Abstract
Autophagy is a process in which cellular components are delivered to lytic vacuoles to be recycled and has been demonstrated to promote abiotic/biotic stress tolerance. Here, we review how the responses triggered by stress conditions can affect autophagy and its signalling pathways. Besides the role of SnRK1 and TOR kinases in regulating autophagy, abscisic acid and its signalling kinase, SnRK2, have emerged as key players to induce autophagy under stress conditions. Furthermore, an interplay between ROS and autophagy is observed, ROS being able to induce autophagy and autophagy able to reduce ROS production. We also highlight the importance of osmotic adjustment for the successful performance of autophagy and discuss the potential role of GABA in plant survival and ethylene-induced autophagy.
Keywords: ROS, TOR, GABA, SnRK1, ATG
Autophagy: a recycling process involved in homeostasis, stress tolerance and senescence
Both abiotic and biotic stress conditions have a negative impact on plant growth, and often threaten agronomical production. In future years, an increased demand for food and a more challenging environment for plant growth is expected [1]. To counteract this, a better understanding of plant resistance to both abiotic and biotic stresses is necessary, and in particular of those processes promoting plant survival on a cellular and organismal level. Plant macroautophagy (here referred to as autophagy, see glossary) is one such process in which macromolecules and cellular components are recycled in lytic vacuoles to be re-used. This recycling is crucial for maintaining cellular homeostasis, acting as a quality control mechanism under non-stressful conditions, and it is stimulated under stress conditions [2].
Autophagy can act either selectively to degrade specific cell components or non-selectively to degrade bulk cytoplasm. In either case, the macromolecules and cellular components to be degraded are encapsulated by a double membrane vesicle (autophagosome), which fuses with the vacuole for recycling of its contents [3]. Autophagy is initiated by the production of an engulfing double-membrane termed a phagophore from the endoplasmic reticulum (ER) [4,5], although other membranes may also contribute to autophagosome formation [6]. The development of the phagophore requires the coordination of different AuTophaGy-related (ATG) proteins, which are highly conserved between plants, yeast, and mammals [7]. Some of these ATG proteins participate in the induction of the phagophore (e.g. ATG1, ATG11 and ATG13), transport of lipids for membrane enlargement (e.g. ATG9), vesicle nucleation (e.g. ATG5 and ATG12), and phagophore expansion and closure (e.g. ATG4, ATG8, ATG3 and ATG7) [8]. After collecting cytosolic components, the phagophore seals, forming an autophagosome, which ultimately fuses with the tonoplast where the cargo is released for its degradation by vacuolar hydrolases [3]. Recent excellent reviews discuss the mechanisms and regulation of autophagosome formation (see [8–11]) and these topics thus will not be covered here in detail.
Autophagy occurs at basal levels in non-stressful conditions [12]. However, stressful conditions, such as nitrogen or carbon starvation, oxidative stress, ER stress, heat, drought, saline, and osmotic stress, sugars excess, and also senescence, induce autophagic flux [13,14]. Autophagy contributes to the recycling and remobilization of nutrients both during organ senescence and in nutrient deficiency [15]. The autophagic recycling process yields amino acids, fatty acids, and sugars which can be used later by the organism as anabolic substrates [16] or for energy production [17,18]. In this way, autophagy can be considered as a process promoting plant survival, particularly during nutrient deficiency (Key Figure 1). In addition, autophagic activity in senescing leaves was observed to contribute to nitrogen remobilization into the seeds [15,19]. In this process of nitrogen remobilization, the selective degradation of chloroplasts by autophagy (chlorophagy) was also shown to be important [20]. The role of other types of selective autophagy, including mitophagy and pexophagy in plant survival under stress conditions remains largely unexplored [21].
Key Figure 1.
Autophagy contributes to cell and plant survival under abiotic and biotic stress conditions. In harsh conditions, cellular organelles can be damaged and their dysfunction increases the generation of reactive oxygen species (ROS) and oxidative damage. At the whole plant level this can lead to plant senescence. Damaged molecules, and even organelles such as mitochondria, chloroplasts and peroxisomes, can be recycled through autophagy. The resulting breakdown products can be used for the de novo synthesis of molecules and organelle biogenesis, thus promoting stress tolerance at both cellular and whole plant levels.
Autophagy is critical for plant tolerance of a wide range of stress conditions. For instance, the growth of Arabidopsis atg mutants, which have impaired autophagy, was shown to be dramatically reduced when subjected to drought and saline stress [22]. When exposed to oxidative stress, autophagy-defective plants become chlorotic [23]. Moreover, autophagic mutants cannot survive under long periods of poor nitrogen and/or carbon conditions, and even in optimal conditions exhibit premature leaf senescence [20]. At a cellular level, abiotic and biotic stresses lead to the overproduction of reactive oxygen and nitrogen species (ROS and RNS) which can damage organelles and biomolecules, affecting their functionality (see box 1 for further detail). In autophagy mutants, these damaged components accumulate due to impaired degradation [17], reducing survival and leading to hypersensitivity to stress conditions.
Box 1. Reactive oxygen and nitrogen species are generated under biotic and abiotic stress.
Stress conditions lead to the overproduction of ROS, including superoxide (O2.−), hydrogen peroxide (H2O2), singlet oxygen (1O2) and hydroxyl radical (.OH). Superoxide can be produced either by electron leakage of the mitochondrial and chloroplastic electron transport chains under stress conditions, or by the respiratory burst oxidase homologue (Rboh, also known as NADPH oxidase). Stress conditions, such as cold, salinity, heat and high light, were shown to induce the expression of Rboh as a rapid systemic response [135]. Mittler et al. proposed the concept of a ROS wave [123], in which each cell senses the wave of superoxide and responds by expressing its own Rboh to propagate the wave through the apoplast. This wave is suggested to be coordinated with a calcium wave and promoted by phytohormones such as ABA [136]. In turn, superoxide yields H2O2, mainly via SOD activity. Given that H2O2 is considerably more stable and biological membranes are not as impermeable to it as for superoxide, it can act as a signalling molecule diffusing between different cell compartments. In fact, hundreds of genes are differentially expressed when plants are exposed to H2O2 [137]. It can also lead to the generation of the most reactive ROS, the hydroxyl radical, in the presence of Fe3+ by the so-called Fenton reaction. This radical reacts with biomolecules in a diffusion-limited rate. The accumulated molecules under stress conditions, such as sugars and proline, react with hydroxyl radicals as a strategy to avoid damage to more essential biomolecules [40,99,100]. Singlet oxygen is mainly produced by the chloroplast when an excited chlorophyll (3Chl) reacts with molecular oxygen. High-light stress and stresses affecting the fluidity of thylakoid membranes enhance its production. In this way, damaged chloroplasts are an important source of ROS, and autophagy (or more specifically, chlorophagy) can contribute to the recycling of these organelles. Nitric oxide (.NO) is a considerably reactive molecule that can diffuse across membranes and act also as a signalling molecule, attenuating or triggering biological processes such as stomatal closure, germination and root elongation [138–141]. Under stress conditions, .NO can be overproduced [142,143], reacting with ROS to generate more reactive species such as peroxynitrite (ONOO−) and nitrogen dioxide (.NO2), collectively known as RNS. The regulatory role of .NO and other RNS is mainly exerted by interfering with critical components in the signalling cascade of phytohormones [144]. In this work, we discuss potential effects of .NO on plant autophagy.
In the last few years, many findings linking plant autophagy with different signalling pathways under abiotic and biotic stress have emerged [13,24–26]. Here we discuss these findings focusing first on the hormonal and metabolic changes that are produced by stress conditions, and then integrating this with the signalling pathways involved in autophagy activation.
Abiotic and biotic stresses induce major metabolic changes
In this section we discuss major metabolic changes occurring under both abiotic and biotic stresses and how these changes can interact with the autophagic process. Cellular events such as alterations in carbohydrate and amino acid fluxes are fundamental features in a plant’s capacity to successfully cope with major biotic and abiotic stresses, directing resources away from growth pathways and towards stress responses. Moreover, perception and modulation of sugar status plays a prominent role in cell survival capacity under stress. This is coordinated mainly through the central energy-sensing SnRK1 (SNF-related kinase 1) kinase, which acts upstream of Target of Rapamycin (TOR) upon sugar-phosphate perception, to induce major changes at the cellular level, regulating plant growth and development, cell cycle progression, induction of stress responses, and autophagy [14,25,27–29]. Besides the signalling function of sugars, their capacity to control osmotic potential, hydrate membranes, scavenge ROS, and their protection of the photosynthetic apparatus make them key molecules in the cellular responses to chilling, freezing, heat, and drought stress [30–32].
