Abstract
The gene arrangements of Drosophila have played a prominent role in the history of evolutionary biology from the original quantification of genetic diversity to current studies of the mechanisms for the origin and establishment of new inversion mutations within populations and their subsequent fixation between species supporting reproductive barriers. This review examines the genetic causes and consequences of inversions as recombination suppressors and the role that recombination suppression plays in establishing inversions in populations as they are involved in adaptation within heterogeneous environments. This often results in the formation of clines of gene arrangement frequencies among populations. Recombination suppression leads to the differentiation of the gene arrangements which may accelerate the accumulation of fixed genetic differences among populations. If these fixed mutations cause incompatibilities, then inversions pose important reproductive barriers between species. This review uses the evolution of inversions in Drosophila pseudoobscura and D. persimilis as a case study for how inversions originate, establish, and contribute to the evolution of reproductive isolation.
Keywords: Drosophila, Chromosomal Inversions, Population Genomics, Recombination, Linkage Disequilibrium, Reproductive Isolation, Speciation
Introduction
Chromosomal inversions were first discovered in 1913 by Alfred Sturtevant as strong crossover modifiers segregating in natural populations of Drosophila melanogaster (Sturtevant, 1917). Because visible markers were required to demonstrate inverted gene order, the analysis of chromosomal inversions remained restricted for the next two decades to D. melanogaster and the closely related D. simulans (Sturtevant, 1921; Sturtevant & Plunkett, 1926). The development of cytogenetic methods that visualized the repeatable patterns of bands and puffs on Drosophila polytene chromosomes (Painter, 1934) opened up a more direct method of analysis of structural variation that exists within populations (Dobzhansky & Sturtevant, 1938) and between species (Sturtevant & Novitski, 1941; Sturtevant & Tan, 1937). Single chromosomal rearrangements generated by spontaneous inversions were already known to exist as fixed differences between closely related species of Drosophila (Sturtevant, 1921), but as more Drosophila species were examined, the degree of gene rearrangement was also found to vary among species., Some species have only one gene order on each chromosome, while have an extensive array of inversion polymorphisms segregating on some or all major chromosomal arms (Dobzhansky & Sturtevant, 1938; Dubinin, Sokolov, & Tiniakov, 1936; Sperlich & Pfriem, 1986).
Careful examination of polytene chromosomes and fixed inversion differences between closely related species were used to develop the first species phylogenies based on genetic characters (Ashburner & Lemeunier, 1976; Dobzhansky, 1944; Sturtevant & Dobzhansky, 1936; Wasserman, 1960). In this case, fixed inversion differences were assumed to be neutral taxonomic characters to discriminate among species. Maps of chromosomal banding patterns were also found to vary among individuals of the same species. Within species inversion polymorphisms were also initially considered neutral variants because only gene order and not gene content differs between arrangements, and thus the geographic distribution of arrangements were thought to be governed by non-selective mechanisms (Dobzhansky & Queal, 1938). The collaboration between Theodosius Dobzhansky and Sewall Wright (Dobzhansky & Wright, 1941, 1943; Provine, 1986; Wright & Dobzhansky, 1946; Wright, Dobzhansky, & Hovanitz, 1942) led to a selective paradigm that emerged from pioneering research from observation of the spatio-temporal distributions of inversions in nature to the subsequent use of population cage experiments to estimate fitnesses of karyotypess (Dobzhansky, 1955; Lewontin, 1974; Provine, 1986).
Despite extensive survey data that quantified levels of variation within and between populations as well as structural differences between species, no clear consensus emerged for the evolutionary genetic mechanisms responsible for (1) the origin of inversion mutations, (2) their subsequent establishment within populations, or (3) their ultimate fixation or elimination from populations (Dobzhansky, 1943, 1948b). This highly productive research program fell out of favor as new genetic markers, such as allozymes, restriction fragment length polymorphisms, nucleotide sequences, and microsatellites were used to address questions of genic and genomic evolutionary processes. Beginning in the 2000s, the application of DNA sequencing technology in natural populations led to the discovery of inversions in organisms other than Drosophila and a renaissance of both empirical and theoretical research on inversion polymorphism ensued (Jones et al., 2012; Kirkpatrick & Barton, 2006; Le Poul et al., 2014; Lowry & Willis, 2010; Stefansson et al., 2005).
Inversion polymorphisms have been implicated in a wide variety of evolutionary processes including chromosomal evolution, complex trait evolution, local adaptation, and speciation. Inversions alter the structure of genomes at different rates, e.g., (genome rearrangement in platyfish Amores et al., 2014; and mosquitos Neafsey et al., 2015). Inversions are observed to capture allelic combinations involved in polygenic traits (sperm morphology and swimming speed in songbirds Kim et al., 2017; reproductive morphotypes in ruffs Küpper et al., 2015; reproductive output in humans Stefansson et al., 2005). Inversions capture suites of genes underlying traits involved in local adaptation (life history traits in mosquitos Cheng et al., 2012; monkey flowers in this issue Coughlan & Willis; flowering time and annual/perennial life history strategies in monkey flowers Lowry & Willis, 2010; altitude and dispersal in teosinte Pyhäjärvi, Hufford, Mezmouk, & Ross-Ibarra, 2013; migratory behavior in Atlantic cod Sinclair‐Waters et al., 2018; neurotransmitters in honey bees Wallberg, Schöning, Webster, & Hasselmann, 2017). Inversions have also been shown to capture genes involved in reproductive isolation between species (Cheng et al., 2012; rats Engelbrecht, Taylor, Daniels, & Rambau, 2011; monkey flowers Fishman, Stathos, Beardsley, Williams, & Hill, 2013; sticklebacks Jones et al., 2012; Drosophila Lohse, Clarke, Ritchie, & Etges, 2015; Lowry & Willis, 2010; Drosophila Noor, Grams, Bertucci, & Reiland, 2001; Pyhäjärvi et al., 2013; Sinclair‐Waters et al., 2018; Stefansson et al., 2005; Wallberg et al., 2017).
Karyotypes and karyotypic frequencies cannot adequately test hypotheses about the common ancestor of arrangements, the age of arrangements, the role that selection plays in their maintenance, and the gene targets of selection; all of which are critical in defining how inversions arise, are established, maintained, and are fixed in populations. The advent of high throughput sequencing has inspired a renewed interest in testing hypotheses about the role genetic changes within inverted and non-inverted regions play in the evolution of populations and the divergence of species. Nucleotide and genome sequence data can better identify breakpoints, infer phylogenies, infer the history of different regions of the genome, and detect regions with signatures of selection. Recent genomic investigations of structural variation in the Drosophila genus have provided important new insights into how inversions shape adaptation and speciation, including that inversion breakpoints are influenced by selection (Corbett-Detig, 2016), inversions can capture multiple genes that are differentially expressed (Fuller, Haynes, Richards, & Schaeffer, 2016; Lavington & Kern, 2017; Said et al., 2018) and are potential targets of selection (Corbett-Detig & Hartl, 2012; Fuller, Haynes, Richards, & Schaeffer, 2017; Kapun, Fabian, Goudet, & Flatt, 2016; Pool, Braun, & Lack, 2017; Rane, Rako, Kapun, Lee, & Hoffmann, 2015; Simões & Pascual, 2018), rearrangements are fixed extensively between species (Bhutkar et al., 2008), and they can prevent the spread of reproductive isolation genes between sibling species (Kulathinal, Stevison, & Noor, 2009; Lohse et al., 2015; McGaugh & Noor, 2012; Noor, Grams, Bertucci, & Reiland, 2001; Sanchez-Flores et al., 2016).
The prevailing view prior to the availability of nucleotide and genome sequence data was that inversion polymorphisms were adaptive, but the mechanisms for the establishment and maintenance of the rearrangements were unclear. Stable clines of inversion frequencies correlated with environmental gradients provided convincing albeit circumstantial evidence that inversion polymorphisms were adaptive, especially when latitudinal clines are recapitulated in the northern and southern hemispheres on multiple continents (Balanya, Huey, Gilchrist, & Serra, 2009; Hoffmann, Sgro, & Weeks, 2004; Kennington & Hoffmann, 2013). The long-term stability of such clines, and concomitant stable seasonal cycling of inversion frequencies (Dobzhansky, 1943; Knibb, 1982; Stalker, 1980), provides some of the strongest evidence of local adaptation to spatio-temporally varying selection (Cogni et al., 2017; Endler, 1977). In a particularly well-characterized example, the relative frequencies of third chromosome arrangements of Drosophila pseudoobscura form an east-west cline across the Southwestern United States, a distribution that has remained stable since its initial description by Dobzhansky nearly 80 years ago (240 generations) (Anderson et al., 1991; Dobzhansky, 1944), while loci on the other autosomes show no evidence for clinal variation (Kovacevic & Schaeffer, 2000; Table 26 in Lewontin, 1974; Riley, Hallas, & Lewontin, 1989; Schaeffer & Miller, 1992), suggesting that the clines are maintained despite the homogenizing effect of gene flow. Population cage experiments in the laboratory also aided in understanding the forces acting on gene arrangement polymorphisms (Dobzhansky, 1948c, 1950; Pavlovsky & Dobzhansky, 1966; Wright & Dobzhansky, 1946).
Population cage experiments showed that two inversions, say A and B, collected from population X reached stable equilibria over several generations (Wright & Dobzhansky, 1946), but the equilibrium points reached differed for the A and B arrangements drawn from localities X and Y (Dobzhansky, 1948c). Mixing pairs of arrangements from different geographic populations, say arrangement A from locality X (AX) and arrangement B from locality Y (BY), failed to reach stable equilibria. Yet, stable equilibria were restored when homokaryotypes had chromosomes from different populations (AX/AY) and the heterokaryotypes had chromosomes from the same population (AX/BX or AY/BY) (Dobzhansky, 1950). Dobzhansky favored overdominance being responsible for the stable equilibria, while Wright’s calculations found that frequency dependent selection fit the frequency change data better (see pages 807–809 in Lewontin, Moore, Provine, & Wallace, 1981; Pavlovsky & Dobzhansky, 1966; Wright & Dobzhansky, 1946). Dobzhansky’s overall model was that the inversions captured coadapted genes not only among the different arrangements, but also among the same arrangement from different populations, however, this hypothesis is inconsistent with nucleotide sequence data that fails to observe differentiation of homosequential inversions among localities (Fuller et al., 2017; Schaeffer et al., 2003).
Inversions are often thought to be locally adaptive because of their ability to maintain associations among multiple alleles that confer higher fitness in particular environments (Kirkpatrick & Barton, 2006). Heterozygosity for chromosomal inversions severely reduces the rate of recombination through multiple mechanisms reviewed below, thereby preventing multiple beneficial alleles and/or epistatically interacting loci from recombining. Polygenic adaptation to heterogeneous environments is sensitive to recombination rates. Typically, rates of adaptation are thought to increase in the presence of recombination (McDonald, Rice, & Desai, 2016) presumably because beneficial alleles can become unlinked from deleterious alleles and new positive epistatic combinations of alleles with respect to fitness can form more rapidly. On the other hand, recombination will also generate maladaptive combinations and beneficial associations can be broken apart (Charlesworth & Barton, 1996). Although inversions are widely thought to be favored because they suppress recombination in the heterozygous state, the potential evolutionary forces that favor their establishment, then either maintain them at appreciable frequencies or drive them to fixation or loss remain unclear in many cases.
Here, we present a review of case studies from the obscura subgroup of Drosophila of how spontaneous inversions originated and were established in D. pseudoobscura populations, then once established how inversions contributed to adaptation, and ultimately what role inversions may have played in the formation of new species. Recent data from Drosophila support the hypothesis that inversions are established because of their indirect effects on fitness through recombination suppression that promotes local adaptation to heterogeneous environments. The reduction of recombination within inverted regions also leads to the accumulation of genetic differences within the inverted segments. We review recent findings about the role that inversions play in establishing species boundaries. We focus our analyses on D. pseudoobscura and D. persimilis, which are model systems for understanding the population genetics of inversions within species as well as the evolutionary genetics of differences between species.
