Abstract
A major limitation of targeted cancer therapy is the rapid emergence of drug resistance, which often arises through mutations at or downstream of the drug target or through intrinsic resistance of subpopulations of tumor cells. Medulloblastoma (MB), the most common pediatric brain tumor, is no exception, and MBs that are driven by sonic hedgehog (SHH) signaling are particularly aggressive and drug-resistant. To find new drug targets and therapeutics for MB that may be less susceptible to common resistance mechanisms, we used a developmental phosphoproteomics approach in murine granule neuron precursors (GNPs), the developmental cell of origin of MB. The protein kinase CK2 emerged as a driver of hundreds of phosphorylation events during the proliferative, MB-like stage of GNP growth, including the phosphorylation of three of the eight proteins commonly amplified in MB. CK2 was critical to the stabilization and activity of the transcription factor GLI2, a late downstream effector in SHH signaling. CK2 inhibitors decreased the viability of primary SHH-type MB patient cells in culture and blocked the growth of murine MB tumors that were resistant to currently available Hh inhibitors, thereby extending the survival of tumor-bearing mice. Because of structural interactions, one CK2 inhibitor (CX-4945) inhibited both wild-type and mutant CK2, indicating that this drug may avoid at least one common mode of acquired resistance. These findings suggest that CK2 inhibitors may be effective for treating patients with MB and show how phosphoproteomics may be used to gain insight into developmental biology and pathology.
INTRODUCTION
A key challenge in developing cancer therapeutics is the identification of a target protein that is essential to the growth, survival, or metastasis of a tumor. One path to such proteins is to test developmental regulators that operate in normal cells from which the tumor is derived. A prime example is medulloblastoma (MB), the most common malignant pediatric brain tumor. Developmental, genetic, and transcriptional analyses have established a clear parallel between sonic hedgehog (SHH)–subtype MB and granule neuron precursors (GNPs) (1–3). During normal cerebellar development, GNPs proliferate extensively in response to hedgehog (Hh) signaling (2) before differentiating into granule neurons, the most abundant type of neuron in the brain (Fig. 1A) (4). In mice, this period begins at postnatal day 1 (P1), peaks at P7, and is largely complete by P14, at which point remaining GNPs have stopped dividing and have begun differentiating (5) into granule neurons (2, 6). Sustained Hh target gene activity in GNPs, due to reduced function of pathway components that negatively regulate Hh signal transduction, such as Patched (PTCH1) (1), or to heightened function of activating Hh signal transducers or effectors, such as GLI2 (3), results in continued proliferation of GNPs beyond P14 and eventual rise of SHH-type MB.
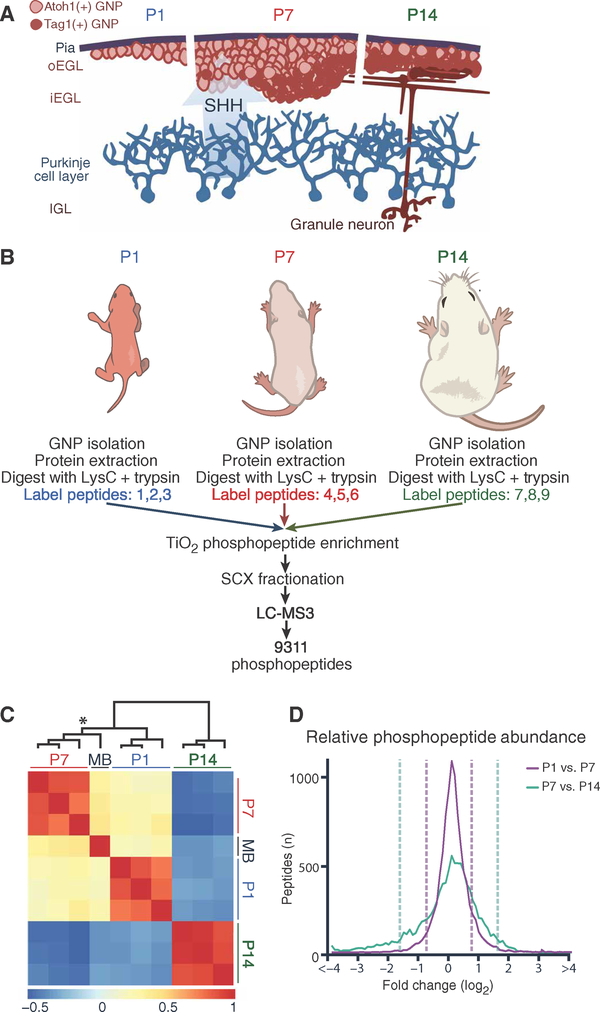
Fig. 1. Quantitative mapping of the phosphoproteome during GNP development.
(A) Schematic of early postnatal proliferation and differentiation of GNPs. Pink, Atoh1-positive proliferative GNPs; red, postmitotic GNPs. oEGL/iEGL, outer/inner external granule layer; IGL, internal granule layer. (B) Experimental scheme for the phosphoproteomic assays. (C) Heat map representing relative phosphopeptide abundance and undirected clustering among three biological replicates and sample types (P1, P7, and P14 GNPs and Ptch+/− MB). Color axis = R2. *Cluster branch of P7 GNPs versus MB sample. (D) Distribution of relative phosphopeptide abundance at developmental transitions. Dashed lines = 1.5 SDs. Mass spectrometry (MS) was performed in n = 3 biological replicates per experimental time point (14 to 40 mice per replicate at each time point).
To date, drugs for SHH-type MB inhibit Smoothened (SMO), a transmembrane protein that acts early in the Hh signal transduction pathway. Patients treated with SMO inhibitors initially have dramatic tumor regression but eventually develop resistance due to mutations in SMO or in genes encoding downstream components of the Hh pathway (7). Worse, at the time of diagnosis, 49% of infants and 59% of children have mutations downstream of SMO; hence, these tumors are resistant to SMO inhibitors from the start (8). Despite the great potential of Hh pathway inhibitors, children with MBs continue to receive multiple nontargeted therapies and, consequently, sustain long-term neurological and cognitive problems. There is a pressing need to identify novel drug targets that affect Hh signal transduction downstream of SMO, preferably at late steps in the pathway. Ideally, inhibition of this target using a specific drug would be refractory to single mutations of Hh pathway components.
To identify important candidate drug targets for MBs, and to explore regulation of Hh transduction, we performed a proteome-wide analysis of in vivo phosphorylation events occurring in murine GNPs during the initiation, peak, and completion of Hh-driven proliferation. Unlike genome-wide transcription assays, phosphoproteomics provided measurement of the modified state of proteins at each developmental stage and thereby inferred which kinases have stage-specific changes in activity. Kinases that are specifically active during the MB-like P7 stage of GNP development may also be required for MB growth. Kinases are critical regulators of Hh signaling (9, 10) and are attractive drug targets, susceptible to inhibition by competitive inhibitors, small molecules that interfere with the binding of adenosine 5′-triphosphate (ATP) (11–13). If a kinase has multiple substrates that are important for Hh transduction and/or survival of MB cells, then the drug will have multiple restraining influences on tumor growth, and mutation of any single substrate is unlikely to cause drug resistance.
RESULTS
Proliferating and postmitotic GNPs have distinct phosphoproteomes
Using a protocol summarized schematically in Fig. 1B, we identified 9311 unique phosphorylation events in GNPs purified from P1, P7, and P14 mouse cerebellum. The false discovery rate (FDR) was <1% (table S1). All samples were prepared in biological triplicate, and the results were strongly reproducible (R2 = 0.76; Fig. 1C and fig. S1). A single MB sample generated from Ptck1+/− mice (1, 14) was included for comparison (Fig. 1C and table S1). In keeping with previous literature (5), P1 and P7 Atoh1-positive cells were highly proliferative, whereas P14 GNPs were almost universally postmitotic (fig. S2). Many changes in phosphorylation distinguished proliferating (P1 and P7) from early postmitotic (P14) GNPs (Fig. 1, C and D). Only 4.9% of the phosphorylated peptides changed in relative phosphorylation level more than twofold from P1 to P7 (3.6% increased, 1.3% decreased); in contrast, 21.4% of phosphorylated peptides changed more than two-fold between P7 and P14 (18.2% decreased, 3.2% increased; Fig. 1D). Strikingly, the pattern of protein abundance and phosphorylation in P7 GNP samples was more similar to that of MB cells than to that of either P1 or P14 GNPs, demonstrating strong similarity between peak proliferating GNPs and MB even at the minute-to-minute kinetics of phosphorylation (Fig. 1C, asterisk). Peptides more highly phosphorylated in P1, P7, and MB relative to P14 included known pro-proliferative phosphorylation sites of the cell cycle regulator retinoblastoma 1 (RB1) (fig. S2) and the proteins encoded by three of eight genes that commonly undergo copy number aberrations in human MBs (Gab1, Irs2, and Yap-1) (15).
CK2 substrates are more highly phosphorylated in GNPs isolated at P7
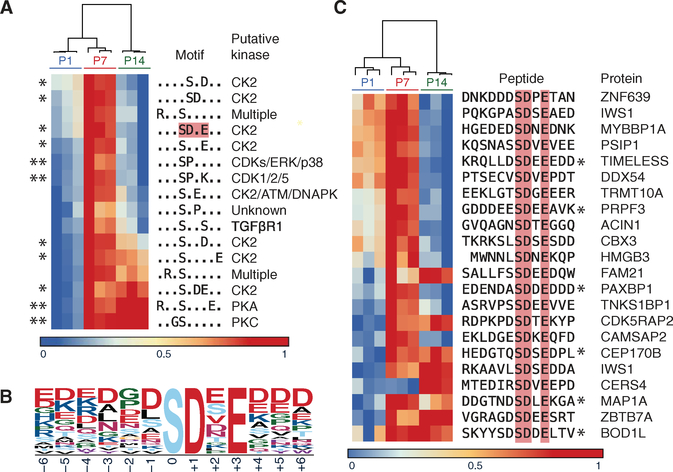
Motif analysis of phosphorylation events in GNPs that changed in frequency (>1.5 SD) over time suggested candidate kinases whose activity might be different during proliferation versus differentiation. Sixteen phosphorylation consensus motifs were prominent among 1522 protein sequences that had phosphorylation changes (Fig. 2A). Four observed motifs [for cyclin-dependent kinases (CDKs), protein kinase A (PKA), and PKC] were consistent with past studies (4, 10, 16). Of the remaining motifs, 7 of 12 matched those targeted by the protein kinase CK2 (casein kinase 2)—the target motifs of which have acidic residues C-terminal to a phosphorylated serine or threonine (Fig. 2, A to C, and table S2) (17). Notably, of 278 phosphorylated peptides with possible CK2 target motifs, 83.8% were more highly phosphorylated in P7 compared to P1 or P14. CK2 itself was not more highly expressed at P7 compared to P1 or P14 in terms of either protein or transcript abundance (18), which is likely why CK2 has not been previously identified in large-scale transcriptional assays of cerebellar development and MB (18, 19). In keeping with this, many CK2 substrates did not change in phosphorylation abundance (table S1). Our findings therefore likely reflect regulated and localized change specifically in CK2 activity, which could have resulted from changes in kinase localization and/or trafficking of its subunits, co-regulator binding, phosphatase abundance, or accessibility of substrate motifs or docking sites (20). This highlights the importance of investigating kinase activity at its terminal readout: proteome-wide assessment of phosphorylation.
