As a member of the Orthomyxoviridae family, the polymerase complex of the influenza C virus structurally resembles its influenza A and influenza B virus counterparts, but the nucleoprotein differs by possessing an extra C-terminal region. We have characterized this region in view of nuclear import and interaction with the importin α protein family. Our results demonstrate the functional significance of a previously uncharacterized region on Orthomyxoviridae nucleoprotein (NP). Based on this work, we propose that importin α binding to influenza C virus NP is regulated by a long bipartite nuclear localization signal. Since the sequence of influenza D virus NP shares high homology to that of the influenza C virus, this work will also shed light on how influenza D virus NP functions.
KEYWORDS: importin alpha, influenza, nuclear localization signal, NLS, nucleoprotein, NP
ABSTRACT
The influenza C virus (ICV) is a human-pathogenic agent, and the infections are frequently identified in children. Compared to influenza A and B viruses, the nucleoprotein of ICV (NPC) has an extended C-terminal region of which the functional significance is ill defined. We observed that the nuclear localization signals (NLSs) found on the nucleoproteins of influenza A and B virus subtypes are absent at corresponding positions on ICV. Instead, we found that a long bipartite nuclear localization signal resides at the extended C-terminal region, spanning from R513 to K549. Our experimental data determined that the KKMK motif within this region plays important roles in both nuclear import and polymerase activity. Similar to the influenza A viruses, NPC also binds to multiple human importin α isoforms. Taken together, our results enhance the understanding of the virus-host interaction of the influenza C virus.
IMPORTANCE As a member of the Orthomyxoviridae family, the polymerase complex of the influenza C virus structurally resembles its influenza A and influenza B virus counterparts, but the nucleoprotein differs by possessing an extra C-terminal region. We have characterized this region in view of nuclear import and interaction with the importin α protein family. Our results demonstrate the functional significance of a previously uncharacterized region on Orthomyxoviridae nucleoprotein (NP). Based on this work, we propose that importin α binding to influenza C virus NP is regulated by a long bipartite nuclear localization signal. Since the sequence of influenza D virus NP shares high homology to that of the influenza C virus, this work will also shed light on how influenza D virus NP functions.
INTRODUCTION
Influenza A, B, C, and D viruses (IAV, IBV, ICV, and IDV, respectively) belong to the Orthomyxoviridae family and contain segmented negative-sense RNA genomes. IAV and IBV mainly cause infection at the respiratory tract in humans and sometimes lead to a fatal outcome. Human infections of zoonotic IAV have also been continuously recorded, including the avian H5N1, H7N9, and swine-origin pandemic H1N1 viruses (1). Similar to IBV, ICV also mainly infects humans, but it may occasionally transmit to canine and swine (2). Although ICV causes only mild upper respiratory symptoms in adults, it was found that the virus can cause lower respiratory tract infection and prolong the duration of hospitalization in pediatric cases (3, 4).
Both IAV and IBV contain eight segments, and each of their RNA genomes is wrapped around a protein core which is composed of the trimeric polymerase complex PA/P3, PB1, PB2, and multiple copies of nucleoprotein (NP), forming a viral ribonucleoprotein complex (vRNP). It was suggested that seven vRNPs are coordinated around the remaining one forming a “7 plus 1” pattern within the virion (5). Recent results demonstrated that the genomes of ICV and IDV also adopt this pattern, although they only encompass seven gene segments (6).
NP is the most abundant component in a ribonucleoprotein complex. It performs multiple structural and functional roles, including encapsidation of viral RNA (vRNA), stabilization of replication intermediates by forming complementary ribonucleoprotein (cRNP) (7), maintenance of vRNP structure by homo-oligomerization (reviewed in references 8 and 9), and interactions with various host factors. At the early stage of infection, NPs are imported into the nucleus as a component of vRNP. The newly synthesized progeny NPs in the cytoplasm are then imported into the nucleus to support transcription and replication and take part in the assembly of cRNP and vRNP (10–12). In IAV, the nuclear import event of NP alone or vRNP were both found to exploit the importin α/β pathway (13–15). NPA contains at least two nuclear localization signals (NLSs), located at residues M1 to M13 and K198 to R216 (12, 16, 17). The crystal structure of NLS1 complexed with mouse importin α1 demonstrated extensive interaction at the minor groove but not the major groove (18). Recently, Wu et al. proposed that NLS1 and NLS2 may bind importin α synergistically (19). The interaction between nucleoprotein and importin α is crucial for other functions, such as modulation of RNP activity and contribution to host tropism (20, 21).
The structures of both NPA and NPB have been solved by protein crystallography (22–24). The structures resemble each other, and their secondary structure elements generally align well. Interestingly, NPB is 72 residues more extended at its N terminus than NPA. An NLS has been mapped from K44 to R47 that resembles a conventional monopartite NLS (25, 26).
Our understanding of the influenza C virus is far less than of the IAV/IBV. Nonetheless, it has been shown that there is actually a high seroprevalence against ICV in the human population, suggesting that the impact of this virus is largely underestimated (27). Moreover, the virus-host interaction of the influenza C virus is poorly characterized. By sequence alignment, we found that NPC has an extended C terminus compared to those of NPA and NPB. In this study, we specifically characterized the nuclear localization signal identified in this region and further examined the functional importance of this extended domain.
RESULTS
Sequence alignment of influenza virus NPs reveals distinctive structural features of NPC.
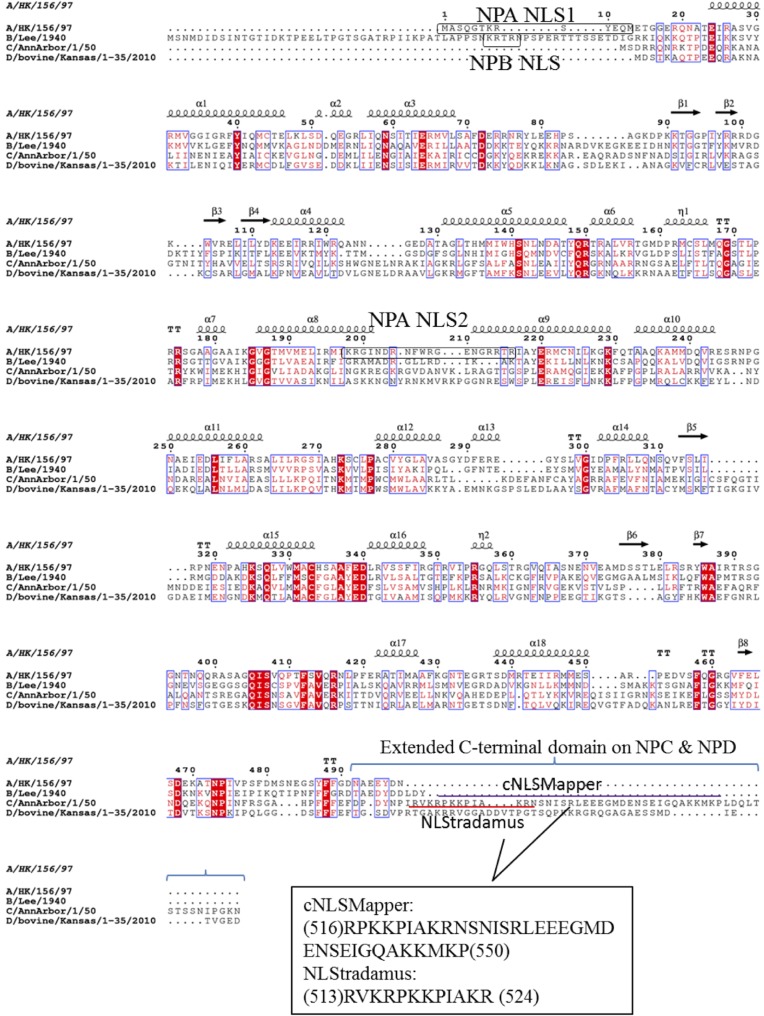
The NPs of IAV, IBV, and ICV (NPA, NPB, and NPC) have 498, 560, and 565 amino acid residues, respectively. The NP sequences of A/HK/156/97 (H5N1), B/Lee/1940, C/AnnArbor/1/50, and a provisional IDV D/bovine/Kansas/1-35/2010 were aligned with ClustalO (Fig. 1). The N terminus of NPC appears relatively shorter than NPA and NPB, but it contains an extended C-terminal domain (CTD) (54 and 51 amino acids longer than NPA and NPB, respectively). The nuclear localization signals on NPA or NPB could not be identified at the corresponding positions on NPC (Fig. 1).
