Abstract
Disruption of virus capsid assembly has compelling antiviral potential that has been applied to Hepatitis B Virus (HBV). HBV core protein assembly can be modulated by heteroaryldihydropyrimidines (HAPs), such molecules are collectively termed core protein allosteric modulators (CpAMs). Though the antiviral effects of CpAMs are acknowledged, the mechanism of action remains an open question. Challenging aspects of characterizing misdirected assembly are the large size and non-uniform nature of the final particles. In this study of HBV assembly, we observed a competition between normative and CpAM-induced aberrant assembly with electron microscopy and single particle nanofluidic techniques. This competition was a function of the strength of the association energy between individual core proteins, which is proportional to ionic strength. With strong association energy, assembly reactions primarily yielded morphologically normal HBV capsids, despite the presence of HAP. At weak association energy, HAPs led to increased assembly product size and disrupted morphology. The smallest particles were T = 4 icosahedra, whereas the larger particles were defective spheres, ellipsoids, and bacilliform cylinders, with regions of T = 4 geometry interspersed with flat regions. Deviation from the spherical T = 4 geometry progressively increased with particle size, which is consistent with the interpretation of a competition between two alternative assembly pathways.
Graphical Abstract
Introduction
Chronic Hepatitis B Virus (HBV) infection is a global health concern.1 Because the established treatment strategies are not curative, new therapeutic strategies are under investigation to identify a cure. One antiviral approach is the development of molecules which target the assembly of virus capsids, the protective container of the viral genome.2–3 The HBV core protein (Cp), the shell of the viral core, is implicated in virus functionality at nearly every step of the viral lifecycle, which explains why Cp is a particularly attractive antiviral target. During viral infection, core protein dimers assemble to form ordered spherical capsids with T = 3 and T = 4 icosahedral geometry, consisting of 90 and 120 dimers, respectively. T = 3 capsids have a diameter of 31.3 nm, whereas T = 4 capsids have a diameter of 35.7 nm, measured by cryogenic electron microscopy (cryo-EM). Under physiological conditions, the T = 4 capsid is the predominant form.4
In vitro assembly of T = 3 and T = 4 capsids, including the preference for T = 4 capsids, can be recapitulated with purified core protein. The core protein is a small, homodimeric protein comprised of an assembly domain, residues 1–149, and C-terminal nucleic acid-binding domain, residues 150–183. The C-terminal domain is necessary for binding and packaging nucleic acid but is dispensable for self-assembly of an empty capsid.5 In the context of a T = 4 capsid, Cp dimers are either AB or CD protein pairs, with five A subunits forming icosahedral fivefold vertices and two sets of B-C-D repeats forming quasi six-fold vertices (Figure 1a, inset).6–7 The interface between dimers is largely hydrophobic, and the per contact association energy is weak,8 ~ 3.6 kcal/mol (6 KBT), which facilitates assembly and minimizes kinetic traps. In vitro, this association energy becomes stronger in solutions of high ionic strength, which can lead to kinetically trapped intermediate species. In vitro assembly has been modeled as a nucleated cascade of second order reactions.8–9 Once assembled, capsids are the structural core of infectious virions and provide a scaffold for the lipid envelope and a container for reverse-transcription of viral RNA into DNA.10 Thus, proper self-assembly of core protein into capsids is imperative for a chronic infection.
Figure 1. Assembly product morphology from competition between dimer-dimer association energy and HAP-TAMRA-induced assembly.
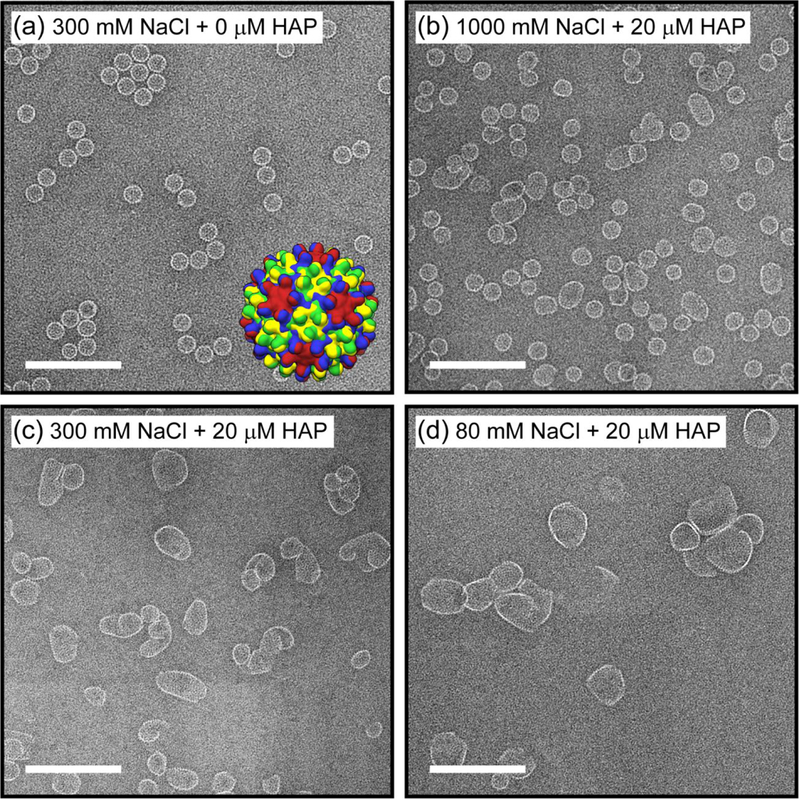
(a) Negative stain transmission electron microscope (TEM) images of an assembly reaction of 5 μM core protein (Cp) dimer with 300 mM NaCl in 50 mM HEPES (pH 7.5) but without HAP-TAMRA (negative control). The inset shows a low resolution rendering of a T = 4 capsid structure, the major product of the control reaction. Each Cp monomer is colored according to its position in an icosahedral asymmetric unit (A: red, B: blue, C: green, D: yellow). Negative stain TEM images of assembly reactions of 5 μM Cp dimer with 20 μM HAP-TAMRA and (b) 1000, (c) 300, and (d) 80 mM NaCl in 50 mM HEPES (pH 7.5). The scale bars are 200 nm. At high ionic strength, most of the particles have sizes close to T = 4 capsids despite the presence of HAP-TAMRA, whereas at low ionic strength, larger and more aberrant structures are formed.
Molecules that target the self-assembly of virus capsids are collectively termed core protein allosteric modulators (CpAMs). The motivation for this study is to gain an understanding of how CpAMs alter virus capsid assembly, towards the ultimate goal of an explicit molecular mechanism of action. CpAMs exhibit a variety of effects by altering assembly kinetics, the stability of dimer-dimer interactions, or the geometry of dimer-dimer interactions.11–12 One of the well-characterized classes of CpAMs consists of the heteroaryldihydropyrimidines (HAPs), which were initially identified with the capability to inhibit viral replication13 and cause in vitro assembly of large aberrant structures.14 Core protein directed antivirals represent an orthogonal approach to nucleoside analog inhibitors, enabling CpAMs to retain antiviral activity even against nucleoside analog resistant mutants.15 Several CpAMs have progressed to the clinical stage and show promise as part of a curative strategy for chronic infection.16
HAP molecules increase assembly rate and extent, and the products can be large and heterogeneous, but the precise mechanism of action is still being elucidated. Despite their heterogeneity, there is evidence that the products consist in part of a repeating hexameric structure.14 Hexameric repeats were recently confirmed with a HAP derivative that facilitated the assembly of Cp cylinders.17 In the majority of reported cases, however, the HAP-induced assembly produces a diverse ensemble of structures. This heterogeneity precludes a comprehensive understanding of the reaction products.
