Abstract
Membrane-embedded transporters are crucial for the stability and performance of microbial production strains. Apart from engineering known transporters derived from model systems, it is equally important to identify transporters from nonconventional organisms that confer advantageous traits for biotechnological applications. Here, we transferred genes encoding fluoride exporter (FEX) proteins from three strains of early-branching anaerobic fungi (Neocallimastigomycota) to Saccharomyces cerevisiae. The heterologous transporters are localized to the plasma membrane and complement a fluoride-sensitive yeast strain that is lacking endogenous fluoride transporters up to 10.24 mM fluoride. Furthermore, we show that fusing an amino-terminal leader sequence to FEX proteins in yeast elevates protein yields, yet inadvertently causes a loss of transporter function. Adaptive laboratory evolution of FEX proteins restores fluoride tolerance of these strains, in one case exceeding the solute tolerance observed in wild type S. cerevisiae; however, the underlying molecular mechanisms and cause for the increased tolerance in the evolved strains remain elusive. Our results suggest that microbial cultures can achieve solvent tolerance through different adaptive trajectories, and the study is a promising step towards the identification, production, and biotechnological application of membrane proteins from nonconventional fungi.
Keywords: Neocallimastigomycota, Anaerobic gut fungi, Membrane proteins, Microbial engineering, Fluoride export proteins
Highlights
-
•
First report describing the heterologous production of functional ion transport proteins sourced from anaerobic gut fungi.
-
•
Codon-optimization enables production of functional, gut fungal membrane proteins in S. cerevisiae but not in E. coli.
-
•
Addition of an N-terminal leader peptide elevates membrane protein yields yet diminishes cellular activity.
-
•
Adaptive laboratory evolution restores cellular fluoride export activity in yeast to levels exceeding native tolerance.
1. Introduction
The performance of engineered microbial production strains can be dictated by membrane-embedded transporter proteins (Boyarskiy and Tullman-Ercek, 2015; Kell et al., 2015). For instance, product inhibition and toxicity is often mitigated via overexpression of drug exporters in biofuel-producing microbes (Dunlop et al., 2011; Foo et al., 2014; Teixeira et al., 2012). Directed evolution and rational protein engineering of known transporters offer a path to improve membrane protein function and ultimately strain performance (Farwick et al., 2014; Fisher et al., 2014; Foo and Leong, 2013; Mingardon et al., 2015). However, to increase the known transporter repertoire, it is valuable to mine transporters with potentially novel and improved functions from non-model organisms. Such approaches have identified transporters from various fungi and plants that were heterologously expressed in Saccharomyces cerevisiae to engineer strains capable of utilizing sucrose (Marques et al., 2018), xylose (Runquist et al., 2010; Saloheimo et al., 2007), and cellobiose (Sadie et al., 2011), and these heterologous transporters have been further improved by classic directed evolution strategies (Wang et al., 2016; Young et al., 2012).
Anaerobic gut fungi (Neocallimastigomycota) are an underexplored source of biotechnologically interesting enzymes and membrane transporters with direct application to lignocellulosic breakdown and hydrolysate fermentation. These early diverging fungi inhabit the digestive systems of large herbivores, where they secrete powerful cellulolytic enzymes that aid in the digestion of recalcitrant, lignin-rich plant fibers (Gordon and Phillips, 1998; Solomon et al., 2016; Theodorou et al., 1996). Because of their unusual habitat and diverse enzymatic secretome, it is likely that anaerobic fungi harbor specialized membrane transporters that allow them to utilize biohydrolysates and thrive in the rumen microbiome. This is corroborated by the recent genomic and transcriptomic sequencing of four different gut fungal strains, revealing a large repertoire of putative membrane-embedded receptors and transporters that are likely involved in their unusual lifestyle (Haitjema et al., 2017; Henske et al., 2017; Seppälä et al., 2016). However, owing to their complex native environment and handling requirements, anaerobic gut fungi are challenging to maintain in the laboratory (Haitjema et al., 2014), and their membrane proteome has not been functionally explored or tested. Apart from the expression of genes encoding ADP/ATP carriers from Neocallimastix sp. in Escherichia coli and S. cerevisiae more than a decade ago, no gut fungal membrane proteins have been heterologously produced or characterized (Haferkamp et al., 2002; van der Giezen et al., 2002; Voncken et al., 2002). Therefore, a wealth of potentially industrially relevant gut fungal membrane proteins remain untested, resulting in a knowledge gap regarding their relative activity and potential for use in model systems.
Here, as proof-of-principle, we show that bioinformatically identified fluoride transporters from three strains of anaerobic fungi can be transferred to a model yeast system and restore fluoride tolerance in a fluoride-sensitive knockout yeast. Fluoride is a ubiquitous xenobiotic that adversely affects several cellular processes (Barbier et al., 2010; Zuo et al., 2018). Consequently, most organisms require fluoride ion exporters to maintain intracellular fluoride concentrations below toxic levels (Baker et al., 2012; Barbier et al., 2010). In this study, we identify and produce functional fluoride exporter (FEX) proteins from Neocallimastix californiae, Piromyces finnis and Anaeromyces robustus in S. cerevisiae. Upon codon optimization, the putative FEX genes are highly expressed, and the membrane proteins are targeted to the endoplasmic reticulum and trafficked to the plasma membrane. The addition of an amino-terminal leader peptide improves total protein titers but reduces tolerance to fluoride. Subsequent evolutionary engineering of strains harboring heterologous FEX proteins not only restores, but improves both protein yields and fluoride tolerance through apparently divergent evolutionary paths. In one case, cellular fluoride tolerance is increased to a level greater than that observed in wildtype yeast. These results are the first to demonstrate heterologous production of a plasma membrane-embedded ion transporter from anaerobic gut fungi, and demonstrate the utility of these transporters for strain engineering.
2. Materials and methods
2.1. Identification and cloning of fluoride exporter (FEX) genes
Genes encoding FEX proteins were identified in transcriptomic and genomic data collected from 3 strains of anaerobic gut fungi (Haitjema et al., 2017; Seppälä et al., 2016; Solomon et al., 2016). The topology of the proteins was predicted using TMHMM (Krogh et al., 2001). Alignments were made using PSI/TM-Coffee and the ExPAsy Boxshade tool (Artimo et al., 2012; Chang et al., 2012). The genes were synthesized by Genewiz (South Plainfield, NJ, USA); the P. finnis and A. robustus FEX genes were codon optimized for expression in E. coli whereas the N. californiae FEX gene was synthesized both in a codon-optimized version and a non-codon optimized version. The genes were subsequently cloned into the yeast centromeric pYC2/CT vector, using restriction enzymes EagI and XhoI and T4 DNA ligase (New England Biolabs, Ipswich, MA, USA). In the pYC vector, the cloned gene is downstream of a GAL1 promoter and 3′-terminally fused to green fluorescent protein followed by a decahistidine tag. In a subset of pYC vectors, the gut fungal FEX gene is preceded by a 5′ Preproleader sequence, which, including a 3’ cloning scar, translates into the following peptide: MKVLIVLLAIFAALPLALAQPVISTTVGSAAEGSLDKREARPDV (Clements et al., 1991). The gene encoding Fex1p was amplified from the S. cerevisiae genome using primers 1 and 2 (Supplementary Table S3) and subcloned into pYC2/CT. DNA- and PCR-purification kits were from Zymo Research (Irvine, CA). Primers were from Eurofins MWG Operon, KY, USA. Plasmids were verified by Sanger sequencing (Genewiz, South Plainfield, NJ, USA).
