Abstract
Chromosomal inversions shape recombination landscapes, and species differing by inversions may exhibit reduced gene flow in these regions of the genome. Though single-crossovers within inversions are not usually recovered from inversion heterozygotes, the recombination-barrier imposed by inversions is nuanced by non-crossover gene conversion. Here, we provide a genome-wide empirical analysis of gene conversion rates both within species and in species hybrids. We estimate that gene conversion occurs at a rate of 1 × 10−5 to 2.5 × 10−5 converted sites per bp per generation in experimental crosses within Drosophila pseudoobscura and between D. pseudoobscura and its naturally-hybridizing sister species D. persimilis. This analysis is the first direct empirical assessment of gene conversion rates within inversions of a species hybrid. Our data show that gene conversion rates in interspecies hybrids are at least as high as within-species estimates of gene conversion rates, and gene conversion occurs regularly within and around inverted regions of species hybrids, even near inversion breakpoints. We also found that several gene conversion events appeared to be mitotic rather than meiotic in origin. Finally, we observed that gene conversion rates are higher in regions of lower local sequence divergence, yet our observed gene conversion rates in more divergent inverted regions were at least as high as in less divergent collinear regions. Given our observed high rates of gene conversion despite the sequence differentiation between species, especially in inverted regions, gene conversion has the potential to reduce the efficacy of inversions as barriers to recombination over evolutionary time.
Keywords: gene conversion, recombination, speciation, inversions
INTRODUCTION
Chromosomal inversions influence meiotic recombination and can shape how populations evolve and how new species arise. In heterokaryotypes, inversions can dramatically alter crossing over both by reducing the recovery of crossover products in inverted regions and by increasing crossover rates on other chromosomes through the interchromosomal effect (Schultz & Redfield 1951). Reduction in crossing over in heterokaryotypes can help maintain haplotypes in linkage-disequilibrium (LD), which affects local adaptation, speciation, and the persistence of hybridizing species (e.g., Noor et al. 2001b; Hoffmann & Rieseberg 2008; Fishman et al. 2013; Kirkpatrick & Barrett 2015).
Inversions are often associated with locally adapted alleles in natural populations, and genetic differences between hybridizing populations may be more likely to persist when recombination barriers help maintain LD (see reviews in Hoffmann & Rieseberg 2008; Kirkpatrick 2017). While many studies document the effects of inversions on crossing over, less is known about effects of inversions on non-crossover gene conversion. Non-crossover gene conversion is a distinct form of recombination with its own unique evolutionary implications (reviewed in Korunes & Noor 2017). Understanding the prevalence, distribution, and size of gene conversion events, particularly in heterokaryotypes, is necessary for understanding how recombination shuffles alleles and affects evolutionary processes such as gene flow.
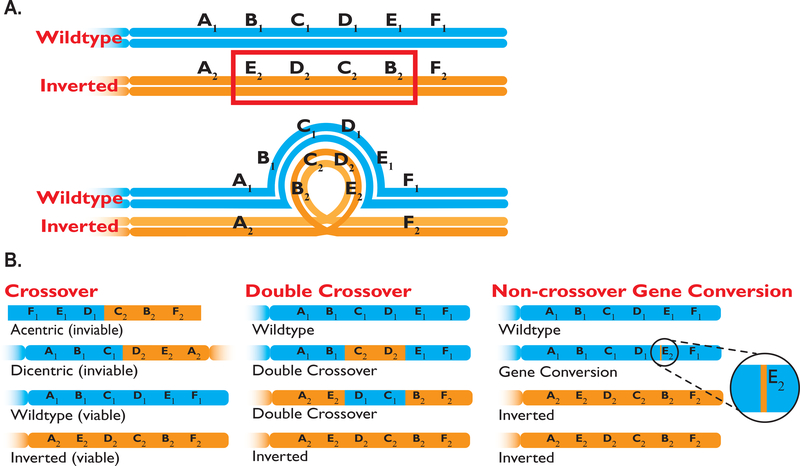
Chromosomal rearrangements can affect chromosome pairing during meiosis in individuals that are heterozygous for such arrangements. Even when rearranged chromosomes pair successfully and recombination initiates normally, the outcomes of recombination are affected (Gong et al. 2005). When double strand breaks (DSBs) initiate homologous recombination, possible outcomes include crossing over and non-crossover gene conversion (Figure 1). Individuals that are heterokaryotypic for inversions experience reduced recovery of recombinants from crossing over, since single crossovers within paracentric inversions can yield inviable acentric and dicentric products. Double crossovers within inversions yield viable chromosomes, but evidence suggests that double crossover rates within inversions are lower than rates in homokaryotypes or in collinear regions that are several megabases away from an inversion (Levine 1956; Ishii & Charlesworth 1977; Stump et al. 2007; Stevison et al. 2011; Manzano-Winkler 2015). Further, double crossovers are unlikely to occur near inversion breakpoints (Navarro et al. 1997). In contrast to crossing over, gene conversion copies a homologous piece of template DNA at the site of the DSB, resulting in a non-reciprocal exchange of DNA, and may have very different properties with respect to inversions (see below).
Figure 1. Outcomes of Recombination Within Inverted Regions.
(A) During meiosis, chromosomes of an inversion heterozygote form an inversion loop, and consequences of recombination in the inverted region (B) include single crossovers (inviable products), double crossovers, and gene conversion.
Gene conversion can occur in genomic regions where meiotic recombination is otherwise suppressed, including centromeric regions (Liebman et al. 1988; Shi et al. 2010; Talbert & Henikoff 2010; Miller et al. 2016) and near other recombination events (Mancera et al. 2008; Miller et al. 2016). Gene conversion may also occur throughout inverted regions in heterokaryotypes, as suggested by within-species population genetic data and LD-based estimates of the frequency of gene conversion within inversions (Rozas & Aguadé 1994; Schaeffer & Anderson 2005; Nóbrega et al. 2008). However, LD-based estimates of gene conversion are necessarily complicated by various assumptions (see Betrán et al. 1997). The occurrence of gene conversion in inverted regions is further supported by limited direct empirical assessments, such as Chovnick’s (1973) study of the rosy locus in Drosophila melanogaster, which showed that gene conversion occurs at similar levels within and outside of an inversion. A recent within-species analysis of heterokaryotypes in D. melanogaster suggests that gene conversion rates may even increase within inversions (Crown et al. 2018). The pervasiveness of gene conversion in areas where crossovers are rare indicates that, despite the redistribution of crossing over in inversion heterozygotes, gene conversion may still work to degrade LD throughout inverted regions.
In addition to potentially happening more homogeneously across the genome than crossing over, gene conversions have other properties that differentiate their evolutionary effects. First, the tracts copied by gene conversion are short, typically not more than a few thousand nucleotides at the longest (Liskay et al. 1987; Judd & Petes 1988; Waldman & Liskay 1988; Jeffreys & May 2004), while crossovers often result in megabase-scale exchanges of sequences. Second, unlike crossing over, if gene conversion occurs in a heterozygous stretch of DNA, it also alters allele frequencies by overwriting one allele with another and can subsequently result in a non-Mendelian 3:1 segregation of alleles. Thus, gene conversion is unique in its immediate effects on nucleotide diversity.
