Significance
Human prion disease is a rapidly fatal and incurable neurodegenerative disease. Reduction of prion protein in the brain is a well-supported therapeutic hypothesis, and antisense oligonucleotides with this mechanism of action are currently in development. To facilitate clinical testing of prion protein-lowering drugs in prion disease, we show that with proper sample handling, brain prion protein levels can be monitored in cerebrospinal fluid, using existing tools, and exhibit suitable short-term stability for drug-dependent decreases to be reliably measured. Cerebrospinal fluid prion protein levels thus may usefully serve as a pharmacodynamic biomarker. This biomarker may open new paths for informative clinical trials in presymptomatic individuals who harbor high-risk mutations for genetic prion disease.
Keywords: prion protein, cerebrospinal fluid, biomarker, human prion disease, Creutzfeldt-Jakob disease
Abstract
Reduction of native prion protein (PrP) levels in the brain is an attractive strategy for the treatment or prevention of human prion disease. Clinical development of any PrP-reducing therapeutic will require an appropriate pharmacodynamic biomarker: a practical and robust method for quantifying PrP, and reliably demonstrating its reduction in the central nervous system (CNS) of a living patient. Here we evaluate the potential of ELISA-based quantification of human PrP in human cerebrospinal fluid (CSF) to serve as a biomarker for PrP-reducing therapeutics. We show that CSF PrP is highly sensitive to plastic adsorption during handling and storage, but its loss can be minimized by the addition of detergent. We find that blood contamination does not affect CSF PrP levels, and that CSF PrP and hemoglobin are uncorrelated, together suggesting that CSF PrP is CNS derived, supporting its relevance for monitoring the tissue of interest and in keeping with high PrP abundance in brain relative to blood. In a cohort with controlled sample handling, CSF PrP exhibits good within-subject test–retest reliability (mean coefficient of variation, 13% in samples collected 8–11 wk apart), a sufficiently stable baseline to allow therapeutically meaningful reductions in brain PrP to be readily detected in CSF. Together, these findings supply a method for monitoring the effect of a PrP-reducing drug in the CNS, and will facilitate development of prion disease therapeutics with this mechanism of action.
Prion disease, a fatal and incurable neurodegenerative disease, is caused by misfolding of the prion protein (PrP), encoded by the gene PRNP (1). PrP is a well-validated drug target for prion disease: knockout animals are invulnerable to prion infection (2), heterozygous knockouts have delayed onset of disease (3), and postnatal depletion of PrP can delay or prevent prion disease (4, 5). Total knockout is tolerated in mice (6, 7), cows (8), and goats (9, 10), and healthy humans with one loss-of-function allele of PRNP have been identified (11). Further, PrP serves as a common target uniting all subtypes of human prion disease, including Creutzfeldt-Jakob disease, fatal familial insomnia, and Gerstmann-Straussler-Scheinker disease (12). Therefore, therapeutic development efforts have sought to lower PrP in the brain (13–16), and antisense oligonucleotides with this mechanism of action are currently in development (17). Similar approaches are being explored in other neurodegenerative diseases, with promising preliminary clinical results (18, 19).
Clinical trials of PrP-lowering therapies will be enhanced by early determination of whether PrP is indeed being lowered effectively at a tolerated dose. The brain is the target tissue for any prion disease therapeutic, but is difficult to monitor directly. Cerebrospinal fluid (CSF) is produced by the choroid plexus of the ventricles, flows in and around the spinal cord, and is in intimate contact with interstitial fluid of brain parenchyma. CSF more closely reflects the biochemistry of the brain than blood or any other accessible tissue, and is obtainable through a minimally invasive lumbar puncture. PrP levels in CSF range from tens to hundreds of nanograms per milliliter, within the range of standard protein detection assays. Multiple groups have reported successful detection of PrP in human CSF, using ELISAs, including the one currently commercially available human PrP ELISA kit, the BetaPrion ELISA (20–24) (Analytik Jena, Leipzig, Germany). The assay is best described as measuring total PrP, which is the variable of interest for PrP-lowering therapeutics (Discussion).
Informed by US Food and Drug Administration’s 2013 Draft Guidance on Bioanalytical Method Validation (25), we assessed the technical performance of the BetaPrion ELISA across 225 human CSF samples spanning a range of diagnoses. We then used this assay to investigate the biological suitability of CSF PrP as a pharmacodynamic biomarker for PrP-reducing therapeutics.
Results
The BetaPrion Human PrP ELISA Quantifies Total CSF PrP Reproducibly, Precisely, Sensitively, and Selectively.
We assessed the assay’s precision, sensitivity, selectivity and reproducibility by analyzing 225 human CSF samples from patients with symptomatic prion disease, presymptomatic prion disease mutation carriers, patients with nonprion dementia, and patients with normal pressure hydrocephalus, as well as other nonprion controls (SI Appendix, Table S1), across 41 plates. The results broadly support the technical suitability of this assay for reliable quantification of CSF PrP (Table 1 and SI Appendix, Fig. S1).
Table 1.
