Significance
Noise-induced hearing loss is a highly prevalent form of sensorineural hearing impairment affecting individuals of all ages. Noise exposure triggers a rapid increase in reactive oxygen species (ROS) levels, causing oxidative damage to auditory hair cells and resulting in hearing loss. Here we describe a novel mechanism of protection against noise-induced auditory hair cell damage. In response to sound exposure, pejvakin, a peroxisome-associated protein, acts as an ROS sensor and recruits the autophagy machinery to trigger the degradation of peroxisomes (pexophagy) damaged by oxidative stress. We characterized the pejvakin interactions involved in this process. We provide evidence that pejvakin-mediated pexophagy plays a key role in controlling peroxisome proliferation and protecting auditory hair cells against noise-induced damage.
Keywords: pejvakin, noise-induced hearing loss, LC3B, pexophagy, intracochlear viral transduction
Abstract
Noise overexposure causes oxidative stress, leading to auditory hair cell damage. Adaptive peroxisome proliferation involving pejvakin, a peroxisome-associated protein from the gasdermin family, has been shown to protect against this harmful oxidative stress. However, the role of pejvakin in peroxisome dynamics and homeostasis remains unclear. Here we show that sound overstimulation induces an early and rapid selective autophagic degradation of peroxisomes (pexophagy) in auditory hair cells from wild-type, but not pejvakin-deficient (Pjvk−/−), mice. Noise overexposure triggers recruitment of the autophagosome-associated protein MAP1LC3B (LC3B; microtubule-associated protein 1 light chain 3β) to peroxisomes in wild-type, but not Pjvk−/−, mice. We also show that pejvakin–LC3B binding involves an LC3-interacting region within the predicted chaperone domain of pejvakin. In transfected cells and in vivo transduced auditory hair cells, cysteine mutagenesis experiments demonstrated the requirement for both C328 and C343, the two cysteine residues closest to the C terminus of pejvakin, for reactive oxygen species-induced pejvakin–LC3B interaction and pexophagy. The viral transduction of auditory hair cells from Pjvk−/− mice in vivo with both Pjvk and Lc3b cDNAs completely restored sound-induced pexophagy, fully prevented the development of oxidative stress, and resulted in normal levels of peroxisome proliferation, whereas Pjvk cDNA alone yielded only a partial correction of the defects. Overall, our results demonstrate that pexophagy plays a key role in noise-induced peroxisome proliferation and identify defective pexophagy as a cause of noise-induced hearing loss. They suggest that pejvakin acts as a redox-activated pexophagy receptor/adaptor, thereby identifying a previously unknown function of gasdermin family proteins.
Noise overexposure increases reactive oxygen species (ROS) levels, causing oxidative damage to auditory hair cells and resulting in hearing loss (1, 2). Excessive ROS production can be counteracted by the antioxidant activities of ROS-metabolizing organelles, mitochondria, and peroxisomes (3, 4). Diverse metabolic pathways, including the β-oxidation of branched and very-long-chain fatty acids and hydrogen peroxide (H2O2) metabolism, occur in peroxisomes. The number, size, shape, and molecular content of these organelles continually adapt to the changing needs of the cell and external conditions (5, 6). The balance between peroxisome biogenesis and degradation is crucial for redox cell homeostasis and may be dysregulated in peroxisomal biogenesis disorders (7). Peroxisome biogenesis can occur de novo or through the proliferation of existing peroxisomes (5, 6), and damaged peroxisomes are degraded by selective autophagy (pexophagy) in yeast and plant vacuoles and in mammalian lysosomes (8).
We recently showed that the physiological response to noise overexposure includes peroxisome proliferation (1). We found that defects of pejvakin, underlying an autosomal recessive form of deafness in humans (DFNB59) (9), cause hypervulnerability to sound in humans and mice (1) associated with high levels of oxidative stress in auditory hair cells and neurons. Pejvakin is a member of the gasdermin protein family, which has six members in humans (gasdermins A–E and pejvakin), four of which (gasdermins A, B, D, and E) trigger pyroptosis, a form of programmed lytic cell death initiated by inflammasome activation (10). These gasdermins have an autoinhibited two-domain architecture, with an N-terminal pore-forming domain in the plasma membrane and a C-terminal repressor domain, separated by a linker region (11–14). Their pyroptotic pore-forming activity results from inflammatory caspase cleavage at the linker domain, separating the N-terminal pore-forming domain from the C-terminal repressor domain. Pejvakin has a markedly different structure than that of other gasdermins, with a divergent shorter C-terminal domain and no identifiable cleavable linker domain (11, 15).
Pejvakin is associated with peroxisomes and is required for the sound-induced proliferation of these organelles (1). However, its role in peroxisome dynamics, in peroxisome proliferation itself, or at other steps in peroxisome turnover, and the direct or indirect nature of its action, remain unknown. In this study, we addressed these issues. We first studied sound-induced peroxisome proliferation in both wild-type and Pjvk−/− mice. We then characterized the role of pejvakin in pexophagy and in protection of the auditory system against noise-induced oxidative stress.
Results
Early Pexophagy Precedes the Proliferation of Peroxisomes Induced by Sound Overexposure.