The Nature of Recombination Suppression in Inversions
Chromosomal inversions alter gene order, but not gene content, such that the major genetic effect of inversion heterozygosity is strong recombination suppression. The direct fitness effects of inversion heterozygosity in Drosophila could be minimal and confined to the potential for gene disruption or gene expression alteration at breakpoints (Fuller et al., 2016, 2017). Alternatively, the population genetic consequences of forming large blocks of tightly linked allelic variants may confer indirect fitness effects on gene rearrangements that ultimately govern the establishment phase of chromosomal inversion evolution. Thus, in a population genetic framework, natural selection drives the evolution of common chromosomal inversions and recombination suppression is the trait selected upon (but not necessarily the trait selected for). There are three mechanisms that can suppress recombination in inversion heterozygotes, and the relative importance of these mechanisms can vary between taxonomic groups. In reverse chronological order they are: (1) a selective process where crossing-over inside of inverted regions causes gross gametic aneuploidy and therefore zygotic lethality, e.g., as observed in maize and mice (Koehler et al., 2002; McClintock, 1941), (2) a mechanical process where crossing-over inside of inverted regions cause dicentric chromosomes that segregate to the polar bodies in female meiosis and are never transmitted, e.g., as observed in Drosophila and Sciara (Carson, 1946; Sturtevant & Beadle, 1936), and (3) crossover modification processes where the distribution of crossover events is shifted away from inverted regions (Crown, Miller, Sekelsky, & Hawley, 2018; Lucchesi & Suzuki, 1968; Sturtevant & Beadle, 1936). Although the transmission genetic consequence of these three mechanisms is the same (namely suppressed recombination), the population genetic consequences are drastically different. It is therefore critical for evolutionary models to accurately reflect the different cause(s) of recombination suppression.
In the heterozygous state, pairing and synapsis of chromosomal inversions and the standard arrangement occur normally throughout the inverted region (Gong, McKim, & Hawley, 2005), to achieve this, a characteristic “inversion loop” is formed. The consequence of crossing-over inside this inversion loop is the duplication and deletion of large regions of the chromosome (Figure 1). If the inversion loop does not encompass the centromere, then in addition to the duplications and deletions the crossover products will either be acentric or dicentric (containing no or two centromeres) which causes segregation abnormalities. The consequences of these forms of gross gametic aneuploidy are almost invariably zygotic lethality, with any viable offspring being non-recombinants giving the population-level appearance of recombination suppression. When this mechanism operates it is a form of strong fitness underdominance (i.e., where heterozygotes are less fit than either homozygote), with the basic population genetic expectation that inversions (as the minority allele) would rapidly be eliminated from the population (Wright, 1941). Although this form of recombination suppression was first observed in maize (McClintock, 1931, 1941) and operates in a wide range of organisms, there are some unique features of dipteran meiosis that make this mechanism largely irrelevant for the evolution of Drosophila inversion polymorphism.
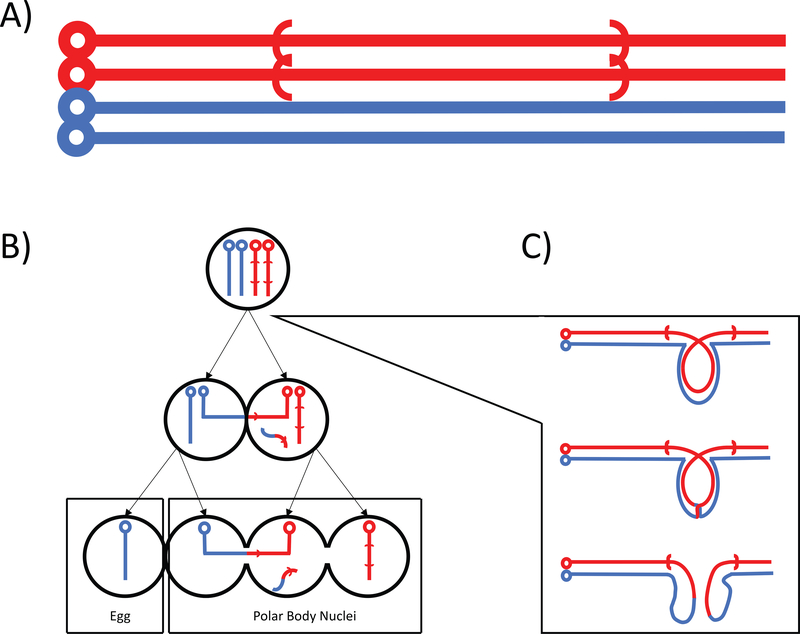
Figure 1.
Drosophila female meiosis in an inversion heterozygote. A) Chromosomes of an inversion heterozygote as an unpaired four strand bundle, red chromosome carry an inverted arrangement illustrating breakpoints with parentheses. B) The progression of asymmetric (female) meiosis contingent on a crossover event causing acentric/dicentric chromosomes. Dicentric chromosome is selectively eliminated by confinement to internal positions, and therefore polar bodies nuclei, in the linear array of meiotic products. C) Formation of the inversion loop illustrating the consequences of crossing-over in the inverted region, showing only two chromatids for simplicity.
All brachycerous Diptera have achiasmate male meiosis, which means crossing over is limited to females (see Gethmann, 1988 for rare exceptions). Female meiosis is asymmetric, that is for every one functional egg there are three polar body nuclei formed. In Drosophila, these form a linear array with one of the terminal nuclei always becoming the gamete (Huettner, 1924). In a process mathematically equivalent to gametic selection there can be competition among homologous chromosomes, or recombinant chromosomes, to be in a terminal position included in the functional egg; this process is termed true meiotic drive and is potentially a very strong evolutionary force (Lindholm et al., 2016; Sandler & Novitski, 1957). There are several forms of meiotic drive in female meiosis of Drosophila melanogaster (Ashburner, Golic, & Hawley, 2005), but perhaps the most important is the preferential segregation of dicentric products of crossing-over in inversion loops to the central position in the linear array of meiotic products. As a result of this meiotic drive, the aberrant chromosomes that would produce lethal zygotes are efficiently sequestered in polar bodies, such that there is no substantial underdominance realized for paracentric inversions in Drosophila species (Sturtevant & Beadle, 1936). Although this effectively eliminates direct fitness effects due to segregation problems, the mechanical process does not apply to pericentric inversions, which in turn is likely responsible for their rarity in natural populations of Drosophila (Coyne, Aulard, & Berry, 1991; Coyne, Meyers, Crittenden, & Sniegowski, 1993; Krimbas & Powell, 1992a; Navarro, Betrán, Barbadilla, & Ruiz, 1997). Even though the operation of the mechanical process appears simple at first, the molecular biology and biophysics for segregation and breakage of dicentric chromosomes is not completely understood and is an active area of research (Hill & Golic, 2015; Koehler et al., 2002; Lopez et al., 2015).
Although the transmission genetic and population genetic consequences of the chromosome mechanics have been well-studied and are at the theoretical core of inversion evolution, the preceding mechanisms are limited to explaining only one quarter of recombination suppression inside inverted regions (Novitski & Braver, 1954), which corresponds to 10–20% of the total chromosome-wide recombination suppression observed in initial studies (Sturtevant & Beadle, 1936), later work (Grell, 1962; Roberts, 1962), and recent investigation of the interference hypothesis (Koury, 2017). Therefore, the vast majority (80–90%) of recombination suppression due to inversion heterozygosity is determined by a third mechanism that shifts the distribution of crossing-over away from inversions – collectively referred to as crossover modifying processes. Unfortunately, almost nothing is known about the causes of these processes and they remain poorly described despite being the major determinant of the selected effect of segregating inversions. In what follows, an outline of the basic components of these effects are described and the interference hypothesis for recombination suppression is introduced as a conceptual framework for future research into the crossover modifying processes.
Just like recombination rates, there are both genetic and environmental sources of variation for recombination suppression and it is a process affected by proximity to structural features of chromosomes (e.g., centromeres, telomeres, boundary sites, heterochromatin, etc.) (Comeron, Ratnappan, & Bailin, 2012; Coyne et al., 1993; Hunter, Robinson, Aylor, & Singh, 2016; Koury, 2017). Recombination suppression, however, has an added degree of complexity because its relative strength is dependent on genetic distance to the heterozygous inversion breakpoints. Therefore, it is useful to conceptualize recombination suppression as an interference effect in the broad sense, i.e., as an altered probability of crossing-over conditional on distance to a heterozygous inversion breakpoint. In this theoretical framework, it is possible to account for strong local and diffuse global suppression, both inside and outside the inverted region, as well as position dependent effects of inversion heterozygosity on recombination landscapes (Koury, 2017; McPeek & Speed, 1995; Navarro et al., 1997).
Evidence for interference effects also comes from the reduced rates of single and double crossover events inside the inversion loop (Krimbas & Powell, 1992b). Interestingly, the rates of gene conversion are not as strongly affected (Chovnick, 1973; Korunes & Noor, 2017; Schaeffer & Anderson, 2005), which is consistent with Gong et al.’s (2005) observation that the distribution of crossover precursors (such as double strand breaks) are approximately normal for inversion heterozygotes and the suppression is achieved by preventing the crossover maturation of chiasmata. Using experimental constructs to avoid the difficulties associated with dicentric chromosomes, or using viability reduction with pericentric inversions as an indirect measure, the crossover rate inside heterozygous inversions is estimated to be about 25% of wildtype (Coyne et al., 1993; Navarro & Ruiz, 1997; Novitski & Braver, 1954). Thus, the lack of recombination both internal and external to inverted regions is largely determined by the interference effects of heterozygous breakpoints as crossover modifiers.
In comparative studies of recombination suppression by heterozygous inversions, size and position dependent effects have often been noted (Krimbas & Powell, 1992b). Smaller inversions distally placed on chromosomes tend to suppress recombination stronger than larger or more proximal inversions near the centromere, although analysis of the relative importance of size versus position is limited by the irregular distribution of natural variants with incompletely known recombination profiles. The size effect, especially in terms of double crossover events inside inversions, have long been known to be a result of crossover interference (Krimbas & Powell, 1992b; Navarro et al., 1997; Sturtevant & Beadle, 1936). More surprisingly, similar results in terms of interference and position effects have been shown for reciprocal translocations (Hawley, 1980; Roberts, 1970, 1972; Sherizen, Jang, Bhagat, Kato, & McKim, 2005), demonstrating these effects are not due to inversions per se, but rather due to breakpoint heterozygosity from any type of chromosomal rearrangement. Several evolutionary arguments for selection acting on inversion polymorphisms are based on the effects of suppressed recombination, and these have variously (and often conflictingly) claimed that distal inversions are favored (Novitski, 1946), smaller inversions are favored (Krimbas & Powell, 1992b; Olvera et al., 1979), larger inversions are favored (Caceres, Barbadilla, & Ruiz, 1999; van Valen & Levins, 1968), or that inversions disrupting boundary sites are necessary for normal crossing-over (Corbett-Detig, 2016). However, all of the selective explanations of inversion polymorphism make simplifying assumptions about recombination suppression (the selected character) based on the mechanical process. Clearly the majority of the phenotypic effect of heterozygous inversions is neither caused by this mechanism, nor is it limited to the inverted region of the chromosome (Fuller et al., 2016; Lavington & Kern, 2017).
Considering how little is mechanistically known in the model organism D. melanogaster, it is hard to speculate about other less studied species. Inversion heterozygosity suppresses recombination in the limited number of Drosophila species studied thus far (Carson, 1953; Dobzhansky & Epling, 1948; Singh & Singh, 1987), and does so via the same mechanisms, yet the relative strength and importance of these mechanisms is unknown (Krimbas & Powell, 1992b). Given that both recombination maps and crossover interference are known to vary between species, it is reasonable to expect the magnitude and extent of recombination suppression to differ as well (Brand, Cattani, Kingan, Landeen, & Presgraves, 2018; Cáceres, Ranz, Barbadilla, Long, & Ruiz, 1999; True, Mercer, & Laurie, 1996). While this expectation is fulfilled for rates of double crossovers inside inversions, interestingly work in D. subobscura (a species with a longer genetic map) shows the effect is in the opposite direction (lower) of expectations (Spurway & Philip, 1952). Indeed, interspecific variation in the distribution of chromosomal inversions, recombination rates, and crossover interference is likely to provide important evolutionary insight to the importance of meiotic phenotypes, and is particularly fruitful ground for further exploration of the interference hypothesis for recombination suppression (Caceres et al., 1999; Gong et al., 2005; Koury, 2017; True et al., 1996).
In summary, it is likely the indirect fitness effects of chromosomal inversions determine their evolutionary fate, and those fitnesses are the combined direct effects of alleles in strong linkage disequilibrium within inverted segments. This linkage is caused by recombination suppression observed for inversion heterozygotes. There are three possible causes of recombination suppression, two of which could generate strong fitness underdominance as a direct effect of inversion heterozygosity. However, the details of meiosis in higher Diptera allow Drosophila species to escape the negative consequences of crossover induced chromosomal aberrations (aneuploidy, dicentric chromosomes, etc.). In the mechanical process, single crossover events generate acentric and dicentric recombinant products that are never included in the functional egg via a meiotic drive mechanism unique to asymmetric female meiosis only known from studies of Diptera and not other taxa (Carson, 1946; Madan, Seabright, Lindenbaum, & Bobrow, 1984; McClintock, 1941; Sturtevant & Beadle, 1936). Despite the mechanical process being more thoroughly studied than the poorly known interference effects, the former mechanism’s operation is limited to the inverted regions and accounts for only 10 to 20 percent of total chromosome-wide recombination suppression (Grell, 1962; Koury, 2017; Roberts, 1962; Sturtevant & Beadle, 1936). In contrast, the crossover modifying process operates both inside and outside inversions, extends chromosome-wide, and accounts for 80 to 90 percent of recombination suppression. Future experimental work, empirical analysis, and evolutionary modelling should focus on the chromosome-wide interference effects caused by the crossover modifying process as the major determinant of recombination suppression for inversion heterozygotes.