Fig. 2. Phosphoproteomic data indicate increased phosphorylation at CK2 motifs at P7.
(A and B) The 16 motifs enriched in the fractions of altered phosphopeptide abundance (>1.5 SDs) in the phosphoproteomic analysis of GNPs purified from mice harvested on P1, P7, and P14 (A). The predicted kinase of each motif is noted down the right. *CK2 target motifs. **Known regulators of GNP development. Color axis: Fraction of maximum enrichment. Red box: CK2 motif characterized by motif logo (B). (C) Enriched phosphopeptides, and the corresponding protein, contributing to the SD.E motif-enriched phosphopeptides described in (A). Proteins include known (*) and currently uncharacterized CK2 substrates. Data are n = 3 biological replicates per experimental time point (14 to 40 mice per replicate at each time point).
CK2 inhibition results in decreased GNP proliferation
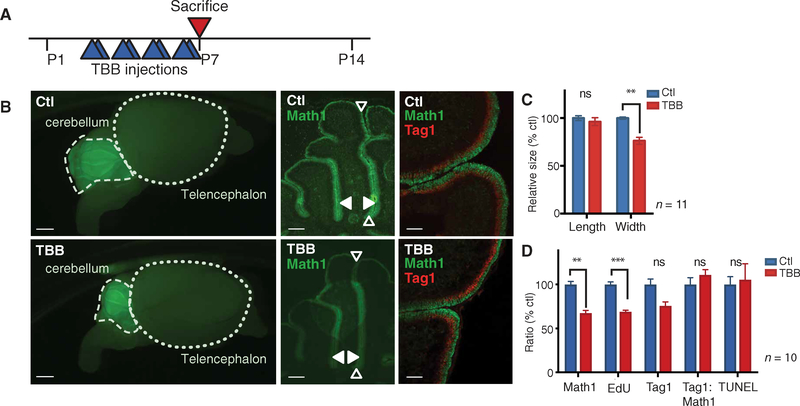
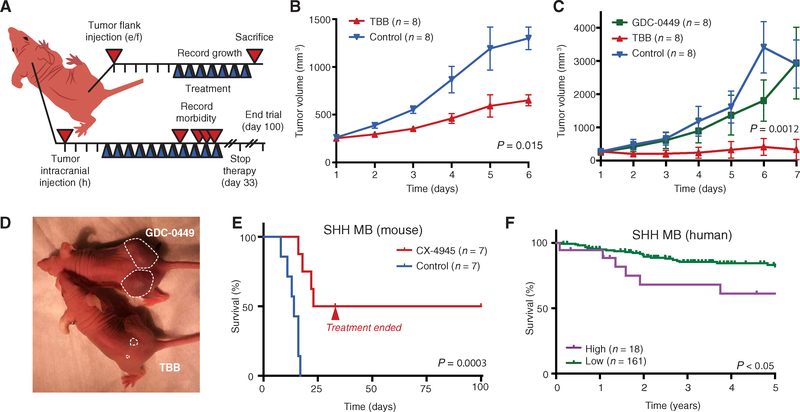
Many peptides with CK2 consensus sites were more highly phosphorylated at the peak proliferative period (P7), suggesting that CK2 activity is important for GNP proliferation. We tested the role of CK2 in cerebellar development by injecting mice with the CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) (Fig. 3A) (21). Although several CK2 inhibitors exist, TBB was used because it is fairly selective (22), is cell-permeable (23), lacks short-term cytotoxicity in vivo (24), and is effective at preventing seizure activity in mice (24). Its potency suggests blood-brain barrier (BBB) permeability. Mice were treated twice a day, from P3 to P7, with TBB [30 mg/kg, intra-peritoneally (ip)] (25) or vehicle control [dimethyl sulfoxide (DMSO)]. Four hours after the final injection, mice were sacrificed and their cerebella were fixed. TBB-treated mice had no change in cerebellar folia length, which is affected by non–GNP-derived neurons (Fig. 3, B and C) (26). In contrast, folia width, which depends on the number of proliferating GNPs and their progeny, granule neurons, was significantly reduced by TBB treatment (P < 0.01; Fig. 3, B and C). The number of Atoh1-positive (P < 0.01) and proliferating GNPs at P7 (P < 0.001) was significantly reduced in treated mice (Fig. 3, B and D). No significant change in the rate of apoptosis or the ratio of proliferation to differentiation was detected (Fig. 3, B and D). Similarly, GNPs cultured in the presence of SHH and CX-4945, a CK2 inhibitor that is highly specific and structurally distinct from TBB, had a dose-dependent decrease in proliferation and a correlating decrease in total Atoh1-positive cells at 24 hours (fig. S3), as assessed by single-cell immunofluorescence imaging. The decrease in folia width was likely to be due to reduced GNP proliferation in response to inhibition of CK2 during the critical postnatal period.
Fig. 3. CK2 is required for cerebellar development.
(A) Experimental scheme for CK2 inhibitor (TBB) studies in early postnatal mice. (B and C) Immunofluorescent imaging of whole brains and sagittal cerebellar sections from control (DMSO) and CK2 inhibitor-treated Math1/nGFP transgenic mice. Green, Math1 (proliferating GNP marker); red, Tag1 (postmitotic GNP marker). Empty arrow: Folia length. Solid arrow: Folia width. (D) Ratio of Math1-expressing GNPs, early differentiating GNPs (Tag1), proliferation [5-ethynyl-2′-deoxyuridine (EdU)], and apoptosis [terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL)] in TBB-treated mice relative to control mice. Data are means ± SD from n = 10 to 11 mice, as indicated. ns = P > 0.05, **P < 0.01, ***P < 0.001, two-tailed t test. Scale bars, 1 mm (left), 0.1 mm (middle), and 0.05 mm (right).
CK2 facilitates Hh signal transduction
Because GNP proliferation in neonatal mice is driven by Hh signaling, we investigated whether CK2 affects the response to SHH. Hh signal transduction is initiated when SHH ligand binds and inhibits the cell surface receptor PTCH1 (Fig. 4A) (1, 27). SHH inhibition of PTCH1 unleashes the G protein (heterotrimeric GTP-binding protein)-coupled receptor SMO (28, 29), which then inhibits a protein called suppressor of fused (SUFU), in turn an inhibitor of the transcription factor GLI2 (30). Inhibition of SUFU thus allows transcription of GLI2 target genes, such as GLI1. Note that CK2 is an acronym derived from the misnomer “casein kinase-2” and is unrelated in both structure and function to the known GLI2 regulator CK1.
Fig. 4. CK2 is required for Hh signal transduction.
(A) Canonical Hh signaling pathway, for reference in subsequent panels. (B and C) Fold change in Gli1 mRNA expression in NIH3T3 cells exposed to murine SHH (3 μg/ml) for 6 hours after knockdown of CSNK2A1, CSNK2A2, or CSNK2B (B) or when treated with the CK2 inhibitor TBB (50 μM) or CX-4945 (10 μM) or SMO inhibitor GDC-0449 (100 nM) (C) relative to controls. (D and E) Gli1 mRNA expression after exposure to TBB (as indicated) or GDC-0449 (100 nM) in NIH3T3 cells pretreated with SMO agonist SAG (100 nM) (D) and in Sufu−/− MEFs (E). (F) Immunoblotting for protein abundance of endogenous GLI2 (GLI2FL, full length) and exogenous constitutively active GLI2 (GLI2ΔN) in NIH3T3 cells treated with various concentrations of TBB for 6 hours. Blots are from a representative of three experiments. (G) Gli1 mRNA expression after exposure to TBB in NIH3T3 cells expressing GLI2ΔN. Data in (A) to (D) and (G) were obtained by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and are shown as means ± SD from n = 3 experiments. ns = P > 0.05, *P < 0.05, **P < 0.01, ****P < 0.0001, two-tailed t test.
Genetic and small-molecule inhibitor studies in NIH3T3 cells (spontaneously immortalized murine embryonic fibroblasts) confirmed a role for CK2 in Hh signal transduction. NIH 3T3 cells are routinely used to assay the Hh pathway because they contain all canonical components of the Hh transduction pathway and have a robust transcriptional response to SHH ligand in culture. In contrast, GNPs in culture exit the cell cycle and rapidly differentiate into non-SHH-responsive granule neurons. A change in Gli1 transcript expression in GNPs can result from perturbation of the Hh pathway or from non–Hh-dependent perturbations, such as cell cycle inhibitors or changes in ion homeostasis, so NIH3T3 cells are used instead.
CK2 is a heterotetrameric holoenzyme made up of four subunits: two catalytic α subunits (either two CSNK2A1 subunits or one CSNK2A1 subunit and one CSNK2A2 subunit) and two regulatory β subunits (CSNK2B). Genetic loss in mice of either CSNK2A1 or CSNK2B results in early embryonic death (31, 32). CSNK2A2 can generally be replaced by CSNK2A1, and knockout of Csnk2a2 in mice results in infertility in males but otherwise has no apparent phenotypic consequence (31).
Small interfering RNA (siRNA)-mediated knockdown of the essential catalytic α subunit, CSNK2A1, in NIH3T3 cells strongly reduced the induction of Gli1 expression by SHH (Fig. 4B). As expected, the effect of knockdown of only the nonessential α prime subunit, CSNK2A2, on Gli1 transcription was not significant. Knockdown of CSNK2B resulted in a significant but modest decrease in Gli1 induction, consistent with previous evidence (32) that even low CSNK2B protein abundance, as in Csnk2b knockdown cells (fig. S4), is sufficient to make substantial amounts of CK2 holoenzyme. Each of the mRNA knockdowns was subunit-specific and did not reduce expression of the non-Hh target Smo (fig. S4).
Two structurally unrelated CK2 inhibitors, CX-4945 and TBB (21, 33), each significantly reduced the expression of the SHH-induced GLI2 targets (Gli1, Gli2, and Ptchl) in NIH3T3 cells as early as 4 hours after drug administration (Fig. 4C). CK2 inhibition did not decrease the transcript abundance of the non-Hh target gene Smo (fig. S5). The rapid influence of CK2 inhibitors on Hh pathway target gene expression suggests that the slower (72-hour) effects of CSNK2A1 knockdown in NIH3T3 cells were due to direct action on Hh signal transduction rather than indirect effects from prolonged CK2 inhibition.
CK2 facilitates Hh signal transduction at late steps in the pathway
When NIH3T3 cells were stimulated with an SMO agonist “SAG” (100 nM) (34), the CK2 inhibitor TBB blocked the increase in Gli1 transcript that normally occurs in response to SAG, working as well as the SMO antagonist GDC-0449 (100 nM; also known as vismodegib) (Fig. 4D) (35). Therefore, CK2 regulates Hh signal transduction at or downstream of SMO. Treatment of Sufu−/− mouse embryonic fibroblasts (MEFs) (36) with TBB also strongly blocked transcription of Gli1 (Fig. 4E), indicating that CK2 is needed at late steps of the Hh signal transduction pathway, at or downstream of SUFU. Treatment with the SMO antagonist GdC-0449 (100 nM) did not block transcription of Gli1 in Sufu−/− cells, as expected, because SMO is upstream of SUFU in the pathway.