2.2. Construction of FEX deletion mutants
The fex1:GSHU fex2Δ deletion strain was generated in the background of S. cerevisiae BJ5465 (MATa ura3-52 trp1 leu2Δ1 his3Δ200 pep4:HIS3 prb1Δ1.6R can1 GAL) (ATCC, 208289) using the delitto perfetto method, as described previously (Stuckey and Storici, 2013). The fex1:GSHU mutation was constructed by integrating the GSHU gene cassette into the FEX1 locus using primers 3 and 4. The fex2 knockout was constructed by integrating the CORE-Kp53 gene cassette into the FEX2 locus using primers 3 and 5 and subsequently removing the cassette using primers 6 and 7. Gene insertions and deletions were verified by PCR and Sanger sequencing (Genewiz, South Plainfield, NJ, USA).
2.3. Transformation and FEX expression in yeast
pYC2/CT plasmids were transformed into S. cerevisiae BJ5465 fex:GSHU fex2Δ using the lithium-acetate/PEG method (Gietz, 2014). Transformants were selected on SD-URA plates (20 g/L D-glucose, 6.7 g/L yeast nitrogen base (Becton, Dickinson & Co., Sparks, MD, USA), 5 g/L casamino acids (Becton, Dickinson & Co., Sparks, MD, USA), 40 mg/L Tryptophan, 40 mg/L Adenine, 16.25 g/L sodium citrate dihydrate, 4.2 g/L citric acid monohydrate, 20 g/L agar (Becton, Dickinson & Co., Sparks, MD, USA)). Genes were expressed essentially as described in Drew et al., with the following modifications (Drew et al., 2008). Briefly, individual colonies were used to inoculate 3 mL overnight cultures in SD-URA medium (20 g/L D-glucose, 6.7 g/L yeast nitrogen base (Becton, Dickinson & Co., Sparks, MD, USA), 5 g/L casamino acids (Becton, Dickinson & Co., Sparks, MD, USA), 40 mg/L Tryptophan, 40 mg/L Adenine, 16.25 g/L sodium citrate dihydrate, 4.2 g/L citric acid monohydrate) and grown at 30 °C, 225 rpm, for ∼16 h. The overnight cultures were used to seed an expression culture in SR-URA medium (same composition as SD-URA, except with 20 g/L raffinose instead of glucose) to a starting OD600 of 0.12. The culture was grown at 30 °C, 225 rpm for 6–7 h or until OD600 ∼0.6–1, when gene expression was induced with 2% w/v galactose for 22–24 h (30 °C, 225 rpm).
2.4. Confocal microscopy
0.1 ODU of induced yeast culture was centrifuged and resuspended in 1X PBS buffer. Samples were transferred to wells of Nunc Lab-Tek chambered slides coated with 0.1% (w/v) poly-L-lysine (Sigma-Aldrich, St. Lous, MO, USA). Microscopy was performed using an Olympus Fluoview 1000 Spectral confocal microscope with a 60x/NA 1.3 objective. GFP was visualized using a 488 nm laser and 530/20 nm bandpass filter. In order to more clearly display protein trafficking patterns, the laser intensity was adjusted for each strain to bring fluorescence intensities near saturating levels.
2.5. Fluorescence-activated cell sorting (FACS)
FEX gene expression was induced as described above. An equivalent of 0.2 ODU was pelleted at 3000 g for 30s and resuspended in 200 μL 1X PBS prior to FACS analysis. Scatter and fluorescence intensity were measured for approximately 60,000 singlet cells from each sample using a BD FACSARIA I flow cytometer. Fluorescence intensity from GFP was determined using a 488 nm laser and 530/30 nm bandpass filter.
2.6. In vivo fluoride tolerance assay and adaptive laboratory evolution
2 ODU of induced yeast culture was collected by centrifugation at 3000×g, 2 min, and resuspended in 40 μL 1x PBS buffer. 3 μL of the cell suspension (0.15 ODU) was streaked on an SG-URA plate (20 g/L galactose, 6.7 g/L yeast nitrogen base (Becton, Dickinson & Co., Sparks, MD, USA), 5 g/L casamino acids (Becton, Dickinson & Co., Sparks, MD, USA), 40 mg/L Tryptophan, 40 mg/L Adenine, 16.25 g/L sodium citrate dihydrate, 4.2 g/L citric acid monohydrate, 20 g/L agar (Becton, Dickinson & Co., Sparks, MD, USA)) supplemented with 50 μM NaF. Plates were incubated at 30 °C for approximately 48 h. For liquid assay, cultures were induced to trigger FEX expression as described above. The induced cultures were used to seed 3 mL cultures in SRG-URA medium (20 g/L raffinose, 20 g/L galactose, 6.7 g/L yeast nitrogen base (Becton, Dickinson & Co., Sparks, MD, USA), 5 g/L casamino acids (Becton, Dickinson & Co., Sparks, MD, USA), 40 mg/L Tryptophan, 40 mg/L Adenine, 16.25 g/L sodium citrate dihydrate, 4.2 g/L citric acid monohydrate)), supplemented with increasing concentrations of NaF (0–81.96 mMNaF), at a starting OD600 = 1. The cultures were incubated at 30 °C, 225 rpm, for 22–24 h. Growth was determined by measuring the optical density at 600 nm. For adaptive laboratory evolution, induced cultures were used to seed 3 mL of SRG-URA medium supplemented with 0.020 mM and 0.040 mM fluoride, at a starting OD600 = 1. The cultures were incubated at 30 °C, 225 rpm. After 16–48 h, OD600 was measured and the cultures were used to seed 3 mL SRG-URA medium supplemented with the highest tolerated concentration of fluoride at which growth could be detected, as well 3 mL SRG-URA medium supplemented with twice the highest tolerated concentration of fluoride, in both cases at a starting OD600 = 1. This serial seeding into increasingly higher concentrations of fluoride was repeated over a period of ∼21 days.
2.7. Plasmid extraction and curing of evolved strains
Plasmids were extracted from the evolved yeast strains using a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research (Irvine, CA)). Isolated plasmids were propagated and extracted from E. coli DH5α and verified by Sanger sequencing (Genewiz, South Plainfield, NJ, USA). In order to cure evolved yeast strains of plasmid, cells were plated onto SD plates (20 g/L D-glucose, 6.7 g/L yeast nitrogen base (Becton, Dickinson & Co., Sparks, MD, USA), 5 g/L casamino acids (Becton, Dickinson & Co., Sparks, MD, USA), 40 mg/L Tryptophan, 40 mg/L Adenine, 40 mg/L uracil, 16.25 g/L sodium citrate dihydrate, 4.2 g/L citric acid monohydrate, 20 g/L agar (Becton, Dickinson & Co., Sparks, MD, USA)) containing the auxotrophic marker (uracil) encoded in the pYC plasmid backbone. Colonies were replica-plated onto SD and SD-URA plates to identify those unable to grow in the absence of uracil due to loss of plasmid. The Ura‒ phenotype was verified by confirming growth or lack of growth after overnight incubation in 3 mL SD and SD-URA cultures, respectively.