The Evolutionary Mechanisms That Establish Inversions in Drosophila Populations- A case history in D. pseudoobscura
Different species of Drosophila offer increasing levels of complexity in the study of inversion polymorphisms in nature. The inversion polymorphisms of D. melanogaster are relatively simple in that two major cosmopolitan arrangements are segregating on the large chromosomal arms (X, 2L, 2R, 3L, and 3R) with a large number of minor rare endemic arrangements (Lemeunier & Aulard, 1992). Corbett-Detig and Hartl (2012) showed that the derived arrangements have putative selective targets distributed across the inverted regions. The inversion polymorphisms of D. robusta and D. subobscura are the most complex in that non-overlapping inversions are found to be in linkage disequilibrium within and between chromosomal arms (Carson, 1958; Krimbas, 1992; Levitan, 1958; Levitan, 1992), which require future studies to understand how each inversion arises, and why the combinations are non-randomly associated with each other. Here, we focus on the third chromosome gene arrangement polymorphism of D. pseudoobscura, a system slightly more complex than D. melanogaster where a single chromosomal arm segregates for more than 30 different overlapping inversions (Dobzhansky & Sturtevant, 1938), many reaching appreciable frequencies and wide geographic distributions.
D. pseudoobscura has served as model system for testing how the effects of recombination suppression lead to the establishment and maintenance of inversions in natural populations, as there is a wealth of well characterized gene arrangements on its third chromosome that were generated through overlapping inversion mutations that strongly suppress recombination (Dobzhansky & Epling, 1948; Dobzhansky & Sturtevant, 1938; Levine, 1956; Levine & Levine, 1954, 1955). D. pseudoobscura is native to the western United States from the Rocky Mountains in the east to the Pacific coast in the west and from British Columbia in the north to Guatemala in the south (Figure 2). A subspecies of D. pseudoobscura is found in the highlands near Bogota, Colombia (D. pseudoobscura bogotana not shown), that has served as a model for the early stages of speciation (Orr, 1989; Phadnis, 2011; Phadnis & Orr, 2009).
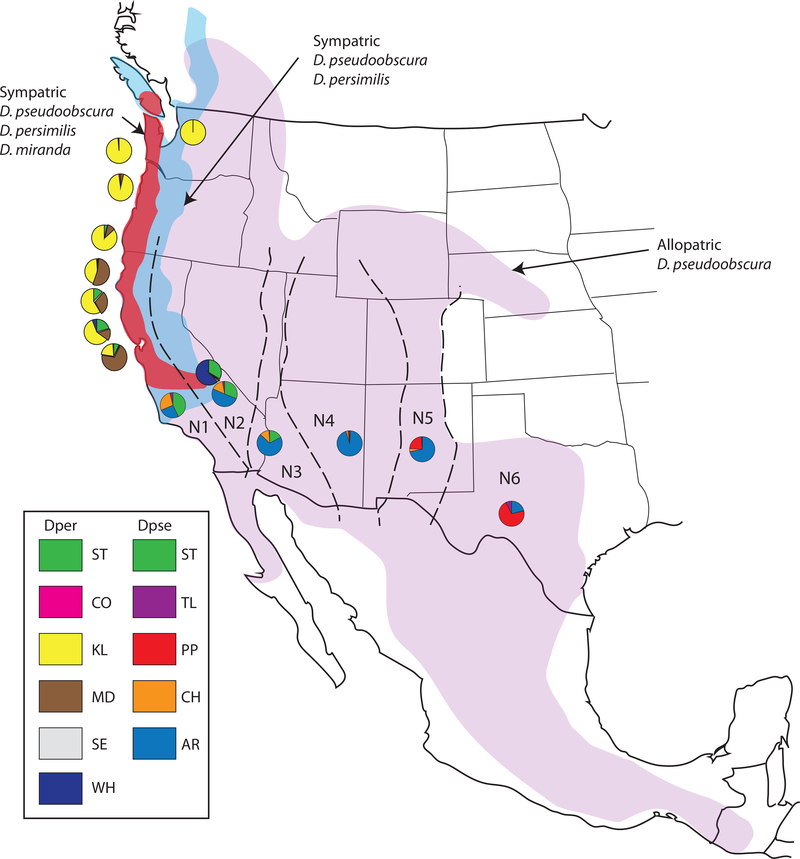
Figure 2.
Distribution of the members of the North American obscura group species, Drosophila pseudoobscura, D. persimilis, and D. miranda. The pie diagrams show the frequencies of the major gene arrangements of D. pseudoobscura and D. persimilis. The gene arrangement frequencies of D. pseudoobscura in six niches (N1 to N6, see Schaeffer, 2008) are indicated by the six pie diagrams at the bottom of the niche while all other pie diagrams shown above are for D. persimilis. Only the Standard (ST) gene arrangement is found in D. pseudoobscura and D. persimilis.
A large body of evidence suggests that the arrangements of the third chromosome are selected. First, the third chromosome’s gene arrangement frequencies are found in a geographical cline that has been stable for at least eighty years (Anderson et al., 1991; Dobzhansky, 1944; Schaeffer et al., 2003) with the major transitions in frequencies occurring at the boundaries of major physiographic provinces (Lobeck, 1948) (Figure 2). The gradient of gene arrangement frequencies exists despite the homogenizing effect of extensive migration (4Nem > 1) among D. pseudoobscura populations (Fuller et al., 2017; Kovacevic & Schaeffer, 2000; Riley et al., 1989; Schaeffer et al., 2003; Schaeffer & Miller, 1992) suggesting that strong selection counters the effects of gene flow. In addition, the gene arrangements cycle in frequency over seasons (Dobzhansky, 1943).
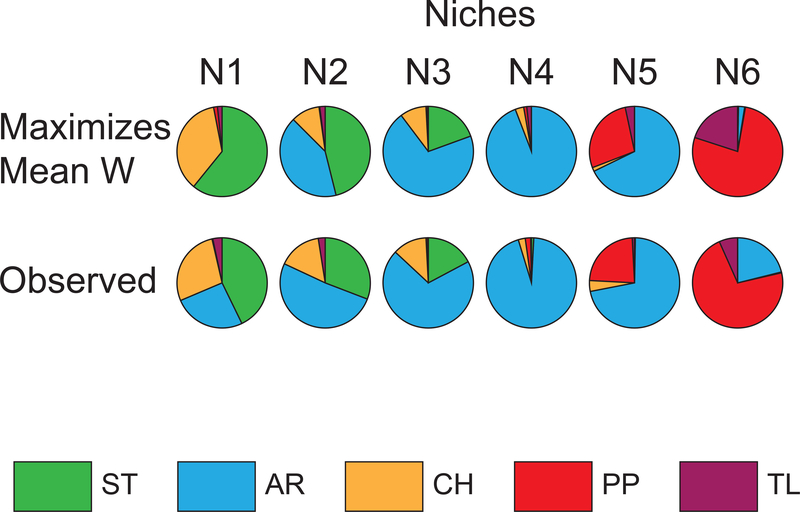
Schaeffer (2008) used gene arrangement frequencies in natural populations to infer karyotypic fitnesses as opposed to the population cage approach of Wright and Dobzhansky (Dobzhansky, 1948c; Wright & Dobzhansky, 1946). The inversion frequency cline can be divided into six niches where the gene arrangement frequencies are similar within niche, but are different between niches. Schaeffer (2008) developed a model of migration-selection balance that estimated karyotypic fitnesses within each niche necessary to transform estimated migrant frequencies to the observed local frequencies. The karyotypic fitness estimates in the six niches do not predict the stable maintenance of all arrangements in a single niche, but instead that they are maintained as a stable protected polymorphism across all niches (Levene, 1953; Levins & MacArthur, 1966). The application of Levins and MacArthur (1966) fitness set analysis suggests that the environments of adjacent niches are similar, but that the climates of non-adjacent niches are different (see supplemental information). In addition, the gene arrangement frequencies that would maximize mean fitness in each niche differ from the observed gene arrangement frequencies suggesting that migration creates a genetic load in each niche (Figure 3).
Figure 3.
Gene arrangements frequencies in the six different niches. The top line of pie diagrams shows the randomly chosen gene arrangement frequencies that maximize the mean fitness (W) within the niche. The bottom line shows the observed gene arrangement frequencies in the six different niches.
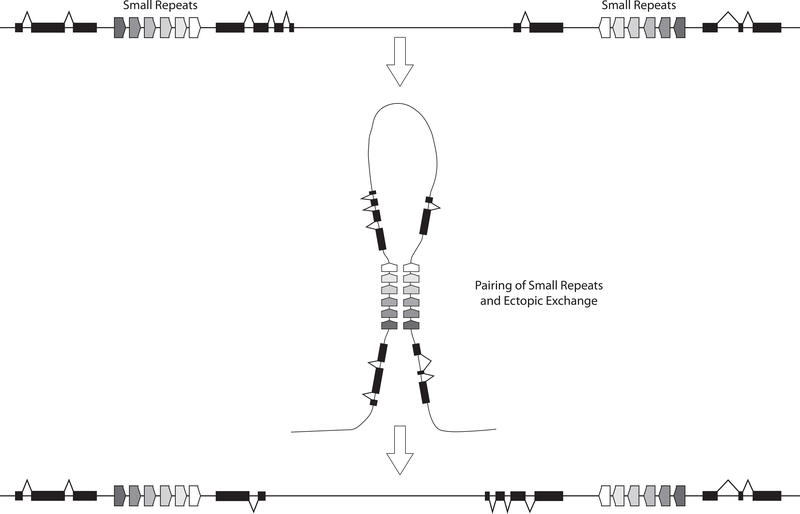
Although the fitness of each arrangement can be estimated from their frequency distribution in space, the genetic basis of their origin, whether selection plays a role, and if so, what the genetic basis for the selection has remained elusive. Genomic and transcriptomic analysis of the D. pseudoobscura third chromosomal gene arrangements have provided clues about how inversions arose and the genetic forces that led to their establishment in D. pseudoobscura populations. The discovery of transposable elements (TEs) suggested that inversion mutations may occur through ectopic exchange between TEs in reverse orientation (Lim & Simmons, 1994). In D. pseudoobscura, no known TEs were found at inversion breakpoints (Richards et al., 2005). Instead, sets of small repeat sequences approximately 100 – 200 bp in length were found at the proximal and distal inversion breakpoints in reverse orientation suggesting that these repeats paired and generated the reversal of the region between the repeats (Figure 4). This differs from the structure of breakpoints in other Drosophila species, where random breaks have been typically observed (Wesley & Eanes, 1994) or staggered cuts in the breakpoint region results in duplication of genes adjacent to the breaks (Calvete, Gonzalez, Betran, & Ruiz, 2012; Matzkin, Merritt, Zhu, & Eanes, 2005; Puerma, Orengo, & Aguadé, 2016; Ranz et al., 2007). The D. pseudoobscura third chromosomal arrangements result from single mutational events based on the monophyly of the major inversions (Fuller et al., 2017; Wallace, Detweiler, & Schaeffer, 2011). A central question is how the D. pseudoobscura arrangements were established in populations after they arose.
Figure 4.
Mechanism to invert chromosomes in D. pseudoobscura that uses small repeats sequences (100–200 bp) in reverse orientation that pair and undergo ectopic exchange (center). Repeats are located between coding regions (black filled in boxes separated by intron sequences).
Kirkpatrick and Barton (2006) summarized the major mechanisms that could establish new inversion mutations in populations. The first possibility is that inversions are neutral and their frequency increases or decreases via genetic drift (Kimura, 1968; Lande, 1984; Lande, 1985). Under the drift model, the current frequency cline in D. pseudoobscura represents a transient phase of gene arrangement polymorphism. This explanation is not likely, however, given the effective population sizes of D. pseudoobscura are estimated to be between 1.9 to 4.5 × 106 based on the mutation (4Neμ) and recombination (4Nec) parameters (Schaeffer, 1995), which is much larger than drift models require (Lande, 1984). The second explanation is that a new inversion mutation is driven to higher frequency because it captured a single sweeping beneficial allele (Maynard Smith & Haigh, 1974).
