Significance
Development of small-molecule lead medicines that potently and specifically modulate RNA function is challenging. We designed a small molecule that cleaves r(CUG)exp, the RNA repeat expansion that causes myotonic dystrophy type 1. In cells and in an animal model, the small-molecule cleaver specifically recognizes the 3-dimensional structure of r(CUG)exp, cleaving it more selectively among transcripts containing short, nonpathogenic r(CUG) repeats than an oligonucleotide that recognizes RNA sequence via Watson-Crick base pairing. The small molecule broadly relieves disease-associated phenotype in a mouse model. Thus, small molecules that recognize and cleave RNA structures should be considered a therapeutic strategy for targeting RNA in vivo.
Keywords: nucleic acids, RNA, chemical biology, genetic disease, RNA splicing
Abstract
Myotonic dystrophy type 1 (DM1) is an incurable neuromuscular disorder caused by an expanded CTG repeat that is transcribed into r(CUG)exp. The RNA repeat expansion sequesters regulatory proteins such as Muscleblind-like protein 1 (MBNL1), which causes pre-mRNA splicing defects. The disease-causing r(CUG)exp has been targeted by antisense oligonucleotides, CRISPR-based approaches, and RNA-targeting small molecules. Herein, we describe a designer small molecule, Cugamycin, that recognizes the structure of r(CUG)exp and cleaves it in both DM1 patient-derived myotubes and a DM1 mouse model, leaving short repeats of r(CUG) untouched. In contrast, oligonucleotides that recognize r(CUG) sequence rather than structure cleave both long and short r(CUG)-containing transcripts. Transcriptomic, histological, and phenotypic studies demonstrate that Cugamycin broadly and specifically relieves DM1-associated defects in vivo without detectable off-targets. Thus, small molecules that bind and cleave RNA have utility as lead chemical probes and medicines and can selectively target disease-causing RNA structures to broadly improve defects in preclinical animal models.
RNA is an important target of therapeutics and chemical probes of function, as it has diverse molecular mechanisms that can cause or contribute to disease. The most common approaches to target a disease-causing RNA include oligonucleotides (1) and CRISPR (2), both of which target RNA sequence. Although these methods are powerful tools, they can have nonnegligible off-target effects (3), and the use of small molecules could be advantageous. Historically, small molecules that target RNA structure have been limited to the bacterial ribosome (4, 5) and riboswitches (6). However, recent efforts have afforded chemical probes that target other RNAs with extensive structure (7), in particular RNA repeat expansions that cause >40 neuromuscular disorders including amyotrophic lateral sclerosis (ALS), Huntington’s disease (HD), and myotonic dystrophy types 1 (DM1) and 2 (DM2).
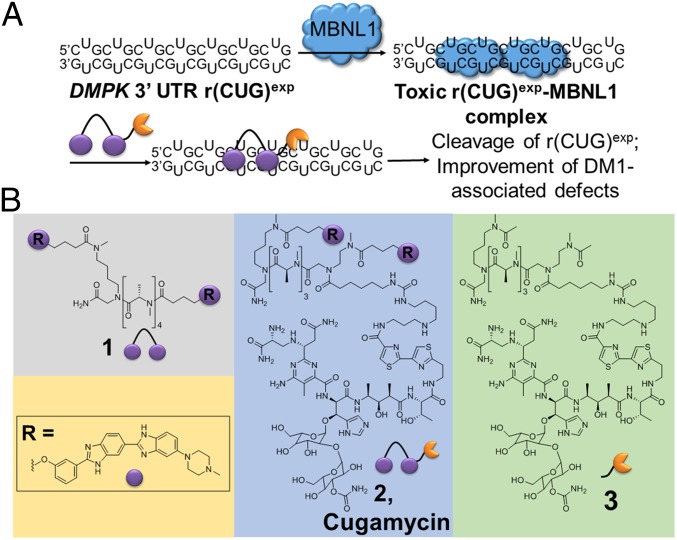
DM1 is the most common cause of adult-onset muscular dystrophy. Although there are no existing treatments, decades of research have elucidated molecular mechanisms driving disease. In DM1, a CTG repeat expansion in the 3′ untranslated region (UTR) of the dystrophia myotonica protein kinase (DMPK) gene is transcribed into RNA, yielding transcripts containing r(CUG)exp (8). This r(CUG)exp is the causative agent of DM1, binding to and sequestering proteins such as Muscleblind-like 1 (MBNL1), a regulator of pre-mRNA splicing (Fig. 1A). Functional loss of MBNL1 by its sequestration in r(CUG)exp-containing nuclear foci (9) causes systemwide pre-mRNA splicing defects (10) that contribute to disease phenotypes (11). Thus, preventing the r(CUG)exp-MBNL complex from forming would constitute a potential route to a therapy.
Fig. 1.
Design of small molecules that cleave r(CUG)exp. (A) r(CUG)exp in the 3′ UTR of the DMPK mRNA folds into a hairpin structure and sequesters MBNL1, resulting in pre-mRNA splicing defects. Compound 2 binds to r(CUG)exp, displaces MBNL1, and cleaves the toxic RNA repeat, improving DM1-associated defects. (B) Chemical structures of 1, 2, and 3.
In DM1, r(CUG)exp folds into a hairpin with an array of repeating 1 × 1 nucleotide UU internal loops (Fig. 1A). This disease-causing RNA has been targeted with antisense oligonucleotides (12–14), CRISPR-based approaches (15), and small molecules with various modes of action. Some of these small molecules bind to the RNA repeat, displace MBNL1, and relieve DM1-associated defects, and have been identified by various methods including screening (16) and rational design (17). There has also been an interest in reducing the levels of r(CUG)exp to elicit a therapeutic response, and this has been achieved through DNA-binding small molecules that inhibit transcription (18) and compounds with multiple modes of inhibition (17, 19, 20).