3. Results and discussion
3.1. Identification and heterologous expression of anaerobic gut fungal FEX homologs
Genomic and transcriptomic data suggest that each of the early diverging fungal strains N. californiae, P. finnis, and A. robustus express one gene encoding a FEX protein homolog that is predicted to extrude fluoride ions from cells (Fig. 1A) (Haitjema et al., 2017; Seppälä et al., 2016). The gut fungal FEX proteins share ∼65% of their amino acid composition and are only ∼30% identical to the S. cerevisiae homologs Fex1p and Fex2p, which are virtually identical to each other (Supplementary Figure S1). Similar to other eukaryotic FEX proteins, the gut fungal proteins are predicted to have 9 transmembrane helices and two Fluc domains (PF02537) (Supplementary Figure S2) (Li et al., 2013; Rapp et al., 2006; Smith et al., 2015; Stockbridge et al., 2013).
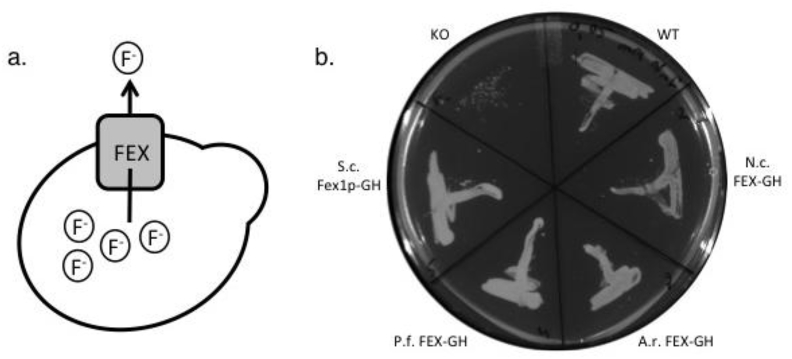
Fig. 1.
Fex proteins extrude fluoride from cells. (a) Cartoon depiction of a cell producing a fluoride transporter (FEX). (b) Anaerobic fungal FEX transporters restore fluoride tolerance in a yeast knockout (KO) strain. 0.15 ODU of induced yeast cultures were harvested and plated on SG-URA supplemented with 2% w/v galactose and 50 μM NaF. Streaked plates were incubated at 30 °C for approximately 48 h. KO = BJ5465 fex1:GSHU Δfex2 carrying the empty pYC vector. WT = BJ5465 carrying the empty pYC vector. N. c. FEX-GH = KO with pYC-N. californiae FEX-GH. A. r. FEX-GH = KO with pYC-A. robustus FEX-GH. P. f. FEX-GH = KO with pYC-P. finnis FEX-GH. S. c. Fex1p-GH = KO with pYC-S. cerevisiae Fex1p-GH.
Three gut fungal FEX genes identified from N. californiae, P. finnis and A. robusts transcriptomes were synthesized and cloned into the centromeric yeast vector pYC2/CT with a carboxy (C)-terminal GFP-His10 (GH) tag to determine whether heterologous FEX transporters could be produced in yeast. Compared to most model organisms, gut fungal genomes are extremely AT-rich (Haitjema et al., 2017; Nicholson et al., 2005; Youssef et al., 2013): the GC-content of the FEX genes is 25–28%, necessitating codon optimization prior to expression in S. cerevisiae (Supplementary Table S1). The genes were initially optimized for expression in E. coli, but upon induction the growth of the bacterial cultures was severely impaired implying that the FEX proteins were toxic to E. coli possibly due to improper targeting, folding, or post-translational modification of the eukaryotic membrane proteins (not shown) (Wagner et al., 2007).
SDS-PAGE analysis of yeast membranes suggests that the gut fungal FEX proteins are produced in the Pep4 protease-deficient yeast strain BJ5465 with minimal degradation or aggregation (Supplementary Figure S3). The expected sizes of the proteins, including the C-terminal GFP-His10 tag, are: 64.6 kDa (N. californiae FEX-GH), 63.5 kDa (P. finnis FEX-GH) and 63.9 kDa (A. robustus FEX-GH), but as often noted for hydrophobic, α-helical membrane proteins, the FEX proteins migrate somewhat faster than expected compared to conventional molecular weight standards (Rath et al., 2009).
3.2. Gut fungal FEX homologs complement a fluoride sensitive yeast strain
S. cerevisiae has two endogenous genes encoding FEX proteins (YOR39OW, Fex1p; and YPL279C, Fex2p); the proteins are virtually identical and both genes must be knocked out for yeast to exhibit fluoride sensitivity (Li et al., 2013). To assess whether the gut fungal proteins are functional in S. cerevisiae, we constructed a fex knock out (KO) strain (BJ5465 fex1:GSHUΔfex2), which fails to grow when exposed to μM concentrations of fluoride (Fig. 1B). The KO strain was subsequently transformed with plasmids encoding gut fungal FEX genes and plated on induction medium containing 50 μM NaF. All three gut fungal proteins restore fluoride tolerance in the KO strain (Fig. 1B).
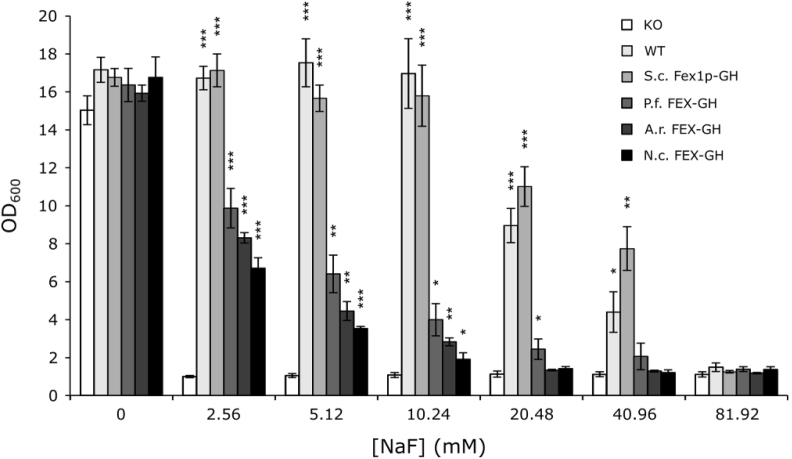
To quantify the degree of fluoride tolerance, we tested the viability of each FEX-producing yeast strain (both endogenous and heterologous) in liquid cultures with increasing concentrations of fluoride. Similar to previous reports, the KO strain exhibits high sensitivity to fluoride resulting in insignificant growth in the lowest NaF concentration tested (Li et al., 2013). In contrast, the wild type strain and the KO strain producing plasmid-borne S. cerevisiae Fex1p-GH demonstrate unimpaired growth in the presence of up to 10.24 mM NaF and attenuated growth in up to 40.96 mM NaF (Supplementary Table S2 and Fig. 2). The activity observed for S. cerevisiae Fex1p-GH contradicts a previous report in which a C-terminal GFP tag significantly reduced the activity of Fex1p (Smith et al., 2015). The activity observed in this work may reflect differences in gene copy number and promoter strength as Smith et al. expressed FEX1 from the native locus under the control of the native promoter, whereas our system utilizes the strong, inducible PGAL1 promoter and a centromeric plasmid with an expected copy number of 1–2 (Clarke and Carbon, 1980). As a result of increased copy number and transcription, functional protein yield may be increased in our system. All strains producing gut fungal FEX-GH proteins exhibit attenuated growth in the presence of 5.12–10.24 mM NaF demonstrating restoration of fluoride tolerance in the KO strain, albeit to levels below that of the WT strain.
