressed genes than either did with the eye (Fig. 1D), perhaps because they are both predominantly composed of epithelial tissue.
When considering the total number of genes that are expressed in each organ, depending on the host developmental stage and symbiotic state (SI Appendix, Fig. S3A), on average juvenile samples expressed 20% more detectable genes than their adult counterparts. Only 7,464 genes were expressed in all three organs, from both juveniles and adults, and in both the SYM and APO state (SI Appendix, Fig. S3B), suggesting that these genes encode core or “housekeeping” functions (for more details of transcriptomic patterns, see Dataset S2). In contrast, when we determined tissue-specific genes (i.e., those that were expressed at least eightfold higher in one organ relative to the other two), a total of 5,587 genes were identified (SI Appendix, Fig. S2E and Dataset S2). Not surprisingly, gene ontology (GO) terms enriched for the eye were related to visual perception or synaptic signaling, while for the gill were linked to gas exchange or pH regulation; similarly, the light organ was enriched in GO terms related to the expected activities of oxidative stress (49, 50) and chitin-associated processes (51) (SI Appendix, Fig. S2 B–D and Dataset S3). Furthermore, we validated the RNA-seq dataset by two methods, qRT-PCR, and the NanoString nCounter XT platform (SI Appendix, Fig. S4 and Dataset S4), which all had a high degree of congruity. These results provide strong evidence that the transcriptional patterns in response to colonization are robust, and clearly differentiate the three tissue types and their developmental states.
Adult Gene Expression in the Light Organ and Remote Tissues Responded Uniquely to Symbiont Colonization.
To identify whether and how the light organ, eye, and gill responded to colonization of the light organ, we compared the gene-expression patterns of these three organs when sampled from adult APO and SYM squid (Fig. 1A). According to the values for the differentially expressed transcripts, the samples clustered by condition (APO or SYM) within each organ (Fig. 2A). Transcripts having expression levels that differed significantly between APO and SYM were identified as up- or down-regulated by symbiosis (Fig. 2B). Unsurprisingly, the light organ, which harbors the symbionts, had the strongest transcriptional response to its colonization, with a total of 206 genes significantly differentially regulated, which clustered into five distinct expression profiles (Fig. 2 and SI Appendix, Fig. S5). Although they are in anatomically remote organs (Fig. 1B), the transcriptomes of both eye and gill also responded to colonization of the light organ. Because of the greater similarity between the number of total transcripts in the light organ and gill (Fig. 1 C and D and SI Appendix, Fig. S2A), and because both of these organs respond to bacteria as part of their normal function, we anticipated that, compared with the eye, more symbiotically responsive genes would be detected in the gill, and they would overlap more significantly with the light organ. However, the eye had twice as many symbiotically regulated genes as the gill (84 vs. 42) (Fig. 2B, SI Appendix, Fig. S5, and Dataset S5). Furthermore, each organ had a distinctive transcriptional response to light-organ colonization: only one gene (annotated as angiotensin-converting enzyme or ACE) was up-regulated in two of the organs (eye and gill).
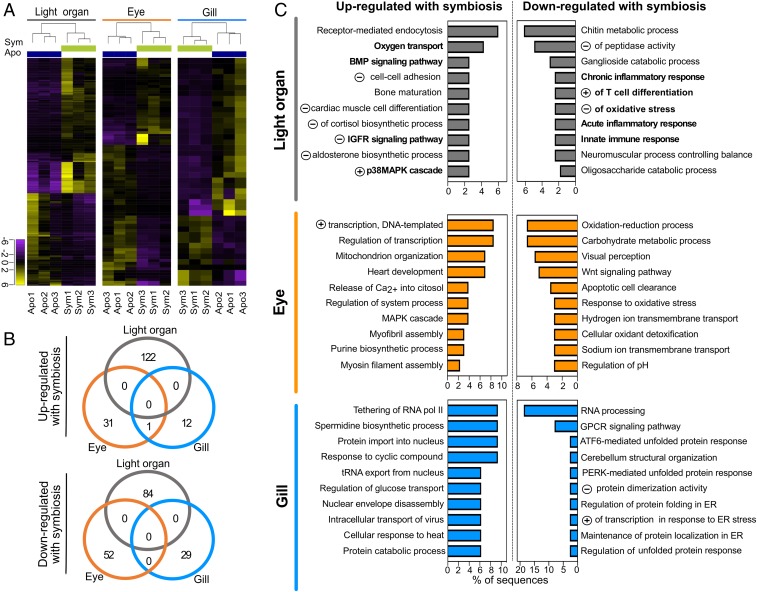
Fig. 2.
Impact of light-organ symbiosis on gene expression in different adult organs. (A) Heat map of expression values, log2-transformed and median centered, for genes significantly differentially expressed (>twofold, Padj < 0.05) in adult light organ, eye, and gill. Apo: aposymbiotic, in dark blue; Sym: symbiotic (colonized by wild-type V. fischeri) in green. (B) Venn diagrams indicating the numbers of significantly differentially expressed genes (>twofold, Padj < 0.05) in response to symbiosis. (C) Functional annotation of symbiosis-responsive genes in remote tissues. The differentially expressed genes were enriched in functional categories based on GO annotation. The top 10 enriched biological processes are shown ordered by percentages of sequences with that function and by its significance level (Fisher’s exact test, FDR < 0.05). “Negative (or positive) regulation of…” is abbreviated by a circled minus (or plus) symbol. Complete GO-term names and codes are in Dataset S6. Bold lettering indicates GO terms described in Results.
To further analyze not only the possible functions of these symbiosis-responsive genes, but also whether there were shared functions (if not genes) among the three organs, we conducted a GO-enrichment analysis for all of the differentially expressed genes. This analysis identified overrepresented terms in each organ, using the entire transcriptome as the background for the enrichment analysis. In APO animals, we found 40, 32, and 29 overrepresented functions in eye, gill, and light organ, respectively. In contrast, overrepresented functions in response to symbiosis were highest in the light organ, followed by gill and eye (Dataset S6). In addition, each of the three organs expressed genes within a unique set of top 10 enriched biological processes, in a symbiosis-dependent manner (Fig. 2C), a further indication of the distinct ways in which they react to the presence of bacteria in the light organ. For example, in the light organ itself, the three major responses to symbiosis, encompassing 9 of the 20 enriched biological functions (Fig. 2C), could be associated with: (i) vascularization and an increased oxygen demand driven by the symbiont’s bioluminescence; (ii) tissue stress from the presence of the symbionts; and (iii) an easing of innate immune responses once the organ is colonized. All of these functions are consistent with previous studies (40, 52, 53). In contrast, in the eye, light-organ colonization resulted in an up-regulation of genes encoding structural proteins, and down-regulation of genes encoding elements of sensory perception and oxidative stress, while the gill exhibited an increased expression of genes encoding stress responses and transcriptional regulation.