Resistive-pulse sensing is a conductivity-based technique that detects individual particles as they translocate through constrictions (e.g., pores) with dimensions comparable to those of the particles.18–22 Resistive-pulse devices are excellent tools for studying virus assembly due to their compatibility with typical assembly buffers, their single-particle sensitivity, and because measurements take place in solution where biomolecules possess their native state.23 Nanofluidic resistive-pulse sensors have already been used to characterize assembly of HBV capsids under a wide range of conditions,24–27 providing a good frame of reference for understanding reactions that become further complicated in the presence of CpAMs.
To understand the HAP action mechanism, we examined individual particles and placed them in context of a larger population. To analyze such a non-uniform ensemble of particles, we based our analysis on two complementary single-particle analysis techniques, electron microscopy (EM) and resistive-pulse sensing. Resistive-pulse sensing counts and sizes each particle to produce a quantitative measure of the particle distribution, whereas EM provides an assessment of particle morphology. The presence of HAP-TAMRA, a HAP CpAM that favors aberrant assembly, allowed us to detect a switch between normative and drug-regulated assembly pathways, two paths which appear to be in direct competition.
Results and Discussion
Characterization of Assembly Products with Electron Microscopy (EM).
To investigate the effect of HAP-TAMRA on the assembly mechanism, we needed to perturb a critical feature of assembly. This perturbation was accomplished by adjusting ionic strength, which predictably alters association energy. We chose concentrations of 80, 300, and 1000 mM NaCl, which correspond to pairwise Cp-Cp association energies of >−3.1, −3.7, and <−4.0 kcal/mol and pseudo-critical concentrations of assembly of >14, 1.9, and 0.5 μM Cp dimer, respectively.8 The pseudo-critical concentration refers to the dimer concentration above which all additional subunits assemble to capsids. At the lowest ionic strength tested (80 mM NaCl), assembly of 5 μM Cp dimer is almost negligible in the absence of CpAM. At the highest ionic strength tested (1000 mM NaCl), assembly is nearly quantitative above the pseudo-critical concentration.
HAP-TAMRA was recently the subject of a cryo-EM reconstruction that determined the position where this modulator binds to pre-assembled HBV capsids.28 In that study, HAP-TAMRA deformed pre-assembled T = 3 and T = 4 capsids, but did not cause a change in the overall icosahedral geometry. In the experiments reported here, the initial concentrations of the HAP-TAMRA and dimeric core protein were 20 μM and 5 μM, respectively, (4 to 1 molar ratio) for all the reaction conditions tested; thus, two active HAP enantiomers were present per dimer.
A control reaction of 5 μM dimer assembled in 300 mM NaCl yielded morphologically normal T = 4 capsids almost exclusively (Figure 1a). Assembly reactions of 5 μM dimer with 20 μM HAP-TAMRA at each salt concentration produced dramatically different product distributions as visualized by EM (Figure 1b-d). Assembly of dimer and HAP in 1000 mM NaCl resulted in an abundance of spherical particles, consistent in diameter with T = 4 capsids (Figure 1b). Assembly in 300 mM NaCl produced a reduced percentage of morphologically normal particles accompanied by an increase in larger aberrant structures (Figure 1c). The influence of HAP was visually evident by the presence of some larger ellipsoidal assemblies, but their abundance was dramatically lower compared to assembly in the lowest salt concentration. A further reduction of salt to 80 mM NaCl produced even larger particles and very few T = 4 capsids (Figure 1d). Molar ratios of HAP-TAMRA to dimer less than 4:1 yielded a smaller percentage of aberrant structures when assembled in 300 and 80 mM NaCl, whereas molar ratios of HAP-TAMRA to dimer greater than 4:1 did not produce a significantly larger fraction of aberrant structures in 1000, 300, or 80 mM NaCl, which suggests at a 4:1 molar ratio HAP-TAMRA was saturating the assembly reaction.
Because assembly in 300 mM NaCl with HAP-TAMRA produced a breadth of particle diversity, we sought to further characterize these products. To identify dominant features among the particles, images of approximately 2000 individual particles were manually extracted from electron micrographs and subjected to 2D class averaging (Figure 2). As Figure 2c indicates, all the class averages have regions with curvature that are visually similar to the curvature of T = 4 capsids. These regions suggest that the aberrant particles are not assembled into a completely novel geometry, but rather particles have regions of T = 4-like capsid geometry interspersed with flat regions. These flat regions are consistent with hexameric repeats of Cp observed previously.14,17 We note that negative staining and dehydration can alter actual shapes and structures, especially of misassembled particles, which we suspect are more flexible as they are not spherical. Despite these limitations, EM and resistive-pulse sensing gave comparable results for the characterization of regular HBV assemblies.25
Figure 2. Favored local geometries for a T = 4 capsid or hexagonal repeat.
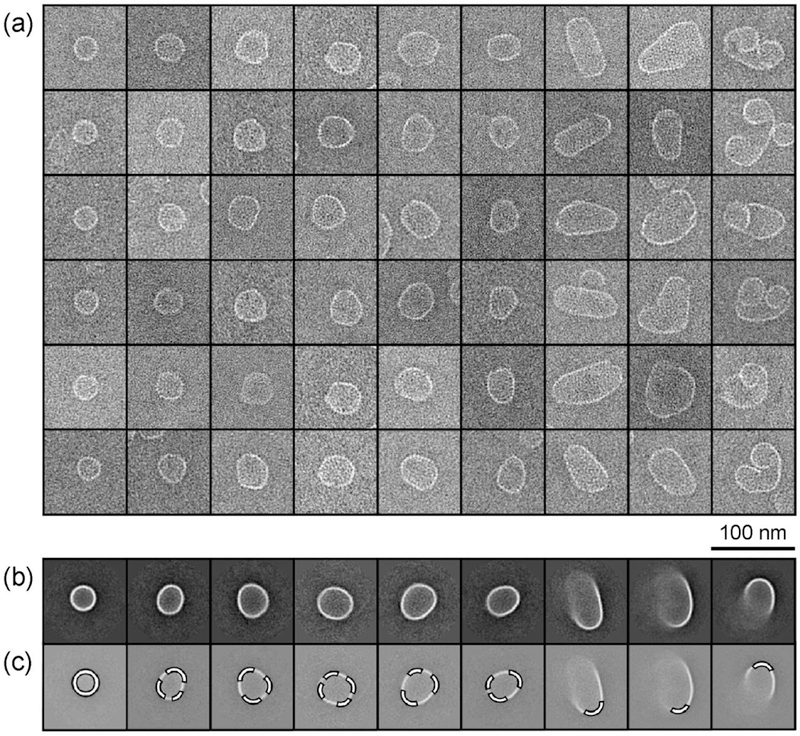
(a) Gallery of negative stain TEM images. Particles in each vertical column were clustered together during a reference-free 2D class averaging procedure. Each column is a representative sample of a class with more particles. (b) The corresponding class averages sorted approximately by ascending size. Particles with a diameter of 35 nm, expected for normal T = 4 capsids, appear on the far left. Larger aberrant structures observed from assembly with HAP-TAMRA include larger, roughly spherical structures, ellipsoidal structures, and structures that appear to have shapes of tubes and spirals. (c) A common feature among all class averaged structures is the highlighted regions with a curvature of 35 nm diameter, comparable to that T = 4 capsids. Particles were assembled from 5 μM Cp dimer with 20 μM HAP-TAMRA and 300 mM NaCl in 50 mM HEPES (pH 7.5).
Resistive-Pulse Measurements of Particle Size Distributions.
