Short abstract
In humans, homozygous mutations in the TPP1 gene results in loss of tripeptidyl peptidase 1 (TPP1) enzymatic activity, leading to late infantile neuronal ceroid lipofuscinoses disease. Using a mouse model that targets the Tpp1 gene and recapitulates the pathology and clinical features of the human disease, we analyzed end-stage (4 months) transcriptional changes associated with lack of TPP1 activity. Using RNA sequencing technology, Tpp1 expression changes in the forebrain/midbrain and cerebellum of 4-month-old homozygotes were compared with strain-related controls. Transcriptional changes were found in 510 and 1,550 gene transcripts in forebrain/midbrain and cerebellum, respectively, from Tpp1-deficient brain tissues when compared with age-matched controls. Analysis of the differentially expressed genes using the Ingenuity™ pathway software, revealed increased neuroinflammation activity in microglia and astrocytes that could lead to neuronal dysfunction, particularly in the cerebellum. We also observed upregulation in the production of nitric oxide and reactive oxygen species; activation of leukocyte extravasation signals and complement pathways; and downregulation of major transcription factors involved in control of circadian rhythm. Several of these expression changes were confirmed by independent quantitative polymerase chain reaction and histological analysis by mRNA in situ hybridization, which allowed for an in-depth anatomical analysis of the pathology and provided independent confirmation of at least two of the major networks affected in this model. The identification of differentially expressed genes has revealed new lines of investigation for this complex disorder that may lead to novel therapeutic targets.
Keywords: neuronal ceroid lipofuscinoses, transcriptome, lysosomal tripeptidyl peptidase 1, pediatric neurodegeneration, neuroinflammation, circadian rhythm
Introduction
Neuronal ceroid lipofuscinoses (NCLs) are among the most common neurodegenerative diseases affecting the pediatric population, with an incidence estimated at 2 to 4 per 100,000 live births (Kollmann et al., 2013; Mink et al., 2013). Different forms of the disease were originally classified on the basis of the time of onset: infantile NCLs, late infantile NCLs, juvenile NCLs (JNCLs), and adult NCLs (Mink et al., 2013). Shared pathologies of the NCLs include progressive visual deterioration resulting in blindness, intellectual and motor decline, epilepsy, ataxia, spasticity, and premature death (Anderson et al., 2013; Schulz et al., 2013; Radke et al., 2015). The ultrastructural appearance of autofluorescent storage material found in patient tissues distinguishes different types of NCLs and has long been used as a diagnostic tool (Palmer et al., 2002, 2013). In the past 20 years, mutations in over a dozen genes have been characterized in families diagnosed with NCLs, and all but one type (Kufs disease) are inherited in an autosomal recessive manner (Carcel-Trullols et al., 2015; Mole and Cotman, 2015). NCLs are considered lysosomal storage diseases because most of the affected proteins have an endo/lysosomal network localization. However, some of these proteins are localized in other cellular compartments such as Golgi, endoplasmic reticulum, cytosol, plasma membrane, and even in the extracellular matrix (Carcel-Trullols et al., 2015) making it difficult to explain how all these gene defects lead to similar clinical symptoms and neurodegeneration. Furthermore, several of the affected genes are lysosomal hydrolases or peptidases, but their cellular substrates are incompletely elucidated, and the primary functions of the membrane proteins mutated in NCLs remain uncertain. Most of the advances in understanding the molecular pathology of NCLs have come from studies on patients’ fibroblast and lymphoblast cell lines and, increasingly, genetic animal models (dogs, sheep, mice, etc.; Cooper et al., 2006; Palmer et al., 2013; Cooper et al., 2015). Several hypotheses have emerged regarding potential cellular triggers for the disease: Evidence for cellular changes, including lysosomal dysregulation, endoplasmic reticulum stress, mitochondrial dysfunction, and compromised autophagy to name a few, have all been implicated (Palmer et al., 2013; Marotta et al., 2017). However, so far, none of these integrate all the genetic origins to a common biological pathway (Cooper et al., 2006; Palmer et al., 2013; Cooper et al., 2015; Marotta et al., 2017). Thus, it is conceivable that the various gene mutations implicated could alter different cellular processes that ultimately trigger a common chronic response that in turn is responsible for the neurodegenerative outcome.
