Abstract
Salmonella enterica serovar Wangata is an important cause of salmonellosis in the state of New South Wales, Australia. Standard surveillance has not identified a common food source and cases have been attributed to an unknown environmental or wildlife reservoir. Investigation of the spatial distribution of cases may provide valuable insights into local risk factors for infection and the potential role of the environment and wildlife.
Using conditional autoregressive analysis, we explored the association between laboratory-confirmed cases of S. Wangata reported to the New South Wales Department of Health and human socio-demographic, climate, land cover and wildlife features. For comparison, a model was also fitted to investigate the association of cases of Salmonella enterica serovar Typhimurium, an established foodborne serotype, with the same features. To determine if cases of S. Wangata were associated with potential wildlife reservoir species, additional variables were included in the S. Wangata model that indicated areas of high suitability for each species.
We found that cases of S. Wangata were associated with warmer temperatures, proximity to wetlands and amphibian species richness. In contrast, cases of S. Typhimurium were associated with human demographic features (proportion of the population comprising children <5 years old), climate (mean annual precipitation and mean annual temperature) and land cover (proportion comprising urban and evergreen broadleaf forest). These findings support the hypothesis that S. Wangata is likely to be associated with an environmental source. Whilst we expected S. Typhimurium to be associated with the human socio-demographic feature, the significance of the land cover features was surprising and might suggest the epidemiology of S. Typhimurium in Australia is more complex than currently understood.
Keywords: Salmonella, Spatial analysis, Landscape epidemiology, Zoonosis, Environmental Salmonella
1. Introduction
Salmonella enterica is a major zoonotic pathogen responsible for a substantial global burden of gastroenteritis [1]. In Australia, salmonellosis is the second most commonly reported cause of gastroenteritis [2] and is responsible for the majority of foodborne gastroenteritis related deaths [3]. The notification rate of salmonellosis in Australia in 2014 was 69.3 per 100,000 people; compared to data from the same year this is more than four and five times the incidence reported in the United States (15.45 per 100,000) [4] and the United Kingdom (12.63 per 100,000) [5], respectively. Salmonella enterica serovar Typhimurium is the most commonly reported serotype in Australia, accounting for 48% of cases [2]. It is a well-recognised foodborne pathogen, accounting for 92% of foodborne outbreaks of salmonellosis in Australia in 2011 [2]. Nonetheless, other serotypes contribute to the total number of salmonellosis cases reported to health authorities with varying geographic distribution [2,6]. Understanding the features that contribute to this geographic variation helps determine infection risk [7,8].
Salmonella enterica serovar Wangata is a commonly reported serotype in New South Wales (NSW) [9] but is otherwise only rarely reported in other states and countries [10]. We recently conducted an epidemiological investigation into human cases of S. Wangata to explore the hypothesis that cases were primarily associated with a wildlife or environmemntal reservoir [11]. In that study, several wildlife species, namely swans, pelicans, sea turtles, brush turkeys, bandicoots, flying-foxes and amphibians, were highlighted as potential reservoirs of the serotype based on microbiological or epidemiological evidence [11].
Understanding the unique epidemiology of serotypes associated with an environmental source is challenging. Studies investigating environmental Salmonella serotypes in Australia have used environmental sampling, case-control interviews or a combination of both [[11], [12], [13], [14], [15], [16], [17]]. Field studies are useful in identifying potential hosts [13,17] however this involves collecting a large number of samples from a wide range of hypothesised reservoirs which is both costly and time consuming. Whilst case-control interviews have identified key risk factors they can also fail to completely identify environmental transmission mechanisms [15].
An additional approach is to investigate the spatial distribution of cases in relation to landscape features, which allows a more complete evaluation of the environmental space that contextualises disease occurrence. It has the additional advantage of leveraging existing surveillance data with the ecological niche of wildlife species to incorporate speculative hosts that may be challenging to sample or difficult to specify in a survey during outbreak investigations (e.g. differentiating between specific bird species or identification of a nocturnal cryptic species).
Using conditional autoregressive (CAR) models, this study describes the spatial distribution of cases of S. Wangata with respect to human socio-demographics, climate, land cover, and wildlife features. For comparison, a similar approach was used to model the spatial distribution of cases of S. Typhimurium, an established foodborne serotype. It was hypothesised that cases of S. Wangata would have an association with wildlife and environmental features whereas cases of S. Typhimurium would primarily be associated with human socio-demographics.
2. Methods
2.1. Data sources
Laboratory-confirmed human cases of S. Wangata and S. Typhimurium occurring in NSW between 1 January 2001 and 31 December 2015 were extracted from the Notifiable Conditions Information Management System (NCIMS) maintained by the NSW Department of Health. The NCIMS captures notifiable diseases reported by pathology laboratories, general practitioners and hospitals in NSW. Cases were excluded if they were suspected to be overseas-acquired based on travel history (S. Wangata n = 50 [8.9%], S. Typhimurium n = 216 [1.1%]). Cases of each serotype were aggregated by postal area (POA) [18]. No cases were missing POA information.
Human footprint (HFP) data were downloaded from the Socioeconomic Data and Applications Centre (SEDAC), which is part of the National Aeronautics and Space Agency's (NASA) Earth Observing System Data and Information System. HFP represents the Human Influence Index (HII) that has been normalized by biome and realm and is made up of 9 global layers characterising human population pressure, land use and infrastructure, and access (e.g. via road or train) [19]. This serves as an indicator of anthropogenic influence and thus the degree of human accessibility. The data were obtained at a resolution of 30 arc sec, which is approximately 1 km2.
Net Human Migration data from 1990 to 2000 were also downloaded from SEDAC and represented as a 30 arc sec raster. Net human migration data represents movement of people into an area minus the migration out of an area [20]. This includes movements both internationally and domestically and can represent, for example, increasing urbanisation in a region.
The population count by age for each POA was extracted from the 2011 Australian Census of Population and Housing [21]. These data were used to estimate population density by dividing the total population by NSW POA area as determined by the 2011 POA shapefile developed by the Australian Bureau of Statistics (ABS) Australian Statistical Geography Standard (ASGS) [18]. The rate of salmonellosis notification is highest in children <5 years old [2], therefore to determine if a high proportion of children was associated with increased cases, an additional variable was created by dividing the count of children <5 years old by the total population of that POA.