Our experiments demonstrating action of CK2 at a late step of the Hh pathway are consistent with previous work showing that the CK2 inhibitor TBB affects GLI2 protein stability, likely through regulation of GLI2 ubiquitination (37, 38). In keeping with this, TBB reduced the amount of endogenous full-length GLI2 expressed in NIH3T3 cells (Fig. 4F), as previously shown (37). TBB did not affect the abundance of an N-terminal-deleted form of Gli2 (HA-Gli2ΔN, hereafter simply Gli2ΔN; Fig. 4F), stably transfected into NIH3T3 cells (39). Gli2ΔN lacks sites required for ubiquitin-dependent degradation (40), remains constitutively nuclear, and normally causes constitutive induction of the GLI2 target gene Gli1. However, treatment with TBB still substantially lowered Gli1 mRNA expression in Gli2ΔN-transfected cells (Fig. 4G). Together, the results suggest that CK2 regulates two steps in the Hh pathway: to stabilize GLI2 itself and to enhance the ability of GLI2 to activate its transcriptional targets. Our phosphoproteomic data point to several potential candidate substrates that might mediate effects of CK2 on GLI2-mediated transcription. For example, phosphorylated CK2 sites were found within enhancer-looping proteins [such as RING finger protein 1 (RING1), nipped-B–like protein (NIPBL), and mediator complex subunit 1 (MED1)] and epigenetic readers and writers [such as Polycomb repressive complex 1 (PRC1), PRC2, and histone deactylase (HDAC) components]. In two cases, the CK2 sites are well-established activating phosphorylation sites (in HDAC1 Ser393/421 and HDAC2 Ser294/422). CK2 may regulate accessibility of GLI2 target gene promoters or their interactions with enhancers.
CK2 inhibitors restrict the growth of mouse and human SHH MB cells in culture
SHH MBs occur because of sustained Hh target gene activity and require persistent action of the Hh signal transduction pathway for survival. Therefore, we reasoned that CK2 inhibitors may be effective at restricting the growth of this MB subtype. Unfortunately, most SHH MB cell lines suppress Hh pathway activity when propagated in vitro and are therefore not useful models of MB. Several previously established SHH MB cell lines that we tested had suppressed SHH pathway activity as indicated by low expression of the Hh target Gli1 (fig. S6). After careful screening, we chose to use the cell lines MB21, MB53, and MB55 (41) in further tests. These murine MB lines retain key characteristics of primary MB tumors that arise in Ptch+/− mice, including sustained, high amounts of Gli1 transcription that is inhibited by SMO inhibitors both in our hands (fig. S6) and in published accounts (41, 42). Hh signaling activity and expression of Hh pathway components in these cell lines are in keeping with primary mouse SHH MB tumors, and when cells are implanted into the flank or cerebellum of mice, they grow into tumors with SHH-subtype histology (41). Gene expression patterns also strongly correlate with both primary mouse SHH MB cells and human SHH MBs, but not human non-SHH subgroup MBs (WNT, group 3, or group 4) (41).
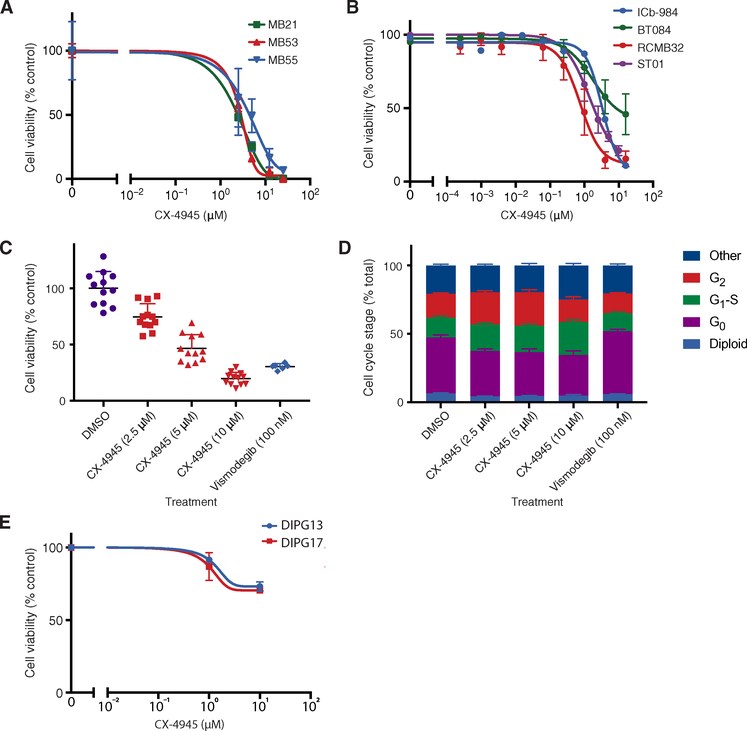
Treatment of MB21, MB53, and MB55 Ptch+/− mouse MB cell lines in culture with the CK2 inhibitor CX-4945 reduced Gli1 expression after 6 hours (fig. S6), suggesting inhibition of Hh pathway activity. The cells underwent dose-dependent cell death after 3 days of treatment [median inhibitory concentration (IC50) = 2.5 to 5.3 μM, relative, normalized to control (DMSO)–treated cells] (Fig. 5A). Three human SHH MB patient-derived xenograft (PDX) lines passaged only in mouse cerebellum were assayed in vitro and similarly demonstrated dose-dependent cell death after 3 days of treatment (Fig. 5B). A human SHH MB tumor cell line with the same activating mutation as the mouse MB cells (RCMB32, Ptch+/−) had even greater sensitivity to CX-4945 (IC50 = 0.76 μM) than MB21, MB53, or MB55. Two other human SHH MB cell lines have TP53 mutations and SHH pathway-activating mutations downstream of SMO, so they are resistant to SMO inhibitors [ICb-984, IC50 = 3.3 μM (GLI2;MYCN-amplified; TP53−/−; chromothripsis); BT084, IC50 = 1.8 μM (GLI2;MYCN;TERT-amplified; TP53−/−; chromothripsis)] (43, 44). These cell lines represent some of the most aggressive forms of human SHH MB. Primary human SHH MB cells (ST01), obtained directly from the operating room and confirmed by pathology to be of the SHH subgroup, also had a dose-dependent decrease in cell viability when treated with CX-4945 for 3 days (IC50 = 1.4 μM; Fig. 5, B and C). As expected, treatment with the SMO inhibitor vismodegib also reduced cell viability and at lower concentrations than CX-4945 (100 nM versus 10 μM), reflecting the favorable ligand abundance, binding accessibility, and/or drug-substrate interactions compared to CX-4945. Notably, cells in G0, which are commonly resistant to chemotherapy and are a cause of SHH MB recurrence (45), were the most sensitive cell population to treatment with CX-4945 (11.1% relative decrease in G0 cells) but the least sensitive population to treatment with vismodegib (5.3% relative increase in G0 cells; Fig. 5D). Control cell lines derived from diffuse intrinsic pontine glioma (DIPG), another aggressive pediatric brain tumor that is not Hh pathway-dependent, did not die in response to CX-4945 (Fig. 5E).
Fig. 5. Efficacy of CK2 inhibitors against MB in vitro.
(A and B) Cell viability after 72 hours of exposure to CX-4945 (10 μΜ), assessed by luminescent cell viability assays, in murine Ptch+/− (MB21 and MB55), Ptch+/−;Tpr53−/− (MB53) MB lines in culture (A) and in human MB PDX lines (RCMB32, BT084, ICb-984, and ST01) assayed in culture (B). (C and D) Cell viability and cell cycle stage of human SHH MB cells cultured in DMSO, CX-4945, or vismodegib for 24 hours, as assessed by high-throughput single-cell imaging. (E) Cell viability after 72 hours of exposure to CX-4945 (10 μM), assessed by luminescent cell viability assays, in pediatric glioma (DIPG) cells. Data are means ± SD from n = 3 experiments.
CK2 inhibitors are effective against SHH MB models in vivo
Ptch+/−;Tpr53−/−;SmoD477G mouse MB cells, which have a mutation in SMO that causes resistance to the U.S. Food and Drug Administration (FDA)-approved Hh (Smo) inhibitor vismodegib (GDC-0449), were injected into the flanks of mice, and the resulting tumors were allowed to grow to 250 to 300 mm3 (Fig. 6A). A parallel experiment was done using implants of Ptch+/−; Tpr53−/− MB cells, which lack the activating SMO mutation and are therefore susceptible to vismodegib. Preclinical work for the two FDA-approved Hh inhibitors (GDC-0449 and LDE225) relied on mouse flank allografts using the same cell lines as those used here (40, 46,47). These mouse cells reportedly closely resemble human MB cells with respect to gene expression (41), response to treatment (40, 47–49), and even the spontaneous occurrence of treatment-induced, drug-resistant mutations (35, 46). Treatment of mice harboring flank allografts with the CK2 inhibitor TBB [30 mg/kg, ip twice daily (BID)] significantly inhibited MB growth as early as 2 days after initiating treatment and, in vismodegib-resistant tumors, near-complete cessation of tumor growth (Fig. 6, B to D).
Fig. 6. Efficacy of CK2 inhibitors against MB in vivo.
(A) Experimental scheme for flank and cerebellar tumor trials. Blue triangle, treatment. (B and C) Relative tumor growth of Ptch+/−;Tpr53−/− (B) and Ptch+/−;Tpr53−/−;SmoD477G (C) MB flank allografts. Data are means ± SD; P values by a two-tailed t test. (D) Representative picture of mice treated with TBB versus GDC-0449. Dashed line demarcates the tumor. (E) Kaplan-Meier survival analysis of mice with Ptch+/−;Tpr53−/−;SmoD477G MB cerebellar allografts treated with CX-4945 or vehicle control. n = 7 mice in each treatment group. (F) Kaplan-Meier analysis of CK2 gene expression and overall survival in human patients (n = 179) with SHH-subtype MB. P values in (E) and (F) were determined by a log-rank Mantel-Cox test.
CK2 inhibition prolonged survival in mice with cerebellar MB. Eighty thousand Ptch+/−;Tpr53−/−;SmoD477G mouse MB cells per mouse were injected into cerebella of NSG mice (Fig. 6A). Seventy-two hours after tumor cells were injected, a collaborator who was blinded to treatment randomized the mice into two groups and began administering either the CK2 inhibitor CX-4945 (37.5 mg/kg) or vehicle control (DMSO). CX-4945 was used because it is the only CK2 inhibitor optimized for clinical use, is provided orally, and has been shown to be safe in humans with solid tumors (50). In agreement with previous trials using these cells (51), 100% of control mice died because of tumors by day 17; in contrast, 43% of mice treated with CX-4945 survived past 100 days, despite termination of treatment at day 30 (Fig. 6E). CX-4945 was well tolerated, because body weights were similar between cohorts throughout the duration of treatment (fig. S7).