FIG 1.
Alignment of NPB, NPC, and NPD sequences with reference to NPA shows that nuclear localization signals on NPA and NPB (as indicated by boxes) are absent at the corresponding positions on NPC. Sequences were aligned using ClustalO (50) and visualized by Esprit 3.0 (51). Two partially overlapping sequences on NPC were predicted as potential NLS sequences by NLStradamus and cNLSMapper servers, which are indicated by red and purple lines, respectively. Point mutations or deletions were then made on these two sequences, and the subcellular localization of variants was investigated.
The extended C terminus of NPC is responsible for nuclear localization.
The NPC sequence was analyzed by two online servers, NLStradamus (28) and cNLSMapper (29), for the prediction of putative nuclear localization signals on proteins. Two overlapping sequences at the C-terminal domain were reported. The former server reported a sequence of 12 amino acids in length, whereas the latter server reported a bipartite sequence with a long linker.
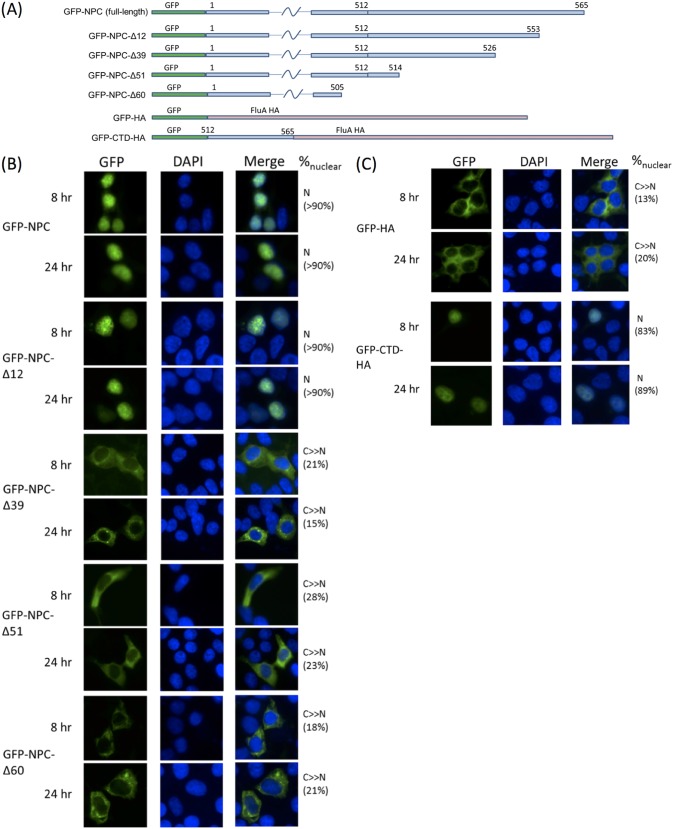
We first tested if the extended C terminus in NPC actually contributes to nuclear localization. Subcellular localizations of wild-type NPC and its C-terminal truncations (Fig. 2A) were compared by fusing them to an N-terminal green fluorescent protein (GFP) tag. At both 8 h and 24 h posttransfection (p.t.), wild-type NPC was predominantly nuclear. In contrast, NPC-Δ60, NPC-Δ51, and NPC-Δ39 were mainly cytoplasmic. Hence, deletion of the extended C terminus rendered NPC cytoplasmic. The localization pattern of the NPC-Δ12 mutant largely resembled that of the wild type. A major NLS likely lies within residues 527 to 553.
FIG 2.
(A) Construction of different NPC truncations in the pEGFP-C1 vector. Numbers represent corresponding amino acid residues on the protein. Constructs are not drawn to scale. (B and C) Constructs in panel A were transfected into 293T cells preseeded on glass slides. At the indicated time points, cells were harvested and fixed for visualization of the GFP signal using a Nikon E80i microscope. A deletion of 12 amino acids from the C terminus of NPC did not affect localization, but deletions of 39, 51, and 60 amino acids impaired nuclear targeting. (C) C-terminal domain (CTD) alone was also able to cause nuclear import. Nuclei were stained with DAPI. Subcellular localization was classified as N (exclusively nuclear), N+C (distributed), or C≫N (mainly cytoplasmic). Percentage values reported represent average percentages of nuclear signal for 20 cells counted.
In order to confirm that the CTD of NPC actually contains an NLS but does not function by other mechanisms such as masking nuclear export signals, two fusion proteins were generated and their localization patterns were compared. The last 60 amino acids of NPC were fused to the C terminus of GFP and at the N terminus of IAV hemagglutinin (GFP-CTD-HA). This construct was compared against a GFP-HA fusion. Hemagglutinin is a virion spike protein and was not found to enter the host nucleus. Indeed, our GFP-HA showed an almost exclusively cytoplasmic pattern at both 8 h and 24 h p.t. (Fig. 2C). GFP-CTD-HA had a mainly nuclear localization at both time points, indicating that CTD alone is sufficient to cause nuclear import.
Influenza C virus NP binds multiple importin α isoforms.