In a similar way, the content of specific amino acids is increased upon stress conditions and is associated with abiotic stress tolerance. For instance, GABA (γ-amino butyric acid) accumulates in plant tissues in response to several stresses and is thought to play a critical role in stomatal closure and water retention [33,34]. Proline is the amino acid with the greatest increase in concentration upon abiotic stress and some authors have associated this response with tolerance to drought, salt, cold and heavy metal stresses [35]. Similar to sugars, proline was suggested to have an important role as a compatible osmolyte and ROS scavenger, in the protection of membranes and proteins, and the maintenance of photosynthetic activity [35,36]. Nonetheless, proline is not an effective scavenger of most ROS [37,38], but is able to protect against hydroxyl radicals [39,40]. In addition, proline metabolism also acts as a redox shuttle, transferring electrons from the cytosol/chloroplast to the mitochondria [41]. Through this pathway, proline catabolism was demonstrated to have direct implications for autophagy in animals. In particular, high proline dehydrogenase (ProDH) activity increases ROS-dependent autophagy, enhancing cell viability [42–44]. However, a link between proline metabolism and autophagy has not yet been explored in plants. This is of particular interest because in normal conditions at night, or after stress conditions, proline catabolism becomes more active and could contribute to cell recovery by increasing autophagic activity. Furthermore, increased levels of amino acid-derived compounds, like glycine-betaine and polyamines, may also contribute to abiotic stress tolerance in several plant species [45,46]. In our view, the osmotic adjustment produced by the accumulation of these compatible osmolytes (sugars and amino acids) in the cytosol also could contribute to the equilibration of the sudden osmotic changes produced in the cell during autophagy (discussed further in box 2). Besides their osmoprotectant role, GABA and polyamines have been proposed as signalling molecules, mainly through the interplay with hydrogen peroxide (H2O2) and nitric oxide (.NO), and by affecting Ca2+ influx [34,47]. Below, we will also discuss their potential to affect autophagy-related signalling.
Box 2. Coordination of autophagy with osmotic adjustment.
The breakdown of more complex structures to monomers in the vacuole during the autophagic process results in increased vacuolar osmolality. Different permeases allow the transport of these recycled compounds back to the cytosol, contributing to the compensation of the osmotic gradient. However, under intense autophagic activity a transient osmotic gradient can be generated, putting the stability of the tonoplast at risk. When the tonoplast is broken, the vacuolar hydrolases are released into the cytoplasm, degrading its content. This process is known as mega-autophagy, and usually results in programmed cell death [8]. To avoid this, it is logical that plant cells perform de novo synthesis of certain metabolites (such as proline) in the cytosol to balance the increased vacuolar osmolality (Figure I). While for most of the recycled compounds, a higher concentration in the vacuole would ensure their transport from the vacuole to the cytosol through the permeases, some other compounds should have an opposite gradient to compensate for the osmolality. For instance, in potato plants under stress conditions proline concentrations were estimated to be 83 mM in the cytosol but 4 mM in the vacuole [145]. In conclusion, any autophagic activity contributing to increased vacuolar osmolality should be coordinated with the accumulation of cytosolic osmo-active compounds (Figure I).
Likewise, the interaction of plants with pathogens has a profound impact on the plant metabolome [48]. The importance of primary metabolism modulation is linked to its role in supplying energy to cells during pathogen attack, enhancing viability of attacked cells [48]. The current opinion tends to accept that metabolic investment in defence disadvantages plant growth in a trade-off process [49]. In this respect, a contribution from biotic stress-induced autophagy (discussed below) to stimulate nutrient recycling and counteract disease progression is a fascinating hypothesis for future investigation. Among the responses to abiotic/biotic stress, the effect of ROS, sugars and polyamines on autophagy is better understood and will be elaborated below. Whether the accumulated amino acids have a role in autophagy remains elusive; however, we suggest here that their cytosolic accumulation should contribute to avoiding mega-autophagy (box 2).
Abiotic stress-induced autophagy
Here, we integrate information on the role of abiotic stress-induced autophagy and the main mechanisms controlling this process, with a goal of contributing to our understanding of whether and how autophagy should be manipulated to enhance abiotic stress tolerance.
Diverse abiotic stresses have been shown to induce autophagy, including osmotic and salt stress, improving plant resistance [22,25]. Moreover, autophagy is relevant for the tolerance of oxidative stress [23], a condition associated with most environmental stresses. The induction of autophagy upon abiotic stress is relatively fast, for example the expression of ATG18a in arabidopsis (Arabidopsis thaliana) was induced within a few hours after exposure to NaCl or mannitol [22]. Also in arabidopsis, the overexpression of ATG5 and ATG7 increases autophagic flux and results in a greater tolerance to oxidative stress [24]. Less is known about the effect of autophagy on priming strategies, in which subjecting plants to a mild stressor induces responses that prepare the plants for a future more severe stress. Because priming is usually also associated with temporal ROS and oxidative stress responses [50,51], it is possible that priming with certain molecules, such as NaCl and H2O2, also induces autophagy. In fact, it was recently shown that thermopriming induces autophagy in arabidopsis [52]. The induction of autophagy by priming could contribute to stress tolerance, as in stress conditions autophagy is usually considered to promote cell survival [3]. However, in this case, autophagy was shown to degrade the heat shock proteins that were induced by the thermopriming to prevent future susceptibility to heat [52], therefore resetting the priming machinery.
Among the proteins coordinating autophagy in abiotic stress, TOR seems to be of particular importance because its overexpression is enough to block starvation-, salt-, and drought-induced autophagy [26]. TOR is a master regulator of growth in response to nutrient availability and its activity was shown to be key for auxin signalling [53–55]. In particular, auxins induce TOR activity through the small GTPase ROP2, which interact with and phosphorylate TOR to activate it [56]. Once active, TOR associates with polysomes to induce the content of upstream open reading frames-mRNA of many auxin response factors (ARF) [57]. In this way, auxin activates TOR which in turn boosts the translation of ARF, enhancing auxin signalling. Concordantly, the auxin analogue 1-naphthaleneacetic acid (NAA) induces TOR activity, resulting in the inhibition of autophagy through a reduction in the number of autophagosomes formed [26]. Sugar phosphates are known to affect TOR activity through the modulation of SnRK1, which can inhibit TOR activity [58,59]. In particular, trehalose-6-phosphate (T6P) inhibits SnRK1 activity [60,61] and inhibits autophagy in response to abiotic stress [25]. In arabidopsis, the catalytic subunit of SnRK1, KIN10 (sometimes referred as AKIN10), was shown to act upstream of TOR (Figure 2) [12]. However, autophagy can also be controlled by TOR-independent mechanisms under certain conditions such as oxidative and endoplasmic reticulum stress [26]. One of the suggested candidates for this regulatory role on autophagy is inositol- requiring enzyme 1b (IRE1b), involved in the regulation of ER-stress related genes. Arabidopsis ire1b mutants exhibited a decreased number of autophagosomes in response to misfolded protein accumulation when compared to wild type (WT) plants [62]. Furthermore, SnRK1 could also induce autophagy independently of TOR, as it was shown to directly interact with ATG1, enhancing its function possibly through phosphorylation (Figure 2) [63].
Figure 2.
Potential effect of abiotic and biotic stress response on plant autophagy. Arrows indicate induction or promotion of a process or product. T-bars indicates inhibition of a process or molecule. Dashed connectors indicate that the process is suggested but not completely known in plants. ABA, abscisic acid; CRKs, Cysteine-rich receptor-like kinases; ET, ethylene; GPXLs, glutathione peroxidase-like proteins; H2O2, hydrogen peroxide; MAPK, mitogen activated protein kinases; O2.-, superoxide; Rboh, respiratory burst homologue; ROS, reactive oxygen species; SnRK1, SNF-related kinase 1; SnRK2, SNF-related kinase 2; TOR, target of rapamycin; TPXs, thiol peroxidases.
Autophagy is also important in submergence stress, as autophagy mutants are hypersensitive to this condition [64]. Moreover, flooding is an environmental stress that generally leads to a hypoxic state in root cells, and arabidopsis KIN10 overexpressing lines can induce tolerance to hypoxia treatments by increasing autophagy [63]. Thus, the evidence points to a positive role of autophagy in flooding, but little is known about the mechanisms by which autophagy enhances tolerance of flooding and hypoxia. More is known in animals, where the expression of hypoxia responsive genes, such as hypoxia induced factor-1 (HIF-1), triggers the expression of proteins belonging to the Bcl-2 family such as BNIP3 and BNP3L, both required to activate hypoxia-induced autophagy [65]. Interestingly, if glucose is provided in the media, the hypoxia-induced autophagy occurs without cell death even at 0.1% pO2 [65]. However, when the levels of glucose are low, hypoxia produces HIF-1-independent autophagy, which seems to be controlled by TOR activity, and results in cell death [65].