The third possibility is that the breakpoints create variation that selection acts upon, which is a direct effect of the inversion mutation. This could take several forms such as disrupting the structure of genes (Puerma et al., 2016) or altering gene expression by separating the coding sequence from its upstream cis regulatory sequences (Puig, Caceres, & Ruiz, 2004). The disruptions caused by breakpoints could have more regional effects if they occur within regions of coordinated gene expression, i.e., topologically associated domains (TADs) (Hou, Li, Qin, & Corces, 2012). Another direct effect of an inversion could be to set up conditions for meiotic drive where one arrangement has an advantage over another based on differences in length of non-sister chromatids (Novitski, 1951). Crossing-over in overlapping inversion heterozygotes generates asymmetry in non-sister chromatids in Meiosis II, which causes meiotic drive favoring the more distally positioned inversions (Koury, 2017). No evidence for meiotic drive was observed when comparing transmission of arrangements in heterokaryotypes in females versus males (chiasmate versus achiasmate segregation) (Meisel & Schaeffer, 2007). Interestingly, the historical sequence of inversions in D. pseudoobscura is consistent with a high fitness of distal inversions on the third chromosome (Koury, 2017). The success of particular gene arrangements could also result from the proximity of breakpoints to pairing sensitive sites that promote crossing over (Corbett-Detig, 2016).
A fourth mechanism to establish new inversions is an indirect effect, where gene rearrangements are established because of their ability to suppress recombination in heterozygotes (Sturtevant & Beadle, 1936). Inversions would be favored if the fitness of a particular gametic combination is greater than the mean fitness of the population (Charlesworth & Charlesworth, 1973). Inversions might also be favored to increase if they capture regions relatively devoid of deleterious recessive mutations (Nei, Kojima, & Schaffer, 1967). Dobzhansky suggested that inversions would increase in frequency if the mutation captured alleles at multiple loci that act epistatically together (Dobzhansky, 1950). Ohta and Kojima (1968) mathematically modeled this idea using a time heterogeneous branching process to incorporate the fitness decay of inversions as deleterious mutations accumulate, and were able to demonstrate that the ultimate fate of inversions is the selective elimination from populations unless there is unique epistasis associated with alleles inside the inversion. Kirkpatrick and Barton (2006) suggested that inversions could be favored to increase if they captured locally adapted alleles in a species where migration allowed maladapted gene combinations to be transported between habitats.
These different scenarios are expected to result in different signatures at the sequence level depending on the age of the inversion (Guerrero, Rousset, & Kirkpatrick, 2012). It is generally assumed that a new derived inversion mutation will occur in a single individual from an ancestral arrangement and increase in frequency either through neutral or selective forces. The derived arrangement will capture a multi-site genotype from one of the segregating ancestral chromosomes. If the new arrangement is neutral, then over time, genetic flux (gene conversion and double cross overs) among arrangements will homogenize variation within the central inverted regions with the most differentiated regions at the breakpoints (Navarro, Barbadilla, & Ruiz, 2000; Navarro et al., 1997). The degree of homogenization depends on the age of the arrangement, with older arrangements being more homogenized (see the dashed line in Figure 1 of Guerrero et al., 2012). On the other hand, if a new arrangement captures one or more selected genes, then genetic flux will homogenize variation in non-selected regions and as the inversion increases in age, selected genes will become increasingly differentiated (see the right hand side of Figure 1 in Guerrero et al., 2012). It should be noted that the Guerrero et al. (2012) do not incorporate new mutations in their model.
Vann (1966) showed that artificially induced inversions are able to establish within laboratory cultures in eight to nine generations, but the young age of these new arrangements would prevent one from distinguishing among establishment hypotheses with nucleotide sequence data. One would expect that all copies of these induced arrangements would be nearly identical in sequence.
The D. pseudoobscura inversion polymorphism is relatively old, with the different arrangements ranging in age from 0.51 to 1.38 million years or equivalently 1.5 to 4.14 million generations ago (Wallace et al., 2011) based on three generations per year (Aquadro, Weaver, Schaeffer, & Anderson, 1991). The majority of naturally occurring inversion heterozygotes differ by at least two inversion events such that most genetic flux in heterokaryotypes will be in the form of gene conversion, estimated to occur at a rate of 3.4 × 10−6 per generation, which is 100 times greater than the mutation rate in this species (Schaeffer & Anderson, 2005), but is 3 to 6 times lower than the single generation gene conversion estimate (Korunes & Noor, 2019). The frequency of heterokaryotypes among niches varies from 10 to 70%, but the mean frequency is expected to be near 40% across all niches based on the extensive migration observed among populations (Kovacevic & Schaeffer, 2000; Riley et al., 1989; Schaeffer & Miller, 1992). Given the age of the inversions and level of genetic flux among arrangements, differentiation is predicted to be significantly decreased in the central regions of the inversion (Guerrero et al., 2012).
Fuller et al. (2016, 2017) used evidence from genomic and transcriptomic analyses of the third chromosome of D. pseudoobscura to address these hypotheses. Fuller et al. (2017) mapped the locations of seven pairs of inversion breakpoints and found no evidence of direct position effect mutations that disrupted the structure of genes near the inversion breakpoints (Fuller et al., 2017) or altered gene expression of loci in the immediate proximity of the breakpoints (Fuller et al., 2016). The expression data, however, did not rule out position effects that disrupt transcriptional variation at a larger genomic scale due to alteration of TADs.
Fuller et al. (2017) sequenced the complete genomes of 54 D. pseudoobscura individuals, representing six different third chromosome arrangements, to detect loci with an abundance of fixed derived mutations (Derived Allele Frequency test - DAF) or significantly long arrangement specific branch lengths (Population Specific Branch Length test - PSBL). A total of 277 outlier loci were discovered with the DAF and PSBL tests which also harbored arrangement specific fixed amino acid changes.
Fuller et al. (2016) quantified gene expression levels among gene arrangements to test whether any loci contained within the inversions were differentially expressed in different life stages (Fuller et al., 2016). They detected 45, 45, and 187 genes on the third chromosome that were differentially expressed in larvae, females, or males, respectively, for a total of 205 genes. The vast majority of differentially expressed genes (95%) were located within the overlapping inverted regions. More recently, Lavington and Kern (2017) and Said et al. (2018) also demonstrated that gene arrangements in D. melanogaster captured differentially expressed genes within inverted regions. Said et al. (2018) ruled out position effects by showing in an elegant synthetic inversion experiment, that an inversion mutation by itself does not lead to widespread gene expression changes within an inversion, suggesting that linked genetic variation captured by the inversion is responsible for transcriptional differences rather than the direct structural effects introduced by the inversion. Their data suggest that the effects of suppressed recombination between inversions in natural D. melanogaster populations maintain arrangement specific patterns of gene expression with potential phenotypic differences and selectable variation. Functional genomic experiments are necessary to demonstrate that transcriptional differences lead to changes in phenotype and fitness, and thus are possible targets of selection. Moreover, similar synthetic inversions are yet to be tested in D. pseudoobscura to exclude the position effect hypothesis.
Taken together, multiple outlier loci are located within the inverted regions of chromosome three and these loci have potentially selectable variation at the amino acid or transcriptional levels. In the majority of cases, this variation exists as changes to encoded proteins rather than gene expression differences. If we assume that outlier loci have adaptive alleles, two non-mutually exclusive models could explain the multiple outlier loci: (1) the initial inversion captured a single sweeping allele and other adaptive alleles were added successively to the arrangement later or (2) the initial inversion events captured suites of adaptive alleles at the outset. The first model of successive addition of adaptive alleles seems inconsistent with the observed data especially when you consider that the Pikes Peak has 127 outlier loci. Successive sweeps would be expected to continually remove genetic variation from the inverted region making it difficult to recover genetic diversity across the inverted region. None of the D. pseudoobscura arrangements show low levels of genetic diversity. The second model suggests that the suite of outlier alleles existed prior to the inversion event. Once captured within the inversion, gene conversion in heterokaryotypes begins to homogenize regions within the inversion except for those that affect fitness. Over time, only the adaptive loci will be differentiated among arrangements. The D. pseudoobscura arrangements are 0.5 million years old or older (Wallace et al., 2011) and the rates of gene conversion in heterokaryotypes are sufficiently high (Korunes & Noor, 2019; Schaeffer & Anderson, 2005) to reveal signatures of adaptation if they exist. Functional genetic studies are needed to rule out selective neutrality of the outlier alleles as an explanation for the inversion polymorphism. In addition, new theoretical models are needed to address what the pattern and organization of genetic diversity will be under these two models. Therefore, we conclude that the D. pseudoobscura inversions were most likely established and maintained due to the indirect effects of inversions as recombination suppressors in response to local adaptation or epistastic selection among loci. Potential functions of the outlier genes that may drive selection are sensory or detoxification activities (Fuller et al., 2016, 2017). An open question about the molecular genetics of inversions is: are there interactions among loci in inversion heterozygotes? Selection does not act on single inversions, but on the individual or inversion karyotype. Transcriptome data show that the majority of loci show additive effects in heterozygous individuals with only a few loci showing underdominant or overdominant expression (Fuller et al., 2016), which suggests that there are few interactions among loci at the expression level. Again, functional genetic experiments are needed to determine if these gene expression differences have effects on fitness.
We now know that selection in heterogeneous environments plays a prominent role in the establishment of the third chromosome inversions of D. pseudoobscura. In addition, selection in heterogeneous environments has led to clinal variation in the frequency of gene rearrangements in space. What happens if populations become isolated?
The Role of Fixed Inversions in the Speciation Process-A case history from D. pseudoobscura and D. persimilis
The establishment of inversion polymorphisms within heterogeneous environments may well set the stage for the formation of new species. Interest in speciation genetics traces its origins to the 1930s. At the time, D. melanogaster was the overwhelming favorite model species and its closest relative for genetic studies of reproductive isolation was D. simulans. D. melanogaster and D. simulans were not an idealas a model for speciation genetics because crosses between these species yielded only dead or sterile hybrids (Sturtevant, 1920). D. obscura Race A and B (Lancefield, 1929), later renamed D. pseudoobscura and D. persimilis (Dobzhansky & Epling, 1944; Frolowa & Astaurow, 1929), were found to be morphologically indistinguishable and yielded sterile males and fertile females in reciprocal genetic crosses. With fertile hybrid female offspring, crosses between the two species allowed Dobzhansky (1936) and later Orr (Orr, 1987) to attribute hybrid male sterility to at least nine genes distributed across the genome with genes on the X chromosome having the largest effect.
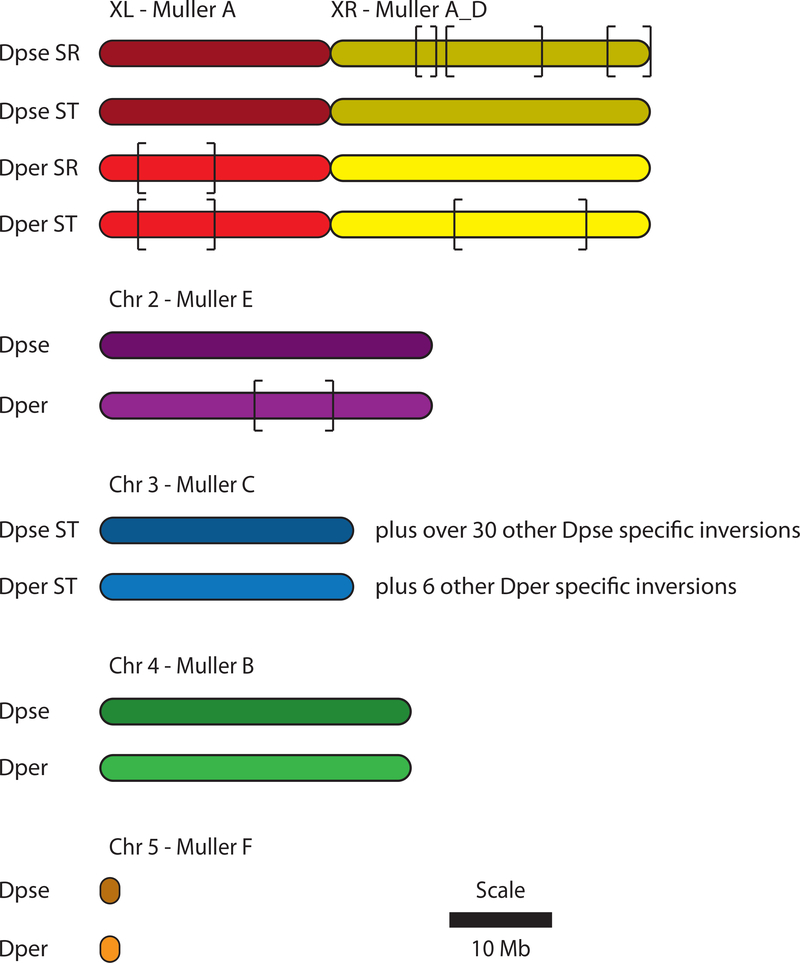
Figure 5 shows the genomic rearrangement differences on the six chromosome arms of D. pseudoobscura and D. persimilis. The X chromosome of the two species is comprised of two arms, XL and XR, which correspond to the A and D Muller (Muller, 1940) elements. XL has a fixed paracentric inversion between the two species, while XR is segregating for two gene arrangements within D. persimilis: Standard and Sex Ratio (SR). D. persimilis males that carry the SR arrangement produce over 95% daughters. The D. persimilis SR chromosome is homosequential and identical by descent with the D. pseudoobscura XR Standard chromosome gene arrangement (Fuller, Leonard, Young, Schaeffer, & Phadnis, 2018). D. pseudoobscura is also segregating for an independent Sex Ratio chromosome, which differs from the Standard arrangement by three non-overlapping inversions. The second chromosome (Muller E) has a fixed inversion difference between the two species. The third chromosome (Muller C) is polymorphic for paracentric inversions in both species, but only one gene arrangement is shared in common, the relatively older Standard (note this naming refers to the third chromosome) arrangement (Figure 6). Meanwhile, chromosomes four (Muller B) and five (Muller F) have the same gene arrangement, i.e., are collinear in both species.