A 3-pore device fabricated for resistive-pulse analysis of HBV assembly in the presence of HAP-TAMRA is illustrated in Figure 3a,b and described in detail in the Methods. The relatively large dimensions of the pores (~100 nm wide and 100 nm deep) allowed the translocation of large protein structures formed in the presence of HAP-TAMRA. For the resistive-pulse measurements, we opted to measure the particles under high ionic strength buffer conditions (e.g., 50 mM HEPES with 1000 mM NaCl) to return the highest signal-to-noise ratio and, more importantly, to compare the particle size distributions quantitatively. Thus, the assembly reaction mixtures were purified by size exclusion chromatography to remove unassembled dimer and excess drug and to stop the assembly reaction prior to adjusting the salt concentration for resistive-pulse sensing.
Figure 3. Resistive-pulse device and pulse-amplitude measurements.
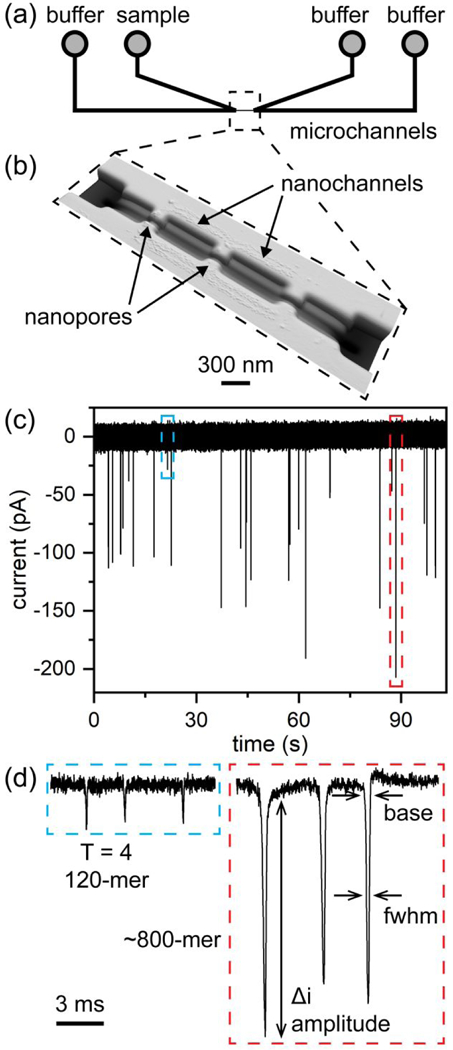
(a) Two V-shaped microchannels are connected through a series of nanochannels and nanopores, indicated with the dashed box. (b) Atomic force microscope (AFM) image of the nanochannels and nanopores. The nanopores are 100 nm wide, 100 nm deep, and 280 nm long. (c) Typical current trace of an assembly reaction of 5 μM Cp dimer with 20 μM HAP-TAMRA and 300 mM NaCl. The current baseline (~17 nA) was subtracted from the signal. Each of what appears as a single pulse on the timescale of the plot in panel (c) consists of (d) three discrete pulses. In panels (c) and (d), the blue dashed line indicates detection of an individual T = 4 capsid, and the red dashed line indicates detection of one of the largest protein particles formed. The arrows in panel (d) indicate the pulse width at a fixed position (3σ or 4σ) below the baseline (base) and pulse width at half-height (i.e., full width at half-maximum, fwhm).
In the resistive-pulse experiments, the negatively charged HBV particles were electrokinetically driven through the pores with an applied potential. A typical current trace in Figure 3c shows pulses with a large range of amplitudes, demonstrating the non-uniform products generated from the assembly reaction with HAP-TAMRA. On the same devices, the much smaller, fully formed T = 3 and T = 4 HBV capsids were well resolved (Figure S1a in the Supporting Information). To correlate the pulse amplitude with the number of dimers in a particle and the pulse width with the particle length (Figure 3d), solutions of T = 3 and T = 4 capsids were used as calibration standards. Because HBV capsids are porous, they displace electrolyte equal to volume of the protein (and not the volume of the capsid shells);23 consequently, the pulse amplitudes can be correlated with the number of dimers in the particle. Well-characterized, larger non-spherical structures composed of Cp dimer are not available as calibration standards.
Histograms of pulse amplitudes from assembly products with HAP-TAMRA and 1000, 300, and 80 mM NaCl are shown in Figure 4a. For higher ionic strength assembly reactions, e.g., 1000 and 300 mM NaCl, the histograms contain broad Gaussian distributions that correspond to particles with the size of T = 4 capsids and larger particles. These distributions are followed by exponential tails suggesting Poisson distributions of products. However, the histogram of assembly products with HAP-TAMRA and 80 mM NaCl is clearly bimodal consisting of two Gaussian distributions centered at ~120–140 dimers and ~600 dimers. Notably, as the ionic strength decreased and HAP-TAMRA influenced particle formation more substantially, the fractions of T = 4 capsids or similarly sized particles in the histograms in Figure 4a decrease and are 55%, 10%, and 5% of the particle populations for 1000, 300, and 80 mM, respectively. To further complicate interpretation of assembly with 20 μM HAP-TAMRA and 1000 mM NaCl, no significant amounts of T = 3 capsids were detected, whereas 20%–30% T = 3 capsids were observed under similar reaction conditions without HAP-TAMRA (Figure S1b). One possible explanation of this observation is that formation of T = 3 capsids requires early intermediates disfavored by HAP-TAMRA, which binds preferentially to quasi-sixfold vertices.25,29
Figure 4. Bimodal distribution of assembly products from competition of two alternative assembly pathways.
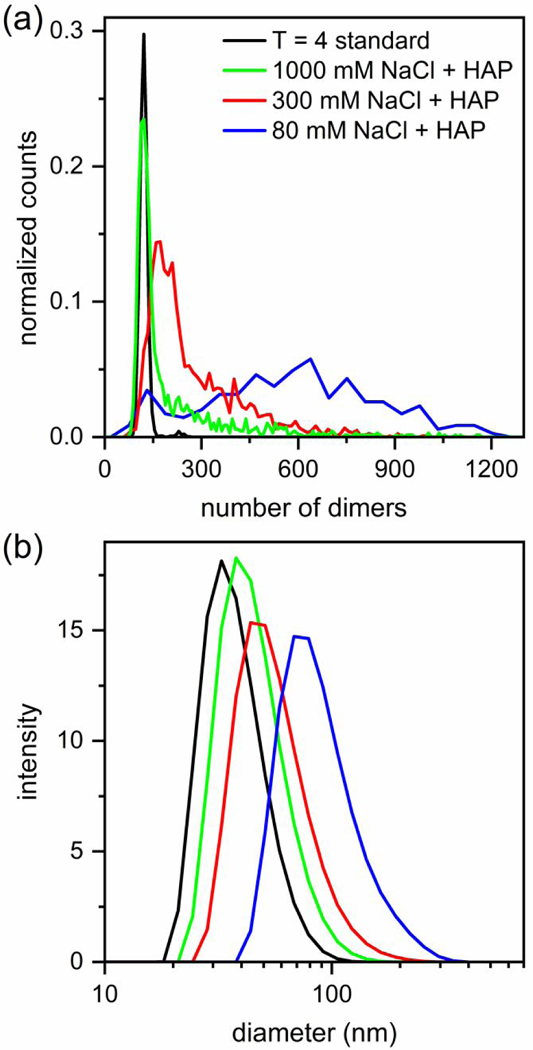
Histograms of assembly reactions of 5 μM Cp dimer with 20 μM HAP-TAMRA and 1000, 300, and 80 mM NaCl in 50 mM HEPES (pH 7.5). Assembly at low ionic strength, 80 mM NaCl, yields a bimodal distribution with centers at ~120–140 and ~600 dimers. The number of measured particles is ~200 for assembly at 80 mM NaCl and > 900 for 300 and 1000 mM NaCl. The histograms were normalized to the same area, and then, scaled for clarity. (b) Dynamic light scattering (DLS) data with mean diameters of 35 nm for T = 4 capsids, and 40, 50, and 80 nm for assembly reactions in 20 μM HAP-TAMRA with 1000, 300, and 80 mM NaCl, respectively. DLS data provide a view that contains less information than the resistive-pulse sensing, but the conclusions remain broadly consistent.