The technical performance of the BetaPrion human PrP ELISA supports reliable quantification of PrP in human CSF
| Experiment | Results |
| Within-plate technical replicate reproducibility (same dilution) | CV = 8% |
| Within-plate technical replicate reproducibility (all dilutions) | CV = 11% |
| Between-plate technical replicate reproducibility | CV was 22% in an interplate control sample run on 17 plates on different days (SI Appendix, Supplementary Discussion). |
| Sensitivity | LLOQ is 3–5× the blank signal, depending on the plate reader used. |
| Selectivity | Nonreactive for recombinant mouse PrP, rat CSF, and cynomolgus monkey CSF [consistent with one amino acid mismatch in the reported detection antibody epitope (18)], artificial CSF, and protease-digested CSF. |
| Dilution linearity | Linear across two samples and five dilutions. See SI Appendix, Fig. S1A. |
| Spike recovery | Using AAA-quantified recombinant human PrP23-230 as a standard, spike recovery of recombinant PrP in CSF was 90% across five concentrations. Titration of a high PrP CSF sample into a low PrP sample resulted in linear recovery. See SI Appendix, Fig. S3. |
| Standard curve reproducibility | CV was <10% at all six nonzero standard curve points across five replicates. See SI Appendix, Fig. S1. |
CV, coefficient of variation; LLOQ, lower limit of quantification.
In assessing within-plate variability, we discerned plate position effects for control samples, with a mild but significant downward trend from upper left to lower right (SI Appendix, Fig. S2). Comparison of the kit standard curve to a standard curve made from recombinant human prion protein quantified by amino acid analysis (AAA) yielded systematic differences, with implications for kit use for absolute versus relative quantification of PrP (SI Appendix, Fig. S3B and Discussion).
Standardized Storage and Handling Are Essential to Reliable Quantification of CSF PrP.
PrP was measurable by ELISA in all 225 CSF samples analyzed, including in CSF from individuals with 13 different genetic prion disease mutations (SI Appendix, Fig. S4 A and B and Table S1). Across all CSF samples analyzed, PrP levels varied by more than two orders of magnitude (SI Appendix, Fig. S4A), ranging from 1.9 to 594 ng/mL. PrP was reduced in individuals with symptomatic prion disease, as previously reported (20, 21, 23, 24, 26). Within matched cohorts containing individuals with prion disease, however, diagnostic category (nonprion, presymptomatic genetic, symptomatic genetic, and sporadic prion disease) explained only a minority of variance in CSF PrP level (adjusted R2 = 0.23; P < 1 × 10−7, linear regression). After excluding individuals with symptomatic prion disease, PrP still differed significantly between the various cohorts included in our study, and within-cohort variation was also dramatic (SI Appendix, Fig. S4C; mean ∼20-fold difference between highest and lowest sample within a cohort). These observations led us to search for other factors that might contribute to either biological or preanalytical variability. CSF PrP was correlated with age (SI Appendix, Fig. S4D), but among our samples, age is confounded with cohort, diagnosis, and likely many unobserved variables, making it unclear whether this correlation is biologically meaningful. CSF PrP did not differ according to sex (SI Appendix, Fig. S4E), and exhibited no lumbar–thoracic gradient over serial tubes collected from the same lumbar puncture (SI Appendix, Fig. S4 F and G). After noticing that PrP levels appeared lower in smaller aliquots of the same CSF sample (SI Appendix, Fig. S5A), we hypothesized that differences in sample handling might be one major source of variability in observed CSF PrP levels.
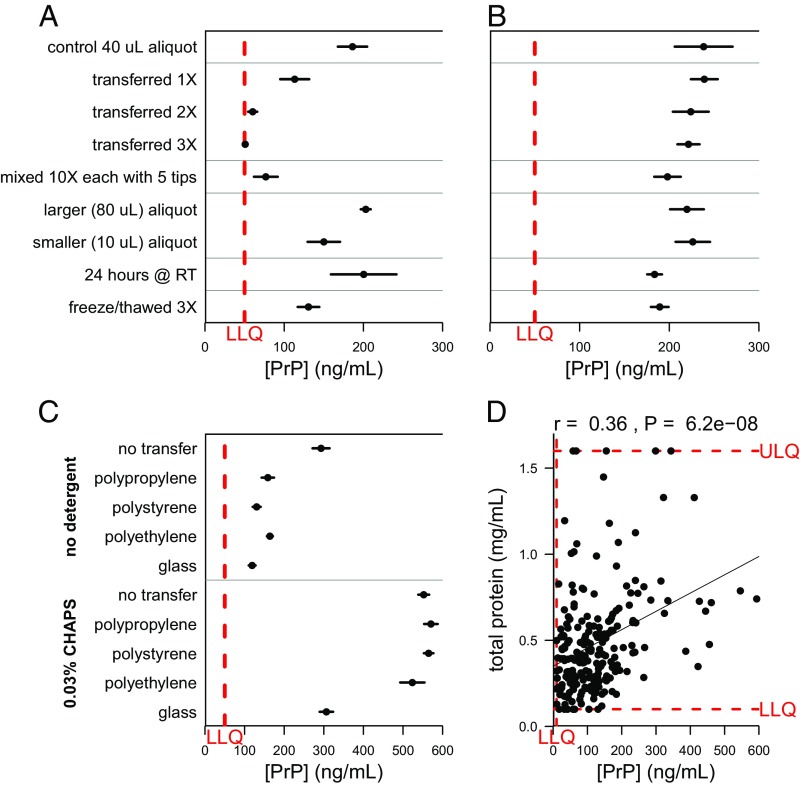
It is known that other neurodegenerative disease-associated amyloidogenic proteins have a high affinity for plastics (27–29), but PrP’s stability under different handling conditions has not previously been systematically investigated. To assess PrP’s susceptibility to differential CSF sample handling, we subjected aliquots of a single CSF sample to variations in number of transfers between polypropylene storage tubes, amount of exposure to polypropylene pipette tips, storage aliquot size, storage temperature, and number of freeze–thaw cycles (Fig. 1A). Strikingly, increased plastic exposure in the first three conditions dramatically reduced measurable PrP in solution (Fig. 1A). To promote PrP solubility in our samples, we experimented with adding small amounts of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate (CHAPS), a common zwitterionic surfactant known to enhance protein solubility in multiple contexts (30–32). Addition of 0.03% CHAPS before aliquoting minimized PrP loss to plastic across most manipulations (Fig. 2B). For instance, transferring a CSF sample to a new microcentrifuge tube three times eliminated at least 73% of detectable PrP (P < 1 × 10−6, two-sided t test) without CHAPS, but only 7.1% (P = 0.37) of PrP was lost in the presence of 0.03% CHAPS. Addition of CHAPS also increased total PrP recovery, presumably by preventing loss to the single polypropylene tube and tips used for plating samples (SI Appendix, Fig. S5), and was effective against loss to multiple plastics, but not glass (Fig. 1C). Storing CSF at room temperature for 24 h or subjecting samples to three freeze–thaw cycles had a less dramatic effect on PrP that did not appear to be affected by CHAPS (Fig. 1 A and B and SI Appendix, Fig. S5 D and E).