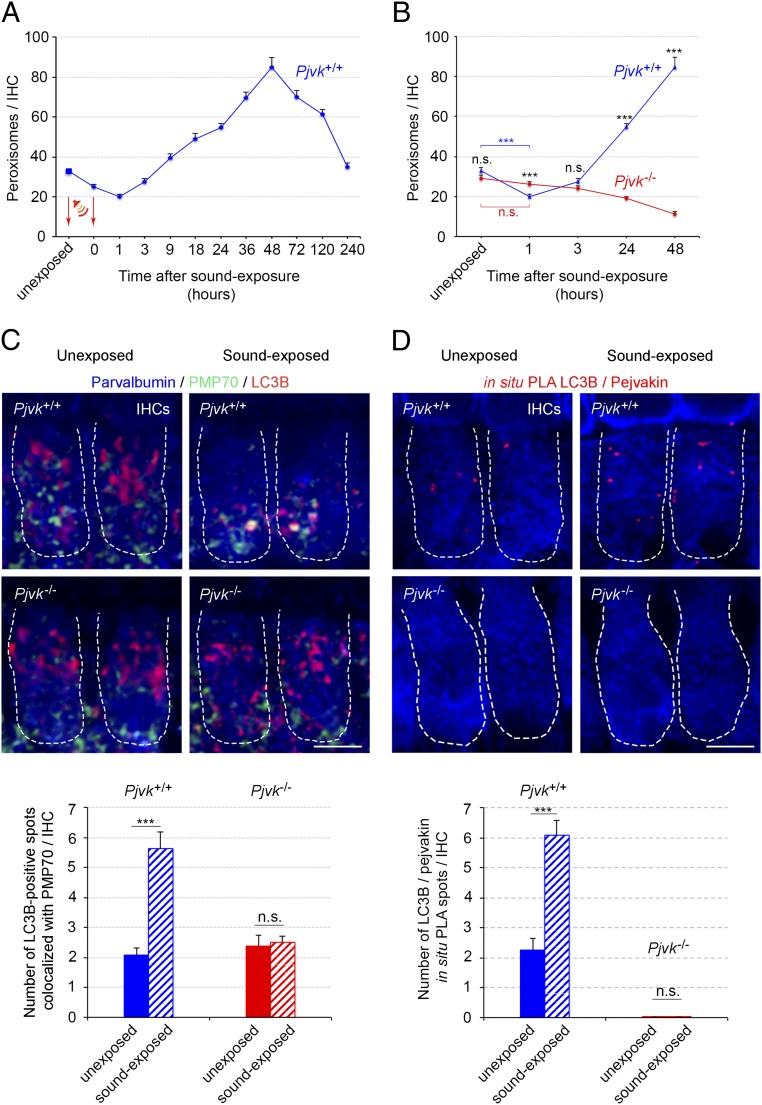
We studied peroxisome kinetics in response to sound in the inner hair cells (IHCs), the genuine sensory cells of the cochlea, the auditory sensory organ, in wild-type (Pjvk+/+) and Pjvk−/− mice. Surprisingly, Pjvk+/+ mice were found to have fewer peroxisomes per IHC than unexposed mice (mean ± SEM, 32.8 ± 1.5 peroxisomes per IHC), 24% lower (24.8 ± 1.4 peroxisomes per IHC; P = 0.0002) at the end of the 1 h of exposure to loud broadband sound (5–40 kHz, 105 dB SPL), and 39% lower at 1 h after the end of sound exposure (20 ± 1.2 peroxisomes per IHC; P < 0.0001) (Fig. 1A). Two hours later, the number of peroxisomes had started to increase, peaking at 48 h at a level 4.3 times that at 1 h after the end of the stimulation (85.3 ± 4.2 peroxisomes per IHC; 2.6 times that before stimulation). It decreased slowly thereafter, reaching baseline values within 10 days (Fig. 1A). In contrast, the number of peroxisomes remained unchanged at 1 h after the end of sound exposure in Pjvk−/− IHCs (mean ± SEM, 29.3 ± 1.3 and 26.1 ± 1.2 peroxisomes per IHC before and after sound exposure, respectively; P = 0.08) and decreased gradually thereafter, by 56% (mean ± SEM, 12.8 ± 1.8; P < 0.0001) over 48 h (Fig. 1B). Therefore, sound-induced peroxisome proliferation is preceded by an early rapid pejvakin-dependent degradation of peroxisomes (pexophagy).
Fig. 1.
Pejvakin promotes pexophagy in response to sound in auditory hair cells. (A) Changes in peroxisome number in P21 Pjvk+/+ IHCs (n = 60; four mice per time point) after sound exposure (5–40 kHz, 105 dB SPL, 1 h). The arrows indicate the sound stimulation period (1 h). (B) Impaired early elimination and proliferation of peroxisomes in Pjvk−/− IHCs in response to sound (n = 60 IHCs; four mice per time point). (C) LC3B-mediated pexophagy is altered in Pjvk−/− IHCs. The number of coimmunolabeled LC3B/PMP70 spots increased at 1 h after sound exposure in Pjvk+/+ IHCs, but not in Pjvk−/− IHCs. (D) The number of LC3B/pejvakin PLA spots (in red) was higher at 1 h after sound exposure in Pjvk+/+ IHCs. No LC3B/pejvakin PLA spots were observed in Pjvk−/− IHCs. The bar charts quantify LC3B/PMP70 coimmunolocalization in IHCs from Pjvk+/+ and Pjvk−/− mice (C) and LC3B/pejvakin PLA spots in Pjvk+/+ and Pjvk−/− IHCs (D) before and at 1 h after sound exposure (n = 40 IHCs; four mice per genotype and condition). Data are mean ± SEM. ***P < 0.001; n.s., not significant, unpaired Student’s t test. (Scale bars: 5 μm.)
We then investigated the possible involvement of pejvakin in coupling of the autophagosome machinery to peroxisomes, by studying the recruitment of LC3B, an autophagosomal marker, to these organelles (16, 17). At the time of maximal peroxisome degradation (1 h after the end of sound exposure), the number of spots double-immunolabeled for LC3B and peroxisome membrane protein 70 (PMP70) had increased by a factor of 2.7 in Pjvk+/+ IHCs (Fig. 1C) but remained unchanged in Pjvk−/− IHCs (Fig. 1C). Moreover, at 24 h after sound exposure, peroxisome aggregates, a hallmark of impaired pexophagy (18), were detected in Pjvk−/− IHCs but not in Pjvk+/+ IHCs (SI Appendix, Fig. S1).
We next investigated the possible recruitment of LC3B by pejvakin in an in situ proximity ligation assay (PLA) (19, 20), a highly sensitive method in which each pair of proteins is visualized as a fluorescent spot. The number of LC3B/pejvakin PLA spots had also increased by a factor of 2.7 in Pjvk+/+ IHCs at 1 h after sound exposure (Fig. 1D). The lack of pejvakin in Pjvk−/− mice did not modify the number of spots double-immunolabeled for LC3B and PMP70 observed under basal conditions, indicating the existence of a pejvakin-independent interaction between peroxisomes and LC3B in unstressed IHCs. Overall, these results suggest that pejvakin triggers the autophagic degradation of noise-induced oxidative stress-damaged peroxisomes by recruiting LC3B.
Pejvakin Promotes Stress-Induced Pexophagy Through the Direct Recruitment of LC3B.