Taken all together, the current view is that autophagy has a clear beneficial effect under abiotic stresses, and the kinases SnRK1 and TOR play a key regulatory role in abiotic stress-induced autophagy. More research is needed to understand how priming strategies can be applied to manipulate autophagy and whether this results in a positive effect for a subsequent stress.
Biotic stress-induced autophagy
Besides abiotic stresses, biotic stresses are also able to influence autophagic events. In this section we discuss the relationship between autophagy and plant-pathogen interactions. Autophagy is a well-established component of the metazoan immune system [66]. In plants, autophagy activation upon pathogen attack has been shown to lead to different outcomes, depending on the pathogen lifestyle [67]. Similarly, the plant immune system is a complex and sophisticated machinery that relies on multiple layers of specificity to optimize defence responses [68]. One of the most well-studied plant defence mechanisms is the hypersensitive response (HR), a form of programmed cell death (PCD) which has a role in restricting pathogen invasion [69]. Importantly, necrotrophic pathogens can take advantage of the HR response, and some of them can manipulate the HR machinery to facilitate infection spreading [70]. Several studies highlighted a tight connection between autophagy and HR regulation during plant immune responses. Pioneering reports in this field provided divergent data regarding the role of autophagy in inducing or restricting HR [71,72] in arabidopsis leaves infected by the bacterium Pseudomonas syringae. Such discrepancies are probably due to the different age of the plants used in the experiments, which can be a critical factor in view of the importance of autophagy in senescence mechanisms and salicylic acid (SA) signalling [73]. In the case of necrotrophs, autophagy execution was often associated with restriction of the HR response, thus contributing to a resistance phenotype [74,75]. By contrast, the same HR restriction has been shown to increase susceptibility to biotrophs [76]. Further research demonstrated an essential role for autophagy in the induction of HR [73,77], adding more layers of complexity. It was proposed that autophagy can favour HR in infection sites and restrict it in surrounding tissues [73]. This model fits well with evidence indicating a role for autophagy in contributing to the elimination of ROS generated during the HR response [78]. Several authors have hypothesized that the role of autophagy may vary according to the specific pathogen considered [74,79], and evidence has emerged that some pathogens have evolved strategies to modulate autophagy to their advantage [80–82]. In conclusion, autophagy can play different roles in plant-pathogen interactions, and such differences appear to be related to the pathogen’s lifestyle. Future research in this field is expected to shed more light on the selectivity and specificity of autophagic events during immune responses.
Effect of (a)biotic stress-induced phytohormones and metabolites on autophagy
In this section we aim to connect the different phytohormones and the metabolic changes that occur under (a)biotic stress to autophagy. In particular, we assess whether the induction of phytohormones by stress conditions can alter the SnRK1-TOR pathway to permit autophagy even when sugars are available. We also examine whether the accumulated metabolites can suppress or enhance the signalling pathways affecting autophagy.
Regarding the potential crosstalk between phytohormones and autophagy under stress, ABA seems to be one of the most promising phytohormones. ABA is suggested to act as an endogenous messenger under abiotic and biotic stresses [83]. Although the most visible effects of ABA are in the leaves, such as the reduction in leaf expansion and stomatal closure, this phytohormone accumulates in all plant organs once the plant senses a reduction in water availability [84]. Thus, ABA is considered to be part of the plant systemic responses. In biotic stress, ABA-induced stomatal closure is proposed to be essential to avoid pathogen entrance by the stomata as a physical mechanism of defence [85]. ABA was known to inhibit the activity of plant TOR, which could lead to the induction of autophagy under stress, but the molecular mechanism was unknown. However, very recently it was demonstrated that one of the key kinases acting downstream of ABA, SnRK2, phosphorylates RAPTOR and inactivates the TOR complex [86] (Figure 2). Previously, TOR was suggested to be primarily inhibited by SnRK1 when sugars were scarce (carbon depletion). However, autophagy is activated during stress even when sugars are abundant [14], suggesting that an alternative pathway should occur. The results from Wang et al. [86] provide this alternative pathway, which is activated in response to ABA and has the SnRK2 kinase as a central player (Figure 2). Moreover, ABA could contribute to the establishment of autophagy via the activation of the respiratory burst oxidase homolog (Rboh, NADPH oxidase in animals, Figure 2).
Another phytohormone of relevance under stress is ethylene (ET). This phytohormone also increases in response to extracellular pathogens through the activation of both mitogen-activated protein kinase (MAPK) signalling pathways and Ca2+ dependent protein kinases, which increase ACS (1-aminocyclopropane-1-carboxylic acid synthase) gene expression [87] (Figure 2). Multiple reports demonstrated that intact ET signalling is required for the Rboh-dependent accumulation of ROS necessary to trigger tolerance to both biotic and abiotic stimuli [88–90], and several lines of evidence showed that ET is able to activate antioxidant systems. In particular, oxidative stress treatments were able to induce ERF1 expression, and ERF6 was shown to upregulate antioxidant enzymes under biotic and abiotic stresses [91]. ET-dependent ROS scavenging is also active during heavy metal and cold stress responses [92,93]. In fact, the role of ERFs as inducers of resistance to many abiotic stresses (salinity, cold, drought, freezing, heat, heavy metal and oxidative stress) is well documented [94]. However, a link between ERF and autophagy is still unknown, and it deserves exploration, in particular because jasmonic acid and SA are now known to regulate ERFs. On the other hand, ABA negatively regulates ERF1 and ERF6 induction; yet ERF1 induces ABA accumulation in arabidopsis [91,95]. Given the importance of ERFs in development and stress responses, and their capacity to converge signals from different phytohormones, it would be interesting to see in future research whether ERF mutants can perform autophagy at the same rate as WT plants.
As discussed above, the accumulation of polyamines, proline, sugars and GABA is one of the most common responses of plants to milder (a)biotic stresses. Once the stress is established, the catabolism of accumulated polyamines by the enzymes polyamine oxidase and diamino oxidase results in the production of H2O2 [96,97]. In this sense, an active catabolism of polyamines can contribute to the H2O2-induced autophagy (discussed below). In fact, exogenous application of physiological concentrations of spermine was shown to induce autophagosome formation in root cells of wheat seedlings [98]. Besides the osmoprotective role discussed in box 2, the accumulated osmolytes are able to reduce ROS levels by reacting with hydroxyl radicals and in this way prevent oxidative stress-induced cell death [99,100]. Also, GABA was shown to induce ET production which in turn can induce ATG8 expression and thus autophagy. This evidence, together with the capacity of GABA to contribute to the TCA cycle through the GABA shunt, suggests that GABA can act at different levels to promote cell survival (box 3).
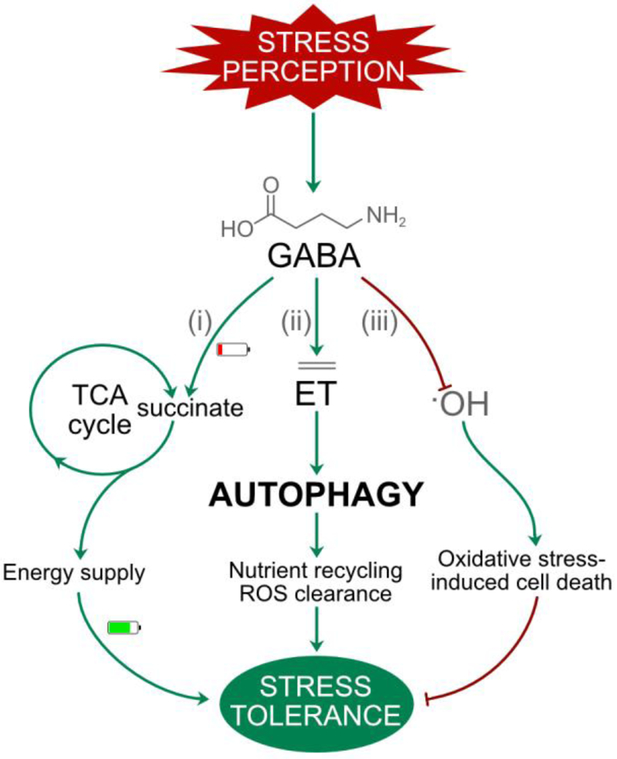
Box 3. GABA promotes cell survival at different levels.
