SUMMARY
The aim of this study is to determine the significance of programmed death-ligand 1 (PD-L1 or CD274) methylation in relation to PD-L1 expression and survival in melanoma. Despite the clinical importance of therapies targeting the PD-1/PD-L1 immune checkpoint in melanoma, factors regulating PD-L1 expression, including epigenetic mechanisms, are not completely understood. In this study, we examined PD-L1 promoter methylation in relation to PD-L1 expression and overall survival in melanoma patients. Our results suggest that DNA methylation regulates PD-L1 expression in melanoma, and we identify the key methylated CpG loci in the PD-L1 promoter, establish PD-L1 methylation as an independent survival prognostic factor, provide proof-of-concept for altering PD-L1 expression by hypomethylating agents, and uncover that PD-L1 methylation is associated with an interferon-signaling transcriptional phenotype. Based on our findings, measuring and altering PD-L1 promoter DNA methylation may have potential prognostic and therapeutic applications in melanoma.
MAIN TEXT
Despite recent breakthroughs in immune checkpoint therapy (Hodi et al., 2010; Larkin et al., 2015; Robert et al., 2015), treatment of metastatic melanoma remains a clinical challenge (Gershenwald et al., 2017). Immune checkpoint blockade therapies have demonstrated promising efficacy, inducing high rates of anti-melanoma immune response (Wolchok et al., 2017), and even robust complete responses in some cases (Schadendorf et al., 2015; Wolchok et al., 2013). However, most patients do not experience a durable response through primary or acquired resistance mechanisms which are incompletely understood (Larkin et al., 2015; Sharma, Hu-Lieskovan, Wargo, & Ribas, 2017). Understanding the mechanisms affecting treatment response, identifying patients most likely to benefit from anti-PD1/PD-L1 therapy, and developing treatments that could potentially expand the efficacy of immune checkpoint therapies to current non-responders remain a significant challenge.
Expression of PD-L1 in treatment naïve melanoma tumor biopsies has been associated with treatment response to anti-PD-1, longer progression-free survival, and longer overall survival (Daud et al., 2016; Tumeh et al., 2014). Despite the common clinical use of PD-L1 expression as a marker of potential response to anti-PD1 therapy (Liu, Wang, & Bindeman, 2017), factors regulating PD-L1 expression are incompletely understood. DNA methylation is an epigenetic modification that plays important roles in regulating gene expression, tumor suppressor silencing, genomic stability, and is commonly dysregulated in melanoma (Micevic, Theodosakis, & Bosenberg, 2017). DNA methylation has been reported to regulate expression of PD-L1 in other malignancies, including acute myeloid leukemia (Goltz, Gevensleben, Grunen, et al., 2017), head and neck squamous cell carcinoma (Goltz, Gevensleben, Dietrich, Schroeck, et al., 2017), myelodysplasias (Yang et al. Myelodysplasias 2004), and colorectal cancer (Goltz, Gevensleben, Dietrich, & Dietrich, 2017), but has not been investigated in melanoma. The aim of this study is to determine whether DNA methylation of the programmed death-ligand 1 (PD-L1 or CD274) promoter occurs in melanoma and whether it is associated with PD-L1 expression and overall survival of melanoma patients.
We analyzed the variation in DNA methylation at all CpG probes (cg15837913, cg02823866, cg14305799, cg1347877, and cg19724470) associated with the PD-L1 promoter across the melanoma patient cohort (Fig. 1A, n=473) from The Cancer Genome Atlas (TCGA). Two CpG loci, cg15837913 and cg19724470, exhibited a large range of methylation across the cohort (Fig. 1A), suggesting a potentially functional role for DNA methylation at these loci. We next analyzed the association of DNA methylation at each of the five CpG loci with PD-L1 mRNA expression (Fig. 1B-F). We found that methylation at cg15837913 (Fig. 1B, p<0.0001) and cg19724470 (Fig. 1F, p<0.0001) significantly negatively correlated with PD-L1 expression. The strongest relationship was seen between methylation of CpG locus cg19724470 and PD-L1 gene expression (Pearson’s r=−0.54, p=2.2E-16). The remaining CpG loci had a modest range of methylation in the patient cohort (Fig. 1C, 1D and 1E).