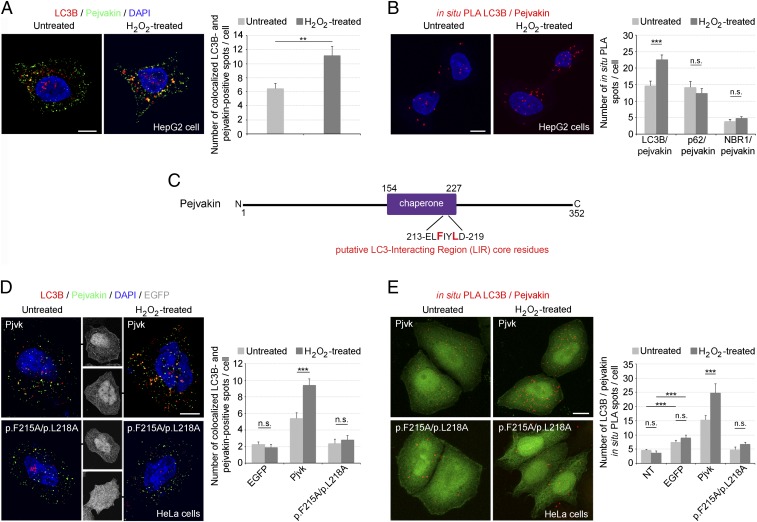
Most selective organelle autophagy pathways in mammalian cells require specific receptors to target organelles for autophagic degradation. The key players and regulators of pexophagy in vertebrates are largely unknown. However, the autophagic receptor/adaptor complex, composed of p62/SQSTM1 (hereinafter referred to as p62) (21) and NBR1 (22), has been shown to induce pexophagy in mammals by binding the targeted peroxisome to a phagophore, particularly in conditions of oxidative stress. Both p62 and NBR1 bind LC3B via their LC3-interacting region (LIR) motifs and ubiquitinated lysine residues of peroxisomal membrane proteins via their ubiquitin-associated (UBA) domains (3). Therefore, we investigated whether the binding of LC3B to pejvakin induced by oxidative stress also involves a direct interaction of pejvakin with p62 or NBR1. HepG2 human hepatoblastoma cells, which are enriched in peroxisomes and strongly express pejvakin, displayed an increase in the numbers of both LC3B/pejvakin double-immunolabeled spots and LC3B/pejvakin PLA spots in response to H2O2-induced oxidative stress (Fig. 2 A and B). In contrast, the number of p62/pejvakin and NBR1/pejvakin PLA spots was unaffected by H2O2 treatment (Fig. 2 A and B). Thus, there was no evidence for a direct interaction of pejvakin with p62/NBR1 in the ROS-induced recruitment of LC3B.
Fig. 2.
Pejvakin recruits LC3B directly via its LIR, promoting autophagy in response to oxidative stress. (A) Coimmunolabeling of pejvakin and LC3B in untreated and H2O2-treated HepG2 cells. The number of LC3B-positive spots colocalizing with PMP70 labeling increased after oxidative stress (bar charts; n = 20 cells per condition). (B) Interactions between LC3B and pejvakin, detected as in situ PLA spots, in untreated and H2O2-treated HepG2 cells. The bar charts show an increase in the number of LC3B/pejvakin PLA spots after oxidative stress but no change in the number of p62/pejvakin or NBR1/pejvakin PLA spots (n = 20 cells per condition). (C) Schematic representation of the putative LIR motif (core residues in red) in the predicted pejvakin chaperone domain. (D and E) Interactions between pejvakin and LC3B in untransfected HeLa cells (NT) and transfected HeLa cells producing EGFP alone, EGFP and wild-type pejvakin (Pjvk), or EGFP and pejvakin with mutated core residues. p.F215A and p.L218A were detected by double-immunolabeling (D) and in situ PLA (E) for pejvakin and LC3B before and after oxidative stress. The bar charts in D and E show that H2O2 treatment increases LC3B recruitment to pejvakin in transfected HeLa cells producing EGFP and wild-type pejvakin but not in cells producing mutated pejvakin or untransfected cells (n = 20 cells per condition). Data are mean ± SEM. **P < 0.01; ***P < 0.001; n.s., not significant, unpaired Student’s t test. (Scale bars: 5 μm.)
An analysis of the murine pejvakin sequence with the Phyre2 web portal (23) found no predicted UBA domain. A predicted chaperone domain was detected between amino acids 154 and 227 (76% confidence), with a putative LIR motif, FIYL, between amino acids 215 and 218 (Fig. 2C). The LIR motif contains a core consensus motif, [W/F/Y]X1X2[L/I/V] (24, 25), either containing or adjacent to (X1, X2) negatively charged residues. In pejvakin, this motif is flanked by the charged acidic residues E213 and D219, positioned N- and C-terminal to the aromatic residue F215, and I216 is one of only five amino acids acceptable in position X1 relative to F215 in F-type LIRs (24, 25). This putative LIR motif has been strongly conserved throughout evolution (SI Appendix, Fig. S2). We assessed its functional relevance by investigating whether mutations of its key residues, F215 and L218, affected pejvakin binding to LC3B under H2O2-induced oxidative stress. After H2O2 treatment, the numbers of LC3B/pejvakin-coimmunolabeled spots and LC3B/pejvakin PLA spots increased in transfected HeLa cells expressing EGFP and normal murine pejvakin (Pjvk-IRES-EGFP), but not in transfected cells expressing EGFP and a mutated form of pejvakin (mutPjvk-IRES-EGFP) carrying the p.F215A and/or p.L218A amino acid substitutions or expressing EGFP alone (Fig. 2 D and E). However, in basal conditions, the number of LC3B/pejvakin PLA spots was larger in transfected cells producing EGFP alone than in untransfected cells, suggesting that transfection induced a stress that triggered the recruitment of LC3B by endogenous pejvakin (Fig. 2E). Five other putative LIR motifs were identified outside the predicted chaperone domain of pejvakin. Site-direct mutagenesis, performed as above, showed that none of these motifs was involved in LC3B recruitment by pejvakin in response to oxidative stress (SI Appendix, Fig. S3). Overall, these findings suggest that oxidative stress-induced pexophagy is dependent on the direct interaction of the defined LIR motif of pejvakin with LC3B.
The Two Most C-Terminal Cysteine Residues of Pejvakin Regulate Stress-Induced Binding to LC3B.