CK2 expression correlates with 5-year survival in patients with SHH MB
Human clinical data were consistent with the idea of using CK2 inhibition to treat MB. Kaplan-Meier survival analysis has revealed that patients with SHH-MB who had low expression of mRNA encoding the main CK2 catalytic subunit α (CSNK2A1) in their tumors had a mean 82% 5-year survival, whereas patients with high expression of CSNK2A1 had a mean 61% 5-year survival (Fig. 6F). No correlation between CSNK2A1 expression and patient survival was observed in other MB subgroups (group 3, group 4, and WNT; fig. S8), which have been previously shown to not require Hh pathway activity for tumor growth (3).
Single mutation in CK2 causes resistance to TBB but not CX-4945
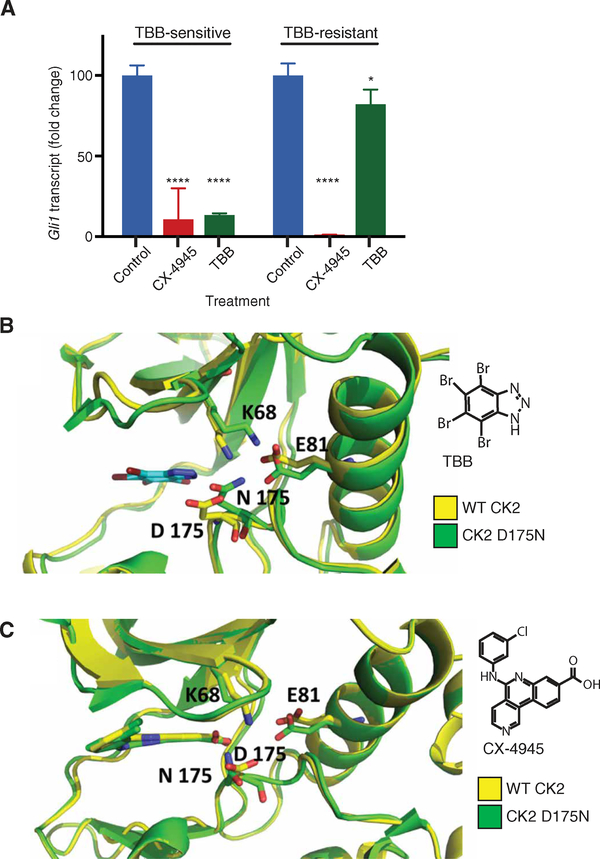
To test the potential for rapid emergence of resistance to CK2 inhibitors, four Ptch+/− mouse MB cell lines (MB21, MB53, MB55, and MB56) were serially passaged for 1 month in the presence of TBB (50 μM), CX-4945 (10 μM), or a DMSO control. During this period, one line (MB55) evolved resistance to TBB, although it maintained sensitivity to CX-4945 (Fig. 7A). Sequencing of Csnk2a1 from the TBB-resistant MB55 cell line revealed five missense mutations (fig. S9A), leading to amino acid substitutions [D175N (fig. S9B), R47M, H183R, G199P, and S217N] that were not present in any of the other cell lines, including the parental TBB-sensitive MB55 line.
Fig. 7. TBB resistance results from mutation in Csnk2a1.
(A) Gli1 transcript abundance in parental, TBB-sensitive MB55 cells and serial passage/drug-induced, TBB-resistant MB55 cells treated with CX-4945 or TBB relative to that in control (DMSO-treated) cells. Data are means ± SD from n = 3 experiments. ns = P > 0.05, *P < 0.05, ****P < 0.0001, two-tailed t test. (B and C) MD simulation and docking analysis of TBB (B) and CX-4945 (C) with wild-type (WT) CK2 and mutant (D175N) CK2 in MB55 cells.
Molecular dynamic (MD) simulations and molecular docking experiments were performed and revealed that, of these five mutations, D175N was the most likely candidate for the reduced efficacy of TBB. In the crystal structure of CSNK2A1 (52), alone and with the inhibitor CX-4945 (33) or TBB (53), Asp175 is located in the well-known and highly conserved DFG motif (DWG in CK2) located in the catalytic binding cleft of CK2α. Together with Glu81, Asp175 is important for the correct orientation of the catalytic residue Lys68 through salt bridges. Our MD simulations revealed that an asparagine in the 175 position was unable to form a salt bridge with catalytic residue Lys68. Consequently, Lys68 strengthened the remaining salt bridge, and Asn175 moved toward Glu81 to protect itself from the hydrophobicity of the surrounding residues (especially Ile174). The resulting displacement of Lys68 and Asn175 increased the ATP-binding cavity volume by about 137 Ä. In wild-type CK2, the binding of TBB requires interaction with both the hinge region and Lys68 (53). Our substrate-specific docking experiments revealed that, when TBB was inside the enlarged cavity of the D175H mutant protein, TBB was too small to efficiently interact with both the hinge region and Lys68 (Fig. 7B). In contrast, CX4945 spanned the entire catalytic site and maintained the same crystallographic binding pattern as in the wild-type protein (Fig. 7C), anchoring both the hinge region at 2.94 Å and to Lys68 at 2.80 Å through hydrophilic interactions. These data indicate that the efficacy of CX-4945 in blocking growth of the MB cell lines—with wild-type and mutant CK2 alike—may be through direct binding-mediated inhibition of CK2.
DISCUSSION
By screening the phosphoproteome during developmental periods when GNPs switch from proliferation to differentiation, we identified CK2 as having increased activity on multiple substrates, specifically in the proliferative, MB-like, P7 stage. The work presented here has already directly led to a phase 1/2 study investigating the use of the CK2 inhibitor CX-4945 in patients with SHH MB. The rapid translation from a phosphoproteomic screen to a clinical trial reflects favorable aspects of the approach to cancer drug target identification. Traditional approaches to discovering novel drug targets often rely on identification of mutations that appear within tumors. However, tumor cells carrying any mutation that is advantageous to cell growth and survival will come to predominate, even if that advantage is small. A great challenge is to determine the biological function of each mutated protein in the context of any one tumor and then distinguishing proteins with activity only slightly advantageous to tumor growth from those that are essential to survival of the tumor. Drugs that target the former have only incremental benefit to patient survival, whereas the latter would be potentially clinically transformative. In contrast, a better understanding of proteins that regulate proliferation versus differentiation in the developmental lineage from which the tumor arose, as provided by our developmental phosphoproteomic approach, can identify important tumor drivers and their likely biological function in proliferation. The critical role of CK2 in GNP development may be the reason why CK2 inhibition resulted in robust tumor stasis and long-term regression of SHH MB but had less robust, often transient, benefit in many other cancer types (54, 55).
Another reason for the rapid path to clinic was the identification of a drug target that is readily conducive to small-molecule inhibition. Kinases, as would be identified through our phosphoproteomic approach, are often readily inhibited by small molecules that compete with ATP binding in the activation cleft. CK2 had many commercially available small-molecule inhibitors and a human-ready compound. This allowed us to bypass lengthy and costly drug development work as would be required for many other protein classes that have undesirable features of a drug target (presence of metal ions, lack of hydrogen bond donors and acceptors, need for adaptive changes in conformation, lipophilicity, and featureless binding sites) (56).
The many potential roles of CK2 in MB, and its influence on two late steps in SHH signal transduction, make it unlikely that a mutation in any one substrate will result in drug resistance, which is another common limitation of many targeted therapies. As we demonstrated, mutations within CK2 itself can still cause resistance. Yet, unlike off-target resistance resulting from mutation of any one of many proteins, predictable resistance due to a mutation in CK2 itself can be tackled through combinations of CK2 inhibitors and/or rational drug design. The quantitative developmental phosphoproteomics approach that we used enabled identification of a previously unknown regulatory mechanism of the Hh signaling pathway and a potential new treatment for this devastating pediatric cancer. Similar approaches may work for other cancers where the developmental cell of origin can be found.
MATERIALS AND METHODS
Animal experiments: Statistics, blinding, and randomization
Given that CK2 promotes Hh signal transduction, we estimated sample size on the basis of previous work performed with Hh inhibitors. Given the dramatic SHH MB tumor regression previously demonstrated with the Hh inhibitor vismodegib (57), we conservatively predicted that CK2 inhibitors would have an effect size of at least 2.2. Therefore, if α error = 0.05 and power = 0.85, then five animals are required per condition. To allow for potential exclusion of one to two animals per condition because of poor tumor engraftment, we used seven to eight animals per condition. Ultimately, we had 100% engraftment [and which met the inclusion criteria (tumor = 250 to 300 mm3)]; thus, all animals were included in the analyses of the tumor growth experiments. Mice were randomized into control or treatment groups daily. Daily randomization ensured that quicker growing tumors were equally represented in both groups. For cerebellar tumor studies, mice were randomized by a blinded collaborator 72 hours after implantation. A blinded collaborator treated and recorded weight, morbidity, and survival. Prism statistical analysis software was used (GraphPad Software) to calculate significance as determined by two-tailed t tests for each paired data point (Fig. 6, B and C) or log-rank Mantel-Cox test (Fig. 6E) for survival studies. All studies were performed in compliance with institutional regulations.
A previous study investigating the effect of Hh inhibition in the developing cerebellum showed that mice with inactive Hh signaling have about 50% reduction in the EGL size, with an effect size of about 5.7 (control: about 80, SD = 7; treatment: 40, SD = 7) (6). Therefore, we conservatively predicted that CK2 inhibitors would be at least 25% as potent as Hh inhibitors, giving an effect size of 1.4. Therefore, with α error = 0.05 and power = 0.85, we expected that we would need and therefore used 10 animals per condition for the developmental experiments. Mice were randomized to treatment or control group at P3, with each litter being equally divided between the two groups. Images were randomized, and all measurements (length, width, Atoh-1, Tag1, and TUNEL staining) were performed by a blinded collaborator. Cerebellar folia length was compared between matched sections of control and treated mice by measuring the maximal length from the mid-base to the tip of the culmen (lobule IV/V). Culmen width was measured 600 pm from the base of the folia between lobules V and VI. Prism statistical analysis software was used (GraphPad Software) to calculate significance as determined by two-tailed t tests. For the Hh epistasis assays specifically, all measurements were taken in biological triplicate, and Prism statistical analysis software was used (GraphPad Software) to calculate significance as determined by two-tailed t tests.
Human Kaplan-Meier curves
Kaplan-Meier survival curves on the human MB data sets were generated using GraphPad Prism, and significance was measured by log-rank Mantel-Cox test.
GNP isolation
Wild-type CD1-Elite mice (strain 482, Charles River) were sacrificed at P1, P7, and P14. Thirty-five to 40 mice were used per P1 and P14 biological replicate, and 14 to 17 mice were used per P7 biological replicate. Cerebella were dissected, and GNPs were isolated and purified using a Percoll gradient, as previously described (58,59). This approach results in a population that is 95 to 99% GNPs (58). Briefly, cerebella were minced and then placed in a digestion buffer consisting of 1× Hanks’ balanced salt solution (HBSS) (14185, Thermo Fisher Scientific), papain (10 U/ml; LSOO3126, Worthington Biochemical), and deoxyribonuclease (DNase) (250 U/ml; D4627, Sigma). Papain solution was aspirated and replaced with 1× HBSS containing ovomucoid (8 mg/ml; LK003182, Worthington Biochemical), bovine serum albumin (BSA) (8 mg/ml; Sigma), and DNase (250 U/ml) and was then triturated using a Pasteur pipette to obtain a single-cell suspension. Cells were passed through a 70-μm nylon cell strainer (21008–952, VWR International) and purified using a step gradient of 35 and 65% Percoll (P4937, Sigma). Pellets were flash-frozen in liquid nitrogen and kept at −80°C until all samples were collected for MS.