To potently and selectively target r(CUG)exp, a dimeric small molecule, 1, was designed to bind adjacent 1 × 1 UU internal loops in the structure of the disease-causing RNA (Fig. 1A) (17). A small molecule with the ability to bind and cleave r(CUG)exp was developed by coupling 1 to bleomycin A5, affording compound 2 (named Cugamycin; Fig. 1 A and B) (17). Bleomycin A5 is an anticancer natural product known to cleave DNA and RNA through the production of a radical species by the N-terminal metal binding core (21). The Hecht group has pioneered investigations into the potential of bleomycin and modified bleomycin analogs to cleave RNA (22). Attachment of bleomycin A5 to an RNA-binding small molecule allows for cleavage of a specific RNA target, in this case r(CUG)exp. This small-molecule cleaver has been shown to cleave r(CUG)exp in vitro and in DM1 patient-derived fibroblasts, resulting in a rescue of DM1-associated splicing defects at lower concentrations than parent compound 1 (17). Herein, we advance 2 as a preclinical candidate by evaluating its activity and selectivity in DM1 patient-derived myotubes, a superior model to fibroblasts because they more closely model the degree of dysregulation of MBNL1-regulated splicing events in disease-affected tissues of a mouse model and DM1 patients (23), and also by evaluating its activity and selectivity in a DM1 mouse model. These studies demonstrate that selective small-molecule degradation of a disease-causing RNA can be achieved, as shown by a rigorous study of on- and off-targeting events, both in cells and in vivo.
Results and Discussion
In Vitro Selectivity of Small-Molecule Cleavers.
To assess the molecular recognition properties of 2, binding to an r(CUG)exp model and DNA was studied. Binding was observed only to r(CUG)exp (EC50 = 365 nM) and not to the DNA hairpin (SI Appendix, Fig. S1). The ability of 2 to cleave these nucleic acids was then assessed in vitro. While 2 has been shown to cleave r(CUG)10 more robustly than a control compound that lacks RNA-binding modules (3; Fig. 1B) (17), the small molecule’s effect on DNA cleavage has not been investigated. In vitro, 2 and 3 cleaved DNA less efficiently than bleomycin A5; 250 nM of 2 or 3 cleaved only ∼15% and ∼20%, respectively, of the DNA, while bleomycin A5 cleaved ∼60% (SI Appendix, Fig. S2). Previous studies of bleomycin A2’s interactions with DNA have shown that elimination of the positive charge on the C terminus of bleomycin reduces DNA cleavage efficiency by 10-fold (21), and structural studies of bleomycin bound to DNA have elucidated important interactions of the C-terminal end with the negatively charged phosphate backbone (24). Thus, it is not surprising that attachment of a small molecule to the C-terminal amine of bleomycin A5 results in less efficient DNA cleavage, as a key point of interaction with DNA has been replaced with a high-affinity RNA-binding module. Notably, cleavage of DNA by 3 or bleomycin was unaffected by addition of r(CUG)10 (SI Appendix, Fig. S2). Thus, in vitro binding and cleavage experiments showed that attachment of bleomycin to an RNA-binding module ablates DNA binding and cleavage while endowing the conjugate with affinity for and the ability to cleave RNA.
Studies of Compounds That Cleave r(CUG)exp in DM1 Patient-Derived Myotubes.
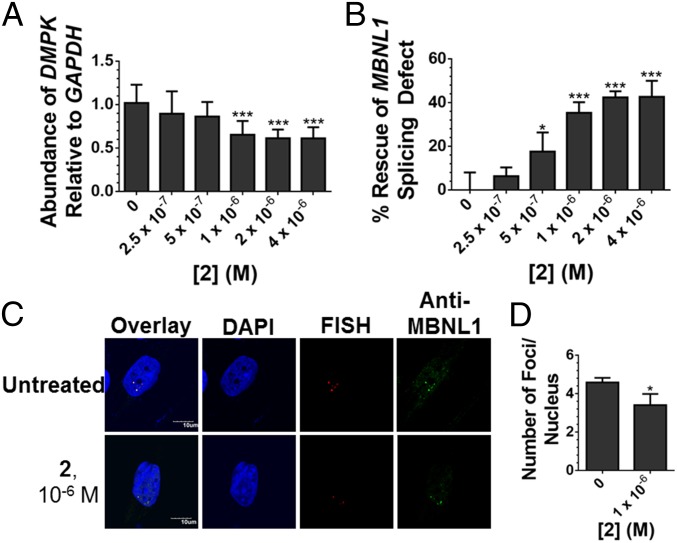
Because of these favorable properties, the activity of 2 was assessed in DM1 patient-derived myotubes (23). Fluorescence microscopy studies revealed that 2 is indeed cell-permeable, localizing in the nucleus (SI Appendix, Fig. S3), where the mutant DMPK allele is also sequestered in foci (9). The ability of 2 to cleave r(CUG)1300 in the 3′ UTR of DMPK was then measured by RT-qPCR. The small molecule cleaved ∼40% of total DMPK mRNA (Fig. 2A), with an EC25 of ∼785 nM, and had no effect on DMPK levels in myotubes derived from healthy patients (SI Appendix, Fig. S4). Thus, cleavage is specific to disease-affected cells expressing mutant DMPK with toxic r(CUG) repeat expansions, and not healthy cells that express wild-type (WT) DMPK containing short, nontoxic, r(CUG) repeats. If we assume that only the mutant allele is cleaved by 2 and estimate that mutant DMPK comprises ∼50–70% of total DMPK abundance in DM1-affected cells (25), then 2 maximally cleaved ∼60–80% of mutant DMPK.
Fig. 2.
