Respiratory viral infections, like IAV and respiratory syncytial virus (RSV) infections, impose great challenges to public health. Alveolar macrophages (AM) are lung-resident immune cells that play important roles in protecting the host against IAV and RSV infections. However, the underlying molecular mechanisms by which AM modulate host inflammation, disease development, and tissue recovery are not very well understood. Here we identify that PPAR-γ expression in AM is crucial to suppress pulmonary inflammation and diseases and to promote fast host recovery from IAV and RSV infections. Our data suggest that targeting macrophage PPAR-γ may be a promising therapeutic option in the future to suppress acute inflammation and simultaneously promote recovery from severe diseases associated with respiratory viral infections.
KEYWORDS: alveolar macrophage, inflammation, influenza, PPAR-γ, RSV, repair
ABSTRACT
Alveolar macrophages (AM) play pivotal roles in modulating host defense, pulmonary inflammation, and tissue injury following respiratory viral infections. However, the transcriptional regulation of AM function during respiratory viral infections is still largely undefined. Here we have screened the expression of 84 transcription factors in AM in response to influenza A virus (IAV) infection. We found that the transcription factor PPAR-γ was downregulated following IAV infection in AM through type I interferon (IFN)-dependent signaling. PPAR-γ expression in AM was critical for the suppression of exaggerated antiviral and inflammatory responses of AM following IAV and respiratory syncytial virus (RSV) infections. Myeloid PPAR-γ deficiency resulted in enhanced host morbidity and increased pulmonary inflammation following both IAV and RSV infections, suggesting that macrophage PPAR-γ is vital for restricting severe host disease development. Using approaches to selectively deplete recruiting monocytes, we demonstrate that PPAR-γ expression in resident AM is likely important in regulating host disease development. Furthermore, we show that PPAR-γ was critical for the expression of wound healing genes in AM. As such, myeloid PPAR-γ deficiency resulted in impaired inflammation resolution and defective tissue repair following IAV infection. Our data suggest a critical role of PPAR-γ expression in lung macrophages in the modulation of pulmonary inflammation, the development of acute host diseases, and the proper restoration of tissue homeostasis following respiratory viral infections.
IMPORTANCE Respiratory viral infections, like IAV and respiratory syncytial virus (RSV) infections, impose great challenges to public health. Alveolar macrophages (AM) are lung-resident immune cells that play important roles in protecting the host against IAV and RSV infections. However, the underlying molecular mechanisms by which AM modulate host inflammation, disease development, and tissue recovery are not very well understood. Here we identify that PPAR-γ expression in AM is crucial to suppress pulmonary inflammation and diseases and to promote fast host recovery from IAV and RSV infections. Our data suggest that targeting macrophage PPAR-γ may be a promising therapeutic option in the future to suppress acute inflammation and simultaneously promote recovery from severe diseases associated with respiratory viral infections.
INTRODUCTION
Acute respiratory viral infections, such as influenza A virus (IAV) and respiratory syncytial virus (RSV) infections, cause severe morbidity and mortality and are among leading causes of death in children and the elderly (1, 2). Particularly, IAV infection kills ∼500,000 people globally and up to 50,000 people in the United States each year (3). In addition to seasonal outbreaks, pandemic IAVs occasionally emerge and can cause catastrophic illness and widespread death. Current strategies for IAV prevention and treatment include yearly vaccination and antiviral drugs. However, frequent changes in the surface antigens of IAV due to antigenic shift and drift can allow IAV to escape antibody (Ab)-mediated immunity following vaccination (4, 5). Antiviral treatment is generally effective only during a very short time period early after IAV infection. Furthermore, many circulating IAV strains have developed resistance to the current antiviral drugs (6). Thus, there is an urgent need to better understand the pathophysiology and the protective immune responses to IAV infection for the development of future preventive and therapeutic means.
The disease pathogenesis associated with IAV infection results from a combination of the deleterious effects of virus replication and the host innate and adaptive immune responses associated with control and, ultimately, clearance of virus (7, 8). The major contribution of the host response to lung injury during IAV infection is exemplified by the immune-mediated lung inflammation and injury associated with infections with the 1918 pandemic IAV or the highly pathogenic H5N1 avian IAV. The inability to control the host responses in these infections results in excessive inflammatory cell infiltration into the lungs and overproduction of proinflammatory mediators (9, 10).
As important components of innate immunity, tissue macrophages and monocyte populations are heterogeneous multifunctional immune sentinel cells important in modulating tissue homeostasis, inflammation, injury, and repair (11–15). The main macrophage population in the respiratory tract is alveolar macrophages (AM), which play important roles in lung homeostasis and pulmonary antimicrobial defense (16, 17). Compared to other tissue macrophages, monocytes, and monocyte-derived cells, AM have distinct functions and phenotypes, which include high autofluorescence, low-level CD11b expression, and high-level expression of CD11c and Siglec F (16, 18). AM precursors develop mainly from fetal monocytes, which seed the lung prior to birth and massively expand and develop into mature macrophages in response to granulocyte-macrophage colony-stimulating factor (GM-CSF) and transforming growth factor β (TGF-β) after birth (18–20). A number of factors, including PPAR-γ, mTORC1, the phosphoinositide kinase PIKfyve, and L-plastin, were also recently shown to be important in AM development and function (19, 21–24). Interestingly, AM appear to be essential for protection against IAV and other respiratory viral infections (25–31). To this end, AM were identified as a major cellular source of the antiviral cytokines type I interferons (IFNs) (29). Furthermore, AM can phagocytize virus and virus-infected cells, clear cellular debris and exudates, and protect alveolar type I (ATI) cells from infection, thereby suppressing the development of lethal inflammation and injury during IAV infection (25–31). AM, particularly AM undergoing alternative polarization (M2), have also been implicated in the repair of damaged tissues following IAV infection (32). However, the underlying molecular mechanisms regulating the protective function of AM against respiratory viral infections remain to be fully elucidated.
PPAR-γ is a nuclear transcription factor, usually forming a heterodimer with RXR (retinoid X receptor), which recruits different coactivators or corepressors to form a complex binding to PPAR-responsive regulatory elements in the genome to modulate the expression of genes involved in adipogenesis, lipid metabolism, and inflammation (33). PPAR-γ has been shown to be vital for M2 polarization and the restriction of excessive production of inflammatory factors (34, 35), although the roles of PPAR-γ in regulating macrophage inflammatory responses against viral infections have not been explored. AM constitutively express high levels of PPAR-γ (19). Mice with loxP-flanked alleles encoding PPAR-γ (Ppargfl/fl) and with CD11c-driven expression of Cre recombinase (Cd11c-cre) that is efficiently expressed in fetal monocytes exhibit severe defects in the AM compartment, suggesting that PPAR-γ is essential for AM development from fetal monocytes (19). Interestingly, prophylactic or therapeutic treatment of mice with natural or synthetic ligands that activate PPAR-γ resulted in diminished host morbidity and mortality during IAV infection (36–40). However, the cellular and molecular mechanisms by which PPAR-γ agonists promote host protection against IAV infection have not been defined. In addition, the physiological and cell type-specific functions of PPAR-γ in response to endogenous ligands during IAV infection are currently unknown.
In this report, we demonstrate that PPAR-γ was downregulated in AM via IFNs following IAV infection. PPAR-γ repressed macrophage proinflammatory responses and promoted the expression of wound healing gene programs independent of M2 polarization, thereby modulating lung inflammation, host morbidity, and tissue repair. We further show that PPAR-γ expression and function in AM were likely important in dictating host diseases and recovery from respiratory viral infection.
RESULTS
IAV infection downregulates PPAR-γ expression in macrophages through IFNs.
