Abstract
In this study we examined potential associations of HPV infection with the cervical microbiota. Cervical samples were collected from 87 HIV-seronegative reproductive-age Black South African women. Microbiota were characterized by Illumina sequencing of the V3-V4 hypervariable regions of the bacterial 16S rRNA gene. Thirty seven (42.5%) and 30 (34.5%) of the women had prevalent HPV and high-risk (HR)-HPV, respectively. Only 23 women (26.4%) had cervical microbiota dominated by a single Lactobacillus species (L. crispatus (2/87 (2.3%)), L. jensenii (2/87 (2.3%)), and L. iners (19/87 (21.8%)). The majority of the women (56/87 (64.4%)) had diverse cervical microbiota consisting of mainly bacterial vaginosis-associated bacteria. The remaining women (8/87 (9.2%)) had microbiota dominated by Aerococcus, Streptococcus, Chlamydia or Corynebacterium. Women with HR-HPV had significantly higher relative abundances of Aerococcaceae, Pseudomonadaceae and Bifidobacteriaceae compared to those with low-risk (LR)-HPV or no HPV-infection (LDA score >2.0, p < 0.05, q < 0.2). Gardnerella, Sneathia, and Atopobium were also found at greater relative abundances in HR-HPV-infected women compared to those with low-risk (LR)-HPV or no HPV-infection (LDA score >2.0, p < 0.05), although the difference was not significant after FDR-adjustment (q > 0.2). Further investigations of the bacterial taxa significantly enriched in HR-HPV-infected women are warranted.
Keywords: High-risk HPV (HR-HPV), Cervical microbiota, Reproductive-age, African, Potential biomarkers
Highlights
-
•
Majority of participants (74%) had cervical microbiota not dominated by Lactobacillus.
-
•
Lactobacillus was not enriched in HPV-negative women compared to HPV-positive women.
-
•
There was no correlation between cervical microbiota diversity and HPV infection.
1. Introduction
Black South African women have a high burden of HPV, including high-risk (HR) genotypes [1,2] that cause cervical cancer [3]. Bacterial vaginosis, a common dysbiosis of the vaginal microbiota especially among reproductive-age women (18–44 years) [4], and high vaginal pH (>4.5) are associated with higher HPV prevalence [5,6]. High vaginal pH has also been correlated with severity of cervical neoplasm [7]. It has been shown that there are racial/ethnic disparities in vaginal pH [[7], [8], [9]], with Black (African) women having less acidic vaginal pH (>4.5) than White, Asian and/or Hispanic women [8,9]. Less acidic vaginal pH may not be protective against vaginal infections [6] and cervical neoplasms [7]. Elevated vaginal pH (≥5), for example, was associated with a 10–20% increased risk of HPV infection in a large population-based study of Costa Rican women aged 18–54 years [6]. Mitra and co-workers (2016) [10] proposed that this may be related to the susceptibility of the HPV E5 oncoprotein to acidic pH [11]. High vaginal pH is consistent with the lack of keystone Lactobacillus spp. such as L. crispatus, L. gasseri, L. iners, and L. jensenii in the cervical and vaginal microbiota (CVM) of reproductive-age women [9,12]. These Lactobacillus spp. are regarded as i) biomarkers for a healthy cervicovaginal milieu and ii) agents that employ a variety of antagonistic mechanisms to promote a protective cervicovaginal milieu [13]. While controversies abound regarding whether the diversity and composition of the microbiota of the different cervicovaginal milieus (vaginal introitus, mid-vagina, lateral vaginal wall, posterior fornix, ectocervical, endocervical sites) is homogeneous (highly concordant) [14,15] or heterogeneous [16,17] within women, most investigators would agree that the Lactobacillus spp. are inherently common bacteria in the CVM [9,12,[14], [15], [16], [17], [18]].
Even though it is emerging that high CVM diversity and depletion of Lactobacillus spp. in CVM are associated with higher HPV prevalence [[19], [20], [21], [22]], some of these findings are conflicting [7,19,20,23,24]. For example, Shannon and colleagues (2017) [20] found an inverse association between relative abundance of L. gasseri and prevalent HPV whereas Reimers and colleagues (2016) [23] found no association. Gao and colleagues (2013) [24] observed that L. gasseri was significantly more frequently detected in women with HPV versus without HPV. Literature is also rapidly expanding to show that specific taxa in the CVM may be potential biomarkers for HPV infection and natural history [7,21,25,26]. While common Lactobacillus spp. such as L. crispatus and L. gasseri are inversely associated with HPV infection [19,23], bacteria such as Sneathia spp. have been positively associated with HPV infection [25] as well as elevated vaginal pH [7] and precancerous cervical lesions [7,[27], [28], [29]]. However, no studies have yet examined these associations on the high HPV-burden population in South Africa.
Evidence from large cohort studies demonstrates that ethnicity/race influences the vaginal microbiota [9,[30], [31], [32]]. The majority of the CVM studies have been performed on Western populations and those of European ancestry which predominantly constitutes Caucasian (White) women [9,12,[30], [31], [32]]. Studies have found that over 70% of reproductive-age White women have vaginal microbiota that are dominated by Lactobacillus spp., particularly L. crispatus [9,[30], [31], [32]]. In contrast, a considerable proportion of reproductive-age African women (23–63%) have been reported to harbour CVM that are Lactobacillus-deficient and are characterized by diverse and heterogeneous populations of mainly BV-associated bacteria [9,18,22,26,[30], [31], [32], [33]]. This has led some investigators [31] to suggest that this could be the reason why African women are more prone to persistent vaginal polybacterial disturbances and invasion by pathogens that include STIs. To date, the diversity of CVM remains largely unexplored and unexplained especially among Black South African women.
Herein, we characterized the composition and diversity of cervical microbiota of reproductive-age Black South African women and examined their associations with HPV infections.
2. Materials and method
2.1. Ethics statement
Participants of this study were women from the HPV Couples Cohort Study [2] who had consented to have their cervical samples used for future research. Both the parent study and this research were reviewed and approved by the Human Research Ethics Committee of the University of Cape Town in South Africa (references 258/2006 and 580/2014, respectively).
2.2. Study samples, study design and study population characteristics
This was a cross-sectional study nested within the 2-year longitudinal HPV Couples Cohort Study that was designed to investigate the prevalence and concordance of genital HPV among 254 heterosexually active Black South African couples with and without HIV infection. The parent study has been previously described in detail [2]. Briefly, with regards to women, all women were aged 18–65 years and were recruited from Gugulethu, Cape Town, South Africa. Endocervical samples were collected using Digene cervical samplers and stored in Digene transport medium at −80 °C until DNA extraction using the MagNA Pure Compact System and the MagNA Pure Compact Nucleic Acid Isolation kit (Roche Molecular Diagnostics, Mannheim, Germany). Detection of 37 HPV types (13 HR and 24 low-risk (LR)) was also performed using the Roche Linear Array HPV genotyping test (Roche Molecular Diagnostics, Mannheim, Germany) as described elsewhere [2].
In the present study, inclusion criteria were: baseline DNA samples with positive human β-globin results (a measure of sample adequacy), sufficient archived sample volume for cervical microbiota analyses, samples from reproductive-age HIV-seronegative women, and information on baseline HPV status. Self-reported menstruation at the time of sampling, being a woman aged <18 or >44 years, and positive HIV status were considered as exclusion criteria. Samples from 91 women were eligible for this study. Findings suggestive of BV, as indicated on the Pap smear results, was abstracted from the participant database.
Of the 91 enrolled women, only 87 (95.6%) were finally included as each had ≥12,161 reads, as outlined in the results section, for 16S rRNA sequence analysis. The baseline characteristics of the 87 reproductive-age heterosexual women are summarized in Table 1.