Figure 1. PD-L1 methylation is associated with PD-L1 expression.
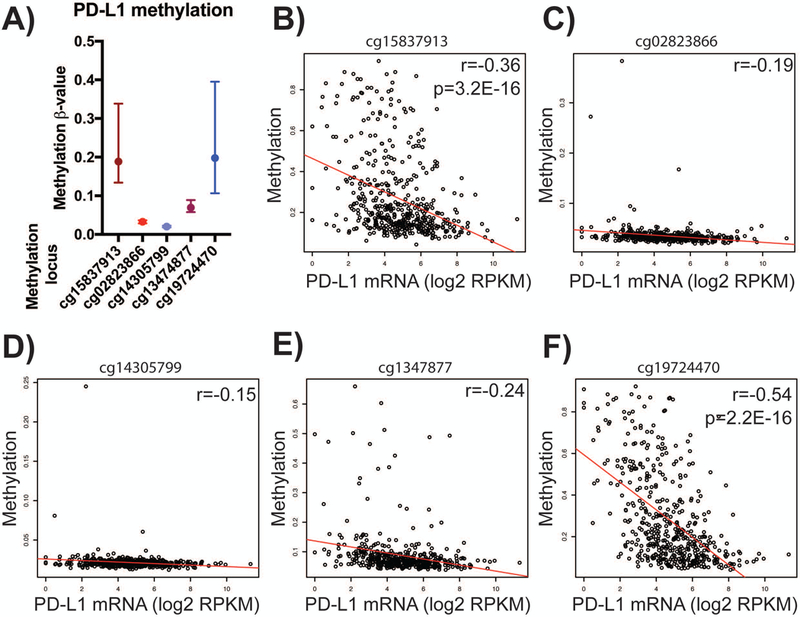
A) Distribution of methylation ß-values (y-axis) across five PD-L1-associated CpG loci (x-axis) in a cohort of 473 melanoma clinical samples from TCGA. B-F) Correlation of individual CpG locus methylation and PD-L1 mRNA expression. Gene expression (log 2 RPKM) shown on x-axis, methylation ß-value on y-axis. B) cg15837913, r = −0.36, p = 3.2E-16. C) cg02823866, r = −0.19. D) cg14305799, r = −0.15. E) cg13474877, r = −0.24. F) cg19724470, r = −0.54, p = 2.2E-16. Pearson’s r and Wilcoxon rank-sum tests used for statistical analysis, respectively. Median with interquartile range shown.
To investigate whether PD-L1 methylation differs between melanoma and normal skin, we analyzed PD-L1 methylation levels in melanoma (n=376) and normal skin samples (n=14). We found that PD-L1 was significantly hypermethylated in melanoma samples compared to normal tissue (Fig. 2A, p<0.005). Based on DNA methylation of promoter CpG elements cg15837913 and cg19724470, we hypothesized that treating melanoma cells with the hypomethylating agent 5-azacytidine would lead to transcriptional de-repression and increased PD-L1 expression. We measured PD-L1 mRNA levels by qPCR upon treating human metastatic melanoma cell lines (YUGEN8, YUSIT1 and YUSAC2, courtesy of Yale SPORE) with 5-azacytidine (in DMSO) for 72 hours and found a significant increase in PD-L1 mRNA expression relative to control treatment (DMSO) in each of the cell lines tested (Figure 2B, p<0.001).
Figure 2. PD-L1 methylation is an independent predictor of melanoma survival.