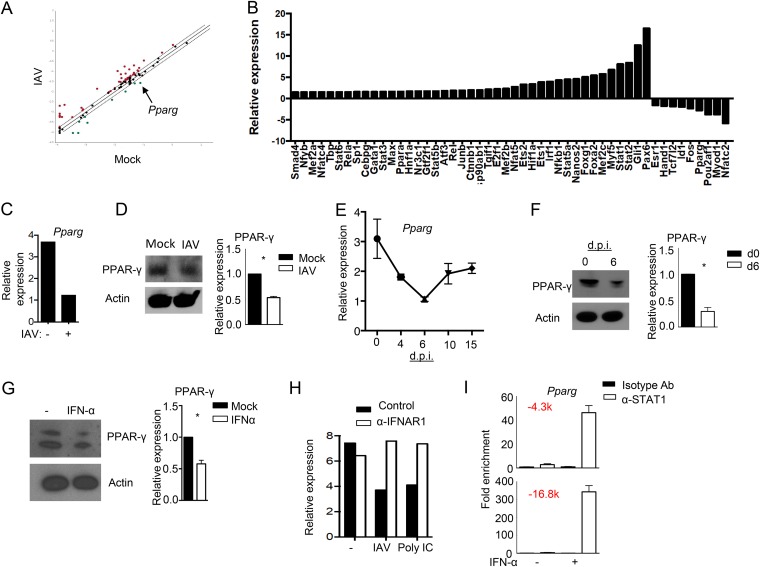
AM are important in regulating antiviral immunity and injury. However, the molecular mechanisms regulating AM responses to viral infection are still not well understood. To explore the transcriptional regulation of AM responding to viral infection, we infected wild-type (WT) AM with the IAV A/PR8/34 strain (here IAV) (multiplicity of infection [MOI] of 10) in vitro and then determined the expression levels of 84 transcription factors (TFs) following culture overnight using a Qiagen RT2-PCR array. We found that a number of TFs involved in antiviral innate immunity were upregulated, while several TFs, including Pparg, were downregulated in AM following IAV infection (Fig. 1A and B). Quantitative PCR (qPCR) results also showed that Pparg was downregulated in AM following IAV infection (Fig. 1C). Western blot analysis confirmed decreased PPAR-γ protein levels in IAV-infected AM (Fig. 1D). To determine whether IAV infection downregulates Pparg in AM in vivo, we sorted AM (CD11c+/Siglec F+) from the lungs of uninfected mice (day 0) or IAV-infected mice (4, 6, 10, or 15 days postinfection [dpi]) and examined Pparg expression by real-time reverse transcription-PCR (RT-PCR) (Fig. 1E). We found that IAV infection diminished Pparg expression in AM, particularly at 6 dpi (Fig. 1E). Western blot analysis confirmed that AM isolated from IAV-infected mice (6 dpi) exhibited decreased PPAR-γ protein levels compared to AM isolated from uninfected mice (Fig. 1F). IAV infection triggers the production of the antiviral cytokines type I IFNs by AM (6). We next examined whether type I IFNs were involved in the regulation of PPAR-γ expression in AM. We found that IFN-α treatment suppressed PPAR-γ expression in AM (Fig. 1G). Next, we infected AM with IAV and then blocked type I IFN signaling with the inclusion of an IFNAR1-blocking antibody in culture. We found that anti-IFNAR1 treatment abolished IAV-induced suppression of Pparg expression in AM (Fig. 1H). Similarly, anti-IFNAR1 treatment abolished poly(I·C)-induced suppression of Pparg expression in AM (Fig. 1H). Together, these data suggest that IAV infection inhibited PPAR-γ expression in AM through IFN signaling. Consistent with this notion, we found that STAT1 could bind to the Pparg locus following IFN-α treatment, suggesting that STAT1 activation following IFN signaling may directly modulate Pparg transcription in AM (Fig. 1I).
FIG 1.
IAV downregulates PPAR-γ expression in AM. (A) Comparison of the expression levels of 84 transcription factors in AM (isolated and pooled from at least 3 mice) with or without IAV infection overnight in vitro by using an RT2 profiler PCR array. Dotted line, fold cutoff of gene expression (1.5-fold); red dots, genes upregulated following IAV infection; green dots, genes downregulated following IAV infection. (B) List of up- or downregulated transcription factors in AM (isolated and pooled from at least 3 mice) following IAV infection in vitro overnight determined by using an RT2 profiler PCR array. (C) Relative expression levels of Pparg in AM (isolated and pooled from at least 3 mice) with or without IAV infection overnight in vitro determined by qRT-PCR. (D) Western blot analysis of PPAR-γ levels in AM (isolated and pooled from at least 3 mice) with or without IAV infection overnight. The bar graph represents the relative density of the PPAR-γ band pooled from three independent experiments. (E) Relative expression levels of Pparg in sorted AM isolated from noninfected (day 0) or IAV-infected mice at 4, 6, 10, or 15 dpi. (F) Western blot analysis of PPAR-γ expression ex vivo in AM (isolated and pooled from at least 3 mice) from noninfected (day 0) or IAV-infected (6 dpi) lungs. The bar graph represents the relative density of the PPAR-γ band pooled from three independent experiments. (G) Western blot analysis of PPAR-γ expression in AM (isolated and pooled from at least 3 mice) with or without IFN-α treatment overnight. The bar graph represents the relative density of the PPAR-γ band pooled from three independent experiments. (H) Relative expression of Pparg in AM (isolated and pooled from at least 3 mice) with or without IAV infection in the presence or absence of anti-IFNAR1 overnight in vitro determined by qRT-PCR. (I) STAT1 binding to the Pparg locus in AM following overnight IFN-α treatment in vitro determined by ChIP analysis (data pooled from >20 mice). Numbers in red are distances of the binding sites to the start codon. Data are representative of results from two to three independent experiments. *, P < 0.05.
PPAR-γ suppresses antiviral inflammation but does not regulate M2 genes following infection.
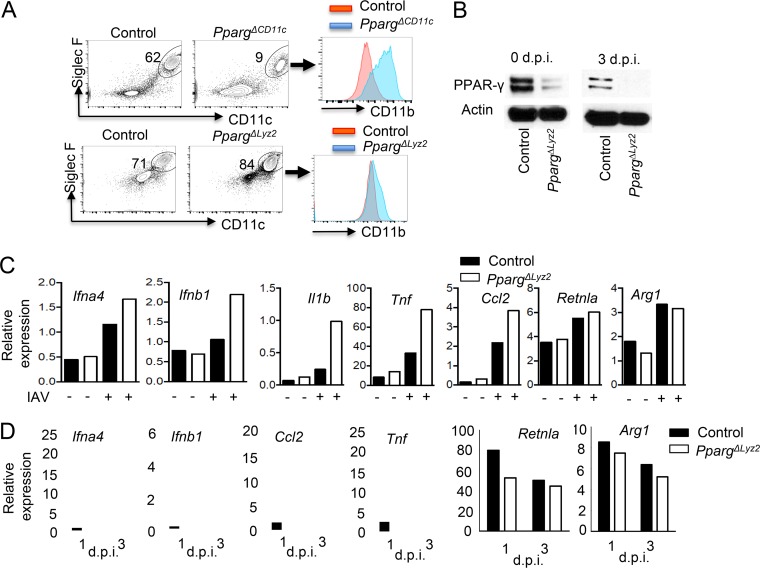
PPAR-γ is required for AM development because the deletion of PPAR-γ in CD11c+ cells (PpargΔCD11c) resulted in impaired AM generation (Fig. 2A) (19). However, compared to CD11c-cre, Lyz2-cre expression in fetal monocytes is incomplete (19). As a result, Lyz2-cre-driven PPAR-γ deficiency (PpargΔLyz2) in AM resulted in relatively normal AM development (Fig. 2A) (19). In comparison to the severe defects of AM development and maturation (evidenced by a dramatic increase of CD11b [19]) observed in PpargΔCD11c mice, PpargΔLyz2 mice had comparable percentages of AM and only slightly increased CD11b expression compared to those of control mice, suggesting that AM development and maturation were relatively normal in PpargΔLyz2 mice (Fig. 2A). Nevertheless, Lyz2-cre is able to mediate gene recombination in adult AM compartments, and AM from adult PpargΔLyz2 mice exhibited impaired PPAR-γ expression (Fig. 2B) (30). We therefore used AM from littermate control (Ppargfl/fl [WT]) or PpargΔLyz2 mice for our further analysis of the roles of PPAR-γ in regulating AM function during respiratory viral infections. We first isolated AM from uninfected control or PpargΔLyz2 mice and infected the AM with IAV in vitro (Fig. 1). Following infection, WT and PpargΔLyz2 AM showed relatively comparable levels of viability (data not shown). We then examined the expression of type I IFNs, inflammatory cytokines, and M2 genes in control or PPAR-γ-deficient AM following IAV infection. We found that PPAR-γ deficiency enhanced the expression of Ifna4, Ifnb1, Tnf, Il1b, and Ccl2 but did not affect the expression of Retnla (encoding RELM-α protein) and Arg1 (encoding Arginase 1 protein) (Fig. 2C). These data suggest that PPAR-γ suppressed AM antiviral and inflammatory responses but did not change macrophage polarization following IAV infection. We next infected control or PpargΔLyz2 mice with IAV and then sorted AM from the lungs of infected mice at 1 or 3 dpi. We found that PPAR-γ-deficient AM exhibited enhanced type I IFN and inflammatory gene expression but showed similar levels of Retnla and Arg1 expression compared to those of control AM at 3 dpi (Fig. 2D). These data suggest that PPAR-γ functioned to inhibit antiviral and inflammatory responses but did not regulate M2 polarization following IAV infection.