Activity of 2 in DM1 myotubes. (A) Effect of 2 on r(CUG)exp-containing DMPK levels in DM1 myotubes as determined by RT-qPCR. Error bars represent SD, n = 3 biological replicates, ***P < 0.001 (one-way ANOVA). (B) Ability of 2 to rescue the MBNL1 exon 5 splicing defect. Error bars represent SD, n = 3 biological replicates, *P < 0.05, ***P < 0.001 (one-way ANOVA). (C) Representative images of r(CUG)exp-MBNL1 foci in DM1 myotubes treated with 2. (D) Quantification of r(CUG)exp-MBNL1 foci/nucleus. Error bars represent SD, n = 3 biological replicates, 40 nuclei counted per replicate, *P < 0.05 (t test).
The ability of 2 to relieve DM1-associated defects was further investigated, including reduction of r(CUG)exp-containing nuclear foci (9) and rescue of pre-mRNA splicing defects caused by functional loss of MBNL1 sequestered in foci (10). MBNL1 self-regulates alternative splicing of its exon 5. Thus, MBNL1 exon 5 is dysregulated in DM1-affected cells (26), manifesting as increased exon inclusion in DM1 myotubes (∼50% inclusion) compared with WT (∼15% inclusion). At a 1-µM dose, 2 shifts MBNL1 exon 5 splicing toward WT, with only 35% included, or rescue of about 40% (Fig. 2B and SI Appendix, Fig. S5A). Notably, the EC25 for rescue of the MBNL1 exon 5 splicing defect is ∼840 nM, which is similar to the EC25 for cleaving the mutant DMPK transcript. No effect was observed on the inclusion of MAP4K4 exon 22a, a NOVA alternative splicing regulator 1-dependent (27), upon treatment with 2 (SI Appendix, Fig. S5 B and C). Further, RNA-seq analysis revealed that 2 specifically rescues DM1-associated splicing events, vide infra. The small molecule also diminished r(CUG)1300-MBNL1 nuclear foci, another hallmark of DM1 pathology, when cells were treated with 1 μM of 2 while 3 did not (Fig. 2 C and D and SI Appendix, Figs. S6 and S7).
Our RT-qPCR data indicate that 2 (1 μM) maximally cleaved ∼60–80% of mutant DMPK transcripts in DM1 patient-derived myotubes. At the same concentration, we observed that 2 rescued the MBNL1 exon 5 splicing defect by ∼40% and reduced the number of r(CUG)exp-MBNL1 foci by ∼30%. The reasons for this difference are likely highly complex and include various factors such as the time it takes for MBNL1 to be released after cleavage of r(CUG)exp by 2 and the half-life of the DMPK RNA, among others.
Molecular Recognition of RNA Structure by Cugamycin and RNA Sequence by Oligonucleotides in DM1-Patient Derived Myotubes.
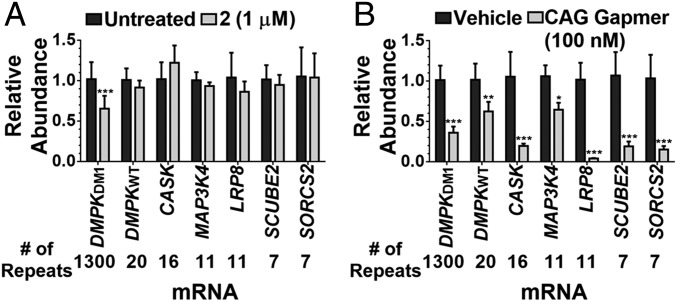
Engendering small molecules with the ability to cleave RNA targets allows for facile profiling of their cleaved targets in cells via RT-qPCR. Because antisense oligonucleotides have this property intrinsically, their cleaving capacity can be used as a proxy for molecular recognition in cells (28). It has been shown that small molecules can selectively cleave disease-driving r(CUG)exp in DM1 fibroblasts; that is, they do not cleave RNAs with short repeats of r(CUG) (17). Therefore, the levels of mRNAs containing nonpathogenic r(CUG) repeats quantifiable in DM1 myotubes were measured upon treatment with 1 μM of 2, a dose that rescues DM1-associated pre-mRNA splicing defects by ∼40%. These six RNAs (including WT DMPK) have repeat lengths of 7–20. Regardless of expression level, these RNAs were unaffected by small-molecule 2 treatment, and only the levels of the disease-causing r(CUG)1300 were reduced (Fig. 3A). The calcium/calmodulin dependent serine protein kinase (CASK) mRNA was upregulated by ∼22%; however, these effects do not meet a >95% confidence interval (P = 0.059, t test). Evidently, repeat length drives the cleavage of RNA by compound 2. Analogously, the effect of a 16-nucleotide-long antisense LNA gap-mer complementary to r(CUG)exp on the same mRNAs was measured (sequence provided in SI Appendix). For these studies, doses were used that decreased DMPK mRNA levels and rescued DM1-associated pre-mRNA splicing defects by ∼40% such that direct comparisons to 2 can be made (between 10 nM–100 nM; SI Appendix, Fig. S8). The levels of all six mRNAs containing r(CUG) repeats, including WT DMPK in healthy myotubes, were reduced at 100 and 10 nM concentrations (Fig. 3A and SI Appendix, Figs. S8 and S9).
Fig. 3.
Recognition of r(CUG)exp by 2 and an oligonucleotide at a dose where they improve DM1-associated splicing defects similarly. (A) Effect of 2 (1 µM) on RNAs containing more than six r(CUG) repeats expressed in DM1 myotubes as determined by RT-qPCR; dark gray bars indicate untreated cells and light gray bars indicate cells treated with 2. (B) Effect of an r(CUG) repeat-targeting antisense oligonucleotide (100 nM) on RNAs containing more than six r(CUG) repeats expressed in DM1 myotubes as determined by RT-qPCR; dark gray bars indicate vehicle-treated cells and light gray bars indicate cells treated with antisense oligonucleotide. Error bars represent SD, n = 3 biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001 (t test).