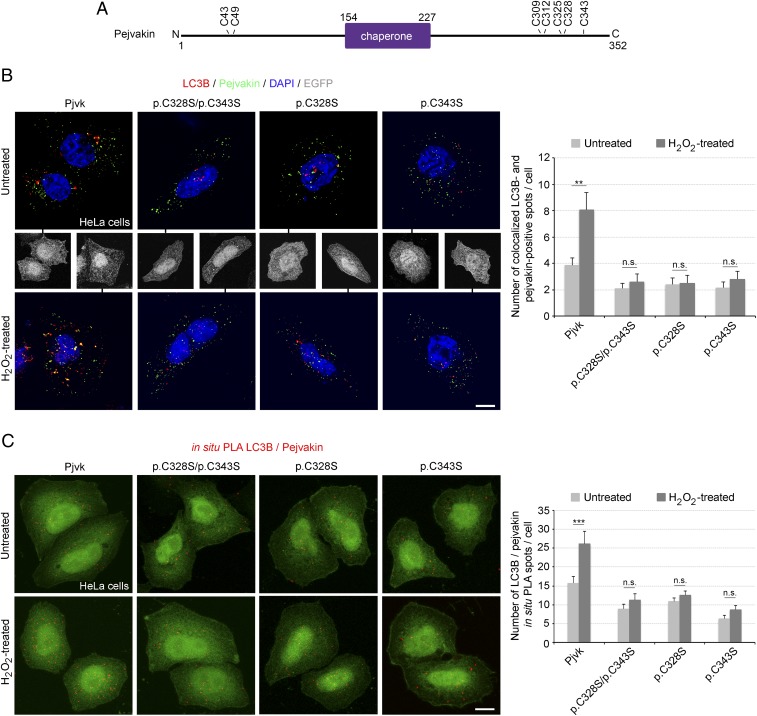
The pejvakin-mediated pexophagy elicited by oxidative stress raised the possibility of redox-regulated LC3B recruitment by pejvakin. The functional LIR motif of pejvakin is part of the largest putative protein-binding pocket detected by the Fpocket tool (26) within the predicted chaperone domain (SI Appendix, Fig. S4), and chaperone protein activity is often regulated by the redox state of the cell (27). Cysteine oxidation releases the compact conformation of the protein, exposing high-affinity binding sites (27–29). The C terminus of pejvakin contains a cysteine-rich region (residues 309–343). We assessed the pejvakin-LC3B binding elicited by H2O2 in HeLa cells transfected with mutated forms of pejvakin carrying cysteine-to-serine substitutions by coimmunolabeling and in situ PLA. Mutations of either of the two cysteine residues closest to the C terminus, p.C328S and p.C343S (a deafness-causing missense mutation in humans) (30), abolished this interaction (Fig. 3). No mutation of the other C-terminal (p.C309S, p.C312S, and p.C325S) or N-terminal (p.C43S and p.C49S) cysteine residues had such an effect (SI Appendix, Fig. S5). Thus, the C328 and C343 residues are essential for ROS-induced pejvakin–LC3B interaction.
Fig. 3.
Stress-induced pejvakin binding to LC3B is regulated by pejvakin C328 and C343 residue oxidation. (A) Schematic representation of cysteines N- or C-terminal positions to the predicted chaperone domain of pejvakin. (B and C) Interactions between pejvakin and LC3B in transfected HeLa cells producing EGFP together with wild-type pejvakin (Pjvk) or the mutated forms of pejvakin, C328S and C343S, detected by double-immunolabeling (B) and in situ PLA (C) for pejvakin and LC3B before and after oxidative stress. The bar charts in B and C show that H2O2 treatment increases LC3B recruitment by pejvakin in transfected HeLa cells producing EGFP and wild-type pejvakin but not in cells producing mutated pejvakin (n = 20 cells per condition). Data are mean ± SEM. **P < 0.01; ***P < 0.001; n.s., not significant, unpaired Student’s t test. (Scale bars: 5 μm.)
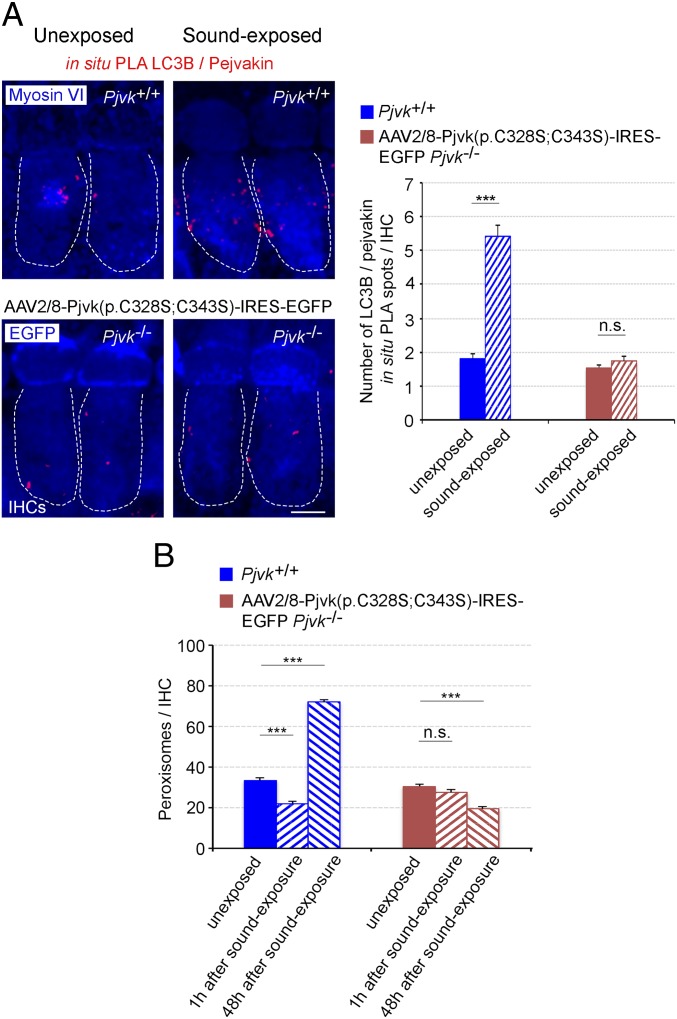
We then investigated the role of these two cysteine residues in vivo in sound-induced LC3B recruitment by pejvakin and pexophagy. We performed a rescue experiment in Pjvk−/− mice, involving the adeno-associated virus 2/8 (AAV2/8) vector-mediated transfer of the p.C328S/p.C343S mutated form of pejvakin [AAV2/8-Pjvk(p.C328S;C343S)-IRES-EGFP] into the cochlea of Pjvk−/− mice. Before sound stimulation, the basal interaction between pejvakin and LC3B detected in the form of LC3B/pejvakin PLA spots (Fig. 4A) was similar in transduced Pjvk−/− and control Pjvk+/+ IHCs. In contrast, at 1 h after the end of sound exposure, no increase in LC3B-mutated pejvakin interaction could be detected in transduced Pjvk−/− IHCs, whereas these interactions had increased by a factor of three in Pjvk+/+ IHCs (Fig. 4A). Moreover, neither pexophagy nor peroxisome proliferation following sound exposure was restored in Pjvk−/− IHCs transduced with the vector expressing the mutated form of pejvakin (Fig. 4B). Thus, the pejvakin C328 and C343 residues play a key role in noise-induced pexophagy, and pejvakin, which displays redox-regulated binding to LC3B, likely acts as a ROS sensor.