Sample preparation for quantitative MS analysis
GNP samples were prepared as previously described (60), with the following modifications. All solutions are reported as final concentrations. Lysis buffer [8 M urea, 1% SDS, 50 mM tris (pH 8.5), protease and phosphatase inhibitors from Roche] was added to the cell pellets to achieve a cell lysate with a protein concentration between 2 and 8 mg/ml. A micro-BCA (bicinchoninic acid) assay (Pierce) was used to determine the final protein concentration in the cell lysate. Proteins were reduced and alkylated as previously described. Proteins were precipitated using methanol/chloroform. In brief, four volumes of methanol were added to the cell lysate, followed by one volume of chloroform, and finally three volumes of water. The mixture was vortexed and centrifuged to separate the chloroform phase from the aqueous phase. The precipitated protein was washed with one volume of ice-cold methanol. The washed precipitated protein was allowed to air dry. Precipitated protein was resuspended in 4 M urea, 50 mM tris (pH 8.5). Proteins were first digested with LysC (1:50; enzyme/protein) for 12 hours at 25°C. The LysC digestion was diluted down to 1 M urea, 50 mM tris (pH 8.5) and then digested with trypsin (1:100; enzyme/protein) for another 8 hours at 25°C. Peptides were desalted using a C18 solid-phase extraction cartridges, as previously described. Dried peptides were resuspended in 200 mM EPPS [4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid] (pH 8.0). Peptide quantification was performed using the micro-BCA assay (Pierce). The same amount of peptide from each condition was labeled with tandem mass tag (TMT) reagent (1:4; peptide/TMT label) (Pierce). The 6- and 10-plex labeling reactions were performed for 2 hours at 25°C. Modification of tyrosine residue with TMT was reversed by the addition of 5% hydroxyl amine for 15 min at 25°C. The reaction was quenched with 0.5% trifluoroacetic acid, and samples were combined at a 1:1 ratio. Combined samples were desalted and offline-fractionated into 24 fractions, as previously described.
Liquid chromatography-MS3 spectrometry
Twelve fractions (every other one) of the 24-peptide fraction from the basic reverse-phase step were analyzed with a liquid chromatography (LC)-MS3 data collection strategy on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) equipped with a Proxeon EASY-nLC 1000 for online sample handling and peptide separations. About 5 μg of peptide resuspended in 5% formic acid + 5% acetonitrile (ACN) was loaded onto a 100-μm-inner diameter fused-silica microcapillary with a needle tip pulled to an internal diameter less than 5 μm. The column was packed in-house to a length of 35 cm with a C18 reverse-phase resin (GP118 resin 1.8 μm, 120 Å; Sepax Technologies). The peptides were separated using a 120-min linear gradient from 3 to 25% buffer B (100% ACN + 0.125% formic acid) equilibrated with buffer A (3% ACN + 0.125% formic acid) at a flow rate of 600 nl/min across the column. The scan sequence for the Orbitrap Fusion began with an MS1 spectrum [Orbitrap analysis, resolution 120,000 (400 to 1400 m/z scan range); AGC target, 2 × 105; maximum injection time, 100 ms (dynamic exclusion of 75 s)]. “Top speed” (1 s) was selected for MS2 analysis, which consisted of collision-induced dissociation [quadrupole isolation set at 0.5 Da and ion trap analysis; AGC, 4 × 103; normalized collision energy (NCE), 35; maximum injection time, 150 ms]. The top 10 precursors from each MS2 scan were selected for MS3 analysis (synchronous precursor selection), in which precursors were fragmented by high-energy collisional dissociation before Orbitrap analysis [NCE, 55; maximum AGC, 5 × 104; maximum injection time, 150 ms; isolation window, 2.5 Da; resolution, 60,000 (10-plex experiments) or 15,000 (6-plex experiments)].
LC-MS3 data analysis
A suite of in-house software tools was used for .RAW file processing and controlling peptide and protein-level FDRs, assembling proteins from peptides, and protein quantification from peptides, as previously described. MS/MS spectra were searched against a UniProt human database (accessed February 2014), with both the forward and reverse sequences. Database search criteria are as follows: tryptic with two missed cleavages, a precursor mass tolerance of 50 parts per million (ppm), fragment ion mass tolerance of 1.0 Da, static alkylation of cysteine (57.02146 Da), static TMT labeling of lysine residues and N termini of peptides (229.162932 Da), and variable oxidation of methionine (15.99491 Da). TMT reporter ion intensities were measured using a 0.03-Da window (6-plex) or 0.003-Da window (10-plex) around the theoretical mass/charge ratio (m/z) for each reporter ion in the MS3 scan. Peptide spectral matches with poor-quality MS3 spectra were excluded from quantitation (<100 summed signal-to-noise ratio across 6 channels and <0.5 precursor isolation specificity for 6-plexes or <200 summed signal-to-noise ratio across 10 channels and <0.5 precursor isolation specificity for 10-plexes).
LC-MS3 sample comparison
Only peptides identified with full confidence were included in the analysis (binomial probability threshold of P < 10−6, occurrence threshold = 20). Each biological replicate was median-adjusted to 10 to account for differences in sample loading, resulting in the following changes: proteomic data set: P1A, −0.0362; P1B, −0.0879; P1C, +0.20476; P7A, +0.03374; P7B, −0.2761; P7C, −0.2865; P14A, +0.78605; P14B, +0.65244; P14C, +0.71771; MB, +0.04692; phosphoproteomic data set: P1A, −0.040163003; P1B, −0.124877212; P1C, −0.009654128; P7A, −0.170361415; P7B, −0.341821765; P7C, −0.117111143; P14A, +1.114888992; P14B, +1.060676918; P14C, +0.955615374; MB, +0.43299154. Log2-transformed P1:P7 and P7:P14 ratios were determined, and phosphopeptide changes were normalized to protein changes. Values outside of 1.5 SD were considered “significant changers.” Motif analysis of significantly changing phosphopeptides was performed using Motif-X, as previously described (61, 62). Briefly, Motif-X is an iterative statistical approach to identifying protein phosphorylation motifs in large-scale phosphoproteomic data sets built on a greedy recursive search of the sequence space to identify highly correlated residue/position pairs with the lowest P values. Here, a binomial probability threshold of P < 10−6 and occurrence threshold of 20 were used.
GNP proliferation
To determine relative proliferation of Atoh1(+) GNPs at P1, P7, and P14, cerebella were dissected from Atohl/nGFP transgenic mice (63) 1 hour after EdU injection [50 mg/kg using a stock (5 mg/ml) diluted in phosphate-buffered saline (PBS)]. Brains were dissected and fixed in 4% paraformaldehyde (PFA) overnight and then transferred into 20% sucrose for 24 hours. Fixed whole cerebella were mounted in optimal cutting temperature (OCT) embedding medium, sectioned at 20 μm, and then air-dried for 20 min before storing at −20°C for up to 2 months. Sections were blocked in 0.2% Triton X-100 and 2% goat serum diluted in sterile PBS for 1 hour at room temperature. EdU staining was performed as per the manufacturer’s instructions (Click-iT Plus EdU Alexa Fluor 647 Imaging Kit, catalog no. C10640, Life Technologies) and counterstained with Hoechst 33258 (final concentration, 1 μl/ml in PBS) at room temperature for 10 min. Images were taken on a Leica TCS SP8 confocal microscope. Total EdU signal within Atoh1(+) regions was quantified using ImageJ (six cerebella per time point, four to seven folia per cerebellar section) after background subtraction via rolling ball radius (50 pixels). Prism statistical analysis software was used (GraphPad Software) to calculate significance as determined by two-tailed t tests.
GNP culture
GNPs from CDl-Elite mice (strain 482, Charles River) were cultured in Neurobasal-A with B27, sodium pyruvate, penicillin/streptomycin, Glutamax, glucose (0.6 mg/ml), and KCl (25 mM). Recombinant SHH (SHH 461–54/CF, R&D Systems) was added to media at 3 μg/ml to promote cell division.
Single-cell immunofluorescence imaging (GNP and MB)
Before cell culture, 96-well glass imaging plates (Cellvis P96–1.5H-N) were incubated with poly-D-lysine (A-003-E, Millipore) at 100 μg/ml for 2 hours followed by laminin (CC095, Millipore) at 10 μg/ml overnight at 37°C. GNPs or MB cells were plated at 1 × 105 cells per well and grown at 37°C. After treatment, the cells were fixed with 4% PFA for 10 min at room temperature. The cells were then blocked using 5% donkey serum, 1% BSA, and 0.2% Triton X-100 for 1 hour at room temperature. Primary antibodies (Abs) were incubated overnight at 4°C. The primary Abs used are as follows: anti-NeuN mouse monoclonal at 1:200 (MAB377, Millipore) and phospho-Rb (Ser807/811) rabbit monoclonal at 1:1000 (clone D20B12, 8516, Cell Signaling Technology). Cells were costained with rhodamine phalloidin (R415, Molecular Probes). Donkey anti-immunoglobulin G (IgG) secondary Abs against mouse and rabbit conjugated to Alexa Fluor 488 and Alexa Fluor 647 at 1:500 (Jackson ImmunoResearch) were used. All cell imaging was performed using the ImageXpress Micro XLS Widefield High Content Screening System (Molecular Devices) using 20× (0.45 or 0.75 numerical aperture) Nikon objectives. The intensity of fluorescence in each cell was automatically calculated using custom MATLAB scripts: Nuclei were segmented using 4′,6-diamidino-2-phenylindole, as previously described (64); downstream analysis was performed in R; cell cycle classification was performed using Gaussian finite mixture modeling with the mclust package (65); and cell cycle clustering analysis in GNPs used the log10-transformed median intensity of each cell, measuring Math1-GFP, phosphorylated Rb, and NeuN, as well as the area of the cell. All studies were done with six individual cultures.