Fig. 1.
Storage and handling can dramatically reduce the amount of PrP detected in CSF samples unless appropriate measures are taken. In A–C, dots represent mean and line segments represent 95% confidence intervals across four to seven aliquots of the same sample, each measured in duplicate at a 1:50 dilution. (D) Dots represent mean of measurements within dynamic range, among 2 dilutions with 2 technical replicates each. (A) Increased polypropylene exposure substantially reduces detectable PrP. (B) Addition of 0.03% CHAPS detergent to samples increases PrP recovery and consistently mitigates PrP loss to plastic. (C) Addition of CHAPS (Bottom) increases total PrP recovery and shows similar rescue across plastics, but substantial PrP loss is still observed on storage in glass. (D) Across 217 CSF samples, total protein levels and PrP levels were modestly correlated (Spearman’s rank correlation coefficient = 0.36; P = 6.2 × 10−8). (A–C) Dots represent mean and line segments represent 95% confidence intervals across four to seven aliquots of the same sample, each measured in duplicate at a 1:50 dilution. (D) Dots represent mean of measurements within dynamic range, among two dilutions with two technical replicates each.
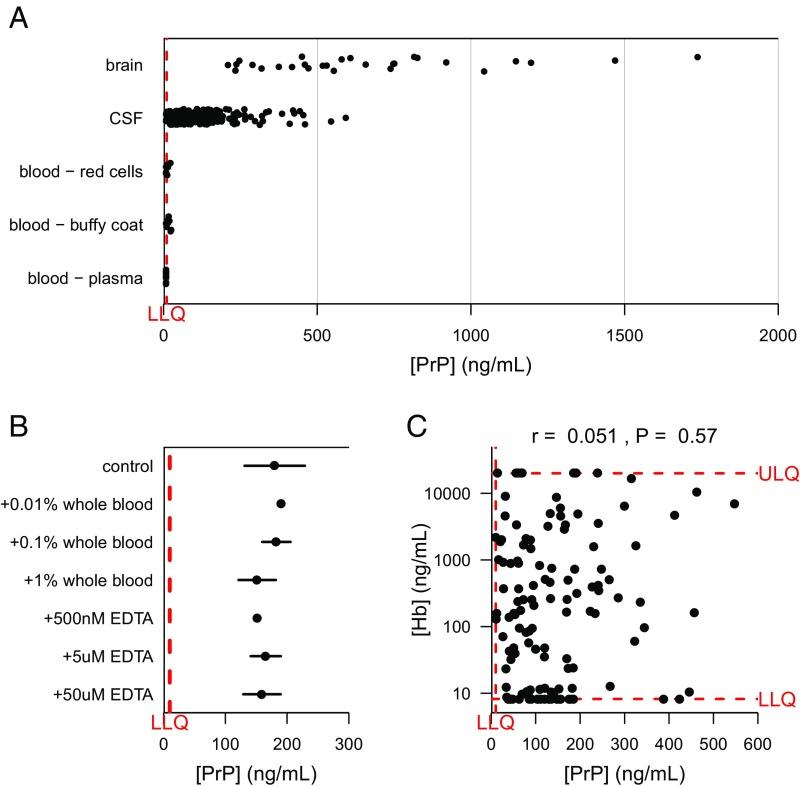
Fig. 2.
Blood PrP contributes negligibly to the PrP detected in CSF. (A) PrP levels were compared by ELISA in 28 postmortem human brain samples, three blood fractions from eight individuals each, and all (n = 225) CSF samples analyzed in the present study. PrP is abundant in a range of human brain regions, undetectable in human plasma, and detectable in the red cell and buffy coat fractions only at low levels compared with PrP in CSF. (B) Spiking whole blood into CSF up to 1% by volume does not affect measured PrP. (C) Across 128 CSF samples spanning multiple cohorts and diagnostic categories, hemoglobin and PrP levels in CSF are uncorrelated. In A and C, dots represent mean of measurements within dynamic range, among 2 technical replicates per dilution. In A–C, dots represent mean and line segments represent 95% confidence intervals across two to three aliquots of the same sample.