A) PD-L1 is hypermethylated in melanoma (red) compared to control normal skin (green) (**p<0.005, Student’s t-test). B) Treatment of human metastatic melanoma cell lines YUGEN8 (shown in red), YUSIT1(blue) and YUSAC2 (green) in vitro with hypomethylating agent 5-azacitidine leads to increased PD-L1 expression (**p < 0.005, Student’s t-test, three technical replicates per experiment). Metastatic melanoma cell lines were obtained from the Yale SPORE, further described in Materials and Methods. C) Survival analysis of melanoma patients (TCGA) stratified by methylation of PD-L1. Cohort was split into high PD-L1 promoter methylation (top quartile, red) versus low PD-L1 promoter methylation (bottom quartile, blue). D) Survival analysis of Stage III melanoma patients only (*p < 0.05, ***p<0.001, Log rank test). E) Summary of Cox-proportional hazard multivariable analysis. PD-L1 methylation is associated with increased risk of death (HR =22.1, 95% CI = 3.51–139.9) in melanoma patients from TCGA cohort. Age, Breslow depth and stage IV disease were also significant predictors of survival (n= 334, number of events= 159). Relapse-free survival (not shown) HR=14.92 (95% CI 1.2–185, n=258, number of events= 72).
We next investigated whether PD-L1 methylation is associated with survival in melanoma. We stratified the overall survival of melanoma patients from The Cancer Genome Atlas (TCGA) based on PD-L1 methylation into a mPD-L1 low (hypomethylated) and mPD-L1 high (hypermethylated) cohort. Clinical characteristics of the cohort are shown in Table S1. Kaplan-Meier analysis demonstrated that hypomethylation of PD-L1 was associated with significantly longer overall survival in melanoma patients of all stages (Figure 2C, p<0.0005), as well as in a subset analysis of Stage III patients only (Figure 2D, p<0.05).
We further investigated whether PD-L1 methylation is an independent prognostic factor of melanoma overall survival. In the univariate Cox-proportional hazard analysis, PD-L1 methylation was significantly associated with overall survival (data not shown). In a multivariate analysis, using age, gender, ulceration status and Breslow thickness as co-variates, PD-L1 methylation remained significantly associated with melanoma overall survival (Figure 2E, p=0.0009).
To define the transcriptome-wide phenotype of PD-L1 hypomethylated tumors (associated with longer survival), we performed a genome-wide differential expression analysis of 20,530 genes between the PD-L1 hypomethylated (n=108) and PD-L1 hypermethylated (n=108) tumor cohorts (Figure 3A). The rank list of all differentially expressed genes in the comparison are in Supplementary Table S2. Among the top differentially expressed genes, many have known immunologic functions, such as PDCD1 (PD-1), IFNG, CD8A, CD8B, TIGIT, TAP1, GZMB, CD3D, IRF1. CD274 (PD-L1) was ranked as the most differentially expressed gene in the PD-L1 hypomethylated cohort, consistent with the strong association between PD-L1 methylation and expression we observed earlier (Figure 1B and 1F). To investigate potentially overrepresented pathways, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005). GSEA of the PD-L1 hypomethylated cohort (Figure 3B top) revealed that the most enriched pathways were allograft rejection (Figure 3C), interferon gamma response (Figure 3D), interferon alpha response (Figure 3E), inflammatory response, JAK/STAT signaling, TNF-α signaling, and apoptosis (Table S3). In the PD-L1 hypermethylated cohort, the only significantly enriched gene set was Myc targets (Figure 3B bottom). These findings suggest that hypomethylation of PD-L1 is associated with a transcriptomic phenotype of immune activation and expression of interferon signaling pathway genes.
Figure 3. PD-L1 methylation is a marker of a transcriptomic phenotype enriched for interferon signaling.

A) Differential transcriptomic analysis between the PD-L1 hypermethylated cohort (n=108, top quartile by methylation) versus the PD-L1 hypomethylated cohort (n=108, bottom quartile by methylation) comparing the expression of 20,530 genes from RNA-seq data. Complete results are available in Supplementary Table S3. B) Top: Gene set enrichment analysis (GSEA) of the PD-L1 hypomethylated transcriptional phenotype uncovered many immunologic pathways, such as interferon signaling. Significantly enriched gene sets are shown in bold. Complete results are available in Supplementary Table S3. Bottom: GSEA of the PD-L1 hypermethylated transcriptional phenotype was significant only for targets of Myc. Complete results are available in Supplementary Table S3. C) Allograft rejection was the most significantly over-represented gene set in the PD-L1 hypomethylated cohort, with an absolute enrichment score of 2.59, and multiple comparison adjusted p-value of 0. D) Interferon gamma and E) Interferon alpha response genes were also enriched in the PD-L1 hypomethylated cohort, with enrichment score of 2.53 and 2.42, respectively and adjusted p-values of 0.