FIG 2.
PPAR-γ suppresses antiviral inflammation but does not regulate M2 genes following infection. (A) Airway AM percentages and CD11b expression on AM from control (Ppargfl/fl), PpargΔCD11c, and PpargΔLyz2 mice. (B) Western blot analysis of PPAR-γ expression in sorted AM (isolated and pooled from 2 to 3 mice) from control (Ppargfl/fl) or PpargΔLyz2 mice at 0 and 3 dpi. (C) qRT-PCR analysis of Ifna4, Ifnb1, Il1b, Tnf, Ccl2, Retnla, and Arg1 expression in AM (isolated and pooled from 3 mice) from control (Ppargfl/fl) or PpargΔLyz2 mice following IAV infection in vitro overnight. (D) Control or PpargΔLyz2 mice were infected with IAV, and Ifna4, Ifnb1, Tnf, Ccl2, Retnla, and Arg1 gene expression levels in AM (isolated and pooled from 2 to 3 mice) from control or PpargΔLyz2 mice were determined at days 1 and 3 postinfection. Data are representative of results from at least two independent experiments.
Myeloid PPAR-γ suppresses lung inflammation and host morbidity and mortality.
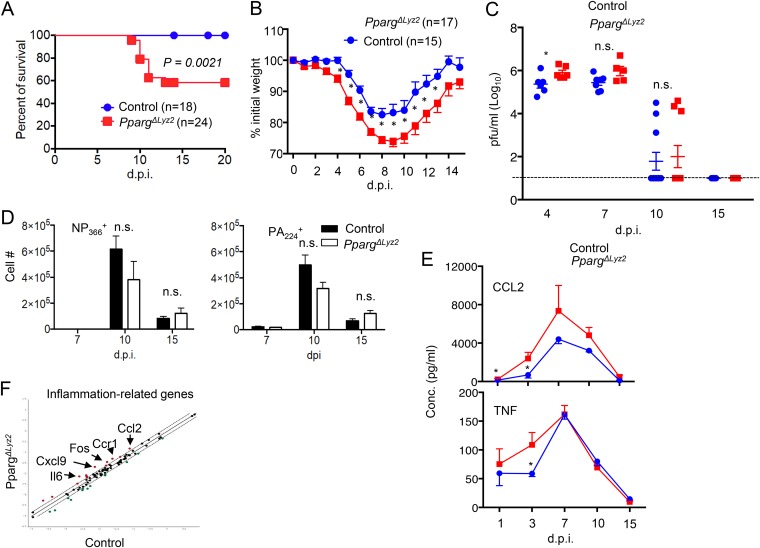
To explore PPAR-γ expression in macrophages in regulating host antiviral responses and disease development following IAV infection, we infected control or PpargΔLyz2 mice with IAV and examined host mortality and morbidity, viral replication, and inflammatory responses at different days postinfection. Compared to control mice, PpargΔLyz2 mice had enhanced host mortality and morbidity and delayed weight recovery following IAV infection (Fig. 3A and B). We examined the kinetics of IAV replication in the respiratory tract using a PFU assay and found that PpargΔLyz2 mice exhibited significantly increased virus titers in the early days following IAV infection (4 dpi) compared to control mice (Fig. 3C). However, PpargΔLyz2 mice had comparable viral titers at 7 dpi, and most of the mice cleared their infectious virus at around 10 dpi (3 out of 11 mice exhibited detectable viruses in control or PpargΔLyz2 bronchoalveolar lavage [BAL] fluid) (Fig. 3C). Thus, PpargΔLyz2 mice showed similar viral clearance kinetics as control mice, which suggests that the enhanced morbidity and mortality observed in PpargΔLyz2 mice were not merely due to the failure of viral clearance. Consistent with the viral clearance data, we found that PpargΔLyz2 mice exhibited comparable levels of IAV-specific CD8+ T cell responses (both H2db NP366–374 tetramer positive and H2db PA224–233 tetramer positive) at 7, 10, and 15 dpi (Fig. 3D).
FIG 3.
Myeloid PPAR-γ suppresses host mortality, morbidity, and pulmonary inflammation. Control or PpargΔLyz2 mice were infected with IAV. (A) Host mortality (percent survival). (B) Host morbidity (percent initial weight). (C) Airway IAV titers (PFU assay) determined at day 4, 7, 10, or 15 postinfection. (D) Lung IAV-specific PA224 and NP366 tetramer-positive CD8+ T cells at days 7, 10, and 15 postinfection. (E) CCL2 and TNF-α levels in BAL fluid, quantified by an ELISA at day 1, 3, 7, 10, or 15 postinfection. (F) Comparison of expression levels of 84 inflammation-related genes in lungs from control or PpargΔLyz2 mice at day 10 postinfection by using an RT2 profiler PCR array. Dotted line, 1.5-fold difference cutoff; red dots, genes upregulated in the lungs of PpargΔLyz2 mice; green dots, genes downregulated in the lungs of PpargΔLyz2 mice. Data are representative of results from at least two independent experiments (n = 3 to 6 mice per group) except for panels A to C (pooled data from 2 to 6 experiments). *, P < 0.05; n.s., not significant.
Next, we measured lung inflammatory cytokine (CCL2 and tumor necrosis factor alpha [TNF-α]) levels in the BAL fluid at different days following IAV infection to determine whether Pparg expression in myeloid cells regulates pulmonary inflammation. We found that PpargΔLyz2 mice had significantly higher CCL2 and TNF-α levels at early days after IAV infection (i.e., 1 or 3 dpi) (Fig. 3E). Notably, although the differences did not reach statistical significance, PpargΔLyz2 mice showed a trend of increased CCL2 protein levels in the BAL fluid at 7, 10, or 15 dpi, indicating that PpargΔLyz2 mice may have modestly increased pulmonary inflammation at later days postinfection. To this end, we used a more sensitive approach to examine inflammatory gene expression in the lungs of control or PpargΔLyz2 mice by a Qiagen RT2-PCR array. We found that lungs of PpargΔLyz2 mice exhibited altered expression of inflammation-related genes, including higher expression levels of a number of proinflammatory genes (such as Il6, Cxcl1, and Fos), at day 10 postinfection (Fig. 3F). Taken together, these data suggest that myeloid PPAR-γ deficiency leads to enhanced early viral replication, an exuberant inflammatory reaction, and increased severity of host sickness.
Myeloid PPAR-γ inhibits inflammation and morbidity during RSV infection.