We also investigated the relationship between measured PrP and total protein in 217 samples, using the DC total protein assay. Across all samples analyzed, a modest correlation (r = 0.36; Spearman rank test, P < 1 × 10−7) between PrP and total protein was observed (Fig. 1D), which may reflect either a biological phenomenon or simply the ability of higher ambient protein levels to serve a blocking function that partially offsets PrP loss by adsorption. In support of the latter interpretation, addition to CSF of 1 mg/mL BSA increased recovery of PrP (SI Appendix, Fig. S5F), although it was less effective than CHAPS at preventing loss resulting from transfers.
PrP in CSF Is CNS Derived and Unlikely To Be Confounded by Blood Contamination.
CSF PrP is an informative tool in prion disease only insofar as it is a faithful proxy for PrP levels in the CNS, the relevant target for any future therapeutic. CSF proteins derive from two major sources, CNS and blood, with proportional contribution driven by relative tissue abundance of a given protein (33, 34). Blood proteins may enter CSF either through passive diffusion as CSF flows along the spinal cord (35), or artifactually if blood from a traumatic lumbar puncture contaminates the collected CSF. To assess the contribution of blood-derived PrP to overall CSF PrP, we compared PrP levels across brain samples and red blood cell, buffy coat, and plasma fractions of blood from nonneurodegenerative disease control individuals versus all the CSF samples in our study (Fig. 2A). Among blood fractions, PrP was most consistently detected in buffy coat, in keeping with reports that blood PrP emanates chiefly from platelets (36, 37); we also detected PrP above the lower limit of quantification in some red cell samples, but never in plasma. As the average PrP concentration in all three blood fractions was still well below that in brain and was lower than that in 96% of CSF samples analyzed, the risk of confounding signal from blood-derived PrP appears negligible. Consistent with this conclusion, spiking whole blood into CSF at up to 1% (vol/vol) did not increase the detected PrP (Fig. 2B). Finally, as a proxy for blood contamination, we measured hemoglobin levels in 128 CSF samples and observed no correlation between CSF hemoglobin and CSF PrP (Fig. 2C). Variation in hemoglobin levels also failed to confound the test–retest reliability of CSF PrP (SI Appendix, Fig. S6). From these lines of evidence, we conclude that the PrP detected in CSF is overwhelmingly derived from the CNS.
CSF PrP Levels in Individuals Are Stable on Short-Term Test–Retest.
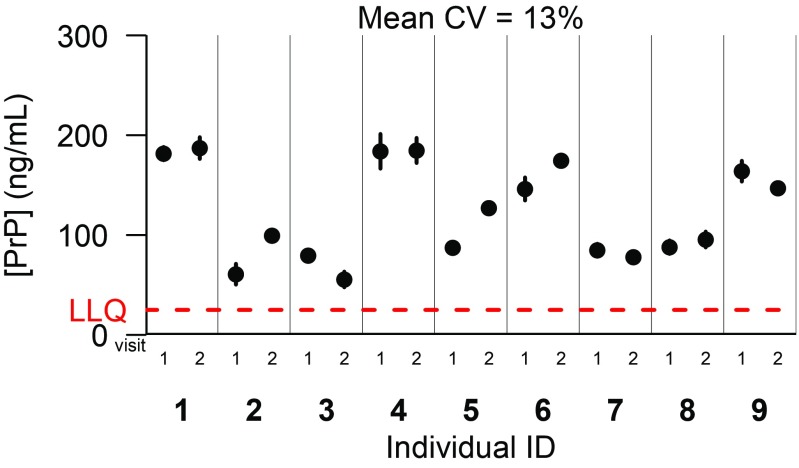
For CSF PrP levels to serve as a meaningful biomarker, they must be stable enough in one individual over time that a drug-dependent reduction could be reliably detected. We quantified PrP in pairs of CSF samples collected from nine individuals (placebo-treated controls with nonprion dementia) who had undergone two fasting morning lumbar punctures at 8- to 11-wk intervals in the context of a clinical trial (38). Lumbar punctures were performed according to a standardized protocol by a single investigator, and samples were subsequently processed uniformly. Under these highly controlled conditions, the mean CV between points for a given participant was reasonably low, at 13% (Fig. 3). Higher CVs of 33–41% were observed in three other cohorts in which sample handling appears to have been less uniform (SI Appendix, Supplementary Discussion and Fig. S7), consistent with PrP’s susceptibility to preanalytical perturbations (Fig. 1).
Fig. 3.
Test–retest stability of CSF PrP. Uniformly processed CSF samples were provided from a past clinical trial, from placebo-treated individuals with mild, nonprion cognitive impairment. Fasting morning lumbar punctures were performed by one investigator on nine individuals and then repeated at an interval of 8–11 wk. Dots represent means, and line segments 95% confidence intervals, of measurements within dynamic range among two dilutions with two technical replicates each.
Discussion
Our data support the use of CSF PrP quantification as a pharmacodynamic biomarker for clinical trials of PrP-lowering therapeutics. CSF PrP is CNS derived, rather than blood derived, so it should respond to PrP lowering in the brain. With appropriate protocols, it can be measured reproducibly and with favorable test–retest reliability.