Fig. 2.
KO-strains harboring anaerobic fungal FEX proteins show attenuated growth in up to 10.24 mM NaF. OD600values of indicated yeast strains were measured after ∼22 h growth in the presence of inducer (2% w/v galactose) and increasing concentrations of NaF. Wildtype (WT) and a KO strain expressing S. cerevisiae Fex1p-GH exhibit growth in up to 40.96 mM NaF. In contrast, KO strains expressing gut fungal homologs show reduced fluoride tolerance exhibiting growth in up to 10.24 mM NaF. Indicated is the standard error of the mean of three biological replicates. Statistically significant differences in growth between FEX-producing strains and the KO control strain were determined using a one-tailed Student's t-test: *** = p-value<0.001, ** = 0.001<p-value<0.01, * = 0.01<p-value<0.05. KO = BJ5465 fex1:GSHU Δfex2 carrying the empty pYC vector. WT = BJ5465 carrying the empty pYC vector. S. c. Fex1p-GH = KO with pYC S. cerevisiae Fex1p-GH. P. f. FEX-GH = KO with pYC P. finnis FEX-GH. A. r. FEX-GH = KO with pYC A. robustus FEX-GH. N. c. FEX-GH = KO with pYC N. californiae FEX-GH.
3.3. Gut fungal FEX homologs are trafficked to the plasma membrane
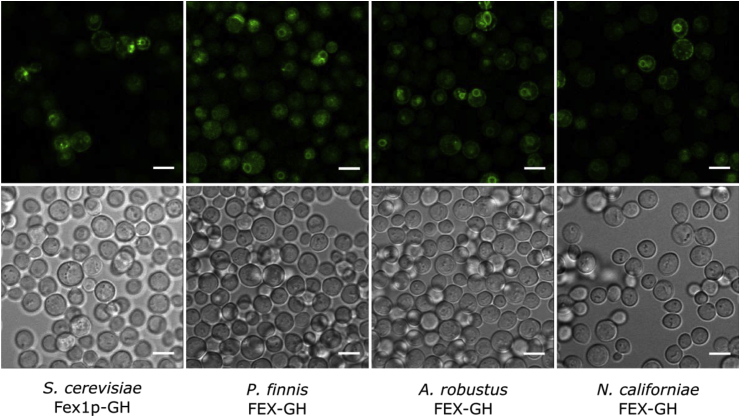
Yeast strains were analyzed using confocal microscopy and flow cytometry to assess the impacts of membrane protein trafficking and yield on the fluoride tolerance of the strains. In S. cerevisiae, targeting to cytoplasmic proteasomes (via the ERAD pathway) is often a consequence of protein misfolding and stacking in the endoplasmic reticulum (ER) (Delic et al., 2013). Confocal microscopy reveals that all gut fungal FEX-GH proteins are targeted to the perinuclear ER and the cell periphery in a pattern similar to the native S. cerevisiae Fex1p-GH (Fig. 3). Additionally, FEX-GH proteins may partially reside in the cortical ER as supported by the observation of punctuate fluorescent structures at the cell periphery. Although protein retention in the cortical ER is plausible, the cellular activity demonstrated in growth assays is indicative of properly trafficked, active FEX proteins at the cell membrane.
Fig. 3.
Confocal micrographs show localization of GFP-tagged FEX proteins produced in a S. cerevisiae knockout strain. Yeast knockout strains producing the indicated FEX homologs exhibit localization of fluorescent protein to the endoplasmic reticulum and plasma membrane signifying proper membrane protein folding and targeting. GFP (top) and transmission (bottom) micrographs were obtained in the absence of fluoride after ∼22–24 h induction of FEX gene expression in raffinose- and galactose-based medium. Microscopy was performed using an Olympus Fluoview 1000 Spectral confocal microscope equipped with a 60x/NA 1.3 objective. Laser intensities were adjusted for each strain to bring fluorescence intensities near saturating levels. All scale bars represent 5 μm.
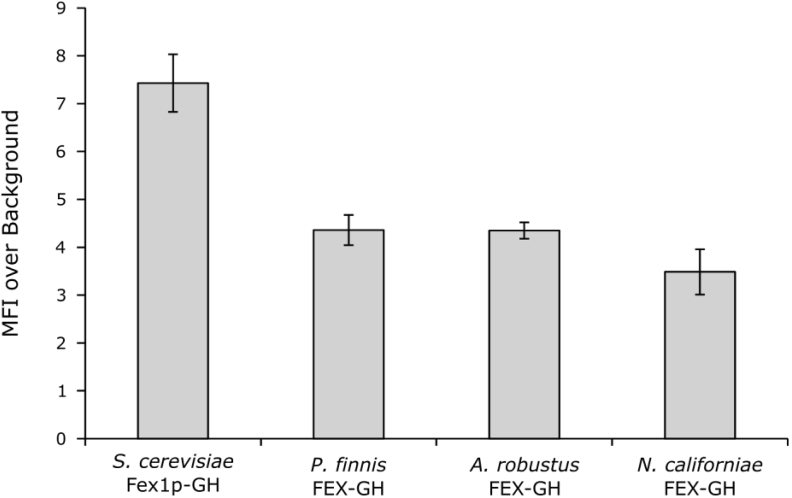
Analysis of recombinant S. cerevisiae strains using flow cytometry demonstrates appreciable yields of each gut fungal homolog (Fig. 4). All three strains exhibit mean fluorescence intensities (MFI) ∼4 fold over background autofluorescence, approximately half of the value observed upon expression of S. cerevisiae FEX1-GH from the same plasmid backbone. The decreased MFI is indicative of a reduction in total protein yield compared to S. cerevisiae Fex1p-GH. Given the similar trafficking patterns, the reduced protein yields may underlie the lower fluoride tolerance associated with strains producing gut fungal FEX proteins.
Fig. 4.
Knockout strains producing gut fungal FEX-GH proteins exhibit decreased total protein yield compared to a strain producing S. cerevisiae Fex1p-GH. Analysis of strains using fluorescence-activated cell sorting (FACS) provides a proxy measurement of total protein yield. Compared to a knockout strain expressing the native S. cerevisiae Fex1p-GH from a low copy vector, strains expressing gut fungal homologs from the same vector backbone display lower cellular mean fluorescence intensities (MFI). Strains were cultured and gene expression was induced in the absence of fluoride. Indicated is the standard deviation from the mean of three biological replicates. For each replicate, fluorescence data was collected for ∼40,000 single cells.