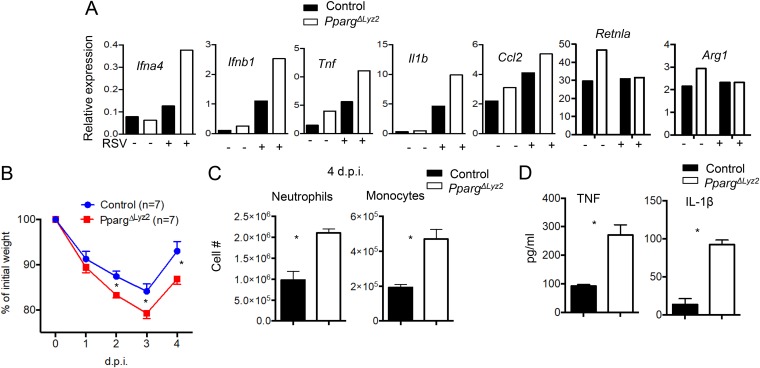
To examine whether PPAR-γ controls AM inflammatory responses to other virus infections, we infected AM isolated from control or PpargΔLyz2 mice with RSV, a virus that affects millions of children. Similar to what we observed following IAV infection (Fig. 2C), we found that PPAR-γ deficiency enhanced Ifna4, Ifnb1, Tnf, Il1b, and Ccl2 expression following RSV (strain line 19; MOI of 10) infection in vitro, suggesting that PPAR-γ also controls antiviral and inflammatory responses against RSV infection (Fig. 4A). We then infected control or PpargΔLyz2 mice with RSV (line 19) and examined host morbidity and lung inflammatory responses. We found that myeloid PPAR-γ deficiency increased weight loss following RSV infection (Fig. 4B). We also found that PpargΔLyz2 mice had enhanced inflammatory innate immune cells (neutrophils and monocytes) present in the lungs at 4 dpi (Fig. 4C), suggesting that PpargΔLyz2 mice had a higher level of pulmonary inflammation than control mice. Consistently, BAL fluid of PpargΔLyz2 mice had higher TNF-α and interleukin-1β (IL-1β) levels than those of control mice at 4 dpi (Fig. 4D). Thus, myeloid PPAR-γ was required for the suppression of exuberant host inflammation and exaggerated morbidity following RSV infection. These data suggest that macrophage PPAR-γ may restrict host disease development in a broad spectrum of respiratory viral infections.
FIG 4.
Myeloid PPAR-γ suppresses pulmonary inflammation during RSV infection. (A) qRT-PCR analysis of Ifna4, Ifnb1, Il1b, Tnf, Ccl2, Retnla, and Arg1 expression in AM (isolated and pooled from at least 3 mice) from control (Ppargfl/fl) or PpargΔLyz2 mice following RSV infection (MOI of 10) in vitro overnight. (B to D) Control (Ppargfl/fl) or PpargΔLyz2 mice were infected with RSV. (B) Host morbidity (percent initial weight) monitored daily. (C) numbers of lung neutrophils or monocytes at 4 dpi. (D) BAL fluid TNF and IL-1β concentrations determined by an ELISA at 4 dpi. Data are representative of results from at least two independent experiments (n = 3 to 4 mice per group) except for panel B (pooled data from 2 experiments). *, P < 0.05.
PPAR-γ expression in resident AM is likely required for controlling host disease development.
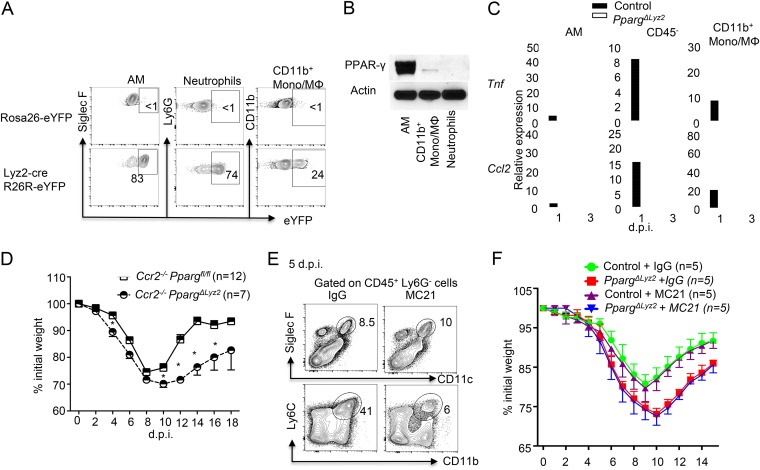
Lysozymes are widely expressed in myeloid cells, including neutrophils, monocytes, and macrophages. We crossed a Lyz2-cre mouse with a cre reporter strain R26R-eYFP mouse to examine Cre deletion in the myeloid compartment. In agreement with data from a previous report (35), we observed Lyz2-cre activity in the majority of alveolar macrophages and neutrophils and partially in CD11b+ monocytes/macrophages (Fig. 5A). Western blot analysis of sorted myeloid cell populations isolated from the lungs revealed that AM expressed high levels of PPAR-γ, and the lung CD11b+ monocyte/macrophage population expressed comparatively low levels of PPAR-γ (Fig. 5B), while neutrophils did not express detectable PPAR-γ, which is consistent with data from a previous report (35) (Fig. 5B). To explore the potential roles of PPAR-γ in regulating inflammation of AM, monocytes/monocyte-derived macrophages, and/or epithelial cells, we sorted AM, CD11b+ monocytes/macrophages, and CD45− cells (mainly epithelial cells) from IAV-infected lungs of control or PpargΔLyz2 mice at 1 and 3 dpi and examined inflammatory cytokine expression. We found that elevated Tnf and Ccl2 expression levels were observed mainly in AM but not in CD11b+ monocytes/macrophages or in CD45− cells (Fig. 5C). We next explored the relative contributions of PPAR-γ in AM and monocytes/monocyte-derived macrophages in controlling host disease development during IAV infection. To this end, we crossed PpargΔLyz2 mice to Ccr2−/− mice to block monocyte traffic to the infected lungs (40–43). We found that, compared to Ccr2−/−/Ppargfl/fl mice, Ccr2−/−/PpargΔLyz2 mice lost more weight and exhibited delayed recovery (Fig. 5D), suggesting that enhanced disease development in PpargΔLyz2 mice is independent of monocytes or monocyte-derived cells. We next assessed whether treatment of anti-CCR2 (monoclonal antibody [mAb] MC21), which selectively depletes recruiting monocytes (44), could affect host morbidity in PpargΔLyz2 mice. As reported previously (43), MC21 treatment greatly decreased monocyte infiltration into the lung (Fig. 5E). However, MC21 treatment did not significantly alter host weight loss in either control or PpargΔLyz2 mice (Fig. 5F), again suggesting that monocytes are dispensable for phenotypes observed in PpargΔLyz2 mice following IAV infection. Taken together, these data suggest that PPAR-γ expression in AM, rather than in monocytes or monocyte-derived cells, is probably responsible for the restriction of exaggerated pulmonary inflammation and the suppression of the development of severe diseases following respiratory viral infection.
FIG 5.
PPAR-γ expression in resident alveolar macrophages is likely required for the suppression of host morbidity. (A) Lyz2-cre gene recombination in AM, neutrophils, and CD11b+ monocytes/macrophages reported as percent enhanced yellow fluorescent protein (eYFP) expression following crossing with R26R-eYFP reporter mice. (B) Western blot analysis of PPAR-γ protein expression in sorted AM, CD11b+ monocytes/macrophages, and neutrophils in the lungs of naive WT mice (pooled from 3 mice). (C) Tnf and Ccl2 expression in the indicated cell populations in the lungs of control (Ppargfl/fl) or PpargΔLyz2 mice at days 1 and 3 postinfection. (pooled from 2 to 3 mice per group). (D) Ccr2−/− Ppargfl/fl and Ccr2−/− PpargΔLyz2 mice were infected with IAV. Host morbidity (percent initial weight) was monitored. (E) WT mice were infected with IAV and treated with control IgG or mAb MC21. Percentages of lung AM (top panel) and monocytes (bottom panel) in CD45+ Ly6G− cells at 5 dpi. (F) Control (Ppargfl/fl) and PpargΔLyz2 mice were infected with IAV and treated with control IgG or MC21 mAb. Host morbidity (percent initial weight) was monitored. Data are representative of results from at least two to three independent experiments, except for panels C (pooled data from 3 experiments) and D. *, P < 0.05.
Macrophage PPAR-γ promotes tissue repair.