A second, less well-resolved view of assembly was determined by dynamic light scattering (DLS), which yielded a rough estimate of the particle diameter assuming a homogenous population of spherical particles (Figure 4b). DLS returned a diameter of 35 nm for T = 4 capsids, which is consistent with values derived from a cryo-EM structural model.28 The mean diameter increased to 40, 50, and 80 nm for assembly with HAP-TAMRA and 1000, 300, and 80 mM NaCl, respectively. The DLS results are consistent with those of resistive-pulse sensing, albeit at much lower resolution. As simple examples, both SEC and DLS are not able to resolve the T = 3 and T = 4 capsid distributions assembled without HAP-TAMRA present or the bimodal distribution of products from assembly with HAP-TAMRA in 80 mM NaCl. Resistive-pulse sensing, however, easily resolves these distributions at the extremes of particle sizes generated in the assembly reactions reported here.
To examine the relative stability and evolution of these large protein particles formed in the presence of HAP-TAMRA, we measured the particle sizes of a sample assembled with 300 mM NaCl over a period of months (Figure 5). We reasoned that large particles would have “frayed” edges, allowing re-equilibration of subunits. Again, to exclude the possible confounding effects of unassembled dimer, samples were purified by size exclusion chromatography to remove free dimer. The histograms in Figure 5 illustrate that over the period of five months, the distribution of particles shifts to larger sizes, which provides evidence that the distribution is governed by kinetics rather than just equilibrium thermodynamics. This change in the size distribution of the particles may be due to the slow dissolution of kinetic traps formed during assembly, rearrangement to form stable complexes, or a snap shot of Ostwald ripening to form larger particles. The presence of geometric imperfections and free polymer ends likely facilitate rearrangements, which can be seen in the aberrant structures but not in well-formed icosahedra. Purified T = 3 and T = 4 capsids remain stable over similar timescales of months as shown by mass spectrometry30 and confirmed by measurements of purified capsid standards with resistive-pulse sensing (Figure S1a). When fully formed T = 3 and T = 4 capsids are exposed to HAP-TAMRA, the capsids become distorted on a much shorter timescale of 24 h, but remain intact.28
Figure 5. Evolution of assembly reaction products over time.
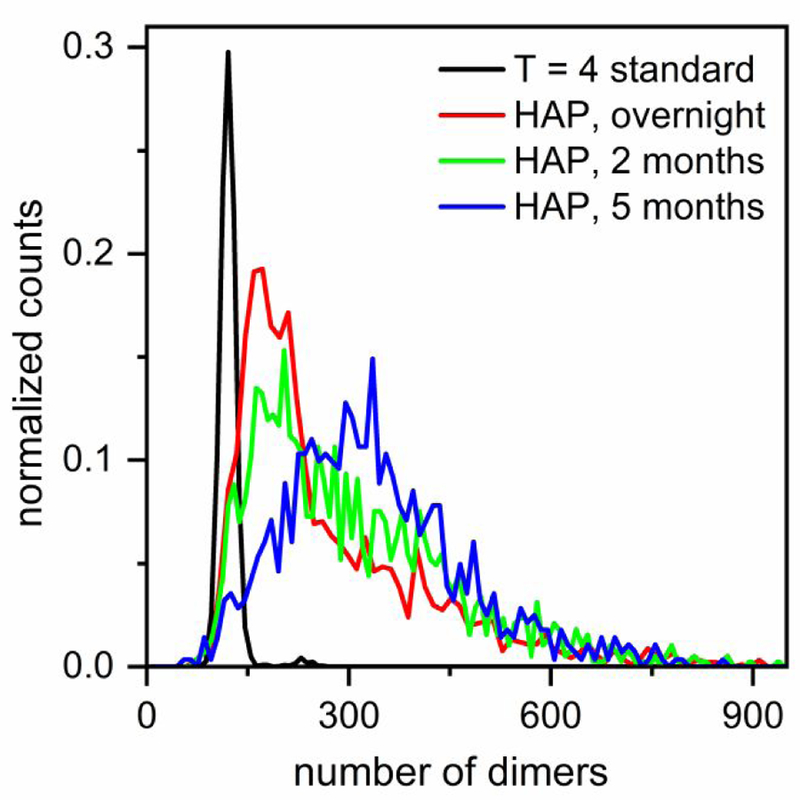
Histograms show the relative stability and evolution of particles assembled from 5 μM dimer with 20 μM HAP-TAMRA and 300 mM NaCl in 50 mM HEPES on the timescale of months and compared to a T = 4 capsid standard. The reaction solution was purified with size exclusion chromatography (SEC) to remove unassembled dimer and excess drug from the solution. Over time, the particles mature to larger sizes, and the distribution becomes more symmetrical. The number of measured particles is >900 for all histograms. For clarity, the y-values for the histograms in the presence of HAP-TAMRA have been multiplied by 4.
Particle Length Estimated from Pulse Width.
Measurements of pulse amplitude contain information about the protein volume which is directly proportional to the number of dimers in the particle.23,26 In addition, the pulse width can be used to estimate the particle length. For particles that are small and can rotate freely as they enter the pore, the pulse width correlates with their average length projected onto the axis of translocation. Particles with one of their dimensions (> 80 nm) approaching the dimensions of the pore (~100 nm) have to orient length-wise to pass through the pores. For these particles, the pulse width correlates with their longer dimension. Although previous studies have reported longer pulse widths for resistive-pulse measurements of larger particles,31 pulse widths have not been used for measuring particle length.
In our experiments, the pores are longer than the length of even the largest particles formed during reactions with HAP-TAMRA. If all particles have the same electrophoretic mobility, the pulse width measured at half-height of the pulse (full width at half-maximum) tracks the movement of the particle center and will not change as function of the particle size. On the contrary, the pulse width measured at the baseline (or equivalently, at a fixed-position below the baseline) contains information about the particle length and increases with particle size. To confirm that particles have equal electrophoretic mobilities, the total time that the particles spent in the three nanopores and two pore-to-pore nanochannels (i.e., the time measured from the start of the first pulse until the end of last pulse), was extracted and, as seen in Figure 6a, does not change with particle size, indicating that all the particles have a similar electrophoretic mobility. At 1000 mM NaCl, the Debye length is ~0.3 nm, which is negligible compared to the >>30 nm particle sizes.
Figure 6. Morphological information extracted from nanochannel data.
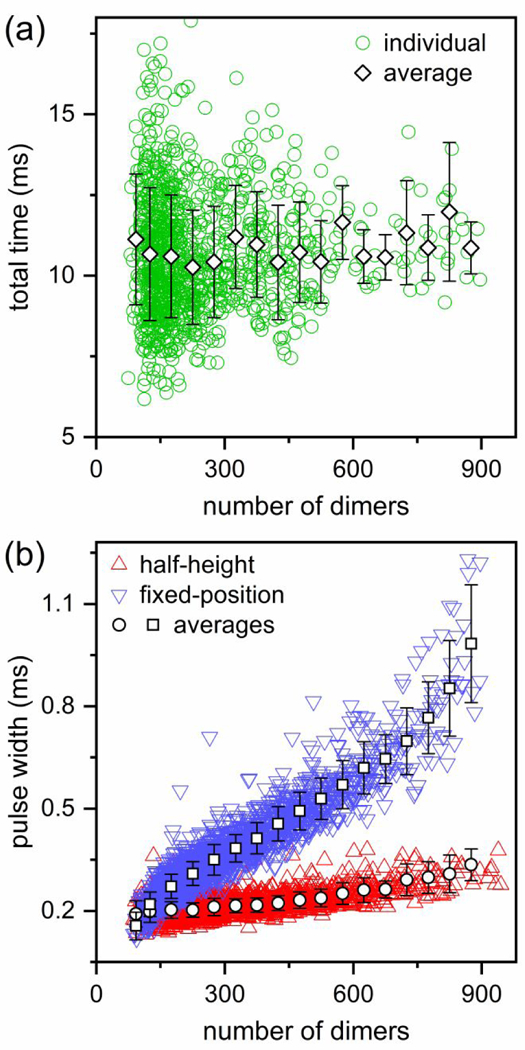
(a) Variation of the total transit time of each particle through the three pores and two pore-to-pore nanochannels with number of dimers. The total time does not change with particle size which indicates that the particles of different sizes have similar electrophoretic mobilities. (b) Variation of average pulse width with number of dimers. The pulse width measured at half-height increases only slightly, whereas the width measured at a fixed position (3σ or 4σ below the baseline current) increases significantly with increased number of dimers. Particles were assembled from 5 μM Cp dimer with 20 μM HAP-TAMRA and 300 mM NaCl in 50 mM HEPES (pH 7.5).