3.4. Addition of a synthetic leader peptide increases FEX protein levels but decreases cellular fluoride tolerance
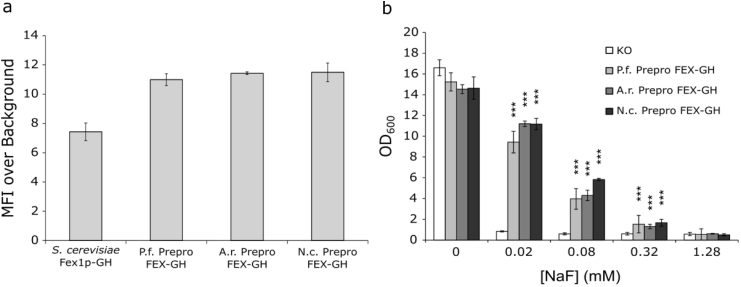
As both the total protein yields and fluoride tolerance were lower for strains producing gut fungal FEX proteins compared to the wild-type yeast Fex1p, we hypothesized that we could improve FEX-GH trafficking, functional yield, and concomitant fluoride tolerance through the addition of an amino-terminal leader peptide. The synthetic Prepro leader peptide, consisting of a consensus eukaryotic signal peptide followed by a linker sequence and a KEX2 protease site, has been successfully used in yeast to promote targeting and trafficking of recombinant proteins to the plasma membrane (Arnold et al., 1998; Clements et al., 1991; O'Malley et al., 2009; von Heijne, 1985). Accordingly, the gene encoding this peptide was cloned upstream of the FEX-GH coding sequences and the resulting constructs were transformed into the KO strain. Analysis of strains producing Prepro-FEX-GH using confocal microscopy suggests trafficking patterns similar to those producing Prepro-less proteins (Supplementary Figure S4). In all strains, fluorescence is localized to the perinuclear ER and cell periphery; however, the relative fluorescence intensities, particularly at the periphery, appear to be greater in these strains. Flow cytometry corroborates the observation of increased cellular fluorescence intensity; yeast strains producing amino-terminally modified FEX proteins exhibit ∼11-fold MFI over background representing ∼3-fold increases compared to strains producing unmodified FEX proteins (Fig. 5a).
Fig. 5.
Addition of Prepro leader peptide to FEX-GH proteins leads to improved total protein yields and reduced cellular fluoride tolerance. (a) Knockout strains producing Prepro-FEX-GH homologs in the absence of fluoride exhibit cellular MFI values significantly greater than the strain producing S. cerevisiae Fex1p-GH. The increase in cellular MFI relative to S. cerevisiae Fex1p-GH is indicative of improved protein yields, suggesting a beneficial effect arises due to addition of Prepro. (b) The strains producing Prepro-FEX-GH homologs exhibit low tolerances to NaF suggesting that the Prepro peptide is deleterious to protein function. Indicated is the standard deviation from the mean of three biological replicates. For each replicate in panel a, fluorescence data was collected for ∼40,000 single cells. In panel b, OD600values of KO strains expressing Prepro-FEX-GH were measured after ∼22–24 h growth in the presence of inducer (2% w/v galactose) and increasing concentrations of NaF. Statistically significant differences in growth between FEX-producing strains and the KO control strain were determined using a one-tailed Student's t-test: *** = p-value<0.001, ** = 0.001<p-value<0.01, * = 0.01<p-value<0.05. KO = BJ5465 fex1:GSHU Δfex2 carrying the empty pYC vector. P. f. FEX-GH = KO with pYC P. finnis FEX-GH. A. r. FEX-GH = KO with pYC A. robustus FEX-GH. N. c. FEX-GH = KO with pYC N. californiae FEX-GH.
While addition of the Prepro peptide improves total protein yields, cellular fluoride tolerance is considerably diminished, and we observe attenuated growth of all strains cultured in media containing up to 0.32 mM NaF (Supplementary Table S2 and Fig. 5b). The Prepro domain is expected to integrate into the ER membrane and undergo subsequent peptidase cleavage (Arnold et al., 1998; Clements et al., 1991; O'Malley et al., 2009; von Heijne, 1985), but SDS-PAGE analysis suggests that the leader peptide preceding the FEX proteins is not processed (the unprocessed Prepro peptide adds ∼4 kDa to the molecular weight of the protein) (Supplementary Figure S3). Therefore, we speculate that the Prepro peptide may interfere with insertion, folding, or function of the downstream protein. Alternatively, the uncleaved signal peptide may be indicative of increased protein retention in the ER, which may reduce functional protein yield in at least two ways. First, the unfolded protein response (UPR) may be induced or exacerbated leading to decreases in functional protein as reported for the overexpression of a heterologous P2 H+/adenosine transporter in S. cerevisiae (Griffith et al., 2003). Second, increased retention in the ER may reduce the amount of protein trafficked to the plasma membrane.
3.5. Evolutionary engineering significantly improves fluoride tolerance of prepro-FEX-GH-producing strains
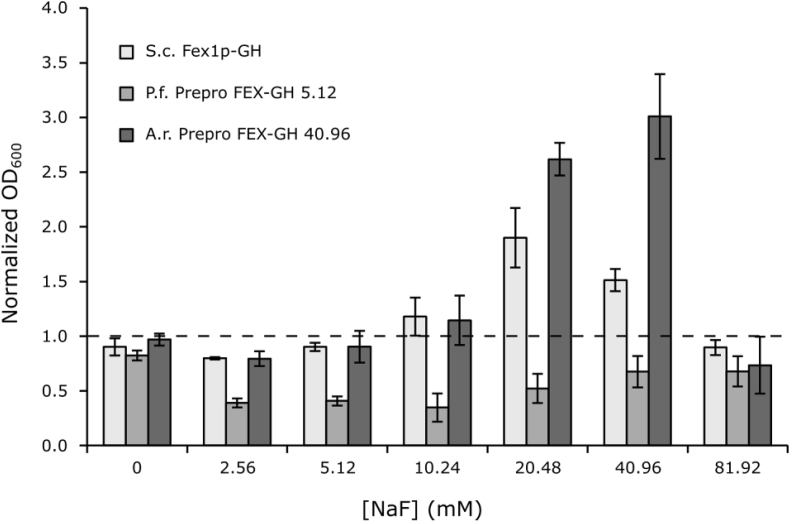
In order to improve cellular fluoride tolerance, we performed an evolutionary engineering experiment wherein strains producing gut fungal Prepro-FEX-GH were propagated in serial batch cultures containing increasing concentrations of fluoride. To challenge the strains to a greater extent and potentially identify mutations resulting in greater phenotypic improvements, the experiment was carried out using strains producing Prepro-FEX-GH, which exhibit high total protein yields but low fluoride tolerance. Starting with a culture supplemented with low (≪ 1 mM) concentrations of fluoride, we serially seeded cultures with increasingly higher concentrations of fluoride and quickly isolated two strains that are able to grow in significantly higher concentrations of fluoride than their parent strains: P. finnis Prepro-FEX-GH 5.12 exhibits attenuated growth in up to 5.12 mM NaF, a 16-fold improvement, whereas A. robustus Prepro-FEX-GH 40.96 tolerates 40.96 mM NaF, which is greater than the concentration toxic to wild type yeast and represents a 128-fold increase in fluoride tolerance (Supplementary Table S2 and Supplementary Figure S5). Remarkably, in 40.96 mM NaF, the evolved strain producing A. robustus Prepro-FEX-GH grows to an OD600 value ∼3-fold greater than WT yeast (Fig. 6). For comparison, a KO strain producing S. cerevisiae Fex1p-GH from the same vector backbone, thus providing a closer comparison with respect to promoter strength and gene copy number, grows to a final OD600 value ∼1.5-fold greater than WT yeast. These results demonstrate the potential value of novel transporters mined from anaerobic fungi and the capacity to evolve improved cellular function through relatively simple methods such as adaptive laboratory evolution.