Table 1.
Baseline demographic, sexual and smoking, behavioural, and clinical information of the 87 heterosexually active Black South African women.
| Characteristic | All participants |
|---|---|
| (N = 87) | |
| Age (years) | 32.0 (25.0–39.0) |
| Age at sexual debut (years)ˆ | 18.0 (16.0–19.0) |
| Number of lifetime sexual partners | 3.0 (2.0–4.0) |
| Number of sex acts with study partner in last monthˆ | 4.0 (2.0–8.0) |
| Currently on hormonal contraception* (% (n/N)) | 42.5 (31/73 |
| HPV infection (% (n/N)) | |
| Any HPV type | 42.5 (37/87) |
| Any high-risk HPV type | 34.5 (30/87) |
| Cervical cytology (% (n/N)) | |
| Normal | 75.6 (62/82) |
| ASCUS | 13.4 (11/82) |
| LSIL | 11.0 (9/82) |
| Experienced vaginal discharge in last 6 months (% (n/N)) | 15.4 (12/78) |
| Experienced genital ulceration in last 6 months (% (n/N)) | 2.3 (2/87) |
| Findings suggestive of BV on Papanicolaou smear (% (n/N)) | 43.7 (38/87) |
| Cigarette use (% (n/N)) | |
| Never smoked | 64.4 (56/87) |
| Ex-smoker | 3.4 (3/87) |
| Current smoker | 32.2 (28/87) |
Abbreviations: HPV – human papillomavirus, ASCUS – atypical cells of undetermined significance, LSIL – low-grade squamous intraepithelial lesion, BV – bacterial vaginosis.
Continuous variables are expressed as medians with interquartile ranges (IQRs, at 25th and 75th percentiles).
ˆData was not available on the age at sexual debut for one woman, lifetime number of sexual partners of one woman, and number of sexual acts with study partner in the last month of two women.
*The hormonal contraceptives included oral pills, norethisterone enanthate, Depo-Provera, and steroids.
Of the 87 women, 37 (42.5%) were HPV-positive. Among the HPV-positive women, 51.4% (19/37) had single HPV infections. A total of 45.9% (17/37) and 81.1% (30/37) of the women were infected with LR- and HR-HPV types, respectively. Among these women, 27.0% (10/37) were infected with both LR- and HR-HPVs. A majority of the women had normal cervical cytology (75.6% (62/82)) while few had experienced vaginal discharge (15.4% 12/78)) or genital ulceration (2.3% (2/87)) in the last 6 months. A total of 43.7% (38/87) of the women had findings suggestive of BV on their Pap smear results. Approximately one-third of the women (32.2%) were currently cigarette smokers.
2.3. Illumina MiSeq V3-V4 16S rRNA metagenomics library preparation and sequencing
The hypervariable V3-V4 16S rRNA region of the bacterial gene was targeted by universal bacterial primer 319F (5’-CCTACGGGNGGCWGCAG-3’) and 806R (5’-GACTACHVGGGTATCTAATCC-3’) as previously described [34]. The 16S rRNA libraries for sequencing were prepared according to the 16S rRNA metagenomics protocol for MiSeq System (Illumina, San Diego, CA, USA) [35], with minor modifications. Here, amplicon PCR was performed in triplicate using TaKaRa Ex Taq® Hot Start Version (Takara Bio Inc., Japan) with the following thermocycling conditions: an initial denaturation at 98 °C for 2 min, followed by 30 cycles of denaturation at 98 °C for 20 s, annealing at 50 °C for 30 s and extension at 72 °C for 45 s, and a final extension step at 72 °C for 10 min. Nuclease free water and Digene specimen transport medium were used as negative and extraction controls, respectively, to check for possible contaminants. The Human Microbiome Project mock communities, HM-782D (even concentration) and HM-783D (staggered concentration) (BEI Resources, Manassas, VA, USA), each comprising of a synthetic mixture of genomic DNA from 20 known bacterial species, were used as positive controls to assess the impact of PCR and sequencing artefacts and optimize the cervical microbiota analyses. Pooled amplicons from each replicate were confirmed by gel electrophoresis (1.5% ethidium bromide-stained agarose gel), purified using Agencourt AMPure XP System (Beckman Coulter, Beverly, MA, USA), and quantified in triplicate using the Quant-iT® PicoGreen dsDNA assay (Thermo Fisher Scientific, Waltham, MA, USA). Next, Index PCR of the purified products was performed using KAPA HiFi HotStart ReadyMix PCR Kit (KAPA Biosystems, Wilmington, MA, USA) under the following PCR conditions: an initial denaturation at 95 °C for 3 min followed by 8 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, with a final extension step at 72 °C for 5 min. Purified libraries were pooled in equimolar concentrations and the final library quantified on a Bioanalyzer High Sensitivity Chip (Agilent Technologies, Santa Clara, CA, USA). The library was sequenced on the Illumina MiSeq using a paired-end 300-bp protocol and v3 reagents by Macrogen Inc. (Seoul, South Korea).
2.4. 16S rRNA gene amplicon sequence data analyses
The sequenced data was analyzed by QIIME v1.8.0 [36] and UPARSE (usearch8.0.1616) [37] using the parameters detailed in Supplementary Table 1. The merged, quality-filtered, globally-trimmed, and dereplicated reads were clustered into operational taxonomic units (OTUs) at 97% sequence similarity threshold followed by picking of the representative sequences, taxonomy assignment using the RDP Naïve Bayesian Classifier [38] (with the Greengenes database (gg13_8 Release) [39]), and creating a phylogenetic tree and OTU table. We modified the conservative filtering strategy described by Narrowe and co-workers (2015) [40] to discard spurious OTUs. Here, the relative abundance (0.02%) of the most abundant false positive (L. iners) in the HM-782D mock community was assumed to represent the level of cross-contamination and was therefore used to filter the OTU table. A rarefaction curve for each sample was generated at multiple sequence depths in order to determine a sufficient subsampling depth necessary to estimate the bacterial diversity. Alpha diversity was computed using chao1, observed_species, Shannon, Simpson, and PD_whole_tree metrics implemented in QIIME. Dominance and Shannon equitability indices were computed using an in-house script in R v3.2.2 (R Core Team 2016). Principal Coordinates Analysis (PCoA) of beta diversity measures (weighted and unweighted UniFrac distances, and Bray-Curtis dissimilarity metric) was used to describe the degree of variations in cervical microbiota composition according to the participants’ metadata. UniFrac distance measures the distance between bacterial communities using phylogenetic information, while the Bray-Curtis metric measures the dissimilarity between communities based on OTU abundance. Differences in beta diversity measures (between sample categories) were tested using permutation multivariate analysis of variance (PERMANOVA) [41], with 999 permutations.
2.5. Hierarchical clustering of cervical microbiota
To identify the community state types (CSTs) of the cervical microbiota, hierarchical clustering (average neighbour linkage) was performed based on Bray-Curtis dissimilarity index calculated using the Vegan R package [42]. Only taxa with relative abundances ≥0.33% were included in the heatmap.
2.6. Identification of potential biomarkers for HR-HPV and HPV infections
Differentially abundant bacterial taxa (potential biomarkers) in women with and without HPV infection, including HR-HPV, were identified by the Linear Discriminant Analysis (LDA) effect size (LefSe) algorithm v1.0 [43]. In this paper, the term biomarker is used to refer to bacterial taxa with significantly different relative abundances between particular groups, as defined by Segata and co-workers (2011) [43]. The alpha value for the factorial Kruskal-Wallis test among classes and the pairwise Wilcoxon signed-rank test between subclasses was 0.05. Given that the LefSe method does not account for multiple comparisons, the p-values were subsequently adjusted based on the false discovery rate (FDR) method [44]. FDR-adjusted p-values (q-values) less than 0.2 were considered significant. This cut-off, frequently applied in microbiome studies [45,46], was selected to ensure important differentially abundant taxa with small effect sizes are identified. Discriminative features were identified at a logarithmic LDA score threshold of 2.0.