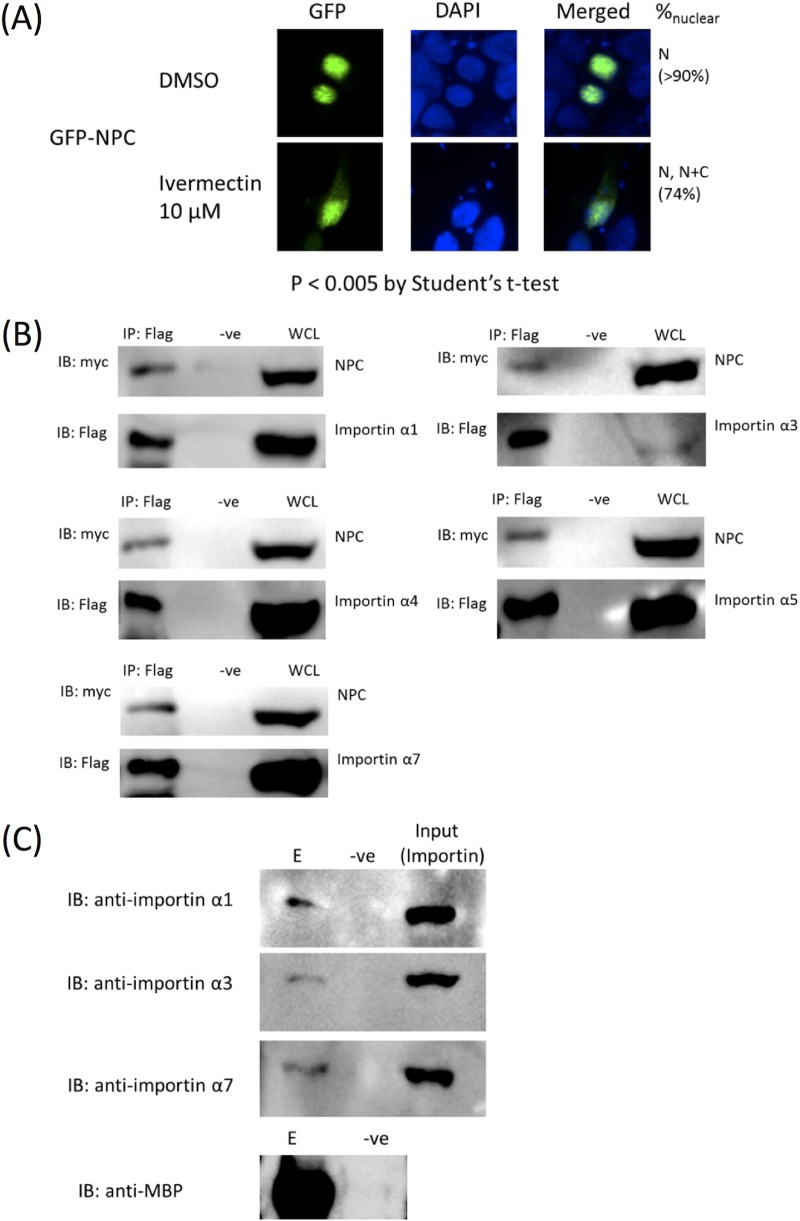
Conventional nuclear import events rely on host importin α/β machinery. The dependence of importin α/β of NPA for nuclear import has been well documented (16, 30, 31). We first tested if this is also true for NPC by investigating the effect of ivermectin on nuclear localization of GFP-NPC. Ivermectin has been shown to inhibit importin α/β-dependent but not importin β1-dependent nuclear import events (32). Nuclear import of NPC was impaired upon treatment with 10 μM ivermectin 2 to 4 h before cell harvest (Fig. 3A). The binding of NPC with host importins α1, α3, α4, α5, and α7 was then tested by co-immunoprecipitation. Importins α6 and α8 were omitted in our experiment, since these isoforms were found to be testis specific or oocyte and embryo specific, respectively (33, 34), and ought not to be the natural targets of NP. Flag-tagged importins α1, α3, α4, α5, and α7 were all able to co-immunoprecipitate a myc-tagged NPC, demonstrating binding of NPC to multiple importin α isoforms (Fig. 3B). We further tested the binding of NPC with endogenous importins α1, α3, and α7 using a copurification approach. Importin α1 is the housekeeping nuclear import adaptor, while importins α3 and α7 were found to be implicated in the pathogenicity of IAV. Maltose-binding protein (MBP)-tagged NPC immobilized on amylose resin was able to copurify importins α1, α3, and α7, further confirming the biological relevance of the interaction (Fig. 3C).
FIG 3.
(A) GFP-NPC-transfected cells were treated with ivermectin before cell harvest. A Student's t test showed a P value of <0.005, indicating significant difference between treated and DMSO groups. Hence, ivermectin enhanced cytoplasmic accumulation of NPC compared to that of the DMSO control, demonstrating the dependence of NPC nuclear import on the importin α/β pathway. (B) NPC interacts with importins α1, α3, α4, α5, and α7. Co-immunoprecipitation experiments were performed using Flag-tagged importin isoforms to precipitate myc-tagged NPC. IP, immunoprecipitation with anti-Flag (Sigma F3165 for α1, α4, α5, and α7; Proteintech 20543-1-AP for α3); -ve, negative control without anti-Flag; WCL, whole-cell lysate. Experiments were performed at least in duplicates, with representative data shown. Each panel represents a separate experiment; hence, it is not appropriate to compare relative binding efficiency across each isoform. (C) HEK293T lysate was allowed to incubate with MBP-tagged NPC which was immobilized on amylose resin. Maltose elution was analyzed by SDS-PAGE and Western blotting. Endogenous importins α1, α3, and α7 were coeluted with MBP-NPC. E. coli lysate without MBP-NPC was used as a negative control. E, elute; -ve, negative control.
The major NLS is a KKMK motif residing on the CTD.
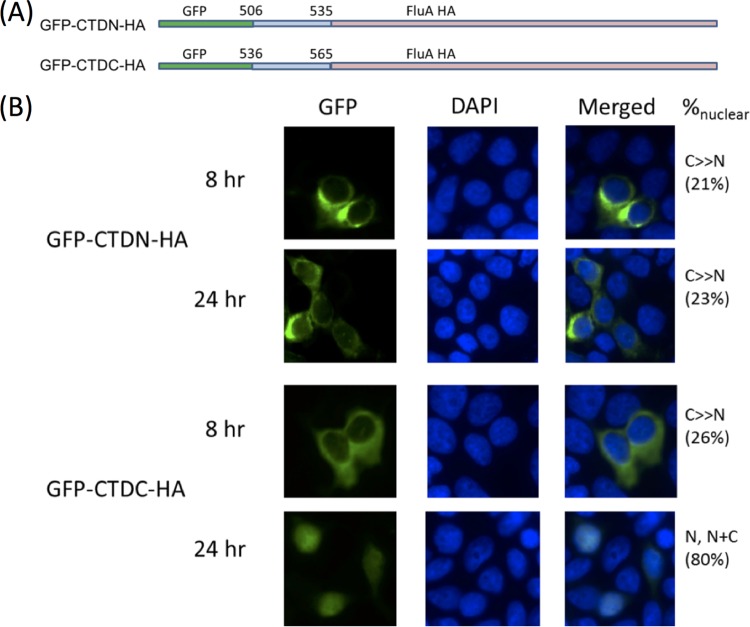
The binding of importin α isoforms to their cargo molecules have been characterized to be via traditional monopartite or bipartite nuclear localization signals. Inspection of the CTD sequence revealed two motifs, namely, 515-KRPKK-519 and 546-KKMK-549, both of which highly resemble a classical monopartite NLS. Two fusion proteins were generated, namely, GFP-CTDN30-HA, which encompassed the N-terminal 30 amino acids of the CTD (i.e., residues 506 to 535 of NPC) between a GFP tag and an IAV hemagglutinin protein, and GFP-CTDC30-HA, which encompassed the C-terminal 30 amino acids of the CTD (i.e., residues 536 to 565 of NPC) between GFP and IAV HA. The GFP-CTDN30-HA construct showed exclusively cytoplasmic localization at 8 h and 24 h p.t. despite a weak signal that leaked into the nucleus at 24 h. Meanwhile, GFP-CTDC30-HA exhibited an intermediate behavior among GFP-HA and GFP-CTD-HA. At 8 h p.t., GFP-CTDC30-HA was mainly cytoplasmic. At 24 h p.t., the majority of the GFP signal observed was nuclear, despite some cells showed a diffused pattern (Fig. 4). Hence, although the kinetics of nuclear import were hampered, residues 536 to 565 were sufficient for nuclear targeting.
FIG 4.