Our experiments suggest best practices for sample handling and assay use. CSF PrP is sensitive to preanalytical factors, but the addition of 0.03% CHAPS detergent mitigates the most dramatic such factor by minimizing PrP loss to plastic. A recommended CSF collection and processing protocol is detailed in SI Appendix, Fig. S8. Also, in light of subtle plate position effects (SI Appendix, Fig. S2), samples intended for comparison could be colocated on the ELISA plate, and/or plate position should be adjusted for using standard curves or control samples. Our comparison of the kit standard curve to an AAA-quantified standard curve suggests that the assay may be most useful for relative rather than absolute quantification of PrP (SI Appendix, Fig. S3B).
Our study has several limitations. First, ELISA might not detect all conformers or fragments of PrP. Although we hypothesize that the BetaPrion assay measures total PrP, we are presently developing a targeted mass spectrometry-based orthogonal method to test this hypothesis. For study of specific PrP isoforms, future ELISA development efforts could leverage additional PrP antibodies to quantify particular subsets of PrP molecules (39). Second, although we have established that CSF PrP is quantifiable in patients with genetic prion disease across a variety of mutations and has good test–retest reliability in a cohort of patients with nonprion dementia, when we embarked on the present study, we did not have access to test–retest samples from presymptomatic genetic prion disease mutation carriers. To address this shortfall, in summer 2017 we launched a longitudinal clinical research study at Massachusetts General Hospital, through which we are collecting serial CSF from PRNP mutation carriers and controls (40). Third, the samples analyzed here were reused after collection for other research or clinical purposes, and in most cases, we cannot fully account for their sample handling history before receipt by our laboratory. Thus, our numbers may exaggerate the interindividual variation in CSF PrP in the population.
PrP levels in CSF as measured by ELISA are reduced by approximately half in patients with symptomatic prion disease (21, 23, 24), a phenomenon reproduced here (SI Appendix, Fig. S4). Multiple plausible biological mechanisms could explain these findings: incorporation of PrP into insoluble plaques (41), internalization of misfolded PrP in the endosomal–lysosomal pathway (42), and posttranslational down-regulation of PrP as a function of disease (43). It is therefore possible that an intrinsic reduction in CSF PrP in the course of symptomatic disease could confound the use of PrP as a biomarker for the activity of PrP-lowering drug tested in a symptomatic population. Although it is important to be aware of this potential limitation, symptomatic patients are not the population most in need of such a biomarker. The rapid progression of prion disease has enabled symptomatic trials to use cognitive or survival endpoints (44–46), and future trials may be further benefit from the use of real-time quaking induced conversion to detect misfolded prion “seeds” in symptomatic patient CSF (47–49).
Instead, this biomarker may have its greatest utility in presymptomatic individuals carrying high-risk genetic prion disease mutations. As trials in symptomatic neurodegenerative disease patients continue to fail or prove uninterpretable, it is increasingly recognized that therapeutic efforts must aim further upstream in the disease process (50). Although identifiable by genetic testing, genetic prion disease mutation carriers appear healthy up to the stark precipice of symptom onset, creating a compelling case for prevention. Because following presymptomatic individuals to a clinical endpoint appears infeasible (51), lowering CSF PrP has been proposed as a surrogate endpoint to enable presymptomatic trials of agents such as the antisense oligonucleotides currently in development.* In this context, CSF PrP may have a near-term opportunity to serve, not just as a pharmacodynamic biomarker but also as a pivotal readout that enables a rational therapeutic to be tested for its ability to extend healthy life, thus honoring the opportunity provided by predictive genetic testing.
Methods
CSF Samples.
De-identified human CSF samples were provided by multiple clinical collaborators and included some previously published samples (38, 52). Samples were shipped on dry ice and stored at −80 °C. Before use, samples were thawed on ice and centrifuged at 2,000 × g (at 4 °C). Ninety percent of the volume was pipetted into a new tube to separate supernatant from cellular or other debris, aliquoted into new polypropylene storage tubes, and refrozen at −80 °C. For indicated samples, 0.03% CHAPS detergent by volume (final concentration, from a 3% CHAPS stock) was preloaded into the supernatant receiving tube before the postcentrifugation transfer, and then mixed into the sample by gentle pipetting before aliquoting.
Quantification of Human PrP in CSF, Brain Tissue, and Blood, Using the BetaPrion Human PrP ELISA Kit.
Across experiments, PrP was quantified using the BetaPrion human PrP ELISA kit (Analytik Jena, cat no. 847–0104000104) according to the manufacturer’s instructions. This sandwich ELISA is configured in 96-well format and relies on an apparently conformational human PrP capture antibody and a horseradish peroxidase-conjugated primary detection antibody to human PrP residues 151–180 (23). In brief, samples were diluted into blocking buffer (5% BSA and 0.05% Tween-20 in PBS, filtered before use) at concentrations ranging from 1:100 to neat, depending on the anticipated PrP content of the sample type. All samples were plated in duplicate. Lyophilized standards and kit reagents were diluted fresh for same-day use, with the exception of wash buffer and blocking buffer, excess of which were stored at 4 °C for reuse within 4 wk. The assay format is 96-well comprised of 12 modular 8-well strips that enabled partial plates to be run in some cases. After all add and incubation steps, the absorption per well was read in either a SpectraMax or FluoStar Optima plate reader at 450 nm, with 620 nm absorbance also monitored as baseline. Data were exported as a text file and analyzed in R.
Unknown CSF samples were run at two dilutions each (typically 1:10 and 1:50). Only one of 225 CSF samples analyzed fell below the range of the assay’s lower limit of detection (1 ng/mL final) at a 1:10 dilution, and was rerun neat, yielding a result of 1.9 ng/mL. Except where noted, samples were run in technical duplicate at two dilutions, and error bars represent 95% confidence intervals around the mean.