From studies of some of the animal models, it is clear that astrogliosis and microglia activation play major roles in the disease (Pontikis et al., 2004; Pontikis et al., 2005; Parviainen et al., 2017) and precede neuronal cell death, such that neuronal loss is prevalent throughout the brain and at the end stages of the disease. There are specific brain areas of vulnerability at early stages such as the thalamocortical system and cerebellum (Cb; Cooper et al., 2006; Palmer et al., 2013; Cooper et al., 2015). Thus, astrogliosis and microglia activation are better predictors than the autofluorescent storage material in indicating initial areas of neuronal loss in the brains of many CLN models (Haltia et al., 2001; Tyynela et al., 2004; Anderson et al., 2013). Interestingly, no direct correlation has been found between the pattern of neuronal loss and the accumulation of storage material, and neither is the degree of neurodegeneration correlated with the amount of intracellular autofluorescent storage bodies accumulated (Cooper et al., 2006; Palmer et al., 2013; Cooper et al., 2015). Furthermore, it has been suggested that the accumulation of storage bodies can be neuroprotective because the few surviving neurons in the end stage of the disease are vastly distended with storage material (Tyynela et al., 2004; Cooper et al., 2015). Although NCLs have a strong neuronal toxicity component, the function of astroglia and microglia in disease progression is still unclear. Recently, stimulation of CLN3 deficient microglia and astrocytes in culture was shown to attenuate their ability to transform morphologically and alter their protein-secretion profiles (Parviainen et al., 2017). Also, there is evidence of aberrant astrocyte CX43 hemichannel activity in JNCL (Burkovetskaya et al., 2014).
We focused on the classical late infantile form of NCL (cLINCL), which in humans is caused by mutations in the TPP1 gene (Sleat et al., 1997). The gene is also known as CLN2 (ceroid lipofuscinosis neuronal 2), GIG1, LPIC, and SCAR7 (NCBI Gene database, www.ncbi.nlm.nih.gov/gene). This disease variant is usually diagnosed between 2 and 4 years of age with onset of seizures followed by the severe neurological deterioration common to all NCLs, and most patients die between 7 and 15 years of age (Sleat et al., 1997; Fietz et al., 2016). The only currently available treatment is a recent FDA-approved enzyme replacement therapy in which a recombinant form of human TPP1 (Brineura® [cerliponase alfa]) is infused into the cerebral spinal fluid of patients. This treatment appears to reduce the loss of walking ability in human trials (clinical trial NCT01907087), but it is expensive and may lead to many side effects due to the prolonged nature of the treatment (Schulz et al., 2018).
A well-established mouse model to study cLINCL was generated and characterized by targeting the mouse gene Tpp1 in the Lobel laboratory (Sleat et al., 2004), resulting in loss of detectable TPP1 activity, progressive neurological phenotypes including ataxia, and increased motor deficiency. Affected mice have a median survival of 19 weeks with motor deficiencies detectable after 10 weeks, thus representing an excellent model of the human disease progression. More recently, Pierce’s laboratory independently made a Tpp1 knockout model with a more severe phenotype (Geraets et al., 2017), but the Lobel model has been better characterized, including its ability to respond to protein replacement therapies (Passini et al., 2006; Chang et al., 2008).
Our laboratory has extensive experience in microglia and astrocyte responses to embryonic and perinatal brain injury (Domowicz et al., 2011; Domowicz et al., 2018). Because these cell types play fundamental roles in neuronal survival and injury repair and express Tpp1 at higher levels in astrocytes (7 times more) and microglia (4 times more; Zhang et al., 2014) than in neurons, they could be major contributors to the pathology of cLINCL. The objective of this study was to perform, for the first time in a cLINCL model, a transcriptome-wide characterization by RNA sequencing (RNA-seq) of two macroareas mostly affected in the 4-month-old Tpp1–/– (Cb and forebrain/midbrain [F/M]). Because we were particularly interested in the microglia and astrocyte response and both cell types have been known to quickly alter their transcriptome profiles in culture (Haimon et al., 2018), we used whole Cb to perform these analyses. We then matched the differentially expressed genes (DEG) in the Tpp1 model to the already reported individual cell transcriptomes (Zhang et al., 2014; Lun et al., 2015; Gosselin et al., 2017; Boisvert et al., 2018; Haimon et al., 2018; Olah et al., 2018). Our results in 4-month-old Tpp1–/– mouse F/M and Cb show strong neurodegenerative inflammatory responses involving microglia and astrocytes, activation of leukocyte extravasation, dysregulation of neurotransmitter production, and upregulation of nitric oxide (NO) and reactive oxygen species (ROS); in contrast, we observed downregulation of transcription factors involved in control of circadian rhythm. This study represents, to our knowledge, the first analysis of this kind made in any animal model for cLINCL.