The long-term consequences of gene conversions include the erosion of genetic differentiation between distinct hybridizing populations or species. Inversions are often invoked as facilitators of speciation, species persistence, and adaptation in the face of hybridization (e.g., Noor et al. 2001b; Hoffmann & Rieseberg 2008; Fishman et al. 2013; Kirkpatrick & Barrett 2015). Despite the recombination-suppressing effects of inversions, theory and limited empirical evidence suggest that gene conversion has the potential to erode inversions’ effects as barriers to recombination over the long-term (Navarro et al. 1997; Andolfatto & Nordborg 1998; Schaeffer & Anderson 2005; Feder & Nosil 2009). If gene conversion rates are sufficiently high and not suppressed by inversions, then the reality of how inversions contribute to maintaining genetic differentiation between species may be quite complex. The recent analysis by Crown et al. observed increased rates of gene conversion in inversions compared to collinear regions (Crown et al. 2018), but this has not yet been studied in an interspecies cross. To our knowledge, no one has examined empirically the extent to which genetic exchange via gene conversion occurs in interspecies hybrids differing by chromosomal inversions. Although the results of Crown et al. (2018) suggest that such gene conversion may be common in such inverted regions, interspecies hybrids often exhibit greater sequence divergence between recombining homologs, and this divergence may decrease the efficiency of recombination (Liskay et al. 1987; Chen et al. 2007). Recombinant genotypes resulting from gene conversion may also be eliminated by natural selection when inversions keep locally adaptive alleles in LD. Through genome-wide analyses of gene conversion, we can now evaluate the extent of recombination to a greater resolution and discuss how resulting genetic exchange contributes to evolutionary processes.
In this study, we assess rates of gene conversion to examine the efficacy of chromosomal inversions as barriers to recombination in a model system: Drosophila pseudoobscura and D. persimilis. We examine gene conversion within rearrangements that differ between these species and within rearrangements that continue to segregate within species. D. pseudoobscura and D. persimilis differ by fixed inversions on chromosome 2 and on the left arm of the × chromosome (each approximately 7 Mb). The divergence times for these fixed inversions are about 1.55 and 1.65 MYA, respectively, which predates the species divergence (Fuller et al. 2018). There is a nearly-fixed inversion difference (approximately 12 Mb in size) on the right arm of the X, and chromosome 3 contains inversion polymorphisms in both species. The inversion on the right arm of the × chromosome diverged from its ancestral arrangement about 1.4 MYA, and the chromosome 3 inversion (“Pikes Peak”) utilized in this study is dated at about 0.75 MY (Fuller et al. 2018). These inversions provide a system in which chromosomal rearrangements differentiate a significant portion of the ~150 Mb genomes of this species pair. Evidence supports the hypothesis that these species persist as distinct at least in part due to reproductive barriers associated with chromosomal inversions. Indeed, nearly all reproductive barriers to gene flow between these species map to the inverted regions (Noor et al. 2001a; b). Ongoing hybridization in wild populations occurs at a rate of ~0.0001 per generation (Dobzhansky 1973; Powell 1983), resulting in gene flow, which is detectable in statistical analyses of polymorphism data (Machado & Hey 2003; Hey & Nielsen 2004). DNA sequence divergence is on average higher in inverted regions than in collinear regions (Table S1; Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009; McGaugh & Noor 2012), and the divergence of collinear regions is lower between sympatric populations than allopatric populations (Kulathinal et al. 2009). Within species, D. pseudoobscura has over 30 arrangements of chromosome 3 (Dobzhansky & Epling 1944; Powell 1992). The genes within these chromosome 3 inversions are likely under selection, as the arrangement frequencies form geographic clines and exhibit seasonal variation (Dobzhansky 1943, 1948). Together, the attributes of these species and their history as a model for the study of speciation make the D. pseudoobscura / D. persimilis system an ideal model for the study of gene exchange in inverted regions and its evolutionary consequences. This system provides an opportunity to examine the permeability of different genetic regions to recombination via gene conversion.
Here, we harness the D. pseudoobscura and D. persimilis model system to obtain the first direct empirical data on rates of gene conversion with respect to chromosomal inversions in species hybrids. Using single-generation crosses allows us to resolve recombination after a single meiosis, revealing individual gene conversion events. We present our analyses of the size, distribution, frequency, and characteristics of gene conversion throughout the genomes of both intra- and interspecies crosses, which provide both homokaryotypic and heterokaryptypic chromosomes. We assess whether the rate of gene conversion varies between differing genomic regions: inside vs outside inversions, near inversion breakpoints, and in regions of differing sequence divergence (Table 1). By addressing gene conversion, this study contributes to the development of a more complete and complex picture of the role that inversions play in population genetics.
Table 1.
Questions Addressed by this Study.
| Question | Dataset |
|---|---|
| 1. Is gene conversion more likely to happen between haplotypes in regions with lower sequence divergence? | Local sequence divergence measured in 1000 bp windows surrounding gene conversion events compared to a control set of genomic windows. |
| 2. Does gene conversion occur within inverted regions? | Whole genome sequencing of two heterokaryotypic crosses, with a total of 5 inversions. |
| 3. Are gene conversions observed at similar rates within inversions compared to collinear regions? | Analysis of the converted proportion of filtered SNPs and maximum likelihood estimation of the rate and tract length. |
| 4. Do the regions surrounding observed gene conversions exhibit patterns in GC content or motif enrichment? | GC content and motif presence in 1000 bp windows surrounding gene conversion events compared to a control set of genomic windows. |
| 5. Does GC-biased gene conversion shape gene conversion in this system? | Direction of gene conversion in AT / GC heterozygotes. |
METHODS
Strains and Crosses
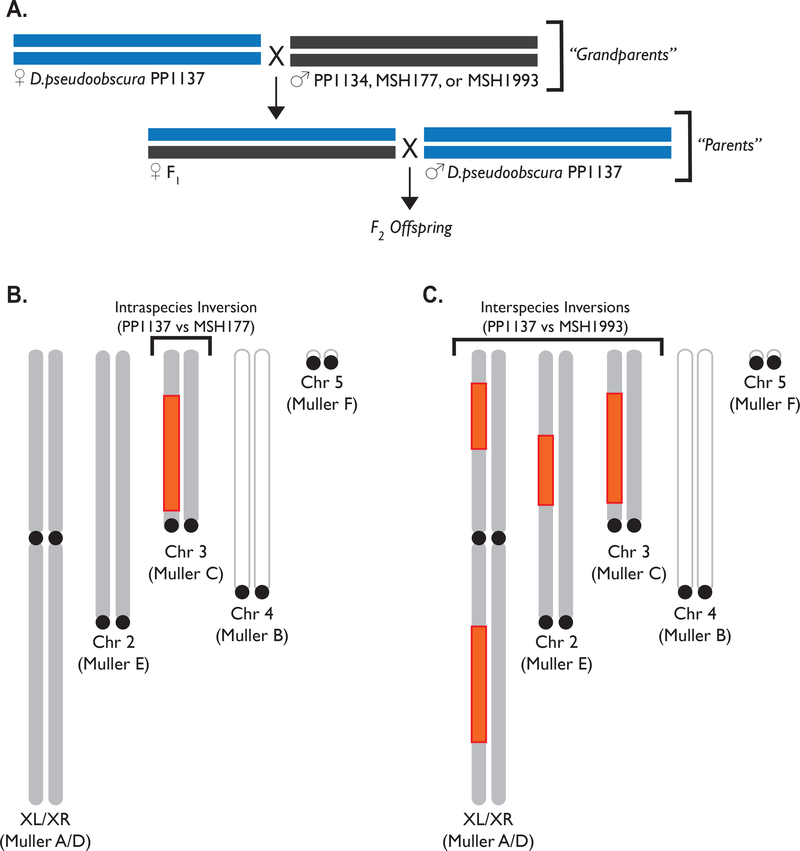
To assess gene conversion within various karyotypes, we generated one interspecies cross and two intraspecies crosses. We used three inbred lines of D. pseudoobscura (PP1137, PP1134, and MSH177), and one line of D. persimilis (MSH1993). The Pikes Peak (PP) inversion segregates on chromosome 3, and PP1137 and PP1134 are homozygous for this inversion. PP1137 and PP1134 were both originally collected from Bosque del Apache National Wildlife Refuge, San Antonio, NM in 2006. The MSH177 line and the D. persimilis MSH1993 line were derived from flies originally collected from Mount St. Helena, CA (MSH), and both are homozygous for the standard inversion arrangement of chromosome 3. In each cross, a PP1137 individual was crossed to the other strain, and a female F1 was then backcrossed to a PP1137 male (Figure 2A). The resulting three crosses had the following karyotypes: PP1137×PP1134 had the same karyotype on all chromosomes; PP1137×MSH177 was heterokaryotypic only on chromosome 3; and PP1137×MSH1993 (the interspecies cross) was heterokaryotypic on chromosome 2, chromosome 3, and both arms of the × chromosome (Figure 2B and 2C; Table S2 for breakpoint coordinates). All heterokaryotypic chromosome arms differ by a single inversion step only. Ten offspring were sequenced from each of the three crosses, including 6 females and 4 males from PP1137×PP1134; 5 females and 5 males from PP1137×MSH177; and 6 females and 4 males from PP1137×MSH1993.