Because a robust systemic response to colonization was observed that included functions associated with light-perception in the squid eye (Fig. 2C), an organ convergent in form and function with the mammalian eye (39), we asked whether and how eyes of another well-studied symbiosis model, the mouse, respond to host colonization; to our knowledge, the impact of the gut microbiota on the transcriptomic profile of the mouse eye has not been reported. We compared, by RNA sequencing, the expression profile of the eye of conventionalized mice (i.e., mice in which the gut microbiota was present) to that of germ-free mice. Adult stage mice and squid were compared to minimize any effects due to differences in their developmental rates. Applying the same level of stringency as used for the squid eye (i.e., an adjusted P < 0.05) only five genes were detected as differentially expressed in the mouse eye in response to conventionalization (Dataset S7). One predicted gene was down-regulated, and four genes were up-regulated, including lactotransferrin, which was previously reported as present in the transcriptome of mouse eye (54), IFN-activated gene 205, a mitochondrial tRNA, and a noncoding RNA of the RIKEN family. Although the evolutionarily convergent eyes of cephalopods and vertebrates share a large number of conserved genes with similar expression levels (55, 56), we detected no shared symbiosis-regulated genes in the eyes of these two organisms.
In summary, in the mature squid symbiosis: (i) functionally distinct and anatomically distant tissues are influenced by the presence of symbiotic bacteria; (ii) unlike the total expression profile for each of the three organs, the transcriptional responses to symbiosis, and their functional annotations, were specific and nonoverlapping (Figs. 1D and 2B); and (iii) although the evolutionarily convergent eyes of cephalopods and vertebrates share a large number of conserved genes with similar expression levels (55, 56), no shared gene regulation was detected within eyes of squid and mouse in response to microbial colonization of distant tissues.
Colonization of Juvenile Hosts Had a Rapid Impact on Gene Expression, Even in Remote Tissues.
In the adult host, transcriptional responses to symbiosis are evident both locally and systemically (Figs. 2 and 3), but how quickly during symbiotic development does this outcome appear? The transcriptional response of the light organ has been reported to occur as early as 3 h following exposure to environmental V. fischeri (52). To determine the manner and timing of symbiosis-specific responses in other organs, more distant from the light-organ symbionts, we compared the RNA-seq gene-expression data of light organ, eye, and gill 24 h after the initiation of symbiosis, when the bacteria have fully colonized and are brightly luminous (28). At this point, we found that the light organ already exhibits a distinct transcriptional response; specifically, when comparing APO and SYM conditions, a total of 1,919 differentially regulated genes were detected, including 17% of the 206 genes characteristic of the adult SYM light-organ response (Fig. 2B). Analysis of this overlapping set of 36 genes revealed that ∼40% are associated with osmoregulatory and immune functions (Datasets S8 and S9). Subclusters 3 and 4 of light-organ differential gene expression comprise highly up-regulated genes in only two of the three analyzed SYM light organs (SI Appendix, Fig. S6), indicating a response whose onset is either variable or transitory. Interestingly, these two subclusters contained genes related to light perception, with significantly enriched functions, such as “structural components of the lens,” “visual perception,” or “phototransduction” (Dataset S9), perhaps reflecting the development of the light organ’s capacity to perceive light (39).
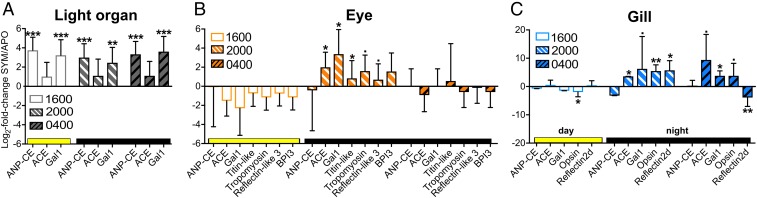
Fig. 3.
Variation of symbiosis-responsive gene expression over the day/night cycle. Gene expression changes in SYM (compared with APO) light-organ (A), eye (B), and gill (C) tissue at different times of day: 0400 hours = 2 h before dawn; 1600 hours = 2 h before dusk; 2000 hours = 2 h after dusk. Juvenile squid were maintained for 3 d under a 12–12 light-dark schedule. Candidate genes where chosen for qRT-PCR based on expression changes observed at 72 h (SI Appendix, Fig. S9 and Table S1 and Dataset S4). BPI3, bactericidal/permeability-increasing protein 3. ***P < 0.001; **P < 0.01; *P <0.05; ·P < 0.1 (SYM vs. APO).
As expected, a smaller number of symbiosis-responsive genes (44 in the eye and 184 in the gill) were detected (SI Appendix, Figs. S6 and S7), and there was essentially no overlap with the adult response in either of these organs. Nevertheless, a trend in which eye (but not gill) genes clustered with colonization state was detected (SI Appendix, Fig. S7). Unlike the light organ, the responses of eye and gill at 24 h were highly variable between samples; thus, we hypothesized that many genes that were differentially regulated in these organs in adults may have not yet become apparent in 24-h juveniles (SI Appendix, Fig. S8A). Thus, we used the NanoString platform to determine whether the patterns of a selected set of 23 genes that were not significantly regulated at 24 h (Dataset S4), but were either trending toward induction at 24 h or would become induced in adult eye and gill had, by 72 h, became significantly differentially regulated by symbiosis. While such a temporal comparison can be made with juvenile eye or gill tissues, changes in the light organ transcriptome are confounded by this organ’s substantial morphogenic transformation between 24 and 72 h (28).
Of the 13 selected adult eye-specific, symbiosis-responsive genes, 4 became clearly differentially regulated in juvenile eye tissue between 24 and 72 h postcolonization (e.g., SI Appendix, Figs. S8A′ and S9), indicating that during this period much of the transcriptional signature of the adult eye was still developing. For the gill, we chose two groups of genes: six that were significantly up-regulated in adults and four that were not, but were trending toward significance in 24-h juveniles. Of the first group, only one (ACE) had become differentially regulated by 72 h, while all four of the 24-h trending genes had. Thus, the data suggest that the gill has a more juvenile-specific response that is not retained in adults. In summary, the analysis of juvenile organs indicates that: (i) a robust transcriptional response to symbiosis appeared in the light organ within 24 h postcolonization, (ii) a smaller systemic response by eye and gill also became apparent, and (iii) by 24 h, the juvenile eye began to show an adult-like response, which became more significant at 72 h.
Expression of Some Symbiosis-Responsive Genes Was Regulated over the Day/Night Cycle.
