Abstract
Copy number variation (CNV) is a major part of the genetic diversity segregating within populations, but remains poorly understood relative to single nucleotide variation. Here, we report on a tRNA ligase gene (Migut.N02091; RLG1a) exhibiting unprecedented, and fitness-relevant, CNV within an annual population of the yellow monkeyflower Mimulus guttatus. RLG1a variation was associated with multiple traits in pooled population sequencing (PoolSeq) scans of phenotypic and phenological cohorts. Resequencing of inbred lines revealed intermediate frequency three-copy variants of RLG1a (trip+; 5/35 = 14%), and trip+ lines exhibited elevated RLG1a expression under multiple conditions. trip+ carriers, in addition to being over-represented in late-flowering and large-flowered PoolSeq populations, flowered later under stressful conditions in a greenhouse experiment (P < 0.05). In wild population samples, we discovered an additional rare RLG1a variant (high+) that carries 250–300 copies of RLG1a totaling ~5.7Mb (20–40% of a chromosome). In the progeny of a high+ carrier, Mendelian segregation of diagnostic alleles and qPCR-based copy counts indicate that high+ is a single tandem array unlinked from the single copy RLG1a locus. In the wild, high+ carriers had highest fitness in two particularly dry and/or hot years (2015 and 2017; both p < 0.01), while single copy individuals were twice as fecund as either CNV type in a lush year (2016: p < 0.005). Our results demonstrate fluctuating selection on CNVs affecting phenological traits in a wild population, suggest that plant tRNA ligases mediate stress-responsive life-history traits, and introduce a novel system for investigating the molecular mechanisms of gene amplification.
Keywords: Structural variant, life history, standing variation, fluctuating selection, balancing selection, flowering time
INTRODUCTION
The standing genetic variation for fitness-related traits is a key determinant of a population’s evolutionary potential, providing the raw material for adaptive evolution when environments change (Barrett & Schluter 2008; Hoffmann & Sgrò 2011; Anderson 2016). However, the abundant genetically-based fitness polymorphism seen within many populations is also paradoxical: at any given locus, efficient directional or stabilizing selection should winnow out all but the variant with highest geometric mean fitness. Balancing selection, in which context-dependence effects of alleles on fitness maintain polymorphism, resolves this problem. Long-term protected polymorphism is robustly predicted under models of heterozygote advantage (overdominance) (Gillespie 1984) and frequency-dependent selection (Wright 1939; Clarke 1979), as well sexual antagonism (Connallon & Clark 2014) and gamete-zygote conflict (Immler et al. 2011; Fishman & Kelly 2015). Empirical examples of such protected polymorphism, often involving multi-gene structural variants with suppressed recombination in heterozygotes, are increasingly common (Joron et al. 2011; Fishman & Kelly 2015; Barson et al. 2015; Llaurens et al. 2017). However, these intrinsic mechanisms of balancing selection likely apply to only a small subset of traits and genes. Spatial and (especially) temporal shifts in selection are more controversial as sources of intra-population polymorphism, but their ubiquity creates abundant opportunities to contribute to the maintenance of functional genetic variation within natural populations (reviewed in Bell 2010; Delph & Kelly 2013). Furthermore, new models that incorporate biologically-realistic factors that buffer against allele loss, such as switches in dominance (Posavi et al. 2014; Wittmann et al. 2017), plasticity (Gulisija et al. 2016) and the storage effect of unselected life-stages (Ellner & Hairston 1994; Svardal et al. 2015), suggest that environmental variation can maintain a large pool of polymorphism. However, although a few recent studies demonstrate fluctuating selection on intermediate-frequency genetic variants (Kerwin et al. 2015; Chakraborty & Fry 2016; Lee et al. 2016; Wittmann et al. 2017; Troth et al. 2018), more work is needed to reveal whether and how environmental fluctuations contribute to functional polymorphism within natural populations.
Genic copy number variants (CNVs), including duplication and deletion of entire genes (Hastings et al. 2009), are likely to be functionally important components of standing variation across diverse taxa (Żmieńko et al. 2014; Marroni et al. 2014; Chain et al. 2014; Zarrei et al. 2015; Salojärvi et al. 2017; Prunier et al. 2017). Changes in copy number are a common form of mutation — in plants, segmental or tandem duplication events occur at a per-gene rate at least as high at the per-site nucleotide mutation rate (Lynch & Conery 2000). Much of this variation appears to persist in populations; for example, diverse maize lines differ in copy number at nearly 35% of genes (Chia et al. 2012) and >15% of Arabidopsis genes are stabilized tandem duplicates (reviewed in Żmieńko et al. 2014). Initially, whole gene duplications or deletions may directly alter expression (Stranger et al. 2007), though various forms of dosage compensation can buffer these immediate effects (Veitia et al. 2013). Duplicates can develop novel or subdivided functions over time (Lynch & Conery 2000), but they may also influence phenotypic variation within and among populations as CNVs on shorter timescales. In plants, CNVs contribute to rapid evolution under experimental thermal stress (DeBolt 2010), underlie the contemporary evolution of glysophate-resistance in weeds (Gaines et al. 2010), and have been implicated in phenological and edaphic (soil stress) adaptation across populations (Rosloski et al. 2010; Maron et al. 2013; Hanikenne et al. 2013; Gordon et al. 2017). Over the longer term, CNV caused by reciprocal deletion of non-tandem duplicates is a common source of epistatic Dobzhansky-Muller incompatibilities (Bomblies et al. 2007; reviewed in Fishman & Sweigart 2018). However, because CNVs are difficult to assay with reference-based short-read resequencing approaches (Hunter et al. 2013; Tiffin & Ross-Ibarra 2014), we are only beginning to understand their contribution to within-population variation, local adaptation, and speciation.
Here, we identify and characterize copy number variants of a gene contributing to the abundant phenological and fitness variation within the Iron Mountain (IM) population (Central Cascades, OR, USA) of yellow monkeyflower (Mimulus guttatus, Phrymaceae). This high-elevation annual population exhibits extremely high levels of nucleotide diversity (πsynonymous = 0.033; Puzey et al. 2017) and quantitative genetic variation (Kelly & Willis 2001; Scoville et al. 2009; Bodbyl Roels & Kelly 2011; Scoville et al. 2011). The abundant variation in fitness-relevant traits (e.g., pollen viability, flower size and seeds/fruit) is largely due to intermediate frequency polymorphism, as expected under local balancing selection rather than mutation-selection or migration-selection balance against unconditionally deleterious variants (Kelly & Willis 2001; Kelly et al. 2013). Spatial and temporal environmental variation are plausible sources of balancing selection at IM, where the depth of winter snowpack and (rare) rain events set the length and lushness of the summer growing season (Mojica et al. 2012; Troth et al. 2018). Furthermore, field measures of fitness in QTL introgression lines (Mojica & Kelly 2010; Mojica et al. 2012) and a genome-wide association mapping panel (Troth et al. 2018) demonstrate fluctuating selection on IM variants that affect flower size via developmental rate. In wetter years, large-flowered, relatively slowly-developing genotypes have high fecundity, whereas in dry years they have a lower probability of living to flower (Mojica & Kelly 2010; Mojica et al. 2012; Troth et al. 2018). Such antagonistic pleiotropy across environments is key to the models for the selective maintenance of standing variation by environmental fluctuations (Wittmann et al. 2017; Brown & Kelly 2018), but the genes that underlie such tradeoffs are almost completely unknown.
In this study, we focus on one gene contributing to the abundant variation in life-history phenotypes within Iron Mountain M. guttatus, and dissect its structural variation, phenotypic associations, and fitness effects in the wild. Previously, a tRNA ligase (Migut.N02091, RLG1a) was identified as a significant genome-wide single nucleotide polymorphism (SNP) outlier in PoolSeq scans for differentiation between early and late flowering cohorts at both IM and the nearby Quarry population (Monnahan & Kelly 2017). This gene is one of two (unlinked) Mimulus homologs of Arabidopsis RLG1 (Englert & Beier 2005), a multi-functional ligase recently implicated in regulation of both growth hormone auxin and the stress-induced unfolded protein response (Leitner et al. 2015; Nagashima et al. 2016). Here, CNV-focused analyses of that IM flowering time dataset, a previously published flower size selection dataset (Kelly et al. 2013), and four new PoolSeq populations with contrasting flowering cues and germination times in the field, reveal that RLG1a coverage varies strongly across phenotypic cohorts. By scanning for coverage variants in a whole-genome resequenced inbred line set (Puzey et al. 2017), we identify an intermediate frequency (5/35 lines) CNV with two additional copies of RLG1a (trip+). We develop a diagnostic marker and use individual genotyping to confirm that trip+ increases in frequency under artificial selection for large flower size and is over-represented in late-flowering plants in 2014 (a dry year). We then use rt-qPCR to test whether trip+ plants exhibited constitutively elevated expression of RLG1a, consistent with a dosage effect (e.g. Gaines et al. 2010), and a conduct a common-garden growout to test for environment-specific life-history effects. We find that trip+ carriers exhibit elevated RLG1a expression and delay flowering under experimentally water-stressed conditions relative to single copy (solo) individuals, indicating that this RLG1a CNV is a stress-responsive contributor to the fast-slow axis of life-history/floral variation at IM.