Likewise, the selectivity of an antisense oligonucleotide complementary to a sequence in DMPK’s coding region (14) was studied (sequence provided in SI Appendix, Fig. S10) such that direct comparisons could be made to 2 and to the LNA gap-mer complementary to the repeats. This oligonucleotide, a 16-nucleotide gap-mer, was evaluated for its effects on other RNAs with partial Watson-Crick base-pairing complementarity (n = 10; 6–13 complementary bases). Out of these 10 RNAs, the levels of 4 were slightly elevated while the levels of 2 containing 12 complementary bases were reduced upon oligonucleotide treatment (SI Appendix, Fig. S10). As only 60% of RNAs were affected by the antisense agent targeting a unique sequence, these studies suggest that targeting a unique sequence in DMPK rather than the CUG repeats is more selective (SI Appendix, Fig. S10).
To gain insights into how small-molecule targeting of an RNA 3D fold could be more selective than an oligonucleotide targeting RNA sequence, an analysis of the structure of each mRNA containing r(CUG)n repeats was completed. These studies modeled the RNA’s structure, its probability of forming, and its evolutionary conservation (29). To model the structures of these r(CUG)n-containing RNAs from the sequence, a method that combines a thermodynamic-based folding algorithm (29) with sequence comparison (30) was used to generate structural models for thermodynamically stable regions. Evolutionary conservation of these structures indicates that they have a common function, thus increasing the confidence of the predicted structure (30). This method has been used to predict evolutionary conserved structures in the influenza A segment-7 viral mRNA (31); these structures were confirmed experimentally by chemical mapping and NMR spectroscopy (32).
Using this RNA structure analysis, we observed that only two mRNAs containing r(CUG) repeats adopt a structure similar to that observed in r(CUG)exp, albeit a much smaller one (SI Appendix, Fig. S11 and Table S1). Further, the computed probability of the two mRNAs forming a structure similar to r(CUG)exp is much lower than that of DMPK mRNA containing r(CUG)1300 (SI Appendix, Table S1). Free energies of structures formed by isolated CUG repeats from each sequence decreased with repeat length, as expected (SI Appendix, Table S1). Collectively, these computational studies provide a rationale for the selectivity of 2 relative to the oligonucleotide and include (i) differences in RNA structure—for example, highly structured regions (with multiple binding motifs) are biased for binding 2 whereas a lack of structure is biased for binding to the oligonucleotide; and (ii) the suprastoichiometric complex between r(CUG)exp and 2, in which a few binding events can effect cleavage and provide optimal selectivity for long repeats whereas the antisense oligonucleotide can be an efficient cleaver of all targets it engages.
Analysis of DNA Cleavage in Cells.
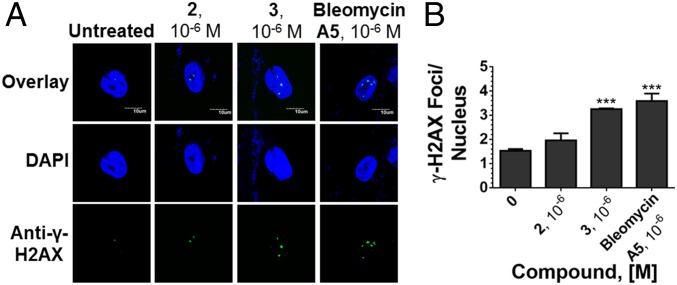
To further examine the selectivity of 2, its effects on DNA cleavage in DM1 myotubes were assessed. Native bleomycin A5 is known to cleave DNA and thereby activate the DNA damage response pathway, resulting in an increase in γ-H2AX foci that occur in response to a DNA double-strand break (33). Based on in vitro binding and cleavage analysis (SI Appendix, Figs. S1 and S2), however, it is believed that 2 would not target DNA as a key positive charge that contributes to DNA binding affinity was replaced with the r(CUG)exp small-molecule binder. Using γ-H2AX immunostaining to quantify the number of foci, both bleomycin A5 and 3 cause a significant increase in the number of γ-H2AX foci/nucleus, while 2 has no effect (Fig. 4 and SI Appendix, Fig. S12). Thus, the appending of RNA-binding modules reprograms bleomycin’s cleavage selectivity both in vitro and in cells.
Fig. 4.
Effects of small molecules on the DNA damage response pathway. (A) Representative images from γ-H2AX immunofluorescence to assess DNA damage in DM1 myotubes upon treatment with 2, 3, or bleomycin A5. (B) Quantification of the number of γ-H2AX foci/nucleus. Error bars represent SD; n = 3 biological replicates, with 40 nuclei counted per replicate; ***P < 0.001 (one-way ANOVA).
Studies of Compounds That Cleave r(CUG)exp in a DM1 Preclinical Mouse Model.
Given these encouraging results, the molecular recognition and cleavage of 2 was further investigated in vivo. Before investigating therapeutic potential, drug metabolism and pharmacokinetic studies were completed to determine the concentration of compound in blood plasma. Mice were treated with 10 mg/kg of 2 via i.p. injection, and the concentration of compound in blood plasma was determined at various times posttreatment. The maximum concentration of compound in plasma peaked at 1 h with a concentration of 2.67 μM, while 2 could be detected at nanomolar concentrations at 24 h (SI Appendix, Fig. S13). The half-life of the compound in blood plasma was determined to be 2.22 h.