Figure 5.
Organization of the D. pseudoobscura and D. persimilis genomes. The brackets indicate inversion differences between the genomes as well as the inversions that are associated with the Sex ratio chromosome on the right arm of the X (XR) in the two species. The D. pseudoobscura chromosomes are colored slightly darker compared to their D. persimilis counterparts.
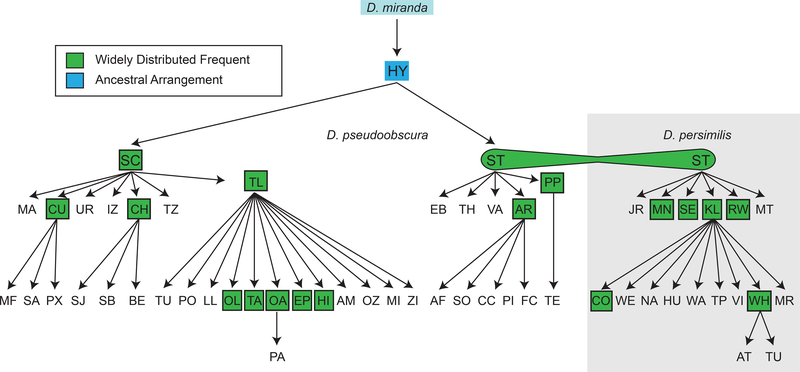
Figure 6.
Phylogeny of the third chromosome gene arrangements of D. pseudoobscura and D. persimilis. The D. pseudoobscura gene arrangements are: AM, Amecameca; AF, American Fork; AR, Arrowhead; BE, Berkeley; CH, Chiricahua; CC, Cochise; CU, Cuernavaca; EB, East Bay; EP, Estes Park; FC, Fort Collins; HI, Hidalgo; HY, Hypothetical; IZ, Iztaccihuatl; LL, Los Lirios; MA, Mammoth; MI, Michoacan; MF, Miraflores; OA, Oaxaca; OL, Olympic; OZ, Ozumba; PA, Patzcuaro; PX, Paxtepec; PP, Pikes Peak; PI, Pinon; PO, Popocatepetl; SA, San Antonio; SJ, San Jacinto; SB, Santa Barbara; SC, Santa Cruz; SO, Sonoita; ST, Standard; TA, Tarasco; TE, Texas; TH, Thomas; TL, Tree Line; TU, Tulancingo; TZ, Tzintzuntzan; UR, Uruapan; VA, Vandeventer; and ZI, Zirahuen (Anderson, Dobzhansky, Pavlovsky, Powell, & Yardley, 1975; Crumpacker & Kastritsis, 1967; Dobzhansky, 1944; Dobzhansky, 1948a; Dobzhansky et al., 1975; Epling & Lower, 1957; Espinoza-Velazquez & Salceda, 1981; Olvera et al., 1979; Olvera et al., 1985; Strickberger & Wills, 1966; Turner & Jeffery, 1977). The D. persimilis gene arrangements are: AT, Atwell Mill; CO, Cowichan; HU, Humboldt; JR, James Reserve; KL, Klamath; MR, Maple Ridge; MT, Mather; MN, Mendocino; NA, Nainaimo; RW, Redwoods; SE, Sequoia; ST, Standard; TP, Thetis Park; TU, Tuolumne; VI, Victoria; WA, Wawona; WE, Weott; and WH, Whitney (Anderson et al., 1975; Dobzhansky, 1944, 1948b; Dobzhansky & Sturtevant, 1938; Moore & Taylor, 1986; Spiess, 1950, 1965). The Hypothetical arrangement was demonstrated to be the ancestral arrangement based on adjacency information of genes that flank inversion breakpoints (see page 3 Supplemental Information, Table S3 and Figures S4-S23, Fuller et al., 2017)
Cytogenetic and molecular genetic data have suggested that D. pseudoobscura and D. persimilis hybridize (Dobzhansky, 1973; Machado & Hey, 2003; Machado, Kliman, Markert, & Hey, 2002; McGaugh & Noor, 2012; Noor, Garfield, Schaeffer, & Machado, 2007; Powell, 1983; Wang & Hey, 1996; Wang, Wakeley, & Hey, 1997). Dobzhansky (1973) observed a single female out of 27,099 examined that was heterozygous for third chromosome gene arrangements specific to either D. pseudoobscura or D. persimilis third chromosome (CH/MN, see Figure 6). Dobzhansky concluded that despite evidence for an interspecific hybrid, reproductive isolation is likely complete because of the absence of widespread sharing of third chromosome gene arrangements between species. Additionally, Powell (1983) observed an unambiguous hybrid rate of 3/30,000 in collected females. It is possible that both of these hybridization rates underestimate the true value, as the species are not equally abundant in sympatric locations, and hence the opportunity for hybridization may vary greatly depending on location. Thus, although hybrids have rarely been observed in nature or in the laboratory, a direct estimate of the level of present gene exchange between the two species is lacking.
Mitochondrial and nuclear genes have shown varying degrees of shared nucleotide polymorphisms or low genetic divergence between D. pseudoobscura and D. persimilis. Shared polymorphisms could result from the persistence of variation from the common ancestral population or they could indicate hybridization or gene flow between the species. Evidence from the study of mitochondrial DNA haplotypes has resulted in conflicting support for the existence of shared haplotypes between D. pseudoobscura and D. persimilis (Hale & Beckenbach, 1985; Powell, 1983; Wang & Hey, 1996). In the nuclear genome, the divergence population genetics approach analyzed nucleotide sequences from multiple loci from the two species and found differentiated as well as undifferentiated loci (Machado & Hey, 2003; Machado et al., 2002; McGaugh & Noor, 2012; Noor et al., 2007; Wang & Hey, 1996; Wang et al., 1997). The differentiated genes map within fixed inversions between the two species and the undifferentiated genes were located in collinear regions of the genome. Furthermore, Noor et al. (2001) showed that genomic regions associated with male hybrid sterility also largely map to the inverted regions of XL and chromosome two. Noor et al. (2001) concluded that inversions play an important role in the speciation process by preventing the spread of incompatibility genes between these two species, while allowing gene flow in collinear regions.
Linkage disequilibrium (LD) based tests of shared derived mutations have been used to identify loci that have experienced gene flow between the two species (Machado et al., 2002), however, it is not clear how these statistics behave when shared polymorphisms are ancestral. Models based on Isolation with Migration (IM) are consistent with gene flow between the two species at loci in collinear regions of the genome, but not within inverted regions (Hey & Nielsen, 2004; Machado & Hey, 2003; Machado et al., 2002). The pattern that emerges is that genes within inverted regions show little evidence for gene flow while collinear regions suggest higher levels of genetic exchange between D. pseudoobscura and D. persimilis (Machado, Haselkorn, & Noor, 2007; Noor et al., 2007). Kulathinal et al. (2009) observed that shared polymorphisms are greater for sympatric (D. pseudoobscura vs. D. persimilis) as opposed to allopatric (D. pseudoobscura pseudoobscura vs. D. pseudoobscura bogotana) species pairs leading the authors to conclude that isolation with gene flow rather than ancestral polymorphism explained the shared polymorphism in collinear regions (Kulathinal et al., 2009). Thus, the IM model proposes that incompatibility genes accumulate throughout the genome, but are selectively removed from collinear regions by natural selection when they are shuffled between two species via gene flow, yet persist within inverted regions due to recombination suppression (Noor, Grams, Bertucci, & Reiland, 2001). However, the ancestral polymorphism hypothesis could not be definitively excluded from the Kulathinal et al. (2009) analysis. (Kulathinal et al., 2009)
When and how did the inversions get fixed between D. pseudoobscura and D. persimilis relative to when gene flow between these two species ceased? There are two contrasting views on this question. On the one hand, genome comparisons have suggested that both of the fixed X chromosome inversion between D. pseudoobscura and D. persimilis originated at the time of divergence from the outgroup species D. miranda, but with the XL inversion fixing prior to the XR inversion (Fuller et al., 2018; McGaugh & Noor, 2012). It is not clear from the IM model, or from invoking post-speciation gene flow, what forces fixed these inversions within isolated populations. Is it that inversions spread initially in a population through a local adaptation mechanism (e.g., see preceding section, Kirkpatrick and Barton (2006)) and subsequently accumulated mutations that generate speciation phenotypes?
On the other hand, a more recent analysis of the SR and Standard XR arrangements of D. persimilis and D. pseudoobscura revealed a pattern of phylogenetic discordance at inversion breakpoints that suggest the inversions arose in the ancestral population (Fuller et al., 2018). Moreover, by estimating sequence divergence between inverted and collinear regions, the evolutionary history of all fixed chromosome differences was reconstructed and provided evidence that, in fact, all inversions distinguishing the two species arose in the ancestral population and were freely segregating for a substantial period of time prior to the beginning of speciation. This model alternatively suggests that inversions may arise through similar mechanisms of local adaptation, yet in the ancestral population. While segregating in this ancestor, the genomic regions spanning the inversion would accumulate genetic divergence by the action of suppressed recombination in heterozygotes. If populations then become allopatric, there may be incomplete lineage sorting of the ancestrally segregating inversions. Here, at the onset of speciation, collinear regions would appear undifferentiated, yet inverted regions would already harbor high levels of divergence and perhaps be predisposed to the formation of incompatible alleles (Fuller et al., 2018). Importantly, these results did not rule out the presence of post-speciation gene flow (Hey & Nielsen, 2004; Kulathinal et al., 2009; Machado et al., 2002; Noor et al., 2007; Noor, Johnson, & Hey, 2000; Wang et al., 1997), but demonstrated that exchange upon secondary contact was not the sole mechanism for observed patterns of increased divergence across fixed inversion differences in D. pseudoobscura and D. persimilis. (Fuller et al., 2017; Fuller et al., 2018; Kirkpatrick & Barton, 2006).
It is intriguing to speculate upon plausible scenarios responsible for the split between D. pseudoobscura and D. persimilis. For example, there is some evidence for an association between altitude and the frequency of the shared Standard (ST) arrangement on the third chromosome in both species in the Sierra Nevada mountains, where ST is found at frequencies of 10–30% (Figure 2)(Dobzhansky, 1944). Furthermore, Dobzhansky had suggested that D. persimilis inhabits higher elevations (>5,000 feet) than D. pseudoobscura (<5,000 feet) (page 14 in Dobzhansky & Epling, 1944). Perhaps the fixed inversion differences observed at present were initially under selection in the ancestral population for adaptation to higher elevation or colder climates. Other phenotypic differences between D. pseudoobscura and D. persimilis have been noted for a number of other ecological factors, including humidity, heat stress, and food availability, among others(Coyne, Bundgaard, & Prout, 1983; Matzkin, Watts, & Markow, 2009). However, without a detailed understanding and characterization of the ecology and life history of current D. pseudoobscura and D. persimilis populations, possible speciation scenarios remain little more than speculation (Dobzhansky, 1948b; Navarro & Barton, 2003).