Materials and Methods
Mice
All animal procedures were performed according to the University of Chicago Institutional Animal Care and Use Committee regulatory policies. Mice were housed under standard conditions, with access to food and water ad libitum, in a 12-hr light and dark cycle. The mouse model for cLINCL, in which Tpp1 is disrupted by gene targeting, has been previously described (Sleat et al., 2004) and is referred in this article as the Tpp1–/– mutant. The mouse line was a gift from Drs. P. Lobel and D. E. Sleat. The line is being maintained by Tpp1+/– heterozygous crosses. In all experiments, Tpp1–/– homozygous mice were compared with heterozygous (Tpp1+/–) or wild-type (Tpp1+/+) siblings of matched age and sex, as indicated.
Gene Expression Analysis
Total RNA was obtained from the Cb and F/M of three 4-month-old Tpp1–/– and wild-type mice by TRIzol (Thermo Fisher Scientific, Waltham, Massachusetts, USA) extraction followed by a QIAGEN RNeasy Kit® cleanup procedure (QIAGEN, Hilden, Germany) which included a DNase digestion step. RNA quality (RIN > 9.5) assessments, cDNA library preparation, and single-end sequencing with 50-nucleotide reads were performed by the University of Chicago Genomics Facility on Illumina platforms. RNA sequence files were transferred to the University of Chicago Center for Research Informatics’ Tarbell High-Performance Computing cluster for analysis.
The quality of raw sequencing data was assessed using FastQC v0.11.5 (Andrews, 2010); Illumina adapter/primer sequences were detected from sequencing reads. All RNA reads were first mapped to the mouse (GRCm38) reference genome using STAR v2.5.2b release with default parameters (Dobin et al., 2013). Picard v2.8.1 (http://broadinstitute.github.io/picard/) was used to collect mapping metrics.
The resulting files from the previous alignment step in the RNA-seq analysis were taken individually as input to quantify transcriptional expression using Cufflinks v2.2.1 (Trapnell et al., 2010; RRID:SCR_014597) and Rsubread:featureCounts v1.26.1 (Liao et al., 2014; RRID:SCR_009803). Afterward, several methods of differential expression analysis (Costa-Silva et al., 2017), including Cuffdiff v2.2.1 (Trapnell et al., 2010; RRID:SCR_014597), edgeR v3.18.1 (Robinson et al., 2010; RRID:SCR_012802), DESeq2 v1.16.1 (Love et al., 2014; RRID:SCR_015687), and limma v3.32.10 (Ritchie et al., 2015; RRID:SCR_010943), were used to discover DE-mRNA genes (fold change ≥ 1.5 and false discovery rate [FDR]) < 0.1 between pairwise groups based on the expression estimate of individual gene mRNAs. To obtain the groups with similar expression trends based on the identified DE-mRNA genes, several in-house scripts were implemented using R (https://www.rproject.org/) and Python (https://www.python.org/) languages.
The identified DE genes were further used as input to functional analysis modules for the identification of enrichment of functional categories and regulatory networks, using Gene Ontology (GO) terms (RRID:SCR_002811) and KEGG-enrichment analyses (RRID:SCR_012773), as well as Ingenuity Pathway Analysis (IPA; RRID:SCR_008653). Pathways significantly enriched in the genes of interest were identified using clusterProfiler (Yu et al., 2012; v3.6.0) at FDR-adjusted p value < .10 (hypergeometric test). Gene Set Enrichment Analysis was performed using clusterProfiler (Yu et al., 2012; v3.6.0; RRID:SCR_016884) as well.
