Abstract
S100 proteins are distinct dimeric EF-hand Ca2+-binding proteins that can bind Zn2+, Mn2+, and other transition metals with high affinity at two sites in the dimer interface. Certain S100 proteins, including S100A7, S100A12, S100A8, and S100A9, play key roles in the innate immune response to pathogens. These proteins function via a “nutritional immunity” mechanism by depleting essential transition metals in the infection that are required for the invading organism to grow and thrive. They also act as damage-associated molecular pattern ligands, which activate pattern recognition receptors (e.g., Toll-like receptor 4 RAGE) that mediate inflammation. Here we present protocols for these S100 proteins for high-level production of recombinant protein, measurement of binding affinities using isothermal titration calorimetry, and an assay of antimicrobial activity.
Keywords: S100 proteins, S100A7, S100A12, S100A8, S100A9, Calprotectin, Nutritional immunity, Metal binding, Host-pathogen interaction, Inflammatory response, Protein expression, Protein purification, Isothermal titration calorimetry, Antimicrobial growth assay
1. Introduction
1.1. S100 Proteins
S100 proteins are a unique subfamily of EF-hand calcium (Ca2+)-binding proteins that have an integrated dimeric structure and can form higher-order oligomers [1]. Although EF-hand proteins are widely known as second messengers that transduce Ca2+ signals, S100 proteins also function in the extracellular milieu [2, 3]. Remarkably, several S100 proteins have been shown to play central roles in the innate immune response to infection by pathogenic organisms, with dual functions via “nutritional immunity” and activation of inflammation [4]. Of the 25 different S100 genes, only S100A7, S100A8, S100A9, and S100A12 have been identified as innate immune modulators.
1.2. Structure and Function of S100 Proteins
S100 proteins contain N-terminal S100-specific and C-terminal canonical, “EF-hand” helix-loop-helix calcium-binding motifs that come together to form the stable four-helix bundle characteristic of all EF-hand proteins. However, S100 proteins are distinct among the EF-hand proteins in that the basic structural unit is a completely integrated, obligate dimer [1]. All S100 proteins except the ancestral and C-terminally truncated S100G form homodimers, and a select few form heterodimers. However, S100A8 and S100A9 are unique in that they preferentially form heterodimers [5].
Ca2+ binding results in a significant shift in the orientation of helix 3 (Fig. 1), exposing a hydrophobic cleft that mediates binding to target proteins [6]. Because the levels of Ca2+ present in the extracellular milieu are so high (in the mM range), when outside cells S100 proteins are constitutively bound to Ca2+ and activated for target binding. Besides binding Ca2+, many S100 proteins also bind transition metal ions (e.g., Mn2+, Cu2+, Fe2+, Ni2+, and Zn2+) [7]. Each S100 dimer has two transition metal-binding sites at symmetrically disposed locations across the dimer interface (Fig. 2) [7, 8]. Binding of transition metals has been characterized for S100A12 [9], S100A7 [10], and S100A8/S100A9 heterodimer (calprotectin, CP) [8]. S100A7 and S100A12 have canonical sites comprised of three histidine and one aspartic acid residue that coordinate Zn2+ or Cu2+ in a tetrahedral fashion [7]. CP has two distinct transition metal-binding sites. One is specific to CP and is comprised of six histidine residues arranged in an octahedral coordination sphere, capable of binding Mn2+, Zn2+, Ni2+, Cu2+ and Fe2+ (S1). The other (S2) is a canonical site that can bind Zn2+ and Cu2+. Although binding of transition metal ions minimally perturbs S100 protein structures, high concentrations of these ions (and also Ca2+) induce the formation of tetramers and higher-order oligomers. In some cases, it appears that these oligomeric states have a key role for function [3, 11].
Fig. 1.
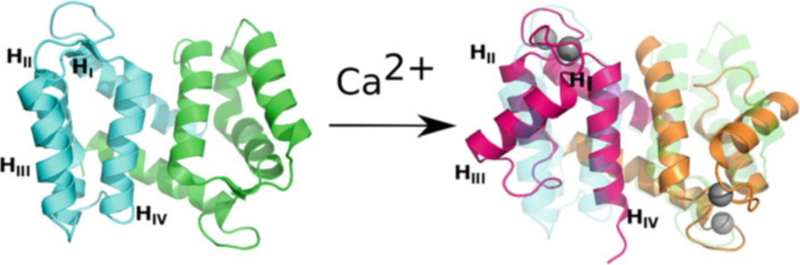
The effect of Ca2+ binding on the structure of S100A12. The Ca2+-free protein (green and cyan) is shown on the left, and the Ca2+-bound state (magenta and orange) is shown on the right, superimposed on the Ca2+-free state. Note the change in position of helix 3 between the Ca2+-free and Ca2+-bound states. PDB ID: 2WCE and 2M9G
Fig. 2.