GABA is suggested to play several roles in plant responses to (a)biotic stresses, and also under starvation or hypoxic conditions [146]. Under non-optimal conditions causing nutrient deficiency, the GABA shunt is activated to produce reducing equivalents through the TCA cycle and feed the cell (Figure I, i). Moreover, GABA has been shown to increase ET levels in sunflower cotyledons [147]. Since ET was shown to promote ATG8 expression in plants [148], contributing to autophagy, we hypothesize that GABA may indirectly promote autophagy (Figure I, ii). Finally, under oxidative stress conditions, GABA is able to reduce ROS levels by reacting with hydroxyl radicals and in this way prevent oxidative stress-induced cell death (Figure I, iii). Thus, GABA promotes cell survival at three different levels, (i) feeding the cells via the GABA shunt under starvation conditions, (ii) promoting plant autophagy by increasing ET signalling, and (iii) inhibiting oxidative stress-induced cell death by scavenging hydroxyl radicals (Figure I).
In addition to the osmoprotective function of sugars, sugars can act as signalling molecules to regulate autophagy. Besides the capacity of intracellular T6P to inhibit SnRK1 and thus autophagy [25], external glucose was demonstrated to induce autophagy via Regulator of G-protein signalling (RGS1) [14,101]. In the coming years, the crosstalk between sugar metabolism and autophagy will likely become clearer. In this regard, trehalose-6-phosphate phosphatase (TPP) seems to be a good candidate linking these processes. The induction and activation of TPP has been correlated with heat stress tolerance in seedlings and developmental progression in bud burst [102,103]. Since TPP activity reduces the levels of T6P, we propose that its role in promoting growth can be related to the attenuation of the T6P-mediated inhibition of autophagy. Additionally, the trehalose produced can have positive implications since exogenous treatments with trehalose were shown to reduce membrane damage and induce tolerance to heat stress [102] and also its catabolism via trehalase was shown to have positive implications for drought tolerance [104].
Thus, the current evidence shows that sugars can both repress (eg. T6P) and induce (eg. glucose) autophagy, but also other metabolites such as polyamines and GABA can contribute to the induction of autophagy by different mechanisms, and this deserves further exploration. In terms of phytohormones, ABA is likely to play a key role in inducing autophagy under stress conditions.
Coordination between autophagy and reactive oxygen and nitrogen species
As ROS usually function in an upstream position in the signalling cascade of many abiotic and biotic stress responses, in this final section we consider the effect of ROS on autophagy, and if autophagy in turn modulates ROS levels. Furthermore, as some proteins involved in the autophagic process were shown to be inhibited by ROS by in vitro experiments or artificial oxidative stress in vivo, we critically revisit this evidence, putting it in the context of what would happen in physiological conditions.
To control ROS levels, plants depend on specific antioxidant systems. Usually, the antioxidant systems are divided into enzymatic and non-enzymatic. The enzymatic antioxidant system includes superoxide dismutase isoforms, catalases and peroxidases, whereas the non-enzymatic system is used to refer to ROS scavengers such as glutathione, ascorbate, tocopherol and also sugars. These can be defined as the canonical antioxidant systems and they act specifically on one type of ROS and on single molecules. Autophagy can promote the degradation of damaged organelles, such as mitochondria, chloroplasts, and peroxisomes, that otherwise would overproduce ROS and RNS. Therefore, autophagy was suggested to also contribute to the antioxidant system [105]. This idea is supported by the observation that autophagy-defective arabidopsis plants have increased H2O2 production [106]. Moreover, silencing of the ATG18a gene in arabidopsis generated hypersensitivity to oxidative stress and increased basal oxidative damage [23,107]. Hence, autophagy can be considered as a non-canonical antioxidant system acting on biomolecules or organelles instead of low molecular weight molecules (Figure 3A). Accordingly, it was shown that entire chloroplasts are transported to the vacuole by autophagy as consequence of irradiation stress [108]. This is of particular interest because the accumulation of oxidized components can lead to cell death [105]. Additionally, as the internal redox state of cells can control cell cycle progression [109,110], it is possible that elimination of oxidized molecules is also essential to determine cell cycle fate.
Figure 3.
Effect of reactive oxygen species on autophagy. (A) Schematic crosstalk between reactive oxygen species and autophagy. (B) Inhibition of redox sensitive proteins involved in the autophagic process or its regulation by ROS (in red). Once the stress is perceived the ROS levels increase contributing to the induction of autophagy. However, if very high intracellular levels of ROS are produced some ATG proteins can be oxidized, attenuating the autophagic process. Although SnRK1 and TOR were shown to be redox sensitive, the evidence suggests that excessive intracellular levels of ROS are required to inhibit their activity and attenuate their control over autophagy. The basal and induced concentrations of H2O2 are merely indicative and were estimated based on their determination in plants [134]. It should be noted that these concentrations are highly variable even within the same cell, as some organelles have much higher rate of production than others. The concentrations of excessive ROS were chosen based on the concentrations used with exogenously applied ROS or in the in vitro assays [23,26,115,117,118,120,121]. The figure also considers the difference between the extracellular and intracellular levels of ROS. When ROS are exogenously applied, only a small fraction reaches the cytoplasm/nucleus to inactivate the discussed proteins as they have to cross the cell wall, membranes, and face the antioxidant system. Arrows indicate induction or promotion of a process or product. T-bars indicate inhibition of a process or molecule. Dashed connectors indicate that the process is suggested but not completely known in plants. ROS, reactive oxygen species; SnRK1, SNF-related kinase 1; TOR, target of rapamycin.
In turn, ROS and RNS could modulate autophagy either by targeting activators or repressors of this process. In this regard, treatment of arabidopsis roots with exogenous H2O2 or methyl viologen were shown to induce autophagy [23]. Since methyl viologen acts on the chloroplastic electron chain, this evidence suggests that ROS produced chloroplastically are able to induce autophagy (Figure 2). This is in line with evidence from Chlamydomonas, in which a deficiency in carotenoid synthesis triggers autophagy in the light, but not in the dark, where the chloroplastic electron transport is inactive [111]. Recently, the inhibition of fatty acid synthase complex in chlamydomonas was shown to affect chloroplast integrity, in particular by hyperstacking of thylakoid membranes, which correlated to a higher autophagic flux in a ROS-independent way [112]. Moreover, it was recently reported that the hydrotropic response of arabidopsis roots requires autophagic activity, evidenced by accumulation of autophagosomes, and the presence of H2O2 [113]. Although H2O2 can directly interact with some regulators of autophagy, more specific mechanisms involving H2O2 sensors might act to mediate H2O2 signalling in autophagy. Cysteine-rich receptor-like kinases, glutathione peroxidase-like proteins or thiol peroxidases are examples of these H2O2 sensors that can mediate more effective H2O2 signalling [114]. It will be interesting to see whether these proteins participate in the regulation of autophagy mediated by H2O2 (Figure 2).
The levels of ROS, site of generation and time of exposure are important factors determining their effect on any biological process. Generally speaking, when ROS are limited, in terms of concentration, time or space, they can trigger a response, which many authors refers to the signalling role of ROS; by contrast, uncontrolled levels of ROS may have a deleterious effect on the process under study. Autophagy is not an exception (Figure 3A). For instance, although exogenous H2O2 is suggested to induce autophagy [23], H2O2 was also shown to oxidise ATG proteins inactivating their function and thus autophagy [115]. However, direct evidence in this regard in plants is still scarce. The first insight comes from mammalian systems, in which Scherz-Shouval et al., [72] showed that H2O2 production is induced upon starvation, inhibiting the delipidating activity of ATG4 by targeting a specific cysteine near the catalytic site through oxidation. Because too much delipidating activity would reduce ATG8-phosphatidylethanolamine conjugation, which is necessary for autophagosome formation, the authors suggested that in this way ROS could induce autophagy [116]. In plants and algae, however, the ATG4 processing activity is also inhibited by H2O2, and thus ROS would abolish autophagosome formation [117,118]. Arabidopsis ATG4s are inhibited to 50% when H2O2 is present at 0.9 mM [117], whereas the protease activity of Chlamydomonas reinhardtii ATG4 is more sensitive to H2O2, being partially inhibited at only 0.1 mM H2O2 [118]. In the latter, the inhibition is produced by the formation of a disulfide bond and very low redox potentials are required for its activation, which is mediated by the cytosolic thioredoxin, TRXh1 [118]. The importance of a functional ATG4 in autophagy is supported by data showing that when ATG4 activity is increased either by phosphorylation or glycosylation, autophagy is activated, and when it is reduced by ubiquitination, autophagy is inhibited [119]. Recently, other two ATG proteins of animals, ATG3 and ATG7, were shown to be more redox sensitive than ATG4, being inhibited by 0.1 mM H2O2 through the oxidation of catalytic thiols, resulting in the inhibition of autophagosome formation [115] (Figure 3B). Moreover, an in vitro study demonstrated that KIN10 activity also depends on its the redox status. In particular, very reducing conditions were shown to produce the highest kinase activity, whereas oxidizing conditions inhibit its activity [120]. These observations demonstrate that redox poise could also modulate autophagy, in a way that extreme oxidizing conditions would attenuate autophagy (Figure 3). In the case of arabidopsis ATG4s and KIN10, the concentration of H2O2 required to inhibit their activity is relatively high (about 1 mM), casting doubts on the physiological relevance of ATG4 and KIN10 regulation by H2O2 in plants.