The index of relative socio-economic advantage and disadvantage was downloaded from the 2011 Socio-Economic Indexes for Areas (SEIFA) develped by the ABS [22]. This index ranks each POA according to its socioeconomic condition relative to the rest of the country. The index is based on census data and is composed of relative socioeconomic advantage and disadvantage which are defined by the ABS in terms of “people's access to material and social resources, and their ability to participate in society.” [23]
Climate data were accessed from the WorldClim Global Climate database [24]. Mean annual temperature, mean temperature of the warmest and coolest quarters, mean annual precipitation, and mean precipitation of the wettest and driest quarters during the period 1950 to 2000 were extracted in 30 arc sec resolution rasters [25].
Land cover was represented by MODIS Land Cover Type (MCD12Q1) and accessed from the United States Geological Survey Land Cover Institute (LCI) [26]. This raster describes the predominant land cover between 2001 and 2010 for each pixel at a resolution of 15 arc sec represented as a value between 0 and 16 [27]. Of the 17 land cover features, 10 were extant throughout the study region. These were: surface water, evergreen broadleaf forest, open shrublands, woody savanna, grassland, permanent wetland, croplands, urban and built up, cropland/natural vegetation mosaic, and barren/sparsely vegetated. QGIS [28] was used to extract raster layers for each of the 10 listed land cover types to make 10 binary rasters. The proximity function was then used to create a distance raster for each type by calculating the distance of each pixel in the geographic extent under study to the given land cover feature. In addition, the mean number of pixels in each POA attributable to evergreen broadleaf forest, open shrublands, grassland, croplands, urban and built up, and cropland/natural vegetation mosaic was calculated to give the proportion of that land cover type per POA.
The MODIS-based maximum green vegetation fraction (MGVF), also accessed from LCI [29], was used to represent the overall vegetation cover at a resolution of 30 arc sec [30]. MVGF is a function of the normalized difference vegetation index which accounts and adjusts for periods of unusual greenness [30]. The 2008 raster was selected as this represents the midpoint for this investigation.
A shapefile of the river systems in Australia was obtained from HydroSHEDS [31]. This file was converted to a binary 30 arc sec raster wherein each pixel represented the presence of river (1) or the absence of river (0). The sum was taken for each POA.
To investigate the association with wildlife, species richness data for mammals [32] and amphibians [33] were downloaded as 30 arc sec grids from the International Union for Conservation of Nature (IUCN) via SEDAC. Data on avian species richness were not available. Species richness represents an aggregation of the number of species from each class present within each grid. These rasters were based on data obtained from the IUCN in 2013 [32,33]. Further, to investigate if human cases were associated with the distribution of putative hosts identified in our previous epidemiological study [11], occurrences of black swans (Cygnus atratus), Australian pelicans (Pelecanus conspicillatus), Australian Brush Turkeys (Alectura lathami), long-nosed bandicoots (Perameles nasuta), black flying foxes (Pteropus alecto), grey-headed flying foxes (Pteropus poliocephalus), and little red flying-foxes (Pteropus scapulatus) observed between 2000 and 2017 were obtained from the Global Biodiversity Information Facility (GBIF) [34]. Only three of the four native species of flying fox in mainland Australia were included in this study because spectacled flying foxes (Pteropus conspicillatus) do not occur in NSW. The ecological niche of each species was then quantified using the Maxent machine learning algorithm described below in the statistical methods.
2.2. Statistical analysis
2.2.1. Ecological niche models (Maxent)
In order to investigate the association between the hypothesised wildlife reservoir species and cases of S. Wangata the distribution of the wildlife species is required. Determining the distribution of a species typically involves presence and absence data [35]. However, where only presence data are available a different approach is required. Therefore, because the distribution of the hypothesised wildlife reservoirs was unknown and no absence data were available, Maximum entropy (Maxent) was used to model the ecological niche of the hypothesised wildlife reservoirs. Maxent is a machine learning algorithm that can estimate the habitat suitability for a species using presence-only data [36].
The fundamental niche of each host species was modelled separately, with the spatial extent equivalent to the presence data of the species to a maximum of the continent of Australia. To model the fundamental niche of a species, the full range of environments that make up the suitable habitat of that species should be sampled [36] meaning maxent should be run over the largest possible spatial extent where species observations occur.
Raster variables were assessed for correlation using a Pearson's correlation test and highly correlated variables were chosen by comparing model performance across different combinations of correlated variables. The variables that resulted in the best model performance (high mean area under the receiver operating characteristic curve (AUC)) were selected for the final model. Landscape features included in each model can be found in Supplementary file 1. Ten thousand background points were sampled separately for each host species' model, weighted by the human footprint to account for the potential spatial sampling bias of presence points in GBIF. HFP was used because it encompasses anthropogenic factors that would facilitate observation accessibility, known to affect reporting effort [37,38]. To minimise overfitting the models, the regularisation multiplier was set to the default of 1.0.
Five-fold cross-validation was performed for each Maxent model to test performance, such that the presence data for each species were randomly divided iteratively into 5 groups; 4 groups were modelled together and then tested against the 5th. This process applies 5 folds wherein each random permutation is used as a test group once across each iteration. The AUC was then used to determine model performance. Similarly, the mean Maxent function was used as the estimate of species habitat suitability per km2 across the region.
The species niche probability function raster was then converted to a binary raster with the value 1 (i.e. suitable habitat) if the probability of species habitat suitability was ≥0.85 and 0 (i.e. unsuitable habitat) if the probability was <0.85. The cut-off of 0.85 was used because this selects for areas where the habitat is highly suitable. The sum of all 1 km2 pixels was then calculated for each POA to quantify the total area of suitable habitat for each species per POA.
Ecological niche modelling produced seven rasters of predicted suitable habitat for each hypothesised S. Wangata host. The rasters of predicted suitable habitat and the model performance metrics are provided in Supplementary file 1.