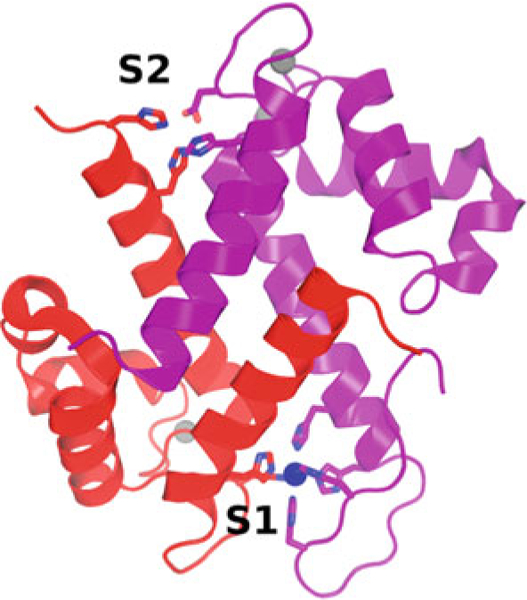
Structure of Ca2+- and Mn2+-bound CP. S100A8 is colored red, and S100A9 is colored purple. Ca2+ ions are gray spheres and the Mn2+ ion is a blue sphere. The side chains chelating transition metal ions in S1 and S2 are shown. PDB ID: 4GGF
1.3. S100 Proteins in Nutritional Immunity
During infections, pathogens acquire nutrient metals from the host, while the host in turn limits the availability of nutrient metals [12, 13]. S100 proteins play a central role in restricting the bio-availability of nutrient transition metals, a mechanism termed “nutritional immunity” [14]. In keeping with this, increasing dietary transition metals can overcome this defense and in turn cause detrimental effects for the host during infection. For example, dietary iron supplementation enhances Mycobacterium avium [15] and Mycobacterium tuberculosis [16] infection, excess dietary zinc heightens Clostridium difficile colonization of the gut [17], and elevated dietary manganese increases susceptibility of the heart to Staphylococcus aureus infection [18]. Consistent with CP playing a critical role during infection, a deficiency in CP enhances susceptibility of the murine host to numerous bacterial and fungal pathogens [19–24]. S100A12 inhibits Campylobacter jejuni [25] and Helicobacter pylori [26] through Zn2+-dependent sequestration. Additionally, S100A12 exhibits antimicrobial activity against multiple parasites, possibly through the binding of copper, which enhances the production of superoxide [27, 28]. In 2007, it was reported that S100A7 and S100A15 have antimicrobial activity toward Escherichia coli during infections of the skin [29, 30]. In addition to limiting metal nutrients, S100A7 can directly adhere to and reduce the survival of E. coli in the skin [31]. These observations suggest that the ability of S100 proteins to bind a variety of nutrient metal ions is critical in establishing nutritional immunity and protection during infections.
1.4. S100 Proteins in the Inflammatory Response
In addition to nutritional immunity, S100 proteins play diverse roles as immunomodulators. S100A7 and S100A15 proteins are expressed in keratinocytes [32, 33], whereas CP and S100A12 are primarily expressed in monocytes, macrophages, and neutrophils [28, 34, 35]. However, during the inflammatory response, CP and S100A12 are also expressed in keratinocyte, endothelial, and epithelial cells [36–39]. Secreted CP can act as a potent chemoattractant for monocytes, macrophages, and neutrophils during inflammation [40, 41]. As a result, CP inhibits immune cell growth or induces apoptosis [17]. In addition, S100 proteins can act as damage-associated molecular patterns by binding pattern recognition receptors including TLR4 [42], RAGE [43, 44], and CD33 [45]. Misregulation of S100 protein expression is implicated in chronic inflammation [46], as overactivation of the pattern recognition receptors TLR4 and RAGE drives NF-κB production and recruitment of neutrophils, monocytes, and macrophages. For example, high levels of calprotectin and S100A12 have been associated with inflammatory bowel disease and rheumatoid arthritis and high levels of S100A7 with inflammatory skin diseases (e.g., psoriasis, atopic dermatitis) [17, 30, 44]. Altered expression of S100 proteins also correlates with multiple autoimmune disorders including systemic lupus erythematosus, Still’s, and Sjogren’s [47–50]. Given the numerous associations to inflammation and infections, S100 proteins have been targeted for potential therapeutic effects [51].
2. Materials
2.1. Expression and Purification of S100 Proteins
2.1.1. Reagents and Solutions
pGEMEX plasmid containing S100A12.
BL21 (DE3) competent cells.
LB/agar plates supplemented with 100 μg/mL ampicillin.
Autoclaved Luria broth (LB) growth media.
100 mg/mL ampicillin solution sterile-filtered prior to use
Ammonium sulfate.
Lysis buffer: 20 mM Tris-HCl (pH 8.0).
Q buffer A: 20 mM Tris-HCl (pH 8.0).
Q buffer B: 20 mM Tris-HCl (pH 8.0), 1 M NaCl.
SEC buffer: 20 mM Tris-HCl (pH 8.0), 100 mM NaCl.
SDS-PAGE buffers and Coomassie stain.
2.1.2. Equipment
Autoclave.
0.45 and 0.2 μm syringe filters.
Plastic syringes.
Appropriate flasks and petri dishes.
Shaking incubator.
Sonication system.
Centrifuges.
Dialysis tubing with a 1 kDa MWCO.
10 kDa MWCO concentrators.
FPLC system.
Q-sepharose column.
S75 column.
SDS-PAGE system and gels.
2.2. Determination of Transition Metal Affinities of S100 Proteins
2.2.1. Reagents and Solutions
CP protein (~2 mL at 10–50 μM) in ITC buffer.