Because the light organ has a well-described daily rhythm of transcriptional regulation (35) that is reflected in crypt-cell ultrastructural remodeling, symbiont luminescence, and metabolic activity in both partners (34, 35), we asked whether the symbiosis-regulated gene expression detected in remote organs also changed over the day (Fig. 1A). Based on the NanoString data for 72-h juveniles at 2000 hours (SI Appendix, Fig. S9), we characterized symbiosis-responsive gene expression from juvenile organs by qRT-PCR at three times: 2 h before dusk (1600 hours, at ∼70-h postcolonization) and 2 h before dawn (0400 hours), both times when the host is quiescent and symbiont luminescence is reduced, compared with 2 h after dusk (2000 hours, at ∼74-h postcolonization) (Fig. 1A), when the host is active and the symbionts are brightly luminous (34).
Expression levels of three genes [atrial natriuretic peptide-converting enzyme (ANP-CE), ACE, and galaxin 1 (Gal1)] were determined across all of the organs. In the light organ, while ACE was not significantly regulated by symbiosis, ANP-CE and Gal1 remained up-regulated in SYM relative to APO at all times tested (Fig. 3A). In contrast, Gal1 and ACE were up-regulated by symbiosis in the eye only at 2000 hours although, in gill, ACE was up at both 2000 and 0400 hours (Fig. 3 B and C). Thus, ANP-CE is specifically regulated in the light organ, as is ACE regulated only in the eye and gill.
Expression of an additional four eye-specific and two gill-specific genes that were symbiosis-regulated at 2000 hours were likewise dependent on time of day. While there are trends of down-regulation of these genes in the eye at 1600 hours, no significant differences appeared beyond 2000 hours (Fig. 3B). Similarly, in gill, opsin is up-regulated at 2000 relative to 1600 hours, while reflectin 2 d becomes down-regulated (relative to APO) at 0400 hours (Fig. 3C). Cephalopods are noted for extraocular photoreceptors (57), but these structures are associated with the surface of the animal, and not with internal organs, such as the gills (58).
In summary, symbiosis-responsive genes that were regulated in one organ at one time of day can be differentially expressed in other organs at a different time, emphasizing the time- and context-dependency of the response. In addition, among the genes examined here, the symbiosis-dependent up-regulation of expression in gill, and especially in eye, was generally most prominent early in the evening (2000 hours), when the host is ecologically active. In contrast, in the light organ, no pattern was observed for these genes (Fig. 3A), although other genes show strong patterns of temporal regulation (35).
Symbiont Luminescence Was the Principal Driver of Transcriptomic Patterns in both the Light Organ and the Eye.
Because symbiosis-induced up-regulation of gene expression occurred at night, coincident with high levels of symbiont luminescence, we asked whether light-emission itself is a factor driving gene expression. To this end, the gene-expression profiles of the juvenile light organ, eye, and gill were compared when the light organ was colonized by either a wild-type, light-producing strain (SYM) or a nonluminous Δlux mutant derivative (SYM-dark). Because such dark mutants can only maintain normal levels of colonization for the first day postinoculation (32), we focused our analyses on 24 h after symbiosis had initiated.
At this time, under normal conditions of SYM colonization, 1,919 genes are regulated in the light organ compared with APO (Fig. 4A). Comparison of the SYM expression profile with that of the SYM-dark animals revealed that at 24 h the light organ has a strong transcriptional response, independent of light production. Specifically, a total of 636 genes were regulated by both strains, over two-thirds of which were down-regulated. The functional annotation of >25% of these down-regulated genes was dominated by GO categories associated with maintenance of ciliary structure and function (Fig. 4A), which is not surprising because both SYM and SYM-dark bacteria induce the cell death and loss of the ciliated surface that mediates initial colonization (28). Also enriched in this shared set were genes up-regulated in immune response and stress (Fig. 4A), which is also not unexpected, as the morphogenesis of the ciliated surface is driven largely by symbiont microbe-associated molecular patterns.
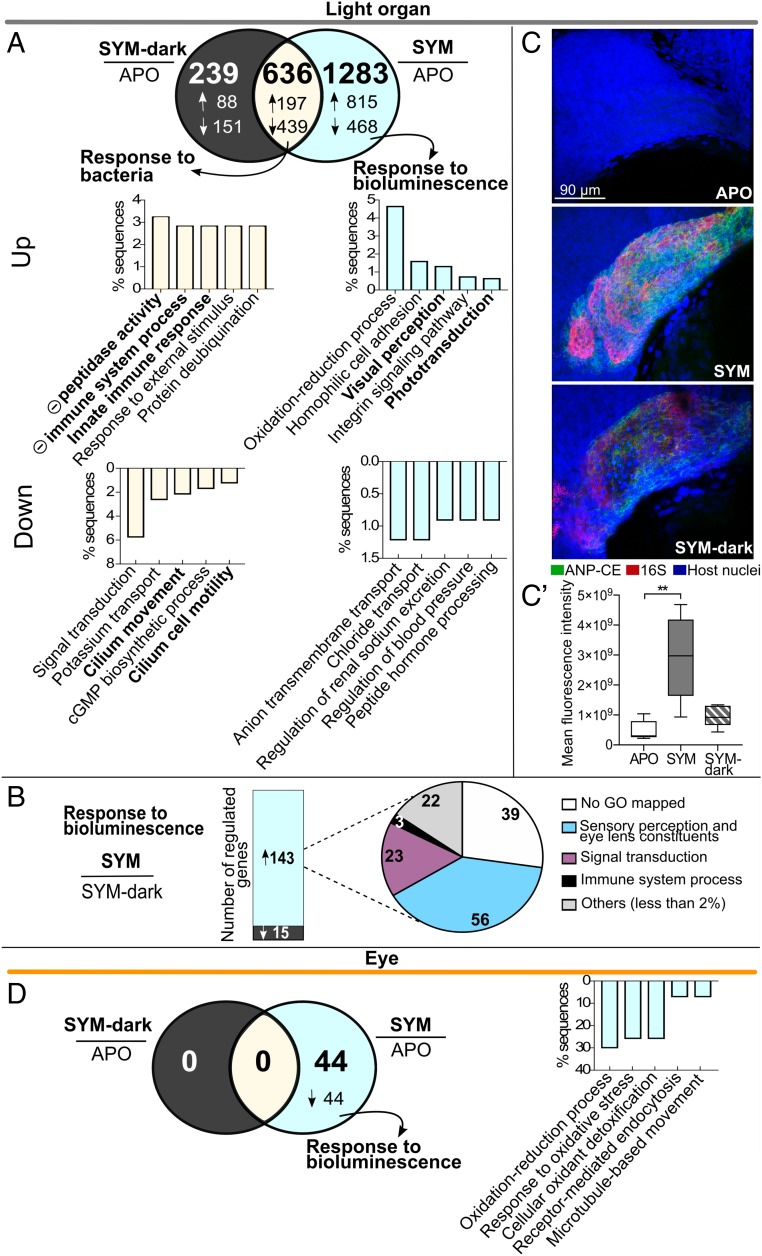
Fig. 4.