Figure 2. Cross Design and Inversion System.
(A) For each of the 3 crosses, a PP1137 individual was crossed to another strain (either PP1134, MSH177, or MSH1993), and a female F1 was then backcrossed to PP1137. The karyotypes present in the resulting crosses are summarized with inverted regions highlighted in orange for the PP1137 × MSH177 cross (B), which was heterokaryotypic on chromosome 3, and for the interspecies cross (C), which was heterokaryotypic on chromosome 2, chromosome 3, and both arms of the × chromosome.. PP1137 × PP1134 had the same karyotype on all chromosomes.
Genome Alignment and SNP Calling
Scripts used for genome alignment, SNP calling, and identification of gene conversion are available on GitHub (https://github.com/kkorunes/2018_GeneConversion_Pipeline). For each of the three single-pair crosses, we obtained whole-genome sequences at an average coverage of 20X from one grandparent, both parents, and 10 F2 offspring. Library preparation and sequencing (Illumina HiSeq 100-bp paired end) was performed by the High-Throughput Sequencing Facility at the University of North Carolina at Chapel Hill. All genome alignments were performed using bwa v0.7.5 with default parameters (Li & Durbin 2009). The PP1137 genome (SRA experiment SRX091311) was aligned to the FlyBase D. pseudoobscura reference genome v3.04 (Gramates et al. 2017) to generate the PP1137 reference genome used for all subsequent alignments, and all coordinates are reported with respect to this assembly. D. pseudoobscura and D. persimilis are closely related, and alignment of D. persimilis line MSH1993 (SRR363439) covers over 95% of positions in our PP1137 reference. The similarity between the species allows for alignment of D. persimilis individuals and D. pseudoobscura × D. persimilis hybrids to the PP1137 reference. To assess the physical location of observed gene conversions, we used the contig order and orientation previously inferred from genetic and physical maps (Schaeffer et al. 2008). To assess gene conversion inside vs outside inverted regions, we used previously published breakpoints (Table S2) for chromosome 3 (Fuller et al. 2017), chromosome 2, and the × chromosome (Machado et al. 2007).
All sequencing data from the above crosses were aligned to the assembled PP1137 genome. SAMtools v1.4 was used to convert to BAM, sort, and index each genome (Li et al. 2009). Picard was used to mark duplicates (http://broadinstitute.github.io/picard), and the Genome Analysis Toolkit (GATK) v3.8 was used for local realignment around indels, followed by SNP calling based on the GATK Best Practices (DePristo et al. 2011; Van der Auwera et al. 2013). Only genotypes with Phred quality scores >=30 were considered. The SNPs in the parent females were subjected to the most stringent set of filters, since the parental SNPs identified in the female F1s were used as the basis of tracking recombination in the offspring of each cross. These F1 female SNPs were further filtered for depth and strand bias (excluding sites with QualByDepth (QD) < 2.0, filtered Depth (DP) < 10, Qual < 30, FisherStrand (FS) > 60, and StrandOddsRatio (SOR) > 3.0). SNPs called in the offspring (Figure 2A) were filtered using the cutoff of DP<10 for all chromosomes in the females and for the autosomes in the males. Since males are hemizygous for the × chromosome, any heterozygous sites on male × chromosomes were discarded, along with any sites with depth less than 5X. These criteria were chosen following confirmation of several putative gene conversion tracts via PCR and Sanger sequencing parents and offspring (see Table S3 for coordinates and primers and Figure S1 for an example). A custom Perl script was applied to add a flag to the filter column of any offspring genotype where a grandparent or parent was unexpectedly heterozygous (i.e., the grandparents and the male parents).
Identification of Gene Conversion
After SNP-calling, custom scripts were used to scan the genomes for gene conversion events. We excluded chromosomes 4 and 5 from these analyses due to poor assembly quality: chromosome 4 is thought to be composed of 14 contigs averaging less than 2 MB each, and chromosome 5 is thought to be composed of 30 contigs averaging less than 0.5 MB each, with uncertain orientation (Schaeffer et al. 2008). Since the parent female in each cross is an F1 of the two strains, every SNP identified in the parent female represents a site that differs between the two strains of the cross. We used these polymorphic sites identified in the parent females as the basis of tracking the inheritance of alleles. In the absence of recombination, each chromosome should be either entirely heterozygous or entirely homozygous, so deviations from this expectation represent possible recombination events. To exclude potential double crossovers, we set an upper limit of 10 kb between the unconverted SNPs flanking a putative gene conversion. Recombination events flanked by SNPs less than 10 kb apart are expected to be gene conversion events and not double crossovers, given the expected size range of gene conversion (Judd & Petes 1988; Waldman & Liskay 1988; Jeffreys & May 2004; Schaeffer & Anderson 2005; Miller et al. 2016) and the effect of crossover interference, which results in the wide spacing of crossovers on the same chromosome (Sturtevant 1913, 1915; Stevison & Noor 2010). For the hemizygous × chromosomes in the males, loci where the inherited allele switched from one parent of origin to the other and back were flagged as putative gene conversion events. For the autosomes and the female × chromosomes, the same logic was applied, except we scanned for loci where the offspring switched from one parent of origin to both parents and back to a single parent (i.e., heterozygosity in an otherwise homozygous background). Scanning for gene conversion events where focal sites are heterozygous according to our pipeline is a more conservative approach than scanning for homozygous sites on a heterozygous background, since whole-genome sequencing cannot inform us whether sequencing biases or insufficient coverage led to the sequencing of only one allele. Only loci with a SNP in the female F1 and a reference allele in the male parent were considered as possible sites of gene conversion, and we required that the alternate allele be supported by at least 3 reads, which must represent at least 25% of total number of reads. Again, these criteria were chosen following confirmation of a subset of the loci via PCR and Sanger sequencing (Table S3). These methods are designed to identify putative non-crossover gene conversion events and are not directed at identifying crossovers or crossover-associated gene conversion.
After identifying putative gene conversions, we performed several additional filtering steps and quality checks. We filtered any candidates that occurred within a 5 bp window on either side of an indel identified in the parent female, since we found indels to be a frequent source of error. Each putative gene conversion was individually examined for accuracy of genotypes using Integrative Genomics Viewer (IGV) v2.4.8 to view the reads and SNP calls for the parents and offspring at the locus of each conversion (Robinson et al. 2011; Thorvaldsdottir et al. 2013). Only visually confirmed putative gene conversions were used in subsequent analyses and reported in gene conversion rates.