2.2.2. CAR model
The global Moran's index (I) identified significant spatial autocorrelation in the POA-aggregated cumulative incidence of both S. Wangata and S. Typhimurium (S. Wangata: Moran I Statistic = 0.221, P-value <.0001. S. Typhimurium: Moran I Statistic = 0.053, P- value = .0183). Initially, a non-spatial Poisson model was used to model the cumulative incidence of S. Wangata and S. Typhimurium per POA. However, the global Moran's I identified significant spatial autocorrelation in the residual deviance for both models (S. Wangata: Moran I Statistic = 0.0914, P-value = .0019, S. Typhimurium: Moran I Statistic = 0.1138, P- value = .0002) so this model was deemed inappropriate. Therefore to account for the spatial autocorrelation inherent to these disease phenomena, conditional autoregressive models (CAR) with random effects were used with the Besag-York-Millie (BYM) prior [39].
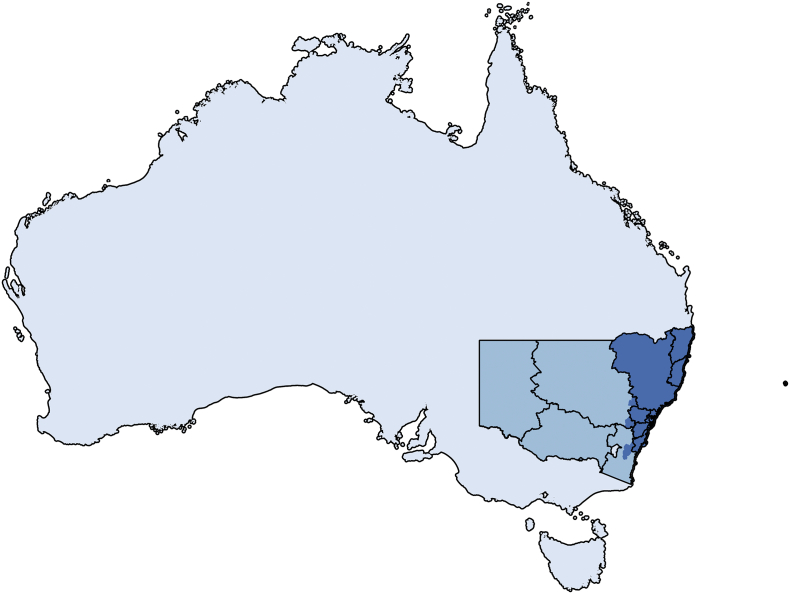
S. Wangata cases were predominately observed in a corridor within NSW, therefore the spatial extent of the CAR models for both serotypes were restricted to the POAs within 11 of the 15 NSW local health districts (LHDs) where cases occurred. Namely: Sydney, South Eastern Sydney, Western Sydney, Northern Sydney, Central coast, Illawarra Shoalhaven, South Western Sydney, Nepean Blue Mountains, Mid North Coast, Northern NSW and Hunter New England LHDs (Fig. 1). Eight POAs fell across the borders of two LHDs and five of these were subsequently excluded from the study extent because the majority of their area was not included in the above LHDs. The 451 POAs from within these local health districts contained 99% of cases of S. Wangata (514/520) reported during the study period. Of the 451 POAs included in the CAR analyses the minimum area was 0.39km2 and the maximum was 8573.49km2, with a median of 15.92km2.
Fig. 1.
The country of Australia (light blue), the state of New South Wales (NSW) (medium blue) and the spatial extent of the CAR analysis (dark blue). The boarders of the local health districts (LHD) of NSW are shown. The CAR extent is comprised of 11 LHDs, namely: Sydney, South Eastern Sydney, Western Sydney, Northern Sydney, Central coast, Illawarra Shoalhaven, Southwestern Sydney, Nepean Blue Mountains, Mid North Coast, Northern NSW and Hunter New England. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For each serotype, the CAR model estimated the number of cases per POA as a function of: human socio-demographics (proportion of the population <5 years old, socioeconomic ranking, mean net human migration), climate (mean annual temperature and precipitation), land cover (mean MGVF, mean distance to wetlands, mean distance to surface water, sum of river pixels, proportion of POA classified as each land cover type (evergreen, grassland, cropland, urban, cropland/natural vegetation mosaic and open shrubland), and wildlife features (mean mammal species richness and mean amphibian species richness). The S. Wangata model also included the total area of suitable habitat per POA for the hypothesised reservoir species as derived from the niche models described above. Maps of the variables included in the CAR analyses are shown in Supplementary file 2.
The CAR models were specified as follows:
where, for each POA k, Xk represents the predictor matrix for human socio-demographics, climate, land cover, and wildlife features, and βk the vector of regression coefficients. φk denotes the random effects and ln(Population Countk * areak) is the offset, which incorporates both POA population and POA area as the offset since log(a) + log(b) = log(a*b).
Bivariate models were fitted for each serotype and predictors that were not significant (95% credible intervals included 0) were excluded from further analysis. It is important to note that these are Bayesian hierarchical models and the inference is based on Markov Chain Monte Carlo simulation. As such the credible intervals represent actual probability distributions of the estimated parameters, wherein the model's regression coefficient for any particular landscape feature is the median of its parameter distribution. This contrasts confidence intervals under conventional frequentist null hypothesis testing, which represent single realisations of random variables under a hypothesised sampling distribution (and are thus absent of parameter distributional information). Therefore, the 95% credible intervals demonstrate the posterior distribution of possible parameter estimates. Significance was taken to be when the 95% credible interval did not include zero, as is the standard used for CAR modelling. All significant predictor variables in the bivariate models were assessed for correlation using a Pearson's test of correlation. For variables with a correlation coefficient >|0.65| the variable with the lower deviance information criterion (DIC) was selected for inclusion in subsequent models.
A full model was then run that included all significant variables. Next, to test the effect of each domain, domain-specific subsets of models were created from significant variables and were compared to the full model. These domains reflect the variables described above and comprised human socio-demographics, climate, land cover, and wildlife, respectively. Model evaluation was based on the DIC. The models with the lowest DIC from each domain were tested in combination with the other domain sub-models. The final model was the one that had the lowest DIC.
For each serotype, the cumulative incidence ratio was derived by exponentiating the regression coefficients and maps were produced to show the predicted number of cases and residual deviance from each final model. The distribution of the residual deviance was then tested for spatial autocorrelation using Moran's I.