In animals, TOR was also observed to be oxidized and agglomerated by H2O2 causing the loss of its functionality [121]. This observation could explain why overexpression of TOR had no effect on oxidative stress-induced autophagy when using high concentrations of H2O2 (5 mM) [26], and suggests that TOR-independent mechanism act to induce autophagy. Yet, 10 min incubations of TOR with 0.5 mM H2O2 were unable to reduce TOR kinase activity [121]. Hence, it is unlikely that in physiological conditions H2O2 directly inactivates TOR to induce autophagy and more specific mechanisms should be involved. For instance, H2O2 was suggested to indirectly activate the protein kinase SnRK2, through the inhibition of the type 2C protein phosphatase HAB1 [122]. In this way H2O2 could enhance the phosphorylation of RAPTOR from the TOR complex by SnRK2, resulting in the inactivation of TOR and induction of autophagy (Figure 2) [86]. It is tempting to speculate that Rboh acts as a specific switch to activate this H2O2-induced autophagy (Figure 2). This mechanism would fit nicely with the ROS wave concept [123], in which Rboh is induced to produce apoplastic superoxide which dismutates to H2O2 and diffuse along the cells transmitting the signal. In fact, Liu et al. [22] showed that Rboh inhibitors were able to suppress autophagy in arabidopsis (Figure 2). Rboh activity is induced during stress by abscisic acid (ABA) and Ca2+ signals [124,125] (Figure 2). Moreover, Rboh is known to induce Ca2+ influx, and cytosolic Ca2+ signals are known to induce autophagy [124,126]. Thus, Rboh would be essential to induce autophagy in the systemic response of plants to biotic and abiotic stresses, whereas the production of mitochondrial and chloroplastic ROS, through an increase in oxidized proteins, would comprise a less specific mechanism acting in particular cells subjected to unfavourable conditions. We propose that more research is needed on how chloroplastic ROS can induce autophagy. This insight has been trailing behind studying the effect of mitochondrial ROS on autophagy, because this is one of the most important sources of ROS in animals. In plants the production of ROS by mitochondria is less relevant, due to the lower respiration rate. Therefore, evidence for mitophagy in plants is limited [13].
RNS have emerged as important molecules that are usually overproduced under stress conditions and have regulatory roles [127]. Recently, under hypoxia .NO was shown to target S-nitrosoglutathione reductase (GSNOR) to the autophagosome by S-nitrosylation [128]. This is of particular interest because GSNOR controls the levels of S-nitrosoglutathione (GSNO), the cellular reservoir of .NO, having a key role in the control of crosstalk between ROS and .NO in plants [129]. The specific targeting of GSNOR to the autophagosome by .NO might also occur under other stresses. For example, in Lotus japonicus roots an induction of .NO content was associated with a reduction of GSNOR activity under drought stress [130]. Besides targeting enzymes to autophagosomes, RNS could inhibit or enhance autophagy by posttranslational modification (protein nitration or S-nitrosylation) of the proteins involved in the autophagic process or its regulation. It is possible that the exposed cysteines of ATG3, ATG4 and ATG7, which are targets of oxidation by H2O2, would also be targets of S-nitrosylation by .NO. This might lead to a mechanism in which .NO could also directly inactivate the autophagic process. Moreover, .NO was shown to induce the expression of genes encoding hydrogen sulfide (H2S)-synthesizing enzymes in tomato plants [131], resulting in higher levels of H2S, which inhibits autophagy by an unknown mechanism [132]. This effect of NO would be less relevant when SnRK1 is more active because SnRK1 was shown to phosphorylate nitrate reductase (NR), inactivating its activity and thus reducing NO production [133].
In conclusion, in abiotic and biotic stress an interplay between ROS and autophagy occurs, in which autophagy can reduce ROS production as a non-canonical antioxidant system, whereas ROS induce autophagy. At excessive levels, ROS could also attenuate autophagy through the oxidation of ATG proteins. Whether the oxidation of these proteins is relevant in a physiological context in plants requires further investigation. In the context of ROS signaling, Rboh is in an upstream position and was demonstrated to be essential to modulate H2O2-induced autophagy. Regarding the effect of RNS on plant autophagy, we consider that more research needs to be done in this field.
Concluding remarks
A steadily growing body of evidence highlights autophagy as a key contributor to abiotic stress tolerance in plants, and the kinases SnRK1 and TOR play a key regulatory role in abiotic stress-induced autophagy. Less clear is the contribution of autophagy to biotic stress resistance as in some cases it was shown to benefit the infection. Sugar starvation-based autophagy is well-known, through the activation of SnK1 inhibiting TOR, an autophagy inhibitor. However, mild abiotic stresses and many biotic stresses lead to sugar accumulation, and in these conditions, it is proposed that ABA signalling inhibits the TOR complex through SnRK2, leading to sugar excess/ ABA mediated autophagy. Furthermore, extracellular glucose is suggested to induce autophagy, suggesting that extracellular and intracellular sugar signalling pathways may differentially affect autophagy. ET is another phytohormone with positive implications in autophagy, whereas auxins were shown to have an inhibitory effect.
The current evidence also shows an interplay between ROS and autophagy, in which autophagy can reduce ROS production, whereas ROS induces autophagy. Rboh seems to be a key player contributing to H2O2-induced autophagy. At very high intracellular levels, ROS could inhibit some ATG proteins compromising autophagy, but the existence of these mechanisms in plants or their relevance under physiological conditions remains to be discovered.
Conversely, little is known in plants about the effect of RNS on autophagy, and because these molecules are usually overproduced under stress conditions we consider that this deserves exploration.
Finally, the accumulation of GABA, proline, and polyamines under stress conditions can indirectly promote autophagy by different pathways, and also contribute to the osmotic adjustment that should be coordinated with the autophagic process to avoid mega-autophagy. Future advances in understanding autophagy dynamics under various stresses (see Outstanding Questions) may lead researchers to develop novel strategies to improve crop resistance under increasingly adverse environments.
Outstanding questions:
Do developmental and spatio-temporal effects control the way SnRK1 is regulated by sugars? How do these differ in sink and source tissues, and does this depend on oxidizing and reducing conditions?
Do plants with enhanced SnRK2 activity have increased autophagy? If yes, is this induction ABA dependent?
How does autophagy contribute to the accumulation of ERF1 under flooding conditions? In turn, do ERFs play a role during induction and establishment of autophagy?
Are endogenously produced ROS able to oxidize TOR and promote autophagy in plants? Alternatively, are endogenously produced ROS able to oxidize SnRK1 and ATG proteins to inhibit autophagy in plants?
What is the link between the autophagosome and H2O2 accumulation in the hydrotropic response of Arabidopsis roots under water stress?
What are the mechanisms by which H2S inhibits autophagy?
Are endogenously produced RNS able to affect autophagy in plants; if so, is it due to protein nitration or nitrosylation, and which proteins are targeted by these posttranslational modifications?
Is the autophagic response affected in plants with impaired accumulation of osmolytes? For instance, are the p5cs1 Arabidopsis mutants (unable to accumulate proline) predisposed to activation of mega-autophagy?
Is targeting of specific components of the autophagic machinery a widespread strategy in plant-pathogen interactions?
(Box2) Figure I.
Coordination of autophagy and osmotic adjustment. When macromolecules or organelles are sent to be degraded in the vacuole (dark arrows in the left-hand side of the figure), the osmolality of the vacuole increases as soon as the degradation starts. (A) If the cell responds by accumulating compatible osmolytes in the cytosol, the osmotic pressure is compensated for. (B) In the absence of this response, a gradient of osmotic pressure (dark-blue arrows) will be generated between the vacuole and the cytosol, producing expansion pressure on the tonoplast (sky-blue arrows). (C) The water flux would result in an increase in vacuolar size or (D) the rupture of the tonoplast. In this last scenario, the vacuolar hydrolases are released into the cytosol resulting in mega-autophagy, which in turn endangers the viability of the cell.