Gene Conversion Rate and Tract Length
To assess rates of gene conversion, we corrected for the proportion of the genome in which we had the power to detect gene conversion. To calculate the proportion of SNPs that were observed to be converted (Converted SNPs / Total SNPs), we determined the denominator using only SNPs that met all filtering criteria used in the gene conversion identification pipeline. As a complementary approach for assessing the rate of gene conversion, we applied the maximum-likelihood method described by Miller et al. 2012 to consider the distribution of the distances separating unconverted SNPs and estimate the average tract length and the rate of DSBs fated to be gene conversions (Miller et al. 2012, 2016). It is important to note the distinction between the rate of DSBs fated to be gene conversions and the proportion of nucleotides converted per generation. The latter accounts for the tract length of each event to give the proportion of converted sites per genome per generation (see Results). We used this maximum-likelihood approach (as per Miller et al. 2012, 2016) to jointly estimate tract length and the rate of gene conversion occurrence by considering tract lengths of 100–500 bp (in increments of 10 bp) and DSB rates ranging from 1 × 10−8 to 3 × 10−7 gene conversion events/bp/gen to identify a local maximum. We then performed additional trials in smaller increments to find the precise local maximum. Due to the differences in our gene conversion detection strategy, we performed this analysis for the hemizygous male × chromosome data, and then we separately analyzed the autosomal chromosomes plus the female × chromosome.
Local Sequence Divergence and Composition
To determine whether gene conversion is associated with local sequence divergence, we calculated divergence in the 1000 bp surrounding each conversion event (500 bp in each direction). We measured divergence using p-distance, the proportion of sites at which two sequences differ: p = nd/n, where nd is the number of different nucleotides, and n is the total number of examined nucleotides (Nei & Kumar 2000). We then took a random sample of 10,000 unconverted SNPs and their flanking 1000 bp from each cross. These 10,000 control regions surrounding unconverted SNPs passed all filtering criteria used for identifying converted SNPs. We quantified differentiation in the control regions using SNPs identified between PP1137 and the other strain of each cross: PP1134, MSH177, and MSH1993. For MSH1993 and PP1134, the genomes available on SRA were used for SNP calling following the same sequence analysis protocol as above (SRX104992 and SRX091323), and for MSH177 we used the male grandparent (20X coverage) of that cross. A Mann-Whitney U test was used to test for differences in the distribution of sequence divergence observed in sampled converted and unconverted regions.
We used a similar strategy to assess the GC content surrounding gene conversion events. We calculated GC content in the 1000 bp surrounding each gene conversion event (500 bp in each direction), and we sampled 10,000 random, unconverted 1000 bp tracts from each cross. Again, a Mann-Whitney U test was applied to test for differences in the distribution of GC content observed in sampled converted and control regions. To test whether AT-to-GC gene conversions and GC-to-AT gene conversions occurred equally in our dataset, we compared the ratio of the observed gene conversions (AT-to-GC/GC-to-AT), where the null expectation is a 1:1 ratio. We used only informative converted positions where there was the opportunity for an AT-to-GC or GC-to-AT gene conversion due to the female parent being an AT / GC heterozygote. Using informative gene conversion tracts (47 out of the 71 gene conversion tracts), we compared the ratio of AT-to-GC/GC-to-AT using a binomial sign test. The parental genotypes and transmission directionality for each gene conversion are included in Table S6.
We also examined the composition of the local regions surrounding gene conversion by checking for overlapping gene annotations using Flybase and searching for motifs using MEME (Bailey et al. 2009). To identify overlap between gene conversion tracts and D. pseudoobscura gene annotations in FlyBase, we determined the maximum possible tract length of each gene conversion event using the flanking, unconverted SNPs. We compared the sequences of these tracts to the collection of D. pseudoobscura gene spans using FlyBase’s BLAST function (see Table S7 for observed overlap and the FlyBase gene symbols). To search for motifs, we used MEME to search the 1000 bp surrounding unique gene conversion events for enrichment of motifs between 6 bp and 12 bp in length. For putative motifs detected at a e-value < 0.05, we used FIMO within the MEME suite (Grant et al. 2011) to search for enrichment of these motifs in 1000 randomly selected control regions per cross, where no gene conversion was detected.
RESULTS
Among the 30 offspring examined across our three crosses, we observed 71 gene conversion products. The observed locations (Figure 3 for their physical distribution, Table S4 for all coordinates) and sizes of gene conversion tracts allow us to assess how much exchange might occur in a single generation due to gene conversion. We used two complementary methods to assess the probability of a nucleotide being transferred via gene conversion in a single generation. First, we directly calculated the proportion of converted sites out of the total observed SNPs for each cross. We found the proportion of converted sites per genome per generation to be ~1 × 10−5 to ~2.5 × 10−5 (Table 2). We also obtained a joint maximum-likelihood estimation of the average tract length and the proportion of DSBs fated to resolve as gene conversions using the method presented in Miller et al. (Miller et al. 2012, 2016). These two complementary methods for assessing the proportion of converted sites per genome per generation both account for the density of SNPs. The maximum likelihood method has the advantage of considering the distribution of the distances separating unconverted SNPs, allowing for an estimation of the average tract length and the rate of DSBs fated to be gene conversions. On the other hand, the calculation of the proportion of SNPs that were observed to be converted (Converted SNPs / Total SNPs) has the advantage of building fewer assumptions into the estimate and potentially being less sensitive to biases due to sample size, which may occur when analyzing subsets of the data such as inverted regions.
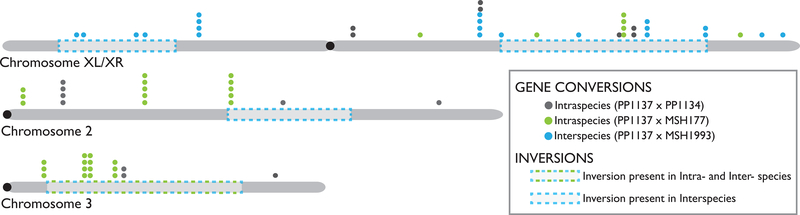
Figure 3. The Physical Distribution of Observed Gene Conversions.
The examined chromosomes are shown in grey with centromere positions shown in black. Inversions are represented with dashed boxes and the approximate relative location of each gene conversion is represented with a dot, colored according to the cross. The interspecies cross was heterokaryotypic on chromosome 2, chromosome 3, and both arms (XL and XR) of the × chromosome. Dashed blue boxes indicate the positions of the inverted regions in the interspecies cross, and blue dots indicate the positions of observed gene conversion tracts in this cross. The inversion on chromosome 3 was also heterokaryotypic in the PP1137 × MSH177 intraspecies cross (inversion shown with blue and green dashed box), and green dots represent the conversion positions in this cross. PP1137 × PP1134 (grey dots) was homokaryotypic.
Table 2. Observed Gene Conversions and Rates.
Each row represents one of the three experimental crosses. PP1137 × MSH1993 is the interspecies cross, PP1137 × MSH177 is the within-species heterokaryotypic cross, and PP1137 × PP1134 is the within-species homokaryotypic cross.
| Conversions by Region | SNPs by Region | Rate (Converted/Total SNPs) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Cross | Inverted | Collinear | Total | Inverted | Collinear | Total | Inverted | Collinear | Total |
| PP1137 × MSH1993 | 11 | 10 | 21 | 815935 | 1110886 | 1926821 | 1.35E-05 | 9.00E-06 | 1.09E-05 |
| PP1137 × MSH177 | 11 | 23 | 34 | 351039 | 2972519 | 3323558 | 3.13E-05 | 7.74E-06 | 1.02E-05 |
| PP1137 × PP1134 | na | 16 | 16 | na | 632503 | 632503 | na | 2.53E-05 | 2.53E-05 |
The maximum likelihood analysis estimates the proportion of converted sites per genome per generation to be in the range of 1.4 × 10−5 - 1.9 × 10−5, which is in agreement with the proportion of converted sites out of the total observed SNPs (reported in Table 2). In the hemizygous male × chromosome, we had the power to detect gene conversion where the inherited genotype switched from one parent of origin to the other in either direction. The maximum-likelihood approach for the hemizygous × chromosome data estimated a tract length of about 390 bp and a rate of gene conversion-fated DSBs of 3.7 × 10−8 gene conversion events/bp/gen. Together, the tract length and rate of gene conversion-fated DSBs suggest the proportion of converted sites per genome per generation to be 1.4 × 10−5. Due to the methodological difference in gene conversion detection, we separately analyzed the autosomes along with the female × chromosomes. In these data, the maximum likelihood estimation of the tract length was 188 bp and the rate of gene conversion-fated DSBs was 4.94 × 10−8 events/bp/gen. Since we searched for gene conversion in only one direction in these data (heterozygous converted SNPs in otherwise homozygous regions), the product of the tract length and twice the estimated rate of gene conversion-fated DSBs gives the estimated proportion of converted sites per genome per generation: 1.9 × 10−5. As expected, this rate is very similar to the rate of 1.4 × 10−5 observed for the hemizygous male × chromosome. The rates presented here (~1 × 10−5 to ~2.5 × 10−5) are comparable to rates observed in D. melanogaster, where the proportion of converted sites per genome per generation likely falls in the range of 9.3 × 10−6 to 1.3 × 10−5 (Blanton et al. 2005; Miller et al. 2012, 2016).