All analyses were performed using the R programming language [40]. Maxent modelling was performed using the maxent function from the dismo package [41]. CAR modelling was performed using the bymCAR.re function available in the CARbayes package [42].
2.2.3. Ethical statement
The use of human data in this study was approved by the NSW Population and Health Services Research Ethics Committee (21 December 2015, LNR 2015/08/038) and The University of Sydney Human Research Ethics Committee (2015/834, 22 October 2015).
3. Results
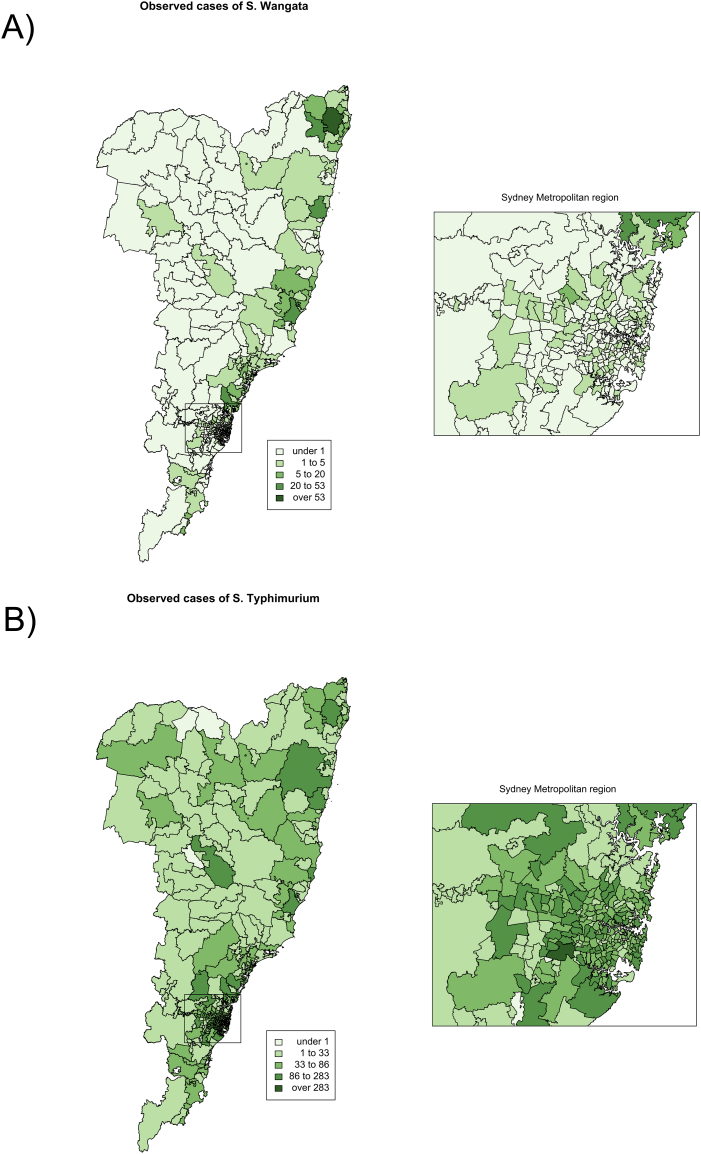
Between 1 January 2001 and 31 December 2015 there were 514 cases of S. Wangata and 18,631 cases of S. Typhimurium reported within the spatial extent under investigation. The number of cases per POA ranged from 0 to 53 cases for S. Wangata and 0 to 283 cases for S. Typhimurium (Fig. 2). Cases of S. Wangata were primarily located in the north east of the state. In contrast, cases of S. Typhimurium were clustered within the metropolitan region of Sydney, along the coast and extending farther west than S. Wangata.
Fig. 2.
Observed cases of S. Wangata (A) and S. Typhimurium (B) in the study area within NSW, Australia between 1 January 2001 and 31 December 2015.
Bivariate analysis (Table 1) revealed that mean annual precipitation, mean annual temperature, mean distance to water, mean distance to wetlands, sum of river length as well as mammal and amphibian species richness were associated with S. Wangata. Of the seven hypothesised reservoir species only brush turkey habitat suitability was significantly associated with cases of S. Wangata. S. Typhimurium was associated with proportion of the populations <5 years old, mean annual precipitation, mean annual temperature, mean MGVF, mean distance to water, mean distance to wetlands, sum of river length, proportion of POA comprising evergreen, proportion of POA comprising grassland, proportion of POA comprising urban/ built as well as amphibian species richness.
Table 1.