Fig. 6.
Evolved Prepro-FEX-producing strains exhibit distinct changes in fluoride tolerance. Following adaptive evolution, the OD600 values of evolved strains were measured and normalized to the OD600 of a wildtype BJ5465 empty vector control grown at each NaF concentration. A KO strain producing S. cerevisiae Fex1p-GH provides a reference controlling for promoter strength and a closer comparison with respect to transporter gene copy number. This strain exhibits comparable or greater growth compared to WT yeast at all but the highest NaF concentrations tested. Similar to S. cerevisiae Fex1p-GH, A. robustus Prepro-FEX-GH 40.96 grows to cell densities comparable to, or greater than, WT at all but the highest NaF concentrations. In contrast, P. finnis Prepro-FEX-GH 5.12 grows to a lower concentration than WT yeast in all cases. OD600 values of evolved KO strains were measured after ∼24 h growth in the presence of inducer (2% w/v galactose) and increasing concentrations of NaF. Indicated is the standard error of the mean of three biological replicates. KO = BJ5465 fex1:GSHU Δfex2 carrying the empty pYC vector. S. c. Fex1p-GH = KO with pYC S. cerevisiae Fex1p-GH.P.f. FEX-GH = KO with pYC P. finnis FEX-GH. A. r. FEX-GH = KO with pYC A. robustus FEX-GH.
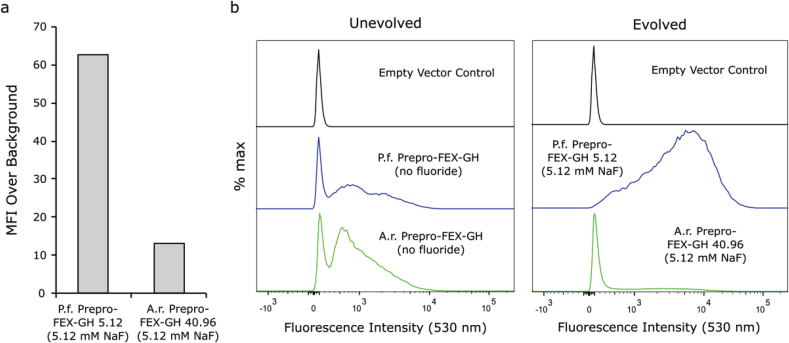
There are several paths that may lead to increased cellular fitness in the presence of fluoride: improved FEX activity, greater yields of functional FEX proteins, altered membrane composition reducing HF influx, etc. Analysis of P. finnis Prepro-FEX-GH 5.12 in the presence of 5.12 mM NaF using flow cytometry reveals a 5.7-fold increase in cellular MFI (Fig. 7a) as well as a significant improvement in phenotypic homogeneity (Fig. 7b). This phenotype is similar in the presence of 10.24 mM NaF (Supplementary Figure S6). These changes suggest that improved protein yield contributes to the apparent improvement in fluoride tolerance compared to the unevolved strain. SDS-PAGE of membranes containing P. finnis Prepro-FEX-GH 5.12 corroborates the elevated protein yields observed using flow cytometry (Supplementary Figures S3). As noted previously, the addition of the Prepro peptide may have exacerbated the cellular UPR possibly resulting in increased FEX proteolysis via the ERAD pathway. It is possible that the cells reduced UPR (e.g., through increased chaperone protein production), resulting in improved protein yield. Additional experiments quantifying the degree of UPR induced in each strain would provide insight on this hypothesis. By sequencing plasmids extracted from the evolved strains, we ruled out beneficial mutations in the FEX coding sequences or flanking regulatory regions as the cause for increased fluoride tolerance. Notably, transformation of plasmid extracted from P. finnis Prepro-FEX-GH 5.12 into the original knock out strain resulted in fluoride tolerance comparable to the unevolved P. finnis Prepro-FEX-GH strain, suggesting that the plasmid in the evolved strain is not the sole cause of the improved phenotype (data not shown). Furthermore, curing the evolved strain of plasmid and re-transforming this cured, evolved strain with the original P. finnis Prepro-FEX-GH plasmid did not result in any elevated fluoride tolerance. We conclude that while the fluoride tolerance of the evolved strain may be a result of genetic mutations outside the regions we screened, they may also be a result of reversible adaptation. For example, Schütz et al. used directed evolution to increase yields of functional G protein-coupled receptor in yeast; however, the yields decreased after repeated culturing or transformation of the receptor into fresh yeast cells (Schütz et al., 2016). In contrast, flow cytometric analysis of A. robustus Prepro-FEX-GH 40.96 in the presence of 5.12 mM NaF suggests an alternative path towards improved cellular fitness. These cells exhibit a 1.1-fold increase in MFI and greater phenotypic heterogeneity despite 2 orders of magnitude improvement in fluoride tolerance (Fig. 7). A similar phenotype is observed in the presence of 10.24 mM NaF (Supplementary Figure S6). Moreover, in the absence of fluoride, A. robustus Prepro-FEX-GH 40.96 exhibits basal fluorescence intensities. The low fluorescence intensity exhibited by A. robustus Prepro-FEX-GH 40.96 suggests decreased production of protein or post-translational proteolysis or misfolding of the GFP tag. Interestingly, in-gel fluorescence suggests that there is a low amount of full-length GFP-tagged protein and no free GFP could be detected (Supplementary Figure S3). Thus, the mechanism by which this strain has acquired its robust tolerance towards fluoride remains unknown.
Fig. 7.
Evolved Prepro-FEX-producing strains exhibit divergent fluorescence phenotypes. (a) P. finnis Prepro-FEX-GH 5.12 exhibits significantly improved MFI relative to its parent strain indicative of increased total protein yield. In contrast, the MFI of A. robustus Prepro-FEX-GH 40.96 is similar to that of the unevolved strain. (b) Histograms reveal phenotypic heterogeneity at the population level. While P. finnis Prepro-FEX-GH 5.12 exhibits complete homogeneity in the presence of NaF, A. robustus Prepro-FEX-GH 40.96 displays increased heterogeneity. Fluorescence data represents the mean fluorescence intensity corresponding to ∼40,000 single cells. KO = BJ5465 fex1:GSHU Δfex2 carrying the empty pYC vector. P. f. FEX-GH = KO with pYC P. finnis FEX-GH. A. r. FEX-GH = KO with pYC A. robustus FEX-GH.