The cell-type specificities of the DEG were determined based on transcriptome data from Zhang et al. (2014; http://web.stanford.edu/group/barres_lab/brain_rnaseq.html). Briefly, genes expressed in excess of fivefold higher in each cell type (neurons, astrocytes, microglia, oligodendrocytes, and endothelial cells) with respect to the bulk expression of all other cell types were considered enriched in that cell type. The individual datasets for each cell type were then matched to the DEG in each brain sample to assign cell-type specificity.
mRNA In Situ Hybridization
Control and Tpp1 mutant 4-month-old brains were fixed in 4% paraformaldehyde and processed for nonradioactive in situ hybridization (ISH) as described previously (Domowicz et al., 2008, 2011). To prepare the digoxigenin (DIG)-labeled RNA probes used for ISH, cDNA fragments from the glial fibrillary acidic protein (Gfap), period1 (Per1), and CD68 antigen (Cd68) genes were generated by PCR and inserted into pCRII dual-promoter vector plasmids (Invitrogen, Waltham, MA, USA). Sequencing of the cloned gene fragments was performed with an ABI PRISM 377XL sequencer (Perkin Elmer, Waltham, MA, USA) by the University of Chicago Cancer Center DNA sequencing facility. Riboprobes incorporating DIG-labeled nucleotides were synthesized from linearized PCR templates with SP6 or T7 RNA polymerase (Roche, Indianapolis, IN, USA). Probed mRNAs were detected after hybridization with an alkaline phosphatase-conjugated anti-DIG antibody (Roche, Indianapolis, IN, USA). Alkaline phosphatase activity was detected (blue color) using BCIP and NBT substrates (Roche, Indianapolis, IN, USA). A virtual library of the images was obtained using a CRi Pannoramic Scan Whole Slide Scanner (Perkin Elmer, Waltham, MA, USA), and final pictures were selected using the Pannoramic Viewer Software (RRID:SCR_014424).
Quantification of the number of CD68+ cells and sizes in Tpp1 mutant and control brains were performed on 10 cortical and thalamic fields using the Image J software package (RRID:SCR_003070). Unfortunately, levels of Cd68 mRNA in wild-type brain were too low to make a reliable quantification.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
RNA purified as described earlier was reverse transcribed with the High-Capacity Reverse Transcription Kit (Applied Biosystems, Waltham, MA, USA), and quantitative real-time reverse transcription polymerase chain reactions (RT-qPCRs) were performed using the SsoAdvanced™ Universal SYBR Green system (Bio-Rad Laboratories, Hercules, CA, USA) in a CFX-96 real-time instrument (Bio-Rad Laboratories, Hercules, CA, USA). For each gene, forward and reverse primers were designed using Primer-BLAST (NCBI; RRID:SCR_003095) and later tested in RT-qPCR; only primer pairs with 90% to 105% efficiency were used. Relative normalized expression values were determined using the ΔΔCt methods for the indicated target genes relative to Gapdh (glyceraldehyde-3-phosphate dehydrogenase) in the wild-type mouse sample. The data are expressed as means ± standard deviations. Determinations were performed in triplicate, and experiments were repeated with independent RNA samples at least three times with consistent results. Statistical significances were evaluated by the paired-samples Student’s t test. Values of p < .05 for the null hypothesis were considered significant.
Results
Tpp1-Mutant Brain Transcriptome Analysis
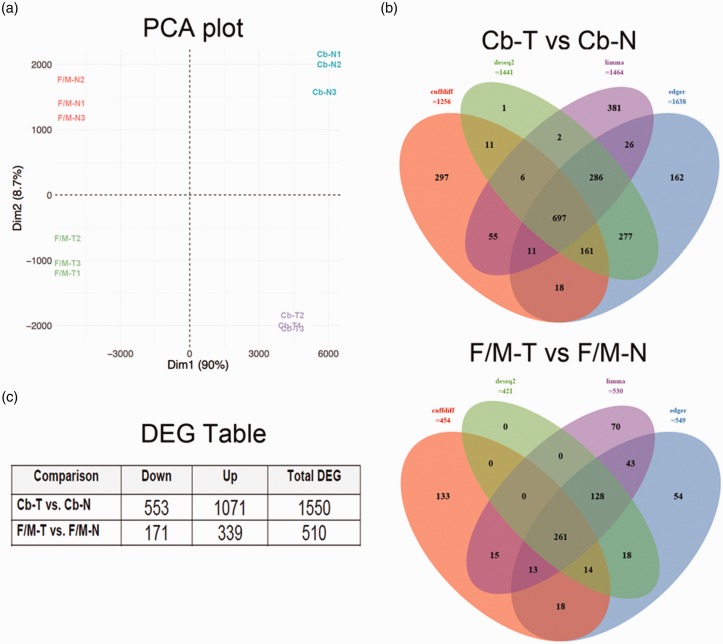