Fig. 4.
Transfer of the p.C328S/p.C343S mutated form of pejvakin into Pjvk−/− IHCs does not restore neither early pexophagy nor peroxisome proliferation after sound exposure. (A) The number of LC3B/pejvakin in situ PLA spots does not increase at 1 h after sound exposure (5–40 kHz, 105 dB SPL, 1 h) in Pjvk−/− IHCs transduced with AAV2/8-Pjvk(p.C328S;C343S)-IRES-EGFP (n = 40 IHCs; four mice per condition). (B) Peroxisome degradation (at 1 h after sound exposure) and proliferation (at 48 h after sound exposure) are not restored in Pjvk−/− IHCs transduced with AAV2/8-Pjvk(p.C328S;C343S)-IRES-EGFP (n = 60 IHCs; four mice per condition). Data are mean ± SEM. ***P < 0.001; n.s., not significant, unpaired Student’s t test. (Scale bar: 5 μm.)
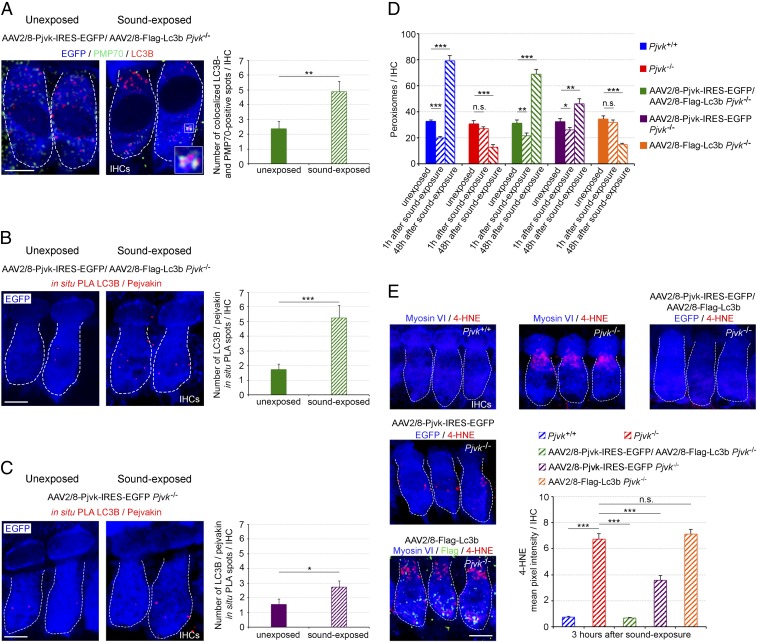
Transfer of the Pjvk and Lc3b Genes Together Restores Pexophagy and Peroxisome Proliferation in Pjvk−/− IHCs.
The foregoing experiments provide strong evidence for a role of pejvakin in pexophagy and the involvement of the pexophagy in peroxisome proliferation. However, we thought that additional in vivo evidence correlating pexophagy, changes in ROS damage to peroxisomes, and peroxisome proliferation might accrue from rescue experiments in Pjvk−/− mice. Therefore, we assessed the effects of pejvakin and LC3B, separately and together, on these processes by injecting AAV2/8-Pjvk-IRES-EGFP, AAV2/8-Flag-Lc3b, or both into Pjvk−/− cochleas. At 1 h after the end of sound exposure, LC3B targeting to peroxisomes in Pjvk−/− IHCs overexpressing both Pjvk and Lc3b cDNAs, as detected by PLA and coimmunolabeling, was similar to that observed in wild-type mice (mean ± SEM, 5.25 ± 0.85 LC3B/pejvakin PLA spots per transduced IHC vs. 6.08 ± 0.5 spots per Pjvk+/+ IHC; P = 0.40 and 4.88 ± 0.85 LC3B/pejvakin-coimmunolabeled spots per transduced IHC vs. 5.63 ± 0.55 spots per Pjvk+/+ IHC; P = 0.40), whereas the reexpression of Pjvk alone led to a smaller increase in the number of these spots (mean ± SEM, 2.73 ± 0.4 LC3B/pejvakin PLA spots per transduced IHC vs. 6.08 ± 0.5 spots per Pjvk+/+ IHC; P < 0.0001) (Fig. 5 A–C).
Fig. 5.