However, because shifts in the frequency of a 3-copy CNV could not explain the >10× coverage differences between some Poolseq population pairs, we hypothesized the existence of rare higher-copy haplotypes shifting in frequency among wild-derived cohorts but absent from the inbred lines. In large wild-derived population samples, we identify two individuals carrying sequence variants common in the high-coverage PoolSeq cohorts; whole genome re-sequencing of these plants (high+ carriers) reveals that they carry~300 ampliconic copies of RLG1. Individual genotyping confirms that high+ alleles significantly decrease in frequency between early and late flowering cohorts in 2014, and thus account for much of the RLG1a coverage variation among pools. We then use genotyping and qPCR on the selfed progeny of one high+ carrier to test whether high+ “alleles” segregate as expected for a single tandemly-duplicated chromosomal array (vs. the alternative hypotheses of transfer to an organelle, transmission as extra-chromosomal DNA, or dispersion across multiple chromosomes). We find that high+ behaves as a presence-absence polymorphism at a single locus unlinked from the single-copy version of RLG1a. Finally, we examine the frequency and fitness effects of RLG1a CNV genotypic categories (solo, trip+, high+) over three years spanning extremes of fecundity and survival in the Iron Mountain population. We document significant (but variable) associations between CNV genotype and fitness in each year, with high+ strongly favored in two low survival and/or fecundity years, and both high+ and trip+ disfavored relative to solo in an intervening lush year. As predicted from relative fitnesses in the preceding year, the frequency of each CNV fluctuates across the three seasons. Together, these analyses suggest that a series of copy number variants of a Mimulus guttatus tRNA ligase influence life-history traits that experience fluctuating selection across years, and thus are likely maintained as polymorphisms by this form of balancing selection.
METHODS
Study System -
The Mimulus guttatus (also known as Erythranthe guttata; Phrymaceae) species complex exhibits tremendous diversity in life history, mating system, and other traits across Western North America. Mimulus guttatus, the most widespread and diverse species (and the likely progenitor of other complex members) is self-compatible but largely outcrossing (Sweigart et al. 1999). We focus here on the well-studied Iron Mountain (IM, Oregon) high-elevation annual population, from which the inbred line (IM62) used to construct the Mimulus guttatus v2.0 reference genome (www.Phytozome.jgi.doe.gov) was derived. Ecologically, the life cycle of the IM population is driven by variable yearly patterns of snow accumulation and melt-out. Germination occurs in both Fall (November, with plants overwintering under snow as small rosettes) and in spring after melt-out (May-June, generally). The maximum summer growing season varies from five to twelve weeks depending on snowpack and other climate variables. This year to year variation in the length, timing and lushness of the growing season creates high temporal variance in mean survival and fecundity (Mojica et al. 2012; Fishman & Kelly 2015; Monnahan et al. 2015; Lee et al. 2016; Troth et al. 2018).
PoolSeq of phenotypic populations -
We re-analyzed PoolSeq datasets from two previous experiments, which examined allele frequency differences among bulk-sequenced phenotypic cohorts, as well as conducting two new PoolSeq experiments (Table 1).
Table 1.
Plant materials (all Mimulus guttatus from Iron Mountain, Oregon, population) used in this study.
| Dataset | Components | Analyses | Source |
|---|---|---|---|
| Flower size selection expt. | High vs. Low vs. Ancestral | PoolSeq, marker genotyping | Kelly et al. 2013 |
| Flowering time cohorts | IM13_Early vs. IM13_Late IM14_Early vs. IM14_Late |
PoolSeq PoolSeq, marker genotyping |
Monnahan & Kelly 2017 |
| Germination time cohorts | Fall vs. Spring (2012/2013) | PoolSeq | this study |
| Vernalization requirement | Bolt vs. Rosette | PoolSeq | this study |
| IM inbred lines (N = 35) |
IM62, IM767, IM664, etc. | whole genome resequencing, trip+ rt-qPCR (N = 11), marker design |
Puzey et al. 2017 Lee et al. 2016 |
| Greenhouse populations | IM13P, IM15P (from outbred wild seeds) | flowering time growout, screens for high+ carriers | this study |
| Individual high+ carriers | IM13P_627c, IM13P_644 IM15P_317 | whole genome resequencing, high+ segregation & qPCR |
this study |
| Wild populations | IM2015, IM2016, IM2017 | Fitness and frequency in field | this study |
Existing datasets:
The High-Low-Ancestral set (Kelly et al. 2013) includes large- and small-flowered populations derived by nine generations of artificial selection with outbreeding (High and Low selection lines; pool N = 49 and 78, respectively), starting from a large outbred IM-derived base population (Ancestral, pool N = 75). A Control population maintained under the same conditions, but without selection, was also generated (Kelly et al. 2013); it was not sequenced, but we use it here for individual genotyping tests. The Early-Late flowering time set consisted of field-collected cohorts of early- and late- flowering plants from IM and nearby Quarry populations in both 2013 and 2014 (Monnahan & Kelly 2017). Because the Quarry population is the product of recent admixture between IM-like annuals and perennial genotypes (Monnahan et al. 2015), we restrict our analyses here to the IM datasets.
Fall-Spring germination time study:
In Fall 2012 and Spring 2013, we collected fall-germinating and spring-germinating seedlings from nine quadrats spaced across the IM site (N= 40 samples per quadrat collected 20/tube, except for quadrant in Spring with only 10 seedlings; total N = 320 and 290, respectively). After DNA extraction, tubes were combined proportionately (equimolar concentrations by sample size) into Fall and Spring pools for library preparation.
Bolt-Rosette vernalization requirement study.
In spring 2012, we grew >600 independent (1 per maternal family) plants derived from 2011 field-collected seeds in University of Montana greenhouse under non-inductive daylengths (12 hour days) for 2 months, and then transitioned to 16 hour days. These conditions induce flowering in IM M. guttatus that do not require vernalization (a cold treatment simulating winter) and inhibit it in those that do (Friedman & Willis 2013; Fishman et al. 2014). About 60% of plants had flowered after 60 days at 16hr daylengths. Individuals were then assigned to Bolt (flowered without vernalization) and Rosette (vernalization-requiring, non-flowering) cohorts, and leaf and/or bud tissue collected in sets of 20 plants per extraction tube. After DNA extraction using a standard Mimulus protocol (Fishman & Kelly 2015), 10 and 15 tubes per population were pooled in equimolar amounts (N = 300 and 200 plants per Bolt and Rosette pools, respectively).
PoolSeq and Inbred Line Resequencing and Analysis —
The High-Low-Ancestral flower size and Early-Late flowering time cohorts were sequenced as described previously (Kelly et al. 2013; Monnahan & Kelly 2017). The Fall-Spring and Bolt-Rosette population pairs were each sequenced with Illumina HiSeq 2500 (PE 100) at the University of Kansas Genomics Core, following the same library preparation protocol as previous pools (Monnahan & Kelly 2017). All eleven PoolSeq populations were processed with the same bioinformatics pipeline (see Table 1 and Supporting Information for details).
For analyses of coverage differences among pooled populations, we restricted our analyses to sites in exons, as extremely high sequence diversity within M. guttatus hampers read-alignment outside of genes (Puzey et al. 2017). We calculated mean read depth at each annotated exon with Mosdepth (Pedersen & Quinlan 2018) and then standardized exon read-depth values by the genome-wide median for a given pool or sequenced individual. After excluding a subset of annotated genes that appeared to be chloroplast-nuclear transfers (or mis-assembled), and a set that appeared to be mis-annotated repetitive DNA (see supplemental methods in Supporting Information), we retained 25019 genes with coverage data from one or more exons. To compare read depths among pools genome-wide, we calculated the absolute value of the difference in standardized read depth between each pair of populations (e.g., High vs. Ancestral, Low vs. Ancestral, Bolt vs. Rosette, Spring vs. Fall, IM13_Early vs. IM13_Late, IM14_Early vs. IM14_Late). For comparisons of mean RLG1a coverage among pools, we restricted estimates to the final 23 exons (spanning ~9 kilobases) of Migut.N02091, as the first three exons showed high variance in coverage depending on whether or not unpaired reads were filtered out and we cannot definitively distinguish additional duplication of this portion of the gene from mis-mapping.
Using a similar bioinformatics pipeline, we examined Migut.N02091 exon coverage in previously resequenced IM inbred lines (1 from Lee et al. 2016; 34 from Puzey et al. 2017) (Table 1). To identify sequence variation among genomic copies in carriers of RLG1a CNV, we also performed metagenome de novo assembly on sequence reads mapping to Migut.N02091 in one inbred line with elevated coverage (IM664). After converting the SAM alignments for extracted reads to fastq format, we used the metagenome assembler MEGAHIT (Li et al. 2015) with default settings. Assembled contigs were mapped back to the Mimulus guttatus v2.0 reference genome with minimap2 (Li 2018) to identify haplotypes diagnostic of duplicated copies of the gene.
RLG1a genetic marker design and genotyping —
For analyses of individual genotype, we designed and amplified an exon-primed length-polymorphic marker (mN2091×16) spanning a 5 amino-acid (5AA; 15 base) insertion-deletion polymorphism in Exon 16 of Migut.N02091, as well as the highly variable Exon16–17 intron (Supporting Information: Table S1). In inbred lines (N = 14 tested) and wild-derived plants, we found multiple (diploid) amplicons corresponding to single-copy sequences (RLG1a_solo), plus two amplicons carrying the Exon 16 5AA deletion: a 426bp fragment was exclusive to IM lines with ~3× coverage of Migut.N02091 (trip+) and a rare 428bp amplicon was uniquely associated with very high coverage individuals (high+) found in wild collections (IM13P, IM15P; Table 1). Both 5AA-deletion variants at mN2091×16 were scored as presence/absence polymorphisms, as the high+ 428bp allele was always “homozygous” due to out-competition of all other alleles in PCR reactions and the trip+ 426bp allele was always found with 2–3 additional amplicons also found in solo lines (see Results).