Concluding remarks
The genus Drosophila, and particularly the sister species of D. pseudoobscura and D. persimilis, is an important model for the study of evolutionary genetics and genomics. Genes have been conserved on chromosomal arms, but gene order has been shuffled extensively both within and between species (Carson, 1992; Dobzhansky, 1944; Levitan, 1982; Throckmorton, 1982; Wasserman, 1982) through the mechanism of chromosomal inversion. While inversions were initially used as a neutral character to discriminate between species, their role as potent drivers of evolutionary change and adaptation in the genus has become increasingly appreciated. If, as we argue, inversions are selected as a result of their recombination suppression in heterozygotes, it is astounding to learn that only 10–20% of this phenotype can be explained by accepted cytogenetic mechanisms. Further research should focus on clarifying the mechanism(s) by which inversion heterozygosity causes recombination suppression, now known to be a form of crossover distribution modification. From empirical, experimental, and theoretical studies ranging from polytene chromosome analysis to high-throughput sequencing, a picture has emerged whereby inversions underlie adaptation across heterogeneous environments in D. pseudoobscura and D. persimilis through their indirect effects as recombination suppressors (Fuller et al., 2018). Further ecological studies are needed to better characterize the heterogeneous environments experienced by both species; for example, the separate challenges faced by a limited dispersal larval phase and an adult phase capable of extensive migration among diverse habitats (Coyne et al., 1982; Coyne, Bryant, & Turelli, 1987). One of the primary reasons that genomic and bioinformatic analysis has been pursued was to obtain valuable clues about the ecology of D. pseudoobscura, which has remained difficult to study. Despite its lengthy history in evolutionary genetics, many basic aspects of its life history remain unknown. Genomic analyses have suggested that D. pseudoobscura may have meaningful interactions with plants in its environment based on detoxication proteins that are enriched among potential selective targets (Fuller et al., 2016, 2017). Adapting polygenic traits for one habitat may be maladaptive for another and evidence suggests that inversions are a potential evolutionary mechanism to both maintain sets of locally adapted loci, while preventing exchange with maladaptive migrant alleles. The ecological diversification of gene arrangements to particular habitats is followed by genetic diversification within inverted regions, which may provide the raw material for incompatibility genes to emerge and isolate populations forming new species. While we call for additional research in this pair of sister species, we also recognize and call attention to the cactophilic species of Drosophila as an additional important model system to understand inversion polymorphism because of the well-known ecology and the extensive inversion polymorphism present (Guillén et al., 2015; Lohse et al., 2015; Wasserman, 1982, 1992).
There is still much work to be done. While inversions present a powerful model for adaptation and speciation, recombination suppression presents challenges for mapping possible Dobzhansky-Muller Incompatibility genes and other loci involved in differences between species and arrangement type (.e.g, cold tolerance, cuticular hydrocarbons, courtship song, among other behavioral traits). It is intriguing to consider that the extent of gene arrangement variation seen within and between Drosophila species is reflective of an ecologically driven process of adaptation to heterogeneous environments and to speculate upon the role of chromosomal inversions in the formation of species.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Emma Berdan, Mohamed A. F. Noor, and two anonymous reviews for comments that improved the manuscript. This work was supported by grants from the National Institute for General Medical Sciences at the National Institutes of Health R01 GM115914 (NP), R01 GM098478 (SWS), a Mario Capecchi endowed assistant professorship (NP), the Pew Biomedical Scholars Program (NP), the Penn State-National Institutes of Health funded Computation, Bioinformatics and Statistics (CBIOS) Predoctoral Training Program to ZLF.
Footnotes
Data Accessibility
Data and Computer Archive: Data was previously published (Fuller et al., 2016, 2017). The computer code is found in a collection within Scholarsphere (https://scholarsphere.psu.edu/collections/h44558f89j) entitled “Computer Code for “How chromosomal rearrangements shape adaptation and speciation in Drosophila” to appear in Molecular Ecology”
References
- Amores A, Catchen J, Nanda I, Warren W, Walter R, Schartl M, & Postlethwait JH (2014). A RAD-Tag Genetic Map for the Platyfish (Xiphophorus maculatus) Reveals Mechanisms of Karyotype Evolution Among Teleost Fish. Genetics, 197(2), 625–U307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WW, Arnold J, Baldwin DG, Beckenbach AT, Brown CJ, Bryant SH,…Moore JA (1991). Four decades of inversion polymorphism in Drosophila pseudoobscura. Proceedings of the National Academy of Sciences USA, 88, 10367–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WW, Dobzhansky T, Pavlovsky O, Powell JR, & Yardley D (1975). Genetics of natural populations. XLII. Three decades of genetic change in Drosophila pseudoobscura. Evolution, 29, 24–36. [DOI] [PubMed] [Google Scholar]
- Aquadro CF, Weaver AL, Schaeffer SW, & Anderson WW (1991). Molecular evolution of inversions in Drosophila pseudoobscura : The amylase gene region. Proceedings of the National Academy of Sciences USA, 88, 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, & Hawley RS (2005). Drosophila: A Laboratory Handbook (2nd ed.). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Ashburner M, & Lemeunier F (1976). Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). I. Inversion polymorphisms in Drosophila melanogaster and Drosophila simulans Proceedings of the Royal Society of London B, 193, 137–157. [DOI] [PubMed] [Google Scholar]
- Balanya J, Huey RB, Gilchrist GW, & Serra L (2009). The chromosomal polymorphism of Drosophila subobscura: a microevolutionary weapon to monitor global change. Heredity, 103(5), 364–367. [DOI] [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo S, Xu M, Smith TF, & Gelbart WM (2008). Chromosomal rearrangement inferred from comparisons of twelve Drosophila genomes. Genetics, 179, 1657–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Cattani MV, Kingan SB, Landeen EL, & Presgraves DC (2018). Molecular evolution at a meiosis gene mediates species differences in the rate and patterning of recombination. Current Biology, 28(8), 1289–1295.e1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres M, Barbadilla A, & Ruiz A (1999). Recombination rate predicts inversion size in Diptera. Genetics, 153(1), 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres M, Ranz JM, Barbadilla A, Long M, & Ruiz A (1999). Generation of a widespread Drosophila inversion by a transposable element. Science, 285(5426), 415–418. [DOI] [PubMed] [Google Scholar]
- Calvete O, Gonzalez J, Betran E, & Ruiz A (2012). Segmental duplication, microinversion, and gene loss associated with a complex inversion breakpoint region in Drosophila. Mol Biol Evol, 29(7), 1875–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson HL (1946). The selective elimination of inversion dicentric chromatids during meiosis in the eggs of Sciara impatiens. Genetics, 31(1), 95–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson HL (1953). The effects of inversions on crossing-over in Drosophila robusta. Genetics, 38(2), 168–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson HL (1958). The Population Genetics of Drosophila robusta In Demerec M (Ed.), Advances in Genetics (Vol. 9, pp. 1–40): Academic Press. [DOI] [PubMed] [Google Scholar]
- Carson HL (1992). Inversions in Hawaiian Drosophila In Krimbas C & Powell JR (Eds.), Drosophila Inversion Polymorphism (pp. 407–439). Boca Raton, FL: CRC Press. [Google Scholar]
- Charlesworth B, & Barton NH (1996). Recombination load associated with selection for increased recombination. Genetical Research, 67(1), 27–41. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, & Charlesworth D (1973). Selection of new inversion in multi-locus genetic systems. Genetical Research, 21(2), 167–183. [Google Scholar]
- Cheng C, White BJ, Kamdem C, Mockaitis K, Costantini C, Hahn MW, & Besansky NJ (2012). Ecological genomics of Anopheles gambiae along a latitudinal cline: A population-resequencing approach. Genetics, 190(4), 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick A (1973). Gene conversion and transfer of genetic information within the inverted region of inversion heterozygotes. Genetics, 75(1), 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogni R, Kuczynski K, Koury S, Lavington E, Behrman EL, O’Brien KR,…Eanes WF (2017). On the Long-term Stability of Clines in Some Metabolic Genes in Drosophila melanogaster. Sci Rep, 7, 42766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Ratnappan R, & Bailin S (2012). The many landscapes of recombination in Drosophila melanogaster. PLoS Genet, 8(10), e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB (2016). Selection on inversion breakpoints favors proximity to pairing sensitive sites in Drosophila melanogaster. Genetics, 204(1), 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, & Hartl DL (2012). Population Genomics of Inversion Polymorphisms in Drosophila melanogaster. PLoS Genet, 8(12), e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan JM, & Willis JH Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex. Molecular Ecology, 0(0). [DOI] [PubMed] [Google Scholar]
- Coyne JA, Aulard S, & Berry A (1991). Lack of underdominance in a naturally occurring pericentric inversion in Drosophila melanogaster and its implications for chromosome evolution. Genetics, 129(3), 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Boussy IA, Prout T, Bryant SH, Jones JS, & Moore JA (1982). Long-distance migration of Drosophila. American Naturalist, 119, 589–595. [Google Scholar]
- Coyne JA, Bryant SH, & Turelli M (1987). Long-distance migration of Drosophila. 2. Presence in desolate sites and dispersal near a desert oasis. American Naturalist, 129, 847–861. [Google Scholar]
- Coyne JA, Bundgaard J, & Prout T (1983). Geographic variation of tolerance to environmental stress in Drosophila pseudoobscura. American Naturalist, 122(4), 474–488. [Google Scholar]
- Coyne JA, Meyers W, Crittenden AP, & Sniegowski P (1993). The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics, 134, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown KN, Miller DE, Sekelsky J, & Hawley RS (2018). Local Inversion Heterozygosity Alters Recombination throughout the Genome. Current Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crumpacker DW, & Kastritsis CD (1967). A new gene arrangement in the third chromosome of Drosophila pseudoobscura. Journal of Heredity, 58, 2–6. [PubMed] [Google Scholar]
- Dobzhansky T (1936). Studies of hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics, 21, 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1943). Genetics of natural populations. IX. Temporal changes in the composition of populations of Drosophila pseudoobscura. Genetics, 28, 162–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1944). Chromosomal races in Drosophila pseudoobscura and Drosophila persimilis. Carnegie Inst. Washington Publ, 554, 47–144. [Google Scholar]
- Dobzhansky T (1948a). Chromosomal variation in populations of Drosophila pseudoobscura which inhabit northern Mexico. The American Naturalist, 82(803), 97–106. [Google Scholar]
- Dobzhansky T (1948b). Genetics of natural populations. XVI. Altitudinal and seasonal changes in certain populations of Drosophila pseudoobscura and Drosophila persimilis. Genetics, 33, 158–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1948c). Genetics of natural populations. XVIII. Experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics, 33, 588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1950). The genetics of natural populations. XIX. Origin of heterosis through natural selection in populations of Drosophila pseudoobscura. Genetics, 35, 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1955). A review of some fundamental concepts and problems of population genetics. Cold Spring Harbor Symposia on Quantitative Biology, 20, 1–15. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T (1973). Is there gene exchange between Drosophila pseudoobscura and Drosophila persimilis in their natural habitats? The American Naturalist, 107(954), 312–314. [Google Scholar]
- Dobzhansky T, & Epling C (1944). Taxonomy, geographic distribution, and ecology of Drosophila pseudoobscura and its relatives In Dobzhansky T & Epling C (Eds.), Contributions to the Genetics, Taxonomy, and Ecology of Drosophila pseudoobscura and its Relatives (pp. 1–46). Baltimore, MD: The Lord Baltimore Press. [Google Scholar]
- Dobzhansky T, & Epling C (1948). The suppression of crossing over in inversion heterozygotes of Drosophila pseudoobscura. Proceedings of the National Academy of Sciences USA, 34, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Felix R, Guzman J, Levine L, Olvera O, Powell JR,…Salceda VM (1975). Population genetics of Mexican Drosophila. I. Chromosomal variation in natural populations of Drosophila pseudoobscura from Central Mexico. Journal of Heredity, 66(4), 203–206. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T, & Queal ML (1938). Genetics of natural populations. I. Chromosome variation in populations of Drosophila pseudoobscura inhabiting isolated mountain ranges. Genetics, 23, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, & Sturtevant AH (1938). Inversions in the chromosomes of Drosophila pseudoobscura. Genetics, 23, 28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, & Wright S (1941). Genetics of natural populations. V. Relations between mutation rate and accumulation of lethals in populations of Drosophila pseudoobscura. Genetics, 26, 23–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, & Wright S (1943). Genetics of natural populations. X. Dispersion rates in Drosophila pseudoobscura. Genetics, 28, 304–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubinin NP, Sokolov NN, & Tiniakov GG (1936). Occurrence and Distribution of Chromosome Aberrations in Nature (Diptera). Nature, 137, 1035. [Google Scholar]