(A) Two GFP-HA-fusion proteins were constructed wherein GFP-CTDN-HA encompasses the N-terminal portion of CTD and GFP-CTDC-HA encompasses the last 30 amino acids of this domain. (B) These constructs were transfected into HEK293T cells and checked for nuclear localization at two time points posttransfection. CTDC retained the ability to drive nuclear import of GFP-HA, showing nuclear signal almost close to an entire CTD at 24 h p.t. CTDN showed little nuclear signal at both 8 and 24 h p.t. Nuclei were stained with DAPI. Subcellular localization was classified and quantified as described in the legend for Fig. 2.
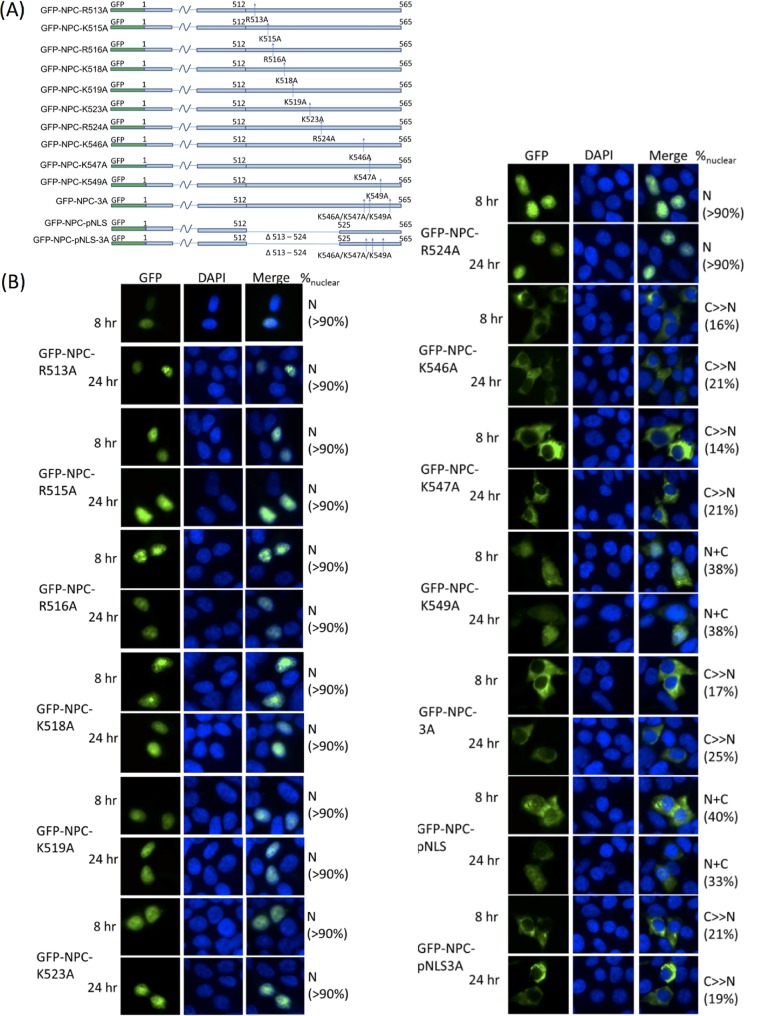
In order to define the residues essential for nuclear import, each and all of the lysine residues in the KKMK motif were then mutated to alanine, and the effect on NPC localization was investigated. Single K-to-A mutations were sufficient to impair nuclear import, with K546 and K547 playing more important roles, accounting for the observation that K546A and K547A each could render NPC cytoplasmic while K549A only led to a diffused NPC distribution. A triple mutant (NPC-3A) 546-AAMA-549 rendered NPC cytoplasmic at both 8 h and 24 h p.t., supporting that this motif is essential for nuclear import of NPC (Fig. 5).
FIG 5.
(A) A series of point and multiple mutants was constructed to test for the importance and relative contribution of positively charged residues on pNLS and KKMK motif. (B) K546A and K547A both abolished nuclear accumulation of NPC. K549 was also essential for nuclear import but to a lesser contribution than K546 and K547. All other point mutants showed minimal effects. Deletion of pNLS was also deficient in nuclear import. NPC-3A harboring 546-AAMA-549 and NPC-pNLS-3A both lost their ability to enter the nucleus. Nuclei were stained with DAPI. Subcellular localization was classified and quantified as described above.
Positive residues R513, K515, R516, K518, K519, K523, and R524 were also systematically mutated to alanines. All of these resides were on a putative nuclear localization signal (pNLS) predicted by NLStradamus. The effect on subcellular localization was revisited on a fluorescence microscope. However, none of these mutations was sufficient to affect the localization of NPC at 8 or 24 h p.t., but a deletion from R513 to R524 (GFP-NPC-pNLS) rendered nuclear localization largely hampered and gave an N+C localization pattern. The effect of pNLS deletion on subcellular localization was less substantial than AAMA mutation. The multiple mutant GFP-NPC-pNLS-3A, wherein pNLS was deleted and all three lysines in the KKMK motif were mutated to alanines, resembled NPC-3A (Fig. 5).
Mutations of the KKMK motif abolish importin binding.
To quantify the effect of AAMA mutations on the major NLS motif, we carried out microscale thermophoresis (MST) experiments to determine the binding affinity of NPC CTD and the AAMA mutant toward human importin α1, which is the housekeeping nuclear import adaptor universally expressed in all cell types. To this end, we expressed and purified maltose-binding protein (MBP) tagged MBP-CTD and MBP-CTD3A. We determined that the Kd (dissociation constant) for CTD toward an autoinhibitory domain-truncated human importin α1 was 48.6 nM. The MBP-CTD3A mutant registered an approximately 10-fold decrease in binding, with a Kd of 533 nM (Fig. 6). This difference supported that the KKMK motif is crucial for importin binding.
FIG 6.
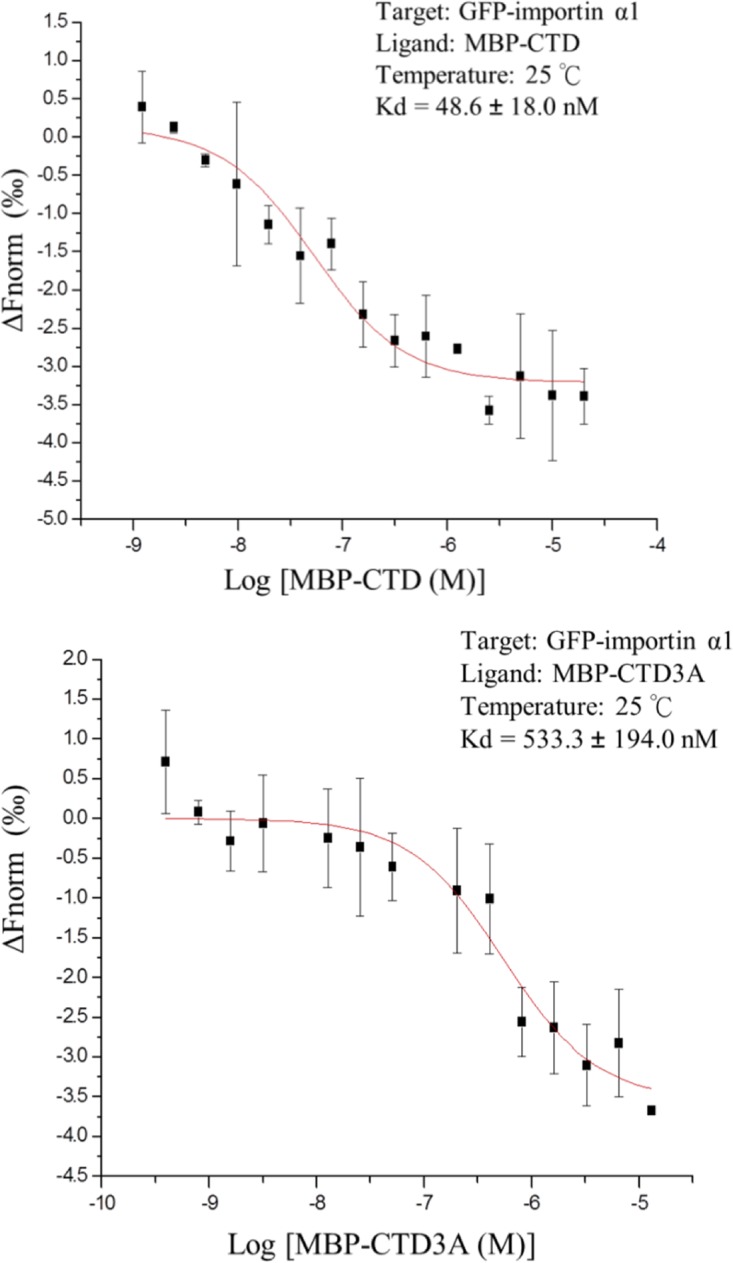
By microscale thermophoresis, the dissociation constants (Kd) of MBP-CTD and MBP-CTD3A with human importin α1 were determined. Autoinhibitory domain-truncated importin α1 was linked to an N-terminal GFP to provide fluorescence signal for MST detection. GFP-importin α1 was titrated against a serial dilution of MBP-CTD or MBP-CTD3A. Experiments were performed at 25°C with 22% LED and 20% MST powers. MST signal at MST-on-time of 1.5 s was analyzed. Kd was calculated with MO.Affinity V2.3 software, and curves were fitted in Origin (MicroCal) using data from three independent measurements. Kd is represented as mean ± standard deviation.