ITC buffer: 20 mM HEPES (pH 7.5), 75 mM NaCl.
Zn(OAc)2 in ITC buffer.
1 M CaCl2.
Equipment
VP-ITC titration calorimeter.
Degassing station.
2.3. Antimicrobial Growth Assay
Reagents and Solutions
Tryptic Soy Agar (TSA) plate.
Tryptic Soy Broth (TSB).
CP protein in CP buffer.
CP buffer: 20 mM Tris (pH 7.5), 100 mM NaCl, 3 mM CaCl2, 10 mM BME, filtered.
Equipment
96-well microtiter plate.
Plastic wrap.
UV/Vis spectrophotometer.
3. Methods
3.1. Expression and Purification of S100 Proteins
A lac operon-based system is used for bacterial expression of S100 proteins. Following standard protocols, Escherichia coli (E. coli) cells are transformed with a plasmid containing the gene of interest, grown to a high optical density, and stimulated to express the protein of interest by the addition of isopropyl β-D-1-thiogalacto-pyranoside (IPTG). After cell lysis, the protein is purified from the soluble lysate. Purification takes advantage of the extraordinary structural stability of S100 proteins. The name S100 is derived from the observation that these proteins remain soluble in 100% ammonium sulfate [52]. Hence, the first step of the purification is an ammonium sulfate precipitation that increases the ionic strength of the solution and results in the precipitation of most of the endogenous E. coli proteins, while S100 remains in solution. The next step is anion-exchange chromatography for further separation of impurities by differences in isoelectric point. The last step of the purification involves size-exclusion chromatography to separate the S100 protein from its impurities by size. Finally, mass spectrometry of the purified product confirms the identity of the protein of interest. Protocols for S100A7, S100A8, S100A9, S100A12, and CP have been published [5, 14, 53–55]. Here we provide a specific detailed example, an optimized 1 L expression and purification protocol for human S100A12.
3.1.1. Procedure
Expression
Transform pGEMEX-S100A12 into BL21 (DE3) chemically competent cells using a standard transformation protocol [56] (see Note 1).
Plate 100 μL of transformation solution on LB/agar plate supplemented with ampicillin, and grow 12–16 h at 37 °C.
Pick a single colony from the freshly transformed plate, and inoculate 25 mL of LB media supplemented with antibiotic.
Grow this culture in a shaking incubator at 250 rpm for 12–16 h at 37 °C.
In the morning, inoculate 10 mL of the culture to inoculate 1 L of LB supplemented with antibiotic (see Note 2).
Grow this culture at 37 °C in a shaking incubator at 250 rpm.
While shaking the culture, monitor cell growth. Once the optical density at 600 nm (O.D.600) is between 0.6 and 0.8 absorbance units, add IPTG to a concentration of 0.5 mM.
Allow the culture to grow at 37 °C in a shaking incubator at 250 rpm for 12–16 h.
Harvest the cells by centrifugation at 5000 × ℊ for 20 min, and freeze the pellets at –80 °C.
Purification
Thaw the frozen cell pellet at room temperature, and resuspend in 30 mL of lysis buffer.
Sonicate on ice at 40% amplitude (5 s on, 5 s off) for 5 min (see Note 3).
Clarify the lysate by centrifugation at 20,000 × ℊ for 25 min at 4 °C.
The S100A12 protein will be in the supernatant (see Note 4). Decant supernatant into 100 mL glass beaker and place the beaker on ice. Add a stir bar and slowly add ammonium sulfate to 60% saturation (see Note 5). Allow the solution to stir in the cold room for 1 h.
Clarify the solution by centrifugation at 20,000 × ℊ for 25 min at 4 °C.
Transfer the supernatant into dialysis tubing. Dialyze against 2 L of Q buffer A at 4 °C. Change the dialysis buffer twice, allowing at least 4 h in between changes (see Note 6).
Using the FPLC system, equilibrate a Q-Sepharose column with 4 column volumes (CV) of Q Buffer A.
Load the protein sample and collect flow through.
Wash the column with four CV of Q buffer A.
Elute proteins with a 0–30% NaCl gradient over 15–20 CV using Q buffer B, collecting 5 mL fractions.
Take an aliquot of each fraction and run SDS-PAGE with Coomassie staining.
Pool the fractions containing S100A12 protein. The protein has a molecular weight of ~10.5 kDa.
Concentrate the fractions to <5 mL using a 10 kDa MWCO concentrator.
Equilibrate an S75 column with 1 CV of SEC buffer (see Note 7).
Load the concentrated S100A12 protein onto the SEC column.
Elute the protein over 1 CV, collecting 5 mL fractions.
Take an aliquot of each fraction, and run SDS-PAGE with Coomassie staining.
Pool together the S100A12 protein fractions, and validate the protein identity using mass spectrometry.
Measure the absorbance of the protein using a spectrophotom¬eter in order to obtain the concentration. The extinction coef¬ficient of S100A12 is 5960 M–1 cm–1 (see Note 8).
Concentrate protein to ~5–10 mg/mL. Concentrated protein will reduce the risk of degradation. Aliquot into 1.5 mL Eppendorf tubes, and flash freeze using liquid nitrogen. Store the protein at –80 °C (see Note 9).