Gene Conversion and Sequence Divergence
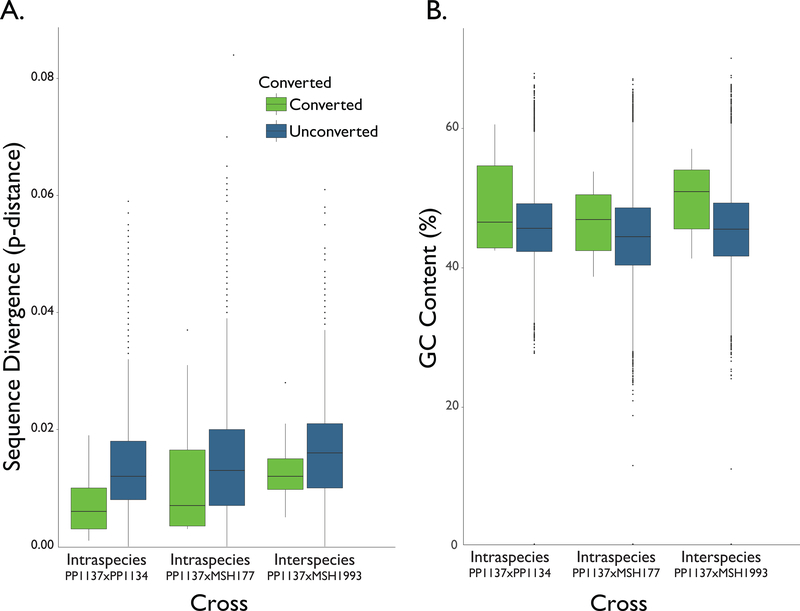
Recombination may be less frequent between more divergent sequences (Liskay et al. 1987; Chen et al. 2007). The effects of local sequence divergence on gene conversion may contribute to how inversions influence rates of genetic exchange, as inversions that are involved in local adaption or speciation often accumulate genetic differences faster than collinear regions. In this system, sequence divergence in inverted regions is on average higher than in collinear regions (Table S1; Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009; McGaugh & Noor 2012). To test whether gene conversion is affected by local sequence divergence, we used the 1000 bp regions surrounding gene conversion events to examine the number of SNPs differentiating the pair of strains involved in each cross. We compared average divergence in the regions surrounding converted SNPs to the distribution of divergence between the strains of each cross in random regions (Figure 4A). Taking the results from all the crosses together, we observed significantly lower local divergence in the regions surrounding gene conversions (p = 0.02, Fisher’s method for combining probabilities (Fisher 1954, Sect 21.1)). The association of gene conversion with regions of lower local sequence divergence suggests that rates of gene conversion may be expected to be lower in more divergent sequences, such as within inverted regions compared to collinear regions. However, our observed gene conversion rates in more divergent inverted regions were at least as high as in less divergent collinear regions (see below). Together, these results suggest that local sequence divergence may negatively affect gene conversion rates, but gene conversion rates are driven by the interaction of multiple factors and cannot be predicted by local divergence alone.
Figure 4. Local Sequence Divergence and GC Content Surrounding Gene Conversions.
In (A), the green boxplots represent local sequence divergence in the 1000 bp windows surrounding gene conversions. The blue boxplots represent local sequence divergence in 10,000 randomly selected 1000 bp windows where no gene conversion was observed. Results are shown for the three crosses: PP1137 × PP1134 (p=0.01), PP1137 × MSH177 (p=0.19), and PP1137 × MSH1993 (p=0.26). (B) shows GC content in the 1000 bp windows surrounding gene conversions (green) and the GC content of 10,000 randomly selected 1000 bp windows (blue) where no gene conversion was observed. Again, results are shown for the 3 crosses: PP1137 × PP1134 (p=0.37), PP1137 × MSH177 (p=0.21), and PP1137 × MSH1993 (p=0.02).
Species Hybrids Experience Gene Conversion within Inversions and Near Breakpoints.
The interspecies cross (PP1137 × MSH1993) presented in this study provides the first direct empirical observation of gene conversion within inverted regions of species hybrids. In this interspecies cross and in the within-species heterokaryotypic cross (PP1137 × MSH177), we observed gene conversion in both inverted and collinear regions. These data provide direct evidence in support of the idea that inversions do not completely suppress recombination and may have no suppressing effects on gene conversion specifically (Chovnick 1973; Rozas & Aguadé 1994; Schaeffer & Anderson 2005; Nóbrega et al. 2008). Unlike crossing over, which is dramatically suppressed within inversions, gene conversion rates in the examined crosses are at least as high within inversions as within collinear regions. In fact, our data suggest that rates within inversions may be higher than within collinear regions within each cross examined (Table 2), though this is not statistically significant (p = 0.35, Fisher’s method for combining probabilities (Fisher 1954, Sect 21.1)). Even outside of inverted regions, the breakpoints of inversions are known to suppress crossing over, so we looked for evidence of suppression of gene conversions near breakpoints. In total, we observed 3 gene conversion events within 1 Mb of a breakpoint. This number of events was not noticeably lower than the number of events observed in similarly sized regions more centrally within inversions or far outside inversions (Table S5). The closest of these gene conversions was about 37 kb from the nearest inversion breakpoint, inside the inversion. All distances of detected conversions from breakpoints are in Table S4.
Recovery of Pre-Meiotic Germ Line Events
We recovered 17 loci where a gene conversion was observed at the same locus in more than one offspring. This observation is consistent with mitotic gene conversion occurring in a germ-line precursor. These gene conversions were all shared within crosses; i.e., no SNP was converted in multiple crosses, as one might expect in the case of a gene conversion “hotspot” driving meiotic gene conversion at a locus. An alternate explanation for the origin of these events are de novo mutations. While this possibility cannot be excluded, it is unlikely that a de novo mutation would occur at the precise position of the parental SNP and match the genotype present in the other parent. When we validated a subset of our putative gene conversions with Sanger sequencing, we included several of these loci. See Figure S1 for an example of a validated mitotic event, where the signature of gene conversion can be observed in multiple offspring at the same locus. Altogether, although we observed 71 gene conversion products among the offspring, these appeared to represent only 32 unique gene conversion events.