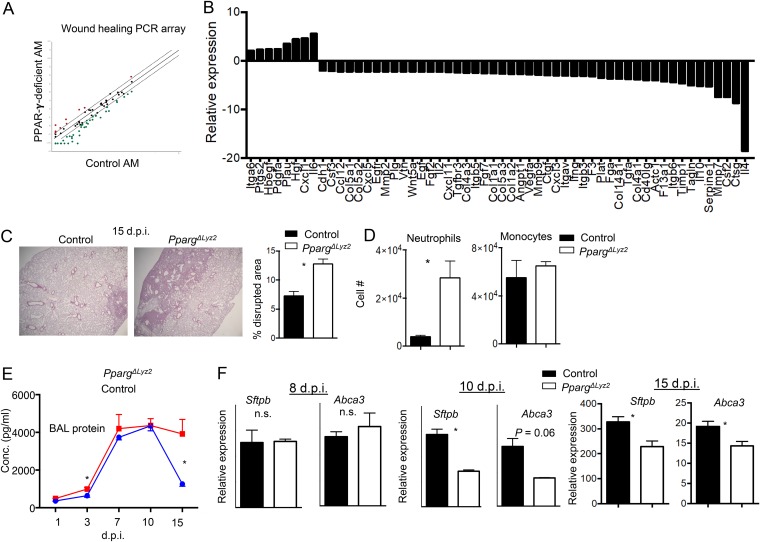
Following the clearance of IAV, the inflammatory responses in the lung resolve, and the damaged tissue undergoes a repair process to restore normal tissue homeostasis. AM are thought to be involved in the tissue repair process following lung injury (32). We therefore examined whether PPAR-γ affects AM tissue repair function. To this end, we isolated control or PPAR-γ-deficient AM from WT or PpargΔLyz2 mice and performed a Qiagen RT2-PCR array to determine wound healing gene expression. We found that PPAR-γ deficiency resulted in impaired expression of large numbers of wound-healing-related genes, including epithelial and endothelial growth factors such as Vegf, Egf, and Fgf7 (Fig. 6A and B). Levels of a number of factors involved in tissue remodeling, including Mmp7, Mmp9, and Timp1, were also decreased in PPAR-γ-deficient AM (Fig. 6A and B). These data suggested that PPAR-γ expression is important in regulating wound healing and tissue repair functions of AM.
FIG 6.
Macrophage PPAR-γ modulates inflammation resolution and tissue repair. (A) Comparison of the expression levels of 84 wound healing genes of AM isolated (pooled from 3 mice) from uninfected control or PpargΔLyz2 mice in vitro. Dotted line, fold cutoff of gene expression (1.5-fold); red dots, genes upregulated in PPAR-γ-deficient AM; green dots, genes downregulated in PPAR-γ-deficient AM. (B) List of up- or downregulated wound healing genes in AM (pooled from 3 mice) from control or PpargΔLyz2 mice determined by using an RT2 profiler PCR array. (C to F) Control (Ppargfl/fl) or PpargΔLyz2 mice were infected with IAV (n = 3 to 4). (C) H&E staining of lung sections of control or PpargΔLyz2 mice at day 15 postinfection. (Left) Representative images; (right) quantification of percentages of inflamed and disrupted alveolar areas in the lungs of control (Ppargfl/fl) and PpargΔLyz2 mice. (D) BAL fluid neutrophil or monocyte numbers enumerated at 15 dpi. (E) BAL fluid total protein concentrations determined at 1, 3, 7, 10, or 15 dpi. (F) Sftpb and Abca3 gene expression in lungs from control or PpargΔLyz2 mice at 8, 10, or 15 dpi. Data are representative of results from at least two independent experiments. *, P < 0.05.
Therefore, we examined whether PpargΔLyz2 mice had impaired tissue recovery in vivo following viral clearance. To this end, we examined lung histopathology by hematoxylin and eosin (H&E) staining of lung sections at 15 dpi, when infectious virus from IAV infection has been cleared (Fig. 3C). We found that PpargΔLyz2 mice still had significantly higher proportions of inflamed and/or damaged areas that were not properly repaired at day 15 postinfection, when mice already recovered most of their lost weight (Fig. 3B and Fig. 6C). To further explore the roles of myeloid PPAR-γ in regulating lung inflammation resolution and tissue repair, we first examined airway inflammatory cell content (monocytes and neutrophils [reflection of lung inflammatory resolution]). We found that PpargΔLyz2 mice exhibited higher neutrophil numbers at 15 dpi, suggesting that PpargΔLyz2 mice had impaired pulmonary inflammation resolution (Fig. 6D). We also measured total protein concentrations in the BAL fluid (reflection of endothelial/epithelial leakage) at different days following IAV infection and observed that PpargΔLyz2 mice had drastically higher protein levels in the BAL fluid than those of control mice at 15 dpi (Fig. 6E). These data indicate that PpargΔLyz2 mice had impaired inflammation resolution and decreased damage repair. In further support of this view, we examined alveolar type II (ATII) epithelial gene expression in the lungs of control and PpargΔLyz2 mice as a surrogate for tissue recovery at 8, 10, or 15 dpi. We found that levels of ATII-specific genes, Sftpb and Abca3, were comparable between control and PpargΔLyz2 lungs at 8 dpi. However, lungs of PpargΔLyz2 mice exhibited lower Sftpb and Abca3 expression levels than those of control mice at 10 or 15 dpi (Fig. 6F), indicating that lungs of infected PpargΔLyz2 mice had diminished ATII cell regeneration and lung recovery during viral clearance. Taken together, these data suggest that PPAR-γ promoted AM tissue repair function and that myeloid deficiency of PPAR-γ resulted in diminished inflammation resolution and impaired tissue recovery following IAV infection.
DISCUSSION
The transcriptional regulation of the lung macrophage responses against respiratory viral infections is largely undefined. Here we identify that PPAR-γ expression in AM is vital for their proper responses during both IAV and RSV infections. PPAR-γ is an anti-inflammatory transcription factor able to antagonize NF-κB-mediated cytokine production constitutively and in response to Toll-like receptor (TLR) ligand stimulation (45). Consistent with this notion, we show that PPAR-γ-deficient AM produced increased levels of both antiviral and proinflammatory cytokines in response to IAV and RSV infections. Notably, AM constitutively express high levels of PPAR-γ, which may help to maintain a tolerogenic environment in the lung during homeostasis. However, AM can also rapidly produce inflammatory cytokines following microbial challenge (17, 46). The downregulation of PPAR-γ in AM may help the AM rapidly respond to certain microbial challenges and provide beneficial functions under certain conditions. Nevertheless, the complete loss of PPAR-γ in macrophages caused an exaggerated release of inflammatory mediators and enhanced disease development in vivo following IAV and RSV infections. These data suggest that PPAR-γ counterregulates the pathogenic inflammatory responses in vivo and acts to ensure the proper function of lung macrophages during respiratory viral infections.
The differential functions of AM and recruited monocyte/macrophage populations during homeostasis and under disease conditions have only begun to be appreciated. During respiratory viral infections, circulating monocytes infiltrate the lungs in a CCR2-dependent manner and can give rise to exudate or inflammatory macrophages at the site of infection (42). These CCR2-dependent inflammatory monocytes and monocyte-derived cells have been associated with the development of immunopathology, although these cells also contribute to normal antiviral responses, as the blockage of their migration to the lung due to CCR2 inhibition or deficiency impaired and/or delayed host viral clearance during RSV and IAV infections (31, 40–42, 44). Notably, CCR2 deficiency or CCR2 blockade did not significantly change overall host morbidity in a PPAR-γ-sufficient or -deficient background in our experimental system compared to what was reported previously (31, 41), in which CCR2 deficiency significantly diminished host morbidity and mortality. Variations in infection schemes, virus stocks, and/or microbiota (46) may contribute to the different results observed. However, our results are supported by the findings of Aldridge et al. (40), in which Ccr2−/− mice exhibited similar morbidity and mortality as WT mice following IAV infection.