Human brain samples were obtained from the Massachusetts Alzheimer's Disease Research Center (n = 26 samples from 5 different control individuals without neurodegenerative disease, with postmortem intervals of 23–72 h, representing diverse cortical and subcortical regions) and from the National Prion Disease Pathology Surveillance Center (n = 2 samples of frontal cortex from nonprion controls) homogenized in PBS with 0.03% CHAPS at 10% weight/vol in 7 mL tubes (Precellys no. KT039611307.7), using a MiniLys tissue homogenizer (Bertin no. EQ06404-200-RD000.0) for three cycles of 40 s at maximum speed. The resulting 10% brain homogenates were diluted 1:10 and 1:100 in blocking buffer for ELISA.
Human blood fractions were obtained from Zen-Bio [three fractions (red blood cell, buffy coat, and plasma) from eight individuals each], 0.03% CHAPS was added, and samples were then mixed either by pipetting up and down or by homogenization in a MiniLys using the same protocol described earlier. Blood fractions were diluted 1:10 in blocking buffer for ELISA.
Negative Controls.
Rat and cynomolgus monkey CSF (BioReclamation IVT; two samples each from two separate animals) and artificial CSF (Tocris no. 3525) were aliquoted and stored at −80 °C. For protease-digested CSF, two CSF samples with 0.03% CHAPS (measured to contain 273 and 643 ng/mL PrP undigested) were digested with 5 µg/mL Proteinase K (WW Grainger Co. cat. no. 5000186667) at 37C for 1 h, after which the digestion was halted with 4 mM PefaBloc (Sigma Aldrich cat. no. 11429868001) immediately before use in ELISA.
Recombinant Prion Protein Purification.
For spike-in experiments and attempted detection of mouse recombinant PrP, in-house purified recombinant full-length human prion protein and mouse prion protein were purified from Escherichia coli, using established vectors (a generous gift from Byron Caughey's laboratory at NIH Rocky Mountain Labs), according to established methods (53, 54). Protein concentration was determined by 280 nm absorbance on a NanoDrop, and by AAA performed in duplicate (New England Peptide) after the addition of 0.03% CHAPS.
Storage and Handling Experiments.
For all storage and handling experiments, each condition was run in parallel on four identical aliquots made from one original CSF sample, and each aliquot was plated in duplicate. For all transfer experiments, 40 µL CSF aliquots were thawed on ice, and then the full volume was transferred to a new 500-µL storage tube the indicated number of times and allowed to sit for a minimum of 15 min in each tube. Where not otherwise indicated, tubes were polypropylene, and sample aliquots were 40 µL.
Total Protein Assay.
The DC total protein assay (Bio-Rad cat. no. 5000111) was used according to the manufacturer’s instructions to measure total protein across 217 CSF samples (all samples in this study except for the n = 8 lumbar-thoracic gradient samples; SI Appendix, Fig. S4 F and G). This assay, similar in principle to a Lowry assay, combines the protein with an alkaline copper tartrate solution and Folin reagent (55). The protein reacts with copper in the alkaline medium, then reduces the Folin reagent to yield species with a characteristic blue color in proportion to abundance of key amino acids, including tyrosine and tryptophan.
Whole Blood Spike-In.
Human whole blood (Zen-Bio) was spiked into parallel aliquots of a single CSF sample containing baseline midrange PrP at 1%, 0.1%, or 0.01% per volume. EDTA spike-ins were performed in parallel to control for EDTA preservative carried in the blood sample. Samples were refrozen after spike-in and then rethawed for use to ensure lysis of cellular fractions before PrP quantification.
Bethyl Laboratories Human Hemoglobin ELISA.
Hemoglobin was quantified in 128 human CSF samples using the Human Hemoglobin ELISA kit (Bethyl Laboratories no. E88-134), according to the manufacturer’s instructions. Samples for this analysis spanned diagnostic categories including normal pressure hydrocephalus, nonprion dementia, and symptomatic genetic and symptomatic sporadic prion disease. Samples were diluted 1:10 and 1:100 for most experiments, and in some cases, 1:20 and 1:100. All samples were plated in duplicate.
Blinding Procedures.
Assay operators were blinded to diagnosis for prion disease CSF cohorts. For test–retest cohorts, assay operators were blinded to test–retest pairing for metformin trial samples and MassGeneral Institute for Neurodegenerative Disease Tissue Bank samples; pairing was known but collection order unknown for University of California, San Francisco, samples; pairing and order were known for sapropterin trial samples.
Statistics, Data, and Source Code Availability.
All statistical analyses were conducted, and figures generated, using custom scripts in R 3.1.2. Raw data from plate readers, associated metadata, and source code sufficient to reproduce the analyses reported here are publicly available at: https://github.com/ericminikel/csf_prp_quantification/.