4. Conclusions
In this work, we produce functional, recombinant gut fungal membrane proteins in S. cerevisiae to enhance ion tolerance. To our knowledge, this is the first study to demonstrate the industrial utility of membrane proteins from the early-diverging Neocallimastigomycota fungi. Further, gut fungal protein yields and cellular ion tolerance exceeded wildtype levels through fusion of an amino-terminal leader peptide combined with adaptive laboratory evolution. Taken together, these results show that heterologous transporters from anaerobic fungi improve the phenotype of an industrially relevant model microorganism. Moreover, this effort represents a critical first step towards the identification, characterization and, ultimately, utilization of membrane proteins from a class of eukaryotic microorganisms that specialize in biomass degradation and possess a wealth of biotechnologically promising yet completely uncharacterized membrane proteins. We foresee that extending these methods to highly valued membrane proteins (e.g., carbohydrate and putative drug transporters) mined from anaerobic fungi and other understudied microorganisms will accelerate strain engineering for a number of industrial applications.
Acknowledgements
The authors acknowledge funding support from the Office of Science (BER), U.S. Department of Energy (DE-SC0010352), the National Science Foundation (MCB-1553721), and the Institute for Collaborative Biotechnologies through grant W911NF-09-0001. The pGSHU and pCORE-Kp53 plasmids were a kind gift from Francesca Storici at the Georgia Institute of Technology. The pYC2/CT plasmid was a kind gift from Patrick Daugherty (UCSB). SS is the recipient of VILLUM Foundation's Young Investigator Programme grant VKR023128 and JIY is supported by a National Science Foundation Graduate Research Fellowship under grant No. 1650114. We acknowledge the use of the NRI-MCDB Microscopy Facility and the Spectral Laser Scanning Confocal supported by the Office of The Director, National Institutes of Health under Award #S10OD010610.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.mec.2019.e00091.
Contributor Information
Susanna Seppälä, Email: sseppala@ucsb.edu.
Justin I. Yoo, Email: justin_yoo@ucsb.edu.
Daniel Yur, Email: dyur@udel.edu.
Michelle A. O'Malley, Email: momalley@ucsb.edu.
Author Contributions
M.A.O., S.S., and J.I.Y. designed the research, analyzed data, and wrote the manuscript. S.S., J.I.Y., and D.Y. performed experiments.
Competing interests
The authors declare no competing interests.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Arnold C.E., Parekh R.N., Yang W., Wittrup K.D. vol 104. 1998. (Leader Peptide Efficiency Correlates with Signal Recognition Particle Dependence in Saccharomyces cerevisiae). [PubMed] [Google Scholar]
- Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., Grosdidier A., Hernandez C., Ioannidis V., Kuznetsov D., Liechti R., Moretti S., Mostaguir K., Redaschi N., Rossier G., Xenarios I., Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J.L., Sudarsan N., Weinberg Z., Roth A., Stockbridge R.B., Breaker R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier O., Arreola-Mendoza L., Del Razo L.M. Molecular mechanisms of fluoride toxicity. Chem. Biol. Interact. 2010;188:319–333. doi: 10.1016/j.cbi.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Boyarskiy S., Tullman-Ercek D. Getting pumped: membrane efflux transporters for enhanced biomolecule production. Curr. Opin. Chem. Biol. 2015;28:15–19. doi: 10.1016/j.cbpa.2015.05.019. [DOI] [PubMed] [Google Scholar]
- Chang J.-M., Di Tommaso P., Taly J.-F., Notredame C. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinf. 2012;13 doi: 10.1186/1471-2105-13-S4-S1. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Clements J.M., Catlin G.H., Price M.J., Edwards R.M. Secretion of human epidermal growth factor from Saccharomyces cerevisiae using synthetic leader sequences. Gene. 1991;106:267–271. doi: 10.1016/0378-1119(91)90209-T. [DOI] [PubMed] [Google Scholar]
- Delic M., Valli M., Graf A.B., Pfeffer M., Mattanovich D., Gasser B. The secretory pathway: exploring yeast diversity. FEMS Microbiol. Rev. 2013;37:872–914. doi: 10.1111/1574-6976.12020. [DOI] [PubMed] [Google Scholar]
- Drew D., Newstead S., Sonoda Y., Kim H., von Heijne G., Iwata S. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 2008;3:784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop M.J., Dossani Z.Y., Szmidt H.L., Chu H.C., Lee T.S., Keasling J.D., Hadi M.Z., Mukhopadhyay A. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farwick A., Bruder S., Schadeweg V., Oreb M., Boles E. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc. Natl. Acad. Sci. 2014;111:5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M. a., Boyarskiy S., Yamada M.R., Kong N., Bauer S., Tullman-Ercek D. Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli AcrB efflux pump to secrete the non-native substrate n-butanol. ACS Synth. Biol. 2014;3:30–40. doi: 10.1021/sb400065q. [DOI] [PubMed] [Google Scholar]
- Foo J.L., Jensen H.M., Dahl R.H., George K., Keasling J.D., Lee T.S., Leong S., Mukhopadhyay A. Improving microbial biogasoline production in Escherichia coli using tolerance engineering. mBio. 2014;5 doi: 10.1128/mBio.01932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo J.L., Leong S.S.J. Directed evolution of an E. coli inner membrane transporter for improved efflux of biofuel molecules. Biotechnol. Biofuels. 2013;6:81. doi: 10.1186/1754-6834-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. Methods in Molecular Biology. Clifton, N.J.; 2014. Yeast transformation by the LiAc/SS carrier DNA/PEG method; pp. 1–12. [DOI] [PubMed] [Google Scholar]
- Gordon G.L., Phillips M.W. The role of anaerobic gut fungi in ruminants. Nutr. Res. Rev. 1998;11:133–168. doi: 10.1079/NRR19980009. [DOI] [PubMed] [Google Scholar]
- Griffith D.A., Delipala C., Leadsham J., Jarvis S.M., Oesterhelt D. A novel yeast expression system for the overproduction of quality-controlled membrane proteins. FEBS Lett. 2003;553:45–50. doi: 10.1016/S0014-5793(03)00952-9. [DOI] [PubMed] [Google Scholar]
- Haferkamp I., Hackstein J.H.P., Voncken F.G.J., Schmit G., Tjaden J. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 2002;269:3172–3181. doi: 10.1046/j.1432-1033.2002.02991.x. [DOI] [PubMed] [Google Scholar]
- Haitjema C.H., Gilmore S.P., Henske J.K., Solomon K.V., de Groot R., Kuo A., Mondo S.J., Salamov A.A., LaButti K., Zhao Z., Chiniquy J., Barry K., Brewer H.M., Purvine S.O., Wright A.T., Hainaut M., Boxma B., van Alen T., Hackstein J.H.P., Henrissat B., Baker S.E., Grigoriev I.V., O'Malley M.A. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017;2:17087. doi: 10.1038/nmicrobiol.2017.87. [DOI] [PubMed] [Google Scholar]
- Haitjema C.H., Solomon K.V., Henske J.K., Theodorou M.K., O'Malley M. a. Anaerobic gut fungi: advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol. Bioeng. 2014;111:1471–1482. doi: 10.1002/bit.25264. [DOI] [PubMed] [Google Scholar]