In contrast, lung-resident AM are often beneficial to the host during respiratory viral infections, as AM depletion impairs host antiviral responses with the concomitant development of severe lung injury during respiratory viral infections (25–31). However, AM release inflammatory mediators following viral infections and thus may contribute to the development respiratory inflammation and/or injury if their responses are not tightly regulated. Multiple lines of evidence presented in this study suggest that PPAR-γ expression in AM rather than in monocytes and/or monocyte-derived cells is important in controlling host inflammation and subsequent disease development. First, AM expressed high levels of PPAR-γ compared to monocytes and monocyte-derived cells. Furthermore, sorted AM rather than monocytes or monocyte-derived cells exhibited increased inflammatory responses. Finally, disruption of monocyte recruitment into the lungs by using anti-CCR2 or genetic CCR2 deletion did not majorly impact the outcome of IAV infection in WT and myeloid PPAR-γ-deficient mice, suggesting that PPAR-γ expression in monocytes may be dispensable for the regulation of the development of severe diseases following respiratory viral infection. Interestingly, PPAR-γ expression also regulated the wound healing function of AM and tissue recovery through the promotion of various growth factors and tissue remodeling factors. Notably, PPAR-γ deficiency did not result in decreased M2 gene expression in AM with or without IAV or RSV infection, suggesting that PPAR-γ may regulate AM repair function independent of M2 polarization. Thus, PPAR-γ is vital for the proper function of AM during respiratory viral infection by restricting their inflammatory features and simultaneously promoting their repair roles.
Type I IFNs are widely recognized as host-beneficial, antiviral cytokines. They lead to the transcription of IFN-stimulated genes that aim to eliminate the virus and prevent its spread by promoting an antiviral state in nearby cells (47). However, type I IFNs are also the key initiators of pulmonary inflammatory responses during respiratory viral infections, and thus, their actions must also be finely balanced to maximize viral clearance while inflicting minimal damage to the tissue (48). Indeed, the exaggerated production of type I IFNs has been implicated in the development of exuberant pulmonary inflammation and severe host morbidity and mortality following respiratory viral infections (44, 49, 50). In this report, enhanced type I IFN production was observed in PPAR-γ-deficient AM, but the absence of PPAR-γ in AM resulted in significantly increased viral titers 4 days following IAV infection, suggesting that the enhanced production of type I IFNs by PPAR-γ-deficient AM was not sufficient to diminish viral replication in the lungs. The exact reasons underlying this phenomenon warrant further investigation. Nevertheless, given the potential inflammatory function of type I IFNs, it is possible that the altered production of type I IFNs along with dysregulated inflammatory cytokine production in PPAR-γ-deficient AM contribute to the severe outcome of IAV infection in myeloid PPAR-γ-deficient mice.
In summary, our findings have uncovered critical roles of PPAR-γ in regulating inflammatory responses of AM, the development of acute host disease, and the proper restoration of tissue homeostasis following respiratory viral infections. Further studies are warranted to examine the therapeutic potential of modalities that can specifically modulate the expression of PPAR-γ in AM for the treatment of severe respiratory viral infections and their associated pathologies.
MATERIALS AND METHODS
Mice and infection.
WT C57BL/6 mice were purchased from the Jackson Laboratory. Lyz2-cre, CD11c-cre, Ppargfl/fl, R26R-eYFP, and Ccr2−/− mice were purchased from the Jackson Laboratory and bred in-house. PpargΔLyz2 mice were generated by crossing Ppargfl/fl mice with Lyz2-cre mice. PpargΔCD11c mice were generated by crossing Ppargfl/fl mice with CD11c-cre mice. Ccr2−/− Ppargfl/fl and Ccr2−/− PpargΔLyz2 mice were generated by crossing Ppargfl/fl or PpargΔLyz2 mice with Ccr2−/− mice. Lyz2-cre R26R-eYFP reporter mice were generated by crossing R26R-eYFP mice with Lyz2-cre mice. All mice were housed in a specific-pathogen-free environment. For IAV infection, the influenza A/PR8/34 virus strain (∼200 PFU/mouse) was diluted in fetal bovine serum (FBS)-free Dulbecco’s modified Eagle’s medium (DMEM) (Corning) on ice and inoculated into anesthetized mice through the intranasal route as described previously (51). Host mortality was determined based on the humane endpoint (more than 30% weight loss or moribund) or deaths before humane sacrifice. For RSV infection, RSV (strain line 19; ∼5 × 106 PFU/mouse) was diluted in FBS-free DMEM (Corning) on ice and inoculated into anesthetized mice through the intranasal route as described previously (52).
AM culture and infection in vitro.
AM were obtained from BAL fluid. Briefly, alveolar lavage fluids were pooled from BAL washes from 3 to 5 mice (phosphate-buffered saline [PBS] with 2 mM EDTA) and stored on ice. Red blood cell lysis was then performed in ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA [pH 7.2]) at room temperature (RT) for 2 min. Freshly isolated cells were rested in complete medium (RPMI 1640, 10% FBS, 1% penicillin-streptomycin) for 4 h at 37°C with 5% CO2. The nonadherent cells were discarded, and the plates were rinsed with warm PBS. For AM infection in vitro, seeded cells were infected with or without IAV PR8 or RSV line 19 at an MOI of 10, as indicated, for 1 h and then cultured overnight. For AM IFN treatment in vitro, 105 AM were plated in a 12-well plate and treated with 50 ng/ml IFN-α (BioLegend) or the vehicle overnight in the presence of recombinant GM-CSF to keep AM alive (10 ng/ml; BioLegend). Cell lysates were analyzed by quantitative RT-PCR (qRT-PCR) or Western blotting.
Quantitative RT-PCR.
mRNA from cultured AM (pooled from multiple mice), in vivo-sorted AM (pooled from multiple mice), or homogenates from individual lungs, as indicated, was isolated with a total RNA purification kit (Sigma) and treated with DNase I (Invitrogen). Random primers (Invitrogen) and Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) were used to synthesize first-strand cDNAs from equivalent amounts of RNA from each sample. RT-PCR was performed with Fast SYBR green PCR master mix (Applied Biosystems). qPCR was conducted in duplicates with QuantStudio3 (Applied Bioscience). Data were generated with the comparative threshold cycle (delta CT) method by normalizing values to the value for hypoxanthine phosphoribosyltransferase (HPRT). Sequences of primers used in the studies are provided as follows: TTCTGGTTCTCCGCTCTGTT and GTACATGAGGGGGATGATGG for Abca3, CAATGAAGAGCTGGCTGGTG and TGAGCATCCACCCAAATGAC for Arg1, GTCACCAAGCTCAAGAGAGAGGTC and CCTACAGAAGTGCTTGAGGTGGTT for Ccl2, CTCCGCCGGCTTCCTCCTCA and ACCTGGTTCATCATCGCTAATC for Hprt, TCCATCAGCAGCTCAATGAC and AGGAAGAGAGGGCTCTCCAG for Ifna4, TCCACCAGCAGACAGTGTT and CTTTGCACCCTCCAGTAATAGC for Ifnb1, GGGCCTCAAAGGAAAGAATC and TACCAGTTGGGGAACTCTGC for Il1b, TGCCAGTTTCGATCCGTAGA and ATGAATCCTTGGCCCTCTGA for Pparg, TGCCCTGCTGGGATGACTGCT and GGACAGTTGGCAGCAGCGGG for Retnla, CTGTGCCAAGAGTGTGAGGA and TTGGGGTTAATCTGGCTCTG for Sftpb, and CATGCGTCCAGCTGACTAAA and TCCCCTTCATCTTCCTCCTT for Tnf.
RT2 profiler PCR array.
Total RNA from lung tissue or AM was extracted as described above. An equal amount of total RNA was used for the synthesis of first-strand cDNA with a kit from Qiagen. First-strand cDNA was mixed with 2× Fast SYBR green master mix (Applied Bioscience) and water in a formula as directed in the manual. Twenty-five microliters of the mixture was added to each well of a 96-well plate provided by the manufacturer. The wells in the plate included different primers in each well to detect 84 target genes, housekeeping genes, and negative- and positive-control genes. qPCR was conducted with QuantStudio3 (Applied Bioscience). The obtained raw data were analyzed with software provided by Qiagen (accessible online on the Qiagen website). Briefly, uploading the raw data, designating the control group, selecting the housekeeping gene to normalize results, and calculating the relative expression level were performed step-by-step according to the manufacturer’s instructions.