Characterization of RLG1a trip+: vernalization, RLG1a expression, and flowering time under stress —
To test whether the patterns observed the PoolSeq results from shifts in RLG1a CNV frequencies, we genotyped individual plants from the flower size selection (High, Low, and Control) populations and from IM14 (Early and Late flowering) populations at mN2091×16 marker. We also performed two separate experiments to assess phenological phenotypes. We grew solo and trip+ inbred lines (inferred from marker genotyping) under the same greenhouse regime as in the Bolt-Rosette PoolSeq experiment in Spring 2015 and sampled rosette leaf tissue one day prior to and six days after the transition from 12hr daylength to16hr daylengths (n = 9 solo lines and 2 trip+ lines, respectively; 2–4 separate individuals per treatment per line; total N = 56). Both trip+ lines (IM664 and IM115) flowered within one month under this regime, consistent with a lack of vernalization requirement, as did three of the 10 solo lines. We extracted RNA, made cDNA, and measured expression of RLG1a (mN2091q primers in Exon 26) relative to two previously developed controls (EF1a and UBQ5) (see Supporting Information for details). Differences in expression were determined from threshold cycle number (Ct) relative to the control (ΔCt = Ct mN2091 - Ct EF1a; averaged over 2 technical reps each) and analyzed using ANOVA with daylength (12hr, 16hr), RLG1a CNV category (solo, trip+), and line (nested within CNV) as main effects and the and daylength × CNV interaction effect.
As part of a separate study on differentiation in plant-soil-microbe interactions, we grew IM outbred plants (from 2013 wild seed) in the University of Montana greenhouse in Summer 2017 in two soil treatments: soil from the Oregon Dunes perennial M. guttatus site (DUN) (Hall & Willis 2006) and from IM, each cut 50:50 with sand. Plants were top-watered daily and fertilized weekly with a low-phosphorus fertilizer. Both treatments mimicked field conditions and produced rapid-flowering phenotypes resembling wild IM plants. However, we infer the DUN soil to be more stressful because it produced a ~2-fold reduction in mean biomass and fruit number at harvest vs. the IM soil (data not shown). We recorded the number of days to first flower, extracted DNA from IM plants, and genotyped individuals at the mN2091×16 marker. Flowering time was analyzed with ANOVA with soil type and genotype (trip+ /solo; three high+ carriers were excluded from this analysis) and their interaction.
Characterization of RLG1a high+ variants: resequencing and qPCR —
To characterize rare high copy (high+) variants of RLG1a, we grew and PCR-genotyped plants from 2013 and 2015 seed collections (IM13P, IM15P; N = 120 maternal families total, 1–4 progeny each). One high+ individual from each year (IM13P_627c and IM15P_317d, respectively) was chosen for whole genome sequencing. Library preparation, sequencing (HiSeq 4000, PE125), and alignment to the M. guttatus v2.0 reference are described in Supporting Information. Median-standardized exon coverage for Migut.N02091 was calculated as with the PoolSeq and line datasets.
In addition, we counted RLG1a genomic copy number by qPCR in two wild-collected (as seed) high+ plants, IM13P_644 and IM13P_627c, as well as trip+ and solo lines. As single-copy control, we used primers within Exon 12 of Migut.G00571 (Isopropyl Malate Isomerase large subunit; mG571q in Table S1). RLG1a ΔCt values (Ct [mN209–Ct[mG571]], were calculated for each sample, averaged cross two technical replicates, and copy number estimated using the equation: 2(Δ Ct solo - Δ Ct high+). To corroborate these estimates in IM13P_644 (which had highest copy number), we created a standard curve by performing 3-fold serial dilutions of template DNA and measuring Ct for each dilution. We then used the linear fit of the standard curve to estimate mN2091q copy number relative to mG571q in the same plant.
Characterization of RLG1a high+ variants: genetic segregation --
To segregate the high copy allele, we selfed IM13P_627c and grew the resultant progeny (which segregate as a pseudo-F2 at loci heterozygous in the parent plant) in the University of Montana greenhouse in Spring 2018. All progeny (N = 192) were genotyped with mN2091×16 to score high+ presence/absence. For tests of location, we also genotyped a linkage-informative (heterozygous in IM13P_627c) marker <100kb from Migut.N02091 (mN2089; Table S1) on a subset. To confirm segregation of the high+ cluster as a single locus, we conducted genomic qPCR (as above) on a subset of progeny (N = 39). If all copies are in tandem, high+ carriers should fall into two distinct categories with a ~2-fold difference in copy number. Individual ΔCt values (calculated as above) were clustered using Normal Mixture methods, the cluster number with the lowest AIC/BIC value chosen, and cluster means compared with ANOVA. Finally, to assess whether expression was elevated in high-copy carriers, we also conducted rt-qPCR on mRNA extracted from floral shoot tissue of a small set of progeny (segregating for 0, 1 or 2 high+ alleles based on the genomic DNA qPCR), using EF1a as a control as in the trip+ analyses. Additional methods in Supporting Information.
Frequency and fitness in the wild —
To evaluate RLG1a CNVs in the field, we genotyped and measured fitness of wild IM plants in 2015–2017. In 2015, we marked spring (cotyledons) and fall (rosette leaves) germinants in May, but almost all of the spring germinants died as seedlings and were lost before DNA could be collected. We marked a second set of flowering plants on two dates a week apart in mid-June; most of these died as well, but we obtained DNA from many non-survivors. Finally, we collected tissue (whole plant except for seeds) for DNA extraction and all seeds from survivors among the marked germinants and flowerers, as well as additional random survivors as in previous studies (Fishman & Kelly 2015; Lee et al. 2016). Thus, we can estimate both survival from flowering to fruit and seedset among survivors, from which we can calculate lifetime fitness conditioned on surviving long enough to be collected. In 2016 and 2017, we obtained DNA and fruit/seed counts of plants that survived to fruit, as in previous studies. Individuals were genotyped at mN2091×16, and coded as trip+, high+ or solo. We analyzed log-transformed lifetime female fitness (survival × seedset +1) from 2015 alone using standard least squares ANOVA. We analyzed seedset and fruitset from 2016 and 2017 together, with GLM analyzes (Poisson, log link) including year, RLG1a genotype, and their interaction.
All statistical analyses of qPCR, genotypic, and phenotypic data were performed in JMP 13 (SAS Institute 1994).
RESULTS
PoolSeq reveals RLG1a coverage variation associated with multiple life history traits
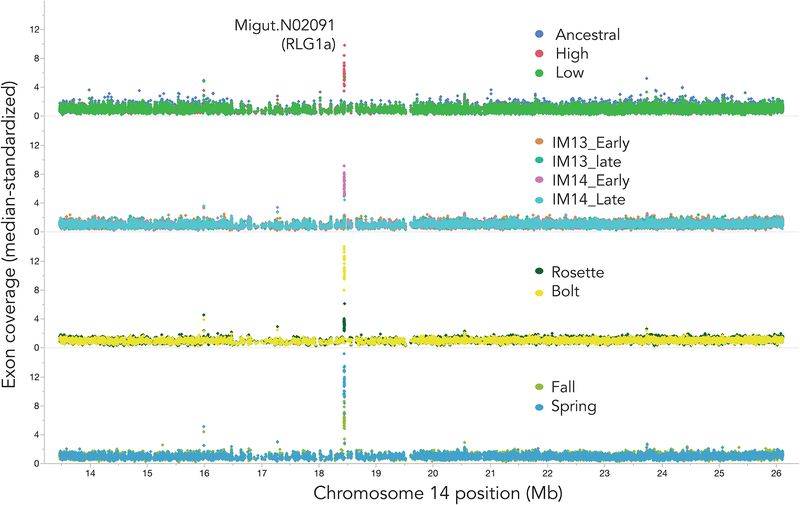
A tRNA ligase gene, Migut.N02091 (RLG1a), was a strong read-depth outlier in four of the six PoolSeq comparisons (Figure 1), including the IM14_Early vs. IM14_Late comparison in which it was previously identified as a SNP outlier (Monnahan & Kelly 2017). The 11 PoolSeq populations varied widely in genome-standardized coverage across Migut.N02091 (Exons 6–28), ranging from near-background in the Ancestral (mean = 1.18) to >10× in the Bolt and Spring germinant pools (means = 11.20 and 11.07, respectively). Like the Ancestral population for flower size selection, both IM13 flowering cohorts, IM14_Late, and the Low flower size selection population exhibited relatively low RLG1a coverage (1.2–1.67× background) indicating few individuals with high-copy genotypes in these pools. The High flower size selection population, Fall germinant, IM14_Early, and Rosette pools were intermediate in coverage (5.82, 5.99, 6.70, and 3.1, respectively),
Figure 1.
Median-standardized exon coverage (read depth) in pooled whole-genome resequencing of 11 Mimulus guttatus phenotypic populations (cohorts), grouped by experiment/dataset (Table 1). Planned contrasts of cohorts from top to bottom: Flower size after artificial selection, flowering time in the field, vernalization requirement, and germination time in the field. Values are shown for 13Mb of Chromosome 14 flanking the focal gene Migut.N02091.