Impact of symbiont bioluminescence on juvenile gene expression. (A) Venn diagram of numbers of differentially expressed genes in the light organ, 24 h after colonization by either SYM (wild-type) or SYM-dark (Δlux) strains, compared with APO (>twofold, Padj < 0.05). Arrows indicate either up (↑) or down (↓) regulation. Bar graphs: functional enrichment of genes significantly up-regulated and down-regulated with symbiosis. For each set, the top five biological process terms are represented (Fisher’s exact test, FDR < 0.01). Notations as in Fig. 2. (B, Left) number of genes significantly up- or down-regulated in SYM compared with SYM-dark colonized light organs; (Right) proportion of annotated biological processes accounting for >2% of up-regulated genes. (C) Visualization of ANP-CE transcript in whole-mount light organs 24 h after colonization. Representative confocal images showing ANP-CE expression in crypt epithelium of APO, SYM, or SYM-dark colonized juvenile squid; merged midsection of z-stack of crypt #1; ANP-CE (green), 16S RNA (symbionts, red), and host nuclei (TOPRO, blue) (SI Appendix, Fig. S10 and Movies S1–S3). (C′) Quantification of ANP-CE signal by fluorescence intensity from z-stacks of crypt #1 in five light organs. P values were calculated using Kruskal–Wallis test and Dunn's multiple comparison test. Error bar: SD (**P < 0.01). (D) Venn diagram of differentially expressed genes in the eye, 24 h after colonization by either SYM (wild-type) or SYM-dark (Δlux) strains, compared with APO (>twofold, Padj < 0.05). Arrow indicates down (↓) regulation. Bar graph: notations as in A (Datasets S8 and S9).
However, it is most striking that over two-thirds of the 1,919 genes regulated by colonization required that the symbionts be luminescent, underscoring how critical a role V. fischeri bioluminescence plays in shaping the symbiosis. In direct contrast to the luminescence-independent response, nearly 70% of the genes of the luminescence-specific response were up-regulated. Notably, the 39 GO categories of up-regulated genes included “visual perception,” “phototransduction,” “photoreceptor activity,” and “structural constituent of eye lens,” all involved with light perception or modulation, as well as homophilic cell adhesion and oxidative-reduction processes (Fig. 4A and Dataset S9). The light organs that were colonized by the dark mutant not only failed to regulate these luminescence-specific genes, but also had an expression signature of their own. The dark mutant regulated 875 genes, only 46% the number regulated by the wild-type strain (1,919), with just over one-quarter of their regulated genes specific to the SYM-dark colonization, compared with the two-thirds of genes specific to the luminous SYM colonization. Furthermore, unlike the SYM animals, the SYM-dark condition resulted principally in down-regulated genes, with no significant functional enrichment in any GO category (Fig. 4A). Finally, when gene regulation was compared directly between SYM and SYM-dark animals, 143 annotated genes were up-regulated in SYM, and only 15 down-regulated (Fig. 4B). Thirty-nine percent of these up-regulated genes were associated with sensory perception of light stimulus or modulation of light (e.g., lens proteins) (Dataset S9).
A particularly interesting difference between colonization conditions was the ∼sevenfold up-regulation in SYM compared with SYM-dark of ANP-CE, which regulates cell volume and inflammation in a variety of systems (59). One of the key developmental features of the light organ is the SYM-induced swelling of the crypt cells with which the bacteria directly associate (Fig. 1A); however, the dark mutant is defective in inducing this phenotype (32). If ANP-CE were involved in such a cell-swelling and inflammation phenotype, we would predict that the transcript for this protein would specifically localize to the crypt epithelium in the SYM host, and be at a higher abundance than in SYM-dark–colonized animals. Using hybridization chain reaction-FISH, we compared the localization of the ANP-CE transcript in light organs at 24-h postinoculation. Abundant transcripts localized specifically to the cytoplasm of the crypt cells in SYM-colonized animals (SI Appendix and Movies S1–S3). In contrast, only low levels were detected in either APO or SYM-dark animals, with no significant difference between these conditions (Fig. 4 C and C′).
Unlike the light organ, no difference in colonization-dependent gene expression was detected between gill tissue of SYM and SYM-dark juveniles at 24 h, perhaps due to this organ’s high variability in development at this time point. However, the eye showed a uniform, down-regulation in the expression of all 44 of the genes that responded in any way to colonization by the luminous symbiont (Fig. 4D); in contrast, no significant change in expression of any of those genes was detected when the symbiont was the dark mutant. These data suggest that, like the light organ, the eye’s principal reaction to symbiotic colonization is mediated by the presence of light production. Interestingly, eye genes down-regulated by symbiont colonization were enriched in biological processes related to oxidation state or tissue reorganization (Fig. 4D and Dataset S9). Only 4 of the 44 genes down-regulated in the eye were shared with the light-organ’s response: specifically, ANP-CE, MAM/LDL-receptor class A domain-containing 2-like, dynein heavy-chain axonemal, as well as the hypothetical protein KGM_03810, which is also regulated in the adult light organ. However, unlike with the juvenile light organ, the first three of these eye genes are up- (not down-) regulated in response to symbiosis. In summary, symbiosis-dependent gene expression in both the light organ and eye was more dependent on the existence of bacterium’s luminescence than on the presence of the bacteria themselves.
Discussion
The data presented here demonstrate that, beginning within hours of the onset of the E. scolopes light-organ symbiosis, localized colonization of tissues by the specific symbiont V. fischeri creates a network of communication across the host’s body that reprograms transcription system-wide. The coordination of this network persists throughout the developmental trajectory of the association, beginning with early symbiosis-induced changes in light-organ form and function, and continuing well into the maturation of the partnership. Furthermore, the network reprograms remote organs to respond to the daily rhythms set by the V. fischeri population within the light organ. Finally, genetic manipulation of the symbiosis revealed that bioluminescence, the principal currency of the symbiont, while having no effect on the gill response, is not only the major symbiosis-dependent driver of transcriptional regulation in the light organ, but also the only driver in the eye.
Transcriptomes of the Host’s Organs Reflect Their Biological Functions Through Development.