Bivariate analysis for S. Wangata and S. Typhimurium. The median coefficient and credible intervals are given. Significant variables are in bold and marked with.a
| Variable | S. Wangata |
DIC | S. Typhimurium |
DIC |
|---|---|---|---|---|
| Median (95% cred. interval) | Median (95% cred. interval) | |||
| Human socio-demographics | ||||
| Proportion < 5 years old (%) | 0.0746 (−0.0824–0.2394) | 826.6365 | 0.0926 (0.0364–0.1456)a | 3085.81 |
| SES score | 0.0018 (−0.0019–0.0057) | 827.0837 | 0.0004 (−0.0007–0.0019) | 3088.221 |
| Mean net migration | 0.0021 (−0.008–0.0051) | 821.0101 | 0.009 (0.0000–0.0022) | 3088.672 |
| Climate | ||||
| Mean annual precipitation (mm) | 0.0028 (0.0014–0.0045)a | 822.2477 | 0.0015 (0.0010–0.0021)a | 3086.083 |
| Mean annual temperature (°C) | 0.3968 (0.1575–0.6788)a | 823.5516 | 0.2871 (0.2001–0.3854)a | 3084.891 |
| Landscape | ||||
| MGVF (%) | −0.0101 (−0.0324–0.0136) | 825.865 | −0.0232 (−0.0299 to −0.0168)a | 3087.073 |
| Distance to wetland (km) | −13.9471 (−20.1996 to −8.2512)a | 816.8665 | −4.8665 (−6.6568 to −3.2084)a | 3090.948 |
| Distance to water (km) | −0.0134 (−0.0200 to −0.0069)a | 819.8108 | −0.0061 (−0.0083 to −0.0040)a | 3089.188 |
| Proportion POA evergreen | −0.5667 (−1.4958–0.3257) | 825.1231 | −1.2930 (−1.6502 to −0.9493)a | 3083.062 |
| Proportion POA grassland | −0.7144 (−2.9826–1.3471) | 829.5128 | −1.3710 (−2.1531 to −0.6182)a | 3088.97 |
| Proportion POA cropland | −0.3070 (−2.7668–2.0996) | 824.3792 | −0.4996 (−1.2711–0.2802) | 3089.431 |
| Proportion POA Urban/built up | 0.8569 (−0.1294–1.8197) | 826.2268 | 1.5309 (1.1493–1.9011)a | 3084.607 |
| Proportion POA cropland/vegetation mosaic | 3.5894 (−1.8161–9.0833) | 827.4662 | 0.7331 (−1.4143–1.9011) | 3089.582 |
| Proportion POA open shrubland | −7.3475 (−20.1413–0.0247) | 827.9412 | −0.6000 (−2.0243–1.0978) | 3089.475 |
| River length total (km) | −0.0028 (−0.0040 to −0.0016)a | 823.6724 | −0.0080 (−0.0011 to −0.0005)a | 3085.947 |
| Wildlife | ||||
| Amphibian species richness | 0.1217 (0.0517–0.2020)a | 817.8239 | 0.0280 (0.0056–0.0476)a | 3087.635 |
| Mammal species richness | 0.1053 (0.0554–0.1707)a | 823.7016 | 0.0127 (−0.0083–0.033) | 3088.437 |
| Black swan (km2) | −0.0253 (−0.1186–0.0208) | 829.345 | ||
| Australian pelican (km2) | 0.0009 (−0.0023–0.0042) | 827.9733 | ||
| Australian brush turkey (km2) | 0.0009 (0.0001–0.0017)a | 822.251 | ||
| Long-nose bandicoot (km2) | −0.0002 (−0.0005–0.0001) | 829.4502 | ||
| Black flying-fox (km2) | 0.0011 (−0.0008–0.0029) | 828.0384 | ||
| Grey-headed flying-fox (km2) | 0.0008 (−0.0002–0.0017) | 825.0789 | ||
| Little red flying fox (km2) | −0.0001 (−0.0002–0.0000) | 827.9556 |
Denotes the variable significantly contributes to bivariate model.
High correlation (>|0.65|) was noted between: 1) mammal and amphibian species richness, 2) mean annual precipitation and distance to wetland, and 3) distance to wetland and distance to water, and 4) MGVF and urban POAs. Among correlated variables, those with lower DIC in the bivariate analysis were selected for multivariable analysis. Therefore S. Wangata models excluded mammal species richness, distance to water and mean annual precipitation, while S. Typhimurium models excluded distance to wetland, MGVF and distance to water. The full model and domain specific sub-models are shown in Supplementary file 3.
The multivariable CAR model that represents the best fit for each Salmonella serotype is shown in Table 2. Cases of S. Wangata infection were associated with warmer temperature, closer proximity to wetlands, and increasing amphibian richness. By comparison, a higher proportion of the population < 5 years old, greater mean annual temperature and rainfall, a lesser proportion of POA comprising evergreen, and a greater proportion of POA comprising urban were associated with S. Typhimurium infections.
Table 2.
Final multivariable analysis for S. Wangata and S. Typhimurium. The models below represent the models with the lowest DIC where all variables significantly contributed to the model.
| Serotype | Variable | Cumulative incidence ratio (95% credible interval) |
|---|---|---|
| S. Wangata | Mean annual temperature (°C) | 1.310 (1.013–1.678) |
| Mean distance to wetland (km) | 5.83 ×106 (1.49 ×108–0.002) | |
| Mean amphibian species richness | 1.095 (1.034–1.172) | |
| S. Typhimurium | Proportion < 5 years old (%) | 1.073 (1.025–1.126) |
| Annual precipitation (mm) | 1.002 (1.002–1.003) | |
| Mean annual temperature (°C) | 1.112 (1.034–1.185) | |
| Sum of river length in POA (km) | 1.000 (0.999–1.000) | |
| Proportion of POA evergreen | 0.309 (0.221–0.447) | |
| Proportion of POA Urban/Built up | 2.711 (2.048–3.605) |
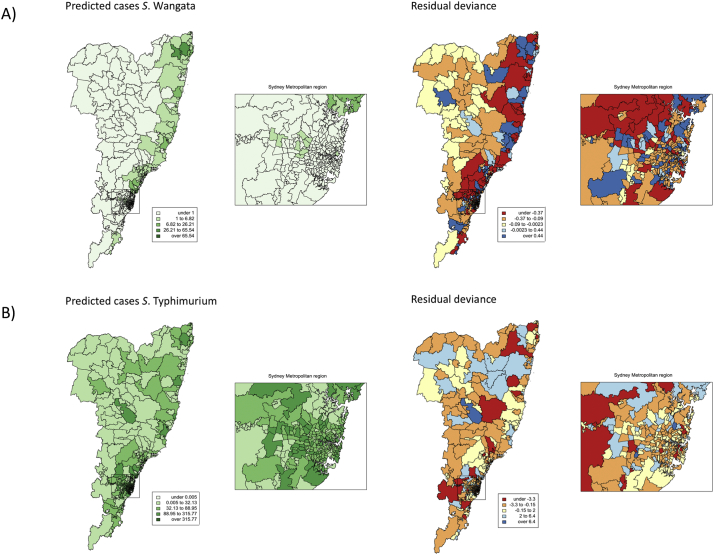
Maps of the predicted cases and residual deviance are shown in Fig. 3. Global Moran's I of the residual deviance for each serotype was non-significant (S. Wangata: Moran I Statistic = −1.0959, P-value = .8634, S. Typhimurium: Moran I Statistic = −4.7719, P- value = 1) suggesting that these two models sufficiently accounted for the spatial autocorrelation observed in S. Wangata and S. Typhimurium cumulative incidence.
Fig. 3.
Predicted cases of A) S. Wangata and B) S. Typhimurium based on the conditional autoregressive models of each serotype. The residual deviance is also shown for each serotype.