Figure 6b shows how the pulse widths at half-height and at a fixed-position below the baseline current (e.g., 3σ or 4σ below the baseline where σ is the standard deviation of the baseline current) varies with the number of dimers in the particles. We chose to measure the pulse widths at a fixed-position below the baseline instead of at the baseline itself to avoid the uncertainty from low frequency noise at the start and end of each pulse. As expected, the pulse widths measured at half-height increased only slightly with the number of dimers. This slight increase is due to the finite data acquisition frequency (e.g., 40 kHz). Generally, for larger particles, the pulse width measured at half-height describes the motion of the center of mass of these particles. Conversely, the pulse widths measured at the fixed position just below the baseline increased more significantly with the number of dimers in a particle because the pulse width captures the leading and trailing edges of the particle entering and exiting the pore, respectively.
The increase in the duration of the pulse width at a fixed position below the baseline is due to the range of velocities experienced by the particles as they enter, transit, and exit the pore. Because the electric field along the pore axis varied, the particle velocity had a low-velocity component at the pore entrance and exit and a high-velocity component in the pore center, which, in turn, led to pulse widths with sufficient duration to be easily measured. We used the average pulse width of T = 3 and T = 4 capsids at the same fixed position below the baseline current to calibrate our measurements. Then, both the length and number of dimers were calculated from measurements of particles assembled with HAP-TAMRA and 300 mM NaCl (Figure 7). Indeed, the average particle length from four different sets of measurements was 53 ± 2 nm, which is in good agreement with the mean diameter from DLS measurements of assembly (Figure 4b).
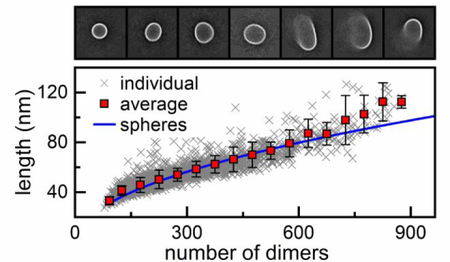
Figure 7. Model of spherical growth consistent with particle morphology.
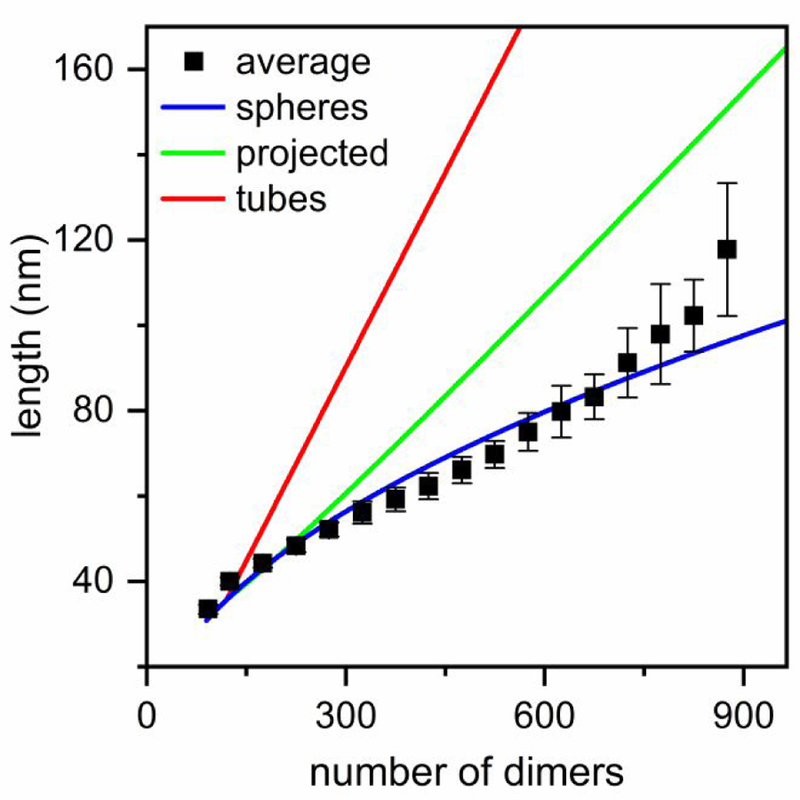
Average particle lengths fitted with models of spherical growth and linear growth of tubes. The pulse width measurements in Figure 5b were converted to particle length with T = 3 and T = 4 capsids as calibration standards. The spherical growth model (blue) yielded the best fit and assumes that particles grow by forming closed spheres, but as particle diameters approach the nanopore dimensions, the model begins to fail. The linear growth of tubes model (red) shows the longer dimension of tube length, whereas the green shows the mean projected length on the translocation axis if the tubes rotate freely as they travel through the pores. Particles were assembled from 5 μM Cp dimer with 20 μM HAP-TAMRA and 300 mM NaCl in 50 mM HEPES (pH 7.5).
To better understand how the particles are formed, we compared two simple, limiting models of particle growth and attempted to fit the particle lengths derived from the resistive-pulse measurements. The first model was spherical growth where particles form closed spheres with a diameter that increases with the addition of dimeric subunits. The second model was linear polymerization where particles form tubes with two hemispherical caps with a diameter equal to that of a T = 4 capsid (35.7 nm). We assumed that the hemispheres formed first and that the length of the cylindrical tube increased with the addition of dimeric subunits. For both models, the diameter of a T = 4 capsid was their only input. For a spherical particle, the length measured by resistive-pulse sensing is simply the diameter (Figure 7-blue). However, the problem becomes more complicated for tubes. The red line in Figure 7 shows how the longer dimension of the tubes changes with the number of dimers in the tube. If a tube is small enough (longer dimension < 80 nm) to rotate during translocation through the pores, then resistive-pulse sensing measures the average projection of the freely rotating particle onto the axis of translocation (Figure 7-green). One important conclusion from Figure 7 is that rotating tubes and ellipsoids with small aspect ratios of the axis lengths (e.g., < 2) yield pulse widths similar to that of spheres. However, tubes and ellipsoids with high aspect ratios (> 2) yield pulse widths different from those of spheres even if they rotate freely as they pass through the pores.