CK2 inhibition in the developing mouse
Math1/nGFP transgenic mice (63) were treated with DMSO or TBB (30 mg/kg, ip BID) (218697, EMD Millipore) from P4 to P7 and then sacrificed 6 hours after their final treatment. Brains were dissected and immediately homogenized in lysis buffer (sc-24948, Santa Cruz Biotechnology) supplemented with protease and phosphatase inhibitors (no. 1861218, Thermo Fisher Scientific) or fixed in 4% PFA overnight and then transferred into 20% sucrose for 24 hours. Lysates were separated on a 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) gel and immunoblotted with pAKT:S129 (no. 13461, Cell Signaling Technology) and AKT (no. 4685, Cell Signaling Technology). Fixed whole cerebella were mounted in OCT, sectioned at 20 μm, and then air-dried for 20 min before storing at −20°C for up to 2 months. Sections were blocked in 0.2% Triton X-100 and 2% goat serum diluted in sterile PBS for 1 hour at room temperature. The primary Abs used were anti-NeuN Ab (1:200, COT: 2459079, EMD Millipore), anti-Tag1 Ab (1:100, Ab derived in the Scott laboratory), TUNEL staining as per the manufacturer’s instructions (In Situ Cell Death Detection Kit, catalog no. 1 684 809, Roche), and EdU staining as per the manufacturer’s instructions (Click-iT Plus EdU Alexa Fluor 647 Imaging Kit, catalog no. C10640, Life Technologies). All sections were incubated in primary Ab overnight at 4°C and counterstained with Hoechst 33258 (final concentration, 1 μl/ml in PBS) at room temperature for 10 min. Images were taken on a Leica TCS SP8 confocal microscope. Images were randomized, and all measurements (length, width, Atoh-1, Tag1, and TUNEL staining) were performed by a blinded collaborator. For each group, two images were taken and measured for each of 10 cerebella. Cerebellar folia length was compared between matched sections of control and treated mice by measuring the maximal length from the mid-base to the tip of the culmen (lobule IV/V). Culmen width was measured (600 μm) from the base of the folia between lobules V and VI. One section was measured from each of 11 cerebella analyzed. Prism statistical analysis software (GraphPad Software) was used to calculate significance as determined by two-tailed t tests.
siRNA studies
Pooled custom Dharmacon ON-TARGETplus specificity enhanced siRNA was created against Csnk2a1, Csnk2a2, and Csnk2b. siRNA sequences were provided by D. Seldin (Boston University School of Medicine) and are listed in table S3. Scramble siRNA was purchased from Dharmacon (D-001810–01-05). NIH3T3 cells or Sufu−/− MEFs were plated in six-well plates at 400,000 cells per well in 10% fetal bovine serum (FBS) media constituted of Dulbecco’s modified Eagle’s medium (DMEM) (11960–069, Invitrogen) supplemented with 10% FBS (SH30070.02, Hyclone Laboratories), penicillin/streptomycin (15140–122, Life Technologies), sodium pyruvate (11360–070, Invitrogen), and Glutamax (35050–061, Invitrogen). Cells were then starved in 0.5% FBS for 24 hours before treatment with 25% SHH-N conditioned media in 0.5% FBS. SHH-N conditioned medium was made using a human embryonic kidney (HEK) 293 cell line that stably secretes Shh-N, as previously described (34).
Hh pathway epistasis studies
Sufu−/− MEFs [conditional deletion of exons 4 to 8 (36)] were provided by P.-T. Chang [University of California, San Francisco (UCSF)]. As in siRNA experiments, NIH3T3 cells or Sufu−/− MEFs were plated in six-well plates at 400,000 cells per well in 10% FBS media (DMEM supplemented with 10% FBS, penicillin/streptomycin, sodium pyruvate, and Glutamax). Cells were then starved in 0.5% FBS for 24 hours before treatment with 25% SHH-N conditioned media in 0.5% FBS ± drug treatment or SAG (100 nM; S7779, Selleckchem). Gli2ΔN cells were treated 48 hours after transfection. Myc-GLI2-DN (17649, pCS2-MT-GLI2-deltaN) plasmid was purchased from Addgene. Plasmid transfection was performed using TurboFect transfection reagent (no. R0531, Thermo Scientific) according to the manufacturer’s instructions, starting by plating NIH3T3 cells in a six-well dish at 400,000 cells per well. Unless otherwise stated, all treatments were applied for 6 hours. Cell lysis and RNA extraction were performed using QIAshredder homogenizers (79654, Qiagen) and RNeasy Mini kits (74104, Qiagen) according to the manufacturer’s instructions. RNA was quantified on a NanoDrop spectrophotometer (ND-2000C, NanoDrop Technologies). Transcript quantification was performed using the TaqMan RNA-to-Ct 1-Step Kit (4392656, Life Technologies) and the Applied Biosystems 7500 RT-PCR system, as per the manufacturer’s instructions. All primers were commercially available TaqMan Gene Expression Assay probes purchased from Life Technologies (csnk2a1, Mm00786779_s1; csnk2a2, Mm01243455_m1; csnk2b, Mm00487216_m1; Gli1, Mm00494654_m1; Gli2, Mm.273292; Ccnd1, Mm.273049; and Ptch, Mm00436047_m1). All studies were done in biological triplicate (cultures).
Western blotting
Gli2ΔN cells were lysed with radioimmunoprecipitation assay buffer (sc-24948, Santa Cruz Biotechnology) supplemented with Halt Protease and Phosphatase Inhibitor (no. 1861218, Thermo Fisher Scientific) for 30 min on ice, and lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4°C. Supernatants were incubated with 6× Laemmli sample buffer (10570021–1, BioWorld) at 95°C for 5 min. The samples were then separated with a 7% SDS-PAGE gel and immunoblotted with anti-Gli2 (AF3635-SP, R&D Systems) and anti-β-tubulin (ab6046, Abcam).
Cell viability
Ptch+/− MB cell lines MB55 and MB21 were gifts from R. Segal [Dana-Farber Cancer Institute (DFCI), Harvard Medical School], and Ptch+/−;p53−/− MB53 cells were gifts from C. Rudin [Memorial Sloan Kettering Cancer Center]. All cell lines were validated for high Gli activity via qRT-PCR and cultured at low density as neurospheres in DMEM/F12 supplemented with B27 and 1% penicillin/streptomycin. Cell viability was assessed using CellTiter-Glo (G7573, Promega) according to the manufacturer’s instructions. Cells were plated at 10,000 cells per well in 96-well plates and treated with drugs as indicated, and data were collected on a TECAN Infinite 200 plate reader. All measurements were done in biological triplicate (cultures). Prism statistical analysis software (GraphPad Software) was used to determine relative IC50s, normalized to control (DMSO)-treated cells [nonlinear regression, log(inhibitor) versus response (variable slope) curve].
Human MB cell lines
The RCMB32 and ICb-984 cell lines were generated in the laboratories of R.J.W.-R. and X.-N.L., respectively. The BT084 cell line was provided by T. Milde (Heidelberg University Hospital). The primary tumor sample (ST01) (Fig. 5, B to D) obtained from the operating room was minced and then placed in a digestion buffer consisting of 1× HBSS (14185, Thermo Fisher Scientific), papain (10 U/ml; LSOO3126, Worthington Biochemical), and DNase (250 U/ml; D4627, Sigma). Papain solution was aspirated and replaced with 1× HBSS containing ovomucoid (8 mg/ml; LK003182, Worthington Biochemical), BSA (8 mg/ml; Sigma), and DNase (250 U/ml) and then triturated using a Pasteur pipette to obtain a single-cell suspension. Cells were pelleted and resuspended in 0.02% HBSS-BSA. Cells were passed through a 70-μm nylon cell strainer (21008–952, VWR International). Of 20 tumors implanted, 5 grew to a sufficient size to resect. Cell viability and cell cycle stage were assayed by high-throughput single-cell immunofluorescence imaging, as described above; 12 individual cultures were assessed at each dose. Gli1 transcript expression was assessed by qRT-PCR, as described above, and three cultures were assessed at each dose. Final pathological report is provided in table S4.
Acquired resistance
MB53, MB55, and MB21 cells were cultured in Neurobasal (10888–022, Life Technologies) + B27 supplement (17504044, Invitrogen) + DMSO (0.1%), CX-4945 (10 μM), or TBB (50 μM). Surviving cells were passaged for 1 month, after which only control (DMSO)-treated and MB55 TBB-treated (50 μM) cells remained. Complementary DNA (cDNA) from the three control lines and MB55 TBB-resistant lines was synthesized using the ABI High-Capacity cDNA Reverse Transcription Kit (4368814, Thermo Fisher Scientific) and amplified using the Q5 High-Fidelity DNA Polymerase (M0491, New England Biolabs) as per the manufacturer’s protocol. Primer sequences are provided in table S3.
In silico analysis of mutant versus wild-type binding ofTBBand CX-4945
All the crystal structures of CK2 were retrieved from the Protein Data Bank (PDB). To perform docking and MD simulations, the crystal structure of human CK2 in complex with CX4945 was used (PDB code: 3PE1). However, to compare the binding motif of TBB and to evaluate its position in the structure 3PE1, the crystallographic complex TBB/CK2 was also considered (PDB code: 1J91). The crystal structures were processed to remove unwanted ligands and water molecules. Hydrogen atoms were added to the protein structure using standard geometries (66). To minimize contacts between hydrogens, the structures were subjected to AMBER99 force-field minimization until the root mean square of conjugate gradient was <0.1 kcal mol−1 Å−1 (1 Å = 0.1 nm), keeping the heavy atoms fixed at their crystallographic positions (66). A similar protocol was used also to minimize the structures subjected to in silico mutagenesis. To strictly calibrate the high-throughput docking protocol, a small database of known CK2 inhibitors was built and a set of docking runs was performed. After the calibration phase, TBB and CX-4945 were docked directly into the ATP-binding site of selected CK2 crystal structures, by using AutoDock software (66). MD simulations of the considered structures (parameterized with AMBER99) were performed with NAMD 2.8 (67) to verify their stability over time; in particular, a 100-ns NPT (isothermal-isobaric) ensemble (1 atm, 300 K) MD simulation was performed after an equilibration phase of 1 ns (positional restraints were applied on carbon atoms to equilibrate the solvent around the protein) (68, 69).
Flank allograft studies
In vivo assays of CK2 inhibitor efficacy were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Stanford University. SmoWT-MB and SmoD477G-MB cells isolated from either parental SmoWT or SmoD477G mouse Ptch+/−;p53−/− MB hind-flank allografts were provided by C. Rudin (Memorial Sloan Kettering Cancer Center). Ptch+/−;Tpr53−/− or Ptch+/−;Tpr53−/−;SmoD477G MB cells were mixed 1:1 with Matrigel, and 107 cells were injected into both flanks of a 6-to 7-week-old Nu/Nu mouse (strain 088, Charles River). Once tumors reached 250 to 300 mm3, mice were randomized to treatment versus control groups. Treatment groups received TBB (30 mg/kg, ip BID; reconstituted in DMSO at 50 μg/μl) or GDC-0449 (37.5 mg/kg, ip BID; reconstituted in DMSO at 62.5 μg/μl) (S1082, Selleckchem). Control mice were treated with an equivalent volume of DMSO. Corn oil was used in all groups to a total volume of 180 ml per injection, and all doses were given with a 28-gauge insulin syringe. Tumors were measured, and mice were weighed daily. Once tumors reached 1.5 cm2 or met institutional euthanasia criteria, mice were sacrificed and tumors were harvested. Growth curves were generated using GraphPad Prism, and significance was measured by two-way analysis of variance (ANOVA).