Cell depletion.
For monocyte depletion, mice were treated intraperitoneally (i.p.) with anti-CCR2 antibody (clone MC21; 25 μg/mouse in 200 μl of PBS) (53) or control IgG daily from day 0 to day 6.
Lung histopathology.
Following euthanasia, mice were perfused with PBS (10 ml) via the right ventricle. Ten percent paraformaldehyde (PF) was then gently instilled into the lung, and the lung was left inflated for 1 min before excising and moving the lobe to 10% PF for 48 h, followed by transfer to ethanol (70%). Samples were shipped to the Mayo Clinic Histology Core Lab (Scottsdale, AZ), where they were embedded in paraffin, and 5-μm sections were cut for hematoxylin and eosin staining. To quantify the percent inflamed or disrupted alveolar area, H&E slides were scanned through the Aperio whole-slide scanning system and exported to image files. Computer-based image analysis was performed using ImageJ software (NIH, Bethesda, MD, USA). We first determined the total lung area by converting the image into grayscale, followed by red highlighting through adjustment of the threshold. For determination of the inflamed and disrupted areas, color images were split into single channels. We then used the green channel, highlighted the inflamed areas in red by adjusting the threshold, and measured the areas based on pixels. The percentages of disrupted and inflamed lung areas were calculated based on the ratio of highlighted disrupted areas to the total lung area in each lung section.
Western blot analysis.
The same numbers of cultured or fluorescence-activated cell sorter (FACS)-sorted AM were lysed in lysis buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, and 10% glycerol) with a protease inhibitor cocktail (Roche). The lysates were then separated by SDS-PAGE and transferred to an Immuno-Blot nitrocellulose membrane (Bio-Rad). The membranes were blocked with 5% nonfat milk in a buffer containing 20 mM Tris (pH 7.5), 0.5 M NaCl, and 0.05% Tween 20 (TBST) for 1 h at RT, followed by incubation with primary Ab against PPAR-γ (1:1,000; Cell Signaling Technology) or β-actin (1:5,000; Santa Cruz Biotechnology) overnight at 4°C. After washing with TBST buffer, membranes were incubated with goat anti-rabbit or anti-mouse secondary Ab (Promega). Peroxidase activity was detected with enhanced chemiluminescence (ECL).
Chromatin immunoprecipitation.
AM were obtained from the lungs of naive WT C57BL/6 mice, using anti-CD169 magnetic beads, as recommended by the manufacturer (Miltenyi Biotec). AM were cultured in complete medium supplemented with 10 ng/ml GM-CSF in the presence of 50 ng/ml IFN-α (BioLegend) or the vehicle overnight. The cells were then subjected to a chromatin immunoprecipitation (ChIP) assay as previously described (54). In brief, 8 × 106 AM were cross-linked for 10 min at 37°C by the addition of 1% freshly made formaldehyde. Fixed cells were pelleted at 4°C and washed with ice-cold PBS. The cells were lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1]) containing protease inhibitors (Roche) on ice for 10 min and sonicated to an average size of 200 to 500 bp. After sonication, samples were centrifuged at 13,000 rpm for 10 min at 4°C, and 5% of the sonicated cell extracts were saved as the input. The resulting whole-cell extract was incubated with protein A/G-agarose (Santa Cruz) for 1 h at 4°C. Precleared extracts were then incubated with 60 μl of protein A/G-agarose (Santa Cruz) for ChIP with 5 μg of the appropriate antibody overnight at 4°C. STAT1 ChIP antibody (clone D1K9Y) was obtained from Cell Signaling. After incubation overnight, beads were washed once with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), once with high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), once with LiCl wash buffer (10 mM Tris-HCl [pH 8.1], 1 mM EDTA, 250 mM LiCl, 1% NP-40), and twice with TE wash buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). DNA was eluted in freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3). Cross-links were reversed by overnight incubation with 5 M NaCl at 65°C. RNA and protein were digested using RNase A and proteinase K (Roche), respectively, and DNA was purified by using a Qiagen MinElute PCR purification kit according to the manufacturer’s instructions. The immunoprecipitated DNA was analyzed by quantitative real-time PCR and normalized relative to the input DNA amount. Primers were designed for a segment that was centered on the PPAR-γ coverage regions. Primers used in this study are as follows: TGGAATGAAAGAATCCTCCAA and GTTGGTGCCACATGGATTTT for Pparg −4.3 kilobases and GCAGATTTGTGCCAAGAACA and TGCAGCCGCTGAATAAATAC for Pparg −16.8 kilobases. Real-time PCR data are represented as fold changes over the control.
ELISA of BAL fluid cytokines.
Fifty microliters of each BAL fluid sample was analyzed by an enzyme-linked immunosorbent assay (ELISA) using commercially available kits for mouse IL-1β, CCL2, and TNF-α (BioLegend) according to the manufacturer’s protocol. A VersaMax microplate reader (Molecular Devices) was used for colorimetric quantification and analysis at a 450-nm wavelength.
BCA protein assay.
A bicinchoninic acid (BCA) protein assay kit was obtained from Thermo Scientific. Two microliters of each BAL fluid sample was used. A VersaMax microplate reader (Molecular Devices) was used for colorimetric quantification and analysis at a 570-nm wavelength.
Plaque assay.
IAV plaque assays were performed as described previously (55). Briefly, MDCK cells were grown in 6-well plates and incubated with a series of dilutions of BAL fluid for 1 h. The plates were then overlaid with low-melting-temperature agarose (0.6%) in minimal essential medium (MEM) with bovine serum albumin (BSA) and trypsin and cultured for 3 days in a 37°C incubator. Plates were then fixed with formaldehyde, and virus plaques were visualized by staining with neutral red.
FACS analysis.
Fluorescence-conjugated FACS Abs were purchased from BioLegend, BD Biosciences, or eBioscience. Ab clones are provided. We defined the following cell populations based on cell surface markers: AM (CD11c+ Siglec F+ CD11blow), neutrophils (CD11b+ Ly6G+), the total CD11b+ monocyte/macrophage population (Ly6G− Siglec F− CD11b+), monocytes (Ly6G− Siglec F− CD11b+ Ly6C+), NP366 tetramer-positive cells (CD8+ NP366-tet+), and PA224 tetramer-positive cells (CD8+ PA224-tet+). Samples were collected on a FACS Attune or FACS Attune NXT flow cytometer (Life Technologies) and analyzed using FlowJo software (TreeStar).
Statistical analysis.
Data are means ± standard errors of the means (SEM) of values from individual mice (in vivo experiments). Unpaired two-tailed Student’s t test (two-group comparison), multiple t tests (weight loss), or a log rank test (survival study) was used to determine statistical significance by using GraphPad Prism software. We consider P values of <0.05 to be significant.
ACKNOWLEDGMENTS
We thank Thomas Braciale and Khashayarsha Khazaie for critical readings of the manuscript. We thank the NIH tetramer facility for providing IAV-specific tetramers and the Mayo flow cytometry core for help with cell sorting.
This work was supported by U.S. National Institutes of Health grants (R01 HL126647, AG047156, AI112844, and R21 AI099753 to J.S.; R01 AI095282 and AI129241 to M.H.K.; R01 HL62150 to A.H.L.; and T32 AG049672 to N.P.G.), a Kogod Aging Center high-risk pilot grant to J.S., and a Huvis Foundation grant to A.H.L.