Consistent with our EM observations, Figure 7 clearly shows the model of spherical growth fits better to the experimental data, especially for smaller particles, but becomes progressively worse as the size of the particles approaches the size of the pores (> 80 nm). Of course, the EM data (Figure 2) show that a simple growth model that predicts the formation of closed spheres or ellipsoids with small aspect ratios is insufficient, because of the many open and heterogeneous structures observed. The predictions of the simple spherical model emphasize a smooth, gradual change of the particle shapes as a function of the particle size, which strongly supports the presence of competing mechanisms where the drug favors formation of kinetically trapped particles. If particles were formed exclusively by one of the two proposed pathways (normative or HAP-regulated assembly), a bimodal distribution consisting of two Gaussians for T = 4 capsids and drug-induced assembly products (Figure 4a-blue) would be observed at all protein association energies. Instead, we observe that aberrant particle formation is bounded on the low end at T = 4 capsids and suggest that the larger particles are formed by disrupting T = 4 geometry. The deviation from spherical growth becomes progressively worse at larger particle sizes which may indicate that formation of flat surfaces (i.e., hexagonal repeats) has a cooperative effect; particles that form flat regions early on are more likely to continue to grow abnormally with extended flat regions than to follow a model like that of spherical growth.
Progress towards a CpAM Mechanism of Action.
Capsid assembly depends on a subunit adopting an assembly active state and then associating with a growing complex. Capsid completion depends on adding subunits faster than the bound subunits dissociate; weak association energy results in a reaction that resembles a one-dimensional drunkard’s walk, whereas strong association results in a unidirectional reaction limited by the on-rate.32 At very low association energy, a subunit with poor interaction geometry is likely to dissociate long before a new subunit adds to the growing capsid.
In low ionic strength, HBV Cp is primarily free dimer with a small amount of transient and unstable oligomer. HAP CpAMs bind poorly, if at all to dimer.33 For this reason, we suspect that HAP-TAMRA will bind to a dimer interface in a small oligomer and will have the immediate effect of stabilizing the pairwise interaction and favoring a geometry consistent with a sixfold or quasi-sixfold vertex. The complex will be stabilized for further association reactions including multivalent interactions that further stabilize the growing complex. Because CpAMs are required to stabilize the earliest complexes, the most probable outcome is that these reactions will favor the CpAM-preferred geometry.
In contrast, at high association energies, even early intermediates are relatively stable. Based on the observation that most capsid assembled in high salt have virus-like geometry, we suggest two possible mechanisms. The on-rate for new dimer may be faster than the rate of binding HAP to a dimer-dimer interface, leading to preferential growth of icosahedral capsid instead of hexagonal sheet. The second possibility, which does not exclude the first, is that strong association energy favors capsid-like geometry, even in the presence of a HAP. In both cases, the most probable assembly product will resemble a normal capsid.
These two extremes incorporate both the predictions that the initial complex(es) formed will influence the final product and that there will be a distribution of results. The bimodal distribution of products seen at low association energies (Figure 4a-blue) provides strong evidence of two alternate modes of assembly proceeding relatively independently of one another. At intermediate association energy (300 mM NaCl), we would expect maximal competition between the available modes of assembly and, consequently, a maximally diverse mixture of icosahedral capsids and aberrant structures. These predictions are consistent with the diverse assembly product distribution (Figures 1c, 2, and 4a-red) for the reaction with intermediate association energy.
Conclusion
We present evidence that the mechanism of action of HAP-TAMRA is to facilitate a dynamic competition between normative and drug-directed assembly. This CpAM was found to be especially disruptive for weak protein-protein association energies by likely stabilizing intermediate oligomers with hexamer structures, incapable of forming capsids with icosahedral geometry. These abnormal intermediates ultimately led to large, non-uniform particles, which are essentially a dead end from the perspective of the virus. Due to the morphology of these particles, we suspect they were derived from a T = 4 geometry disrupted by flat regions and regions of incomplete closure. In contrast to what was observed at low ionic strengths, HAP-TAMRA did not significantly affect the assembly process at very high protein association energies. At high ionic strengths, most intermediate oligomers completed the regular icosahedral geometry, normally seen in the absence of CpAMs. Invoking the same mechanism proposed for low ionic strength assembly, higher protein association energies allow the intermediate structures to force the normal local geometry or because the dimer assembles faster than the time needed for CpAM molecules to be inserted into the edges of the growing capsids. In a normal infection, both empty and nucleic acid filled particles are present. For the latter, assembly is nucleated by a pgRNA-reverse transcriptase complex to yield immature RNA-filled cores, which will mature to infectious virions. The presence of RNA may emulate the results obtained here at higher ionic strengths.34
Experimental Methods
Dimer and Virus Capsids.
Wild type core protein dimers (Cp149 dimer, 34 kDa) were expressed in E. coli and purified.35 T = 4 capsids (abundance > 90%) were obtained by overnight assembly of Cp149 in 300 mM NaCl and 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.5). To purify the reaction solution, the unassembled dimer was removed by size exclusion chromatography through a Superose6 column. After assembly, T = 3 capsids were purified on a 10−40% (w/v) continuous sucrose gradient in 50 mM HEPES with 300 mM NaCl that was centrifuged for 6 h at 150,000 g.36 HAP-TAMRA was synthesized as described previously.28 To study the assembly process, 5 μM Cp149 dimer was assembled with 20 μM HAP-TAMRA and 1000, 300, or 80 mM NaCl in 50 mM HEPES. After overnight assembly at room temperature and prior to single-particle measurements, the reaction solution was purified on a Superose6 size exclusion column equilibrated in 50 mM HEPES with 300 mM NaCl to remove the unassembled dimer and excess drug.
Transmission Electron Microscopy (TEM).
To collect the electron micrographs, the assembly reaction solutions were mixed with ammonium molybdate stain (6% final (w/v)) and trehalose (0.5% final (w/v)). Trehalose preserves the supramolecular structure of the particles. The mixture was immediately applied to glow discharged CF300-CU-TH carbon-mesh copper grids (Electron Microscopy Sciences) and air dried. Images were collected with a JEOL TEM 1400plus microscope operated at 120 kV and 50,000X magnification. The 2D class averages presented in Figure 2 were processed with the RELION software package.37
Dynamic Light Scattering (DLS).
DLS measurements of the reaction solutions were acquired with a Malvern Zetasizer Nano instrument, operated at 22 °C. The intensity measurements were normalized by volume to reveal the distributions of particle diameters.
Fabrication of Nanofluidic Devices.
D263 glass substrates were coated with a 120 nm thick layer of chromium and a 3 μm thick layer of S1813 photoresist. Then microchannels were fabricated by UV lithography and wet chemical etching.26 To fabricate the nanochannels and nanopores, a focused ion beam (FIB) instrument (Auriga 60, Carl Zeiss, GmbH) controlled by the Nano-Pattering and Visualization Engine (NPVE; FIBICS, Inc.) was used. The nanochannels were milled with a 30 kV beam at 50 pA. The side nanochannels were milled with a dose of 1.1 nC/μm2, whereas the bridge and pore-to-pore nanochannels were milled with a dose of 0.7 nC/μm2. To create the nanopores that were 100 nm wide by 100 nm deep, a rectangle was milled with a 30 kV beam at 20 pA and a dose of 0.24 nC/μm2, whereas to create the nanopores that were 120 nm wide by 120 nm deep (control experiments in Supporting Information), the dose was 0.28 nC/μm2. During the FIB milling, an electron flood gun (FG 15/40, SPECS, GmbH), operated at 5 eV and 20 μA, compensated for the build-up of positive charge on the substrate surface.
Dimensions of the nanochannels and nanopores were measured with an atomic force microscope (AFM; MFP-3D, Asylum Research, Inc.) and the scanning electron microscope (SEM) on the FIB instrument. The nanopores were 100 ± 5 nm wide, 100 ± 5 nm deep, and 280 ± 10 nm long. The bridge and pore-to-pore nanochannels were 320 ± 10 nm wide and 180 ± 5 nm deep. The length of the bridge nanochannels was 580 ± 20 nm, whereas the length of the pore-to-pore nanochannels was 810 ± 10 nm. The side nanochannels were 290 ± 10 nm deep and 510 ± 10 nm wide. The V-shaped microchannels were 11 ± 1 μm deep and 35 ± 1 μm wide. To bond the devices, the substrates and No. 1.5 cover slips were cleaned in 1 M NaOH for 12 min, sonicated in ultrapure water for 10 min, and brought into contact with each other while still wet. The devices were dried overnight at 90 °C and annealed at 545 °C for 12 h. To hold samples and buffers and make electrical contact to them, glass reservoirs were epoxied over the access holes.