2.7. Statistical analyses
Statistical analyses were done with STATA v13.0 (StataCorp, College Station, TX, USA) and R v3.2.2 (R Core Team 2016). Associations between the established CSTs and participants’ metadata were calculated using Fisher’s exact/Chi-square tests for categorical variables and Mann-Whitney unpaired nonparametric test for continuous variables, with statistical significance at p < 0.05 (two-tailed).
The alpha diversity between the CSTs was compared using Mann-Whitney unpaired nonparametric test, with p < 0.05 showing significance. More than two-group comparison of the alpha diversity measures of the CSTs was computed by Kruskal-Wallis test.
3. Results
3.1. Data analysis and output
A total of 5,694,432 high-quality reads were finally analyzed (≥12,161 reads per sample) from 87 samples. This cut-off of reads was selected based on rarefaction curves of alpha diversity that showed that for each sample, 12,161 reads could provide a reasonable coverage (subsampling depth) of bacterial diversity (Supplementary Fig. 1). The rarefaction analysis of observed_species and Shannon indices are shown in Supplementary Fig. 1a and Supplementary Fig. 1b, respectively. The rarefaction curves of all the samples had plateaued at 12,161 reads when using the Shannon and Simpson Index. The plots of observed_species, chao1, and PD_whole_tree metrics for many samples did not however plateau at the cut-off point. Metrics such as observed_species and chao1 did not plateau because they are sensitive to rare (low-abundant) species, which are usually incorporated in their calculations [47]. Analyses of the negative and extraction controls indicated that contaminants were nearly absent in the sequenced library, thus, unlikely to affect the cervical microbiota diversity estimation especially after we optimized our analyses using the mock communities.
3.2. Bacterial prevalence and relative abundance in the cervical microbiota
A total of 120 non-spurious OTUs were identified in the 87 samples (27–85 OTUs per sample). These OTUs were classified into a total of eight bacterial phyla and 58 genera. Supplementary Table 2 is a summary of the overall relative abundance and prevalence of the eight phyla detected, the corresponding major genera in each phylum and the most abundant OTUs in these genera. Firmicutes was the most abundant phylum, represented by 45 (37.5%) of the 120 OTUs. Lactobacillus was the major genus in this phylum, represented by six OTUs. The most abundant OTU (classified as L. iners) was found in this genus and was present in all the samples. Other Lactobacillus OTUs (L. jensenii, L. crispatus, L. gasseri, L. reuteri, and an unclassified Lactobacillus sp.) were present at significantly lower relative abundance and prevalence. Bacteroidetes was the second most abundant phylum, with Prevotella being the major genus in this phylum. Prevotella was the most diverse of the 58 genera, with a total of 21 OTUs. Among these, Prevotella.4 was the most abundant. Actinobacteria and Fusobacteria were the next most abundant phyla, with Gardnerella and Sneathia, respectively, being the most abundant genera. Three OTUs were detected in each of these genera, with G. vaginalis.3 and Sneathia.3 being the most abundant. Surprisingly, the G. vaginalis.3 and Sneathia.3 OTUs were present in all the samples (Supplementary Table 2).
3.3. Clustering of the cervical microbiota into distinct community state types
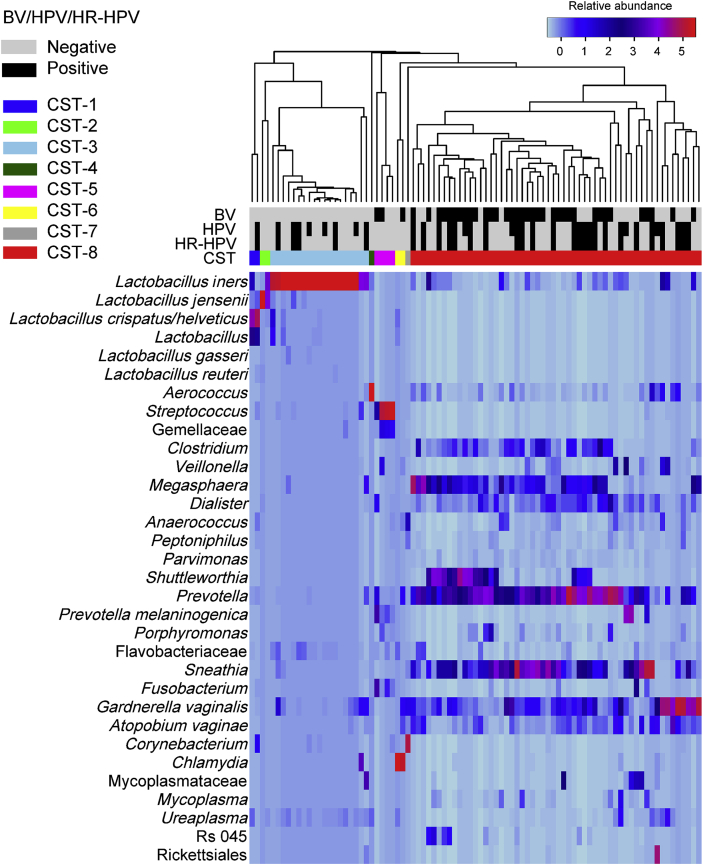
The cervical microbiota were hierarchically clustered by their composition and relative abundances of bacterial taxa. A heatmap of the relative abundances of the 32 major taxa in the 87 women and clustering of the cervical microbiota is shown in Fig. 1.
Fig. 1.
Heatmap of the relative abundances of bacterial taxa found in cervical microbiota of 87 South African reproductive-age women. Rows represent the bacterial taxa (sorted by classification) and columns the samples. The colour key for the relative abundances is indicated in the upper right corner. The human papillomavirus (HPV) and high-risk human papillomavirus (HR-HPV) infection status as well as bacterial vaginosis (BV) findings of the women are indicated. The dendrogram based on average linkage hierarchical clustering of the Bray-Curtis dissimilarity matrix is shown and was used to define the eight community state types (CSTs). Except for L. gasseri and L. reuteri, each with 0.03% relative abundance, the other 30 bacterial taxa included had ≥0.33% relative abundance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The cervical microbiota were clustered into eight CSTs (designated CST 1–8). The percentage relative abundance of the most abundant OTUs in each of the CSTs is provided in Supplementary Table 3 for CSTs 1–7 and Supplementary Table 4 for CST-8.
CSTs 1–3 were dominated by Lactobacillus spp. and included 26.4% (n = 23) of the women. CST-1 (dominated by L. crispatus) and CST-2 (dominated by L. jensenii) were each found in only 2 (2.3%) women whereas CST-3 (dominated by L. iners) was found in 19 women (21.8%). There was no CST dominated by L. gasseri.
CSTs 4–8 were Lactobacillus-deficient. CST-8 was the most prevalent CST (64.4%, n = 56). Women in CST-8 had diverse cervical microbiota, mostly comprising a continuum of BV-associated bacteria such as G. vaginalis, Prevotella, Sneathia, Atopobium, Shuttleworthia, Clostridium, Megasphaera, and Dialister. The remaining CSTs occurred at a low prevalence, with CST-4 (dominated by Aerococcus), CST-5 (Streptococcus), CST-6 (Chlamydia), and CST-7 (Corynebacterium) occurring in only one, four, two and one women, respectively. For ease of reference, these CSTs are referred to by the most dominant bacterial taxa, though each cervical microbiota in these CSTs is colonized by an array of bacterial taxa at varying relative abundances. Moreover, the cervical microbiota were hierarchically clustered according to composition and relative abundances of these bacterial taxa as underscored in subsection 2.5. The identity of these CSTs should therefore be interpreted with caution.