Cerebellar allografts
Cells from Ptck+/−;Tpr53−/−;SmoD477G mice were suspended in sterile PBS to an adjusted final concentration of 80,000 cells per 2-μl injection. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (6 to 7 weeks old; The Jackson Laboratory) were anesthetized and fixed to a stereotactic frame, and the head was sterilized. A small sagittal incision was made in the mouse’s scalp to expose the area surrounding lambda. The skull was sterilized, and a 0.7-mm drill bit was used to create a midline hole located 2 mm down from lambda. A Hamilton syringe with affixed 26-gauge needle was lowered to a depth of ~3 mm and then withdrawn to 2.75 mm before injecting 2-μl (80,000) cells at 0.5 μl/min. The drill hole was closed with a thin slip of bone wax, and the wound was closed. Mice were randomized, and a blinded collaborator began treatment at 72 hours with either CX-4945 (37.5 mg/kg, orally BID; reconstituted in DMSO) or DMSO (equivalent volume to treatment group, dosed ip BID). Corn oil was used as vehicle for both CX-4945 and DMSO. Weight, morbidity, and survival were measured by a blinded collaborator.
Mouse strains
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Stanford University. Male and female wild-type CDl-Elite mice (strain 482, Charles River) were sacrificed at P1, P7, and P14 for initial phosphoproteomic study. Female and male Atohl/nGFP transgenic mice (63) were used for cerebellar development studies. Female Nu/Nu mice (6 to 7 weeks old; strain 088, Charles River) were used for flank studies, and female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (6 to 7 weeks old; strain 005557, The Jackson Laboratory) were used for cerebellar tumor studies.
Supplementary Material
Fig. S1. Relative phosphopeptide abundance between P14 biological replicates.
Fig. S2. Relative proliferation in the P1, P7, and P14 cerebellum.
Fig. S3. GNP proliferation after CX-4945 treatment in culture.
Fig. S4. Knockdown specificity of pooled siRNA constructs targeting Csnk2a1, Csnk2a2, Csnk2b, and Smo.
Fig. S5. Gli1 and Hh-associated transcript expression in NIH3T3 cells after CK2 inhibition.
Fig. S6. Gli1 transcript expression in cultured SHH MB cell lines.
Fig. S7. Weight of control or CX-4945-treated mice with cerebellar SHH MB.
Fig. S8. CSNK2A1 expression and 5-year survival of patients with SHH MB.
Fig. S9. Sequence conservation of Csnk2a1 mutations in TBB-resistant SHH MB cells.
Table S3. Primer and siRNA sequences.
Table S4. Pathological report for the primary human MB sample (ST01).
Table S1. Phosphoproteome of GNPs isolated at P1, P7, and P14.
Table S2. Phosphopeptides of enriched kinase motifs.
Acknowledgments:
We thank R. Segal (DFCI), C. Rudin (Memorial Sloan Kettering Cancer Center), T. Milde [German Cancer Research Center (DKFZ)], P.-T. Chang (UCSF), and D. Seldin (Boston University) for sharing reagents. We thank R. Kunz, S. Gygi, and the Thermo Fisher Center for Multiplexed Proteomics at Harvard Medical School for performing the phosphoproteomics and providing valuable discussion. We thank A. Brown (Stanford University), C. Baker (Stanford University), and the members of the Fuller laboratory, the Scott laboratory, and Cho laboratory for discussions and critical reading of the manuscript. We thank the patients who donated their tumor tissue for the human SHH MB aspects of this study.
Funding: T.P. was supported by the Felix and Heather Baker BioX Stanford Interdisciplinary Graduate Fellowship and the Neurosurgery Research and Education Foundation (NREF) Fellowship. J.P. was supported by the American Brain Tumor Association Basic Research Fellowship and the 2016 B*CURED-NREF Research Grant. The research was supported by Stanford SPARK (T.P.); the Stanford Cancer Institute (T.P.); the Department of Developmental Biology, Stanford School of Medicine (T.P.); the Weston Havens Foundation (T.P.); NIH grant R21HD076367 (M.P.S. and J.E.E.); the Reed-Hodgson Chair in Human Biology at Stanford University (M.T.F.); Fondazione per la ricerca sulla Fibrosi Cistica grant FFC#10/2017 (G.C.); German Cancer Aid (Deutsche Krebshilfe) P-91650709 (J.T.); and Advanced Imaging Research Center grant IG-14180 (L.A.P.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: A provisional patent application entitled “Compositions and methods for treating medulloblastoma” has been submitted to United States Patent and Trademark Office. This provisional patent contains some overlap with parts of this manuscript. The human SHH MB line BT084 was received under a material transfer agreement with X.N.L. The phosphoproteomic data for this study have also been deposited in the Peptide Atlas database (www.peptideatlas.org/; accession ID PASS01240). These and all other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Zurawel RH, Allen C, Wechsler-Reya R, Scott MP, Raffel C, Evidence that haploinsufficiency of Ptch leads to medulloblastoma in mice. Genes Chromosomes Cancer 28, 77–81 (2000). [PubMed] [Google Scholar]
- 2.Wechsler-Reya RJ, Scott MP, Control of neuronal precursor proliferation in the cerebellum by Sonic hedgehog. Neuron 22, 103–114 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho Y-J, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM, Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 123, 465–472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta SK, Meiri KF, Mahfooz K, Bharti U, Mani S, Coordination between extrinsic extracellular matrix cues and intrinsic responses to orient the centrosome in polarizing cerebellar granule neurons. J. Neurosci 30, 2755–2766 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinosa JS, Luo L, Timing neurogenesis and differentiation: Insights from quantitative clonal analyses of cerebellar granule cells. J. Neurosci 28, 2301–2312 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL, Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 131, 5581–5590 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Robinson GW, Orr BA, Wu G, Gururangan S, Lin T, Qaddoumi I, Packer RJ, Goldman S, Prados MD, Desjardins A, Chintagumpala M, Takebe N, Kaste SC, Rusch M, Allen SJ, Onar-Thomas A, Stewart CF, Fouladi M, Boyett JM, Gilbertson RJ, Curran T, Ellison DW, Gajjar A, Vismodegib exerts targeted efficacy against recurrent sonic hedgehog-subgroup medulloblastoma: Results from phase II Pediatric Brain Tumor Consortium studies PBTC-025B and PBTC-032. J. Clin. Oncol 33, 2646–2654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kool M, Jones DTW, Jäger N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Sturm D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, Schmidt C, Rausch T, Shih D, Reimand J, Sieber L, Wittmann A, Linke L, Witt H, Weber UD, Zapatka M, König R, Beroukhim R, Bergthold G, van Sluis P, Volckmann R, Koster J, Versteeg R, Schmidt S, Wolf S, Lawerenz C, Bartholomae CC, von Kalle C, Unterberg A, Herold-Mende C, Hofer S, Kulozik AE, von Deimling A, Scheurlen W, Felsberg J, Reifenberger G, Hasselblatt M, Crawford JR, Grant GA, Jabado N, Perry A, Cowdrey C, Croul S, Zadeh G, Korbel JO, Doz F, Delattre O, Bader GD, McCabe MG, Collins VP, Kieran MW, Cho YJ, Pomeroy SL, Witt O, Brors B, Taylor MD, Schüller U, Korshunov A, Eils R, Wechsler-Reya RJ, Lichter P, Pfister SM; ICGC PedBrain Tumor Project, Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25, 393–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price MA, Kalderon D, Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell 108, 823–835 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Hammerschmidt M, Bitgood MJ, McMahon AP, Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev. 10, 647–658 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Yang PL, Gray NS, Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight ZA, Lin H, Shokat KM, Targeting the cancer kinome through polypharmacology. Nat. Rev. Cancer 10, 130–137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu P, Nielsen TE, Clausen MH, FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci 36, 422–429 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Goodrich LV, Milenkovic L, Higgins KM, Scott MP, Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Northcott PA, Jones DTW, Kool M, Robinson GW, Gilbertson RJ, Cho Y-J, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, Pfister SM, Medulloblastomics: The end of the beginning. Nat. Rev. Cancer 12, 818–834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom J, Cross FR, Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol 8, 149–160 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Meggio F, Pinna LA, One-thousand-and-one substrates of protein kinase CK2? FASEBJ. 17, 349–368 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Zhu X, Girardo D, Govek EE, John K, Mellén M, Tamayo P, Mesirov JP, Hatten ME, Role of Tet1/3 genes and chromatin remodeling genes in cerebellar circuit formation. Neuron 89, 100–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stütz AM, Korshunov A, Reimand J, Schumacher SE, Beroukhim R, Ellison DW, Marshall CR, Lionel AC, Mack S, Dubuc A, Yao Y, Ramaswamy V, Luu B, Rolider A, Cavalli FM, Wang X, Remke M, Wu X, Chiu RY, Chu A, Chuah E, Corbett RD, Hoad GR, Jackman SD, Li Y, Lo A, Mungall KL, Nip KM, Qian JQ, Raymond AG, Thiessen NT, Varhol RJ, Birol I, Moore RA, Mungall AJ, Holt R, Kawauchi D, Roussel MF, Kool M, Jones DT, Witt H, Fernandez-L A, Kenney AM, Wechsler-Reya RJ, Dirks P, Aviv T, Grajkowska WA, Perek-Polnik M, Haberler CC, Delattre O, Reynaud SS, Doz FF, Pernet-Fattet SS, Cho BK, Kim SK, Wang KC, Scheurlen W, Eberhart CG, Fevre-Montange M, Jouvet A, Pollack IF, Fan X, Muraszko KM, Gillespie GY, Di Rocco C, Massimi L, Michiels EM, Kloosterhof NK, French PJ, Kros JM, Olson JM, Ellenbogen RG, Zitterbart K, Kren L, Thompson RC, Cooper MK, Lach B, McLendon RE, Bigner DD, Fontebasso A, Albrecht S, Jabado N, Lindsey JC, Bailey S, Gupta N, Weiss WA, Bognár L, Klekner A, Van Meter TE, Kumabe T, Tominaga T, Elbabaa SK, Leonard JR, Rubin JB, Liau LM, Van Meir EG, Fouladi M, Nakamura H, Cinalli G, Garami M, Hauser P, Saad AG, Iolascon A, Jung S, Carlotti CG, Vibhakar R, Ra YS, Robinson S, Zoilo M, Faria CC, Chan JA, Levy ML, Sorensen PH, Meyerson M, Pomeroy SL, Cho YJ, Bader GD, Tabori U, Hawkins CE, Bouffet E, Scherer SW, Rutka JT, Malkin D, Clifford SC, Jones SJ, Korbel JO, Pfister SM, Marra MA, Taylor MD, Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng H-C, Qi RZ, Paudel H, Zhu H-J, Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011,794089 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarno S, Reddy H, Meggio F, Ruzzene M, Davies SP, Donella-Deana A, Shugar D, Pinna LA, Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’). FEBS Lett. 496, 44–48 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Pagano MA, Bain J, Kazimierczuk Z, Sarno S, Ruzzene M, Di Maira G, Elliott M, Orzeszko A, Cozza G, Meggio F, Pinna LA, The selectivity of inhibitors of protein kinase CK2: An update. Biochem. J 415, 353–365 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Cozza G, Mazzorana M, Papinutto E, Bain J, Elliott M, di Maira G, Gianoncelli A, Pagano MA, Sarno S, Ruzzene M, Battistutta R, Meggio F, Moro S, Zagotto G, Pinna LA, Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2. Biochem. J 421,387–395 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Brehme H, Kirschstein T, Schulz R, Köhling R, In vivo treatment with the casein kinase 2 inhibitor 4,5,6,7-tetrabromotriazole augments the slow afterhyperpolarizing potential and prevents acute epileptiform activity. Epilepsia 55, 175–183 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Ljubimov AV, Caballero S, Aoki AM, Pinna LA, Grant MB, Castellon R, Involvement of protein kinase CK2 in angiogenesis and retinal neovascularization. Invest. Ophthalmol. Vis. Sci 45, 4583–4591 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudarov A, Joyner AL, Cerebellum morphogenesis: The foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Dev. 2, 26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie J, Johnson RL, Zhang X, Bare JW, Waldman FM, Cogen PH, Menon AG, Warren RS, Chen L-C Jr., M. P. Scott, E. H. Epstein, Mutations of the PATCHED gene in several types of sporadic extracutaneous tumors. Cancer Res. 57, 2369–2372 (1997). [PubMed] [Google Scholar]