Transfer of Pjvk and Lc3b into Pjvk−/− IHCs restores pexophagy and abolishes lipid peroxidation after sound exposure. (A) The number of LC3B-positive spots colocalizing with PMP70 labeling was higher at 1 h after sound exposure (5–40 kHz, 105 dB SPL, 1h) in Pjvk−/− IHCs cotransduced with AAV2/8-Pjvk-IRES-EGFP and AAV2/8-Flag-Lc3b. (Inset) LC3B recruitment to peroxisomes. (B) The number of LC3B/pejvakin in situ PLA spots was increased at 1 h after sound exposure in Pjvk−/− IHCs cotransduced with AAV2/8-Pjvk-IRES-EGFP and AAV2/8-Flag-Lc3b. (C) The number of LC3B/pejvakin in situ PLA spots had increased slightly at 1 h after sound exposure in Pjvk−/− IHCs transduced with AAV2/8-Pjvk-IRES-EGFP. The bar charts in A–C quantify LC3B-positive spots colocalizing with PMP70 labeling (A) and LC3B/pejvakin in situ PLA spots (B and C) in treated IHCs before and after sound exposure (n = 40 IHCs; four mice per condition). (D) Peroxisome degradation (at 1 h after sound exposure) and proliferation (at 48 h after sound exposure) were fully restored in Pjvk−/− IHCs cotransduced with AAV2/8-Pjvk-IRES-EGFP and AAV2/8-Flag-Lc3b. AAV2/8-Pjvk-IRES-EGFP transduction of Pjvk−/− IHCs partially rescued early pexophagy and proliferation, whereas the transduction of Pjvk−/− IHCs with AAV2/8-Flag-Lc3b had no effect (n = 40 IHCs from four mice). (E) Lipid peroxidation levels, as assessed by 4-HNE immunolabeling, were higher in Pjvk−/− IHCs than in Pjvk+/+ IHCs at 3 h after sound exposure. Lipid peroxidation was abolished in Pjvk−/− IHCs cotransduced with AAV2/8-Pjvk-IRES-EGFP and AAV2/8-Flag-Lc3b but was significantly increased in both Pjvk−/− IHCs and AAV2/8-Flag-Lc3b Pjvk−/− IHCs. 4-HNE immunoreactivity was weaker in AAV2/8-Pjvk-IRES-EGFP Pjvk−/− IHCs than in Pjvk−/− IHCs (n = 50 cells; four mice per condition). Data are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant, unpaired Student’s t test. (Scale bars: 5 μm.)
Forty-eight hours later, peroxisome numbers had increased by a factor of 2.2 with respect to their prestimulation number in Pjvk−/− IHCs transduced with both AAV2/8-Pjvk-IRES-EGFP and AAV2/8-Flag-Lc3b and by a factor of 2.4 in Pjvk+/+ IHCs, the difference between these two values being nonsignificant (Fig. 5D). In contrast, the transduction of Pjvk−/− IHCs with Pjvk cDNA alone increased peroxisome numbers by a factor of 1.4 (Fig. 5D). The injection of Lc3b cDNA alone neither increased early LC3B-pejvakin interactions nor elicited peroxisome proliferation in Pjvk−/− mice (Fig. 5D).
We then investigated lipid peroxidation in auditory hair cells (as a marker of ROS damage) by assessing immunoreactivity for a byproduct of this process, 4-hydroxy-2-nonenal (4-HNE). The inverse correlation between the level of pexophagy (Figs. 1 C and D and 5 B and C) and that of 4-HNE (Fig. 5E) under the various experimental conditions, with the maximal effect obtained following cotransduction with Pjvk and Lc3b cDNAs, provided evidence for a major role of pejvakin-mediated pexophagy in redox homeostasis and the protection of auditory hair cells against noise-induced damage. This situation is reminiscent of the link between oxidative stress-induced pexophagy and redox homeostasis observed in cell lines (31). Moreover, the parallel changes in the magnitude of pexophagic and peroxisome proliferation responses to sound under the various experimental conditions, together with the observation of the strongest effects when LC3B was coexpressed with pejvakin (Fig. 5), suggested that noise-induced peroxisome proliferation is dependent on pejvakin-mediated pexophagy.
Discussion
Noise overexposure increases ROS levels, causing oxidative damage to auditory hair cells and neurons, resulting in hearing loss (1, 2). The peroxisome is a major source of H2O2. Its production and processing of H2O2 enables it to maintain its own redox balance, but this balance can be overwhelmed, leading to ROS-mediated damage to peroxisomal proteins and membrane lipids and a disturbance in the redox homeostasis of the cell (5). Peroxisomes adapt to the metabolic state of the cell by varying their proliferation and degradation and modifying their enzymatic content and shape (5, 6). We previously showed that peroxisome proliferation occurs at 48 h after noise overexposure, and that this proliferation is impaired in a mouse model of human deafness (DFNB59) lacking pejvakin and characterized by hypervulnerability to sound (1).
We show here that peroxisome proliferation in response to sound overstimulation is preceded by an early autophagic degradation of peroxisomes (pexophagy). The molecular mechanisms of pexophagy have been deciphered in detail in yeast and plants, but they remain poorly understood in mammals (32). Mammalian pexophagy, the principal mechanisms by which damaged or excess peroxisomes are degraded, involves the sequestration of these organelles in autophagosomes and their transfer to lysosomes. Three ubiquitin-dependent pexophagy pathways have been described (3), including one triggered by high levels of ROS. In this recently deciphered pathway, the ataxia telangiectasia mutated kinase (ATM), activated by ROS via the formation of a disulfide cross-linked dimer, is localized to peroxisomes (31) by the peroxisomal import receptor, PEX5 (33, 34). In response to ROS, ATM phosphorylates PEX5, promoting its monoubiquitination. Ubiquitinated PEX5 is recognized by the autophagy receptor/adaptor protein p62, directing the autophagosome to peroxisomes to induce selective pexophagy (31). NBR1 is also involved in this process (22). The p62 and NBR1 proteins have similar domain organizations; p62 has one LIR, and NBR1 has two LIRs, which bind to the LC3 associated with the nascent autophagosome.
We show here that pejvakin plays an essential role in sound-induced pexophagy. Unlike p62 and NBR1, pejvakin, which also contains an LIR domain, has no UBA domain. The lack of detection of a direct interaction of pejvakin with p62 and NBR1 by PLA under oxidative stress conditions does not exclude the involvement of pejvakin in this pathway. However, the absence of a UBA domain in pejvakin suggests that this protein may mediate an ROS-dependent and ubiquitination-independent pexophagy pathway. This situation is reminiscent of the pexophagy induced by nutrient starvation in mammalian cells, which has been shown to involve activation of the peroxisomal membrane protein, PEX14, which binds directly to LC3B (35). In vitro and in vivo experiments identified two cysteine residues of pejvakin, C328 and C343, as playing crucial roles in ROS-induced pejvakin–LC3B interaction and the pexophagy pathway. It remains to be clarified how these cysteine residues regulate pejvakin activity according to their redox state, and whether they are involved in intramolecular or intermolecular disulfide bond formation.