To analyze the pattern of DEG underlying the pathology in this end-stage model of cLINCL, RNA-seq of F/M and Cb RNA from 4-month-old Tpp1–/– (T) and wild-type control (N) brains was conducted. Readouts obtained from RNA-seq (and passing quality control) were aligned to the UCSC Mus musculus mm10 reference genome; 88.3% of the reads were successfully mapped to the mouse reference genome. After eliminating low-expressed-mRNA genes, 15,294 genes out of 52,550 annotated mouse genes were further analyzed. Dimensionality reduction, an informative approach for clustering and exploring the relationships between samples, was conducted with the principal component analysis plot based on the normalized mRNA expression profiles. As presented in Figure 1(a), four obvious groups were separated in terms of tissues and treatments. To identify DE-mRNA genes between pairwise groups, several state-of-the-art tools (Costa-Silva et al., 2017), including Cuffdiff, DEseq2, edgeR, and limma were used. Two pairwise comparisons, F/M-T versus F/M-N and Cb-T versus Cb-N, were conducted using the criteria of fold changes greater than 1.5 and FDR less than 0.1. To identify a more robust collection of intrinsically regulated genes, the overlapped DEG detected by more than one method were considered as the final set of DEG for further analysis. Venn diagrams of overlapping DEG identified from the previously mentioned methods are shown in Figure 1(b). A total of 1,550 and 510 mRNAs were found to be differentially expressed in Cb and F/M areas, respectively (Figure 1(c)).
Figure 1.
Statistical determination of DEG. (a) PCA plot of all samples based on normalized expressions. Control forebrain/midbrain (F/M-N), Tpp1–/– forebrain/midbrain (F/M-T), control cerebellum (Cb-N), and Tpp1–/– cerebellum (Cb-T) of triplicate samples are represented. (b) Venn diagram of differentially expressed mRNA genes identified by four different methods: Cuffdiff, edgeR, DESeq2, and limma. Only genes identified by at least two methods were used for further analysis. (c) Number of up-/downregulated DEG in different comparison(s).
PCA = principal component analysis; DEG = differentially expressed genes.
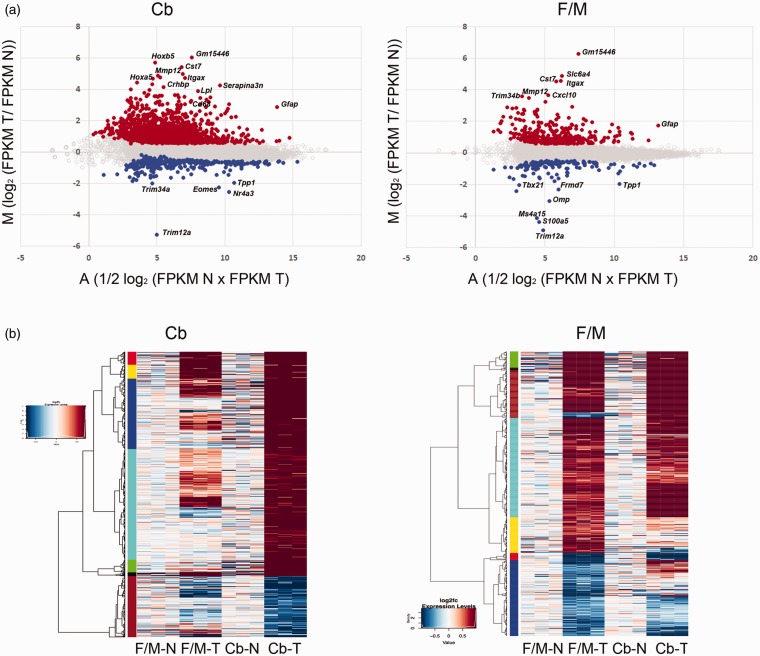
The differential gene expressions in the Tpp1–/– samples compared with controls are visualized in Figure 2, where the MA plots present the ratio of FPKM (Fragments Per Kilobase of transcript per Million mapped reads) expression values between the two conditions. All 15,294 genes are represented in the plot with DEG highlighted in different colors (Figure 2(a)). The heatmap in Figure 2(b) demonstrates the hierarchical clustering between the three controls and Tpp1–/– samples from Cb and F/M areas, thus illustrating differential expression of the significantly regulated genes. For comparison, the levels of expression of the Cb and F/M areas are plotted in both heatmaps. It is noteworthy that a considerable number (299) of the genes whose expressions are significantly changed in Cb also have changed expression (or have a tendency to change; Table S1) in the same direction in the F/M area and vice versa. This finding suggests that many of the networks and pathways implicated in the disease are likely affected in the whole brain but that the Cb is perhaps further along (more affected) in developing the pathology. A full list of DEG is provided in supplemental data (Table S1), and all sequence FASTQ files are available from the Gene Expression Omnibus (accession number # GSE123509).