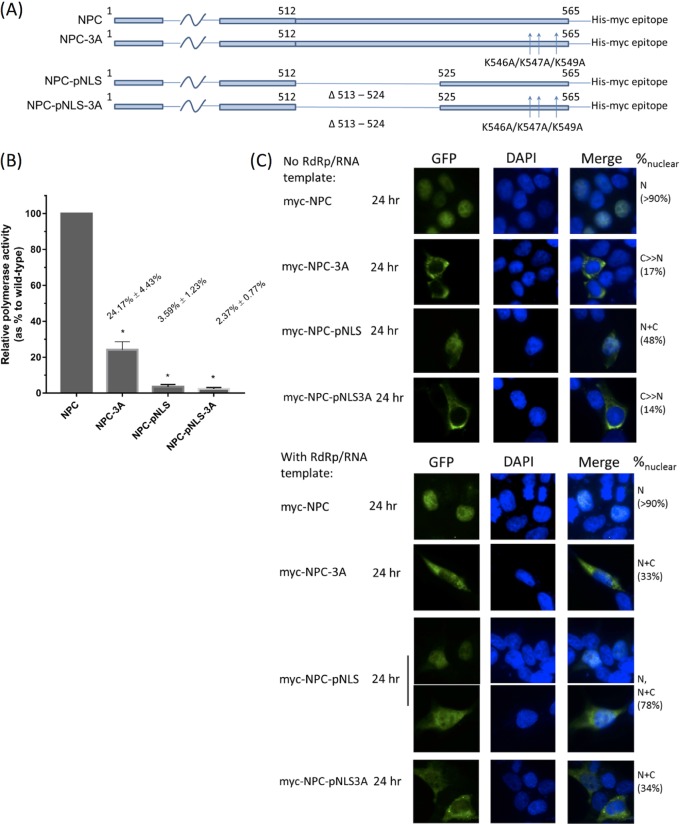
Influenza C virus RNP function was impaired with mutations of the KKMK motif or deletion of pNLS.
We modified a luciferase reporter system, which has been well established for IAV and IBV (25, 35), to test the effect of our NLS triple mutant on RNP activity in ICV. The firefly luciferase gene was cloned in negative sense in a pUC18 backbone (see Materials and Methods) which enabled the transcription into a positive sense firefly luciferase flanked by the ICV promoter sequences in cRNA sense. Luciferase protein was expressed by the viral RNP artificially reconstituted in vivo, hence enabling assessment of activity of the RNP. We employed this luciferase system to test the effect of triple mutations of the KKMK motif on viral RNP activity. Compared to wild-type NPC, the triple 546-AAMA-549 mutant suppressed RNP activity to only around one-fourth compared to that of wild-type NPC, while deletion of pNLS almost suppressed RNP activity to the basal level. Multiple mutant pNLS-3A resembled pNLS in the luciferase reporter assay and gave only basal RNP activity (Fig. 7B). By immunofluorescence, we confirmed that the presence of reconstituted RNP promoted the nuclear import of NPC-3A, NPC-pNLS, and NPC-pNLS3A mutants (Fig. 7C).
FIG 7.
RNP activity of influenza C viruses. (A) Each of the four constructs was cotransfected into HEK293T cells with plasmids expressing the RNP components and a pEGFP-C1 plasmid to control for DNA uptake. Cells were harvested. (B) NPC-3A, NPC-pNLS, and NPC-pNLS-3A mutants all demonstrated diminished polymerase activity as shown by luciferase reporter assay. Normalized results are shown as mean polymerase activity relative to that of the wild type wherein the activity for wild-type NPC was manually set as 100%. *, P < 0.001 by one-way ANOVA followed by Dunnett’s post hoc test (n = 3). Statistical analysis was performed in GraphPad Prism. (C) Subcellular localization of NPC in the context of reconstituted RNP was studied by immunofluorescence. Myc-tagged NPC was transfected with RdRP and luciferase template for generation of RNA with viral promoter. After cell harvest, myc-tagged NPC was detected by mouse monoclonal antibody and Alexa Fluor 488-conjugated anti-mouse antibody. Nuclei were stained with DAPI. Subcellular localization was classified and quantified as described in the legend for Fig. 2. To validate the system, we also tested the localization of myc-NPC, myc-NPC3A, myc-NPCpNLS, and myc-NPCpNLS3A with immunofluorescence, all of which show localizations resembling those by direct fluorescence shown in Fig. 5.
DISCUSSION
In this study, we identified important residues at the extended C-terminal domain of NPC which mediates nuclear import.
Similar to NPA, NPC was also found to bind multiple importin α isoforms, including members from all three karyopherin alpha subfamilies. Previously, Hudjetz and Gabriel (36) showed that NPA binds importins α1, α3, α4, α5, and α7, with higher affinity to α1, α5, and α7. N319K mutation of NPA was found to increase NP binding to importin α1 and was proposed to be responsible for host range adaption (21). Our results suggested that NPs of influenza viruses may commonly utilize multiple importin α isoforms for their transport from the cytoplasm into the nucleus.
Arranz et al. (37) and Moeller et al. (38) individually modeled NPA onto their RNP structure resolved by cryo-electron microscopy (cryo-EM). Although differences were seen comparing the two models, both of them showed that only NLS1 but not NLS2 on NPA was surface exposed. Moreover, the extreme C terminus is also surface exposed. Despite the presence of nuclear localization signals on the polymerase, nuclear import of the vRNP of IAV was found to rely mainly on NLS1 of NP (39). Assuming NPA and NPC adopt a similar orientation when in the form of a vRNP, the CTD of NPC should also be exposed. It is highly probable that the CTD of NPC is responsible for RNP import upon infection of influenza C virus.