(Box 3) Figure I.
Multiple mechanisms by which GABA promotes cell survival and stress tolerance. This scheme illustrates the proposed roles of GABA in maintaining cell viability during (a)biotic stress responses. (i) GABA shunts provide energy required for cell survival. (ii) The induction of ethylene (ET) by GABA can induce autophagy, which contributes to recycling damaged molecules and enhances stress tolerance. (iii) GABA can directly scavenge hydroxyl radicals (.OH) and protect against oxidative stress-induced cell death. Green arrows indicate promotion, whereas red T-bars indicate inhibition.
Highlights.
Autophagy enhances tolerance of many abiotic stresses and oxidative stress conditions.
The energy sensors SnRK1 and TOR control autophagy under energy deficiency, but also under diverse stress conditions.
Independently of the nutritional state of the cells, the stress-responsive SnRK2 emerges as a new player to inhibit TOR and induce autophagy under stress conditions.
Under biotic stress, autophagy can be advantageous to the host as well as being exploited by the pathogen, depending on the pathosystem considered.
Reactive oxygen species (ROS) contribute to the establishment of autophagy, whereas autophagy contributes to ROS scavenging.
Acknowledgments
The authors apologize to all researchers whose work could not be cited due to space limitations. SS thanks the Scientific Research-Flanders (FWO, Belgium) for his postdoctoral fellowship. SS is an associate researcher of the National System of Researchers (SNI) of Uruguay. ŁPT and WVdE are supported by FWO grant no. G0A4915N. DCB is supported by grant no. 1R01GM120316-01A1 from the National Institutes of Health.
Glossary
- Abscisic acid
A plant hormone with important signaling function in development and stress conditions
- Autophagosome
A double membrane compartment that delivers cytoplasmic cargo to be recycled into the vacuole
- Chlorophagy
The selective degradation of chloroplasts via autophagy
- Compatible osmolyte
Organic compounds, usually of low molecular weight, that can be accumulated at high concentration in cells without having toxic effects
- Ethylene
A plant hormone with important signalling functions in development and stress conditions
- Jasmonic acid
A plant hormone with important signalling functions in development and stress conditions
- Macroautophagy
A conserved catabolic process in which part of the cytoplasm, including organelles and damaged molecules, is transferred to the vacuole for degradation
- Mega-autophagy
The massive degradation of the cell contents leading to programmed cell death
- Mitophagy
The selective degradation of mitochondria via autophagy
- Osmotic adjustment
The accumulation of compatible osmolytes to compensate for differing water potentials, within a cell or between the cell and the extracellular environment
- Priming
To induce in a plant a physiological process that prepares it for a faster and/or stronger response in case of a future stress condition
- Pexophagy
The selective degradation of peroxisomes via autophagy
- Phagophore
Double membrane compartment that encloses and isolates cytoplasmic content
- Reactive oxygen species (ROS)
A group of molecules derived from oxygen that are highly reactive and can oxidize other biomolecules
- Reactive nitrogen species (RNS)
A group of small molecules containing nitrogen and oxygen which are highly reactive and can oxidize other biomolecules
- ROS/RNS scavengers
Molecules capable of reacting chemically with ROS/RNS, attenuating their reactivity, at relatively high frequency. This high frequency can be achieved either by high reaction rate constants or by high concentrations of the molecules
- Salicylic acid
A plant hormone with important signalling functions in development and stress conditions
- Systemic response
A generalized response that includes the whole plant and not only the site of stress or infection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tilman D. et al. (2011) Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci 108, 20260–20264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avin-Wittenberg T. et al. (2018) Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot 69, 1335–1353 [DOI] [PubMed] [Google Scholar]
- 3.Bassham DC (2007) Plant autophagy - more than a starvation response. Curr. Opin. Plant Biol 10, 587–593 [DOI] [PubMed] [Google Scholar]
- 4.Zhuang X. et al. (2017) ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci 114, E426–E435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bars R. et al. (2014) ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun 20, 4121. [DOI] [PubMed] [Google Scholar]
- 6.Lamb CA et al. (2013) The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol 14, 759–74 [DOI] [PubMed] [Google Scholar]
- 7.Hayward AP et al. (2009) Autophagy and plant innate immunity: Defense through degradation. Semin. Cell Dev. Biol 20, 1041–1047 [DOI] [PubMed] [Google Scholar]
- 8.Marshall RS and Vierstra RD (2018) Autophagy: The Master of Bulk and Selective Recycling. Annu. Rev. Plant Biol 69, 173–208 [DOI] [PubMed] [Google Scholar]
- 9.Michaeli S. et al. (2016) Autophagy in Plants - What’s New on the Menu? Trends Plant Sci. 21, 134–144 [DOI] [PubMed] [Google Scholar]
- 10.Soto-Burgos J. et al. (2018) Dynamics of Autophagosome Formation. Plant Physiol. 176, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang X. et al. (2018) Autophagosome Biogenesis and the Endoplasmic Reticulum: A Plant Perspective. Trends Plant Sci. 23, 677–692 [DOI] [PubMed] [Google Scholar]
- 12.Inoue Y. et al. (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. DOI: 10.1093/pcp/pcl031 [DOI] [PubMed] [Google Scholar]
- 13.Avin-Wittenberg T. (2018) Autophagy and its Role in Plant Abiotic Stress Management. Plant. Cell Environ DOI: 10.1111/pce.13404 [DOI] [PubMed] [Google Scholar]
- 14.Janse van Rensburg HC et al. (2019) Autophagy in plants: both a puppet and a puppet master of sugars. Front Plant Sci DOI: 10.3389/fpls.2019.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masclaux-Daubresse C. et al. (2017) Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol 39, 8–17 [DOI] [PubMed] [Google Scholar]
- 16.Hirota T. et al. (2018) Vacuolar protein degradation via autophagy provides substrates to amino acid catabolic pathways as an adaptive response to sugar starvation in arabidopsis thaliana. Plant Cell Physiol. 59, 1363–1376 [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N. (2007) Autophagy: Process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. et al. (2013) Autophagy Contributes to Leaf Starch Degradation. Plant Cell 25, 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Berardino J. et al. (2018) Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot 69, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishida H. and Makino A. (2018) Impacts of autophagy on nitrogen use efficiency in plants. Soil Sci. Plant Nutr 64, 100–105 [Google Scholar]
- 21.Broda M. et al. (2018) Mitophagy: A Mechanism for Plant Growth and Survival. Trends Plant Sci. 23, 434–450 [DOI] [PubMed] [Google Scholar]
- 22.Liu Y. et al. (2009) Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963 [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y. et al. (2007) Degradation of Oxidized Proteins by Autophagy during Oxidative Stress in Arabidopsis. Plant Physiol. 143, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minina EA et al. (2018) Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot 69, 1415–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soto-Burgos J. and Bassham DC (2017) SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One 12, e0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pu Y. et al. (2017) TOR-Dependent and -Independent Pathways Regulate Autophagy in Arabidopsis thaliana. Front. Plant Sci 8, 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sablowski R. and Carnier Dornelas M. (2014) Interplay between cell growth and cell cycle in plants. J. Exp. Bot 65, 2703–2714 [DOI] [PubMed] [Google Scholar]
- 28.Dobrenel T. et al. (2013) Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front. Plant Sci 4, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulsmans S. et al. (2016) The SnRK1 Energy Sensor in Plant Biotic Interactions. Trends Plant Sci. 21, 648–661 [DOI] [PubMed] [Google Scholar]
- 30.Tarkowski ŁP and Van den Ende W. (2015) Cold tolerance triggered by soluble sugars: a multifaceted countermeasure. Front. Plant Sci 6, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pommerrenig B. et al. (2018) In concert: orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant Cell Physiol 59, 1290–1299 [DOI] [PubMed] [Google Scholar]
- 32.Sami F. et al. (2016) Role of sugars under abiotic stress. Plant Physiol. Biochem 109, 54–61 [DOI] [PubMed] [Google Scholar]
- 33.Mekonnen DW et al. (2016) Gamma-aminobutyric acid depletion affects stomata closure and drought tolerance of Arabidopsis thaliana. Plant Sci. 245, 25–34 [DOI] [PubMed] [Google Scholar]
- 34.Bown AW and Shelp BJ (2016) Plant GABA: Not Just a Metabolite. Trends Plant Sci. 21, 811–813 [DOI] [PubMed] [Google Scholar]
- 35.Szabados L. and Savouré A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97 [DOI] [PubMed] [Google Scholar]