REFERENCES
- 1.Openshaw PJM, Chiu C, Culley FJ, Johansson C. 2017. Protective and harmful immunity to RSV infection. Annu Rev Immunol 35:501–532. doi: 10.1146/annurev-immunol-051116-052206. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H, Thompson WW, Viboud CG, Ringholz CM, Cheng PY, Steiner C, Abedi GR, Anderson LJ, Brammer L, Shay DK. 2012. Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993–2008. Clin Infect Dis 54:1427–1436. doi: 10.1093/cid/cis211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 4.Doherty PC, Turner SJ, Webby RG, Thomas PG. 2006. Influenza and the challenge for immunology. Nat Immunol 7:449–455. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 5.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. 2006. Cell-mediated protection in influenza infection. Emerg Infect Dis 12:48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. 2017. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist 10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braciale TJ, Sun J, Kim TS. 2012. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Braciale TJ. 2013. Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol 3:425–429. doi: 10.1016/j.coviro.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha DQ, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, Hatta Y, Kim JH, Halfmann P, Hatta M, Feldmann F, Alimonti JB, Fernando L, Li Y, Katze MG, Feldmann H, Kawaoka Y. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 11.Lavin Y, Mortha A, Rahman A, Merad M. 2015. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol 15:731–744. doi: 10.1038/nri3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geissmann F, Mass E. 2015. A stratified myeloid system, the challenge of understanding macrophage diversity. Semin Immunol 27:353–356. doi: 10.1016/j.smim.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. 2010. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol 10:453–460. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginhoux F, Guilliams M. 2016. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Perdiguero EG, Geissmann F. 2016. The development and maintenance of resident macrophages. Nat Immunol 17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussell T, Bell TJ. 2014. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol 14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 17.Kopf M, Schneider C, Nobs SP. 2015. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol 16:36–44. doi: 10.1038/ni.3052. [DOI] [PubMed] [Google Scholar]
- 18.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. 2013. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. 2014. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol 15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, Buttgereit A, Lelios I, Utz SG, Cansever D, Becher B, Greter M. 2017. The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity 47:903.e4–912.e4. doi: 10.1016/j.immuni.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Schneider C, Nobs SP, Heer AK, Hirsch E, Penninger J, Siggs OM, Kopf M. 2017. Frontline science: coincidental null mutation of Csf2ralpha in a colony of PI3Kgamma−/− mice causes alveolar macrophage deficiency and fatal respiratory viral infection. J Leukoc Biol 101:367–376. doi: 10.1189/jlb.4HI0316-157R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng W, Yang J, Lin X, Shin J, Gao J, Zhong XP. 2017. Essential role of mTORC1 in self-renewal of murine alveolar macrophages. J Immunol 198:492–504. doi: 10.4049/jimmunol.1501845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki T, Ito K, Miyata H, Akira S, Kawai T. 2017. Deletion of PIKfyve alters alveolar macrophage populations and exacerbates allergic inflammation in mice. EMBO J 36:1707–1718. doi: 10.15252/embj.201695528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd EM, Zhou JY, Szasz TP, Deady LE, D’Angelo JA, Cheung MD, Kim AHJ, Morley SC. 2016. Alveolar macrophage development in mice requires L-plastin for cellular localization in alveoli. Blood 128:2785–2796. doi: 10.1182/blood-2016-03-705962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laidlaw BJ, Decman V, Ali MA, Abt MC, Wolf AI, Monticelli LA, Mozdzanowska K, Angelosanto JM, Artis D, Erikson J, Wherry EJ. 2013. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog 9:e1003207. doi: 10.1371/journal.ppat.1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider C, Nobs SP, Heer AK, Kurrer M, Klinke G, van Rooijen N, Vogel J, Kopf M. 2014. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog 10:e1004053. doi: 10.1371/journal.ppat.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HM, Lee YW, Lee KJ, Kim HS, Cho SW, van Rooijen N, Guan Y, Seo SH. 2008. Alveolar macrophages are indispensable for controlling influenza viruses in lungs of pigs. J Virol 82:4265–4274. doi: 10.1128/JVI.02602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purnama C, Ng SL, Tetlak P, Setiagani YA, Kandasamy M, Baalasubramanian S, Karjalainen K, Ruedl C. 2014. Transient ablation of alveolar macrophages leads to massive pathology of influenza infection without affecting cellular adaptive immunity. Eur J Immunol 44:2003–2012. doi: 10.1002/eji.201344359. [DOI] [PubMed] [Google Scholar]
- 29.Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, Akira S. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Cardani A, Boulton A, Kim TS, Braciale TJ. 2017. Alveolar macrophages prevent lethal influenza pneumonia by inhibiting infection of type-1 alveolar epithelial cells. PLoS Pathog 13:e1006140. doi: 10.1371/journal.ppat.1006140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goritzka M, Makris S, Kausar F, Durant LR, Pereira C, Kumagai Y, Culley FJ, Mack M, Akira S, Johansson C. 2015. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med 212:699–714. doi: 10.1084/jem.20140825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorski SA, Hufford MM, Braciale TJ. 2012. Recent insights into pulmonary repair following virus-induced inflammation of the respiratory tract. Curr Opin Virol 2:233–241. doi: 10.1016/j.coviro.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. 2013. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med 19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. 2007. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chawla A. 2010. Control of macrophage activation and function by PPARs. Circ Res 106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moseley CE, Webster RG, Aldridge JR. 2010. Peroxisome proliferator-activated receptor and AMP-activated protein kinase agonists protect against lethal influenza virus challenge in mice. Influenza Other Respir Viruses 4:307–311. doi: 10.1111/j.1750-2659.2010.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darwish I, Mubareka S, Liles WC. 2011. Immunomodulatory therapy for severe influenza. Expert Rev Anti Infect Ther 9:807–822. doi: 10.1586/eri.11.56. [DOI] [PubMed] [Google Scholar]
- 38.Cloutier A, Marois I, Cloutier D, Verreault C, Cantin AM, Richter MV. 2012. The prostanoid 15-deoxy-delta12,14-prostaglandin-j2 reduces lung inflammation and protects mice against lethal influenza infection. J Infect Dis 205:621–630. doi: 10.1093/infdis/jir804. [DOI] [PubMed] [Google Scholar]
- 39.Fedson DS. 2013. Treating influenza with statins and other immunomodulatory agents. Antiviral Res 99:417–435. doi: 10.1016/j.antiviral.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Aldridge JR Jr, Moseley CE, Boltz DA, Negovetich NJ, Reynolds C, Franks J, Brown SA, Doherty PC, Webster RG, Thomas PG. 2009. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci U S A 106:5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, Seeger W, Lohmeyer J. 2008. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med 205:3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. 2008. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol 180:2562–2572. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 43.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, Garcia-Sastre A, Stanley ER, Ginhoux F, Frenette PS, Merad M. 2013. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gautier EL, Chow A, Spanbroek R, Marcelin G, Greter M, Jakubzick C, Bogunovic M, Leboeuf M, van Rooijen N, Habenicht AJ, Merad M, Randolph GJ. 2012. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. J Immunol 189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Sastre A, Biron CA. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 48.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. 2011. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teijaro JR. 2015. The role of cytokine responses during influenza virus pathogenesis and potential therapeutic options. Curr Top Microbiol Immunol 386:3–22. doi: 10.1007/82_2014_411. [DOI] [PubMed] [Google Scholar]
- 50.Davidson S, Crotta S, McCabe TM, Wack A. 2014. Pathogenic potential of interferon alphabeta in acute influenza infection. Nat Commun 5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, Madan R, Karp CL, Braciale TJ. 2009. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med 15:277–284. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao S, Jiang L, Moser EK, Jewett LB, Wright J, Du J, Zhou B, Davis SD, Krupp NL, Braciale TJ, Sun J. 2015. Control of pathogenic effector T-cell activities in situ by PD-L1 expression on respiratory inflammatory dendritic cells during respiratory syncytial virus infection. Mucosal Immunol 8:746–759. doi: 10.1038/mi.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mack M, Cihak J, Simonis C, Luckow B, Proudfoot AE, Plachy J, Bruhl H, Frink M, Anders HJ, Vielhauer V, Pfirstinger J, Stangassinger M, Schlondorff D. 2001. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol 166:4697–4704. doi: 10.4049/jimmunol.166.7.4697. [DOI] [PubMed] [Google Scholar]
- 54.Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. 2013. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity 39:833–845. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huprikar J, Rabinowitz S. 1980. A simplified plaque assay for influenza viruses in Madin-Darby kidney (MDCK) cells. J Virol Methods 1:117–120. doi: 10.1016/0166-0934(80)90020-8. [DOI] [PubMed] [Google Scholar]