The efficacy of 2 was studied in the HSALR mouse model of DM1, which contains 250 CTG repeats driven by the human skeletal actin (HSA) promoter and recapitulates aspects of the human disease including dysregulation of MBNL1-dependent splicing events, loss of the muscle-specific chloride ion channel (CLCN1), and myotonia in muscle tissues (34, 35). Compound 2 was delivered i.p. to HSALR mice at 10 mg/kg every other day for 1 week (n = 4 total injections). This dose was well tolerated in mice, and lung fibrosis, a common side effect of bleomycin treatment (36), was not observed at the end of the treatment period (day 8) as measured by lung hydroxyproline content (SI Appendix, Fig. S14). The absence of lung fibrosis was not surprising, as it does not manifest until at least 4 weeks after repeated i.p. injection or systemic delivery of bleomycin (37, 38).
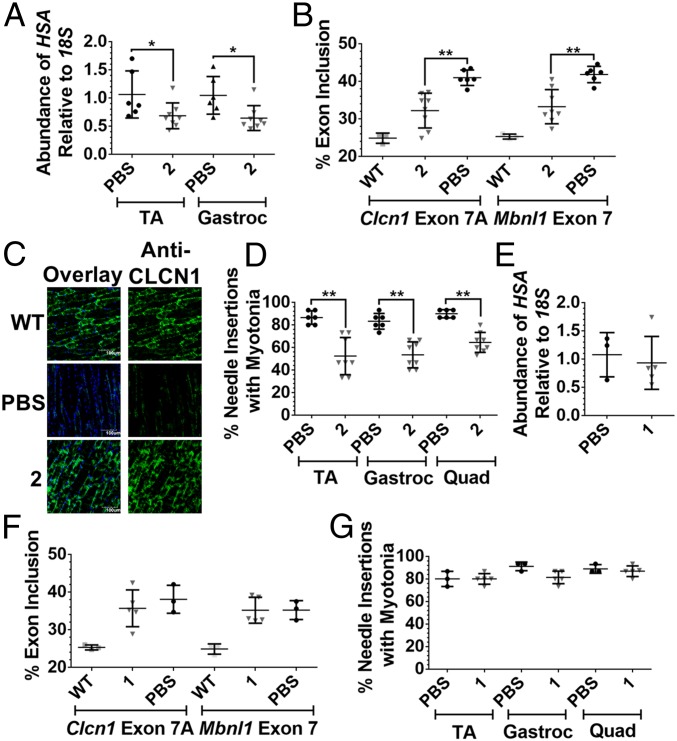
The ability of 2 to improve DM1-associated pre-mRNA splicing, protein defects, and myotonia was measured after 1 week of dosing. Treatment with 2 led to a ∼40% reduction in the r(CUG)250-containing HSA transgene in the tibialis anterior (TA) and gastrocnemius muscles (Fig. 5A and SI Appendix, Fig. S14), indicating that the designer compound is distributed to muscle tissue, binds to r(CUG)exp in vivo, and targets the transcript for destruction. The small molecule’s effect on pre-mRNA splicing patterns in the TA and gastrocnemius muscles was studied. Aberrant splicing of mouse Mbnl1 exon 7 and Clcn1 exon 7A, caused by sequestration of MBNL1 by r(CUG)exp, was rescued, whereas the alternative splicing of Itgb1 exon 17 and Capzb exon 8, two non-MBNL1-regulated splicing events, was not affected (Fig. 5B, and SI Appendix, Fig. S14). Aberrant inclusion of Clcn1 exon 7a results in a premature termination codon that triggers destruction of Clcn1 mRNA via non-sense-mediated decay, leading to a decrease in the CLCN1 protein and manifestation of myotonia (35). It is thought that rescue of Clcn1 splicing is accompanied by an increase in CLCN1 protein levels. Indeed, improvement of Clcn1 splicing patterns upon treatment with 2 restored CLCN1 protein levels in TA muscle sections as determined by immunostaining (Fig. 5C and SI Appendix, Fig. S14).
Fig. 5.
Evaluation of 1 and 2 in HSALR mice. (A) RT-qPCR analysis of the r(CUG)exp-containing HSA transgene in TA and gastrocnemius (gastroc) muscles of vehicle-treated and 2-treated mice; *P < 0.05 (t test). (B) Effect of 2 on Clcn1 exon 7A and Mbnl1 exon 7 splicing in TA muscle of HSALR mice; **P < 0.01 (one-way ANOVA). (C) Representative images of CLCN1 immunostaining in TA muscle sections. (D) Effect of 2 on myotonia in the TA, gastroc, and quadriceps muscles of HSALR mice; **P < 0.01 (t test). Error bars represent SD, n = 3 mice for WT, n = 6 mice for vehicle-treated, n = 8 mice for 2-treated. (E) RT-qPCR analysis of the r(CUG)exp-containing HSA transgene in TA muscle of vehicle-treated and 1-treated mice. (F) Effect of 1 on Clcn1 exon 7A and Mbnl1 exon 7 splicing in the TA muscle of HSALR mice. (G) Effect of 1 on myotonia in the TA, gastroc, and quadriceps muscles of HSALR mice. Error bars represent SD, n = 3 mice for WT, n = 3 mice for vehicle-treated, n = 5 mice for 1-treated.
The CLCN1 protein is a chloride voltage-gated ion channel, and loss of this protein causes various muscle defects, including myotonia, due to of a loss of action potential gradient (35). As expected, based on increased levels of CLCN1, a reduction in myotonia was observed, as measured by the percentage of needle insertions showing a myotonic discharge (80% in untreated versus 50% in treated TA, gastrocnemius, and quadriceps muscles) (Fig. 5D). Interestingly, myotonia is statistically significantly reduced in multiple muscle tissues, indicating that 2 reaches disease-affected tissues. Unfortunately, as there are no effective therapies for DM1, the amount of reduction in myotonia required in a mouse model to translate to alleviation of symptoms in patients has yet to be determined. Cugamycin provides an effective lead compound with which to design preclinical candidates that may be optimized to reduce myotonia to an even greater extent. Collectively, these studies showed that 2 elicits improvement in DM1-associated defects in vivo at the pre-mRNA, protein, and phenotype levels.