A common 3-copy (trip+) variant of RLG1a contributes to flower size and flowering time variation
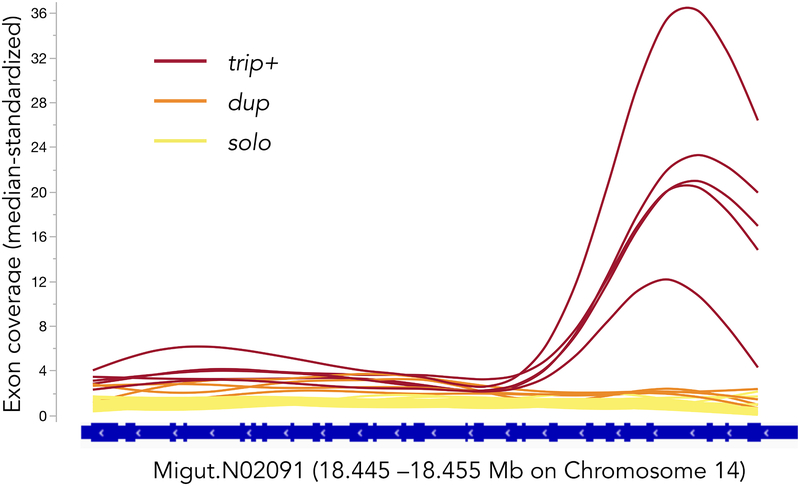
Differences in the standardized coverage of RLG1a between paired population pools (Figure 1) suggest shifts in the frequency of multi-copy haplotypes between cohorts/phenotypes. To identify haplotypes contributing to these PoolSeq patterns, we examined sequence variation and RLG1a exon coverage in a set of 35 whole genome resequenced inbred lines from IM. Four lines (IM664, IM115, IM922, IM239) exhibited a characteristic pattern of ~3× elevated coverage (and heterozygosity) across most of the gene (last 23 exons) plus extremely high coverage (26–47× background) of the first three exons (Figure 2). These lines were uniquely scored as heterozygous for a 15bp (5 amino acid; 5AA) deletion in Exon 16 relative to the reference sequence plus three linked intronic SNPs. Metagenomic assembly of IM664 identified three haplotypes across this section of the gene; one of these contains the 5AA Exon 16 deletion and linked SNPs. One additional line (IM1145) exhibited a similar (but weaker) pattern of elevated coverage gene-wide (mean = 16.4× across first 3 exons and remainder of gene, respectively) and was only heterozygous at the SNPs. We infer it to be a heterozygote of multi- and single-copy genotypes at the time of resequencing. Thus, 1/7 of the inbred lines carry an RLG1a genotype (henceforth trip+) with three distinct paralogs plus (apparently) additional duplications of the 5’ end of the gene. All trip+ lines tested (n = 4, including IM1145) carried a 426bp mN2091×16 amplicon (with 5AA Exon 16 deletion), which as absent from single-copy (solo) lines (n = 9). trip+ lines (which are highly inbred and should be homozygous genome-wide) always had at least two other amplicons, which differed among trip+ plants and overlapped in size with single-copy (solo) amplicons. Therefore, we score trip+ as a dominant (presence/absence) marker. An additional four inbred lines (IM138, IM359, IM549, IM709) exhibited ~2× coverage (Figure2); and dozens of heterozygous sites gene-wide together, this suggests stacking of diverged duplicates rather than retained heterozygosity. Unfortunately, we were unable to identify diagnostic marker alleles for this putative RLG1a_dup genotype; therefore we roll them into the single-copy (solo) genotypic class for analyses of outbred individuals.
Figure 2.
Smoothed exon coverage (standardized by chromosomal median) of Migut.N02091 (RLG1a) in 35 inbred lines of Iron Mountain Mimulus guttatus. Colors indicated inferred copy number category based on gene-wide coverage patterns and “heterozygosity”. The marker mN2091×16 spans exons 16–17 and the qPCR marker mN2091q is in exon 26 (exons are numbered consecutively from right to left). The gene region containing the first three exons shows homology to the housekeeping gene EF1a, and appears to be additionally amplified in trip+ individuals.
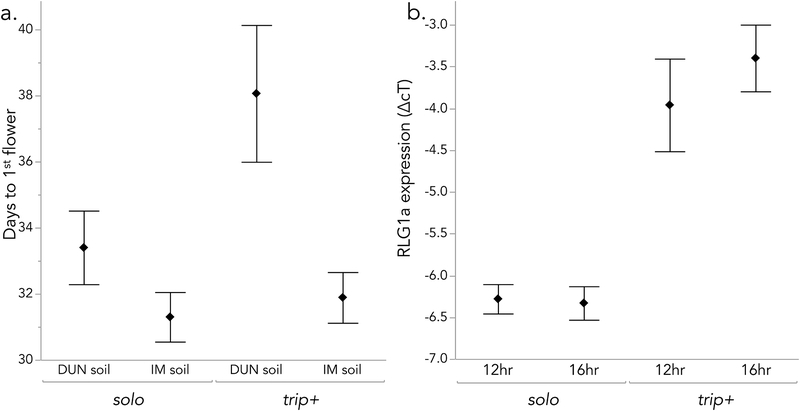
Individual genotyping confirmed a substantial (x22 df = 11.2, P < 0.005) difference in trip+ frequency among the flower size selection populations, with twice as many trip+ plants in the High (36/72 = 50%) vs. Low (13/55 = 26%) and Control (7/29 = 24%) cohorts individually genotyped. Plants carrying trip+ also exhibited late flowering in two independent datasets, one from the field and one under field-mimicking conditions in the greenhouse. First, individual genotyping of the 2014 field PoolSeq collections revealed twice the frequency of trip+ plants (32%) in Late vs. Early (15%) flowering cohorts (χ21 df = 6.3, P = 0.012, N = 179). This may reflect a relatively slow transition to flowering by trip+ plants under the drought-stress conditions experienced in normal-to-dry years at Iron Mountain. Consistent with this inference, trip+ was associated with later flowering in plants grown under two stressful soil conditions in the greenhouse (F 3. 75 = 6.6, P = 0.03 for genotype effect). Although, the interaction with soil type was nonsignificant (P = 0.09), the main genotypic effect was primarily due to delayed flowering of trip+ plants (mean = 38.1 ± 1.3 SE days, n = 17) relative to non-carriers (mean = 33.4 ± 1.3 SE days, n = 18) in the sandier and more drought-stessed DUN soil, with no difference evident in the IM soil (Figure 3a).
Figure 3.
Flowering time and expression of RLG1a genotypes (solo, trip+) under controlled conditions. a) Time to first flower (mean ± 1 SE) for outbred plants grown in two field soils (DUN = drier; see Methods) in greenhouse. Analyzed with ANOVA (overall r2 = 0.21, n = 79). Soil type effect: P < 0.001; RLG1a genotype effect: P = 0.03; interaction: P = 0.09). b. RLG1a expression (relative to EF1a control; mean ± 1 SE) in leaf tissue of solo (N = 9) and trip+ (N =2) inbred lines grown under two daylength conditions. Analyzed with nested ANOVA (overall r2 = 0.78, n = 56): Daylength effect: p = 0.58; RLG1a CNV effect: p < 0.0001; Line nested within genotype effect; p = 0.002)
Consistent with increased mRNA dosage due to increased copy number, two trip+ lines exhibited highly elevated RLG1a expression relative to solo lines (n = 9) under both short (12hr) and long (16hr, after 12hr) daylength conditions (P < 0.00001 for RLG1 genotype effect; Figure 3b). Daylength, non-vernalized flowering phenotype (Bolt/Rosette), and their interaction had no effect on RLG1a expression in leaves (all P > 0.15), but line was highly significant beyond the RLG1a CNV effect (P < 0.002). The RLG1a CNV effect was not a consequence of flowering phenotype; both trip+ lines tested had the Bolt phenotype, but so did two of the solo lines. However, there was hint of interaction between daylength and CNV (P = 0.09), with one of the trip+ lines in particular (IM115) exhibiting increased expression after the shift to 16hr days.
A rare variant with ~300 RLG1a copies (high+) drives PoolSeq coverage patterns and segregates at a single nuclear locus
Even dramatic shifts in the frequency of a 3-copy trip+ haplotype cannot explain the >10× gene-wide coverage of RLG1a in the Spring germinant, Bolt, and IM14_Early cohorts. In genotyping individuals from the IM14 and flower size selection PoolSeq populations, we identified a second amplicon with the 5AA Exon 16 deletion (428bp: 3 each in High and Ancestral, 0 in Low; total N = 162), and in IM14_Early (6/90 = 6.7%) but not IM14_Late (0/95 = 0%). The skew in incidence of the 428bp allele in IM14 was statistically significant, even when trip+ (elevated in IM14_Late; see above) and solo variants were coded as a single category (Pearson χ2 = 6.5, p = 0.01). Despite its rarity, the 428bp amplicon (hereafter high+) was always “homozygous”, suggesting it out-competes other alleles during mN2091×16 PCR-amplification.
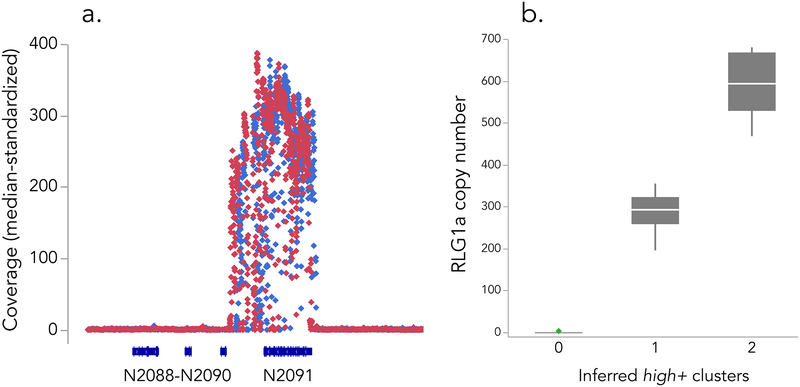
None of the resequenced IM M. guttatus inbred lines (N = 35) had RLG1a coverage greater than trip+ or the indel variation necessary to generate a 428bp mN2091×16 allele. However, two wild-derived 428bp carriers (IM13P_627c and IM15P_617d) exhibited a stack of ~275× (median-standardized) coverage across Migut.N02091 (Figure 4a). The vast majority of stacked reads represented a single haplotype, with the Exon 16 5AA deletion present at >99% (1608/1626 reads had no coverage at the central site of the deletion). Elevated coverage extended across the entire gene as well as the intergenic region between Migut.N02091 and Migut.N02090, spanning ~19 kb. If all copies were arranged in tandem, this high+ RLG1a haplotype would span at least 5.7Mb, or ~22–46% of a M. guttatus chromosome in the current v2 assembly.
Figure 4.