Altogether, our results suggest that hypomethylation of PD-L1 is an independent prognostic factor for better overall survival in melanoma and implicate PD-L1 methylation as a potential marker of an immune transcriptional phenotype, setting the stage for future investigations in larger prospective trials.
DISCUSSION
In this study, we examined the significance of PD-L1 methylation in relation to PD-L1 expression, and overall survival in melanoma. Hypermethylation of PD-L1 is associated with poorer survival in several tumor types (Gevensleben, Dietrich, et al., 2016). In colorectal cancer, hypermethylation of PD-L1 is associated with shorter overall survival, recurrence-free survival, and is an independent prognostic factor of overall survival (Goltz, Gevensleben, Dietrich, & Dietrich, 2017). Similarly, in prostate cancer, PD-L1 methylation is a prognostic factor of biochemical recurrence (Gevensleben, Holmes, et al., 2016). PD-L1 promoter methylation was reported to be associated with transcriptional repression in the setting of HPV infection in head and neck squamous cell carcinoma (Franzen et al., 2018). In non-small cell lung cancer, expression of PD-L1 is regulated by DNA methylation in response to TGF-ß or NF-kB signaling (Asgarova et al., 2018). However, despite the prominent role of anti PD-1 therapies in melanoma, no studies have yet investigated PD-L1 methylation in melanoma. We found that hypermethylation of PD-L1 in melanoma is associated with decreased PD-L1 expression and shorter patient overall survival, in consensus with studies from other tumor types. We identified the key CpG loci regulating PD-L1 expression, and demonstrated that PD-L1 expression can be altered by using DNA hypomethylating agents in human melanoma. Furthermore, we uncovered that PD-L1 methylation is a significant independent prognostic factor of overall survival. Interestingly, transcriptome analysis of PD-L1 hypomethylated tumor samples uncovered that PD-L1 methylation status is associated with a transcriptional phenotype enriched for interferon signaling, and other immunologic pathways, in contrast to PD-L1 hypermethylated tumors. Why PD-L1 is hypermethylated in some melanomas and portends a poor survival is currently unknown. Others have reported the existence of methylation clusters and a “methylator phenotype” in melanoma that are associated with poorer overall survival (Cancer Genome Atlas, 2015; Lauss et al., 2015). Importantly, it remains to be investigated whether PD-L1 methylation is an appropriate marker of an aberrant methylation program that also silences interferon signaling. If such a methylation cluster exists, there may be therapeutic benefit in combining immune checkpoint therapies with hypomethylating agents. Pre-clinical evidence suggests that hypomethylating agents in combination with immune-checkpoint therapy can induce a double-stranded RNA response and improve efficacy (Chiappinelli et al., 2017). Several clinical trials are currently investigating combination therapy in melanoma (reviewed in (Micevic et al., 2017). This report sets the stage for future investigations measuring and targeting PD-L1 methylation, which may have prognostic and therapeutic applications in melanoma.
MATERIALS AND METHODS
Cell culture
Melanoma cell lines YUSIT1, YUGEN8 and YUSAC2 were obtained from the Yale SPORE, are part of the Yale University melanoma cell line collection, and have been described previously (Halaban et al., 1997; Halaban et al., 2009; Theodosakis et al., 2015). They were originally isolated from melanoma specimens collected from adult donors with participants’ informed consent according to Health Insurance Portability and Accountability Act regulations with the Human Investigative Committee protocol at Yale University. Human melanoma cell lines were grown at 37°C and 5% CO2, in Opti-MEM (GIBCO, Grand Island, New York, USA) media supplemented with heat-inactivated fetal bovine serum (Sigma-Aldrich) and penicillin–streptomycin (GIBCO), as described previously (Micevic et al., 2016). For 5-aza-2-deoxycytidine (5-aza-dC) treatment, growth medium was supplemented with 10 μmol/l 5-aza-dC (Sigma-Aldrich) every 24 h over a 72 h period, or as otherwise specific in figure.