To compare in vivo the activity of 2 with that of parent compound 1, which is not capable of cleaving r(CUG)exp, 1 was delivered intraperitoneally to HSALR mice at the same dose of 10 mg/kg every other day for 1 week (4 total injections). After 1 week, the parent small molecule’s effect on r(CUG)exp levels, pre-mRNA splicing defects, and myotonia was assessed. Treatment with 1 under these conditions did not result in a significant change in levels of the r(CUG)exp-containing HSA transgene in TA (Fig. 5E). Thus, reduction of r(CUG)exp levels by 2 is due to cleavage of the RNA and not only binding. Furthermore, treatment with 1 at the same dose as 2 did not result in a significant improvement in DM1-associated splicing defects or myotonia (Fig. 5 and SI Appendix, Fig. S15), suggesting that a stronger rescue of DM1-associated defects can be achieved in vivo through cleavage of the disease-causing RNA. The observation that cleaved compound 2 is more active than 1 at the same dose suggests that cleavage can provide a more prolonged and potent effect in vivo.
Transcriptome-Wide Assessment of On- and Off-Targets in Vivo via RNA Sequencing.
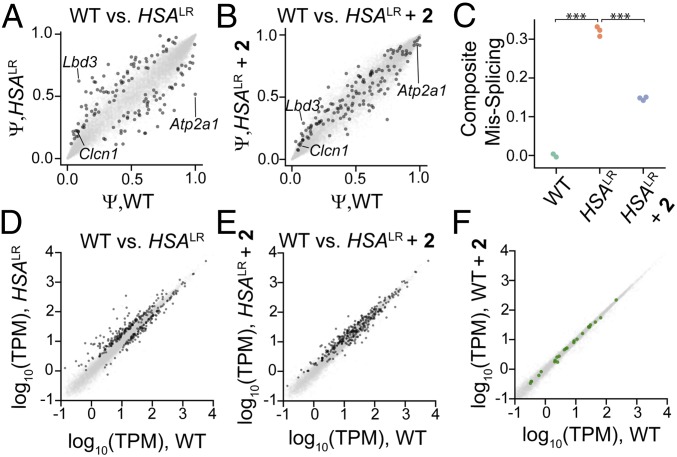
The Mbnl1 and Clcn1 splicing defects, which are both improved∼50% by 2, (Fig. 5B and SI Appendix, Fig. S14) are two of many MBNL1-dependent pre-mRNA splicing events implicated in DM1 and known to be perturbed in the HSALR mouse model. To assess the ability of 2 to rescue DM1-associated splicing defects transcriptome-wide, RNA-seq was performed on RNA isolated from the TA muscle of WT, vehicle-treated HSALR mice, and 2-treated HSALR mice. Percent spliced in (PSI, Ψ) was estimated for alternative exons across the transcriptome by mixture of isoforms (MISO) analysis (39). First, we identified 138 exons that were significantly dysregulated (|ΔΨ| > 0.1, |Z| > 1.4) in HSALR mice relative to WT mice (dark gray points in Fig. 6A); 134 out of 138 of these exons (97%) shifted back toward the WT splicing pattern upon treatment with 2 (Fig. 6B), consistent with the ability of 2 to cleave toxic r(CUG)exp and rescue MBNL-regulated splicing activity. MBNL1 is known to recognize and bind particular RNA structures to regulate splicing outcomes (40). Given this broad improvement in DM1 splicing defects by 2, it became clear that small molecules that recognize the toxic r(CUG)exp’s 3D fold can be selective and do not interfere with the splicing of canonical MBNL1 substrates, whether by affecting MBNL1 itself or by binding to canonical substrates.
Fig. 6.
RNA-seq analysis of gene expression changes upon treatment with 2. (A) Splicing events in vehicle-treated HSALR mice and WT mice. The x-axis denotes Ψ in WT mice and the y-axis denotes Ψ in vehicle-treated HSALR mice. Dark gray spots represent significantly misspliced events. (B) 2 rescues splicing patterns in HSALR mice. The x-axis denotes Ψ in WT mice and the y-axis denotes Ψ in 2-treated HSALR mice. Dark gray spots indicate events that are mis-spliced in HSALR mice; those that move toward the diagonal compared with A are rescued by 2. (C) Composite mis-splicing score across 70 splicing events, indicating rescue of these events upon treatment with 2. *** P < 0.0001, t test. (D) Gene expression changes in vehicle-treated HSALR mice and WT mice. The x-axis denotes gene expression in WT mice and the y-axis denotes gene expression in vehicle-treated HSALR mice. Significantly dysregulated genes are indicated by dark gray spots. (E) Quantification of gene expression in 2-treated HSALR mice (y-axis) compared with WT mice (x-axis). Dark gray points indicate dysregulated genes in HSALR mice, and dark gray points moving toward the diagonal compared with D are rescued by 2. (F) Treatment of WT mice with 2 elicits minimal changes in gene expression. Log2 (vehicle-treated WT) is plotted as a function of log2 (2-treated WT). Genes with >9 contiguous CTG repeats are shown in green.
The extent of rescue achieved for each particular exon may depend on a number of factors—for example, the binding of cis elements or half-life of the mRNA. Thus, the perceived potency of any given therapeutic intervention depends on which subset of exons is interrogated. We chose to define a composite score derived from the most dysregulated exons (70 exons with |ΔΨ| > 0.25% and |Z| > 1.4 when comparing WT and HSALR). A similar composite score has been used (41, 42) and is essentially the average |ΔΨ| across all 70 exons relative to WT, adjusted for whether MBNL regulates inclusion or exclusion of the exon. Upon treatment with 2, the composite missplicing score decreased from 0.3 in vehicle-treated HSALR mice to 0.15 in HSALR mice treated with 2 (Fig. 6C). This observation, combined with the observation of decreased myotonia assessed by electromyography (EMG), suggests that 2 can cleave r(CUG)exp sufficiently to reverse molecular and physiological phenotypes in this model.