Identification and segregation of high+ RLG1a genotype. a. Median-standardized coverage (25bp windows) of high+ carriers IM_627c and IM15P_317d, showing amplification of Migut.N02091 and the Migut.N02090–2091 intergenic region. b. Quantile box plots for RLG1a copy number in selfed progeny of IM13P_627c segregating for 0 (n = 7), 1 (n = 20) or 2 (n = 12) high+ variants. RLG1a copy number was estimated as 2(Δ Ct), where ΔCt is the absolute difference between Ct mN2091q and Ct mG571q for an individual, averaged across two technical replicates.
qPCR of genomic DNA confirmed a massive copy-number expansion in high+ carriers. In two wild-derived carriers, (IM13P_627c and IM13P_644), mN2091q amplified 8.5 and 9.5 cycles earlier than the single copy control gene, respectively. An internally controlled standard curve estimated copy number of the latter to be >500 (502 and 513 in two technical replicates), and the single-cycle difference in ΔCt is consistent with IM13P_627c having one high+ “allele” and IM13P_644 having two). In the PoolSeq populations, capture of fewer than 10 high+ individuals (each with ~250–300 copies of RLG1a), can explain elevated coverage in IM14_Early vs. IM14_Late. Although we could not genotype individuals from the Fall-Spring and Bolt-Rosette experiments, commensurate shifts in frequency (e.g., high+ carriers relatively common in Bolt and Spring, but absent from Rosette and other low coverage pools) plausibly explain their coverage differences.
The extraordinary amplification of RLG1a in high+ plants raises the possibility of transfer of nuclear DNA to an organellar genome (which occur in many copies per cell), rather than duplication within the nuclear genome. Furthermore, even if encoded in the nuclear genome, RLGA1 could be dispersed across many clusters genome-wide (Gaines et al. 2010). Finally, even if tandemly amplified as a single genetic locus, high+ copies need not be located in tandem with the (solo) annotation of Migut.N02091 in the reference genome. To distinguish these possibilities from the working hypothesis that solo and high+ versions of RLG1a are alternative alleles at the same locus, we examined segregation in a pseudo-F2 made by selfing high+ carrier IM13P_627c. The proportion of high+ carriers in the progeny (84% or 146/173) was slightly elevated over the Mendelian expectation of 75% for a dominant marker (χ22 df = 4.58, P = 0.03), but also substantially lower than the two-locus expectation of 93.75% (χ22 df = 7.57, P = 0.006). Genomic qPCR confirm the inference of a single high+ locus: progeny ΔΔ Ct values (N = 39) clustered into three distinct groups (normal mixtures analysis, AIC/BIC lowest for 3 clusters). Solo individuals had ΔΔCt values not different from 0 (mean = 0.10 ± 0.14; n = 7), whereas high+ carriers sorted into hemizygous (mean ΔΔ Ct = 8.195 ± 0.08; n = 20) and homozygous (mean ΔΔ Ct = 9.19 ± 0.11; n = 12) classes with the expected 2-fold difference in inferred copy number (286.5 ± 11.6 SE and 590.5 ±15.9 SE, respectively; Figure 4b. Both marker and qPCR segregation ratios point to a single nuclear locus segregating 0, 1 or 2 high+ RLG1a arrays consisting of ~300 copies of the gene.
To test whether the high+ locus was genetically coincident with Migut.N02091 (i.e., a local tandem expansion on Chromosome 14), we genotyped 62 IM13P_627c progeny at a tightly-linked flanking marker (mN2089). In solo progeny (n = 8), mN2091×16 segregated for two amplicons perfectly associated with segregating mN2089 genotype, indicating that Migut.N02091 is correctly located in the reference genome. However, mN2089 also exhibited Mendelian segregation within the remaining high+ carriers (χ22 df = 1.2, P >0.50 vs. 1:2:1 expectation). Two segregating alleles at Migut.N02091 in solo progeny, plus no linkage between high+ presence and m2089 genotype, demonstrates that RLG1a high+ segregates as a presence/absence polymorphism at an unlinked locus. Thus, RLG1a exhibits multiple levels of structural variation, varying in the number of genetic loci as well as the number of copies per locus.
As a coarse assay of whether high+ carriers exhibited constitutive expression of RLG1a in proportion to copy number, we conducted rt-qPCR on shoot tissue from greenhouse-grown siblings of the two sequenced high+ plants (outbred, so segregating for all possible genotypes). Both high+ (ΔCt = −2.87, n=3) and trip+ (ΔCt = - 2.98; n =2) plants had ~2-fold higher expression than their solo siblings (ΔCt = −3.90; n = 2), but any genotypic differences in expression between solo and multi-copy plants were not significant with this small sample size (ANOVA, P = 0.37). Regardless, there was no evidence of 300-fold higher expression in high+ plants.
Copy number variants at RLG1a experience fluctuating selection across years
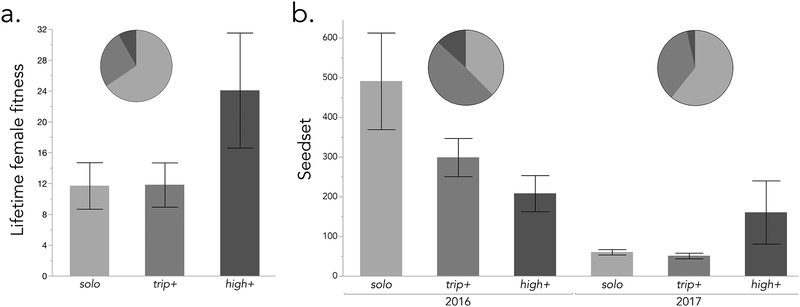
We examined genotype-fitness associations over three years (2015–2017) spanning the extremes of fecundity in the Iron Mountain population (seedset of plants surviving to fruitset = 29.7 ± 2.9 SE, 287.6 ± 32.2 SE, and 63.8 ± 5.5 SE, respectively; all phenotypic N > 350). In 2015, which was a record drought year in the Oregon Cascades (https://wcc.sc.egov.usda.gov) with an unusual April melt-out, we also monitored survival (< 32% overall). We recovered tissue from only ~50% of marked seedlings (N = 243 Spring/Fall germinant pairs) and all 59 Spring germinants recovered (as dead tissue for DNA extraction) died prior to flowering. Additional flowering plants marked one week apart in mid-June also mostly died prior to setting seed (54.6% mortality, N = 185), with those that initiated flowering at the later time significantly more likely to survive (59.7% vs. 38.2%; χ21 df = 7.7, P < 0.01). RLG1a genotype was significantly associated with survival to seedset (χ22 df = 8.56, P = 0.014); 64.7% (11/17) of high+ individuals marked as seedlings or flowerers set seed, whereas only 33.0% (33/100) and 29.8% (54/181) of trip+ and solo plants did. The survival advantage of high+ plants led to significantly higher log-transformed female fitness in monitored plants (ANOVA, P = 0.005, N = 298), as they made (on average) ~2× as many seeds (Figure 5a). There was no significant fecundity effect in the set of random survivors collected at fruiting (P = 0.38, N = 110).
Figure 5.
Fitness of RLG1a copy number variants in the field (means ± 1 SE) and frequencies of each genotype in survivors (pie charts). a. Lifetime female fitness (survival × seedset) in 2015, a severe drought year. b. Seedset (fecundity) of plants that survived to fruit in 2016 and 2017. Note different y-axis scales.
Fecundity varied significantly with RLG1a genotype in 2016 and 2017, but in opposite directions. In lush 2016, which had 5–10× the mean fecundity of flanking years, solo individuals matured 50% more fruits than plants with either CNV (12.4 vs. 7.7 and 8.6 for trip+ and high+ respectively), and, on average, produced twice as many seeds (Figure 5b). In 2017, a heat wave at the end of June/early July sharply ended an otherwise lush flowering season (L. Fishman, pers. obs.). In this truncated season, high+ plants made (on average) twice as many fruits (4.7 vs. 2.4 and 2.2) and 3 times as many seeds as the other two genotypes. These inter-annual shifts result in significant (for fruits; P = 0.016) and marginally non-significant (for seeds; P = 0.07) year × genotype interaction effects in the GLM, along with main effects of RLG1a genotype (both P < 0.005). Along with the 2015 data (and the bias toward early flowering in 2014), shifts in its relative fecundity suggest that high+ influences life history in ways that are beneficial in stressful years (and/or microsites), but deleterious when conditions are good.
Notably, the relative proportions of the CNVs changed as predicted from the previous year’s fitness effects, consistent with real-time responses to fluctuating selection. Following disproportionate high+ survival in 2015 (6.8% in fruiting plants), high+ was maximally common (13%) in 2016 (when it had low seedset), and then dropped to 3.6% in 2017 (Figure 5). Together, these results suggest that RLG1a high+ variants influence life-history traits whose fitness effects vary antagonistically from year to year.
DISCUSSION
Despite the decreasing costs of genome sequencing, connecting phenotype to genotype remains a empirical challenge. Our original intention was to use simple PoolSeq genome-wide association mapping experiments to gain insight into genetic basis of standing variation in floral and life-history traits: the focal Iron Mountain M. guttatus population is a place where selection should be maximally effective (and detectable) at the level of the individual nucleotide, yet also maintains tremendous diversity in fitness-relevant phenotypes. Our results provide a case study for our original question about the processes maintaining polymorphism, but also highlight the challenging complexity of both genome architecture and phenotypic expression in wild populations.