The reduction of the complex compositional data by clustering of the cervical microbiota into CSTs allowed for statistical comparisons as discussed in the subsequent subsection.
3.4. Comparison of characteristics of the women with cervical microbiota in the community state types 3 and 8
The characteristics of the women with cervical microbiota in CST-3 and CST-8 were statistically compared and are shown in Supplementary Table 5. Women with cervical microbiota from the remaining CSTs (CST-1, CST-2, and CST-4 to CST-7) were excluded from this comparison as these groups had low numbers (≤4 women).
None of the women with cervical microbiota in CST-3 had findings suggestive of BV (p < 0.0001) or atypical cells of undetermined significance (ASCUS) (p = 0.055). Similar to the other participants’ metadata, prevalent HPV infections did not statistically differ between CST-3 and CST-8. Finally, it is worthwhile mentioning that an analysis of the baseline bacterial taxa and longitudinal HPV infection outcomes were not possible due to the complexity of HPV infections outcomes and the small sample sizes in each of the multiple categories of HPV infection outcomes (as depicted in Supplementary Table 5).
3.5. Comparison of alpha diversity across CSTs, BV, HPV, and high-risk HPV groups
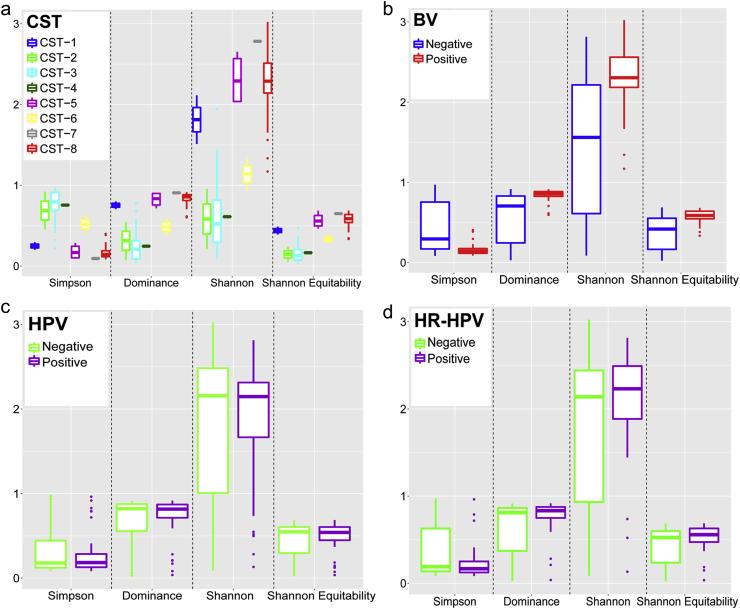
Differences in community composition of the CSTs were assessed by comparing alpha diversity metrics (Fig. 2a). All the alpha diversity measures of the two major CSTs (CST-3 and CST-8) were significantly different, with CST-8 having a greater diversity (less dominance, high richness and evenness) than CST-3. The median Shannon index for instance, for CST-3 versus CST-8 was as follows – Shannon: 0.5 (0.3–0.9) versus 2.3 (2.1–2.5), p < 0.0001.
Fig. 2.
Alpha diversity measures of cervical microbiota. Comparison of the alpha diversity of cervical microbiota grouped by a) community state type (CST), b) bacterial vaginosis (BV) findings, c) human papillomavirus (HPV) infection status and d) high-risk (HR)-HPV infection status. In each plot, the box ranges from the first to the third quartile, with the median represented by the horizontal line. The whiskers extend to the smallest and largest non-outliers and outliers are represented by the dots.
The alpha diversity of the cervical microbiota also significantly varied with BV (Fig. 2b), with women with findings suggestive of BV (Shannon index: 2.3 (2.1–2.5)) having a higher bacterial diversity than those without (1.6 (0.6–2.2)), p < 0.0001.
The alpha diversity of the cervical microbiota of women with HPV (Fig. 2c) and HR-HPV (Fig. 2d) infection did not differ significantly to that in cervical microbiota from uninfected women. The median Shannon index of the cervical microbiota was 2.2 (0.9–2.5) for HPV-negative and 2.1 (1.7–2.4) for HPV-positive women, p = 0.747. For women with and without HR-HPV, median Shannon indices were 2.2 (1.8–2.5) and 2.1 (0.9–2.4), respectively, p = 0.198.
3.6. Comparison of beta diversity across CSTs, BV, HPV, and high-risk HPV groups
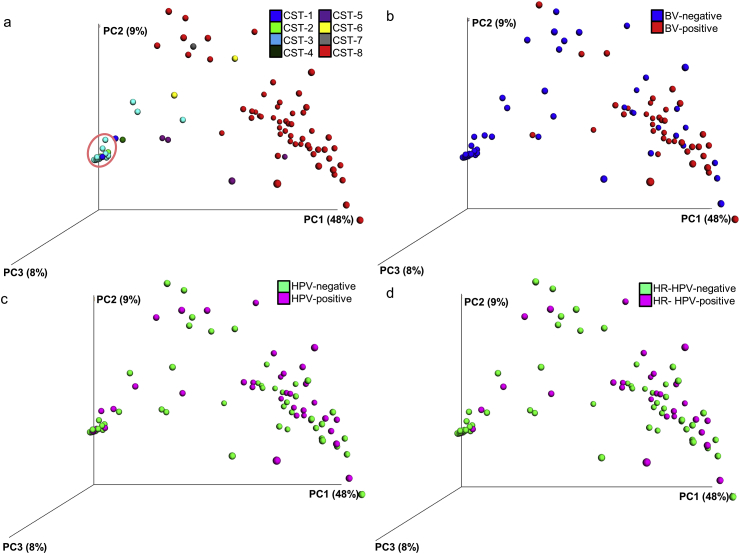
We present the results of the weighted UniFrac distance that was applied to compute beta diversity estimation between the CSTs, BV and HPV infection groups. The PCoA of the weighted UniFrac distance matrix of the 87 cervical microbiota is shown in Fig. 3.
Fig. 3.
Beta diversity of the cervical microbiota. Principal Coordinates Analysis (PCoA) plots of the weighted UniFrac distances of the cervical microbiota coloured according to a) community state type (CST), b) bacterial vaginosis (BV) findings, c) human papillomavirus (HPV) infection status, and d) high-risk (HR)-HPV infection status. The first three principal coordinate (PC) axes and the percentage variation explained by each (PC1: 48%, PC2: 9%, and PC3: 8%) are shown. Each solid point represents a bacterial community.
The CSTs were spatially segregated and therefore compositionally and phylogenetically distinct from each other (pseudo-F statistic = 14.3, p = 0.001). A few samples (21.8%, 19/87), all with Lactobacillus dominance, clustered tightly together (pink-circled cluster in Fig. 3a).
The majority of the cervical microbiota samples from women with no findings suggestive of BV distinctively segregated in the 3D space from cervical microbiota samples from women with findings suggestive of BV (Fig. 3b), pseudo-F statistic = 21.2, p = 0.001.
There was no distinct segregation of the cervical microbiota on the PCoA plot by HPV status (Fig. 3c), pseudo-F statistic = 1.4, p = 0.181; or HR-HPV infection status (Fig. 3d), pseudo-F statistic = 2.2, p = 0.099. The majority of the Lactobacillus-dominated samples (73.7%, 14/19) that clustered tightly together were HPV-negative.