3.2. Determination of Transition Metal Affinities of S100 Proteins by Isothermal Titration Calorimetry
Determining transition metal ion-binding affinities of S100 proteins is one key to understanding their role at the host-pathogen interface as this informs on their ability to function via the nutritional immunity mechanism, i.e., outcompete bacterial metal ion transporters for the nutrient metals [53]. Many techniques have been implemented to determine the dissociation constants (Kd) for S100 proteins, which have been reported in the μM-pM range [7]. Isothermal titration calorimetry (ITC), which measures the amount of heat released or absorbed upon binding, has been used to measure μM-nM transition metal ion-binding affinities of S100 proteins [8, 57, 58]. ITC estimates of the binding affinities of CP for Zn2+ (S1–3 nM, S2–8 nM) and Mn2+ (S1–6 nM) have been reported (Fig. 3). Additional methods for determining metal ion affinities include optical absorption spectroscopy [53], electron paramagnetic resonance spectroscopy [53], competition chelator fluorescence experiments [53, 59], and equilibrium gel filtration [60, 61], each method having its own set of advantages and limitations. Competition chelator fluorescence measurements are the most sensitive and are able to measure Kd values in the pM range, although this method requires transition metal-specific fluorescence dyes and can be complicated by interference from Ca2+ [62]. Here, we provide a protocol for measuring transition metal binding to CP protein using ITC. This is the only experimental method that allows for the complete thermodynamic characterization of a reaction (ΔH, ΔG, and ΔS) and also provides the stoichiometry of the reaction.
Fig. 3.
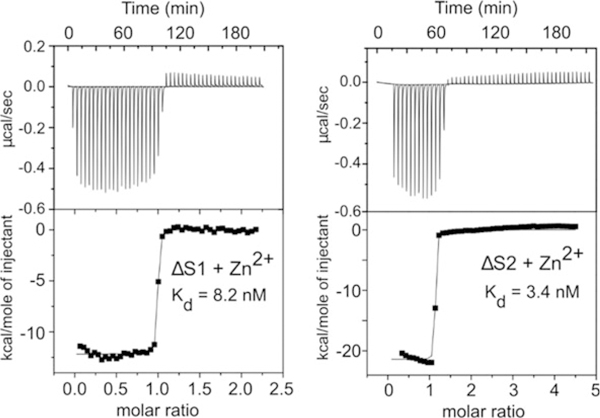
The binding of Zn2+ to CP monitored by ITC. ITC isotherms of CP as Zn2+ are titrated from the syringe into the cell with S1 or S2 knockout mutants (ΔS1, ΔS2). Note the modest differences in Kd values between S1 and S2. These panels were taken from Fig. 1 in the original publication in Damo et al. [8]
3.2.1. Procedure
Dialyze CP protein (10–50 μM) in ITC buffer for Replace with fresh buffer twice for a total of three dialysis exchanges (see Note 10).
Prepare a 100–750 μM Zn(OAc)2 solution using the dialysis ITC buffer. Save the remaining dialysis buffer for rinsing the syringe and sample cell of the ITC instrument (see Note 11).
Add stoichiometric amount of CaCl2 to both the protein and the metal solutions.
Degas the protein and metal ion solutions for 10 min.
Rinse the ITC sample cell three times with ITC buffer.
Add the CP protein to the sample cell of the ITC instrument. Ensure that no air bubbles are present in the syringe.
Load the Zn(OAc)2 solution into the ITC titration syringe.
Titrate the metal ion solution into the protein. We recommend starting with 50 injections, each of 6 μL and 210 s between each injection. The number of injections, injection volumes, and the time between injections must be optimized depending on the metal-binding affinities and the heat absorbed or released upon binding of the system. Before performing another injection, make sure that the heat from each injection is fully dissipated (see Note 12).
To correct for the heat of dilution, perform a metal solution to buffer titration after thoroughly rinsing out the sample cell with the ITC buffer. Repeat steps 2–8, except add only ITC buffer to the sample cell (see Note 13).
To process the data, subtract the buffer isotherm from the protein isotherm. Fit a single-site binding model to the data using the VP-ITC Origin software (Northampton, MA), and obtain the thermodynamic parameters (see Note 14). An example of data depicting the titration of CP mutants with ZnSO4 is shown in Fig. 3.
3.3. Antimicrobial Growth Assay
The ability of S100 proteins to sequester nutrient transition metals inhibits growth of pathogens [20, 22, 26]. Here we describe an antimicrobial growth assay that measures the activity of WT and mutant CP, monitoring the ability of CP to inhibit the growth of Staphylococcus aureus. This assay verifies the ability of CP to sequester transition metal ions. The results of the assay are reported as O.D.600 plotted over time, quantifying bacterial growth in the presence of decreasing concentrations of CP (Fig. 4)[ 20]. Typically, CP at a concentration ≥ 75 μg/mL is sufficient to inhibit the growth of S. aureus in minimal media; however, the inhibition of bacterial growth is also species dependent with different concentrations of CP required for inhibition. Salmonella is greater than 125 μg/mL. Acinetobacter baumannii is greater than 37.5 μg/mL. Helicobacter pylori is greater than 300 μg/mL. Pseudomonas aeruginosa and Clostridium difficile require 1 mg/mL. Inhibition of bacterial growth by CP could vary based on transition metal enrichment of the media chosen to facilitate bacterial growth. This same assay can be implemented to probe the antimicrobial activity of other S100 proteins such as S100A12 [26].