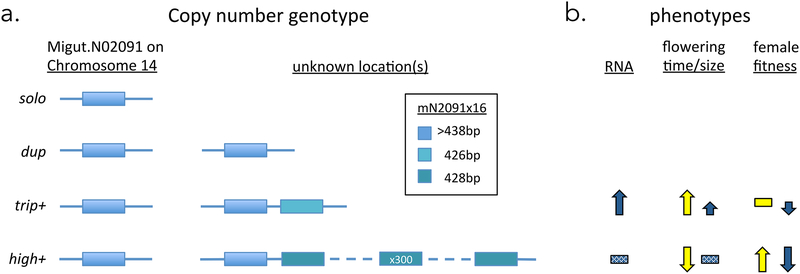
From initial Poolseq scans, we identified a tRNA ligase (RLG1a) as a coverage outlier associated with intra-population variation in flowering time, germination time, flower size, and vernalization requirement. Elevated RLG1a exon coverage in some populations and inbred lines pointed to CNV, and we identified an intermediate frequency (>14%) variant with at least three copies of the gene (trip+) (Figure 6a). Individual genotyping confirmed an assocation of trip+ variants with a “slow” life-history (late flowering under stress and large flower size), and expression analyses suggest that elevated RLG1a dosage in trip+ plants may contribute to its effects (Figure 6b). However, even complete fixation of a 3× variant could not account for observed 10× coverage in three of our population pools, implying the existence of even higher copy-number variants. By targeting rare wild individuals carrying a marker allele sharing a diagnostic exonic indel with trip+, we identified a second CNV (high+) that carries an unprecedented 250–300 haploid copies of RLG1a in a single (presumably tandem) cluster segregating as a presence/absence polymorphism at a nuclear locus unlinked from the single-copy RLG1a (Figure 6a). The presence of even a small number of high+ individuals in a pool was sufficient to grossly bias initial PoolSeq scans, but individual genotyping confirmed significant shifts in CNV frequency in the flower size and 2014 flowering time datasets. Finally, using field fitness measures from three years with very different climatic conditions, we found that high+ experiences fluctuating selection, increasing survival and/or seed production in two drought years, but associated with low fitness relative to non-carriers in a lush year (Figure 6b). Taken together, our discovery and analysis of the trip+ and high+ variants provides insight into the origins of CNV, suggests new candidate functions for an essential but poorly understood plant gene, and supports a role for climatic fluctuation in the maintenance of standing variation for fitness traits.
Figure 6.
Proposed models for a.) the structure of RLG1a copy number variation (CNV) in Mimulus guttatus and b.) the phenotypic effects of trip+ and high+ CNVs relative to solo and dup genotypes. Yellow/light = drought-stress conditions; blue/dark = lush conditions. Solid effects had statistical support in at least one field or greenhouse experiment; stippling indicates sample size was too low for inference.
An embarrassment of riches: detection and dissection of multiple layers of genic CNV
This study of just a single gene, within a single population of monkeyflowers, illustrates both the richness of copy number variation in natural populations and the significant challenges it creates for connecting genotype and phenotype (Hoban et al. 2016). Using genomic resequencing, qPCR, and genetic markers, we define and describe three distinct classes of RLG1a genotype, spanning at least two loci (Figure 6). One locus, Migut.N02091 (RLG1a_1) on LG14, segregates diverse single-copy variants. A second unlinked locus (RLG1a_2) segregates for the ~300-copy high+ variant. The trip+ variant(s), in which one of two inferred copes share a diagnostic 5AA deletion in exon16 with trip+, is likely also an allele at this locus and there are apparently also duplicated (dup) individuals with one copy at each locus. Thus, the history of these variants includes (at minimum) duplication/ insertion of a gene into new chromosomal location, local tandem duplication and sequence divergence, and massively ampliconic tandem duplication, spanning the described mechanisms of CNV generation (Hastings et al. 2009). Although more work will be necessary to fully describe the evolutionary history of this extensive copy number variation, we can propose testable hypotheses about its origins.
The tRNA ligase RLG1 is considered essential (Yang et al. 2017) and is the sole member of its gene family in plants (Englert & Beier 2005). Except for three Brassicaceae and M. guttatus, all 37 Eudicot genomes in Phytozome (ww.Phytozome.org, accessed 07/15/2018) encode just a single copy of this large gene (26 coding exons, >1200AA in M. guttatus). Thus, RLG1 appears highly conserved for copy number despite the multiple rounds of whole genome duplication across Eudicot lineages. Mimulus appears different, even beyond the remarkable high+ amplification. In addition to our focal locus Migut.N02091, a second tRNA ligase homolog (Migut.D02182; RLG1b) is found on Chromosome 4. As annotated, these two genes share 82.6% amino-acid identity, suggesting a recent (most likely within Mimulus) duplication. Retention of two copies of RLG1 in an ancestral Mimulus genome may have been key to further copy number diversification within M. guttatus. Although the functional biology of RLG1 is still poorly understood (see next section), relaxation of the constaints imposed by a single gene performing multiple essential functions (Englert & Beier 2005) may have allowed for neo- and/or sub-functionalization (Lynch & Conery 2000) that then facilitated intraspecific CNV.
The origin of copy number variants at Iron Mountain likely involved whole-gene duplication and deletion deeper in the history of the M. guttatus species complex. Segregation analyses show that the high+ haplotype is present as an an unlinked locus (RLG1a_2 with 250–300 copies in high+ and putatively two copies in trip+; Figure 6a). This nontandem duplication is likely recent, as good agreement between estimates of copy number from qPCR amplification of a highly conserved segment and resequence read-coverage indicate high sequence identity among copies at the two loci. The diagnostic 5AA indel in Exon 16 (captured by our marker mN2091×16) provides some intriguing clues as to how this may have happened. All IM individuals genotyped (including dup, trip+ and high+, as well as the reference gemone and other solo genotypes) have at least one copy with the longer sequence at this position, suggesting that it is ancestral. However, genome alignments from M. nasutus and other “Southern Clade” members of the M. guttatus complex (Brandvain et al. 2014) reveal that the deletion found in trip+ (as one of several copies) and high+ (as 99% of hundreds) is the common single-copy variant elsewhere in the range (L. Fishman, unpublished data). Barring recurrent mutation or gene conversion, this suggests that the IM reference single copy (insertion) and the Southern clade single copy (deletion) are both derived (via reciprocal gene loss) from a duplicated ancestor similar to putative dup IM lines (Figure 6a). Such reciprocal deletion of duplicates has recently been implicated in hybrid lethality in M. guttatus species complex (Zuellig & Sweigart 2018), and is a common mechanism of plant hybrid incompatibility (Fishman & Sweigart 2018), so may have implications beyond phenotypic differences of the alternative genotypes. Testing this scenario against alternatives will require genetic mapping and physical deconvolution (e.g., with long-range sequencing technologies) of high+, trip+ and dup RLG1a haplotypes, as well as molecular evolution analyses of genotypes from throughout the species range.
Finally, how did the massive expansion of RLG1a gene copies in high+ haplotypes occur? The largest genic CNV known in plants is the target gene for glyphosate herbicides, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), which expands to 5–160 copies in Amaranthus palmerii (Gaines et al. 2010). Expansion of EPSPS confers herbicide resistance via gene dosage effects (i.e., increased expression), and is thus strongly favored in sprayed crop fields. Amaranthus EPSPS clusters are dispersed throughout the genome (Gaines et al. 2010) and cytogenetics reveals that EPSPS clusters form extra-chromosomal circular DNA (eccDNA) molecules rather than being integrated into linear chromosomes (Koo et al. 2018). Rolling circle amplification in eccDNA was previously implicated in the origin of the sobo satellite repeat in Solanum bulbocastanum, in which a 360kb (nongenic) monomer amplified to 4.7 Mb in a single (hemizygous) chromosomal location (Tek et al. 2005). RLG1a high+ segregates as the presence/absence of a tandem array, and thus may have originated via rapid amplification in eccDNA and re-insertion (without copy number intermediates) into the nuclear genome (Charlesworth et al. 1994). More work is necessary to evaluate this scenario, and to test the effects of RLG1a2 expansion on the recombination and segregation of linked loci.
This study highlights how much CNVs complicate genome scans for adaptive differentiation, despite being a large proportion of potentially functional variants. Especially in pooled population sequencing (PoolSeq) experiments, CNV generates substantial complications. For example, Migut.N02091 outlier sites were censored (for excess coverage) in SNP scans of the High-Low flower size selection experiment (Kelly et al. 2013), but it appeared to be strong SNP outlier in the IM flowering time study (Monnahan & Kelly 2017). Sites with low coverage are rightly dropped from most analytical pipelines (DePristo et al. 2011), so gene deletions relative to a reference genome (particularly within clusters of paralogs where cross-mapping may occur) are difficult to identify and such loci may be lost from scans. Particularly in PoolSeq studies, the coverage stacks created by duplication can generate false positives via statistical (over-confidence) and biological bias (mis-representation of allele frequencies). Summary statistics of differentiation between pooled populations reflect sample size (coverage) as well as allele frequencies (Kofler et al. 2011; Magwene et al. 2011; Kelly et al. 2013), so even identical duplicates can inflate statistical significance. At the same time, stacking of multiple paralogs with shared SNP/indel differentiation (like trip+ and high+ vs. solo) exaggerates differences among pools. Without the 2bp indel that happens to distinguish trip+ and high+ diagnostic PCR amplicons (and follow-up targeted resequencing, crossing, and qPCR), we would have parsmoniously inferred the existence of a 5-copy variant much more differentiated in frequency among pools. Distortion of PoolSeq SNP scans by CNV adds to the existing arguments for using large sample sizes (>100 individuals) in PoolSeq experiments (Lynch et al. 2014). In addition, validation of results from genome scans (using both individual and pooled sequencing approaches) with independent genotyping is an important step in investigating the genetic basis of adaptive traits.
Our results, along with increasing evidence that CNV contributes to fitness-relevant variation in diverse systems (Girirajan et al. 2011; Maron et al. 2013; Mickelbart et al. 2015; Prunier et al. 2017; Dolatabadian et al. 2017), suggest that explicit CNV surveys should be routine part of genome-wide scans for adaptive loci. A number of new pipelines combine coverage, read-pair information, and other data to infer structural variants from short-read data (Layer et al. 2014; Mohiyuddin et al. 2015), and could fruitfully be used to catalog CNV in (for example) the Iron Mountain inbred line set. The detection of putative 2×-covered dup individuals in that set (Figure 2) is promising, though poor alignment in intergenic regions in diverse M. guttatus limits the utility of read-pair information as corroborative evidence. Without such a reference catalog of variants, however, whole-gene CNVs are likely to often remain a component of the missing heritability (Manolio et al. 2009).