- Henske J.K., Gilmore S.P., Knop D., Cunningham F.J., Sexton J.A., Smallwood C.R., Shutthanandan V., Evans J.E., Theodorou M.K., O'Malley M.A. Transcriptomic characterization of Caecomyces churrovis: a novel, non-rhizoid-forming lignocellulolytic anaerobic fungus. Biotechnol. Biofuels. 2017;10:305. doi: 10.1186/s13068-017-0997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D.B., Swainston N., Pir P., Oliver S.G. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015;33:237–246. doi: 10.1016/j.tibtech.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Li S., Smith K.D., Davis J.H., Gordon P.B., Breaker R.R., Strobel S. a. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19018. doi: 10.1073/pnas.1310439110. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques W.L., Mans R., Henderson R.K., Marella E.R., Horst J. Ter, Hulster E. de, Poolman B., Daran J.-M., Pronk J.T., Gombert A.K., van Maris A.J.A. Combined engineering of disaccharide transport and phosphorolysis for enhanced ATP yield from sucrose fermentation in Saccharomyces cerevisiae. Metab. Eng. 2018;45:121–133. doi: 10.1016/j.ymben.2017.11.012. [DOI] [PubMed] [Google Scholar]
- Mingardon F., Clement C., Hirano K., Nhan M., Luning E.G., Chanal A., Mukhopadhyay A. Improving olefin tolerance and production in E. coli using native and evolved AcrB. Biotechnol. Bioeng. 2015;112:879–888. doi: 10.1002/bit.25511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson M.J., Theodorou M.K., Brookman J.L. Molecular analysis of the anaerobic rumen fungus Orpinomyces - insights into an AT-rich genome. Microbiology. 2005;151:121–133. doi: 10.1099/mic.0.27353-0. [DOI] [PubMed] [Google Scholar]
- O'Malley M.A., Mancini J.D., Young C.L., McCusker E.C., Raden D., Robinson A.S. Progress toward heterologous expression of active G-protein-coupled receptors in Saccharomyces cerevisiae: linking cellular stress response with translocation and trafficking. Protein Sci. 2009;18:2356–2370. doi: 10.1002/pro.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp M., Granseth E., Seppälä S., von Heijne G. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 2006;13:112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- Rath A., Glibowicka M., Nadeau V.G., Chen G., Deber C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runquist D., Hahn-Hägerdal B., Rådström P. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels. 2010;3:5. doi: 10.1186/1754-6834-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadie C.J., Rose S.H., den Haan R., van Zyl W.H. Co-expression of a cellobiose phosphorylase and lactose permease enables intracellular cellobiose utilisation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2011;90:1373–1380. doi: 10.1007/s00253-011-3164-z. [DOI] [PubMed] [Google Scholar]
- Saloheimo A., Rauta J., Stasyk O.V., Sibirny A.A., Penttilä M., Ruohonen L. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl. Microbiol. Biotechnol. 2007;74:1041–1052. doi: 10.1007/s00253-006-0747-1. [DOI] [PubMed] [Google Scholar]
- Schütz M., Schöppe J., Sedlák E., Hillenbrand M., Nagy-Davidescu G., Ehrenmann J., Klenk C., Egloff P., Kummer L., Plückthun A. Directed evolution of G protein-coupled receptors in yeast for higher functional production in eukaryotic expression hosts. Sci. Rep. 2016;6:21508. doi: 10.1038/srep21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä S., Solomon K.V., Gilmore S.P., Henske J.K., O'Malley M.A. Mapping the membrane proteome of anaerobic gut fungi identifies a wealth of carbohydrate binding proteins and transporters. Microb. Cell Factories. 2016;15:212. doi: 10.1186/s12934-016-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.D., Gordon P.B., Rivetta A., Allen K.E., Berbasova T., Slayman C., Strobel S.A. Yeast Fex1p is a constitutively expressed fluoride channel with functional asymmetry of its two homologous domains. J. Biol. Chem. 2015;290:19874–19887. doi: 10.1074/jbc.M115.651976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon K.V., Haitjema C.H., Henske J.K., Gilmore S.P., Borges-Rivera D., Lipzen A., Brewer H.M., Purvine S.O., Wright A.T., Theodorou M.K., Grigoriev I.V., Regev A., Thompson D.A., O'Malley M.A. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science. 2016;351:1192–1195. doi: 10.1126/science.aad1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge R.B., Robertson J.L., Kolmakova-Partensky L., Miller C. A family of fluoride-specific ion channels with dual-topology architecture. Elife. 2013;2 doi: 10.7554/eLife.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey S., Storici F. Methods in Enzymology. first ed. Elsevier Inc; 2013. Gene knockouts, in vivo site-directed mutagenesis and other modifications using the delitto perfetto system in Saccharomyces cerevisiae. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C., Godinho C.P., Cabrito T.R., Mira N.P., Sá-Correia I. Increased expression of the yeast multidrug resistance ABC transporter Pdr18 leads to increased ethanol tolerance and ethanol production in high gravity alcoholic fermentation. Microb. Cell Factories. 2012;11:98. doi: 10.1186/1475-2859-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou M.K., Mennim G., Davies D.R., Zhu W.Y., Trinci A.P., Brookman J.L. Anaerobic fungi in the digestive tract of mammalian herbivores and their potential for exploitation. Proc. Nutr. Soc. 1996;55:913–926. doi: 10.1079/pns19960088. [DOI] [PubMed] [Google Scholar]
- van der Giezen M., Slotboom D.J., Horner D.S., Dyal P.L., Harding M., Xue G.-P., Embley T.M., Kunji E.R.S. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 2002;21:572–579. doi: 10.1093/emboj/21.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences - the limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Voncken F., Boxma B., Tjaden J., Akhmanova A., Huynen M., Verbeek F., Tielens A.G.M., Haferkamp I., Neuhaus H.E., Vogels G., Veenhuis M., Hackstein J.H.P. Multiple origins of hydrogenosomes: functional and phylogenetic evidence from the ADP/ATP carrier of the anaerobic chytrid Neocallimastix sp. Mol. Microbiol. 2002;44:1441–1454. doi: 10.1046/j.1365-2958.2002.02959.x. [DOI] [PubMed] [Google Scholar]
- Wagner S., Baars L., Ytterberg A.J., Klussmeier A., Wagner C.S., Nord O., Nygren P.-A., van Wijk K.J., de Gier J.-W. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteomics. 2007;6:1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang M., Yu C., Zhao H. Directed evolution of xylose specific transporters to facilitate glucose-xylose co-utilization. Biotechnol. Bioeng. 2016;113:484–491. doi: 10.1002/bit.25724. [DOI] [PubMed] [Google Scholar]
- Young E.M., Comer A.D., Huang H., Alper H.S. A molecular transporter engineering approach to improving xylose catabolism in Saccharomyces cerevisiae. Metab. Eng. 2012;14:401–411. doi: 10.1016/J.YMBEN.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Youssef N.H., Couger M.B., Struchtemeyer C.G., Liggenstoffer A.S., Prade R.A., Najar F.Z., Atiyeh H.K., Wilkins M.R., Elshahed M.S. The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 2013;79:4620–4634. doi: 10.1128/AEM.00821-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H., Chen L., Kong M., Qiu L., Lü P., Wu P., Yang Y., Chen K. Toxic effects of fluoride on organisms. Life Sci. 2018;198:18–24. doi: 10.1016/J.LFS.2018.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).