Fig. 4.
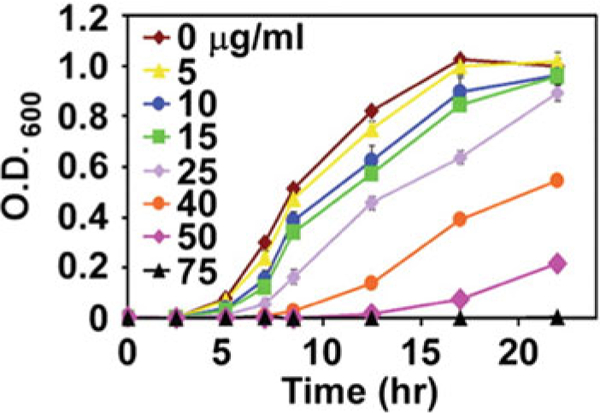
Representative growth assay of S. aureus in the presence of increasing concentrations of recombinant CP. Growth was quantified by 0.D.600. Error bars represent the standard deviation of at least three replicates. Panel is adapted from the original publication in Corbin et al. [20]
Streak a TSA plate with S. aureus, and incubate at 37 °C for 16–18 h (see Note 15).
Select a single colony of S. aureus, start a 5 mL overnight culture in TSB, and incubate at 37 °C on a shaker for 16–18 h at 180 rpm.
Back dilute the overnight culture 1:50 in fresh TSB, and incu¬bate at 37 °C on a roller drum for 1 h at 40 rpm.
- During the incubation, prepare a 96-well microtiter plate as follows:
-
(a)Rows B-H will each contain 62 μL CP buffer and 38 μL TSB (see Note 16).
-
(b)Row A will contain the highest concentration of CP with CP buffer to a total volume of 124 μL. In addition, each well in this row will have 76 μL TSB. For example, if you want 200 μg of protein in row A and have a protein stock that is 10 μg/μL, you would add 20 μL protein stock to 104 μL CP buffer.
-
(c)The CP stock should be thawed and supplemented with 3 mM CaCl2 prior to use (see Note 17).
-
(d)To create a 1:2 serial dilution of CP for this assay, transfer 100 μL from row A into row B. Pipet to mix, and transfer 100 μL from row B to row C. Repeat until you transfer 100 μL from row F into row G. After mixing row G, remove 100 μL from row G so that there is a final volume of 100 μL. Do not transfer row G to row H. Row H should only contain CP buffer and TSB without protein (see Note 18).
-
(e)After adding all the media components to the wells, briefly centrifuge (800 r.c.f., 2 min) the plate to remove bubbles.
-
(a)
After 1 h, add 1 μL of back-diluted culture from step 3 to each well.
Measure initial O.D.600 to establish background and 0 h time point (see Note 19).
Incubate the microtiter plate at 37 °C with shaking. Wrap the microtiter plate in plastic wrap to prevent evaporation (see Note 20).
Take time points every 1–2 h. Vortex the microtiter plate prior to reading the O.D.600 to resuspend any bacteria that have settled to the bottom. Representative data for antimicrobial activity of WT CP against S. aureus are shown in Fig. 4.
4. Notes
S100 proteins may also be purified from E. coli using a lac operon-based system with a plasmid-containing N-terminal 6 × His tag. The protein can then be purified by immobilized metal affinity chromatography (IMAC). This is the case for S100A7 and S100A15 [55]. A plasmid-containing N-terminal 6× His tag has been reported for S100A4 with purification via IMAC followed by anion-exchange chromatography [63].
The use of auto-inducing media in place of LB can significantly increase yields for S100A12 [54, 64]. Whether this is true for other S100 proteins must be experimentally verified.
S100 proteins are stable enough to perform the purification at room temperature with the exception of the sonication, centrifugation, and ammonium sulfate precipitation steps.
Purification of CP requires a protein-refolding step before the anion-exchange column [5, 14, 53], as S100A8–S100A9 are expressed into inclusion bodies rather than in the supernatant.
The optimum ammonium sulfate percentage must be optimized for each protein.
For S100 proteins containing cysteine residues, it may be necessary to include BME (or other reducing agents) to the buffers right before use to prevent disulfide bond formation.
Adding 0.5–5 mM of CaCl2 to the chromatography buffers may increase protein stability and yields. However, this must be removed by the addition of EDTA and dialysis prior to storing the protein, as Ca2+ induces S100 protein oligomerization.
Typical yields for S100 proteins using this protocol are 35–45 mg per 1 L of culture. However, the optimal induction time must be empirically determined for each protein.
To maintain S100 protein activity, avoid repeated freeze-thaw cycles.
The protein and metal ion concentrations have to be accurately determined as this will affect the results. Protein concentrations can be experimentally determined with the use of the extinction coefficients and absorbance at 280 nm.
It is critical that the protein in the sample cell and the metal ion solutions in the syringe have the exact same buffer. The best way to ensure that the buffer is exactly the same is to prepare the syringe solutions in the dialysis buffer that the protein was dialyzed in. If the buffers are not exactly the same, fluctuations in the data will give false binding results.