Statistical analysis
Statistical analyses were performed using paired and unpaired two-tailed Student’s t-test, Mann-Whitney U-test and Log-rank (Mantel-Cox) test in GraphPad Prism using a significance cut-off *p<0.05. GSEA was performed using publicly available software from the Broad Institute. All experiments were conducted in triplicate (three technical replicates). The statistical significance of all variables examined was reached when the two-sided Student’s t-test reached P-value less than 0.05. For multiple comparison adjustment, Benjamini-Hochberg and FDR q-values were used for correction.
PD-L1 RT-PCR
1 μg of total RNA was converted to cDNA using the FirstStrand kit (Roche) with oligo-dT primers per manufacturer’s instructions. RT-PCR was carried using a StepOne Plus thermal cycler (Applied Biosystems) for SYBR green-based quantification. DNA melting profiles were observed to ensure single products for each target gene. Standard curves were also created for each target by serial dilution of a reference sample. Quantification was determined using the ΔΔCt method for comparison to a GAPDH control. RT-PCR primers used: 5′-ATATAAAATAAATAATCATTCTTATACG-3′ and 5′ CGTTTAGGGATTTTGGATTTGTTTAGC-3′. The assays were performed using the TaqMan® Universal Master Mix II on a StepOne (AppliedBiosystems) thermal cycler according to manufacturer’s instructions.
TCGA data analysis
Methylation beta values were obtained from the TCGA SKCM DNA Methylation (Methylation450k) dataset using the UCSC Cancer Genomics Browser. Melanoma patients for which DNA methylation and survival information were available were divided by PD-L1 methylation level into the top quartile - high methylation and bottom quartile - low methylation group. Full cohort is information available in Table S1. Groups were compared using the Mantel-Cox test. PD-L1 expression values and patient clinical information were obtained from the TCGA SKCM Gene Expression (IlluminaHiSeq) dataset. Kaplan-Meier plots were constructed using GraphPad Prism version 6.00
Supplementary Material
SIGNIFICANCE.
This is the first report of PD-L1 methylation as a regulator of PD-L1 expression and as an independent predictor of overall survival in melanoma. PD-L1 methylation has been investigated in other tumor types, where hypermethylation is associated with a poor prognosis. However, despite the important clinical role of PD-1/PD-L1 checkpoint inhibition in melanoma, the role of PD-L1 methylation is currently unclear. We identify the key CpG loci affecting PD-L1 expression in melanoma, and uncover that PD-L1 methylation is part of a wider transcriptional phenotype of immune activation. Our findings implicate PD-L1 methylation as a potential biomarker in melanoma, and set the stage for future investigations measuring and targeting PD-L1 methylation.
ACKNOWLEDGEMENTS
The authors acknowledge the Yale MSTP NIH T32 GM007205 grant. G.M. is also supported by NRSA F30 CA19608901. The results herein are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/ ).
Footnotes
CONFLICT OF INTEREST
MB serves as a consultant for Eli Lilly and Company. Other authors state no conflict of interest.