Covariates and Characteristics of Gene Conversion
To further explore the characteristics of the observed gene conversions and their surrounding genomic regions, we assessed local GC content, GC-bias, and motif enrichment. GC content is positively associated with crossover rates in many taxa including yeast, mammals, and Drosophila (Fullerton et al. 2001; Liu & Li 2008; Kiktev et al. 2018). The local GC content in the regions surrounding our observed gene conversions is slightly higher than the background GC content in each cross (Figure 4B; p = 0.048, Fisher’s method for combining probabilities (Fisher 1954, Sect 21.1)). This observation raises the question of whether GC content in this system is affected by biased transmission of G and C alleles compared to A and T alleles when gene conversion occurs at site that is AT / GC heterozygous. This bias is termed GC-biased gene conversion and occurs widely in many taxa, including mammals, birds, and yeast (Galtier et al. 2001; Webster et al. 2005, 2006; Lesecque et al. 2013; Glémin et al. 2015). We compared the ratio of AT-to-GC gene conversion to GC-to-AT gene conversions (AT-to-GC/GC-to-AT). The null expectation is a 1:1 ratio, while a ratio >1 would be consistent with GC-biased gene conversion. We observed an excess of GC-to-AT gene conversions in the set of 71 gene conversions: the ratio of AT-to-GC/GC-to-AT was 17:30 for the 47 AT/GC heterozygotes (see Table S6), p-value = 0.08, binomial sign test. The directionality of this bias is opposite of the direction expected under GC-biased gene conversion, and this excess of GC-to-AT gene conversions is consistent with other results in Drosophila (Robinson et al. 2014).
Though other studies have reported crossover-associated motifs in Drosophila (Kulathinal et al. 2008; Stevison & Noor 2010; Miller et al. 2012; Singh et al. 2013), our search for motifs in the 1000 bp regions surrounding gene conversions did not return convincing evidence that any motif is significantly enriched near our observed gene conversion tracts. We detected six putative motifs that appear to be significantly enriched in the gene conversion spans at a significance of e-value < 0.05: GTGT[GC]TGTGTGT, TTT[TG]T[TG][GT]TTT[TG], CAGC[GA][GC]CAGC[AG]G, GG[GA]GG[GC]GGGGGG, [GA]AG[AG]G[AG][GC]AGAGA, and [AG][CA]AA[CA][AG][GA][CG]A[AG][CA]A. Each of these six motifs was present in 46–58% of our randomly sampled regions. This rate of occurrence in the background and the low-complexity of the motifs suggests that they are not robustly associated with gene conversion. However, sample size limits power to detect motifs, particularly if multiple motifs are associated with gene conversion, and future studies may reveal DNA motifs associated with gene conversion.
DISCUSSION
The present study provides direct empirical evidence that gene conversion occurs within inversions in species hybrids. These data support other lines of evidence that gene conversion happens regularly within inversions (Chovnick 1973; Navarro et al. 1997; Schaeffer & Anderson 2005; Crown et al. 2018). By analyzing the rates and characteristics of gene conversion, we can better understand the evolutionary effects of this type of recombination. First, these data allow us to assess how much DNA sequence may be affected by gene conversion in a generation, both genome-wide and within inversions. Our estimated gene conversion rates fall within the range of rates observed in other species. Across taxa, conversion rates tend to be on the order of magnitude of 10−4 to 10−6 converted sites per genome per generation (Korunes & Noor 2017). Multiple estimation methods using our data suggest a proportion of converted sites per genome per generation of about 1 × 10−5 to 2 × 10−5 in Drosophila pseudoobscura. Our estimates of average tract length ranged from 188–390 bp, which again falls within the range observed in other studies. For example, in Drosophila, Miller et al. (2016) estimated a tract length of 440 bp which suggests a rate of about 7.5 × 10−6 in D. melanogaster (440 bp × the rate of gene conversion-fated DSBs: 1.7 × 10−8), and Schaeffer and Anderson (2005) gave a conservative tract length estimate of 205 bp in D. pseudoobscura from LD-based data, with a rate of 3.4 × 10−6. Here, we focus on gene conversion rates that have been corrected for SNP-density to avoid bias caused by variation in sequencing coverage among individuals and among chromosomes. Importantly, we did not observe gene conversions on all chromosomes, but we do not claim this as evidence that no gene conversions occurred. Variation in sequencing coverage and quality among individuals limits us in analyzing how the gene conversion landscape varies among individuals. Thus, the data presented here provide an important step towards understanding gene conversion in hybridizing populations and species, and future studies are needed to add greater resolution to this understanding and dissect the correlates of individual variation in gene conversion rates and landscapes.
Gene Conversion is Pervasive Despite Chromosomal Rearrangement and Sequence Divergence
Within inversions, the rates of gene conversion appear to be at least as high or higher than rates in collinear regions. This observation is particularly striking because gene conversions were observed in regions of lower local divergence compared to the background (Figure 4A), but the inverted regions in D. pseudoobscura and D. persimilis exhibit higher sequence divergence compared to collinear regions (e.g., Kulathinal et al. 2009; Stevison et al. 2011; McGaugh & Noor 2012). The relatively high rates of gene conversion within inversions despite greater sequence divergence may be explained by the interchromosomal effect. In D. melanogaster inversion heterozygotes, Crown et al. observed that an increase in crossovers on collinear chromosomes accompanied an increase in gene conversion rates within inversions (Crown et al. 2018). The data presented by Crown et al. suggest that the number of DSBs remains the same in heterokaryotypes, but inversions alter the proportions of crossovers to gene conversions on each chromosome, implicating the interchromosomal effect as a driver of increased gene conversion within inversions. Our results are consistent with their hypothesis.
Meiotic vs. Pre-Germline Mitotic Gene Conversion Events
Unexpectedly, we observed both single meiotic gene conversions and mitotic gene conversions that were shared among multiple offspring within a cross. When considering changes in allele frequencies and sequence divergence over generations, a mitotic gene conversion event may have a greater impact than a meiotic one because multiple offspring immediately inherit the converted tract. Each individual offspring inheriting a mitotic gene conversion contributes to the next generation in a manner equivalent to offspring with a single meiotic gene conversion event. LD-based estimates of gene conversion likely include mitotic events, as these methods cannot distinguish between meiotic gene conversion and mitotic gene conversion that enters the germ-line.
Emerging Covariates and Characteristics of Gene Conversion
One possible explanation for the association of gene conversion with GC content relates to the observation that gene conversion seems to prone to occur in regions of low divergence. Regions of low divergence may be relatively conserved due the presence of coding regions, which tend to be GC rich compared to non-coding regions. Many of our observed gene conversions do overlap with annotated gene spans (Table S7), indicating that this interaction between gene conversion and conserved genomic regions may be an interesting area for future research. Another potential contributor to the landscape of gene conversion is GC-biased gene conversion, which could increase GC content in regions prone to gene conversion. An advantage of our single-generation method over population-based methods is that we can use the parental alleles of each gene conversion to see whether an AT-to-GC or GC-to-AT conversion occurred, rather than inferring GC-biased gene conversion from polymorphism and divergence data. Our results suggest that GC-biased gene conversion did not contribute to patterns of transmission in the examined data. This result is consistent with other results that imply a lack of GC-biased gene conversion in Drosophila. Robinson et al. reported a similar lack of evidence for GC-biased gene conversion based on the ratio of AT-to-GC/GC-to-AT polymorphisms in D. melanogaster, which revealed a significant excess of GC-to-AT (Robinson et al. 2014). Thus, our results lend support to the previously reported population-based inferences. Understanding how gene conversion contributes to genome evolution over time will require future efforts to unravel patterns such as biased gene conversion and the covariation of gene conversion with other genomic variables.
Relative Effects of Gene Conversion and Crossing Over on Decay of Differentiation in Inverted Regions
Though gene conversion occurs regularly within inversions, individual gene conversion events affect relatively small segments of DNA. We estimate that gene conversion tracts within this system tend to be 200–400 bp. A single gene conversion event of 200–400 bp is unlikely to overlap important SNPs within a large inversion spanning several megabases, even if the inversion contains multiple locally adapted loci. If the gene conversion overwrites a locally-adapted allele, the recombinant offspring may face a selective disadvantage. This hypothesis may explain why our observed rate of gene conversion in a single generation is 3–6 times higher than the LD-based estimate of 3.4 × 10−6 from Schaeffer and Anderson’s (2005) study of differing chromosomal arrangements within D. pseudoobscura, particularly given these inversions are locally adapted (see Fuller et al., this issue). If an inversion is selected for its ability to keep locally adapted alleles together, we would expect that double crossovers would be more deleterious compared to gene conversion, given that double crossovers involve much larger stretches of DNA. Thus, gene conversion may be relatively more effective than crossing over at diminishing genetic divergence outside of the locally adapted sites, but double crossovers are more likely to immediately result in maladaptive combinations.