3.7. Potential biomarkers for HPV and high-risk HPV infection
The comparison of the cervical microbiota of HPV or HR-HPV-infected women and -uninfected women indicated that overall cervical microbiota composition (as measured by alpha and beta diversity) did not differ significantly. Next, we used LefSe algorithm [43] to identify potential biomarkers in cervical microbiota associated with HPV and HR-HPV infection. Women with HR-HPV infection comprised of women with any of the HR-HPV genotypes regardless of infection with any of the LR-HPV genotypes whereas women without HR-HPV infection consisted of women that were either negative for HPV or positive for LR-HPV.
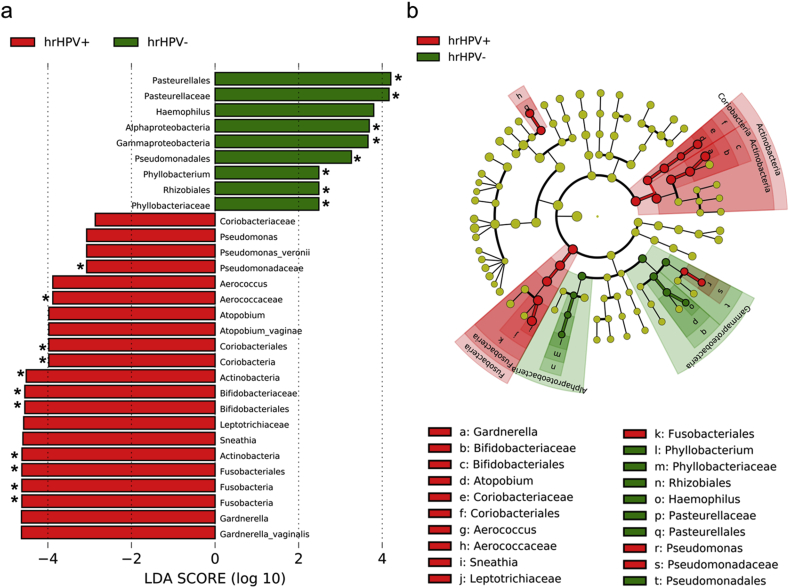
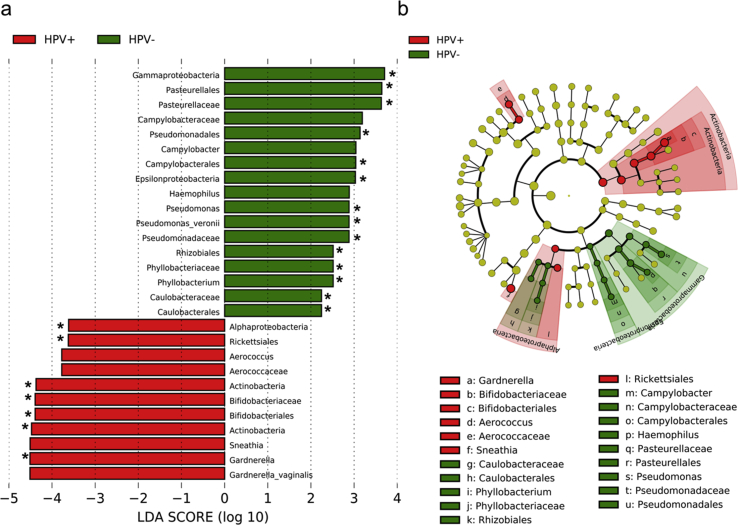
When comparing the cervical microbiota of women with and without HR-HPV, LefSe identified 30 bacterial taxa that were differentially abundant (LDA score >2.0, p < 0.05) (Fig. 4). The relative abundances of the bacterial families Aerococcaceae, Pseudomonadaceae and Bifidobacteriaceae were found to be significantly higher (LDA score >2.0, p < 0.05, q < 0.2) in women with HR-HPV compared to those with low-risk (LR)-HPV or no HPV-infection. Genera Gardnerella, Sneathia, Atopobium, Aerococcus, and Pseudomonas were identified as potential biomarkers for HR-HPV by LefSe (LDA score >2.0, p < 0.05), but significance was lost following adjustment for multiple comparisons (q > 0.2). At species level, the strongest association with HR-HPV infection was found with G. vaginalis, followed by A. vaginae and P. veronii. Haemophilus and Phyllobacterium were the only genera enriched in women without HR-HPV infection, with Phyllobacterium being significantly differentially abundant (q < 0.2).
Fig. 4.
Potential biomarkers for high-risk HPV by LefSe. a) Histogram of differentially abundant taxa in cervical microbiota of women with and without high-risk (HR)-HPV infections, and b) A six-level cladogram with a taxonomic hierarchical structure. Each coloured solid represents a taxon and its diameter is proportional to the taxon’s relative abundance. Red and green solids represent statistically significant taxon ranks in HR-HPV-positive and HR-HPV-negative group, respectively. Only features with logarithmic LDA score >2.0 are shown. Asterisks indicate significantly differentially abundant taxa with q < 0.2 after FDR correction. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
A total of 28 bacterial taxa were found to be differentially abundant (LDA score>2.0, p < 0.05) between the cervical microbiota of HPV-negative and HPV-positive women (Supplementary Fig. 2). Prevalent HPV was strongly associated with higher abundances of Gardnerella, Sneathia, and Aerococcus and concomitantly with lower abundances of Campylobacter, Haemophilus, and Pseudomonas (LDA score >2.0, p < 0.05). Following FDR-adjustment for multiple comparisons, Gardnerella and Pseudomonas were found to be significantly differentially abundant (q < 0.2).
Neither Lactobacillus nor species in this genus were found to be differentially abundant between women with and without HPV or HR-HPV infections.
4. Discussion
In this study, we examined if there was an association between prevalent HR-HPV and cervical microbiota of 87 reproductive-age Black South African women. It is the first study to assess potential biomarkers for genital HPV and HR-HPV infection in South Africa. We began by first characterizing the composition and diversity of the cervical microbiota. By using hierarchical clustering, we found eight CSTs, three of which were dominated by Lactobacillus spp. (CST-1: L. crispatus, CST-2, L. jensenii, and CST-3: L. iners). CSTs 4–7 were less common and were dominated by Aerococcus, Streptococcus, Chlamydia, and Corynebacterium, respectively. CST-8 was a diverse and heterogeneous Lactobacillus-deficient cervical microbiota.
The low prevalence of Lactobacillus-dominated cervical microbiota (26%) corroborates previous findings that Lactobacillus-dominated CVM are less common among African women [9,22,30,31], including Black South Africans [18]. CST-3 (L. iners) was the most prevalent (83%) among the Lactobacillus-dominated CSTs, as has been reported in several other cohorts of reproductive-age HIV-negative women of African descent [9,16,22,26,30,33,48,49]. However, the prevalence of the L. iners-dominated CST was higher than that reported in these studies (31–65%) [9,16,22,26,30,33,48,49]. Compared to other lactobacilli, particularly L. crispatus, L. iners is less protective [13] as it can predispose women to BV [50] and has been associated with reduced protection against a variety of STIs, such as C. trachomatis [22] and HPV [7,19,22], including HR-HPV [22]. Its genome plasticity allows it to survive in dysbiotic cervicovaginal milieu [51]. Unlike the other keystone Lactobacillus spp., L. iners colonization is less dependent on glycogen (the main carbon source in the vagina) [52], which when catabolized leads to production of lactic acid that is responsible for low vaginal pH [53]. Instead, L. iners can utilize non-glycogen sources such as glycerol, producing short-chain fatty acids (e.g., succinate, a biomarker for BV) that are known to increase vaginal pH [51,53]. Furthermore, its metabolite profile is intermediate between that of L. crispatus/L. jensenii and BV-associated bacteria [53].