The transcriptomes examined in this study, which segregated both by organ and by life stage, reflect known functions of the light organ, eye, and gill: that is, control of symbiont luminescence, vision, and respiration, respectively (SI Appendix, Fig. S3 and Dataset S3). The light organ, eye, and gill transcriptomes clustered separately between juveniles and adults (Fig. 1C and SI Appendix, Fig. S2A), a finding that may reflect the dramatic change in the animal’s ecology upon its maturation. Briefly, from hatching to ∼4-wk postcolonization, the juvenile’s behavior is not controlled by a daily rhythm, being either active or quiescent at all times of day and night; however, by about 1 mo, the animal has assumed a profound diel rhythm of burying in the substrate during the day and coming out to forage at night, a behavior that will persist throughout its ∼1-y life (60). This change in lifestyle coincides with a dramatic shift in the daily cycling of host and symbiont metabolism (34), a shift that promotes a brighter luminescence of the bacteria in the evening, when the squid is active and using the light emission of the symbionts to camouflage itself by “counterilluminating” (28, 61). We hypothesize that the eye may be responding transcriptionally not only to its commitment to a diel rhythm of environmental light, but also to the requirement that the eye coordinate its function with the light organ’s emission. Specifically, during counterillumination, the light organ modulates its luminescence in response to the intensity of down-welling moonlight and starlight, which is monitored by the eye, through as yet undefined mechanisms. With such developmental changes in day/night behavior, it is not surprising that the transcriptomes of both the light organ and eye adopt new patterns as these organs mature. Similarly, the respiratory and immune functions of gill tissue, like its transcriptome (Fig. 1C), change between juvenile and adults as the animal begins to bury in the substrate each day by 4 wk of age.
Symbiosis-Induced Changes in Squid–Host Gene Expression Show Similarities with Those Reported for the Mammalian Microbiota.
As with the global patterns of squid gene expression (Fig. 1C), both the light organ and remote tissues reacted robustly to colonization by V. fischeri, and several features of this life-stage and organ-specific transcriptomic response appear to be evolutionarily conserved between the squid light-organ and mammalian-gut symbioses. For example, a comparison of intestinal epithelial cell transcriptomes from germ-free and conventionalized mice found that the response of intestinal epithelial cells to the presence of the microbiota was only a fraction of the genes expressed in these cells and, as in squid, little overlap in this response occurs between juvenile and adult mice (62). In the squid, such largely stage-specific patterns of symbiosis reprogramming apply not only to the colonized tissue (i.e., the light organ), but also to the anatomically remote eye and gill. As yet, we know little about how remote tissues receive information about the colonization state or activity of symbiotic organs, but two modes are possible. The first is a chemical signal, such as a bacterial metabolite, delivered through the circulation (63, 64); one such metabolite, acetate, is generated in the light organ as byproduct of symbiont metabolism (34). For example, the presence of V. fischeri in the light organ has a systemic effect on hemocyte signaling (65). The second mode is neural: cephalopods, in particular, produce both targeted and systemic responses via their nervous system, similar to mammals, where the vagus nerve conveys information about the gut microbiota to the brain (66). However they are delivered, the transcriptional changes appearing during postnatal development in the mouse and squid organs reveal only part of the impact of symbiosis. In fact, many of these effects are deployed during embryogenesis, when symbionts are not yet present: that is, both in the squid–vibrio system and in mammals, the developing host creates specific target-tissue conditions that are poised to respond to the eventual arrival of their symbiotic partners (28). Conversely, in a synoptic study comparing digestive system transcriptomes of four regions of the mouse intestine and liver (67), as with the squid, very little overlap in the transcriptional response to symbiosis occurred across the body.
To our knowledge, an integrated comparison both across tissues and through development has not yet been addressed in the mouse; thus, whether the trajectory of the robust mammalian transcriptional response to symbiont colonization over early development varies among organs, as it did in the squid (i.e., first the light organ, then eye, then gill) (Datasets S4 and S8) remains to be determined. However, taken together, these similarities in life-stage and organ/tissue-specific responses to symbiosis in the two distantly related animal taxa suggest that, rather than having a generalized response to bacteria or their products, as they develop each tissue or organ near to or distant from sites of colonization integrates the partnership into its specific function. For example, in the squid association, we observed an up-regulation of genes involved in vascular development of tissues (Fig. 2C and Dataset S6), which has also been reported for the nutrition-based gut symbioses of mammals (68). The finding that the squid system, wherein bioluminescence and not nutrition is the principal benefit to the host, suggests, not surprisingly, that increased vascularization is important for other aspects of this symbiosis, such as facilitating the support of the bacterial population and their dialogue with host tissues (34, 53).
Symbiotic Regulation of Genes Encoding Specific Functions Occurs on a Diel Cycle.
In addition to sharing life-stage and tissue-specific responses to symbiosis, in both mammalian (69) and squid hosts, there exist diel transcriptomic rhythms in colonized and remote tissues that are influenced by their bacterial partners. For example, although the overall transcriptomes of the three squid organs examined here differed greatly (Fig. 1C), the juvenile light organ shared with eye and gill a symbiosis-dependent diel regulation of two genes: ACE and Gal1. ACE occurs widely among animals, controlling blood pressure and electrolyte balance, although the conserved function of this protein family is immune modulation in both vertebrates (70, 71) and invertebrates (59, 72). Immunity is also likely to be its ancestral role because ACE occurs even in taxa without a closed circulatory system; however, in the squid, whose circulation is closed, this protein may serve both an immune and a vascular function. In contrast, galaxins are invertebrate-specific proteins, first identified in corals; in E. scolopes, Gal1 is an antimicrobial peptide present in the light organ (73). Although the genes encoding ACE and Gal1 were both regulated (either up or down) by symbiosis, each had a different daily rhythm of expression depending on the host organ (Fig. 3).
What biological purposes might underlie the diel regulation of these genes? At all times analyzed—that is, 0400, 1600, and 2000 hours—the light organs were fully colonized (74); however, at 1600 and 0400 hours, the per cell luminescence of the symbionts is relatively low, compared with its maximum at 2000 hours (75). The light organ expressed ACE and Gal1 constitutively throughout the day, whereas ACE and Gal1 expression was up-regulated in the eye only when luminescence was highest (2000 hours). In addition, the light organ had symbiosis-induced expression of ANP-CE, which transforms a propeptide to the edema-related peptide ANP (76). Because ANP-CE and ACE often offset one another’s functions in immunity and vascular homeostasis (77), these findings suggests that an ACE-driven modulation of ANP-CE activity in the light organ releases ANP into the host blood stream, modulating the biochemistry of the eye and gill during the day/night cycle. Taken together, the transcriptional data are consistent with an organ-specific expression of an increased immune potential at particular times of day.
Light Organ and Eye Are More Similar in Their Responses to Symbiosis.
The repertoire of gene expression characteristic of light organ and gill tissues overlapped more in gene identity and number (Fig. 1D), consistent with both their similar relationship to the environment (i.e., both are bathed with bacteria-rich seawater during ventilation) and their shared immune function. In contrast, the interior portions of the eye examined here are protected from any direct exposure to environmental microbes. However, compared with the gill, the differential gene expression in symbiosis responses of the squid’s eye and light organ were more similar in magnitude (Fig. 2B), reinforcing the hypothesis proposed above, that a coordination between these organs facilitates the host’s counterillumination behavior within the ambient light field (61).