Supplementary Material
Acknowledgments
We thank Joanne Kotz of Jnana Therapeutics for discussions. We are also grateful to the patients and their families who contributed samples to this research. We thank the Massachusetts Alzheimer’s Disease Research Center (supported by NIH Grant P50 AG005134) and the MassGeneral Institute for Neurodegenerative Disease Tissue Bank for providing human brain and cerebrospinal fluid samples for this study. This study was approved by the Broad Institute’s Office of Research Subjects Protection (protocols ORSP-3587 and NHSR-4693). The research was supported in part by the National Institute of General Medical Sciences (Award R35GM127045 to S.L.S.). S.M.V. is supported by the National Science Foundation (GRFP Award 2015214731). E.V.M. is supported by the National Institutes of Health (F31 Award AI122592). This work was supported by BroadIgnite and the Next Generation Fund at the Broad Institute of MIT and Harvard, Prion Alliance, anonymous donors, and an anonymous organization. Work in Germany was supported by Robert-Koch-Institute through funds from the Federal Ministry of Health (Grant 1369-341 to I.Z.) and Spanish Ministry of Health-Instituto Carlos III (Grant Miguel Servet-CP16/00041 to F.L.) University of California, San Francisco, sample collection was supported by the NIH/National Institute on Aging (Grants R01 AG-031189 and R01 AG-032289) and the National Center for Advancing Translational Sciences (Grant NIH UCSF-CTSI UL1 RR024131).
Footnotes
Conflict of interest statement: S.L.S. is a member of the Board of Directors of the Genomics Institute of the Novartis Research Foundation ("GNF"); a shareholder and member of the Board of Directors of Jnana Therapeutics; a shareholder of Forma Therapeutics; a shareholder of and adviser to Decibel Therapeutics and Eikonizo Therapeutics; an adviser to Eisai, Inc., the Ono Pharma Foundation, and F-Prime Capital Partners; and a Novartis Faculty Scholar.
*U.S. Food and Drug Administration, Critical Path Innovation Meeting: Genetic Prion Disease, Silver Spring, MD 20903, November 14, 2017.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901947116/-/DCSupplemental.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Büeler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 3.Büeler H, et al. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- 4.Mallucci G, et al. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 5.Safar JG, et al. Prion clearance in bigenic mice. J Gen Virol. 2005;86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- 6.Büeler H, et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 7.Bremer J, et al. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 8.Richt JA, et al. Production of cattle lacking prion protein. Nat Biotechnol. 2007;25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu G, et al. Generation of goats lacking prion protein. Mol Reprod Dev. 2009;76:3. doi: 10.1002/mrd.20960. [DOI] [PubMed] [Google Scholar]
- 10.Benestad SL, Austbø L, Tranulis MA, Espenes A, Olsaker I. Healthy goats naturally devoid of prion protein. Vet Res (Faisalabad) 2012;43:87. doi: 10.1186/1297-9716-43-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minikel EV, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8:322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mead S. Prion disease genetics. Eur J Hum Genet. 2006;14:273–281. doi: 10.1038/sj.ejhg.5201544. [DOI] [PubMed] [Google Scholar]
- 13.White MD, et al. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nazor Friberg K, et al. Intracerebral infusion of antisense oligonucleotides into prion-infected mice. Mol Ther Nucleic Acids. 2012;1:e9. doi: 10.1038/mtna.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silber BM, et al. Novel compounds lowering the cellular isoform of the human prion protein in cultured human cells. Bioorg Med Chem. 2014;22:1960–1972. doi: 10.1016/j.bmc.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn M, et al. Convection-enhanced delivery of AAV2-PrPshRNA in prion-infected mice. PLoS One. 2014;9:e98496. doi: 10.1371/journal.pone.0098496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clancy K. 2019 One couple’s tireless crusade to stop a genetic killer. Wired. Available at https://www.wired.com/story/sleep-no-more-crusade-genetic-killer/. Accessed January 25, 2019.
- 18.Miller TM, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442, and erratum (2013) 12:423. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wild EJ, Tabrizi SJ. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16:837–847. doi: 10.1016/S1474-4422(17)30280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llorens F, et al. PrP mRNA and protein expression in brain and PrP(c) in CSF in Creutzfeldt-Jakob disease MM1 and VV2. Prion. 2013;7:383–393. doi: 10.4161/pri.26416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyne F, et al. Total prion protein levels in the cerebrospinal fluid are reduced in patients with various neurological disorders. J Alzheimers Dis. 2009;17:863–873. doi: 10.3233/JAD-2009-1110. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz M, et al. Codon 129 polymorphism and the E200K mutation do not affect the cellular prion protein isoform composition in the cerebrospinal fluid from patients with Creutzfeldt-Jakob disease. Eur J Neurosci. 2010;31:2024–2031. doi: 10.1111/j.1460-9568.2010.07224.x. [DOI] [PubMed] [Google Scholar]
- 23.Dorey A, et al. Association of cerebrospinal fluid prion protein levels and the distinction between Alzheimer disease and Creutzfeldt-Jakob disease. JAMA Neurol. 2015;72:267–275. doi: 10.1001/jamaneurol.2014.4068. [DOI] [PubMed] [Google Scholar]
- 24.Abu Rumeileh S, et al. Diagnostic accuracy of a combined analysis of cerebrospinal fluid t-PrP, t-tau, p-tau, and Aβ42 in the differential diagnosis of Creutzfeldt-Jakob disease from Alzheimer’s disease with emphasis on atypical disease variants. J Alzheimers Dis. 2017;55:1471–1480. doi: 10.3233/JAD-160740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Food and Drug Administration 2013 Guidance for industry: Bioanalytical method validation. Available at https://www.regulations.gov/document?D=FDA-2013-D-1020-0002. Accessed March 20, 2019.