This ITC procedure can be used for all S100 proteins; however, the number of injections and concentrations might need to be adjusted. It should also be noted that the typical sensitivity of ITC places a lower limit on Kd in the nM range. Methods described above might need to be used if the binding affinities fall outside of this range.
Reducing agents can adversely affect the linearity of the base¬line. Whether your system can tolerate the reducing agent in the buffer must be determined empirically. In general, TCEP is preferable to BME, which is preferable to DTT. Another alternative is to make Cys to Ser mutations in your protein.
Repeat the experiments in triplicate.
This assay can use a variety of different bacterial strains, other than S. aureus. If another strain is used, the appropriate media for the chosen bacteria needs to be implemented.
A lower amount of CP buffer relative to growth media can be used in order to facilitate bacterial growth; however, this will usually increase the concentration of CP necessary to inhibit growth.
Add BME to the CP buffer right before use.
For most assays, a twofold dilution of CP starting at a concentration of 100 μg/100 μLin rows A–G will suffice for S. aureus.
With S. aureus, there will be some initial growth at the highest dose of CP, most likely due to residual metal ions stored within the bacteria.
It is possible to use an automated plate reader to quantify the O.D.600. In this situation, it is recommended for the lid to remain closed to avoid evaporation.
Acknowledgments
This work was supported by the National Institutes of Health R01AI101171 (EPS and WJC) and the National Science Foundation HRD1547757 (SMD).
References
- 1.Nelson MR, Chazin WJ (1998) Structures of EF-hand Ca(2+)-binding proteins: diversity in the organization, packing and response to Ca2+ binding. Biometals 11(4):297–318 [DOI] [PubMed] [Google Scholar]
- 2.Donato R (2003) Intracellular and extracellular roles of S100 proteins. Microsc Res Tech 15:540–551 [DOI] [PubMed] [Google Scholar]
- 3.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL (2013) Functions of S100 proteins. Curr Mol Med 13:24–57 [PMC free article] [PubMed] [Google Scholar]
- 4.Zackular JP, Chazin WJ, Skaar EP (2015) Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem 290:18991–18998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter MJ, Chazin WJ (1998) High level expression and dimer characterization of the S100 EF-hand proteins, migration inhibitory factor-related proteins 8 and 14. J Biol Chem 273:12427–12435 [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya S, Bunick CG, Chazin WJ (2004) Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta 1742 (1–3):69–79 [DOI] [PubMed] [Google Scholar]
- 7.Gilston BA, Skaar EP, Chazin WJ (2016) Binding of transition metals to S100 proteins. Science China 59:792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ (2013) Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A 110 (10):3841–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, Bronstein IB (2009) The crystal structures of human S100A12 in apo form and in complex with zinc: new insights into S100A12 oligomerisation. J Mol Biol 391:536–551 [DOI] [PubMed] [Google Scholar]
- 10.Brodersen DE, Nyborg J, Kjeldgaard M (1999) Zinc-binding site of an S100 protein revealed. Two crystal structures of Ca2+−bound human psoriasin (S100A7) in the Zn2+−loaded and Zn2+−free states. Bio-chemistry 38:1695–1704 [DOI] [PubMed] [Google Scholar]
- 11.Moroz OV, Burkitt W, Wittkowski H, He W, Ianoul A, Novitskaya V, Xie J, Polyakova O, Lednev IK, Shekhtman A, Derrick PJ, Bjoerk P, Foell D, Bronstein IB (2009) Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Bio-chem 10:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood MI, Skaar EP (2012) Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10(8):525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer LD, Skaar EP (2016) Transition metals and virulence in bacteria. Annu Rev Genet 50:67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP (2011) Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10:158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhople AM, Ibanez MA, Poirier TC (1996) Role of iron in the pathogenesis of Mycobacterium avium infection in mice. Microbios 87 (351):77–87 [PubMed] [Google Scholar]
- 16.McDermid JM, Hennig BJ, van der Sande M, Hill AV, Whittle HC, Jaye A, Prentice AM (2013) Host iron redistribution as a risk factor for incident tuberculosis in HIV infection: an 11-year retrospective cohort study. BMC Infect Dis 13:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP (2016) Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22(11):1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juttukonda LJ, Berends ETM, Zackular JP, Moore JL, Stier MT, Zhang Y, Schmitz JE, Beavers WN, Wijers CD, Gilston BA, Kehl-Fie TE, Atkinson J, Washington MK, Peebles RS, Chazin WJ, Torres VJ, Caprioli RM, Skaar EP (2017)Dietary manganese promotes staphylococcal infection of the heart. Cell Host Microbe 22(4):531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besold AN, Gilston BA, Radin JN, Ramsoomair C, Culbertson EM, Li CX, Cormack BP, Chazin WJ, Kehl-Fie TE, Culotta VC (2018) Role of calprotectin in withholding zinc and copper from Candida albicans. Infect Immun 86:e00779–e00717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319 (5865):962–965 [DOI] [PubMed] [Google Scholar]