- 36.Signorelli S. (2016) The fermentation analogy: A point of view for understanding the intriguing role of proline accumulation in stressed plants. Front. Plant Sci 7, 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Signorelli S. et al. (2016) In vivo and in vitro approaches demonstrate proline is not directly involved in the protection against superoxide, nitric oxide, nitrogen dioxide and peroxynitrite. Funct. Plant Biol 43, 870–879 [DOI] [PubMed] [Google Scholar]
- 38.Signorelli S. et al. (2013) Proline does not quench singlet oxygen: Evidence to reconsider its protective role in plants. Plant Physiol. Biochem 64, 80–83 [DOI] [PubMed] [Google Scholar]
- 39.Smirnoff N. and Cumbes QJ (1989) Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 28, 1057–1060 [Google Scholar]
- 40.Signorelli S. et al. (2014) Molecular mechanisms for the reaction between .OH radicals and proline: insights on the role as reactive oxygen species scavenger in plant stress. J. Phys. Chem. B 118, 37–47 [DOI] [PubMed] [Google Scholar]
- 41.Sharma S. et al. (2011) Essential Role of Tissue-Specific Proline Synthesis and Catabolism in Growth and Redox Balance at Low Water Potential. Plant Physiol. 157, 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W. and Phang JM (2012) Proline dehydrogenase (oxidase), a mitochondrial tumor suppressor, and autophagy under the hypoxia microenvironment. Autophagy 8, 1407–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandhare J. et al. (2015) A novel role of proline oxidase in HIV-1 envelope glycoprotein-induced neuronal autophagy. J. Biol. Chem 290, 25439–25451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zabirnyk O. et al. (2010) Oxidized low-density lipoproteins upregulate proline oxidase to initiate ROS-dependent autophagy. Carcinogenesis 31, 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen THH and Murata N. (2011) Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant, Cell Environ. 34, 1–20 [DOI] [PubMed] [Google Scholar]
- 46.Zarza X. et al. (2017) Polyamine oxidase 5 loss-of-function mutations in Arabidopsis thaliana trigger metabolic and transcriptional reprogramming and promote salt stress tolerance. Plant Cell Environ. 40, 527–542 [DOI] [PubMed] [Google Scholar]
- 47.Pál M. et al. (2015) Speculation: Polyamines are important in abiotic stress signaling. Plant Sci. 237, 16–23 [DOI] [PubMed] [Google Scholar]
- 48.Rojas CM et al. (2014) Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci 5, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eichmann R. and Schäfer P. Growth versus immunity-a redirection of the cell cycle? , Curr Opin Plant Biol, 26 (2015) , 106–112 [DOI] [PubMed] [Google Scholar]
- 50.Galletti R. et al. (2008) The AtrbohD-Mediated Oxidative Burst Elicited by Oligogalacturonides in Arabidopsis Is Dispensable for the Activation of Defense Responses Effective against Botrytis cinerea. Plant Physiol. 148, 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Aubel G. et al. (2016) Plant immunity induced by COS-OGA elicitor is a cumulative process that involves salicylic acid. Plant Sci. 247, 60–70 [DOI] [PubMed] [Google Scholar]
- 52.Sedaghatmehr M. et al. (2018) A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. DOI: 10.1111/pce.13426 [DOI] [PubMed] [Google Scholar]
- 53.Weiste C. et al. (2017) The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 13, e1006607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henriques R. et al. (2014) Balancing act: Matching growth with environment by the TOR signalling pathway. J. Exp. Bot 65, 2691–701 [DOI] [PubMed] [Google Scholar]
- 55.Deng K. et al. (2016) Target of Rapamycin Is a Key Player for Auxin Signaling Transduction in Arabidopsis. Front. Plant Sci 7, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schepetilnikov M. et al. (2017) GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 36, 886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schepetilnikov M. et al. (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32, 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nukarinen E. et al. (2016) Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep 6, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lastdrager J. et al. (2014) Sugar signals and the control of plant growth and development. J. Exp. Bot 65, 799–807 [DOI] [PubMed] [Google Scholar]
- 60.Baena-González E. et al. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 [DOI] [PubMed] [Google Scholar]
- 61.Tsai AY-L and Gazzarrini S. (2014) Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci 5, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X. et al. (2016) Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 85, 83–95 [DOI] [PubMed] [Google Scholar]
- 63.Chen L. et al. (2017) The AMP-Activated Protein Kinase KIN10 Is Involved in the Regulation of Autophagy in Arabidopsis. Front. Plant Sci 8, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L. et al. (2015) Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazure N. and Pouysségur J. (2010) Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol 22, 177–180 [DOI] [PubMed] [Google Scholar]
- 66.Deretic V. et al. (2013) Autophagy in infection, inflammation, and immunity. Nat Rev Immunol. 13, 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leary AY et al. (2018) Modulation of plant autophagy during pathogen attack. J. Exp. Bot 69, 1325–1333 [DOI] [PubMed] [Google Scholar]
- 68.Jones JD and Dangl JL (2006) The plant immune system. Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- 69.Coll NS et al. (2011) Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lorang J. et al. (2012) Tricking the guard: Exploiting plant defense for disease susceptibility. Science (80-. ). 338, 659–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patel S. and Dinesh-Kumar SP (2008) Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4, 20–27 [DOI] [PubMed] [Google Scholar]
- 72.Hofius D. et al. (2009) Autophagic Components Contribute to Hypersensitive Cell Death in Arabidopsis. Cell 137, 773–783 [DOI] [PubMed] [Google Scholar]
- 73.Coll NS et al. (2014) The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 21, 1399–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenz HD et al. (2011) ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal. Behav 6, 122–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kabbage M. et al. (2013) Cell Death Control: The Interplay of Apoptosis and Autophagy in the Pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y. et al. (2011) The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal. Behav 6, 1408–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han S. et al. (2015) Cytoplastic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with ATG3 to Negatively Regulate Autophagy and Immunity in Nicotiana benthamiana. Plant Cell 27, 1316–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou S. et al. (2018) Autophagy contributes to regulate the ROS levels and PCD progress in TMV-infected tomatoes. Plant Sci. 269, 12–19 [DOI] [PubMed] [Google Scholar]
- 79.Li Y. et al. (2016) Aspartyl protease mediated cleavage of AtBAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28, 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Popa C. et al. (2016) The effector AWR5 from the plant pathogen Ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep 6, 27058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dagdas YF et al. (2016) An effector of the irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 5, e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Üstün S. et al. (2018) Bacteria Exploit Autophagy for Proteasome Degradation and Enhanced Virulence in Plants. 30, 668–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raghavendra AS et al. (2010) ABA perception and signalling. Trends Plant Sci. 15, 395–401 [DOI] [PubMed] [Google Scholar]
- 84.Zhang J. and Tardieu F. (1996) Relative contribution of apices and mature tissues to ABA synthesis in droughted maize root systems. Plant Cell Physiol. 37, 598–605 [Google Scholar]
- 85.Lim CW et al. (2015) Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci 16, 15251–15270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang P. et al. (2018) Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 69, 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li S. et al. (2018) Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant. Cell Environ 41, 134–147 [DOI] [PubMed] [Google Scholar]
- 88.Jiang C. et al. (2013) An Arabidopsis Soil-Salinity-Tolerance Mutation Confers Ethylene-Mediated Enhancement of Sodium/Potassium Homeostasis. Plant Cell 25, 3535–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamauchi T. et al. (2014) Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot 65, 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mersmann S. et al. (2010) Ethylene Signaling Regulates Accumulation of the FLS2 Receptor and Is Required for the Oxidative Burst Contributing to Plant Immunity. Plant Physiol. 154, 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sewelam N. et al. (2013) Ethylene Response Factor 6 Is a Regulator of Reactive Oxygen Species Signaling in Arabidopsis. PLoS One 8, e70289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu L. et al. (2008) Transcriptional Modulation of Ethylene Response Factor Protein JERF3 in the Oxidative Stress Response Enhances Tolerance of Tobacco Seedlings to Salt, Drought, and Freezing. Plant Physiol. 148, 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schellingen K. et al. (2015) Ethylene signalling is mediating the early cadmium-induced oxidative challenge in Arabidopsis thaliana. Plant Sci. 239, 137–146 [DOI] [PubMed] [Google Scholar]