4. Discussion
We investigated the geographic distribution of cases of the putative environmental serotype S. Wangata, with regard to human socio-demographics, climate, land cover, and wildlife features in NSW, Australia, and compared this to the distribution of the established foodborne serotype, S. Typhimurium. We found cases of S. Wangata were associated with warmer areas with greater overall proximity to wetlands and greater amphibian richness whereas cases of S. Typhimurium were associated with urban areas with a younger population, that are warmer and experience more precipitation, but which are more urbanised and have less evergreen forest. Moreover, the models used to quantify these relationships sufficiently accounted for the spatial dependence associated with the cumulative incidence of each serotype.
The association between cases of S. Wangata and warmer areas that have a higher amphibian species richness and more landscape proximate to wetlands is consistent with the hypothesis that this serotype is linked to a wildlife or environmental reservoir. Cases of S. Wangata are more frequent during the summer months [9] however this is the first study to show that an overall warmer climate is associated with cases. Proximity and/or access to waterways such as rivers, creeks, lakes and beaches was considered as a potential risk factor in our previous epidemiological investigation although no association was found [11]. This might suggest that wetlands specifically, not just surface water, are sources of greater risk in the landscape. The importance of wetlands as a potential environmental reservoir is supported by the cumulative incidence ratio of distance to wetland in the multivariable model (CIR 5.83 ×106, 95% CI [1.49 ×108–0.002]). A study in the United States found that the presence of wetland environments was strongly correlated with cases of salmonellosis, postulating that wetlands may aid the persistence of Salmonella and harbour reservoir hosts [43]. This theory is consistent with our previous investigation where S. Wangata was isolated from a number of wildlife species associated with wetland environments (i.e. swans and pelicans) [11]. This is further supported by the finding of amphibian species richness in the multivariable model. Amphibians are known carriers of Salmonella [44] and indirect exposure to frogs was associated with cases of S. Wangata in our previous epidemiological study although we did not test amphibians in this investigation [11]. Regions of higher amphibian species richness may also be indicative of underlying high biodiversity, as seen in the correlation between this variable and mammal species richness. A number of studies have reported outbreaks of Salmonella associated with wild mammalian populations [14,45,46] and wild birds [47,48] and it is plausible that regions of greater biodiversity, such as wetlands, might promote increased likelihood of spill over events from a range of species into human populations.
None of the predicted host species habitat suitability variables were significant in the final multivariable model for S. Wangata. Based on our previous investigation, we believe S. Wangata is persistent in the environment and cycles through a variety of hosts including occasional spill over to humans, as has been reported for different serotypes in other studies [12,16]. Findings from this study support this hypothesis. Therefore, a clear public health message could be to target controllable interactions between humans and wildlife such as advising against the use of feeding stations which have been linked to wildlife-associated outbreaks [45,49]. Delivering these messages during the warmer months or in and around wetlands may further target at risk populations.
In contrast to S. Wangata where no anthropogenic variables were associated with cases, the final multivariable model for S. Typhimurium included positive associations with population (population <5 years old) and urban (proportion of POA urban) features, in addition to climate and land cover. A higher risk of cases in POAs with larger populations of children <5 years old is consistent with the epidemiology of a foodborne pathogen [50]. Similarly, the observed association between higher temperatures and rainfall has been reported by a number of studies [[51], [52], [53]]. The finding of a higher risk of S. Typhimurium cases in urban environments is less clear. By using the population count in the model offset the cases were modelled per person, per km2 so increased populations in the urban centres should not have influenced this result. Furthermore, a number of studies have found an increased risk of salmonellosis in rural areas [54,55] potentially due to additional exposures such as personal water supplies [56], contaminated soils [54], and contact with infected livestock [57]. In the present study, none of the land cover variables that serve as a proxy for agricultural regions (namely, proportion of POA classed as grassland, cropland, cropland/natural vegetation mosaic and open shrubland) were found to be significant in the bivariate models. The association with highly urban POAs may therefore be due to a reporting bias as there would be a greater abundance of healthcare providers in an urban setting [58]. Moreover, it was not possible to exclude outbreak associated cases from the present study. A number of S. Typhimurium point source outbreaks associated with food outlets have been recorded in the urban areas of Sydney [59,60] which may have inflated the effect of urban POAs. Nevertheless, it cannot be ruled out that there is an alternative reason for the increased risk of cases in an urban environment. The negative relationship between cases of S. Typhimurium and evergreen forest is unlikely to suggest a protective relationship but rather that these features are usually not predominant in highly populated and urban environments.
5. Limitations
A major limitation of this study is aggregation of Salmonella data to the POA level. By aggregating cases by POA there was a loss in the spatial resolution of findings. Zoonotic transmission of a pathogen occurs at the individual level and is therefore best described using a high-resolution spatial unit such as a home address. This information was not available in the current study due to ethics considerations. Inference about individual risk was therefore not possible, however these findings were able to account for spatial variation across the study region and are useful in describing key features of POAs that can form the basis of future investigations. We acknowledge that not all wildlife species that have been identified as hosts of S. Wangata were included as variables in this paper. In particular, S. Wangata has been isolated from a silver gull (Chroicocephalus novaehollandiae) [61] and green sea turtles (Chelonia mydas) [11]. We pursued some specific hypotheses about potential reservoir hosts given findings in our recent epidemiological investigation [11]. Finally, we used population data from the 2011 census which is skewed towards the later years of the case data. POA boundaries changed slightly in 2011, therefore the 2011 census data was required in order to align shapefiles, case data and population data.
6. Conclusions
Salmonella Wangata and Typhimurium manifest divergent geographies of risk in NSW, Australia. S. Wangata – a serotype with a suspected environmental reservoir – was primarily associated with non-anthropogenic variables: warmer temperatures, proximity to wetlands, and amphibian species richness. In contrast, S. Typhimurium – which is the major serotype implicated in foodborne outbreaks of salmonellosis – was predominately associated with anthropogenic variables, as well as climate and areas with less forest. These divergent geographies support the hypothesis that S. Wangata is not predominately transmitted via food but instead via an environmental vector. Control of S. Wangata and other environmental serotypes will therefore require a novel approach beyond microbial reduction in foods, such as targeted public health messaging.