The squid eye also had a stronger relative response to light-organ colonization (84 genes) (Fig. 2B) than the mouse eye did to colonization of the gut by the microbiota (five genes) (Dataset S7). This difference in the scale of transcriptional response was unexpected because, while the mice were an inbred strain, the squid were genetically diverse, reared from wild-caught parents, and such genetic variation should lead to an underestimation of significant transcriptomic differences. However, the relatively weak reprogramming of the mouse by the presence of its gut microbiota may reflect the vertebrate eye’s status as a site of immune privilege (17). Nevertheless, colonization has been reported to influence the mouse eye’s lipid content (16), and metabolomics studies have revealed a gut–retina axis that correlates with a proclivity toward development of macular degeneration (15, 78). In short, the difference between squid and mouse responses may be either due to differences in how the immune system interacts with the eye, or because of a greater need for light-organ/eye coordination in bioluminescent symbiotic associations. The latter hypothesis could be tested by a study of fishes with light-organ symbioses, which have both the vertebrate immune privilege of the eye, and the need to modulate luminescence by the symbiotic organ (79, 80).
Symbiont Luminescence Has a Disproportionately Large Transcriptomic Effect.
A striking feature of symbiont-induced gene expression was discovered during a comparison of the organ transcriptomes of squid colonized with either the wild-type or the Δlux mutant symbiont. The results highlight the remarkable dominance of luminescence in reprogramming gene regulation in not only the symbiont-containing light organ (Fig. 4 A and B), but also the anatomically remote eye (Fig. 4D). In many other associations, studies of such systemic consequences of losing the symbiont’s principal activity have been clouded by the resultant physiological effects on the host. For example, in the rhizobium-legume symbiosis, bacterial mutants defective in nitrogen fixation similarly have a differential affect on transcription in both nodule tissue (81) and other plant organs (82); however, it was difficult to separate functions within the nodule from their general nutritional role. Similarly, animal–microbe interactions within gut tracts and bacteriomes also revolve around the symbiont’s provision of an important nutrition function (83). That is, unlike bioluminescence, these functions directly impact the host’s general physiology and health. Because the light-organ symbiosis plays an ecological role for the squid (i.e., antipredation), under laboratory conditions, the physiology of the host is not negatively affected by carriage of a dark mutant (32). As such, this system serves as a paradigm for studies of the reprogramming of remote-tissue by other microbiota assemblages with nonnutritional functions [e.g., an antibiotic or defensive toxin (84, 85)], like those of the skin and urogenital tract of humans (86).
ANP-CE is one of the most abundant and highly regulated genes in the squid transcriptomes; specifically, it is overexpressed in SYM relative to APO tissues (Fig. 4C), it’s level fluctuates on a day/night cycle in eye and gill (Fig. 3 B and C), and in light organ and eye its expression is induced by the symbiont’s luminescence (Dataset S8). Because ANP-CE plays a role in inducing cell edema (76), and the epithelia lining the symbiont-containing crypts swell when colonized by wild-type, but not dark-mutant, V. fischeri (32, 87), we predicted that ANP-CE expression would be reduced in the epithelium when the symbionts are dark mutants (Fig. 4C). This reduction suggested a direct link between symbiont function and host-tissue response, and may indicate a mechanism by which dark mutants are sanctioned (32, 88), possibly by withholding the nutrients typically delivered by the crypt’s edematous epithelium.
The data presented here demonstrate that different tissues can have different transcriptional drivers in response to symbiosis, and future work will address the array of symbiont features that trigger the systemic response to light-organ colonization. Here we demonstrate that, among several effects on remote tissues, the most remarkable was the role of the symbiont’s product (luminescence), rather than the presence of the symbiont itself, as the sole driver reprogramming the eye’s transcriptome (Fig. 4D). As yet, it is unclear how the eye recognizes the presence of a luminous symbiont: perhaps a signal is delivered indirectly through a humoral or neural signal from the light organ (39). In any case, determining the mechanisms underlying this response presents a rich horizon for discovering shared principles of microbe–organ communication. The transcriptomic responses described here not only document symbiosis-induced molecular networks across the host, but also reveal how these networks may influence the behavior and ecology of the host: for example, mediating the counterillumination antipredatory strategy.
In conclusion, we determined three types of organ-specific transcriptional responses to symbiont colonization over the trajectory of development and over the day/night cycle: (i) a strong reaction to both the microbe and its primary product, luminescence (light organ); (ii) a response to the presence of the symbionts, independent of their luminescence (gill); and (iii) a response triggered solely by the luminescence product (eye). Determining the presence and mechanistic basis of the interorgan network that connects symbiotic and remote tissues, enabling a coordinated response, is a critical area of exploration, and will eventually reveal the degree to which symbioses can influence host health and homeostasis throughout life.
Materials and Methods
Sample Collection.
Animals were collected 24 and 72 h after hatching (juveniles), or ∼5 mo (adults) (Fig. 1A), and anesthetized in seawater containing 2% ethanol; juveniles were stored whole in RNAlater (Sigma-Aldrich) as previously described (52), while adult tissues were dissected before storage. With the exception of day/night cycle studies, all samples were collected at 2000 hours, 2 h after dusk.
RNA-Seq Assembly and Analysis.
A total of 2.2 billion paired-end reads were de novo assembled using the Trinity-v2.4.0 RNA-Seq assembler (89) (Dataset S1), and annotated by BLASTx against the National Center for Biotechnology Information nonredundant protein database. For functional annotation of the reference transcriptome, GO mapping of the transcripts was performed with Blast2GO software (90). To estimate the relative expression value for transcripts, RSEM software (91) was used in combination with the R package edgeR (92) to identify the significantly differentially expressed transcripts. Statistical enrichment of GO terms for differentially expressed genes was performed in Blast2Go using the Fisher’s exact test with a false-discovery rate (FDR) < 0.01. In addition, gene set-weighted enrichment analysis with 500 permutations and FDR < 0.1 was performed on the differentially expressed transcripts (SI Appendix and Dataset S10).
Transcript Quantification by qRT-PCR or NanoString nCounter Analysis.
Changes in host gene expression were measured by qRT-PCR using LightCycler 480 SYBR Green I Master Mix (Roche). Ribosomal protein 19L, serine hydroxymethyl transferase, and heat-shock protein 90 were used to normalize the transcript level (SI Appendix, Table S1). The nCounter Custom CodeSet Kit (NanoString Technologies) was used to detect changes in gene expression (Dataset S4). Assay and spike-in controls were used for normalization based on identical amounts of input RNA. Analysis was performed with nSolverAnalysis software v3.0.