- 21.Gaddy JA, Radin JN, Loh JT, Piazuelo MB, Kehl-Fie TE, Delgado AG, Ilca FT, Peek RM, Cover TL, Chazin WJ, Skaar EP, Scott Algood HM (2014) The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog 10(10):e1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP (2012) Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog 8 (12):e1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP (2013) MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81(9):3395–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A (2009) Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog 5(10):e1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shank JM, Kelley BR, Jackson JW, Tweedie JL, Franklin D, Damo SM, Gaddy JA, Murphy CN, Johnson JG (2018) The host antimicrobial protein Calgranulin c participates in the control of campylobacter jejuni growth via zinc sequestration. Infect Immun 86(6): e00234–e00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haley KP, Delgado AG, Piazuelo MB, Mortensen BL, Correa P, Damo SM, Chazin WJ, Skaar EP, Gaddy JA (2015) The human antimicrobial protein calgranulin C participates in control of Helicobacter pylori growth and regulation of virulence. Infect Immun 83(7):2944–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroz OV, Antson AA, Grist SJ, Maitland NJ, Dodson GG, Wilson KS, Lukanidin E, Bronstein IB (2003) Structure of the human S100A12-copper complex: implications for host-parasite defence. Acta Crystallogr D Biol Crystallogr 59(Pt 5):859–867 [DOI] [PubMed] [Google Scholar]
- 28.Ravasi T, Hsu K, Goyette J, Schroder K, Yang Z, Rahimi F, Miranda LP, Alewood PF, Hume DA, Geczy C (2004) Probing the S100 protein family through genomic and functional analysis. Genomics 84(1):10–22 [DOI] [PubMed] [Google Scholar]
- 29.Buchau AS, Hassan M, Kukova G, Lewerenz V, Kellermann S, Wurthner JU, Wolf R, Walz R, Walz M, Gallo RL, Ruzicka T (2007) S100A15, an antimicrobial protein of the skin: regulation by E. coli through Toll-like receptor 4. J Invest Dermatol 127:2596–2604 [DOI] [PubMed] [Google Scholar]
- 30.Glaser R, Harder J, Lange H, Bartels J, Christophers E, Schroder JM (2005) Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol 6:57–64 [DOI] [PubMed] [Google Scholar]
- 31.Lee KC, Eckert RL (2007) S100A7 (Psoriasin)-mechanism of antibacterial action in wounds. J Invest Dermatol 127:945–957 [DOI] [PubMed] [Google Scholar]
- 32.Boniface K, Bernard F-X, Garcia M, Gurney AL, Lecron J-C, Morel F (2005) IL-22 inhibits epidermal differentiation and induces proin-flammatory gene expression and migration of human keratinocytes. J Immunol 174:3695–3702 [DOI] [PubMed] [Google Scholar]
- 33.Wolf R, Mirmohammadsadegh A, Walz M, Lysa B, Tartler U, Remus R, Hengge U, Michel G, Ruzicka T (2003) Molecular cloning and characterization of alternatively spliced mRNA isoforms from psoriatic skin encoding a novel member of the S100 family. FASEB J 17:1969–1971 [DOI] [PubMed] [Google Scholar]
- 34.Hsu K, Champaiboon C, Guenther BD, Sorenson BS, Khammanivong A, Ross KF, Geczy CL, Herzberg MC (2009) Anti-infective protective properties ofS100 calgranulins. Antiinflamm Antiallergy Agents Med Chem 8(4):290–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagasse E, Clerc RG (1988) Cloning and expression of two human genes encoding calcium-binding proteins that are regulated during myeloid differentiation. Mol Cell Biol 8(6):2402–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goebeler M, Roth J, van den Bos C, Ader G, Sorg C (1995) Increase of calcium levels in epithelial cells induces translocation of calcium-binding proteins migration inhibitory factor-related protein 8 (MRP8) and MRP14 to keratin intermediate filaments. Biochem J 309(Pt 2):419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimbaldeston MA, Geczy CL, Tedla N, Finlay-Jones JJ, Hart PH (2003) S100A8 induction in keratinocytes by ultraviolet A irradiation is dependent on reactive oxygen intermediates. J Invest Dermatol 121(5):1168–1174 [DOI] [PubMed] [Google Scholar]
- 38.Mork G, Schjerven H, Mangschau L, Soyland E, Brandtzaeg P (2003) Proinflammatory cytokines upregulate expression of calprotectin (L1 protein, MRP-8/MRP-14) in cultured human keratinocytes. Br J Dermatol 149(3):484–491 [DOI] [PubMed] [Google Scholar]
- 39.Stoll SW, Zhao X, Elder JT (1998) EGF stimulates transcription of CaN19 (S100A2) in HaCaT keratinocytes. J Invest Dermatol 111 (6):1092–1097 [DOI] [PubMed] [Google Scholar]
- 40.Cornish CJ, Devery JM, Poronnik P, Lackmann M, Cook DI, Geczy CL (1996) S100 protein CP-10 stimulates myeloid cell chemotaxis without activation. J Cell Physiol 166(2):427–437 [DOI] [PubMed] [Google Scholar]
- 41.Lackmann M, Cornish CJ, Simpson RJ, Moritz RL, Geczy CL (1992) Purification and structural analysis of a murine chemotactic cytokine (CP-10) with sequence homology to S100 proteins. J Biol Chem 267(11):7499–7504 [PubMed] [Google Scholar]