- 28.Milenkovic L, Scott MP, Rohatgi R, Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J. Cell Biol 187, 365–374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milenkovic L, Weiss LE, Yoon J, Roth TL, Su YS, Sahl SJ, Scott MP, Moerner WE, Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proc. Natl. Acad. Sci. U.SA 112, 8320–8325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R, The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou DY, Dominguez I, Toselli P, Landesman-Bollag E, O’Brien C, Seldin DC, The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol. Cell. Biol 28, 131–139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchou T, Vernet M, Blond O, Jensen HH, Pointu H, Olsen BB, Cochet C, Issinger OG, Boldyreff B, Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol 23, 908–915 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battistutta R, Cozza G, Fabrice Pierre E. Papinutto, Lolli G, Sarno S, O’Brien SE, Siddiqui-Jain A, Haddach M, Anderes K, Ryckman DM, Meggio F, Pinna LA, Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry 50, 8478–8488 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA, Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. U.S.A 99, 14071–14076 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA, Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 361, 1173–1178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M-H, Wilson CW, Li Y-J, Law KKL, Lu C-S, Gacayan R, Zhang X, Hui C.-c., Chuang P-T, Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910–1928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, Jia J, Casein kinase 2 promotes Hedgehog signaling by regulating both Smoothened and Cubitus interruptus. J. Biol. Chem 285, 37218–37226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Wang Y, Mao J-H, Hsieh D, Kim I-J, Hu L-M, Xu Z, Long H, Jablons DM, You L, Inhibition of CK2a down-regulates Hedgehog/Gli signaling leading to a reduction of a stem-like side population in human lung cancer cells. PLOS ONE 7, e38996 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M, A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum. Mol. Genet 14, 2181–2188 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, Yu Q, Ostrom L, Fordjour P, Anderson DL, Monahan JE, Kelleher JF, Peukert S, Pan S, Wu X, Maira S-M, García-Echeverría C, Briggs KJ, Neil Watkins D, Yao Y.-m., Lengauer C, Warmuth M, Sellers WR, Dorsch M, Interfering with resistance to Smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci. Transl. Med 2, 51ra70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, Li W, Atwood SX, Whitson RJ, Chang ALS, Li J, Oro AE, Chan JA, Kelleher JF, Segal RA, RAS/MAPK activation drives resistance to Smo inhibition, metastasis, and tumor evolution in Shh pathway-dependent tumors. Cancer Res. 75, 3623–3635 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Pak E, Ornell KJ, Pazyra-Murphy MF, MacKenzie EL, Chadwick EJ, Ponomaryov T, Kelleher JF, Segal RA, A transposon screen identifies loss of primary cilia as a mechanism of resistance to SMO inhibitors. CancerDiscov. 7, 1436–1449 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, Maier DL, Udaka YT, Murad N, Morrissy S, Seker-Cin H, Brabetz S, Qi L, Kogiso M, Schubert S, Olson JM, Cho YJ, Li XN, Crawford JR, Levy ML, Kool M, Pfister SM, Taylor MD, Wechsler-Reya RJ, HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell 29, 311–323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brun SN, Markant SL, Esparza LA, Garcia G, Terry D, Huang J-M, Pavlyukov MS, Li X-N, Grant GA, Crawford JR, Levy ML, Conway EM, Smith LH, Nakano I, Berezov A, Greene MI, Wang Q, Wechsler-Reya RJ, Survivin as a therapeutic target in Sonic hedgehog-driven medulloblastoma. Oncogene 34, 3770–3779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, Kushida M, Head R, Morrissy S, Zhu X, Aviv T, Voisin V, Clarke ID, Li Y, Mungall AJ, Moore RA, Ma Y, Jones SJM, Marra MA, Malkin D, Northcott PA, Kool M, Pfister SM, Bader G, Hochedlinger K, Korshunov A, Taylor MD, Dirks PB, Quiescent SOX2+ cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 26, 33–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yauch RL, Dijkgraaf GJP, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, Bazan JF, Kan Z, Seshagiri S, Hann CL, Gould SE, Low JA, Rudin CM, de Sauvage FJ, Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodon J, Tawbi HA, Thomas AL, Stoller RG, Turtschi CP, Baselga J, Sarantopoulos J, Mahalingam D, Shou Y, Moles MA, Yang L, Granvil C, Hurh E, Rose KL, Amakye DD, Dummer R, Mita AC, A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor sonidegib (LDE225) in patients with advanced solid tumors. Clin. Cancer Res 20, 1900–1909 (2014). [DOI] [PubMed] [Google Scholar]
- 48.LoRusso PM, Rudin CM, Reddy JC, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Chang I, Darbonne WC, Graham RA, Zerivitz KL, Low JA, Von Hoff DD, Phase I trial of hedgehog pathway inhibitor vismodegib (GDC-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin. Cancer Res. 17, 2502–2511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robarge KD, Brunton SA, Castanedo GM, Cui Y, Dina MS, Goldsmith R, Gould SE, Guichert O, Gunzner JL, Halladay J, Jia W, Khojasteh C, Koehler MFT, Kotkow K, La H, LaLonde RL, Lau K, Lee L, Marshall D, Marsters JC Jr., Murray LJ, Qian C, Rubin LL, Salphati L, Stanley MS, Stibbard JHA, Sutherlin DP, Ubhayaker S, Wang S, Wong S, Xie M, GDC-0449—A potent inhibitor of the hedgehog pathway. Bioorg. Med. Chem. Lett 19, 5576–5581 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Marschke RF, Borad MJ, McFarland RW, Alvarez RH, Lim JK, Padgett CS, Von Hoff DD, O’Brien SE, Northfelt DW, Findings from the phase I clinical trials of CX-4945, an orally available inhibitor of CK2. J. Clin. Oncol 29, 3087–3087 (2011). [Google Scholar]
- 51.Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Kim J, Chen B, King EM, Borodovsky A, Riggins GJ, Epstein EH, Beachy PA, Rudin CM, Itraconazole and arsenic trioxide inhibit Hedgehog pathway activation and tumor growth associated with acquired resistance to smoothened antagonists. Cancer Cell 23, 23–34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niefind K, Guerra B, Ermakowa I, Issinger O-G, Crystal structure of human protein kinase CK2: Insights into basic properties of the CK2 holoenzyme. EMBOJ. 20, 5320–5331 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarno S, Moro S, Meggio F, Zagotto G, Ben DD, Ghisellini P, Battistutta R, Zanotti G, Pinna LA, Toward the rational design of protein kinase casein kinase-2 inhibitors. Pharmacol. Ther. 93, 159–168 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Bian Y, Han J, Kannabiran V, Mohan S, Cheng H, Friedman J, Zhang L, VanWaes C, Chen Z, Mek inhibitor PD-0325901 overcomes resistance to CK2 inhibitor CX-4945 and exhibits anti-tumor activity in head and neck cancer. Int. J. Biol. Sci 11,411–422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrer-Font L, Villamañan L, Arias-Ramos N, Vilardell J, Plana M, Ruzzene M, Pinna LA, Itarte E, Arús C, Candiota AP, Targeting protein kinase CK2: Evaluating CX-4945 potential for GL261 glioblastoma therapy in immunocompetent mice. Pharmaceuticals 10, E24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surade S, Blundell TL, Structural biology and drug discovery of difficult targets: The limits of ligandability. Chem. Biol 19, 42–50 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, Connelly M, Stewart CF, Gould S, Rubin LL, Curran T, Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/~p53~−~ mice. Cancer Cell 6, 229–240 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Lee HY, Greene LA, Mason CA, Manzini MC, Isolation and culture of post-natal mouse cerebellar granule neuron progenitor cells and neurons. J. Vis. Exp, 990 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ge X, Milenkovic L, Suyama K, Hartl T, Purzner T, Winans A, Meyer T, Scott MP, Phosphodiesterase 4D acts downstream of Neuropilin to control Hedgehog signal transduction and the growth of medulloblastoma. eLife 4, e07068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang ECY, Aicheler R, Murrell I, Wilkinson GWG, Lehner PJ, Gygi SP, Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell 157, 1460–1472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz D, Gygi SP, An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol 23, 1391–1398 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Chou MF, Schwartz D, Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics 35, 15–24 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE, Math1-driven GFP expression in the developing nervous system of transgenic mice. GeneExpr. Patterns 3, 389–395 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T, Irreversible APCCdh1 inactivation underlies the point of no return for cell-cycle entry. Cell 166, 167–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraley C, Raftery AE, Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc 97, 611–631 (2002). [Google Scholar]
- 66.Cozza G, Girardi C, Ranchio A, Lolli G, Sarno S, Orzeszko A, Kazimierczuk Z, Battistutta R, Ruzzene M, Pinna LA, Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: Structural features and pharmacological potential. Cell. Mol. Life Sci 71, 3173–3185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K, Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costa R, Arrigoni G, Cozza G, Lolli G, Battistutta R, Carlos J, Belmonte I, Pinna LA, Sarno S, The lysine-specific demethylase 1 is a novel substrate of protein kinase CK2. Biochim. Biophys. Acta 1844, 722–729 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Venerando A, Franchin C, Cant N, Cozza G, Pagano MA, Tosoni K, Al-Zahrani A, Arrigoni G, Ford RC, Mehta A, Pinna LA, Detection of phospho-sites generated by protein kinase CK2 in CFTR: Mechanistic aspects of Thr1471 phosphorylation. PLOS ONE 8, e74232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Relative phosphopeptide abundance between P14 biological replicates.
Fig. S2. Relative proliferation in the P1, P7, and P14 cerebellum.
Fig. S3. GNP proliferation after CX-4945 treatment in culture.
Fig. S4. Knockdown specificity of pooled siRNA constructs targeting Csnk2a1, Csnk2a2, Csnk2b, and Smo.
Fig. S5. Gli1 and Hh-associated transcript expression in NIH3T3 cells after CK2 inhibition.
Fig. S6. Gli1 transcript expression in cultured SHH MB cell lines.
Fig. S7. Weight of control or CX-4945-treated mice with cerebellar SHH MB.
Fig. S8. CSNK2A1 expression and 5-year survival of patients with SHH MB.
Fig. S9. Sequence conservation of Csnk2a1 mutations in TBB-resistant SHH MB cells.
Table S3. Primer and siRNA sequences.
Table S4. Pathological report for the primary human MB sample (ST01).
Table S1. Phosphoproteome of GNPs isolated at P1, P7, and P14.
Table S2. Phosphopeptides of enriched kinase motifs.