See SI Appendix, SI Materials and Methods for additional experimental details. All experiments involving mice were performed using protocols approved by the University of Wisconsin–Madison Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank members of the M.J.M.-N. and E.G.R. laboratories for helpful discussions, and Dr. Tara Essock-Burns for her assistance with confocal imaging. This work was funded by NIH Grants R37 AI50661 (to M.M.-N) and R01 OD11024 (to E.G.R.). F.E.R. was supported by the Office of the Vice-Chancellor for Research and Graduate Education at the University of Wisconsin–Madison, with funding from the Wisconsin Alumni Research Foundation.
Footnotes
Conflict of interest statement: L.Z. and M.J.M.-N. are coauthors on a 2015 Comment article.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database, the National Center for Biotechnology Information Sequence Read Archive (accession nos. PRJNA473394, PRJNA498343, and PRJNA498345).
See Commentary on page 7617.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819897116/-/DCSupplemental.
References
- 1.Blaser MJ, et al. Toward a predictive understanding of Earth’s microbiomes to address 21st century challenges. MBio. 2016;7:e00714-16. doi: 10.1128/mBio.00714-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch TC, McFall-Ngai MJ. Metaorganisms as the new frontier. Zoology (Jena) 2011;114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]
- 4.Hsu T, et al. Urban transit system microbial communities differ by surface type and interaction with humans and the environment. mSystems. 2016;1:e00018-16. doi: 10.1128/mSystems.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kembel SW, et al. Architectural design drives the biogeography of indoor bacterial communities. PLoS One. 2014;9:e87093. doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyal MS, Venkatesh S, Milbrandt J, Gordon JI, Raichle ME. Feeding the brain and nurturing the mind: Linking nutrition and the gut microbiota to brain development. Proc Natl Acad Sci USA. 2015;112:14105–14112. doi: 10.1073/pnas.1511465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–675. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharon G, Sampson TR, Geschwind DH, Mazmanian SK. The central nervous system and the gut microbiome. Cell. 2016;167:915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider KM, et al. Successful fecal microbiota transplantation in a patient with severe complicated Clostridium difficile infection after liver transplantation. Case Rep Gastroenterol. 2018;12:76–84. doi: 10.1159/000481937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamo T, Akazawa H, Suzuki JI, Komuro I. Novel concept of a heart-gut axis in the pathophysiology of heart failure. Korean Circ J. 2017;47:663–669. doi: 10.4070/kcj.2017.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serino M, Blasco-Baque V, Nicolas S, Burcelin R. Far from the eyes, close to the heart: Dysbiosis of gut microbiota and cardiovascular consequences. Curr Cardiol Rep. 2014;16:540. doi: 10.1007/s11886-014-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppo R. The gut-kidney axis in IgA nephropathy: Role of microbiota and diet on genetic predisposition. Pediatr Nephrol. 2018;33:53–61. doi: 10.1007/s00467-017-3652-1. [DOI] [PubMed] [Google Scholar]
- 13.Budden KF, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 14.Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol. 2015;6:1085. doi: 10.3389/fmicb.2015.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin P. The role of the intestinal microbiome in ocular inflammatory disease. Curr Opin Ophthalmol. 2018;29:261–266. doi: 10.1097/ICU.0000000000000465. [DOI] [PubMed] [Google Scholar]
- 16.Oresic M, Seppänen-Laakso T, Yetukuri L, Bäckhed F, Hänninen V. Gut microbiota affects lens and retinal lipid composition. Exp Eye Res. 2009;89:604–607. doi: 10.1016/j.exer.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Zárate-Bladés CR, et al. Gut microbiota as a source of a surrogate antigen that triggers autoimmunity in an immune privileged site. Gut Microbes. 2017;8:59–66. doi: 10.1080/19490976.2016.1273996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang X, FitzGerald GA. Timing the microbes: The circadian rhythm of the gut microbiome. J Biol Rhythms. 2017;32:505–515. doi: 10.1177/0748730417729066. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L, Zhang C. Microbiome: Keeping rhythm with your gut. Nat Microbiol. 2017;2:16273. doi: 10.1038/nmicrobiol.2016.273. [DOI] [PubMed] [Google Scholar]
- 20.Barr T, et al. Concurrent gut transcriptome and microbiota profiling following chronic ethanol consumption in nonhuman primates. Gut Microbes. 2018;9:338–356. doi: 10.1080/19490976.2018.1441663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury SR, et al. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics. 2007;8:215. doi: 10.1186/1471-2164-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sommer F, Nookaew I, Sommer N, Fogelstrand P, Bäckhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015;16:62. doi: 10.1186/s13059-015-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thaiss CA, et al. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Thion MS, et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 2018;172:500–516.e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Postler TS, Ghosh S. Understanding the holobiont: How microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26:110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFall-Ngai M. Divining the essence of symbiosis: Insights from the squid-vibrio model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFall-Ngai MJ. The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heath-Heckman EA, Foster J, Apicella MA, Goldman WE, McFall-Ngai M. Environmental cues and symbiont microbe-associated molecular patterns function in concert to drive the daily remodelling of the crypt-cell brush border of the Euprymna scolopes light organ. Cell Microbiol. 2016;18:1642–1652. doi: 10.1111/cmi.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamarcq LH, McFall-Ngai MJ. Induction of a gradual, reversible morphogenesis of its host’s epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–785. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartzman JA, et al. The chemistry of negotiation: Rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci USA. 2015;112:566–571. doi: 10.1073/pnas.1418580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wier AM, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carey HV, Duddleston KN. Animal-microbial symbioses in changing environments. J Therm Biol. 2014;44:78–84. doi: 10.1016/j.jtherbio.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery MK, McFall-Ngai MJ. The muscle-derived lens of a squid bioluminescent organ is biochemically convergent with the ocular lens. Evidence for recruitment of aldehyde dehydrogenase as a predominant structural protein. J Biol Chem. 1992;267:20999–21003. [PubMed] [Google Scholar]