Resistive-Pulse Measurements.
To rinse the devices, the two reservoirs in the middle were filled with solution, and vacuum was applied to the reservoirs on the ends. An Axopatch 200B (Molecular Devices, Inc.) was used to apply the potential between the two reservoirs in the middle through Ag/AgCl electrodes and to record the current. Measurements on all the devices were taken with a baseline current of ~17 nA. Data were collected for ~10 min for T = 3 and T = 4 capsid standards and for 0.5 – 1.5 h for assembly in the presence of HAP-TAMRA. The applied potential was in the range of 480 – 580 mV. All data were collected with a sampling frequency of 40 kHz and a filter frequency of 10 kHz.
Data Analysis.
To determine the pulse amplitudes (Δi) and the pore-to-pore times, we imported the raw data into MatLab R2017a (Mathworks, Inc.) and used a modified version of Open Nanopore 1.2.38 To determine the pulse width at fixed position below the baseline and pulse width at half-height, a MatLab program was developed.27 The pore-to-pore times were the criterion to distinguish between pulses correlated in time and uncorrelated pulses from measurements of different particles. Pulse sequences that did not consist of 3 pulses, e.g., a series of current pulses produced by the simultaneous presence of two capsids inside the 3-pore region, were neither analyzed further nor included in the pulse amplitude and width histograms. To calibrate each device and convert the pulse amplitude to number of dimers, a 1:1 mixture of T = 3 and T = 4 capsid standards was measured (Figure S1a in Supporting Information). To eliminate the drift noise, the average pulse amplitude (Δi) of the sequence of three pulses was divided by the baseline current (i). On these devices, the ratio of the average current displacements for T = 3 to T = 4 capsids was 0.73 ± 0.01, which is in good agreement with the ratio of their masses, which is 0.75.
Control Experiments.
Control experiments are described in detail in the Supporting Information. Specifically, SEC does not significantly alter the size distribution of the particles (Figure S2a), resistive-pulse measurements do not affect the distribution of sizes and are not biased against larger particles (Figure S2b), and dilution into high salt of the purified reaction products does not alter the size distributions on the timescale of one day (Figure S3).
Modeling of Spheres and Tubes with Hemispherical Caps on the Sides.
To estimate how the diameter of virus particles changes with the number of dimers, when dimers assemble and form closed spheres of different sizes, a model was developed with the number of subunits and diameter of T = 4 capsids (120 dimers, 35.7 nm in diameter as measured from a cryo-EM structural model) as its only inputs. Equation 1 estimates the surface area of spherical virus particles, Ap(sp), with the assumption that each subunit contributes equally to the particle surface area.
| (1) |
where dp is the diameter of each particle, nd(sp) is the number of dimers in the particle, and Ad is the surface area that corresponds to a single dimer.
The surface area of each dimer was calculated to be ~33.4 nm2 from Equation 1. Then, the diameter, lp(sp), of the spherical virus particles as function of the number of dimers was estimated by Equation 2 as
| (2) |
To model the growth of virus particles consisting of two hemispherical caps on the sides and a cylindrical portion between the caps, we assumed that the two hemispheres have the radius of a T = 4 capsid and consist of 60 dimers each. The length of the cylindrical portion varies with addition of dimers. The surface area of these particles can be estimated by Equation 3.
| (3) |
where Ap(tu) is the surface area of the tube-shaped particles, Ap(hem) is the surface area of each hemispherical cap, Ap(cyl) is the surface area of the cylindrical bacilliform, r is the radius of the hemispheres, and lp(tu) is the total length of the tube.
The number of dimers, nd(tu), is given by Equation 4. Then, Equation 5, which calculates the length of the tubes as a function of the number of subunits, is directly derived from Equation 4.
| (4) |
or equivalently
| (5) |
To estimate the average length of the tubes with hemispherical caps measured by resistive-pulse sensing when small particles can pass through the pores with a random orientation, the shape of these particles was assumed to approximate ellipsoids. Then, the mean projection of these ellipsoidal particles on the axis of their translocation was estimated numerically by Equation 6.
| (6) |
where s is the length of projection and θ is the angle between lp(tu) and the axis of the translocation through the pores. Note that calculation of two times the radius of gyration gives results similar to the projected length. A symbolic solution of Equation 6 was also in close agreement with the numerical solution in Figure 7. For the symbolic solution, Equation 7 was solved on Mathematica 11.2 (Wolfram Research).
| (7) |
where Ε is the complete elliptic integral of the second kind.
Supplementary Material
Acknowledgments.
This work was supported in part by NIH R01 GM129354, NSF CHE-0923064, and the Indiana Clinical and Translational Science Institute (CTSI). The authors thank the Indiana University Nanoscale Characterization Facility and Electron Microscopy Center for use of their instruments.
Footnotes
Conflict of Interest. The authors declare the following competing financial interest: A.Z. reports a financial interest in a company based on core protein allosteric modulators (CpAM).
Supporting Information Available. Control experiments for the effect of size exclusion chromatography (SEC) and resistive-pulse measurements on the particle size distributions. Figures of resistive-pulse measurements of calibration standards and capsid assembly without HAP-TAMRA, impact of size exclusion chromatography and resistive-pulse measurements on particle size distributions, and size distribution of purified products after dilution into high ionic strength buffer. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Gish RG; Given BD; Lai CL; Locarnini SA; Lau JYN; Lewis DL; Schluep T Chronic Hepatitis B: Virology, Natural History, Current Management and a Glimpse at Future Opportunities. Antiviral Res 2015, 121, 47–58. [DOI] [PubMed] [Google Scholar]
- (2).Zlotnick A; Venkatakrishnan B; Tan ZN; Lewellyn E; Turner W; Francis S Core Protein: A Pleiotropic Keystone in the HBV Lifecycle. Antiviral Res 2015, 121, 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pei YM; Wang CT; Yan SF; Liu G Past, Current, and Future Developments of Therapeutic Agents for Treatment of Chronic Hepatitis B Virus Infection. J. Med. Chem 2017, 60, 6461–6479. [DOI] [PubMed] [Google Scholar]
- (4).Crowther RA; Kiselev NA; Bottcher B; Berriman JA; Borisova GP; Ose V; Pumpens P 3-Dimensional Structure of Hepatitis-B Virus Core Particles Determined by Electron Cryomicroscopy. Cell 1994, 77, 943–950. [DOI] [PubMed] [Google Scholar]
- (5).Birnbaum F; Nassal M Hepatitis-B Virus Nucleocapsid Assembly - Primary Structure Requirements in the Core Protein. J. Virol 1990, 64, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Caspar DLD; Klug A Physical Principles in Construction of Regular Viruses. Cold Spring Harbor Symp. Quant. Biol 1962, 27, 1–24. [DOI] [PubMed] [Google Scholar]