The presence of an extended CTD, albeit not found in NPA and NPB, is also observed in the NP of influenza D virus and infectious salmon anemia virus under the Isavirus genus of the Orthomyxoviridae family (40). IDV is a veterinary virus identified in recent years which was found to infect mainly bovine and sometimes swine (41). The genome structure of IDV resembles that of ICV, containing only seven segments (41). By sequence alignment, NPC and NPD share 37.3% protein sequence similarity. The KKMK motif was not found in the CTD of NPD, which instead contains two stretches of positive amino acids, 510-RTGAKRR-516 and 531-KKRGR-535, highly resembling a bipartite NLS. Recently, Donchet et al. (42) reported that the CTD in NPD is intrinsically disordered. They also proposed that a bipartite NLS exists between K514 and R535 on NPD.
Our MST experiment determined the Kd of NPC CTD toward importin α1 to be around 50 nM. This value is roughly in the same range as traditional monopartite and bipartite NLSs to importin α (43, 44) but is 20-fold stronger than NPA NLS1 (1 to 5 μM) (18, 19) and the NPB N-terminal domain (0.8 μM) (45). This value is also similar to the affinity of NPD CTD toward importin α7 determined by Donchet et al. (42). The fact that MBP-CTD3A also registered a Kd of around 500 nM implicated that the pNLS sequence alone still affords considerable binding to importin. This supports that the NLS is a long bipartite sequence.
Our results showed that the KKMK motif itself represents a minimal sequence to facilitate nuclear import (Fig. 3 and 4). Also, point mutations R513A, K515A, R516A, K518A, K519A, K523A, or R524A showed minimal effect on NPC subcellular distribution, but single mutations K546A, K547A, or K549A each can render NPC cytoplasmic to different extents. We believe that the pNLS sequence affects nuclear accumulation of NPC, because an N+C pattern was observed for NPC-pNLS at both 8 h and 24 h p.t., suggesting that NPC retains, at least in part, its ability of nuclear import even after deletion of the pNLS sequence. The pNLS sequence plausibly also modulates binding of NLS to importin α and hence the kinetics of nuclear import. This argument is supported by recent work by Smith et al. (46) who showed that a hydrophobic residue at the “P4” position of a monopartite NLS (such as methionine at the KKMK motif) is generally unfavored, and one of the strategies to remediate such a detrimental effect is the presence of positive charges upstream of the NLS.
Although the KKMK motif shows greater importance on nuclear localization as visualized under the fluorescence microscope, mutating KKMK to AAMA still gave roughly 25% basal polymerase activity (Fig. 5B). We cannot rule out the existence of an auxiliary NLS outside the C-terminal domain which could possibly explain this residual activity. There also exists the possibility that NPC is brought into the nucleus along with a viral or host factor it binds to. In fact, in our immunofluorescence experiment, we showed that the presence of reconstituted RNP partially restored the nuclear import and/or accumulation of NPC NLS mutants. Although NPC-pNLS and NPC-pNLS3A exist to some extent in the nucleus in the context of a reconstituted RNP, these mutants could not support RNP activity. This strongly hints that pNLS sequence is functionally important in viral replication and/or transcription, possibly as in the case of NPA NLS2, which serves also as a nucleolar localization signal (47).
MATERIALS AND METHODS
Biological materials.
HEK293T cells (ATCC) were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (Invitrogen). The NP sequence used for fluorescence microscopy is based on a clinical influenza C virus strain isolated in Hong Kong (Prince Wales Hospital, Shatin, HK). NPC was subcloned into pEGFP-C1 (Clontech) for fluorescence microscopy experiments. DNA Sanger sequencing was performed at BGI Genomics (Shenzhen, China), and the amino acid sequence was deduced thereby as shown in Table S1 in the supplemental material. For co-immunoprecipitation and luciferase reporter assays, NP based on C/AnnArbor/1/50 was subcloned into relevant vectors as described. Importins α1, α3, α4, α5, and α7 were purchased from Harvard Plasmid Bank and subcloned into pCMV-Tag2B (Agilent) for co-immunoprecipitation experiments. Transfection was carried out either by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol or by the polyethyleneimine (PEI) method at a DNA/PEI ratio of 1:2.
Direct fluorescence and immunofluorescence microscopy.
For direct fluorescence, HEK293T cells were seeded on glass slides and transfected with the indicated constructs for 8 h or 24 h using the Lipofectamine 2000 method. For ivermectin (Sigma) treatment, 10 μM ivermectin or an equivalent volume of dimethyl sulfoxide (DMSO) was added to cells 2 to 4 h before harvest. Cells were fixed using 4% paraformaldehyde at room temperature for 5 min and partially lysed by 0.1% Triton X-100 for 5 min. For immunofluorescence, NPC constructs were transfected into preseeded HEK293T cells together with the plasmids encoding the ICV RdRP and an artificial luciferase template, as in the reporter assay setup described below. After 24 h, cells were fixed with 4% paraformaldehyde at room temperature for 5 min, partially lysed by 0.1% Triton X-100 for 5 min, and probed with a mouse monoclonal anti-myc (Cell Signaling) and then an Alexa Fluor 488-coupled anti-mouse secondary antibody (Thermo Fisher Scientific).
In both experiments, nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) for 2 min. Cells were mounted with Dako mounting medium. Fluorescence was visualized by a Nikon E80i fluorescence microscope equipped with respective filters and digital camera at ×40 magnification. Images were selected with contrast and brightness appropriately adjusted to facilitate data presentation using ImageJ software (48). Localization of NPC or mutants was classified into N (exclusively nuclear), N+C (distributed), or C≫N (mainly cytoplasmic). Quantification of the fluorescence signal was performed in ImageJ by counting 20 cells for each construct and calculating with the formula shown below. Cells showing more than 90% nuclear signals were denoted as >90% nuclear to account for ambiguity in defining cytosolic boundary in such cases.
For quantifying the effect of ivermectin, a two-tailed unpaired Student's t test was used.
Co-immunoprecipitation and copurification.
Myc-His-tagged NPC was cotransfected with flag-tagged importin α1, α3, α4, α5, or α7 into HEK293T cells using the PEI method. Cells were harvested 36 to 48 h posttransfection using an IP lysis buffer (50 mM Tris [pH 6.5 to 7.0], 500 mM NaCl, 2 mM EDTA, 0.3% Triton X-100). Protein A-coupled agarose beads (Sigma) were preincubated with anti-flag antibody. For importins α1, α4, α5, and α7, a mouse monoclonal anti-flag from Sigma (F-3165) was used. For importin α3, a rabbit polyclonal anti-flag from Proteintech (20543-1-AP) was used. Antibody-bound beads were subsequently added to lysates to perform immunoprecipitation at room temperature for 1 h. Beads were washed at least 3 times by lysis buffer before SDS-PAGE loading buffer was added. NPC and importins α1, α3, α4, α5, and α7 were immunoblotted by mouse monoclonal anti-myc (2276; Cell Signaling) and anti-flag (F-3165; Sigma), respectively. All co-immunoprecipitation experiments were performed at least twice. Full-sized membrane images for co-immunoprecipitation are available upon request.