In our study, cervical microbiota dominated by L. crispatus (CST-1) and L. jensenii (CST-2) were less common, each with an overall prevalence of 2.3%. The prevalence of CST-1 is much lower than that reported in the CVM of African women (7–22%) whereas that of CST-2 is relatively consistent with previous studies (up to 1% prevalence) [9,16,18,22,31,33,48]. Compared to other keynote Lactobacillus spp., L. jensenii is an uncommon Lactobacillus spp. in cervical samples [12,14,17]. CVM with L. crispatus dominance are known to have the lowest vaginal pH [9], hence thought to be the most protective Lactobacillus spp. against BV [22,50,54], STIs [19,22,23], and cervical disease [29]. We detected L. gasseri and L. reuteri as low-abundant Lactobacillus spp. in the cervical microbiota, but not as distinct CSTs. L. gasseri-dominated CVM are very uncommon among African women (<5%) [9,31] and have not been reported among Black South African women [16,18,33,48]. The paucity of certain Lactobacillus spp. dominating the CVM of Black South African women may be due to genetic [9,30,31,49] and behavioural factors [30]. In addition to these factors, the low prevalence of Lactobacillus-dominated microbiota in our study could have been impacted by our sample type (cervical swabs), although this remains an ongoing debate [14,15,17]. Beyond speculation [15], molecular studies have reported that the cervix and vagina may be differentially colonized with some Lactobacillus spp. [14,16,17]. A study on South African adolescents noted that L. crispatus was less abundant in the endocervix than in the lateral vaginal wall [16].
The predominance of a highly diverse and heterogeneous Lactobacillus-deficient cervical microbiota (CST-8) reinforces earlier findings on African women that have reported a high prevalence (up to about 63%) [9,18,30,31]. Greater community diversity has been associated with BV [20,22,33,48]. Diverse CVM tend to occur in less acidic milieus (pH 4.7–6.2) [9] and to vary in stability and resilience [12,31,55,56]. BV-associated diverse CVM have been associated with heightened genital proinflammatory responses [18,57], abrupt and persistent postpartum microbiota disturbance [56], detectability and delayed clearance of HPV [[19], [20], [21], [22],25,26], neoplastic cervical lesions [27,29], cervical and carcinogenesis [28]. In this study, all women with ASCUS cytology were in CST-8. Diverse vaginal microbiota have been found to be more common in women with ASCUS than in women with normal cervical cytology [29].
Besides CST-8, we also found other Lactobacillus-deficient cervical microbiota that were dominated by other bacteria (CST-4: Aerococcus, CST-5: Streptococcus, CST-6: Chlamydia, and CST-7: Corynebacterium). While CST-5 has been reported [32,58], there are no previous reports on the other CSTs. We are part of the growing number of investigators [26,28,30,31] who have reported unique Lactobacillus-deficient CSTs. This could be an indication that the CVM, especially of Africans, is not yet completely characterized. The predominant taxa in CST-4 (Aerococcus) and CST-5 (Streptococcus) are in the order Lactobacillales and are lactic acid-producers [9,58,59]. Similar to L. iners, these bacteria have potential bidirectional roles in cervicovaginal health [30,55] and disease [9,18,28]. Aerococcus has been associated with genital inflammation in vivo, particularly in women with diverse CVM [18]. Streptococcus and Aerococcus have been reported as two of the bacteria populating the cervicovaginal milieu following depletion of Lactobacillus in women who cleared HPV infections [21]. For CST-6, which included two women, the species was identified as C. trachomatis. These cervical microbiota are likely to be full-blown C. trachomatis infection and not a CST. It is worth mentioning that about 43% of the women (37/87) in our cohort had C. trachomatis infection as detected by sequencing. Recent studies that have used laboratory tests for STI diagnosis have illustrated that there is a great burden of C. trachomatis infection among Black South Africa women, particularly the young (13–43% prevalence) [18,33]. C. trachomatis has been associated with elevated vaginal pH [6]. C. trachomatis have been reported to be more abundant in endocervical samples (similar to the samples our study used) than lateral vaginal wall samples [16]. Lastly, only one woman had Corynebacterium-dominated cervical microbiota (CST-7). Corynebacterium has been found in up to moderate abundances in women with diverse CVM [16,22,32,55] and its high relative abundances has been associated with BV in women of European ancestry [30]. Additional studies are needed to confirm characterize these Lactobacillus-deficient cervical microbiota.
Following characterization of the cervical microbiota in our cohort, we examined the associations between HPV infections and the most common CSTs: CST-3 and CST-8. No associations were observed. CVM/CSTs may not necessarily be associated with STIs [12,18,19,26]. Given the mounting importance of certain cervicovaginal bacterial taxa in genital HPV infection [7,21,23,[25], [26], [27],60,61], we next sought to assess the association between HPV and the relative abundances of cervical bacterial taxa. Higher relative abundances of Sneathia, Gardnerella, Atopobium, Pseudomonas, and Aerococcus were found in women with HR-HPV infections compared to those without, although this was found not to be significant following FDR-adjustment. Gardnerella, Sneathia, and Aerococcus were also associated with any HPV infection, with Gardnerella being significantly (q < 0.2) more abundant in the cervical microbiota of HPV-infected women compared to uninfected women. Sneathia, Atopobium, Gardnerella, and Aerococcus have been reported to be more enriched in African women than White women [30,31,49] and have been associated with BV [9] and found in cases of recurrent BV [62].
Of all the published putative bacteriological markers for HPV/HR-HPV infections, Sneathia is the most widely detected and has been reported in Nigerian [26], Italian [21], Korean [25], Chinese [61], Arizona Hispanic and non-Hispanic [7] cohort studies. None of these studies adjusted their analyses for multiple comparisons. In addition to being identified as a potential biomarker for HPV/HR-HPV infections in women of different races, other studies have demonstrated the pathogenic potential of specific genital Sneathia spp., including S. sanguinegens, and point to the possible mechanisms by which it may play a role in HPV acquisition and persistence. Such Sneathia spp. have been associated with high vaginal pH [7], squamous intraepithelial lesion (SIL) in HPV-positive women [28,29], cervical neoplasm [7], and increased genital inflammation [18,33,57]. S. amnii, a highly similar bacterium to S. sanguinegens, has been reported in mid-vaginal samples of North American women and found to have genes that encode proteins implicated in its invasion, adherence to cervical cells and cytotoxicity [63].
A recent Chinese cohort study found that the abundances of G. vaginalis and A. vaginae were greater in women with HPV than without HPV infection [61]. An Italian study observed that these two bacteria populated the vaginal milieu following depletion of protective Lactobacillus in women with persistent HPV infection [21]. In addition, women with persistent HPV infection were noted to have higher relative abundances of Atopobium than women who cleared HPV infections [21]. Atopobium and Gardnerella have been associated with BV-linked clue cells [49] and increased risk of cervical disease [7,27]. Gardnerella, a likely key driver of biofilm formation in the vagina [64], has been associated with preterm delivery [56], heightened immune activation, disruption of epithelial integrity, and impaired wound-healing [65]. The presence of microwounds in the female genital tract allows HPV to access the basal epithelial cells of the cervical transformation zone [66]. The sialidase-encoding gene from G. vaginalis has been identified as a potential biomarker for HPV persistence [21]. A. vaginae is thought to augment the development of biofilm development since it is predominant in G. vaginalis-dominated biofilm [67].