- (7).Zlotnick A; Cheng N; Conway JF; Booy FP; Steven AC; Stahl SJ; Wingfield PT Dimorphism of Hepatitis B Virus Capsids Is Strongly Influenced by the C-Terminus of the Capsid Protein. Biochemistry 1996, 35, 7412–7421. [DOI] [PubMed] [Google Scholar]
- (8).Ceres P; Zlotnick A Weak Protein-Protein Interactions Are Sufficient to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry 2002, 41, 11525–11531. [DOI] [PubMed] [Google Scholar]
- (9).Zlotnick A; Johnson JM; Wingfield PW; Stahl SJ; Endres D A Theoretical Model Successfully Identifies Features of Hepatitis B Virus Capsid Assembly. Biochemistry 1999, 38, 14644–14652. [DOI] [PubMed] [Google Scholar]
- (10).Dryden KA; Wieland SF; Whitten-Bauer C; Gerin JL; Chisari FV; Yeager M Native Hepatitis B Virions and Capsids Visualized by Electron Cryomicroscopy. Mol. Cell 2006, 22, 843–850. [DOI] [PubMed] [Google Scholar]
- (11).Bourne C; Lee S; Venkataiah B; Lee A; Korba B; Finn MG; Zlotnick A Small-Molecule Effectors of Hepatitis B Virus Capsid Assembly Give Insight into Virus Life Cycle. J. Virol 2008, 82, 10262–10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Li LC; Chirapu SR; Finn MG; Zlotnick A Phase Diagrams Map the Properties of Antiviral Agents Directed against Hepatitis B Virus Core Assembly. Antimicrob. Agents Chemother 2013, 57, 1505–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Deres K; Schroder CH; Paessens A; Goldmann S; Hacker HJ; Weber O; Kramer T; Niewohner U; Pleiss U; Stoltefuss J; Graef E; Koletzki D; Masantschek RNA; Reimann A; Jaeger R; Gross R; Beckermann B; Schlemmer KH; Haebich D; Rubsamen-Waigmann H Inhibition of Hepatitis B Virus Replication by Drug-Induced Depletion of Nucleocapsids. Science 2003, 299, 893–896. [DOI] [PubMed] [Google Scholar]
- (14).Stray SJ; Bourne CR; Punna S; Lewis WG; Finn MG; Zlotnick A A Heteroaryldihydropyrimidine Activates and Can Misdirect Hepatitis B Virus Capsid Assembly. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Billioud G; Pichoud C; Puerstinger G; Neyts J; Zoulim F The Main Hepatitis B Virus (HBV) Mutants Resistant to Nucleoside Analogs Are Susceptible in vitro to Non-Nucleoside Inhibitors of HBV Replication. Antiviral Res 2011, 92, 271–276. [DOI] [PubMed] [Google Scholar]
- (16).Feng S; Gao L; Han XC; Hu TS; Hu YM; Liu HX; Thomas AW; Yan ZP; Yang S; Young JAT; Yun HY; Zhu W; Shen HC Discovery of Small Molecule Therapeutics for Treatment of Chronic HBV Infection. ACS Infect. Dis 2018, 4, 257–277. [DOI] [PubMed] [Google Scholar]
- (17).Liu C; Fan GZ; Wang Z; Chen HS; Yin CC Allosteric Conformational Changes of Human HBV Core Protein Transform Its Assembly. Sci. Rep 2017, 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).DeBlois RW; Bean CP Counting and Sizing of Submicron Particles by Resistive Pulse Technique. Rev. Sci. Instrum 1970, 41, 909–916. [Google Scholar]
- (19).Howorka S; Siwy Z Nanopore Analytics: Sensing of Single Molecules. Chem. Soc. Rev 2009, 38, 2360–2384. [DOI] [PubMed] [Google Scholar]
- (20).Luo L; German SR; Lan WJ; Holden DA; Mega TL; White HS Resistive-Pulse Analysis of Nanoparticles. In Annu. Rev. Anal. Chem, Cooks RG; Pemberton JE, Eds. Annual Reviews: Palo Alto, 2014; Vol. 7, pp 513–535. [DOI] [PubMed] [Google Scholar]
- (21).Harms ZD; Haywood DG; Kneller AR; Jacobson SC Conductivity-Based Detection Techniques in Nanofluidic Devices. Analyst 2015, 140, 4779–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Haywood DG; Saha-Shah A; Baker LA; Jacobson SC Fundamental Studies of Nanofluidics: Nanopores, Nanochannels, and Nanopipets. Anal. Chem 2015, 87, 172–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Harms ZD; Haywood DG; Kneller AR; Selzer L; Zlotnick A; Jacobson SC Single-Particle Electrophoresis in Nanochannels. Anal. Chem 2015, 87, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhou K; Li L; Tan Z; Zlotnick A; Jacobson SC Characterization of Hepatitis B Virus Capsids by Resistive-Pulse Sensing. J. Am. Chem. Soc 2011, 133, 1618–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Harms ZD; Selzer L; Zlotnick A; Jacobson SC Monitoring Assembly of Virus Capsids with Nanofluidic Devices. ACS Nano 2015, 9, 9087–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kondylis P; Zhou JS; Harms ZD; Kneller AR; Lee LS; Zlotnick A; Jacobson SC Nanofluidic Devices with 8 Pores in Series for Real-Time, Resistive-Pulse Analysis of Hepatitis B Virus Capsid Assembly. Anal. Chem 2017, 89, 4855–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zhou J; Kondylis P; Haywood DG; Harms ZD; Lee LS; Zlotnick A; Jacobson SC Characterization of Virus Capsids and Their Assembly Intermediates by Multicycle Resistive-Pulse Sensing with Four Pores in Series. Anal. Chem 2018, 90, 7267–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Schlicksup CJ; Wang JCY; Francis S; Venkatakrishnan B; Turner WW; VanNieuwenhze M; Zlotnick A Hepatitis B Virus Core Protein Allosteric Modulators Can Distort and Disrupt Intact Capsids. eLife 2018, 7, e31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Endres D; Miyahara M; Moisant P; Zlotnick A A Reaction Landscape Identifies the Intermediates Critical for Self-Assembly of Virus Capsids and Other Polyhedral Structures. Protein Sci 2005, 14, 1518–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Uetrecht C; Watts NR; Stahl SJ; Wingfield PT; Steven AC; Heck AJR Subunit Exchange Rates in Hepatitis B Virus Capsids Are Geometry- and Temperature-Dependent. Phys. Chem. Chem. Phys 2010, 12, 13368–13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Platt M; Willmott GR; Lee GU Resistive Pulse Sensing of Analyte-Induced Multicomponent Rod Aggregation Using Tunable Pores. Small 2012, 8, 2436–2444. [DOI] [PubMed] [Google Scholar]
- (32).Elrad OM; Hagan MF Mechanisms of Size Control and Polymorphism in Viral Capsid Assembly. Nano Lett 2008, 8, 3850–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hacker HJ; Deres K; Mildenberger M; Schroder CH Antivirals Interacting with Hepatitis B Virus Core Protein and Core Mutations May Misdirect Capsid Assembly in a Similar Fashion. Biochem. Pharmacol 2003, 66, 2273–2279. [DOI] [PubMed] [Google Scholar]
- (34).Perlmutter JD; Perkett MR; Hagan MF Pathways for Virus Assembly around Nucleic Acids. J. Mol. Biol 2014, 426, 3148–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zlotnick A; Ceres P; Singh S; Johnson JM A Small Molecule Inhibits and Misdirects Assembly of Hepatitis B Virus Capsids. J. Virola 2002, 76, 4848–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Zlotnick A; Palmer I; Kaufman JD; Stahl SJ; Steven AC; Wingfield PT Separation and Crystallization of T = 3 and T = 4 Icosahedral Complexes of the Hepatitis B Virus Core Protein. Acta Crystallogr. Sect. D-Biol. Crystallogr 1999, D55, 717–720. [DOI] [PubMed] [Google Scholar]
- (37).Scheres SHW Relion: Implementation of a Bayesian Approach to Cryo-Em Structure Determination. J. Struct. Biol 2012, 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Raillon C; Granjon P; Graf M; Steinbock LJ; Radenovic A Fast and Automatic Processing of Multi-Level Events in Nanopore Translocation Experiments. Nanoscale 2012, 4, 4916–4924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