- Endler JA (1977). Geographic Variation, Speciation, and Clines. Princeton, NJ: Princeton University Press. [PubMed] [Google Scholar]
- Engelbrecht A, Taylor PJ, Daniels SR, & Rambau RV (2011). Chromosomal polymorphisms in African Vlei Rats, Otomys irroratus (Muridae: Otomyini), detected by banding techniques and chromosome painting: inversions, centromeric shifts and diploid number variation. Cytogenetic and Genome Research, 133(1), 8–15. [DOI] [PubMed] [Google Scholar]
- Epling C, & Lower WR (1957). Changes in an inversion system during a hundred generations. Evolution, 11(2), 248–256. [Google Scholar]
- Espinoza-Velazquez J, & Salceda VM (1981). Inversion polymorphism of a Drosophila pseudoobscura population from Saltillo, Coahuila, Mexico. The Southwestern Naturalist, 25(4), 479–483. [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF, & Hill JP (2013). Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (Monkey flowers). Evolution, 67(9), 2547–2560. [DOI] [PubMed] [Google Scholar]
- Frolowa SL, & Astaurow BL (1929). Die chromosomengarnitur als systematisches merkmal (eine vergleichende untersuchung der russischen und amerikanischen Drosophila obscura Fall.). Zeitschrift fur Zellforschung und Mikroskopische Anatomie, 10, 201–213. [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, & Schaeffer SW (2016). Genomics of natural populations: How differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics, 204, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, & Schaeffer SW (2017). Genomics of natural populations: Evolutionary forces that establish and maintain gene arrangements in Drosophila pseudoobscura. Molecular Ecology, 26, 6539–6562. [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Leonard CJ, Young RE, Schaeffer SW, & Phadnis N (2018). Ancestral polymorphisms explain the role of chromosomal inversions in speciation. Public Library of Science Genetics, 14(7), e1007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gethmann RC (1988). Crossing Over in Males of Higher Diptera (Brachycera). Journal of Heredity, 79(5), 344–350. [DOI] [PubMed] [Google Scholar]
- Gong WJ, McKim KS, & Hawley RS (2005). All Paired Up with No Place to Go: Pairing, Synapsis, and DSB Formation in a Balancer Heterozygote. Public Library of Science Genetics, 1(5), e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell RF (1962). A new model for secondary nondisjunction: The role of distributive pairing. Genetics, 47(12), 1737–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Rousset F, & Kirkpatrick M (2012). Coalescent patterns for chromosomal inversions in divergent populations. Philosophical Transactions of the Royal Society B: Biological Sciences, 367(1587), 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén Y, Rius N, Delprat A, Williford A, Muyas F, Puig M,…Ruiz A (2015). Genomics of ecological adaptation in cactophilic Drosophila. Genome Biology and Evolution, 7, 349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale LR, & Beckenbach AT (1985). Mitochondrial DNA variation in Drosophila pseudoobscura and related species in Pacific northwest populations. Can J Genet Cytol, 27(3), 357–364. [DOI] [PubMed] [Google Scholar]
- Hawley RS (1980). Chromosome sites necessary for normal levels of meiotic recombination. 1. Existence and mapping of the sites. Genetics, 94, 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, & Nielsen R (2004). Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics, 167(2), 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill H, & Golic KG (2015). Preferential breakpoints in the recovery of broken dicentric chromosomes in Drosophila melanogaster. Genetics, 201(2), 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM, & Weeks AR (2004). Chromosomal inversion polymorphisms and adaptation. Trends in Ecology and Evolution, 19(9), 482–488. [DOI] [PubMed] [Google Scholar]
- Hou C, Li L, Qin, Zhaohui S, & Corces, Victor G (2012). Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Molecular Cell, 48(3), 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner AF (1924). Maturation and fertilization in Drosophila melanogaster. Journal of Morphology, 39, 249–265. [Google Scholar]
- Hunter CM, Robinson MC, Aylor DL, & Singh ND (2016). Genetic background, maternal age, and interaction effects mediate rates of crossing over in Drosophila melanogaster females. G3: Genes|Genomes|Genetics, 6(5), 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J,…Kingsley DM (2012). The genomic basis of adaptive evolution in threespine sticklebacks. Nature, 484(7392), 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M, Fabian DK, Goudet J, & Flatt T (2016). Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Molecular Biology and Evolution, 33(5), 1317–1336. [DOI] [PubMed] [Google Scholar]
- Kennington W, & Hoffmann A (2013). Patterns of genetic variation across inversions: geographic variation in the In(2L)t inversion in populations of Drosophila melanogaster from eastern Australia. BMC Evolutionary Biology, 13(1), 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-W, Bennison C, Hemmings N, Brookes L, Hurley LL, Griffith SC,…Slate J (2017). A sex-linked supergene controls sperm morphology and swimming speed in a songbird. Nature Ecology & Evolution, 1(8), 1168–1176. [DOI] [PubMed] [Google Scholar]
- Kimura M (1968). Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genetical Research, 11, 247–269. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, & Barton N (2006). Chromosome inversions, local adaptation and speciation. Genetics, 173(1), 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb WR (1982). Chromosome inversion polymorphisms in Drosophila melanogaster II. Geographic clines and climatic associations in Australasia, North America and Asia. Genetica, 58(3), 213–221. [Google Scholar]
- Koehler KE, Millie EA, Cherry JP, Burgoyne PS, Evans EP, Hunt PA, & Hassold TJ (2002). Sex-specific differences in meiotic chromosome segregation revealed by dicentric bridge resolution in mice. Genetics, 162(3), 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korunes KL, & Noor MAF (2017). Gene conversion and linkage: effects on genome evolution and speciation. Molecular Ecology, 26(1), 351–364. [DOI] [PubMed] [Google Scholar]
- Korunes KL, & Noor MAF (2019). Pervasive gene conversion in chromosomal inversion heterozygotes. Molecular Ecology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury SA (2017). Studies in experimental population genetics of drosophila melanogaster. (Ph.D.), State University of New York at Stony Brook, Stony Brook, NY. [Google Scholar]
- Kovacevic M, & Schaeffer SW (2000). Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics, 156, 155–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas CB (1992). The inversion polymorphism of Drosophila subobscura In Krimbas CB & Powell JR (Eds.), Drosophila Inversion Polymorphism (pp. 127–220). Boca Raton: CRC Press. [Google Scholar]
- Krimbas CB, & Powell JR (1992a). Drosophila Inversion Polymorphism. Ann Arbor, MI: CRC Press. [Google Scholar]
- Krimbas CB, & Powell JR (1992b). Introduction In Krimbas CB & Powell JR (Eds.), Drosophila Inversion Polymorphism (pp. 1–52). Ann Arbor, MI: CRC Press. [Google Scholar]
- Kulathinal RJ, Stevison LS, & Noor MAF (2009). The genomics of speciation in Drosophila: Diversity, divergence, and introgression estimated using low-coverage genome sequencing. Public Library of Science Genetics, 5(7), e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper C, Stocks M, Risse JE, dos Remedios N, Farrell LL, McRae SB,…Burke T (2015). A supergene determines highly divergent male reproductive morphs in the ruff. Nature Genetics, 48, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancefield DE (1929). A genetic study of crosses of two races or physiological species of Drosophila obscura. Zeitschrift für Inductive Abstammungs-und Vererbungslehre, 52, 287–317. [Google Scholar]
- Lande R (1984). The expected fixation rate of chromosomal inversions. Evolution, 38, 743–752. [DOI] [PubMed] [Google Scholar]
- Lande R (1985). The fixation of chromosomal rearrangements in a subdivided population with local extinction and colonization. Heredity, 54, 323. [DOI] [PubMed] [Google Scholar]
- Lavington E, & Kern AD (2017). The Effect of Common Inversion Polymorphisms In(2L)t and In(3R)Mo on Patterns of Transcriptional Variation in Drosophila melanogaster. G3: Genes|Genomes|Genetics, 7(11), 3659–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul Y, Whibley A, Chouteau M, Prunier F, Llaurens V, & Joron M (2014). Evolution of dominance mechanisms at a butterfly mimicry supergene. Nature Communications, 5, 5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier F, & Aulard S (1992). Inversion polymorphism in Drosophila melanogaster In Krimbas CB & Powell JR (Eds.), Drosophila Inversion Polymorphism (pp. 339–405). Boca Raton: CRC Press. [Google Scholar]
- Levene H (1953). Genetic equilibrium when more than one ecological niche is available. American Naturalist, 87, 311–313. [Google Scholar]
- Levine RP (1956). Crossing over and inversions in coadapted systems. American Naturalist, 90, 41–45. [Google Scholar]
- Levine RP, & Levine EE (1954). The genotypic control of crossing over in Drosophila pseudoobscura. Genetics, 39, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP, & Levine EE (1955). Variable crossing over arising in different strains of Drosophila pseudoobscura. Genetics, 40, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R, & MacArthur R (1966). The maintenance of genetic polymorphism in a spatially heterogeneous environment: Variations on a theme by Howard Levene. American Naturalist, 100, 585–589. [Google Scholar]
- Levitan M (1958). Non-Random Associations of Inversions. Cold Spring Harbor Symposia on Quantitative Biology, 23, 251–268. [DOI] [PubMed] [Google Scholar]
- Levitan M (1992). Chromosomal variation in Drosophila robusta Sturtevant In Krimbas CB (Ed.), Drosophila Inversion Polymorphism (pp. 221–338). Boca Raton: CRC Press. [Google Scholar]
- Levitan M (Ed.) (1982). The robusta and melanica groups (Vol. 3b). New York, NY: Academic Press. [Google Scholar]
- Lewontin RC (1974). The Genetic Basis of Evolutionary Change (1 ed.). New York: Columbia University Press. [Google Scholar]
- Lewontin RC, Moore JA, Provine WB, & Wallace B (Eds.). (1981). Dobzhansky’s Genetics of Natural Populations I-XLIII. New York: Columbia University Press. [Google Scholar]
- Lim JK, & Simmons MJ (1994). Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays, 16, 269–275. [DOI] [PubMed] [Google Scholar]
- Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L,…Price TA (2016). The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol, 31(4), 315–326. [DOI] [PubMed] [Google Scholar]
- Lobeck AK (Cartographer). (1948). Physiographic diagram of North America [Google Scholar]
- Lohse K, Clarke M, Ritchie MG, & Etges WJ (2015). Genome-wide tests for introgression between cactophilic Drosophila implicate a role of inversions during speciation. Evolution, 69(5), 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez V, Barinova N, Onishi M, Pobiega S, Pringle JR, Dubrana K, & Marcand S (2015). Cytokinesis breaks dicentric chromosomes preferentially at pericentromeric regions and telomere fusions. Genes & Development, 29(3), 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, & Willis JH (2010). A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. PLOS Biology, 8(9), e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, & Suzuki DT (1968). The interchromosomal control of recombination (Vol. 2, pp. 53–86). [Google Scholar]
- Machado CA, Haselkorn TS, & Noor MA (2007). Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics, 175(3), 1289–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, & Hey J (2003). The causes of phylogenetic conflict in a classic Drosophila species group. Proc Biol Sci, 270(1520), 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, & Hey J (2002). Inferring the history of speciation from multilocus DNA sequence data: The case of Drosophila pseudoobscura and close relatives. Molecular Biology and Evolution, 19(4), 472–488. [DOI] [PubMed] [Google Scholar]
- Madan K, Seabright M, Lindenbaum RH, & Bobrow M (1984). Paracentric inversions in man. Journal of Medical Genetics, 21(6), 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJ, Zhu CT, & Eanes WF (2005). The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics, 170(3), 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Watts TD, & Markow TA (2009). Evolution of stress resistance in Drosophila: interspecific variation in tolerance to desiccation and starvation. Functional Ecology, 23(3), 521–527. [Google Scholar]
- Maynard Smith J, & Haigh J (1974). The hitch-hiking effect of a favorable gene. Genetical Research, 23, 23–35. [PubMed] [Google Scholar]
- McClintock B (1931). The Order of the Genes C, Sh and Wx in Zea Mays with Reference to a Cytologically Known Point in the Chromosome. Proceedings of the National Academy of Sciences, 17(8), 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1941). The stability of broken ends of chromosomes in Zea mays. Genetics, 26(2), 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rice DP, & Desai MM (2016). Sex speeds adaptation by altering the dynamics of molecular evolution. Nature, 531(7593), 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, & Noor MAF (2012). Genomic impacts of chromosomal inversions in parapatric Drosophila species. Philosophical Transactions of the Royal Society B: Biological Sciences, 367(1587), 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, & Speed TP (1995). Modeling interference in genetic recombination. Genetics, 139(2), 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, & Schaeffer SW (2007). Meiotic transmission of Drosophila pseudoobscura chromosomal arrangements. Public Library of Science One, 2, e530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BC, & Taylor CE (1986). Drosophila of southern California. III Gene arrangements of Drosophila persimilis. Journal of Heredity, 77, 313–323. [Google Scholar]
- Muller HJ (1940). Bearings of the ‘Drosophila’ work on systematics In Huxley J (Ed.), The New Systematics (pp. 185–268). Oxford: Clarendon Press. [Google Scholar]