We found that genus Pseudomonas and P. veronii occurred at greater relative abundances in women without HPV infection compared to women with HPV infection (unadjusted and FDR-adjusted analyses). In contrast to this, the genus Pseudomonas and P. veronii were identified as potential biomarkers for prevalent HR-HPV infection (unadjusted analysis). The greater relative abundance of Pseudomonas in women with HR-HPV infection than in women without HR-HPV infection was in agreement with an Italian cohort study [21]. However, after FDR-adjustment, the difference lost its significance. Several species in the genus Pseudomonas are increasingly being identified as potential biomarkers in women with and without HPV/HR-HPV infection. For example, a small pilot study on Mexican cohort observed higher abundances of Pseudomonas oleovorans in HPV-positive women with normal cervical cytology [28]. In a recent study on Chinese cohort, Pseudomonas aeruginosa was associated with baseline HPV negativity [60]. Prospective studies may be needed to determine if Pseudomonas spp. are modifiers of HPV/HR-HPV infection.
Of note, Lactobacillus and L. iners were not found to be differentially abundant in women with and without HPV/HR-HPV infection. Di Paola and colleagues (2017) [21] and Chao and colleagues (2019) [61], similarly found that Lactobacillus was not differentially abundant in the CVM of Italian and Chinese women, respectively, with and without HPV/HR-HPV infection. Several studies, however, have reported a greater relative abundance of lactobacilli, including L. gasseri and L. iners, in the CVM of women without HPV/HR-HPV [20,25,26]. Interestingly, L. iners has been found to be enriched in Arizona Hispanic and non-Hispanic women with HPV [7]. In our study, the relative abundance of lactobacilli was not significantly different in women with versus without HPV infection probably due to the low abundances of Lactobacillus spp., such as L. crispatus and L. gasseri, in the study population.
The main limitations of this study include the relatively small sample size, use of phenotypic trait (black skin colour) to define women of African ancestry, lack of a standard method to diagnose BV, and inadequate information on female sexual behaviour. As a result, it was not possible to examine whether some of the observed associations were confounded by the missing information. Furthermore, the high prevalence of Chlamydia in this cohort may have confounded our ability to detect other microbes associated with HPV. C. trachomatis is associated with HPV prevalence and infection with this STI often results in cervicitis, with this inflammation potentially causing epithelial damage that may predispose women to HPV infection [68]. The small sample size prevented effective control for this potential confounding effect. Despite these limitations, we presented the baseline composition and diversity of the cervical of reproductive-age HIV-seronegative Black South African women and identified bacterial taxa that could be important in HPV and HR-HPV infection.
5. Conclusion
To date, this remains the first study to examine the association between prevalent HPV and cervical microbiota in a Black South African cohort. Further investigations into the role of the cervical and vaginal microbiome in HPV/HR-HPV infections are warranted.
Data statement
The Illumina MiSeq raw sequence data and metadata have been deposited in the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/Traces/sra/) under BioProject PRJNA473351.
Declarations of interest
None.
Funding
This work is based on the research supported in part by the National Research Foundation of South Africa (Grant Number: 64815), Poliomyelitis Research Foundation (PRF), the Cancer Association of South Africa (CANSA), University of Cape Town (UCT) Research Incentive Scheme and the UCT Cancer Research Initiative. Harris Onywera was also a recipient of the UCT International and Refugee Students’ Scholarship and the postgraduate publication incentive (PPI).
Financial and competing interests’ disclosure
The authors declare that no financial or conflict of interest related to this article exists.
Acknowledgements
The authors wish to thank all the study participants and personnel involved in the HPV Couples Cohort Study. Computations were performed using the University of Cape Town’s Division of Information and Communication Technology Services (ICTS) High Performance Computing Facility http://hpc.uct.ac.za.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.pvr.2019.04.006.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
figs1.
figs2.
References
- 1.Bruni L., Barrionuevo-Rosas L., Albero G., Serrano B., Mena M. HPV Information Centre; 2017. Human Papillomavirus and Related Diseases in the World. Summary Report 19 April 2017. ICO Information Centre on HPV and Cancer. [Google Scholar]
- 2.Mbulawa Z.Z.A., Coetzee D., Marais D.J., Kamupira M., Zwane E. Genital human papillomavirus prevalence and human papillomavirus concordance in heterosexual couples are positively associated with human immunodeficiency virus coinfection. J. Infect. Dis. 2009;199:1514–1524. doi: 10.1086/598220. [DOI] [PubMed] [Google Scholar]
- 3.Nobbenhuis M.A.E., Walboomers J.M.M., Helmerhorst T.J.M., Rozendaal L., Remmink A.J. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet. 1999;354:20–25. doi: 10.1016/S0140-6736(98)12490-X. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C., Colebunders R., Crucitti T. The global epidemiology of bacterial vaginosis: a systematic review. Am. J. Obstet. Gynecol. 2013;209:505–523. doi: 10.1016/j.ajog.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Gillet E., Meys J.F.A., Verstraelen H., Bosire C., De Sutter P. Bacterial vaginosis is associated with uterine cervical human papillomavirus infection: a meta-analysis. BMC Infect. Dis. 2011;11:10. doi: 10.1186/1471-2334-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke M.A., Rodriguez A.C., Gage J.C., Herrero R., Hildesheim A. A large, population-based study of age-related associations between vaginal pH and human papillomavirus infection. BMC Infect. Dis. 2012;12:33. doi: 10.1186/1471-2334-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laniewski P., Barnes D., Goulder A., Cui H., Roe D.J. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 2018;8:7593. doi: 10.1038/s41598-018-25879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens-Simon C., Jamison J., McGregor J.A., Douglas J.M. Racial variation in vaginal pH among healthy sexually active adolescents. Sex. Transm. Dis. 1994;21:168–172. doi: 10.1097/00007435-199405000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S.K. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra A., MacIntyre D.A., Marchesi J.R., Lee Y.S., Bennett P.R. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome. 2016;4:58. doi: 10.1186/s40168-016-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straight S.W., Herman B., McCance D.J. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J. Virol. 1995;69:3185–3192. doi: 10.1128/jvi.69.5.3185-3192.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith B.C., McAndrew T., Chen Z., Harari A., Barris D.M. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PLoS One. 2012;7:e40425. doi: 10.1371/journal.pone.0040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrova M.I., Lievens E., Malik S., Imholz N., Lebeer S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol. 2015;6:81. doi: 10.3389/fphys.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y.E., Wang Y., He Y., Ji Y., Wang L.P. Homogeneity of the vaginal microbiome at the cervix, posterior fornix, and vaginal canal in pregnant Chinese women. Microb. Ecol. 2015;69:407–414. doi: 10.1007/s00248-014-0487-1. [DOI] [PubMed] [Google Scholar]