For copurification experiments, MBP-tagged NPC was expressed in BL21(pLysS) Escherichia coli cells. Lysate with MBP-NPC was incubated with amylose resin (New England Biolabs) for 2 h to overnight. After extensive washing with phosphate-buffered saline (PBS), HEK293T lysate was loaded to the resin and was incubated for a further 1 h. Resins were washed with PBS and proteins were eluted with 100 mM maltose. Coeluted proteins were analyzed by SDS-PAGE and Western blotting. To control for nonspecific binding, nontransformed BL21(pLysS) E. coli cell lysate was used as a negative control. Antibodies used for detection were anti-MBP (66003, mouse monoclonal; Proteintech), anti-importin α1 (SC55538, mouse monoclonal; Santa Cruz), anti-importin α3 (PA5-21749, rabbit polyclonal; Invitrogen), and anti-importin α7 (SC390055, mouse monoclonal; Santa Cruz).
Luciferase reporter assay.
P3, PB1, PB2, and NP genes of C/Ann Arbor/1/50 were synthesized (Genscript, Nanjing, China) and subcloned into pcDNA3 and pcDNA3.1mycHisA (Thermo Fisher) vectors to give pCDNA3-CP3, pCDNA3-CPB1, pCDNA3-CPB2 and pcDNA3.1mycHisA-CNP, respectively. A reporter plasmid (pPolI-Luci-CP3-RT) was constructed based on the modified pUC18 backbone encoding a human RNA polymerase I promoter and a hepatitis delta virus ribozyme. (The backbone is a kind gift from Peter Palese, Mount Sinai School of Medicine; see reference 49.) The negative-sense sequence of the firefly luciferase gene was cloned into the cassette flanked by the noncoding sequences of the P3 gene (segment 3). Transfection of each of the plasmids together with a control pEGFP-C1 empty plasmid into 293T cells preseeded on a 24-well plate was achieved by the PEI method. Cells were harvested in luci-lysis buffer (50 mM Tris [pH 7.8], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM dithiothreitol [DTT], 1 mM EDTA) at 48 to 52 h posttransfection, briefly sonicated, and centrifuged. Lysate was separated into two portions. One portion was directly subject to a ClarioStar spectrophotometer for measuring the GFP signal. The other portion was added to firefly luciferase substrate (purchased from Promega), and luciferase signals were then measured also on a ClarioStar spectrophotometer. All chemiluminescence signals were normalized to the GFP signal and, in cases of mutants, normalized with NP expression level. For each mutant, the normalized result was expressed as mean relative polymerase activity to wild-type level (± standard deviation [SD], n = 3, wild-type level set manually to 100%). Statistical comparison was by one-way analysis of variance (ANOVA) and Dunnett’s post hoc test in GraphPad Prism.
Protein expression and purification.
N-terminal autoinhibitory domain-truncated human importin α1 was cloned into a pET28a (Novagen) vector. For purification of GFP-tagged protein, enhanced GFP (EGFP) was then inserted between an N-terminal 6×His tag and importin α1 using BamHI and XhoI restriction sites. The C-terminal domain and its mutant harboring the AAMA mutation were cloned into an in-house pRHisMBP vector, which was a kind gift from K. B. Wong (The Chinese University of Hong Kong), to obtain MBP-CTD and MBP-CTD3A, respectively. All proteins were expressed in a BL21(pLysS) E. coli strain in LB medium supplemented with appropriate antibiotics. Protein expression was induced by 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density (OD) of 0.6 to 0.8 at 16°C. Cells were collected after 12 to 16 h by centrifugation.
Cells expressing GFP-importin α1 were lysed in lysis buffer A (PBS [pH 7], 15% glycerol, and 20 mM imidazole and supplemented with phenylmethylsulfonyl fluoride [PMSF]) by sonication. GFP-importin α1 was then purified on a nickel affinity column according to a standard protocol. Cells expressing MBP-CTD and MBP-CTD3A were lysed in lysis buffer B (PBS pH 7, 5% glycerol, PMSF), and proteins were purified by amylose-affinity columns (New England Biolabs). For use in the MST experiments, all protein samples were buffer exchanged to 20 mM sodium phosphate (pH 7), 50 mM NaCl, and 5% glycerol. MBP-CTD and MBP-CTD3A were concentrated to 0.08 mM and 0.027 mM, respectively.
Microscale thermophoresis.
Tween 20 was added to all protein samples to a final concentration of 0.1% (vol/vol) before the MST experiments. MST experiments were performed on a Monolith NT.115 machine (Nanotemper, Munich, Germany). Five microliters of GFP-importin α1 (target protein) was titrated against a serial dilution of ligand proteins (MBP-CTD or MBP-CTD3A) to make up 16 MST samples at a total volume of 10 μl. MST samples were loaded onto standard-treated capillaries. Measurements were made at 25°C with 22% light-emitting diode (LED) power and 20% MST power. Data analysis was performed on Nanotemper’s MO.Affinity V2.3 software. Thermophoresis was analyzed at an MST-on-time of 1.5 s using the Kd model assuming 1:1 binding. For better data presentation, dose-response curves were fitted by Origin software (MicroCal) using data from three independent measurements. Kd was represented as mean ± standard deviation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Palese, Mount Sinai School of Medicine, for the pPolI-RT backbone plasmid.
The work was supported by a theme-based research grant of the Research Grant Council, Hong Kong SAR (project number T11-705/14N).
We declare no conflicts of interest.
Y.-S.T. designed and performed experiments, analyzed data, and drafted the manuscript; C.-Y.L. designed experiments and analyzed data; C.K.-P.M. designed experiments and analyzed data; P.K.-S.C. collected clinical samples and isolated and identified a strain of ICV; P.-C.S. designed experiments, analyzed data, and provided key scientific advice.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02048-18.