- 26.Torres M, et al. Altered prion protein expression pattern in CSF as a biomarker for Creutzfeldt-Jakob disease. PLoS One. 2012;7:e36159. doi: 10.1371/journal.pone.0036159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewczuk P, et al. Effect of sample collection tubes on cerebrospinal fluid concentrations of tau proteins and amyloid β peptides. Clin Chem. 2006;52:332–334. doi: 10.1373/clinchem.2005.058776. [DOI] [PubMed] [Google Scholar]
- 28.Wild EJ, et al. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J Clin Invest. 2015;125:1979–1986. doi: 10.1172/JCI80743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perret-Liaudet A, et al. Risk of Alzheimer’s disease biological misdiagnosis linked to cerebrospinal collection tubes. J Alzheimers Dis. 2012;31:13–20. doi: 10.3233/JAD-2012-120361. [DOI] [PubMed] [Google Scholar]
- 30.Wetlaufer DB, Xie Y. Control of aggregation in protein refolding: A variety of surfactants promote renaturation of carbonic anhydrase II. Protein Sci. 1995;4:1535–1543. doi: 10.1002/pro.5560040811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cladera J, Rigaud JL, Villaverde J, Duñach M. Liposome solubilization and membrane protein reconstitution using Chaps and Chapso. Eur J Biochem. 1997;243:798–804. doi: 10.1111/j.1432-1033.1997.00798.x. [DOI] [PubMed] [Google Scholar]
- 32.Hjelmeland LM, Chrambach A. Solubilization of functional membrane proteins. Methods Enzymol. 1984;104:305–318. doi: 10.1016/s0076-6879(84)04097-0. [DOI] [PubMed] [Google Scholar]
- 33.You J-S, et al. The impact of blood contamination on the proteome of cerebrospinal fluid. Proteomics. 2005;5:290–296. doi: 10.1002/pmic.200400889. [DOI] [PubMed] [Google Scholar]
- 34.Reiber H. Proteins in cerebrospinal fluid and blood: Barriers, CSF flow rate and source-related dynamics. Restor Neurol Neurosci. 2003;21:79–96. [PubMed] [Google Scholar]
- 35.Aasebø E, et al. Effects of blood contamination and the rostro-caudal gradient on the human cerebrospinal fluid proteome. PLoS One. 2014;9:e90429. doi: 10.1371/journal.pone.0090429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barclay GR, Hope J, Birkett CR, Turner ML. Distribution of cell-associated prion protein in normal adult blood determined by flow cytometry. Br J Haematol. 1999;107:804–814. doi: 10.1046/j.1365-2141.1999.01789.x. [DOI] [PubMed] [Google Scholar]
- 37.MacGregor I, et al. Application of a time-resolved fluoroimmunoassay for the analysis of normal prion protein in human blood and its components. Vox Sang. 1999;77:88–96. doi: 10.1159/000031082. [DOI] [PubMed] [Google Scholar]
- 38.Koenig AM, et al. Effects of the insulin sensitizer metformin in Alzheimer disease: Pilot data from a randomized placebo-controlled crossover study. Alzheimer Dis Assoc Disord. 2017;31:107–113. doi: 10.1097/WAD.0000000000000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linsenmeier L, et al. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. 2018;13:18. doi: 10.1186/s13024-018-0248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prion Alliance 2017 Prion alliance sponsors MGH research study. Available at www.prionalliance.org/2017/07/19/prion-alliance-sponsors-mgh-research-study/. Accessed March 7, 2018.
- 41.Parchi P, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- 42.Caughey B, Raymond GJ, Ernst D, Race RE. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): Implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mays CE, et al. Prion disease tempo determined by host-dependent substrate reduction. J Clin Invest. 2014;124:847–858. doi: 10.1172/JCI72241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto M, et al. Efficacy of flupirtine on cognitive function in patients with CJD: A double-blind study. Neurology. 2004;62:714–718. doi: 10.1212/01.wnl.0000113764.35026.ef. [DOI] [PubMed] [Google Scholar]
- 45.Geschwind MD, et al. Quinacrine treatment trial for sporadic Creutzfeldt-Jakob disease. Neurology. 2013;81:2015–2023. doi: 10.1212/WNL.0b013e3182a9f3b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haïk S, et al. Doxycycline in Creutzfeldt-Jakob disease: A phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2014;13:150–158. doi: 10.1016/S1474-4422(13)70307-7. [DOI] [PubMed] [Google Scholar]
- 47.Atarashi R, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking-induced conversion. Nat Med. 2011;17:175–178. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- 48.McGuire LI, et al. Real time quaking-induced conversion analysis of cerebrospinal fluid in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 2012;72:278–285. doi: 10.1002/ana.23589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foutz A, et al. Diagnostic and prognostic value of human prion detection in cerebrospinal fluid. Ann Neurol. 2017;81:79–92. doi: 10.1002/ana.24833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDade E, Bateman RJ. Stop Alzheimer’s before it starts. Nature. 2017;547:153–155. doi: 10.1038/547153a. [DOI] [PubMed] [Google Scholar]
- 51.Minikel EV, et al. 2018. Age of onset in genetic prion disease and the design of preventive clinical trials. bioRxiv:10.1101/401406.
- 52.Takada LT, et al. Genetic prion disease: Experience of a rapidly progressive dementia center in the United States and a review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2017;174:36–69. doi: 10.1002/ajmg.b.32505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilham JM, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6:e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orrù CD, et al. RT-QuIC assays for prion disease detection and diagnostics. Methods Mol Biol. 2017;1658:185–203. doi: 10.1007/978-1-4939-7244-9_14. [DOI] [PubMed] [Google Scholar]
- 55.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.