- 15.Smith W.L., Hedges S.R., Mordechai E., Adelson M.E., Trama J.P. Cervical and vaginal flora specimens are highly concordant with respect to bacterial vaginosis-associated organisms and commensal Lactobacillus species in women of reproductive age. J. Clin. Microbiol. 2014;52:3078–3081. doi: 10.1128/JCM.00795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balle C., Lennard K., Dabee S., Barnabas S.L., Jaumdally S.Z. Endocervical and vaginal microbiota in South African adolescents with asymptomatic Chlamydia trachomatis infection. Sci. Rep. 2018;8:11109. doi: 10.1038/s41598-018-29320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim T.K., Thomas S.M., Ho M., Sharma S., Reich C.I. Heterogeneity of vaginal microbial communities within individuals. J. Clin. Microbiol. 2009;47:1181–1189. doi: 10.1128/JCM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anahtar M.N., Byrne E.H., Doherty K.E., Bowman B.A., Yamamoto H.S. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–976. doi: 10.1016/j.immuni.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brotman R.M., Shardell M.D., Gajer P., Tracy J.K., Zenilman J.M. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J. Infect. Dis. 2014;210:1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon B., Yi T.J., Perusini S., Gajer P., Ma B. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol. 2017;10:1310–1319. doi: 10.1038/mi.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Paola M., Sani C., Clemente A.M., Iossa A., Perissi E. Characterization of cervico-vaginal microbiota in women developing persistent high-risk human papillomavirus infection. Sci. Rep. 2017;7:10200. doi: 10.1038/s41598-017-09842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borgdorff H., Tsivtsivadze E., Verhelst R., Marzorati M., Jurriaans S. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 2014;8:1–13. doi: 10.1038/ismej.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimers L.L., Mehta S.D., Massad L.S., Burk R.D., Xie X. The cervicovaginal microbiota and its associations with human papillomavirus detection in HIV-infected and HIV-uninfected women. J. Infect. Dis. 2016;214:1361–1369. doi: 10.1093/infdis/jiw374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao W., Weng J., Gao Y., Chen X. Comparison of the vaginal microbiota diversity of women with and without human papillomavirus infection: a cross-sectional study. BMC Infect. Dis. 2013;13:271. doi: 10.1186/1471-2334-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J.E., Lee S., Lee H., Song Y.-M., Lee K. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One. 2013;8:e63514. doi: 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dareng E.O., Ma B., Famooto A.O., Akarolo-Anthony S.N., Offiong R.A. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol. Infect. 2016;144:123–137. doi: 10.1017/S0950268815000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh H.Y., Kim B.S., Seo S.S., Kong J.S., Lee J.K. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin. Microbiol. Infect. 2015;21:674 e671–679. doi: 10.1016/j.cmi.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Audirac-Chalifour A., Torres-Poveda K., Bahena-Roman M., Tellez-Sosa J., Martinez-Barnetche J. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitra A., MacIntyre D.A., Lee Y.S., Smith A., Marchesi J.R. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015;5:16865. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fettweis J.M., Brooks J.P., Serrano M.G., Sheth N.U., Girerd P.H. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology. 2014;160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X., Brown C.J., Abdo Z., Davis C.C., Hansmann M.A. Differences in the composition of vaginal microbial communities found in healthy Caucasian and Black women. ISME J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 32.Borgdorff H., van der Veer C., van Houdt R., Alberts C.J., de Vries H.J. The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lennard K., Dabee S., Barnabas S.L., Havyarimana E., Blakney A. Microbial composition predicts genital tract inflammation and persistent bacterial vaginosis in adolescent South African women. Infect. Immun. Dec 2017;86(1):e00410–e00417. doi: 10.1128/IAI.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Illumina: 16S Metagenomic Sequencing Library Preparation. Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System. 2013. https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf Available from: [Google Scholar]
- 36.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narrowe A.B., Albuthi-Lantz M., Smith E.P., Bower K.J., Roane T.M. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome. 2015;3:6. doi: 10.1186/s40168-015-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 42.Philip D. Computer program review: VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- 43.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 45.The Human Microbiome Project Consortium: structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aagaard K., Petrosino J., Keitel W., Watson M., Katancik J. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–60. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautam R., Borgdorff H., Jespers V., Francis S.C., Verhelst R. Correlates of the molecular vaginal microbiota composition of African women. BMC Infect. Dis. 2015;15:86. doi: 10.1186/s12879-015-0831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srinivasan S., Hoffman N.G., Morgan M.T., Matsen F.A., Fiedler T.L. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verstraelen H., Verhelst R., Claeys G., De Backer E., Temmerman M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macklaim J.M., Fernandes A.D., Bella J.M.D., Hammond J.-A., Reid G. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome. 2013;1:12. doi: 10.1186/2049-2618-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirmonsef P., Hotton A.L., Gilbert D., Burgad D., Landay A. Free glycogen in vaginal fluids is associated with Lactobacillus colonization and low vaginal pH. PLoS One. 2014;9 doi: 10.1371/journal.pone.0102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasan S., Morgan M.T., Fiedler T.L., Djukovic D., Hoffman N.G. Metabolic signatures of bacterial vaginosis. mBio. 2015;6 doi: 10.1128/mBio.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziklo N., Vidgen M.E., Taing K., Huston W.M., Timms P. Dysbiosis of the vaginal microbiota and higher vaginal kynurenine/tryptophan ratio reveals an association with Chlamydia trachomatis genital infections. Front. Cell Infect. Microbiol. 2018;8:1. doi: 10.3389/fcimb.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gajer P., Brotman R.M., Guoyun B., Sakamoto J., Schütte U.M.E. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiGiulio D.B., Callahan B.J., McMurdie P.J., Costello E.K., Lyell D.J. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. U.S.A. 2015;112:11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gosmann C., Anahtar M.N., Handley S.A., Farcasanu M., Abu-Ali G. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29–37. doi: 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Witkin S.S., Mendes-Soares H., Linhares I.M., Jayaram A., Ledger W.J. Influence of vaginal bacteria and D- and L-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer: implications for protection against upper genital tract infections. mBio. 2013;4 doi: 10.1128/mBio.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hickey R.J., Zhou X., Settles M.L., Erb J., Malone K. Vaginal microbiota of adolescent girls prior to the onset of menarche resemble those of reproductive-age women. mBio. 2015;6:1–14. doi: 10.1128/mBio.00097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ritu W., Enqi W., Zheng S., Wang J., Ling Y. Evaluation of the associations between cervical microbiota and HPV infection, clearance, and persistence in cytologically normal women. Cancer Prev. Res. (Phila) 2019;12:43–56. doi: 10.1158/1940-6207.CAPR-18-0233. [DOI] [PubMed] [Google Scholar]
- 61.Chao X.-P., Sun T.-T., Wang S., Fan Q.-B., Shi H.-H. Correlation between the diversity of vaginal microbiota and the risk of high-risk human papillomavirus infection. Int. J. Gynecol. Cancer. 2019;29 doi: 10.1136/ijgc-2018-000032. [DOI] [PubMed] [Google Scholar]
- 62.Srinivasan S., Liu C., Mitchell C.M., Fiedler T.L., Thomas K.K. Temporal variability of human vaginal bacteria and relationship with bacterial vaginosis. PLoS One. 2010;5:e10197. doi: 10.1371/journal.pone.0010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harwich M.D., Jr., Serrano M.G., Fettweis J.M., Alves J.M., Reimers M.A. Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics. 2012;13(Suppl 8):S4. doi: 10.1186/1471-2164-13-S8-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alves P., Castro J., Sousa C., Cereija T.B., Cerca N. Gardnerella vaginalis outcompetes 29 other bacterial species isolated from patients with bacterial vaginosis, using in an in vitro biofilm formation model. J. Infect. Dis. 2014;210:593–596. doi: 10.1093/infdis/jiu131. [DOI] [PubMed] [Google Scholar]
- 65.Zevin A.S., Xie I.Y., Birse K., Arnold K., Romas L. Microbiome composition and function drives wound-healing impairment in the female genital tract. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doorbar J., Quint W., Banks L., Bravo I.G., Stoler M. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 67.Hardy L., Jespers V., Dahchour N., Mwambarangwe L., Musengamana V. Unravelling the bacterial vaginosis-associated biofilm: a multiplex Gardnerella vaginalis and Atopobium vaginae fluorescence in situ hybridization assay using peptide nucleic acid probes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0136658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silva J., Cerqueira F., Medeiros R. Chlamydia trachomatis infection: implications for HPV status and cervical cancer. Arch. Gynecol. Obstet. 2014;289:715–723. doi: 10.1007/s00404-013-3122-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.