One piece of the puzzle: a tRNA ligase and standing variation for life-history traits
The genic CNVs identified in this study are undoubtedly a tiny sample of the variants affecting life-history phenotypes in IM M. guttatus (Troth et al. 2018), but RLG1a is the first (and hopefully worst!) gene to be dissected mechanistically as a direct contributor to fitness phenotypes in this model population. Importantly, despite not knowing the chromosomal locations of the CNVs or full individual genotypes, we can be reasonably confident that RLG1a copy number per se and/or sequence variation within the characterized CNVs underlies the observed phenotypic and fitness associations. In IM M. guttatus, linkage disequilibrium (LD) falls off sharply at distances >1000bp (with the exception of several inversions and/or recent selective sweeps; Puzey et al. 2017). Thus, trip+ and high+ are unlikely to be neutral markers for functionally-distinct loci tightly linked to RLG1a2; in addition, a preliminary scan for associated sites (i.e, scanning for differences in nucleotide diversity within vs. between solo and trip+ inbred line sets; data not shown) did not reveal any strong or extensive LD with other genomic regions. Thus, we discuss the phenotypic and fitness results in the context of RLG1 function.
Why a tRNA ligase CNV? Plant tRNA ligases share little sequence homology with tRNA ligases in other taxa, but retain the core function of splicing the two intron-containing nuclear-encoded tRNAs (tRNATyr and elongator tRNAMet) (Englert & Beier 2005). The single-copy Arabidopsis RLG1 is expressed in most tissues and is embryo-lethal when knocked out (Yang et al. 2017). RLG1 has also been implicated in the joint regulation of the key plant hormone auxin and the endoplasmic reticulum-based unfolded protein response (UPR) in Arabidopsis (Leitner et al. 2015; Nagashima et al. 2016). One arm of the UPR is regulated by cytoplasmic splicing of bZIP60 transcription factor mRNAs; RLG1 performs the key ligation step (Nagashima et al. 2016). Via its regulation of the UPR, RLG1 may play a central role in plant reactivity and resistance to abiotic stressors such as heat and drought, as well as hormonal control of tradeoffs between vegetative growth and reproduction (Deng et al. 2013). In addition, the bZIP60 branch of the UPR is constitutively up-regulated in pollen (which is notoriously vulnerable to heat stress) (Deng et al. 2016) and integral to plant Reponses to viral infection (Zhang et al. 2015). Even beyond its remarkable copy number diversity, M. guttatus (with both RLG1a and RLG1b in everyone) provides a rare opportunity to investigate the multiple functions of plant tRNA ligases without the constraint of an essential single copy.
Our phenotype and fitness results suggest that RLG1a CNVs contribute to stress-responsive variation in the timing of reproduction, along a spectrum from highly reactive to stress (fast) to relatively nonchalant (slow). Interestingly, carriers of the trip+ variant appear “slow” (large flowers, later reproduction in field and under drought stress), whereas high+ carriers appear “fast” (early flowering in 2014, highest survival and/or seedset in drought years). Further analyses of sequence variants and expression patterns will be necessary to determine the physiological mechanisms underlying this pattern. Importantly, the fecundity effects of RLG1a CNV that we see in the field are caused by differences in fruit number rather than seedset/fruit (which did not significantly differ among years or RLG1a genotypes). Thus, fluctuating fitness effects depend on life-history plasticity/stress tolerance traits that affect the timing of flowering and senescence. This parallels work showing fluctuating selection on “fast/small” vs. “slow/large” alleles using QTL introgression lines (Mojica & Kelly 2010; Mojica et al. 2012) and a large genome-wide association mapping set (Troth et al. 2018) planted at a nearby site, but highlights genotype × environment interactions for phenotype as well as fitness. For example, alternative RLG1a genotypes may differ in whether they accelerate bolting when sensing short-term heat stress via the UPR, with the fitness consequences depending on whether heat stress accurately predicts imminent dry-down and death in a given year and microsite. In addition to mediating stress-responsive cues, RLG1a CNVs could also affect stress-tolerance; elevated expression of RLG1a (as in trip+ lines; Figure 3) may increase the efficiency of UPR-upregulation and thus protect against heat or cold damage that shortens life span. Unmeasured effects on male fitness (e.g., via heat tolerance of pollen) may also contribute to the maintenance of alternative RLG1a CNVs. Exploring these intriguing possibilities in the appropriate environmental contexts will require long-term monitoring of natural populations, as well as experimental manipulation of candidate stress regimes.
Temporally fluctuating selection is an appealing but controversial mechanism to maintain fitness variation, as simple models cannot indefinitely maintain multiple alleles when their fitness effects are not perfectly balanced (reviewed in Delph & Kelly 2013). However, plasticity (which we observe) and shifts in dominance (which are implied by high+/solo fitness reversals; Figure 4) can enlarge the parameter space over which temporal variation prevents allele loss (Posavi et al. 2014; Gulisija et al. 2016; Wittmann et al. 2017). In addition, dormant life-stages (e.g. a seedbank) and spatial variation within a population (e.g., unpredictable soil depth) can slow the dynamics of displacement and thus help maintain standing variation over observable time scales (Ellner & Hairston 1994; Svardal et al. 2015). Although we cannot be sure that high+ will not either fix or be driven from the Iron Mountain population in the long term (especially as it may be a relatively new variant not at equilibrium), the observed shifts in relative fitness from year to year plausibly contribute to its maintenance at low to intermediate frequency. This parallels the classic Linathus parryae system, in which temporal and spatial variation in drought stress contributes to the maintenance of a flower color polymorphism (Schemske & Bierzychudek 2001; 2007). In a model inspired by that system, variants with geometric mean fitness < 1 (i.e., ones that should be deterministically lost otherwise) can be maintained if they have highest fitness in “good” years and a seedbank spreads that advantage over time (Turelli et al. 2001). Like other variants contributing to life-history tradeoffs in the IM M. guttatus population (Troth et al. 2018), RLG1a CNVs experience similar dynamics, mediated (in part) by stress-responsive phenological traits. Our results add to the growing evidence that temporally fluctuating selection contributes the abundant quantitative genetic variation seen in many short-lived taxa (Siepielski et al. 2009; Troth et al. 2018).
Supplementary Material
ACKNOWLEDGMENTS
We thank Gloria Goni-McAteer, Hanna McIntosh, John Crandall and Emily Beck for assistance with the field plant collections, and Aurora Bayless, Daniel Crowser, and Katie Zarn for lab and greenhouse help. We appreciate the logistical support for library preparation, qPCR, and marker genotyping provided by Tamara Max and Denghui (David) Xing in the University of Montana Genomics Core. This work was supported by NSF DEB-0846089, DEB-1457763, and OIA-1736249 to L.F. and by NIH R01-GM073990 to J.K.K.
Footnotes
- Whole genome resequence datasets (fastqs or bams): NCBI Sequence Read Archive: Inbred lines - SAMN05852485-SAMN05852522, SAMN04517335; Iron Mountain flowering time pools - PRJNA336318; Other pools - SAMN10169032-SAMN10169038; high+ accessions - SAMN10134433, SAMN10134434.
- Genotype, phenotype, and /or qPCR datasets: doi:10.5061/dryad.m1j24tf
LITERATURE CITED
- Anderson JT (2016) Plant fitness in a rapidly changing world. New Phytologist, 210, 81–87. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Schluter D (2008) Adaptation from standing genetic variation. Trends in Ecology & Evolution, 23, 38–44. [DOI] [PubMed] [Google Scholar]
- Barson NJ, Aykanat T, Hindar K et al. (2015) Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature, 528, 405–408. [DOI] [PubMed] [Google Scholar]
- Bell G (2010) Fluctuating selection: the perpetual renewal of adaptation in variable environments. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 365, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodbyl Roels SA, Kelly JK (2011) Rapid evolution caused by pollinator loss in Mimulus guttatus. Evolution, 65, 2541–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P et al. (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biology, 5, e236–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL (2014) Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genetics, 10, e1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Kelly JK (2018) Antagonistic pleiotropy can maintain fitness variation in annual plants. Journal of Evolutionary Biology, 31, 46–56. [DOI] [PubMed] [Google Scholar]
- Chain FJJ, Feulner PGD, Panchal M et al. (2014) Extensive copy-number variation of young genes across stickleback populations. PLoS Genetics, 10, e1004830–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Fry JD (2016) Evidence that environmental heterogeneity maintains a detoxifying enzyme polymorphism in Drosophila melanogaster. Current Biology, 26, 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature, 371, 215–220. [DOI] [PubMed] [Google Scholar]
- Chia J-M, Song C, Bradbury PJ et al. (2012) Maize HapMap2 identifies extant variation from a genome in flux. Nature Genetics, 44, 803–807. [DOI] [PubMed] [Google Scholar]
- Clarke BC (1979) The evolution of genetic diversity. Proceedings of the Royal Society of London. Series B, Biological sciences, 205, 453–474. [DOI] [PubMed] [Google Scholar]
- Connallon T, Clark AG (2014) Balancing selection in species with separate sexes: insights from Fisher’s geometric model. Genetics, 197, 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S (2010) Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biology and Evolution, 2, 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph LF, Kelly JK (2013) On the importance of balancing selection in plants. New Phytologist, 201, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proceedings of the National Academy of Sciences USA, 110, 19633–19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Quilichini TD et al. (2016) IRE1, a component of the Unfolded Protein Response signaling pathway, protects pollen development in Arabidopsis from heat stress. The Plant Journal. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics, 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatabadian A, Patel DA, Edwards D, Batley J (2017) Copy number variation and disease resistance in plants. Theoretical and Applied Genetics, 130, 2479–2490. [DOI] [PubMed] [Google Scholar]
- Ellner S, Hairston NG Jr. (1994) Role of overlapping generations in maintaining genetic variation in a fluctuating environment. American Naturalist, 143, 403–417. [Google Scholar]
- Englert M, Beier H (2005) Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Research, 33, 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Kelly JK (2015) Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution, 69, 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Sweigart AL (2018) When two rights make a wrong: the evolutionary genetics of plant hybrid incompatibilities. Annual Review of Plant Biology, 69, 701–737. [DOI] [PubMed] [Google Scholar]