REFERENCES
- 1.WHO. 2018. Influenza update–324. WHO, Genevea, Switzerland. [Google Scholar]
- 2.Shaw ML, Palese P. 2013. Orthomyxoviridae: the viruses and their replication, p 1691–1740. In Howley PM, Knipe DM (ed), Fields virology 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Principi N, Scala A, Daleno C, Esposito S. 2013. Influenza C virus–associated community-acquired pneumonia in children. Influenza. Influenza Other Respir Viruses 7:999–1003. doi: 10.1111/irv.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu Y, Abiko C, Ikeda T, Mizuta K, Matsuzaki Y. 2015. Influenza C virus and human metapneumovirus infections in hospitalized children with lower respiratory tract illness. Pediatr Infect Dis J 34:1273–1275. doi: 10.1097/INF.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 5.Noda T, Murakami S, Nakatsu S, Imai H, Muramoto Y, Shindo K, Sagara H, Kawaoka Y. 2018. Importance of the 1 + 7 configuration of ribonucleoprotein complexes for influenza A virus genome packaging. Nat Commun 9:54. doi: 10.1038/s41467-017-02517-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatsu S, Murakami S, Shindo K, Horimoto T, Sagara H, Noda T, Kawaoka Y. 2018. Influenza C and D viruses package eight organized ribonucleoprotein complexes. J Virol 92:02084-17. doi: 10.1128/JVI.02084-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vreede FT, Ng AKL, Shaw PC, Fodor E. 2011. Stabilization of influenza virus replication intermediates is dependent on the RNA-binding but not the homo-oligomerization activity of the viral nucleoprotein. J Virol 85:12073–12078. doi: 10.1128/JVI.00695-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo CY, Tang YS, Shaw PC. 2018. Structure and function of influenza virus ribonucleoprotein, p. 95–128. In Harris J, Bhella D (ed), Virus protein and nucleoprotein complexes. Subcellular biochemistry, vol 88 Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Tang YS, Shaw PC. 2014. Structure and function of nucleoproteins from Orthomyxoviruses. BioDesign 2:91–99. [Google Scholar]
- 10.Hutchinson E, Fodor E. 2012. Nuclear import of the influenza A virus transcriptional machinery. Vaccine 30:7353–7358. doi: 10.1016/j.vaccine.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson E, Fodor E. 2013. Transport of the influenza virus genome from nucleus to nucleus. Viruses 5:2424–2446. doi: 10.3390/v5102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J Virol 71:9690–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Neill RE, Palese P. 1995. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology 206:116–125. doi: 10.1016/S0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 14.Wu WW, Panté N. 2009. The directionality of the nuclear transport of the influenza A genome is driven by selective exposure of nuclear localization sequences on nucleoprotein. Virol J 6:68. doi: 10.1186/1743-422X-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu WWH, Weaver LL, Panté N. 2007. Ultrastructural analysis of the nuclear localization sequences on influenza A ribonucleoprotein complexes. J Mol Biol 374:910–916. doi: 10.1016/j.jmb.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, Palese P, O’Neill RE. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol 71:1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber F, Kochs G, Gruber S, Haller O. 1998. A classical bipartite nuclear localization signal on thogoto and influenza A virus nucleoproteins. Virology 250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 18.Nakada R, Hirano H, Matsuura Y. 2015. Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci Rep 5:15055. doi: 10.1038/srep15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu W, Sankhala RS, Florio TJ, Zhou L, Nguyen NLT, Lokareddy RK, Cingolani G, Panté N. 2017. Synergy of two low-affinity NLSs determines the high avidity of influenza A virus nucleoprotein NP for human importin α isoforms. Sci Rep 7:11381. doi: 10.1038/s41598-017-11018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, Sauter M, Shmidt T, Rother F, Baumgarte S, Keiner B, Hartmann E, Bader M, Brownlee GG, Fodor E, Klenk HD. 2011. Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat Commun 2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin α1 is a determinant of host range of influenza A virus. PLoS Pathog 4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng AKL, Zhang H, Tan K, Li Z, Liu JH, Chan PKS, Li SM, Chan WY, Au SWN, Joachimiak A, Walz T, Wang JH, Shaw PC. 2008. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J 22:3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng AKL, Lam MKH, Zhang H, Liu JH, Au SWN, Chan PKS, Wang J, Shaw PC. 2012. Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein. J Virol 86:6758–6767. doi: 10.1128/JVI.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Q, Krug RM, Tao YJ. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 25.Liu M, Lam MKH, Zhang Q, Elderfield R, Barclay WS, Shaw PC. 2015. The functional study of the N-terminal region of influenza B virus nucleoprotein. PLoS One 10:e0137802. doi: 10.1371/journal.pone.0137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanitchang A, Narkpuk J, Jongkaewwattana A. 2013. Nuclear import of influenza B virus nucleoprotein: involvement of an N-terminal nuclear localization signal and a cleavage-protection motif. Virology 443:59–68. doi: 10.1016/j.virol.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki Y, Sugawara K, Furuse Y, Shimotai Y, Hongo S, Oshitani H, Mizuta K, Nishimura H. 2016. Genetic lineage and reassortment of influenza C viruses circulating between 1947 and 2014. J Virol 90:8251–8265. doi: 10.1128/JVI.00969-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. 2009. NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinformatics 10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kosugi S, Hasebe M, Tomita M, Yanagawa H. 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melén K, Fagerlund R, Franke J, Köhler M, Kinnunen L, Julkunen I. 2003. Importin α nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem 278:28193–28200. doi: 10.1074/jbc.M303571200. [DOI] [PubMed] [Google Scholar]
- 31.Boulo S, Akarsu H, Lotteau V, Müller CW, Ruigrok RWH, Baudin F. 2011. Human importin alpha and RNA do not compete for binding to influenza A virus nucleoprotein. Virology 409:84–90. doi: 10.1016/j.virol.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. 2012. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J 443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köhler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. 1997. Cloning of two novel human importin-α subunits and analysis of the expression pattern of the importin-α protein family. FEBS Lett 417:104–108. doi: 10.1016/S0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Park KE, Koser S, Liu S, Magnani L, Cabot RA. 2012. KPNA7, an oocyte- and embryo-specific karyopherinαsubtype, is required for porcine embryo development. Reprod Fertil Dev 24:382. doi: 10.1071/RD11119. [DOI] [PubMed] [Google Scholar]
- 35.Ng AKL, Chan WH, Choi ST, Lam MKH, Lau KF, Chan PKS, Au SWN, Fodor E, Shaw PC. 2012. Influenza polymerase activity correlates with the strength of interaction between nucleoprotein and PB2 through the host-specific residue K/E627. PLoS One 7:e36415. doi: 10.1371/journal.pone.0036415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudjetz B, Gabriel G. 2012. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-α1 and -α7. PLoS Pathog 8:e1002488. doi: 10.1371/journal.ppat.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 38.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. 2012. Organization of the influenza virus replication machinery. Science 338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cros JF, García-Sastre A, Palese P. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6:205–213. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- 40.Zheng W, Olson J, Vakharia V, Tao YJ. 2013. The crystal structure and RNA-binding of an orthomyxovirus nucleoprotein. PLoS Pathog 9:e1003624. doi: 10.1371/journal.ppat.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su S, Fu X, Li G, Kerlin F, Veit M. 2017. Novel influenza D virus: epidemiology, pathology, evolution and biological characteristics. Virulence 8:1580–1591. doi: 10.1080/21505594.2017.1365216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donchet A, Oliva J, Labaronne A, Tengo L, Miloudi M, Gerard F, Mas C, Schoehn G, Ruigrok RW, Ducatez M, Crépin T. 2019. The structure of the nucleoprotein of influenza D shows that all Orthomyxoviridae nucleoproteins have a similar NPCORE, with or without a NPTAIL for nuclear transport. Sci Rep 9:600. doi: 10.1038/s41598-018-37306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Catimel B, Teh T, Fontes MRM, Jennings IG, Jans DA, Howlett GJ, Nice EC, Kobe B. 2001. Biophysical characterization of interactions involving importin-α during nuclear import. J Biol Chem 276:34189–34198. doi: 10.1074/jbc.M103531200. [DOI] [PubMed] [Google Scholar]
- 44.Fanara P, Hodel MR, Corbett AH, Hodel AE. 2000. Quantitative analysis of nuclear localization signal (NLS)-importin α interaction through fluorescence depolarization. J Biol Chem 275:21218–21223. doi: 10.1074/jbc.M002217200. [DOI] [PubMed] [Google Scholar]
- 45.Labaronne A, Milles S, Donchet A, Jensen MR, Blackledge M, Bourhis JM, Ruigrok RWH, Crépin T. 2017. Structural analysis of the complex between influenza B nucleoprotein and human importin-α. Sci Rep 7:17164. doi: 10.1038/s41598-017-17458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith KM, Di Antonio V, Bellucci L, Thomas DR, Caporuscio F, Ciccarese F, Ghassabian H, Wagstaff KM, Forwood JK, Jans DA, Palù G, Alvisi G. 2018. Contribution of the residue at position 4 within classical nuclear localization signals to modulating interaction with importins and nuclear targeting. Biochim Biophys Acta Mol Cell Res 1865:1114–1129. doi: 10.1016/j.bbamcr.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. 2007. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:3671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pleschka S, Jaskunas R, Engelhardt OG, Zürcher T, Palese P, García-Sastre A. 1996. A plasmid-based reverse genetics system for influenza A virus. J Virol 70:4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.