- 38.Peyer SM, Heath-Heckman EAC, McFall-Ngai MJ. Characterization of the cell polarity gene crumbs during the early development and maintenance of the squid-vibrio light organ symbiosis. Dev Genes Evol. 2017;227:375–387. doi: 10.1007/s00427-017-0576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong D, et al. Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gestal C, Castellanos-Martínez S. Understanding the cephalopod immune system based on functional and molecular evidence. Fish Shellfish Immunol. 2015;46:120–130. doi: 10.1016/j.fsi.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Bose JL, Rosenberg CS, Stabb EV. Effects of luxCDABEG induction in Vibrio fischeri: Enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol. 2008;190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casaburi G, Goncharenko-Foster I, Duscher AA, Foster JS. Transcriptomic changes in an animal-bacterial symbiosis under modeled microgravity conditions. Sci Rep. 2017;7:46318. doi: 10.1038/srep46318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kremer N, et al. Persistent interactions with bacterial symbionts direct mature-host cell morphology and gene expression in the squid-vibrio symbiosis. mSystems. 2018;3:e00165-18. doi: 10.1128/mSystems.00165-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albertin CB, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524:220–224. doi: 10.1038/nature14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belcaid M, et al. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc Natl Acad Sci USA. January 11, 2019;116:3030–3035. doi: 10.1073/pnas.1817322116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alon S, et al. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife. 2015;4:e05198. doi: 10.7554/eLife.05198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.da Fonseca RR, et al. Next-generation biology: Sequencing and data analysis approaches for non-model organisms. Mar Genomics. 2016;30:3–13. doi: 10.1016/j.margen.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Bose JL, et al. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 50.Ruby EG, McFall-Ngai MJ. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- 51.Schwartzman JA, Ruby EG. A conserved chemical dialog of mutualism: Lessons from squid and vibrio. Microbes Infect. 2016;18:1–10. doi: 10.1016/j.micinf.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremer N, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rageh AA, et al. Lactoferrin expression in human and murine ocular tissue. Curr Eye Res. 2016;41:883–889. doi: 10.3109/02713683.2015.1075220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogura A, Ikeo K, Gojobori T. Comparative analysis of gene expression for convergent evolution of camera eye between octopus and human. Genome Res. 2004;14:1555–1561. doi: 10.1101/gr.2268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida MA, et al. Molecular evidence for convergence and parallelism in evolution of complex brains of cephalopod molluscs: Insights from visual systems. Integr Comp Biol. 2015;55:1070–1083. doi: 10.1093/icb/icv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingston AC, Kuzirian AM, Hanlon RT, Cronin TW. Visual phototransduction components in cephalopod chromatophores suggest dermal photoreception. J Exp Biol. 2015;218:1596–1602. doi: 10.1242/jeb.117945. [DOI] [PubMed] [Google Scholar]
- 58.Mäthger LM, Roberts SB, Hanlon RT. Evidence for distributed light sensing in the skin of cuttlefish, Sepia officinalis. Biol Lett. 2010;6:600–603. doi: 10.1098/rsbl.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takei Y. Does the natriuretic peptide system exist throughout the animal and plant kingdom? Comp Biochem Physiol B Biochem Mol Biol. 2001;129:559–573. doi: 10.1016/s1096-4959(01)00366-9. [DOI] [PubMed] [Google Scholar]
- 60.McFall-Ngai MJ, Ruby EG. Sepiolids and vibrios: When first they meet. Bioscience. 1998;48:257–265. [Google Scholar]
- 61.Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- 62.Pan WH, et al. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018;10:27. doi: 10.1186/s13073-018-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pluznick JL, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujisaka S, et al. Diet, genetics, and the gut microbiome drive dynamic changes in plasma metabolites. Cell Rep. 2018;22:3072–3086. doi: 10.1016/j.celrep.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAnulty SJ, Nyholm SV. The role of hemocytes in the Hawaiian bobtail squid, Euprymna scolopes: A model organism for studying beneficial host-microbe interactions. Front Microbiol. 2017;7:2013. doi: 10.3389/fmicb.2016.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonaz B, Bazin T, Pellissier S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front Neurosci. 2018;12:49. doi: 10.3389/fnins.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mardinoglu A, et al. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol Syst Biol. 2015;11:834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tahara Y, et al. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep. 2018;8:1395. doi: 10.1038/s41598-018-19836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Vito P. Atrial natriuretic peptide: An old hormone or a new cytokine? Peptides. 2014;58:108–116. doi: 10.1016/j.peptides.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 71.Moskowitz DW, Johnson FE. The central role of angiotensin I-converting enzyme in vertebrate pathophysiology. Curr Top Med Chem. 2004;4:1433–1454. doi: 10.2174/1568026043387818. [DOI] [PubMed] [Google Scholar]
- 72.Salzet M, Verger-Bocquet M. Elements of angiotensin system are involved in leeches and mollusks immune response modulation. Brain Res Mol Brain Res. 2001;94:137–147. doi: 10.1016/s0169-328x(01)00229-7. [DOI] [PubMed] [Google Scholar]
- 73.Heath-Heckman EA, et al. Shaping the microenvironment: Evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-Vibrio symbiosis. Environ Microbiol. 2014;16:3669–3682. doi: 10.1111/1462-2920.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: Description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 75.Boettcher KJ, Ruby EG, McFall-Ngai MJ. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol A. 1996;179:65–73. [Google Scholar]
- 76.Theilig F, Wu Q. ANP-induced signaling cascade and its implications in renal pathophysiology. Am J Physiol Renal Physiol. 2015;308:F1047–F1055. doi: 10.1152/ajprenal.00164.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boudoulas KD, Triposkiadis F, Parissis J, Butler J, Boudoulas H. The cardio-renal interrelationship. Prog Cardiovasc Dis. 2017;59:636–648. doi: 10.1016/j.pcad.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 78.Rinninella E, et al. The role of diet, micronutrients and the gut microbiota in age-related macular degeneration: New perspectives from the gut-retina axis. Nutrients. 2018;10:E1677. doi: 10.3390/nu10111677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gould AL, Dougan KE, Koenigbauer ST, Dunlap PV. Life history of the symbiotically luminous cardinalfish Siphamia tubifer (Perciformes: Apogonidae) J Fish Biol. 2016;89:1359–1377. doi: 10.1111/jfb.13063. [DOI] [PubMed] [Google Scholar]
- 80.Haygood MG. Light organ symbioses in fishes. Crit Rev Microbiol. 1993;19:191–216. doi: 10.3109/10408419309113529. [DOI] [PubMed] [Google Scholar]
- 81.Barnett MJ, Toman CJ, Fisher RF, Long SR. A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci USA. 2004;101:16636–16641. doi: 10.1073/pnas.0407269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Rourke JA, et al. An RNA-seq based gene expression atlas of the common bean. BMC Genomics. 2014;15:866. doi: 10.1186/1471-2164-15-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Douglas AE. The Symbiotic Habit. Princeton Univ Press; Princeton, NJ: 2010. [Google Scholar]
- 84.McCutcheon JP. Genome evolution: A bacterium with a Napoleon complex. Curr Biol. 2013;23:R657–R659. doi: 10.1016/j.cub.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 85.Sharp KH, Davidson SK, Haygood MG. Localization of ‘Candidatus Endobugula sertula’ and the bryostatins throughout the life cycle of the bryozoan Bugula neritina. ISME J. 2007;1:693–702. doi: 10.1038/ismej.2007.78. [DOI] [PubMed] [Google Scholar]
- 86.Meisel JS, et al. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018;6:20. doi: 10.1186/s40168-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sycuro LK, Ruby EG, McFall-Ngai M. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol. 2006;267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 88.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol. 2014;23:1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.