Widespread gene expression changes also occur in HSALR relative to WT mice as a result of toxic r(CUG)exp expression and MBNL sequestration. Gene expression estimates and changes were assessed as described by Kallisto (43) and Sleuth (44), respectively, affording 326 genes that are significantly dysregulated in HSALR (dark gray points in Fig. 6D). Treatment with 2 resulted in normalization of the transcriptome such that 177 of the 326 genes were no longer dysregulated (Fig. 6E).
Any change in gene expression caused by 2 can reflect either desirable “on-target” normalizing effects, or undesirable “off-target” effects. Because these normalizing effects observed with 2 in HSALR mice can preclude accurate assessment of off-target effects, we differentiated observed changes as on- and off-target by comparing RNA-seq data obtained from vehicle-treated WT mice and 2-treated WT mice. Across 15,818 genes assayed, no significant changes were observed (Fig. 6F). Furthermore, no trend in gene expression changes related to the number of contiguous CTG repeats within transcripts was observed (Fig. 6F). Thus, 2 cleaves r(CUG)exp in vivo with high selectivity, resulting in release of MBNL1 and normalization of the transcriptome. The observation that a transcriptome lacking the putative target remains unperturbed upon treatment with a small-molecule RNA cleaver bodes well for our approach to be generalized for many other disease-causing structures.
Collectively, the small molecule developed here is a promising chemical probe and lead medicine. Indeed, 2 broadly rescues DM1-associated defects with no detectable off-target effects, suggesting that small molecules that selectively recognize a disease-causing 3D fold are ideal targets compared with those that target regions not associated with disease. One of the many advantages of non-oligonucleotide-based strategies is the ability to synthesize many derivatives. Derivatization is limited for oligonucleotides, as only modest changes to the backbone, sugar, or bases can be tolerated to retain activity. As modification is more flexible for compounds like 2, derivative compounds with improved properties can be easily developed. All of the analyses for Cugamycin reported herein, however, indicate that is has limited liabilities as a preclinical candidate.
Of general importance is that these studies have allowed for RNA target profiling in cells and in vivo via RNA-seq. Several approaches have been developed to study RNAs that are recognized by small molecules in cells, including those that employ cross-linking (17, 45). Cleavage can also be used to profile small-molecule recognition whether by direct cleavage, as demonstrated here, or by recruitment of cellular nucleases (46). Indeed, we have shown not only that 2 binds and cleaves r(CUG)exp in vivo, validating it as the therapeutic target, but also that 2 is highly specific.
Conclusions and Outlook
These studies demonstrate that small molecules can be designed to selectively cleave RNAs in a human disease model and improve phenotype. Furthermore, simple appendage of the bleomycin warhead at a free amine can allow programmable control over its targets, moving it away from DNA and toward a desired RNA target. This approach is likely applicable to many RNA-mediated diseases, particularly microsatellite disorders. Because 2 has broad selectivity, targeting 3-dimensional RNA folds with a small molecules may have some advantages over sequence-based targeting. Importantly, this study suggests that several approaches can be used to target and cleave RNAs and do so selectively.
The studies described here may represent a benchmark by which to assess the selectivity of small molecules designed to purposely affect RNA. Evidently, small molecules with varying modes of action, ranging from binding to cleavage, should be considered as potential chemical probes and lead medicines. The number of targets to which such approaches can be applied will likely expand as a more comprehensive understanding of the molecular recognition of RNA by small molecules emerges (7).
Methods
General Methods.
General experimental procedures are given in the SI Appendix.
Cell Lines.
Compounds were tested in two cell lines that could be differentiated into myotubes (23): (i) a DM1 (1,300 CUG repeats) conditional MyoD-fibroblast cell line and (ii) a WT conditional MyoD-fibroblast cell line.
Cell Culture, Compound Treatment, and Oligonucleotide Transfection.
Conditional MyoD-fibroblast cell lines (23) were grown in DMEM growth medium (Corning), 10% FBS (Sigma), 1% antibiotic, antimycotic (Corning), and 1% Glutagro (Corning). Once cells reached ∼80% confluency, fibroblasts were differentiated in DMEM differentiation medium (Corning), 1% antibiotic, antimycotic (Corning), 0.01% iron-transferrin, 0.001% insulin (Life Technologies), and 2 µg/mL doxycycline (Sigma) for 24 h. For small-molecule treatment, cells were treated with small molecules in medium (DMEM, 1% antibiotic, antimycotic, 0.01% insulin). Antisense oligonucleotides were transfected in medium using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s protocol.
Evaluation of Nuclear Foci.
Fluorescence in situ hybridization (FISH) was used to determine the small molecules’ effect on formation and disruption of nuclear foci as previously described (17). Detailed methods are provided in the SI Appendix. The number of r(CUG)exp-MBNL1 foci were counted in 40 nuclei/replicate (120 total nuclei counted over 3 replicates).
Mouse Studies.
HSALR, WT (FVB), and C57BL/6 mice were housed in the Scripps Florida vivarium, and all experiments using live animals were approved by the Scripps Florida Institutional Animal Care and Use Committee. FVB and C57BL/6 mice were purchased from the Jackson Laboratory. Please see SI Appendix, SI Methods for a description of all mouse studies.
RNA Sequencing.
All sequencing data detailed in this paper are deposited in the Gene Expression Omnibus (accession no. GSE127809) (47).