- Fishman L, Sweigart AL, Kenney AM, Campbell S (2014) Major quantitative trait loci control divergence in critical photoperiod for flowering between selfing and outcrossing species of monkeyflower (Mimulus). New Phytologist, 201, 1498–1507. [DOI] [PubMed] [Google Scholar]
- Friedman J, Willis JH (2013) Major QTLs for critical photoperiod and vernalization underlie extensive variation in flowering in the Mimulus guttatus species complex. New Phytologist, 199, 571–583. [DOI] [PubMed] [Google Scholar]
- Gaines TA, Zhang W, Wang D et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proceedings of the National Academy of Sciences USA, 107, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH (1984) Pleiotropic overdominance and the maintenance of genetic variation in polygenic characters. Genetics, 107, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE (2011) Human copy number variation and complex genetic disease. Annual Review of Genetics, 45, 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Woods DP et al. (2017) Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nature Communications, 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulisija D, Kim Y, Plotkin JB (2016) Phenotypic plasticity promotes balanced polymorphism in periodic environments by a genomic storage effect. Genetics, 202, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Willis JH (2006) Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution, 60, 2466–2477. [PubMed] [Google Scholar]
- Hanikenne M, Kroymann J, Trampczynska A et al. (2013) Hard selective sweep and ectopic gene conversion in a gene cluster affording environmental adaptation. PLoS Genetics, 9, e1003707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G (2009) Mechanisms of change in gene copy number. Nature Reviews Genetics, 10, 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE et al. (2016) Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. American Naturalist, 188, 379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM (2011) Climate change and evolutionary adaptation. Nature, 470, 479–485. [DOI] [PubMed] [Google Scholar]
- Hunter B, Wright KM, Bomblies K (2013) Short read sequencing in studies of natural variation and adaptation. Current Opinion in Plant Biology, 16, 85–91. [DOI] [PubMed] [Google Scholar]
- Immler S, Arnqvist G, Otto SP (2011) Ploidally antagonistic selection maintains stable genetic polymorphism. Evolution, 66, 55–65. [DOI] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT et al. (2011) Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature, 477, 203–U102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK, Willis JH (2001) Deleterious mutations and genetic variation for flower size in Mimulus guttatus. Evolution, 55, 937–942. [DOI] [PubMed] [Google Scholar]
- Kelly JK, Koseva B, Mojica JP (2013) The genomic signal of partial sweeps in Mimulus guttatus. Genome Biology and Evolution, 5, 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwin R, Feusier J, Corwin J et al. (2015) Natural genetic variation in Arabidopsis thaliana defense metabolism genes modulates field fitness. eLife, 4, 2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Orozco-terWengel P, De Maio N et al. (2011) PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. (Kayser M, Ed,). PloS one, 6, e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D-H, Molin WT, Saski CA et al. (2018) Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proceedings of the National Academy of Sciences USA, 115, 3332–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM (2014) LUMPY: a probabilistic framework for structural variant discovery. Genome biology, 15, R84–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YW, Fishman L, Kelly JK, Willis JH (2016) A segregating inversion generates fitness variation in yellow monkeyflower (Mimulus guttatus). Genetics, 202, 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Retzer K, Malenica N et al. (2015) Meta-regulation of Arabidopsis auxin responses depends on tRNA maturation. Cell Reports, 11, 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 31, 1674–1676. [DOI] [PubMed] [Google Scholar]
- Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 3, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaurens V, Whibley A, Joron M (2017) Genetic architecture and balancing selection: the life and death of differentiated variants. Molecular Ecology, 26, 2430–2448. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science, 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M, Bost D, Wilson S, Maruki T, Harrison S (2014) Population-genetic inference from pooled-sequencing data. Genome Biology and Evolution, 6, 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene PM, Willis JH, Kelly JK (2011) The statistics of bulk segregant analysis using next generation sequencing. PLoS computational biology, 7, e1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron LG, Guimarães CT, Kirst M et al. (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proceedings of the National Academy of Sciences USA, 110, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroni F, Pinosio S, Morgante M (2014) Structural variation and genome complexity: is dispensable really dispensable? Current Opinion in Plant Biology, 18, 31–36. [DOI] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nature Reviews Genetics, 16, 237–251. [DOI] [PubMed] [Google Scholar]
- Mohiyuddin M, Mu JC, Li J et al. (2015) MetaSV: an accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics, 31, 2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica JP, Kelly JK (2010) Viability selection prior to trait expression is an essential component of natural selection. Proceedings of the Royal Society London B, 277, 2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica JP, Lee YW, Willis JH, Kelly JK (2012) Spatially and temporally varying selection on intrapopulation quantitative trait loci for a life history trade-off in Mimulus guttatus. Molecular Ecology, 21, 3718–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnahan PJ, Kelly JK (2017) The genomic architecture of flowering time varies across space and time in Mimulus guttatus. Genetics, 206, 1621–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnahan PJ, Colicchio J, Kelly JK (2015) A genomic selection component analysis characterizes migration-selection balance. Evolution, 69, 1713–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Iwata Y, Mishiba K-I, Koizumi N (2016) Arabidopsis tRNA ligase completes the cytoplasmic splicing ofbZIP60 mRNA in the unfolded protein response. Biochemical and Biophysical Research Communications, 470, 941–946. [DOI] [PubMed] [Google Scholar]
- Pedersen BS, Quinlan AR (2018) Mosdepth: quick coverage calculation for genomes and exomes. (Hancock J, Ed,). Bioinformatics, 34, 867–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posavi M, Gelembiuk GW, Larget B, Lee CE (2014) Testing for beneficial reversal of dominance during salinity shifts in the invasive copepod Eurytemora affinis, and implications for the maintenance of genetic variation. Evolution, 68, 3166–3183. [DOI] [PubMed] [Google Scholar]
- Prunier J, Caron S, Lamothe M et al. (2017) Gene copy number variations in adaptive evolution: The genomic distribution of gene copy number variations revealed by genetic mapping and their adaptive role in an undomesticated species, white spruce (Picea glauca). Molecular Ecology, 26, 5989–6001. [DOI] [PubMed] [Google Scholar]
- Puzey JR, Willis JH, Kelly JK (2017) Population structure and local selection yield high genomic variation in Mimulus guttatus. Molecular Ecology, 26, 519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloski SM, Jali SS, Balasubramanian S, Weigel D, Grbic V (2010) Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics, 186, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salojärvi J, Smolander O-P, Nieminen K et al. (2017) Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nature Genetics, 49, 904–912. [DOI] [PubMed] [Google Scholar]
- Institute SAS (1994) JMP user’s guide. Version 3.0.2. [Google Scholar]
- Schemske DW, Bierzychudek P (2001) Evolution of flower color in the desert annual Linanthus parryae: Wright revisited. Evolution, 55, 1269–1282. [DOI] [PubMed] [Google Scholar]
- Schemske DW, Bierzychudek P (2007) Spatial differentiation for flower color in the desert annual Linanthus parryae: was Wright right? Evolution, 61, 2528–2543. [DOI] [PubMed] [Google Scholar]
- Scoville AG, Lee YW, Willis JH, Kelly JK (2009) Contribution of chromosomal polymorphisms to the G-matrix of Mimulus guttatus. New Phytologist, 183, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville AG, Lee YW, Willis JH, Kelly JK (2011) Explaining the heritability of an ecologically significant trait in terms of individual quantitative trait loci. Biology Letters, 7, 896–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepielski AM, DiBattista JD, Carlson SM (2009) It’s about time: the temporal dynamics of phenotypic selection in the wild. Ecology Letters, 12, 1261–1276. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M et al. (2007) Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science, 315, 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svardal H, Rueffler C, Hermisson J (2015) A general condition for adaptive genetic polymorphism in temporally and spatially heterogeneous environments. Theor. Popul. Biol, 99, 76–97. [DOI] [PubMed] [Google Scholar]
- Sweigart AL, Karoly K, Jones A, Willis J (1999) The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity, 83, 625–632. [DOI] [PubMed] [Google Scholar]
- Tek AL, Song J, Macas J, Jiang J (2005) Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. Genetics, 170, 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Ross-Ibarra J (2014) Advances and limits of using population genetics to understand local adaptation. Trends in Ecology & Evolution, 29, 673–680. [DOI] [PubMed] [Google Scholar]
- Troth A, Puzey JR, Kim RS, Willis JH, Kelly JK (2018) Selective trade-offs maintain alleles underpinning complex trait variation in plants. Science, 361, 475–478. [DOI] [PubMed] [Google Scholar]
- Turelli M, Schemske DW, Bierzychudek P (2001) Stable two-allele polymorphisms maintained by fluctuating fitnesses and seed banks: protecting the blues in Linanthus parryae. Evolution, 55, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA (2013) Gene dosage effects: nonlinearities, genetic interactions, and dosage compensation. Trends in Genetics, 29, 385–393. [DOI] [PubMed] [Google Scholar]
- Wittmann MJ, Bergland AO, Feldman MW, Schmidt PS, Petrov DA (2017) Seasonally fluctuating selection can maintain polymorphism at many loci via segregation lift. Proceedings of the National Academy of Sciences USA, 114, E9932–E9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S (1939) The distribution of self-sterility alleles in populations. Genetics, 24, 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KJ, Guo L, Hou XL, Gong HQ, Liu CM (2017) ZYGOTE-ARREST 3 that encodes the tRNA ligase is essential for zygote division in Arabidopsis. Journal of integrative plant biology, 59, 680–692. [DOI] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW (2015) A copy number variation map of the human genome. Nature Reviews Genetics, 16, 172–183. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen H, Brandizzi F, Verchot J, Wang A (2015) The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genetics, 11, e1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuellig MP, Sweigart AL (2018) Gene duplicates cause hybrid lethality between sympatric species of Mimulus. PLoS Genetics, 14, e1007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żmieńko A, Samelak A, Kozłowski P, Figlerowicz M (2014) Copy number polymorphism in plant genomes. Theoretical and Applied Genetics, 127, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.