- 94.Müller M. and Munné-Bosch S. (2015) Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 169, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng M-C et al. (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different cis-Acting Elements in Response to Different Stress Signals. Plant Physiol 162, 1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen J. et al. (2016) Hydrogen Sulfide-Mediated Polyamines and Sugar Changes Are Involved in Hydrogen Sulfide-Induced Drought Tolerance in Spinacia oleracea Seedlings. Front. Plant Sci 7, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cona A. et al. (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci. 11, 80–88 [DOI] [PubMed] [Google Scholar]
- 98.Dmitrieva SA et al. (2018) Spermine Induces Autophagy in Plants: Possible Role of NO and Reactive Oxygen Species. Biochem. Biophys. Mol. Biol 483, 341–343 [DOI] [PubMed] [Google Scholar]
- 99.Signorelli S. et al. (2015) Connecting proline and γ-aminobutyric acid in stressed plants through non-enzymatic reactions. PLoS One 10, e0115349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matros A. et al. (2015) Sugars as hydroxyl radical scavengers: Proof-of-concept by studying the fate of sucralose in Arabidopsis. Plant J. 82, 822–839 [DOI] [PubMed] [Google Scholar]
- 101.Yan Q. et al. (2017) Endocytosis of AtRGS1 Is Regulated by the Autophagy Pathway after D-Glucose Stimulation. Front. Plant Sci 8, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li ZG et al. (2014) Involvement of trehalose in hydrogen sulfide donor sodium hydrosulfide-induced the acquisition of heat tolerance in maize (Zea mays L.) seedlings. Bot. Stud 55, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Signorelli S. et al. (2018) Roles for light, energy and oxygen in the fate of quiescent axillary buds. Plant Physiol. 176, 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Van Houtte H. et al. (2013) Overexpression of the Trehalase Gene AtTRE1 Leads to Increased Drought Stress Tolerance in Arabidopsis and Is Involved in Abscisic Acid-Induced Stomatal Closure. Plant Physiol 161, 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Minibayeva F. et al. (2012) Oxidative stress-induced autophagy in plants: The role of mitochondria. Plant Physiol. Biochem 59, 11–19 [DOI] [PubMed] [Google Scholar]
- 106.Yoshimoto K. et al. (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiong Y. et al. (2007) Disruption ol autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 3, 257–258 [DOI] [PubMed] [Google Scholar]
- 108.Izumi M. et al. (2017) Entire Photodamaged Chloroplasts Are Transported to the Central Vacuole by Autophagy. Plant Cell 29, 377–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vivancos PD et al. (2010) Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 64, 825–838 [DOI] [PubMed] [Google Scholar]
- 110.Velappan Y. et al. (2017) Cell cycle arrest in plants: What distinguishes quiescence, dormancy and differentiated G1? Ann. Bot 120, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Perez-Perez ME et al. (2012) Reactive Oxygen Species and Autophagy in Plants and Algae. Plant Physiol 160, 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heredia-Martínez LG et al. (2018) Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas. Plant Physiol. DOI: 10.1104/pp.18.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiménez-Nopala G. et al. (2018) Autophagy mediates hydrotropic response in Arabidopsis thaliana roots. Plant Sci. 272, 1–13 [DOI] [PubMed] [Google Scholar]
- 114.Smirnoff N. and Arnaud D. (2018) Hydrogen peroxide metabolism and functions in plants. New Phytol. In press, [DOI] [PubMed] [Google Scholar]
- 115.Frudd K. et al. (2018) Oxidation of Atg3 and Atg7 mediates inhibition of autophagy. Nat. Commun 9, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Scherz-Shouval R. et al. (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woo J. et al. (2014) Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc. Natl. Acad. Sci 111, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pérez-Pérez ME et al. (2016) Control of Autophagy in Chlamydomonas Is Mediated through Redox-Dependent Inactivation of the ATG4 Protease. Plant Physiol. 172, 2219–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maruyama T. andNoda NN (2018) Autophagy-regulating protease Atg4: structure, function, regulation and inhibition. J. Antibiot. (Tokyo) 71, 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wurzinger B. et al. (2017) Redox state-dependent modulation of plant SnRK1 kinase activity differs from AMPK regulation in animals. FEBS Lett. 591, 3625–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oka SI et al. (2017) Thioredoxin-1 maintains mechanistic target of rapamycin (mTOR) function during oxidative stress in cardiomyocytes. J. Biol. Chem 292, 18988–19000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sridharamurthy M. et al. (2014) H2O2 inhibits ABA-signaling protein phosphatase HAB1. PLoS One 9, e113643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mittler R. et al. (2011) ROS signaling: the new wave? Trends Plant Sci. 16, 300–9 [DOI] [PubMed] [Google Scholar]
- 124.Görlach A. et al. (2015) Calcium and ROS: A mutual interplay. Redox Biol. 6, 260–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pel ZM et al. (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734 [DOI] [PubMed] [Google Scholar]
- 126.Bootman MD et al. (2018) The regulation of autophagy by calcium signals: Do we have a consensus? Cell Calcium 70, 32–46 [DOI] [PubMed] [Google Scholar]
- 127.Del Río LA (2015) ROS and RNS in plant physiology: An overview. J. Exp. Bot 66, 2827–2837 [DOI] [PubMed] [Google Scholar]
- 128.Zhan N. et al. (2018) S-Nitrosylation Targets GSNO Reductase for Selective Autophagy during Hypoxia Responses in Plants. Mol. Cell 71, 142–154 [DOI] [PubMed] [Google Scholar]
- 129.Lindermayr C. (2017) Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Radic. Biol. Med 122, 110–115 [DOI] [PubMed] [Google Scholar]
- 130.Signorelli S. et al. (2013) Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus. Plant Sci. 201–202, 137–146 [DOI] [PubMed] [Google Scholar]
- 131.da-Silva CJ et al. (2017) NO, hydrogen sulfide does not come first during tomato response to high salinity. Nitric Oxide - Biol. Chem 76, 164–173 [DOI] [PubMed] [Google Scholar]
- 132.Laureano-Marín AM et al. (2016) Negative regulation of autophagy by sulfide in Arabidopsis thaliana is independent of reactive oxygen species. Plant Physiol. 171, 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jossier M. et al. (2009) SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59, 316–328 [DOI] [PubMed] [Google Scholar]
- 134.Chakraborty S. et al. (2016) Quantification of hydrogen peroxide in plant tissues using Amplex Red. Methods 109, 105–113 [DOI] [PubMed] [Google Scholar]
- 135.Miller G. et al. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal 2, ra47. [DOI] [PubMed] [Google Scholar]
- 136.Evans MJ et al. (2016) A ROS-Assisted Calcium Wave Dependent on the AtRBOHD NADPH Oxidase and TPC1 Cation Channel Propagates the Systemic Response to Salt Stress. Plant Physiol. 171, 1771–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Desikan R. et al. (2000) Hydrogen peroxide-induced gene expression in Arabidopsis thaliana. Free Radic. Biol. Med 28, 773–778 [DOI] [PubMed] [Google Scholar]
- 138.García Mata C. and Lamattina L. (2001) Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 126, 1196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lombardo MC et al. (2006) Nitric oxide functions as a positive regulator of root hair development. Plant Signal. Behav 1, 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bethke PC et al. (2006) Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot 57, 517–526 [DOI] [PubMed] [Google Scholar]
- 141.Prado AM et al. (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131, 2707–2714 [DOI] [PubMed] [Google Scholar]
- 142.Rodríguez-Ruiz M. et al. (2018) Arsenate disrupts ion balance, sulfur and nitric oxide metabolisms in roots and leaves of pea (Pisum sativum L.) plants. Environ. Exp. Bot DOI: 10.1016/j.envexpbot.2018.06.028 [DOI] [Google Scholar]
- 143.Signorelli S. et al. (2018) Drought stress triggers the accumulation of NO and SNOs in cortical cells of Lotus japonicus L. roots and the nitration of proteins with relevant metabolic function. Environ. Exp. Bot DOI: 10.1016/j.envexpbot.2018.08.007 [DOI] [Google Scholar]
- 144.Signorelli S. and Considine MJ (2018) Nitric oxide enables germination by a four-pronged attack on ABA-induced seed dormancy. Front. Plant Sci 9, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Büssis D. and Heineke D. (1998) Acclimation of potato plants to polyethylene glycol-induced water deficit. II. Contents and subcellular distribution of organic solutes. J. Exp. Bot 49, 1361–1370 [Google Scholar]
- 146.Miyashita Y. and Good AG (2008) Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 49, 92–102 [DOI] [PubMed] [Google Scholar]
- 147.Kathiresan A. et al. (1997) y-Aminobutyric Acid Stimulates Ethylene Biosynthesis in Sunflower. Plant Physiol. 115, 129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shibuya K. et al. (2013) Pollination induces autophagy in petunia petals via ethylene. J. Exp. Bot 64, 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]