REFERENCES
- Asgarova A, Asgarov K, Godet Y, Peixoto P, Nadaradjane A, Boyer-Guittaut M, . . . Hervouet E. (2018). PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology, 7(5), e1423170. doi: 10.1080/2162402X.2017.1423170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N (2015). Genomic Classification of Cutaneous Melanoma. Cell, 161(7), 1681–1696. doi: 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, . . . Strick R. (2017). Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell, 169(2), 361. doi: 10.1016/j.cell.2017.03.036 [DOI] [PubMed] [Google Scholar]
- Daud AI, Wolchok JD, Robert C, Hwu WJ, Weber JS, Ribas A, . . . Hamid O. (2016). Programmed Death-Ligand 1 Expression and Response to the Anti-Programmed Death 1 Antibody Pembrolizumab in Melanoma. J Clin Oncol, 34(34), 4102–4109. doi: 10.1200/JCO.2016.67.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen A, Vogt TJ, Muller T, Dietrich J, Schrock A, Golletz C, . . . Dietrich D. (2018). PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget, 9(1), 641–650. doi: 10.18632/oncotarget.23080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, . . . Discovery P. (2017). Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin, 67(6), 472–492. doi: 10.3322/caac.21409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevensleben H, Dietrich D, Golletz C, Steiner S, Jung M, Thiesler T, . . . Kristiansen G. (2016). The Immune Checkpoint Regulator PD-L1 Is Highly Expressed in Aggressive Primary Prostate Cancer. Clin Cancer Res, 22(8), 1969–1977. doi: 10.1158/1078-0432.CCR-15-2042 [DOI] [PubMed] [Google Scholar]
- Gevensleben H, Holmes EE, Goltz D, Dietrich J, Sailer V, Ellinger J, . . . Kristiansen G. (2016). PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget, 7(48), 79943–79955. doi: 10.18632/oncotarget.13161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D, Gevensleben H, Dietrich J, & Dietrich D (2017). PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology, 6(1), e1257454. doi: 10.1080/2162402X.2016.1257454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D, Gevensleben H, Dietrich J, Schroeck F, de Vos L, Droege F, . . . Dietrich D. (2017). PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget, 8(25), 41011–41020. doi: 10.18632/oncotarget.17354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltz D, Gevensleben H, Grunen S, Dietrich J, Kristiansen G, Landsberg J, & Dietrich D (2017). PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia, 31(3), 738–743. doi: 10.1038/leu.2016.328 [DOI] [PubMed] [Google Scholar]
- Halaban R, Cheng E, Zhang Y, Moellmann G, Hanlon D, Michalak M, . . . Hebert DN. (1997). Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc Natl Acad Sci U S A, 94(12), 6210–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R, Krauthammer M, Pelizzola M, Cheng E, Kovacs D, Sznol M, . . . Enghild JJ (2009). Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS One, 4(2), e4563. doi: 10.1371/journal.pone.0004563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, . . . Urba WJ (2010). Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med, 363(8), 711–723. doi: 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, . . . Wolchok JD (2015). Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med, 373(1), 23–34. doi: 10.1056/NEJMoa1504030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauss M, Ringner M, Karlsson A, Harbst K, Busch C, Geisler J, . . . Jonsson G. (2015). DNA methylation subgroups in melanoma are associated with proliferative and immunological processes. BMC Med Genomics, 8, 73. doi: 10.1186/s12920-015-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wang S, & Bindeman W (2017). Clinical applications of PD-L1 bioassays for cancer immunotherapy. J Hematol Oncol, 10(1), 110. doi: 10.1186/s13045-017-0479-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevic G, Theodosakis N, & Bosenberg M (2017). Aberrant DNA methylation in melanoma: biomarker and therapeutic opportunities. Clin Epigenetics, 9, 34. doi: 10.1186/s13148-017-0332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, . . . Ascierto PA (2015). Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med, 372(4), 320–330. doi: 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, . . . Wolchok JD (2015). Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J Clin Oncol, 33(17), 1889–1894. doi: 10.1200/JCO.2014.56.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, & Ribas A (2017). Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell, 168(4), 707–723. doi: 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, . . . Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 102(43), 15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosakis N, Held MA, Marzuka-Alcala A, Meeth KM, Micevic G, Long GV, . . . Bosenberg MW (2015). BRAF Inhibition Decreases Cellular Glucose Uptake in Melanoma in Association with Reduction in Cell Volume. Mol Cancer Ther, 14(7), 1680–1692. doi: 10.1158/1535-7163.MCT-15-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, . . . Ribas A. (2014). PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature, 515(7528), 568–571. doi: 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, . . . Larkin J. (2017). Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med, 377(14), 1345–1356. doi: 10.1056/NEJMoa1709684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, . . . Sznol M. (2013). Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med, 369(2), 122–133. doi: 10.1056/NEJMoa1302369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