- 42.Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, Nacken W, Foell D, van der Poll T, Sorg C, Roth J (2007) Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med 13 (9):1042–1049 [DOI] [PubMed] [Google Scholar]
- 43.Foell D, Wittkowski H, Vogl T, Roth J (2007) S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol 81(1):28–37 [DOI] [PubMed] [Google Scholar]
- 44.Leclerc E, Fritz G, Vetter SW, Heizmann CW (2009) Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta 1793 (6):993–1007 [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Eksioglu EA, Zhou J, Zhang L, Djeu J, Fortenbery N, Epling-Burnette P, Van Bijnen S, Dolstra H, Cannon J, Youn JI, Donatelli SS, Qin D, De Witte T, Tao J, Wang H, Cheng P, Gabrilovich DI, List A, Wei S (2013) Induction of myelodysplasia by myeloid-derived suppressor cells. J Clin Invest 123 (11):4595–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bresnick AR, Weber DJ, Zimmer DB (2015) S100 proteins in cancer. Nat Rev 15:96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi IY, Gerlag DM, Holzinger D, Roth J, Tak PP (2014) From synovial tissue to peripheral blood: myeloid related protein 8/14 is a sensitive biomarker for effective treatment in early drug development in patients with rheumatoid arthritis. PLoS One 9(8):e106253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frosch M, Ahlmann M, Vogl T, Wittkowski H, Wulffraat N, Foell D, Roth J (2009) The myeloid-related proteins 8 and 14 complex, a novel ligand of toll-like receptor 4, and interleukin-1beta form a positive feedback mechanism in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum 60 (3):883–891 [DOI] [PubMed] [Google Scholar]
- 49.Haga HJ, Brun JG, Berntzen HB, Cervera R, Khamashta M, Hughes GR (1993) Calprotectin in patients with systemic lupus erythematosus: relation to clinical and laboratory parameters of disease activity. Lupus 2 (1):47–50 [DOI] [PubMed] [Google Scholar]
- 50.Nordal HH, Brun JG, Halse AK, Madland TM, Fagerhol MK, Jonsson R (2014) Calprotectin (S100A8/A9), S100A12, and EDTA-resistant S100A12 complexes (ERAC) in primary Sjogren’s syndrome. Scand J Rheumatol 43 (1):76–78 [DOI] [PubMed] [Google Scholar]
- 51.Pietzsch J (2011) S100 proteins in health and disease. Amino Acids 41:755–760 [DOI] [PubMed] [Google Scholar]
- 52.Moore BW (1965) A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun 19:739–744 [DOI] [PubMed] [Google Scholar]
- 53.Brophy MB, Hayden JA, Nolan EM (2012) Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J Am Chem Soc 134:18089–18100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson E, Little S, Franklin DS, Gaddy JA, Damo SM (2017) Expression, purification, and antimicrobial activity of S100A12. J Vis Exp 123:6–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray JI, Tonkin ML, Whiting AL, Peng F, Farnell B, Cullen JT, Hof F, Boulanger MJ (2012) Structural characterization of S100A15 reveals a novel zinc coordination site among S100 proteins and altered surface chemistry with functional implications for receptor binding. BMC Struct Biol 12:16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Froger A, Hall JE (2007) Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 6:e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sivaraja V, Kumar TK, Rajalingam D, Graziani I, Prudovsky I, Yu C (2006) Copper binding affinity of S100A13, a key component of the FGF-1 nonclassical copper-dependent release complex. Biophys J 91:1832–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilder PT, Varney KM, Weiss MB, Gitti RK, Weber DJ (2005) Solution structure of zinc- and calcium-bound rat S100B as determined by nuclear magnetic resonance spectroscopy. Biochemistry 44:5690–5702 [DOI] [PubMed] [Google Scholar]
- 59.Koch M, Bhattacharya S, Kehl T, Gimona M, Vasak M, Chazin W, Heizmann CW, Kroneck PM, Fritz G (2007) Implications on zinc binding to S100A2. Biochim Biophys Acta 1773:457–470 [DOI] [PubMed] [Google Scholar]
- 60.Schäfer BW, Fritschy JM, Murmann P, Troxler H, Durussel I, Heizmann CW, Cox JA (2000) Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J Biol Chem 275:30623–30630 [DOI] [PubMed] [Google Scholar]
- 61.Sturchler E, Cox JA, Durussel I, Weibel M, Heizmann CW (2006) S100A16, a novel calcium-binding protein of the EF-hand super-family. J Biol Chem 281:38905–38917 [DOI] [PubMed] [Google Scholar]
- 62.Cuden LS, Gaillard A, Nolan EM (2016) Calcium ions tune the zinc-sequestering properties and antimicrobial activity of human S100A12. Chem Sci 7:1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ecsédi P, Kiss B, Gόgl G, Radnai L, Buday L, Koprivanacz K, Liliom K, Leveles I, Vértessy B, Jeszenὄi N, Hettinyi C, Schlosser G, Katona G, Nyitray L (2017) Regulation of the equilibrium between closed and open conformations of Annexin A2 by N-terminal phosphorylation and S100A4-binding. Structure 25:1195–1207 [DOI] [PubMed] [Google Scholar]
- 64.Studier FW (2014) Chapter 2: Stable expression clones and auto-induction for protein production in E. coli. Methods Mol Biol 1091:17–32 [DOI] [PubMed] [Google Scholar]