Our results reveal that pejvakin-mediated pexophagy precedes the proliferation of peroxisomes and protects the auditory hair cells against oxidative damage. Rescue experiments in Pjvk−/− mice stimulated with sound revealed a strong correlation between levels of pexophagy induction and the restoration of peroxisome proliferation. From the finding that adding LC3B to pejvakin increased not only pexophagy, but also peroxisome proliferation, we infer that ROS-induced pexophagy is involved in controlling peroxisome proliferation. The role of pejvakin in pexophagy can itself account for the reported lack of peroxisome proliferation in Pjvk−/− mice (1), but we cannot formally exclude the possibility of an additional direct role of pejvakin in peroxisome proliferation.
Overall, our findings reveal the key role of pexophagy in maintaining redox homeostasis and protecting the auditory system against noise-induced hearing loss. Peroxisomes and mitochondria are closely related organelles with similar clearance mechanisms (32, 36). Therefore, it would be interesting to investigate whether mitochondrial autophagy (mitophagy) also protects the hearing system against noise-induced damage.
Finally, our results establish that the DFNB59 form of deafness is a pexophagy disorder. Several peroxisomal membrane proteins have been implicated in pexophagy, and dysfunctions of pexophagy are being increasingly reported in peroxisomal disorders (37). The role of pejvakin in oxidative stress-induced pexophagy reported here demonstrates another function for gasdermin family proteins, which for most of them triggers pyroptosis (11–14).
Materials and Methods
Animals.
All animal experiments were performed in accordance with French and European legislation on the care and protection of laboratory animals (EC Directive 2010/63, French Law 2013–118, February 6, 2013) and the regulations of the Institut Pasteur Animal Care Committee. Pjvk−/− mice (approved allele designation Pjvktm2.1Ugds; MGI accession ID 5908028) were produced in a C57BL/6–129/Sv mixed genetic background (1).
Housing of Mice in an Acoustically Quiet Environment.
Because Pjvk−/− mice are hypervulnerable to the natural acoustic environment (1), we split pups from the same litter and placed in isolated boxes. The pups were separated before P10, corresponding to several days before onset of hearing in mice. The boxes were kept in quiet booths, shielded from the sounds emanating from other cages. Each cage contained four mice and a foster mother.
Acoustic Overexposure.
Three-week-old animals were exposed for 1 h to broadband white noise subjected to bandpass filtering over the 5–40 kHz frequency interval and applied at an intensity of 105 dB SPL. The white noise signal was generated with in-house MATLAB software (MathWorks) and was delivered by an amplifier to a set of four Ultrasonic Vifa speakers (Avisoft Bioacoustics).
Treatment of HepG2 and HeLa Cells with H2O2.
The cells were treated with 0.5 mM H2O2 at 37 °C for 6 h under normoxic conditions (95% air) and then fixed in 4% paraformaldehyde (PFA).
Plasmids and DNA Transfection.
The full-length pejvakin cDNA was obtained by RT-PCR, as described previously (1). The mutant pejvakin clones were prepared from the wild-type pejvakin clone with the QuikChange Site-Directed Mutagenesis kit (Stratagene). HeLa cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Immunofluorescence Studies.
For whole-mount immunolabeling analyses, inner ears were fixed in 4% PFA in PBS, and the cochlear sensory areas (organ of Corti) were microdissected. The tissues were rinsed twice in PBS, then permeabilized and blocked by incubation in 20% normal goat serum and 0.3% Triton X-100 in PBS for 1 h at room temperature. For GFP or Flag detection, whole-mount cochleas were incubated with a chicken anti-GFP antibody (1:100; Abcam ab13970) or a rabbit anti-Flag antibody (1:100; Sigma-Aldrich F7425) in 1% BSA in PBS. Anti-parvalbumin (1:200; Abcam Ab32895) and anti-myosin VI (1:100; Santa Cruz Biotechnology sc-23568) antibodies were used to delimit the contours of IHCs and outer hair cells (OHCs). Anti-PMP70 (1:100; Sigma-Aldrich SAB4200181) and anti-LC3B (1/100; Abcam ab51520) antibodies were used to label peroxisomes and autophagosomes, respectively.
For immunocytofluorescence analyses, HeLa and HepG2 cells were fixed by incubation in 4% PFA in PBS for 15 min, washed in PBS, and incubated in 50 mM NH4Cl and 0.2% Triton X-100 for 15 min at room temperature. The cells were then washed and incubated in 20% normal goat serum in PBS for 1 h, then incubated with the primary antibody in 1% BSA in PBS for 1 h. Antibodies against peroxisome membrane protein 70 (PMP70, 1:100; Sigma-Aldrich SAB4200181) and LC3B (1:100; Abcam ab51520) were used to label peroxisomes and autophagosomes, respectively. The mouse monoclonal antibody against pejvakin (Pjvk-G21) (1) was used at a concentration of 100 μg/mL. Cells were washed in PBS and incubated with the appropriate secondary antibody for 1 h at room temperature.
For immunofluorescence studies, we used Atto-647–conjugated goat anti-rabbit IgG (1:500; Sigma-Aldrich 40839), Atto-550–conjugated goat anti-mouse IgG (1:500, Sigma-Aldrich 43394), and Alexa Fluor 488-conjugated goat anti-chicken IgG (1:500; Invitrogen A11039) as secondary antibodies. DAPI (1:7,500, Sigma-Aldrich D9542) was used to label cell nuclei. Images were acquired with a Zeiss LSM700 Meta confocal microscope (Carl Zeiss MicroImaging). For whole-mount cochlea experiments, we analyzed the auditory hair cells from the middle part of the cochlea.
In Situ PLA.
The Duolink in situ PLA was performed according to the manufacturer’s protocol (https://www.sigmaaldrich.com/technical-documents/protocols/biology/duolink-fluorescence-user-manual.html) to detect the interaction between pejvakin and Lc3b, NBR1, or p62 in HepG2, HeLa, and cochlear hair cells. Cells were immunolabeled with a mouse monoclonal antibody against pejvakin (Pjvk-G21; 1:50) and rabbit polyclonal antibodies against LC3B (1:100; Abcam ab51520), NBR1 (1:100; Abcam ab219862), and p62 (1:100; Abcam ab91526). Anti-mouse and anti-rabbit secondary antibodies with PLA probes were supplied in the Duolink kit (Olink; DUO92101-1KT). DAPI was used to label cell nuclei.
For GFP detection, whole-mount cochleas were incubated with a chicken anti-GFP antibody (1:100; Abcam ab13970). Goat anti-myosin VI antibody (1:100; Santa Cruz Biotechnology sc-23568) was used to delimit the contours of IHCs and OHCs. Cells were observed by confocal microscopy, and interactions were detected as a fluorescent signal (red spot).