Supplementary Material
Acknowledgments
We thank Prof. Denis Furling [Centre de Recherche en Myologie (UPMC/Inserm/CNRS), Institut de Myologie], for his generous gift of cell lines used in this paper. We thank Expansion Therapeutics for providing funding to M.D.D. to complete distribution metabolism and pharmacokinetics studies. This work was funded by the US National Institutes of Health (Grant DP1NS096898) and the Muscular Dystrophy Association (Grant 380467). A.J.A. was partially supported by a diversity supplement from the US National Institutes of Health (Grant DP1NS096898-02S1). S.G.R. was partially supported by a postdoctoral fellowship from the Myotonic Dystrophy Foundation.
Footnotes
Conflict of interest statement: M.D.D. and E.T.W. are consultants for Expansion Therapeutics. S.G.R. is a current employee of Expansion Therapeutics.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data detailed in this paper are deposited in the Gene Expression Omnibus (accession no. GSE127809).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901484116/-/DCSupplemental.
References
- 1.Kole R, Krainer AR, Altman S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stojic L, et al. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 2018;46:5950–5966. doi: 10.1093/nar/gky437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 5.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 6.Howe JA, et al. Selective small-molecule inhibition of an RNA structural element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- 7.Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brook JD, et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 9.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 11.Nakamori M, et al. Splicing biomarkers of disease severity in myotonic dystrophy. Ann Neurol. 2013;74:862–872. doi: 10.1002/ana.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jauvin D, et al. Targeting DMPK with antisense oligonucleotide improves muscle strength in myotonic dystrophy type 1 mice. Mol Ther Nucleic Acids. 2017;7:465–474. doi: 10.1016/j.omtn.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batra R, et al. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell. 2017;170:899–912.e10. doi: 10.1016/j.cell.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamori M, Taylor K, Mochizuki H, Sobczak K, Takahashi MP. Oral administration of erythromycin decreases RNA toxicity in myotonic dystrophy. Ann Clin Transl Neurol. 2015;3:42–54. doi: 10.1002/acn3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rzuczek SG, et al. Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat Chem Biol. 2017;13:188–193. doi: 10.1038/nchembio.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siboni RB, et al. Actinomycin D specifically reduces expanded CUG repeat RNA in myotonic dystrophy models. Cell Rep. 2015;13:2386–2394. doi: 10.1016/j.celrep.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen L, et al. Rationally designed small molecules that target both the DNA and RNA causing myotonic dystrophy type 1. J Am Chem Soc. 2015;137:14180–14189. doi: 10.1021/jacs.5b09266. [DOI] [PubMed] [Google Scholar]
- 20.Guan L, Disney MD. Small-molecule-mediated cleavage of RNA in living cells. Angew Chem Int Ed Engl. 2013;52:1462–1465. doi: 10.1002/anie.201206888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boger DL, Cai H. Bleomycin: Synthetic and mechanistic studies. Angew Chem Int Ed Engl. 1999;38:448–476. doi: 10.1002/(SICI)1521-3773(19990215)38:4<448::AID-ANIE448>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 22.Carter BJ, et al. Site-specific cleavage of RNA by Fe(II).bleomycin. Proc Natl Acad Sci USA. 1990;87:9373–9377. doi: 10.1073/pnas.87.23.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arandel L, et al. Immortalized human myotonic dystrophy muscle cell lines to assess therapeutic compounds. Dis Model Mech. 2017;10:487–497. doi: 10.1242/dmm.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu W, Vanderwall DE, Turner CJ, Kozarich JW, Stubbe J. Solution structure of Co·Bleomycin A2 green complexed with d(CCAGGCCTGG) J Am Chem Soc. 1996;118:1281–1294. [Google Scholar]
- 25.Wojciechowska M, et al. Quantitative methods to monitor RNA biomarkers in myotonic dystrophy. Sci Rep. 2018;8:5885. doi: 10.1038/s41598-018-24156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 27.Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 28.Childs-Disney JL, Disney MD. Approaches to validate and manipulate RNA targets with small molecules in cells. Annu Rev Pharmacol Toxicol. 2016;56:123–140. doi: 10.1146/annurev-pharmtox-010715-103910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz R, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seemann SE, et al. The identification and functional annotation of RNA structures conserved in vertebrates. Genome Res. 2017;27:1371–1383. doi: 10.1101/gr.208652.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss WN, Priore SF, Turner DH. Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA. 2011;17:991–1011. doi: 10.1261/rna.2619511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang T, Kennedy SD, Moss WN, Kierzek E, Turner DH. Secondary structure of a conserved domain in an intron of influenza A M1 mRNA. Biochemistry. 2014;53:5236–5248. doi: 10.1021/bi500611j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 34.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 35.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 36.Blum RH, Carter SK, Agre K. A clinical review of bleomycin–A new antineoplastic agent. Cancer. 1973;31:903–914. doi: 10.1002/1097-0142(197304)31:4<903::aid-cncr2820310422>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 37.Moore B, et al. Animal models of fibrotic lung disease. Am J Respir Cell Mol Biol. 2013;49:167–179. doi: 10.1165/rcmb.2013-0094TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adamson IY, Bowden DH. The pathogenesis of bleomycin-induced pulmonary fibrosis in mice. Am J Pathol. 1974;77:185–197. [PMC free article] [PubMed] [Google Scholar]
- 39.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. doi: 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner SD, et al. Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 2016;12:e1006316. doi: 10.1371/journal.pgen.1006316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 44.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 45.Guan L, Disney MD. Covalent small-molecule-RNA complex formation enables cellular profiling of small-molecule-RNA interactions. Angew Chem Int Ed Engl. 2013;52:10010–10013. doi: 10.1002/anie.201301639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costales MG, Matsumoto Y, Velagapudi SP, Disney MD. Small molecule targeted recruitment of a nuclease to RNA. J Am Chem Soc. 2018;140:6741–6744. doi: 10.1021/jacs.8b01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Angelbello AJ, et al. 2019 Precise small molecule cleavage of a r(CUG) repeat expansion in a myotonic dystrophy mouse model. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse127809. Deposited March 22, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.