Conflict of interest
The authors declare they have no conflicts of interest.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2019.100092.
Contributor Information
Kelly M.J. Simpson, Email: kelly.simpson@sydney.edu.au.
Siobhan M. Mor, Email: siobhan.mor@liverpool.ac.uk.
Michael P. Ward, Email: michael.ward@sydney.edu.au.
Michael G. Walsh, Email: michael.walsh1@sydney.edu.au.
Appendix A. Supplementary data
Output of ecological niche models and maps of variables used.
Maps of variables used in conditional autoregressive model.
Domain-specific sub-models.
References
- 1.Majowicz S.E. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50(6):882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.OzFoodNet Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: Annual report of the OzFoodNet network, 2011. Commun. Dis. Intell. 2015;39(2):E236–E264. doi: 10.33321/cdi.2015.39.22. [DOI] [PubMed] [Google Scholar]
- 3.A.G.D.o. Health . 2014. Foodborne Illness in Australia: Annual Incidence Circa 2010. Canberra, Australia. [Google Scholar]
- 4.Crim S.M. Preliminary incidence and trends of infection with pathogens transmitted commonly through food — Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2014. Morb. Mortal. Weekly Rep. 2015;64(18):495–498. [PMC free article] [PubMed] [Google Scholar]
- 5.Public Health England . PHE Publications; 2018. Salmonella data from 2007 to 2016. [Google Scholar]
- 6.OzFoodNet Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet Network, 2009. Commun. Dis. Intell. 2010;34(4):396–426. doi: 10.33321/cdi.2010.34.40. [DOI] [PubMed] [Google Scholar]
- 7.Varga C. Evaluating area-level spatial clustering of Salmonella Enteritidis infections and their socioeconomic determinants in the greater Toronto area, Ontario, Canada (2007 - 2009): a retrospective population-based ecological study. BMC Public Health. 2013;13(Nov):1078. doi: 10.1186/1471-2458-13-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varga C. Spatial-temporal epidemiology of human Salmonella Enteritidis infections with major phage types (PTs 1, 4, 5b, 8, 13, and 13a) in Ontario, Canada, 2008–2009. BMC Public Health. 2015;15(Dec):1247. doi: 10.1186/s12889-015-2592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.NNDSS . 2019. Salmonella Public Data set.http://www9.health.gov.au/cda/source/pub_salmo.cfm Available from: [Google Scholar]
- 10.Ballal M. Fatal case of diarrhea with acute kidney injury and hemiplegia due to Salmonella enterica serovar wangata: the first report from the Indian subcontinent. Jpn. J. Infect. Dis. 2015;68(6):530–531. doi: 10.7883/yoken.JJID.2014.474. [DOI] [PubMed] [Google Scholar]
- 11.Collins J. A One Health investigation of Salmonella enterica serovar Wangata in north-eastern New South Wales, Australia, 2016–2017. Epidemiol. Infect. 2019;147(e150):1–11. doi: 10.1017/S0950268819000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashbolt R., Kirk M.D. Salmonella Mississippi infections in Tasmania: the role of native Australian animals and untreated drinking water. Epidemiol. Infect. 2006;134(6):1257–1265. doi: 10.1017/S0950268806006224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ball A. University of Tasmania; 1991. The Epidemiology of Salmonella Serovars in Tasmania. Masters Thesis. [Google Scholar]
- 14.Staff, M Salmonellosis outbreak traced to playground sand, Australia, 2007–2009. Emerg. Infect. Dis. 2012;18(7):1159. doi: 10.3201/eid1807.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beard F. Risk factors for sporadic Salmonella Birkenhead infection in Queensland and northern New South Wales: a case control study. New South Wales Public Health Bull. 2004;15(10):172–177. doi: 10.1071/nb04037. [DOI] [PubMed] [Google Scholar]
- 16.Williams S. Individual and household-level risk factors for sporadic salmonellosis in children. J. Inf. Secur. 2016;72(1):36–44. doi: 10.1016/j.jinf.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Williams S. Salmonella in the tropical household environment – everyday, everywhere. J. Inf. Secur. 2015;71(6):642–648. doi: 10.1016/j.jinf.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Australian Bureau of Statistics, Australian Statistical Geography Standard (ASGS) Canberra; Australia: 2011. Volume 3-Non ABS Structures. [Google Scholar]
- 19.WCS and CIESIN . 2005. Last of the Wild Project, Version 2, Global Human Footprint Dataset (Geographic). 4-May-2018. Available from: [Google Scholar]
- 20.de Sherbinin A. 2015. Global Estimated Net Migration Grids by Decade: 1970–2000. [Google Scholar]
- 21.Australian Bureau of Statistics, Census Datapacks, Basic Community Profile, Table B04, Australian Bureau of Statistics. 2011. [Google Scholar]
- 22.Australian Bureau of Statistics, Socio-economic Indexes for Areas (SEIFA), Data Cube, Australian Bureau of Statistics. 2011. [Google Scholar]
- 23.Australian Bureau of Statistics . 2011. Technical Paper: Socio-Economic Indexes for Areas (SEIFA). Canberra, Australia. [Google Scholar]
- 24.WorldClim—Global Climate, Data for current Conditions (∼1950–2000) | WorldClim—Global Climate Data., WorldClim—Global Climate Available from: http://www.worldclim.org/current.
- 25.Hijmans R.J. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25(15):1965–1978. [Google Scholar]
- 26.USGS Land Cover Institute, 0.5 Km MODIS-Based Global Land Cover Climatology, USGS Land Cover Institute, Available at: https://landcover.usgs.gov/global_climatology.php.