To assess the relative amount of recombination via gene conversion in inversions, we must also consider the amount of recombination facilitated by double crossovers. A screen of 9739 D. pseudoobscura × D. persimilis hybrids revealed just one double recombinant within the 12 Mb inversion on the right arm of the × chromosome (Stevison et al. 2011). If the double crossover spanned roughly a megabase, the proportion of exchanged sites per genome per generation would be 1/12 × 1/9739, or just under 1 × 10−5. Double crossovers of this size would account for only slightly lower rates of exchange, averaged per site per generation, than our estimated rate of gene conversion. The balance tips in the favor of double crossovers outweighing the effects of gene conversion when double crossovers are larger than 1 megabase. However, double crossover rates are affected by the size of the inversion. Chromosomal inversions experience both crossover interference (reviewed in Berchowitz & Copenhaver 2010) and crossover suppression near inversion breakpoints, and these forces diminish the likelihood that double crossovers will occur at all (or at best, constrain where they can occur) within smaller inversions. In contrast, gene conversion occurs near inversion breakpoints, as we observed a gene conversion as close as 37 kb from a breakpoint. Together with our direct observations of high gene conversion rates within inversions, our results provide direct empirical support for the idea that gene conversion may dominate as the mechanism of genetic exchange within smaller inversions (Navarro et al. 1997; Andolfatto & Nordborg 1998). As our understanding of gene conversion within inversions improves, we will be able to assess the interaction of the effect of inversion length with other variables such as inversion age and the rates and timing of migration. Assessing the time-scale at which gene conversion affects genetic divergence will require in-depth population genetic analyses that incorporate these variables.
To understand the relative contributions of these different outcomes of recombination, we need to work towards more thoroughly developed theory that incorporates analyses of migration rates, variation in gene conversion landscapes, and variation in inversion characteristics such as length and age. The direct evidence that gene conversion occurs within inversions shows that speciation facilitated by chromosomal rearrangement may be more complex than often assumed. These results reinforce the utility of models of gene flow within inversions that account for gene conversion (e.g., Guerrero et al. 2012). Since some theoretical frameworks support the idea that even a modest level of gene flow within a region can shape genetic differentiation over generations (Navarro et al. 1997; Feder & Nosil 2009), neglecting the role of gene conversion within inversions oversimplifies the role that inversions play in genome evolution.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Castillo, J. Coughlan, K. Samuk, and the three reviewers for helpful comments on this manuscript, J. Sekelsky for advice, and N. Crown and D. Miller for sharing their manuscript pre-publication. S. Schaeffer kindly provided us with several of the Drosophila lines used and confirmed their inversion arrangement. This work was supported by the National Science Foundation Graduate Research Fellowship Program (GRFP to KLK) under Grant No. DGE 1644868, Sigma Xi (Grant-in-Aid of Research to KLK), National Institutes of Health grant GM086445 to MAFN, and National Science Foundation grant DEB-1545627 to MAFN.
Footnotes
DATA ACCESSIBILITY
All genome sequence data will be publicly available through NCBI’s Short Read Archive under Project PRJNA492790 at the time of publication. The custom scripts written to analyze the data are publicly available on GitHub (https://github.com/kkorunes/2018_GeneConversion_Pipeline).
REFERENCES
- Andolfatto P, Nordborg M (1998) The Effect of Gene Conversion on Intralocus Associations. Genetics, 148, 1397–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C et al. (2013) From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline In: Current Protocols in Bioinformatics, p. 11.10.1–11.10.33. John Wiley & Sons, Inc., Hoboken, NJ, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske F a. et al. (2009) MEME Suite: Tools for motif discovery and searching. Nucleic Acids Research, 37, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP (2010) Genetic interference: don’t stand so close to me. Current genomics, 11, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E, Rozas J, Navarro A, Barbadilla A (1997) The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics, 146, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton HL, Radford SJ, McMahan S et al. (2005) REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS genetics, 1, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-M, Cooper DN, Chuzhanova N, Férec C, Patrinos GP (2007) Gene conversion: mechanisms, evolution and human disease. Nature reviews. Genetics, 8, 762–75. [DOI] [PubMed] [Google Scholar]
- Chovnick A (1973) Gene Conversion and Transfer of Genetic Information within the Inverted Region of Inversion Heterozygotes. Genetics, 75, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown KN, Miller DE, Sekelsky J, Hawley RS (2018) Local Inversion Heterozygosity Alters Recombination throughout the Genome. Current biology : CB, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R et al. (2011) A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nature Genetics, 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1943) Genetics of Natural Populations IX. Temporal Changes in the Composition of Populations of Drosophila Pseudoobscura. Genetics, 28, 162–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1948) Genetics of natural populations; experiments on chromosomes of Drosophila pseudoobscura from different geographic regions. Genetics, 33, 588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T (1973) Is there Gene Exchange between Drosophila pseudoobsura and Drosophila persimilis in Their Natural Habitats? The American Naturalist, Vol. 107, No. 954, pp. 312–314. [Google Scholar]
- Dobzhansky T, Epling C (1944) Contributions to the Genetics, Taxonomy, and Ecology of Drosophila Pseudoobscura and its relatives. Carnegie Institute of Washington, 544, 1–46. [Google Scholar]
- Feder JL, Nosil P (2009) Chromosomal Inversions and Species Differences: When Are Genes Affecting Adaptive Divergence and Reproductive Isolation Expected to Reside Within Inversions? Evolution, 63, 3061–75. [DOI] [PubMed] [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF, Hill JP (2013) Chromosomal Rearrangements and the Genetics of Reproductive Barriers in Mimulus (Monkey Flowers). Evolution, 67, 2547–60. [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW (2017) Genomics of Natural Populations: Evolutionary Forces that Establish and Maintain Gene Arrangements in Drosophila pseudoobscura. Molecular Ecology, 26, 6539–6562. [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Leonard CJ, Young RE, Schaeffer SW, Phadnis N (2018) Ancestral Polymorphisms Explain the Role of Chromosomal Inversions in Speciation. PLOS Genetics, 14, e1007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG (2001) Local Rates of Recombination Are Positively Correlated with GC Content in the Human Genome. Molecular Biology and Evolution, 18, 1139–1142. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L (2001) GC-Content Evolution in Mammalian Genomes: The Biased Gene Conversion Hypothesis. Genetics, 159, 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Arndt PF, Messer PW et al. (2015) Quantification of GC-biased gene conversion in the human genome. Genome research, 25, 1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WJ, McKim KS, Hawley RS (2005) All Paired Up with No Place to Go: Pairing, Synapsis, and DSB Formation in a Balancer Heterozygote. PLoS Genetics, 1, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, dos Santos G et al. (2017) FlyBase at 25: Looking to the Future. Nucleic Acids Research, 45, D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics, 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Rousset F, Kirkpatrick M (2012) Coalescent Patterns for Chromosomal Inversions in Divergent Populations. Philosophical transactions of the Royal Society B, 367, 430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R (2004) Multilocus Methods for Estimating Population Sizes, Migration Rates and Divergence Time, with Applications to the Divergence of Drosophila pseudoobscura and D. persimilis. Genetics, 167, 747–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH (2008) Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annual review of ecology, evolution, and systematics, 39, 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Charlesworth B (1977) Associations between Allozyme Loci and Gene Arrangements Due to Hitch-hiking Effects of New Inversions. Genetics Research, 30, 93–106. [Google Scholar]