- Navarro A, Barbadilla A, & Ruiz A (2000). Effect of inversion polymorphism on the neutral nucleotide variability of linked chromosomal regions in Drosophila. Genetics, 155, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, & Barton NH (2003). Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution, 57(3), 447–459. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, & Ruiz A (1997). Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics, 146, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, & Ruiz A (1997). On the Fertility Effects of Pericentric Inversions. Genetics, 147(2), 931–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE,…Besansky NJ (2015). Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science, 347(6217). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kojima K-I, & Schaffer HE (1967). Frequency changes of new inversions in populations under mutation-selection equilibria. Genetics, 57, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Garfield DA, Schaeffer SW, & Machado CA (2007). Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics, 177, 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Almendarez Y, Reiland J, & Smith KR (2001). The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution, 55(3), 512–521. [DOI] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, & Reiland J (2001). Chromosomal inversions and the reproductive isolation of species. Proceedings of the National Academy of Sciences USA, 98(21), 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Johnson NA, & Hey J (2000). Gene flow between Drosophila pseudoobscura and D. persimilis. Evolution, 54(6), 2174–2175; discussion 2176–2177. [DOI] [PubMed] [Google Scholar]
- Novitski E (1946). Chromosomal variation in Drosophila athabasca. Genetics, 31, 508–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski E (1951). Non-random disjunction in Drosophila. Genetics, 36, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski E, & Braver G (1954). An analysis of crossing over within a heterozygous inversion in Drosophila melanogaster. Genetics, 39, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, & Kojima KI (1968). Survival probabilities of new inversions in large populations. Biometrics, 24(3), 501–516. [PubMed] [Google Scholar]
- Olvera O, Powell JR, De La Rosa ME, Salceda VM, Gaso MI, Guzman J,…Levine L (1979). Population genetics of Mexican Drosophila VI. Cytogenetic aspects of the inversion polymorphism in Drosophila pseudoobscura. Evolution, 33, 381–395. [DOI] [PubMed] [Google Scholar]
- Olvera O, Rockwell RF, de la Rosa ME, Gaso MI, Gonzalez F, Guzman J, & Levine L (1985). Chromosomal and behavioral studies of Mexican Drosophila. III. Inversion polymorphism of D. pseudoobscura. J Hered, 76(4), 258–262. [DOI] [PubMed] [Google Scholar]
- Orr HA (1987). Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and D. persimilis. Genetics, 116(4), 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA (1989). Genetics of sterility in hybrids between two subspecies of Drosophila. Evolution, 43, 180–189. [DOI] [PubMed] [Google Scholar]
- Painter TS (1934). A new method for the study of chromosomal aberrations and the plotting of chromosomal maps in Drosophila melanogaster. Genetics, 19, 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovsky O, & Dobzhansky T (1966). Genetics of natural populations. XXXVII. The coadapted system of chromosomal variants in a population of Drosophila pseudoobscura. Genetics, 53, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N (2011). Genetic Architecture of Male Sterility and Segregation Distortion in Drosophila pseudoobscura Bogota-USA Hybrids. Genetics, 189(3), 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, & Orr HA (2009). A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science, 323(5912), 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Braun DT, & Lack JB (2017). Parallel Evolution of Cold Tolerance within Drosophila melanogaster. Mol Biol Evol, 34(2), 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR (1983). Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: Evidence from Drosophila. Proceedings of the National Academy of Sciences USA, 80, 492–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine WB (1986). Sewall Wright and Evolutionary Biology. Chicago, IL: University of Chicago Press. [Google Scholar]
- Puerma E, Orengo DJ, & Aguadé M (2016). Multiple and diverse structural changes affect the breakpoint regions of polymorphic inversions across the Drosophila genus. Sci Rep, 6, 36248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Caceres M, & Ruiz A (2004). Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc Natl Acad Sci U S A, 101(24), 9013–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhäjärvi T, Hufford MB, Mezmouk S, & Ross-Ibarra J (2013). Complex patterns of local adaptation in Teosinte. Genome Biology and Evolution, 5(9), 1594–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane RV, Rako L, Kapun M, Lee SF, & Hoffmann AA (2015). Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Molecular Ecology, 24(10), 2423–2432. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, Grotthuss M. v., Hillier LW, Roote J,…Bergman CM (2007). Principles of genome evolution in the Drosophila melanogaster species group Public Library of Science Biology, 5, 1366–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R,…Gibbs RA (2005). Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene and cis-element evolution. Genome Research, 15, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Hallas ME, & Lewontin RC (1989). Distinguishing the forces controlling genetic variation at the Xdh locus in Drosophila pseudoobscura. Genetics, 123, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P (1962). Interchromosomal Effects and the Relation between Crossing-over and Nondisjunction. Genetics, 47(12), 1691–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PA (1970). Screening for x-ray-induced crossover suppressors in Drosophila melanogaster: prevalence and effectiveness of translocations. Genetics, 65(3), 429–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PA (1972). Differences in synaptic affinity of chromosome arms of Drosophila melanogaster revealed by differential sensitivity to translocation heterozygosity. Genetics, 71(3), 401–415. [DOI] [PubMed] [Google Scholar]
- Said I, Byrne A, Serrano V, Cardeno C, Vollmers C, & Corbett-Detig R (2018). Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions. Proceedings of the National Academy of Sciences, 115, 5492–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Flores A, Peñaloza F, Carpinteyro-Ponce J, Nazario-Yepiz N, Abreu-Goodger C, Machado CA, & Markow TA (2016). Genome Evolution in Three Species of Cactophilic Drosophila. G3: Genes|Genomes|Genetics, 6(10), 3097–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, & Novitski E (1957). Meiotic drive as an evolutionary force. American Naturalist, 91, 105–110. [Google Scholar]
- Schaeffer SW (1995). Population genetics in Drosophila pseudoobscura : A synthesis based on nucleotide sequence data for the Adh gene In Levine L (Ed.), Genetics of Natural Populations: The Continuing Importance of Theodosius Dobzhansky (pp. 329–352). New York, NY: Columbia University Press. [Google Scholar]
- Schaeffer SW (2008). Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution, 62, 3082–3099. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, & Anderson WW (2005). Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics, 171, 1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, Goetting-Minesky P, Kovacevic M, Peoples J, Graybill JL, Miller JM,…Anderson WW (2003). Evolutionary genomics of inversions in Drosophila pseudoobscura: Evidence for epistasis. Proceedings of the National Academy of Sciences USA, 100, 8319–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, & Miller EL (1992). Estimates of gene flow in Drosophila pseudoobscura determined from nucleotide sequence analysis of the alcohol dehydrogenase region. Genetics, 132, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherizen D, Jang JK, Bhagat R, Kato N, & McKim KS (2005). Meiotic recombination in Drosophila females depends on chromosome continuity between genetically defined boundaries. Genetics, 169, 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões P, & Pascual M (2018). Patterns of geographic variation of thermal adapted candidate genes in Drosophila subobscura sex chromosome arrangements. BMC Evolutionary Biology, 18(1), 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair‐Waters M, Bradbury IR, Morris CJ, Lien S, Kent MP, & Bentzen P (2018). Ancient chromosomal rearrangement associated with local adaptation of a postglacially colonized population of Atlantic Cod in the northwest Atlantic. Molecular Ecology, 27(2), 339–351. [DOI] [PubMed] [Google Scholar]
- Singh BN, & Singh AK (1987). The effects of heterozygous inversions on crossing-over in Drosophila ananassae. Genome, 29(5), 802–805. [Google Scholar]
- Sperlich D, & Pfriem P (1986). Chromosomal polymorphism in natural and experimental populations In Ashburner M, Carson HL, & Thomson JN (Eds.), The Genetics and Biology of Drosophila 3e (Vol. 3e, pp. 257–309). New York, NY: Academic Press. [Google Scholar]
- Spiess EB (1950). Experimental Populations of Drosophila persimilis from an Altitudinal Transect of the Sierra Nevada. Evolution, 4(1), 14–33. [Google Scholar]
- Spiess EB (1965). A Discovery and Rediscovery of Third Chromosome Arrangements in Drosophila persimilis. The American Naturalist, 99(908), 423–425. [Google Scholar]
- Spurway H, & Philip U (1952). Genetics and cytology of Drosophila subobscura. VII. Abnormal gene arrangements in element A. Journal of Genetics, 51, 198–215. [Google Scholar]
- Stalker HD (1980). Chromosome Studies in Wild Populations of Drosophila melanogaster. II. Relationship of Inversion Frequencies to Latitude, Season, Wing-Loading and Flight Activity. Genetics, 95(1), 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J,…Stefansson K (2005). A common inversion under selection in Europeans. Nature Genetics, 37(2), 129–137. [DOI] [PubMed] [Google Scholar]
- Strickberger MW, & Wills CJ (1966). Monthly frequency changes of Drosophila pseudoobscura third chromosome gene arrangements in a California locality. Evolution, 20(4), 592–602. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH (1917). Genetic factors affecting the strength of genetic linkage in Drosophila. Proceedings of the National Academy of Sciences USA, 3, 555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH (1920). Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics, 5, 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH (1921). A case of rearrangement of genes in Drosophila. Proceedings of the National Academy of Sciences USA, 7, 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, & Beadle GW (1936). The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics, 21, 544–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, & Dobzhansky T (1936). Inversions in the third chromosome of wild races of Drosophila pseudoobscura, and their use in the study of the history of the species. Proceedings of the National Academy of Sciences USA, 22, 448–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, & Novitski E (1941). The homologies of the chromosome elements in the genus Drosophila. Genetics, 26, 517–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, & Plunkett CR (1926). Sequence of corresponding third-chromosome genes in Drosophila melanogaster and Drosophila simulans. Biological Bulletin, Wood’s Hole, 50, 56–60. [Google Scholar]
- Sturtevant AH, & Tan CC (1937). The comparative genetics of Drosophila pseudoobscura and D. melanogaster. Journal of Genetics, 34, 415–432. [Google Scholar]
- Throckmorton LH (1982). The virilis species group In Ashburner M, Carson HL, & Thompson JN (Eds.), The Genetics and Biology of Drosophila (Vol. 3b, pp. 227–296). New York, NY: Academic Press. [Google Scholar]
- True JR, Mercer JM, & Laurie CC (1996). Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics, 142, 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner ME, & Jeffery DE (1977). Drosophila pseudoobscura of the Great Basin I. A new inversion in chromosome 3. Journal of Heredity, 68(4), 256–257. [Google Scholar]
- van Valen L, & Levins R (1968). The origins of inversion polymorphisms. American Naturalist, 102, 5–24. [Google Scholar]
- Vann E (1966). The Fate of X-Ray Induced Chromosomal Rearrangements Introduced into Laboratory Populations of Drosophila melanogaster. The American Naturalist, 100(914), 425–449. [Google Scholar]
- Wallace AG, Detweiler D, & Schaeffer SW (2011). Evolutionary history of the third chromosome gene arrangements of Drosophila pseudoobscura inferred from inversion breakpoints. Molecular Biology and Evolution, 28, 2219–2229. [DOI] [PubMed] [Google Scholar]
- Wallberg A, Schöning C, Webster MT, & Hasselmann M (2017). Two extended haplotype blocks are associated with adaptation to high altitude habitats in East African honey bees. Public Library of Science Genetics, 13(5), e1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, & Hey J (1996). The speciation history of Drosophila pseudoobscura and close relatives: inferences from DNA sequence variation at the period locus. Genetics, 144(3), 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Wakeley J, & Hey J (1997). Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics, 147, 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman M (1960). Cytological and phylogenetic relationships in the repleta group of the genus Drosophila. Proceedings of the National Academy of Sciences USA, 46, 842–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman M (1982). Evolution of the repleta group In Ashburner M, Carson HL, & Thompson JN (Eds.), The Genetics and Biology of Drosophila 3b (Vol. 3b, pp. 61–139). New York: Academic Press. [Google Scholar]
- Wasserman M (1992). Cytological evolution of the Drosophila repleta species group In Krimbas CB (Ed.), Drosophila Inversion Polymorphism (pp. 455–552). Boca Raton: CRC Press. [Google Scholar]
- Wesley CS, & Eanes WF (1994). Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proceedings of the National Academy of Sciences USA, 91(8), 3132–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S (1941). On the Probability of Fixation of Reciprocal Translocations. The American Naturalist, 75(761), 513–522. [Google Scholar]
- Wright S, & Dobzhansky T (1946). Genetics of natural populations. XII. Experimental reproduction of some of the changes caused by natural selection in certain populations of Drosophila pseudoobscura. Genetics, 31, 125–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Dobzhansky T, & Hovanitz W (1942). Genetics of natural populations. VII. The allelism of lethals in the third chromosome of Drosophila pseudoobscura. Genetics, 27, 363–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.