- 27.Broxton P.D. A global land cover climatology using MODIS data. J. Appl. Meteorol. Climatol. 2014;53(6):1593–1605. [Google Scholar]
- 28.QGIS Development Team . 2018. QGIS Geographic Information System. [Google Scholar]
- 29.USGS Land Cover Institute b. 1 Km MODIS-Based Maximum Green Vegetation Fraction. https://landcover.usgs.gov/green_veg.php Available from:
- 30.Broxton P.D. A MODIS-based global 1-km maximum green vegetation fraction dataset. J. Appl. Meteorol. Climatol. 2014;53(8):1996–2004. [Google Scholar]
- 31.Lehner B., Verdin K., Jarvis A. New global hydrography derived from spaceborne elevation data. Trans. Am. Geophys. Union. 2008;89(10):93–94. [Google Scholar]
- 32.IUCN and CIESIN . 2015. Gridded Species Distribution: Global Mammal Richness Grids, 2015 Release. [Google Scholar]
- 33.IUCN and CIESIN . 2015. Gridded Species Distribution: Global Amphibian Richness Grids, 2015 Release. 20180508. [Google Scholar]
- 34.Global Biodiversity Information Facility . 2017. GBIF Occurrence Download, Global Biodiversity Information Facility. [Google Scholar]
- 35.Corsi F., Duprè E., Boitani L. A large-scale model of wolf distribution in Italy for conservation planning. Conserv. Biol. 1999;13(1):150–159. [Google Scholar]
- 36.Phillips S.J., Anderson R.P., Schapire R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006;190(3):231–259. [Google Scholar]
- 37.Phillips S.J. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl. 2009;19(1):181–197. doi: 10.1890/07-2153.1. [DOI] [PubMed] [Google Scholar]
- 38.Phillips S.J., Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31(2):161–175. [Google Scholar]
- 39.Besag J., York J., Mollie A. Bayesian image restoration with two applications in spatial statistics (with discussion) Ann. Inst. Stat. Math. 1991;43(1):1–59. [Google Scholar]
- 40.R Core Team . 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 41.Hijmans R.J., Elith J. 2013. Species Distribution Modeling with R. R Package Version 0; pp. 8–11. [Google Scholar]
- 42.Lee D. CARBayes: an R package for Bayesian spatial modeling with conditional autoregressive priors. J. Stat. Softw. 2013;55(13):1–24. [Google Scholar]
- 43.Huang J.Y. Association between wetland presence and incidence of Salmonella enterica serotype Javiana infections in selected US sites, 2005-2011. Epidemiol. Infect. 2017;145(14):2991–2997. doi: 10.1017/S0950268817001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribas A., Poonlaphdecha S. Wild-caught and farm-reared amphibians are important reservoirs of Salmonella, a study in north-East Thailand. Zoonoses Public Health. 2017;64(2):106–110. doi: 10.1111/zph.12286. [DOI] [PubMed] [Google Scholar]
- 45.Handeland K. Prevalence of Salmonella Typhimurium infection in Norwegian hedgehog populations associated with two human disease outbreaks. Epidemiol. Infect. 2002;128(3):523–527. doi: 10.1017/s0950268802007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lawson B. Salmonella Enteritidis ST183: emerging and endemic biotypes affecting western European hedgehogs (Erinaceus europaeus) and people in Great Britain. Sci. Rep. 2018;8(1):2449. doi: 10.1038/s41598-017-18667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tauni M., Österlund A. Outbreak of Salmonella Typhimurium in cats and humans associated with infection in wild birds. J. Small Anim. Pract. 2000;41(8):339–341. doi: 10.1111/j.1748-5827.2000.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 48.Bloomfield S.J. Genomic analysis of Salmonella enterica serovar Typhimurium DT160 associated with a 14-year outbreak, New Zealand, 1998-2012. Emerg. Infect. Dis. 2017;23(6):906–913. doi: 10.3201/eid2306.161934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warnken J. The localized environmental degradation of protected areas adjacent to bird feeding stations: a case study of the Australian brush-Turkey Alectura lathami. J. Environ. Manag. 2004;70(2):109–118. doi: 10.1016/j.jenvman.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Ford L. Increasing incidence of Salmonella in Australia, 2000-2013. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0163989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akil L., Ahmad H.A., Reddy R.S. Effects of climate change on Salmonella infections. Foodborne Pathog. Dis. 2014;11(12):974–980. doi: 10.1089/fpd.2014.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Bi P., Hiller J.E. Climate variations and Salmonella infection in Australian subtropical and tropical regions. Sci. Total Environ. 2010;408(3):524–530. doi: 10.1016/j.scitotenv.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y., Bi P., Hiller J.E. Projected burden of disease for Salmonella infection due to increased temperature in Australian temperate and subtropical regions. Environ. Int. 2012;44(Sep):26–30. doi: 10.1016/j.envint.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Lal A. The epidemiology of human salmonellosis in New Zealand, 1997–2008. Epidemiol. Infect. 2012;140(9):1685–1694. doi: 10.1017/S0950268811002470. [DOI] [PubMed] [Google Scholar]
- 55.Dore K. Risk factors for Salmonella Typhimurium DT104 and non-DT104 infection: a Canadian multi-provincial case-control study. Epidemiol. Infect. 2004;132(3):485–493. doi: 10.1017/s0950268803001924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Said B. Outbreaks of infectious disease associated with private drinking water supplies in England and Wales 1970–2000. Epidemiol. Infect. 2003;130(3):469–479. [PMC free article] [PubMed] [Google Scholar]
- 57.Baker M. A recurring salmonellosis epidemic in New Zealand linked to contact with sheep. Epidemiol. Infect. 2007;135(1):76–83. doi: 10.1017/S0950268806006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ford M.W. A descriptive study of human Salmonella serotype Typhimurium infections reported in Ontario from 1990 to 1997. Can. J. Infect. Dis. Med. Microbiol. 2003;14(5):267–273. doi: 10.1155/2003/936084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norton S. A large point-source outbreak of Salmonella Typhimurium linked to chicken, pork and salad rolls from a Vietnamese bakery in Sydney. Western Pacific Surveillance Response. 2012;3(2):16. doi: 10.5365/WPSAR.2012.3.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mannes T. A large point-source outbreak of Salmonella Typhimurium phage type 9 linked to a bakery in Sydney, march 2007. Communicable Diseases Intelligence Q. Rep. 2010;34(1):41. doi: 10.33321/cdi.2010.34.6. [DOI] [PubMed] [Google Scholar]
- 61.Dolejska M. High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J. Antimicrob. Chemother. 2016:dkv306. doi: 10.1093/jac/dkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Output of ecological niche models and maps of variables used.
Maps of variables used in conditional autoregressive model.
Domain-specific sub-models.