AAV-Pjvk and AAV-Lc3b Viral Constructs and Intracochlear Transduction.
AAV2/8 expressing either wild-type pejvakin or a mutated form of pejvakin (p.C328S;C343S), and AAV2/8-Flag-Lc3b were obtained by inserting the murine wild-type or mutated pejvakin cDNAs flanked by an IRES-EGFP reporter gene and Lc3b cDNA flanked by an Flag reporter gene into the multiple cloning site of the pENN.AAV.CB7.CI.RBG vector (PennVector PL-C-PV1044; Penn Medicine Vector Core, University of Pennsylvania School of Medicine). The viruses were produced and titrated by Penn Medicine Vector Core.
Intracochlear viral transduction was performed in P3 Pjvk−/− mice as described previously (1). A fixed volume (2 μL) of a solution containing AAV2/8-Pjvk-IRES-EGFP and AAV2/8-Flag-Lc3b alone or together (1013 viral genomes/mL), or AAV2/8-Pjvk(p.C328S;C343S)-IRES-EGFP alone, was carefully injected into the perilymphatic compartment of the cochlea through the round window.
Quantification of Lipid Peroxidation.
Lipid peroxidation in the cochlea was detected by immunohistofluorescence with an antibody against 4-hydroxy-2-nonenal (4-HNE) (1:200; Abcam ab48506). Images were acquired with a Zeiss LSM700 Meta confocal microscope under identical settings to allow comparisons. The mean pixel intensity of 4-HNE immunolabeling in cochlear hair cells was quantified with ImageJ software.
Prediction Tools.
The Phyre2 web portal was used to predict and analyze pejvakin structure and function (23). Fpocket, an open-source platform, was used to detect the ligand-binding pocket (26). The pejvakin protein sequences were multiply aligned using the MAFFT web server.
Statistical Analyses.
Quantitative data are presented as mean ± SEM, and statistical analyses were performed with GraphPad Prism. Data were analyzed using the unpaired Student t test and, for multiple comparisons, by the t test with Bonferroni correction. The differences between groups were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank M. Ricchetti and J. Boutet de Monvel for their critical reading of the manuscript. This work was supported by the Louis-Jeantet Foundation, Fondation Bettencourt Schueller, Fondation Agir pour l’Audition, Humanis Novalis-Taitbout, Réunica-Prévoyance, BNP Paribas, and the French Investissements d’Avenir program (ANR-10-LABX-65, to C.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821844116/-/DCSupplemental.
References
- 1.Delmaghani S, et al. Hypervulnerability to sound exposure through impaired adaptive proliferation of peroxisomes. Cell. 2015;163:894–906. doi: 10.1016/j.cell.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Ohlemiller KK, Wright JS, Dugan LL. Early elevation of cochlear reactive oxygen species following noise exposure. Audiol Neurotol. 1999;4:229–236. doi: 10.1159/000013846. [DOI] [PubMed] [Google Scholar]
- 3.Walker CL, Pomatto LCD, Tripathi DN, Davies KJA. Redox regulation of homeostasis and proteostasis in peroxisomes. Physiol Rev. 2018;98:89–115. doi: 10.1152/physrev.00033.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015;22:207–218. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrader M, Bonekamp NA, Islinger M. Fission and proliferation of peroxisomes. Biochim Biophys Acta. 2012;1822:1343–1357. doi: 10.1016/j.bbadis.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Waterham HR, Ferdinandusse S, Wanders RJ. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta. 2016;1863:922–933. doi: 10.1016/j.bbamcr.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Delmaghani S, et al. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet. 2006;38:770–778. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
- 10.Kovacs SB, Miao EA. Gasdermins: Effectors of pyroptosis. Trends Cell Biol. 2017;27:673–684. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 12.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 13.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 14.Rogers C, et al. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Op de Beeck K, et al. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur J Hum Genet. 2011;19:965–973. doi: 10.1038/ejhg.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 18.Shibata M, et al. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013;25:4967–4983. doi: 10.1105/tpc.113.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauthier T, Claude-Taupin A, Delage-Mourroux R, Boyer-Guittaut M, Hervouet E. Proximity ligation in situ assay is a powerful tool to monitor specific ATG protein interactions following autophagy induction. PLoS One. 2015;10:e0128701. doi: 10.1371/journal.pone.0128701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 21.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deosaran E, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 23.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alemu EA, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: Sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birgisdottir AB, Lamark T, Johansen T. The LIR motif: Crucial for selective autophagy. J Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 26.Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway ME, Lee C. The redox switch that regulates molecular chaperones. Biomol Concepts. 2015;6:269–284. doi: 10.1515/bmc-2015-0015. [DOI] [PubMed] [Google Scholar]
- 28.Jang HH, et al. Two enzymes in one: Two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, et al. Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a′. J Biol Chem. 2012;287:1139–1149. doi: 10.1074/jbc.M111.303149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mujtaba G, Bukhari I, Fatima A, Naz S. A p.C343S missense mutation in PJVK causes progressive hearing loss. Gene. 2012;504:98–101. doi: 10.1016/j.gene.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anding AL, Baehrecke EH. Cleaning house: Selective autophagy of organelles. Dev Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apanasets O, et al. PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic. 2014;15:94–103. doi: 10.1111/tra.12129. [DOI] [PubMed] [Google Scholar]
- 34.Ma C, Hagstrom D, Polley SG, Subramani S. Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J Biol Chem. 2013;288:27220–27231. doi: 10.1074/jbc.M113.492694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L, Hara-Kuge S, Yamashita S, Fujiki Y. Peroxin Pex14p is the key component for coordinated autophagic degradation of mammalian peroxisomes by direct binding to LC3-II. Genes Cells. 2015;20:36–49. doi: 10.1111/gtc.12198. [DOI] [PubMed] [Google Scholar]
- 36.Lismont C, Nordgren M, Van Veldhoven PP, Fransen M. Redox interplay between mitochondria and peroxisomes. Front Cell Dev Biol. 2015;3:35. doi: 10.3389/fcell.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho DH, Kim YS, Jo DS, Choe SK, Jo EK. Pexophagy: Molecular mechanisms and implications for health and diseases. Mol Cells. 2018;41:55–64. doi: 10.14348/molcells.2018.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.