- Jeffreys AJ, May CA (2004) Intense and Highly Localized Gene Conversion Activity in Human Meiotic Crossover Hot Spots. Nature Genetics, 36, 151–6. [DOI] [PubMed] [Google Scholar]
- Judd SR, Petes TD (1988) Physical Lengths of Meiotic and Mitotic Gene Conversion Tracts in Saccharomyces cerevisiae. Genetics, 118, 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Sheng Z, Lobachev KS, Petes TD (2018) GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 115, E7109–E7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M (2017) The Evolution of Genome Structure by Natural and Sexual Selection. Journal of Heredity, 108, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barrett B (2015) Chromosome Inversions, Adaptive Cassettes and the Evolution of Species’ Ranges. Molecular Ecology, 24, 2046–55. [DOI] [PubMed] [Google Scholar]
- Korunes KL, Noor MAF (2017) Gene Conversion and Linkage: Effects on Genome Evolution and Speciation. Molecular Ecology, 26, 351–364. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MAF (2008) Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proceedings of the National Academy of Sciences of the United States of America, 105, 10051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF (2009) The Genomics of Speciation in Drosophila: Diversity, Divergence, and Introgression Estimated Using Low-Coverage Genome Sequencing. PLoS genetics, 5, e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesecque Y, Mouchiroud D, Duret L (2013) GC-biased gene conversion in yeast is specifically associated with crossovers: molecular mechanisms and evolutionary significance. Molecular biology and evolution, 30, 1409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP (1956) Crossing Over and Inversions in Coadapted Systems. The American Naturalist, Vol. 90, No. 850 41–45. [Google Scholar]
- Li H, Durbin R (2009) Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman S, Symington L, Petes T (1988) Mitotic Recombination within the Centromere of a Yeast Chromosome. Science, 241, 1074–1077. [DOI] [PubMed] [Google Scholar]
- Liskay RM, Letsou A, Stachelek JL (1987) Homology Requirement for Efficient Gene Conversion Between Duplicated Chromosomal Sequences in Mammalian Cells. Genetics, 115, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li H (2008) The Correlation Between Recombination Rate and Dinucleotide Bias in Drosophila melanogaster. Journal of Molecular Evolution, 67, 358–367. [DOI] [PubMed] [Google Scholar]
- Machado CA, Haselkorn TS, Noor MAF (2007) Evaluation of the Genomic Extent of Effects of Fixed Inversion Differences on Intraspecific Variation and Interspecific Gene Flow in Drosophila pseudoobscura and D. persimilis. Genetics, 175, 1289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Hey J (2003) The causes of phylogenetic conflict in a classic Drosophila species group. Proceedings of the Royal Society B: Biological Sciences, 270, 1193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM (2008) High-Resolution Mapping of Meiotic Crossovers and Non-Crossovers in Yeast. Nature, 454, 479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano-Winkler B (2015) Suppressed double crossovers in D. pseudoobscura inversion heterozygotes. Drosophila Information Service, 57–59. [Google Scholar]
- McGaugh SE, Noor MAF (2012) Genomic Impacts of Chromosomal Inversions in Parapatric Drosophila species. Philosophical Transactions of the Royal Society B: Biological Sciences, 367, 422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Smith CB, Yeganeh Kazemi N et al. (2016) Whole-Genome Analysis of Individual Meiotic Events in Drosophila melanogaster Reveals that Noncrossover Gene Conversions are Insensitive to Interference and the Centromere Effect. Genetics, 203, genetics.115186486-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Takeo S, Nandanan K et al. (2012) A Whole-Chromosome Analysis of Meiotic Recombination in Drosophila melanogaster. G3, 2, 249–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Betrán E, Barbadilla A, Ruiz A (1997) Recombination and Gene Flux Caused by Gene Conversion and Crossing Over in Inversion Heterokaryotypes. Genetics, 146, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S (2000) Molecular Evolution and Phylogenetics. Oxford University Press. [Google Scholar]
- Nóbrega C, Khadem M, Aguadé M, Segarra C (2008) Genetic Exchange Versus Genetic Differentiation in a Medium-Sized Inversion of Drosophila: the A2/Ast Arrangements of Drosophila subobscura. Molecular Biology and Evolution, 25, 1534–43. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Garfield DA, Schaeffer SW, Machado CA (2007) Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics, 177, 1417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA et al. (2001a) The Genetics of Reproductive Isolation and the Potential for Gene Exchange Between Drosophila pseudoobscura and D. persimilis via Backcross Hybrid Males. Evolution, 55, 512–521. [DOI] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J (2001b) Chromosomal Inversions and the Reproductive Isolation of Species. Proceedings of the National Academy of Sciences, 98, 12084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR (1983) Interspecific Cytoplasmic Gene Flow in the Absence of Nuclear Gene Flow: Evidence from Drosophila. Proceedings of the National Academy of Sciences, 80, 492–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR (1992) Inversion Polymorphisms in Drosophila pseudoobscura and Drosophila persimilis. Drosophila Inversion Polymorphism, 73–126. [Google Scholar]
- Robinson MC, Stone EA, Singh ND (2014) Population genomic analysis reveals no evidence for GC-biased gene conversion in Drosophila melanogaster. Molecular biology and evolution, 31, 425–33. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W et al. (2011) Integrative Genomics Viewer. Nature Biotechnology, 29, 24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Aguadé M (1994) Gene Conversion is Involved in the Transfer of Genetic Information between Naturally Occurring Inversions of Drosophila. Proceedings of the National Academy of Sciences, 91, 11517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, Anderson WW (2005) Mechanisms of Genetic Exchange within the Chromosomal Inversions of Drosophila pseudoobscura. Genetics, 171, 1729–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, Bhutkar A, McAllister BF et al. (2008) Polytene Chromosomal Maps of 11 Drosophila Species: The Order of Genomic Scaffolds Inferred from Genetic and Physical Maps. Genetics, 179, 1601–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Redfield H (1951) Interchromosomal Effects on Crossing Over in Drosophila. Cold Spring Harbor Symposia on Quantitative Biology, 16, 175–97. [DOI] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM et al. (2010) Widespread Gene Conversion in Centromere Cores. PLoS biology, 8, e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh ND, Stone EA, Aquadro CF, Clark AG, Corley-Smith G (2013) Fine-scale heterogeneity in crossover rate in the garnet-scalloped region of the Drosophila melanogaster × chromosome. Genetics, 194, 375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison LS, Hoehn KB, Noor MAF (2011) Effects of Inversions on Within- and Between-Species Recombination and Divergence. Genome Biology and Evolution, 3, 830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison LS, Noor MAF (2010) Genetic and Evolutionary Correlates of Fine-Scale Recombination Rate Variation in Drosophila persimilis. Journal of Molecular Evolution, 71, 332–345. [DOI] [PubMed] [Google Scholar]
- Stump AD, Pombi M, Goeddel L et al. (2007) Genetic Exchange in 2La Inversion Heterokaryotypes of Anopheles gambiae. Insect molecular biology, 16, 703–9. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH (1913) A third group of linked genes in Drosophila ampelophila. Science, 37, 990–992. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH (1915) The behavior of the chromosomes as studied through linkage. Zeitschrift für Induktive Abstammungs- und Vererbungslehre, 13, 234–287. [Google Scholar]
- Talbert PB, Henikoff S (2010) Centromeres Convert but don’t Cross. PLoS biology, 8, e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Briefings in Bioinformatics, 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman AS, Liskay RM (1988) Dependence of Intrachromosomal Recombination in Mammalian Cells on Uninterrupted Homology. Molecular and Cellular Biology, 8, 5350–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Axelsson E, Ellegren H (2006) Strong regional biases in nucleotide substitution in the chicken genome. Molecular biology and evolution, 23, 1203–16. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NGC, Hultin-Rosenberg L, Arndt PF, Ellegren H (2005) Male-driven biased gene conversion governs the evolution of base composition in human alu repeats. Molecular biology and evolution, 22, 1468–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.