Figure 2.
Visualization of DEG. (a) MA plot representing the ratio of FPKM expression values between Tpp1–/– cerebellum (Cb) and forebrain/midbrain (F/M) areas and their corresponding control areas (N) plotted against their average. The statistically significant genes with >1.5 fold change and false discovery rate of less than 0.1 are plotted in orange color (upregulated genes) and blue (downregulated genes). The rest of the genes are represented with gray open circles. (b) Heat map displaying the 1,550 and 510 genes that are differentially expressed between Wt (N) versus Tpp1–/– (T) cerebellum (Cb) and forebrain/midbrain (F/M), respectively. Representation of the same genes in complementary samples is also included for comparison. Each column represents an individual triplicate. Red color represents relative increase in abundance, blue color represents relative decrease, and white color represents no change as measured by log2fc (Log2 of T vs. N ratio) and represented in the inset. The rows are organized by hierarchical clustering (represented at the left).
FPKM= Fragments Per Kilobase of transcript per Million mapped reads.
DEG Cell Type Profile Analysis
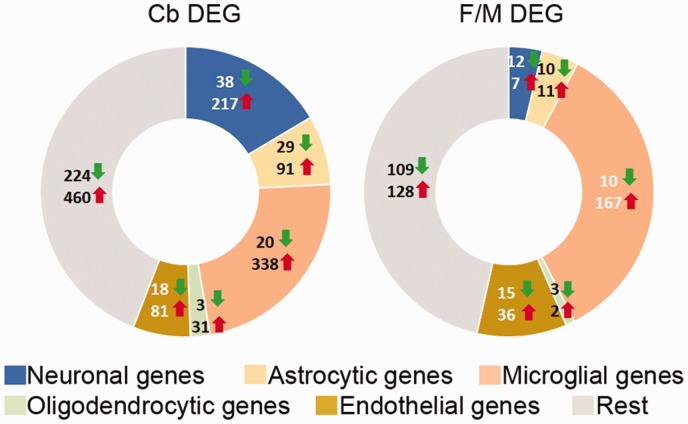
We also determined the cell-type enrichment profile for both sets of DEG based on reported cell-type transcriptomes (Zhang et al., 2014; Figure 3). Even though 44% to 46% of the genes are unassigned (probably because they are expressed in several cell types), these data indicate that the microglial transcriptome is highly altered in the Tpp1–/– animals, followed in descending order of alteration by those of neurons, astrocytes, endothelial cells, and the least-affected cell types, oligodendrocytes. The most strongly upregulated genes in astrocytes from both F/M and Cb were Serpina3n, Gfap, C4b, and Aspg, which were all found to be expressed in reactive astrocytes after lipopolysaccharide injection or middle cerebral artery occlution brain injury (Zamanian et al., 2012) and are also elevated in aging astrocytes (Clarke et al., 2018). Many genes were associated with activated microglia; among the largest changes are Itgax, Ccl3, C3, and Rgs1, which have also been shown to be increased in human aging microglia (Olah et al., 2018).
Figure 3.
Identification of cell-type-specific DEG in wt and Tpp1–/– brain regions. Distribution of cell-type-specific genes that are commonly enriched (upregulated: red up arrow; downregulated green down arrow) in between DEG from Cb-T versus Cb-N and F/M-T versus F/M-N comparisons. Cell-type specificity of DEG was established from data imputed from Zhang et al. (2014; http://web.stanford.edu/group/barres_lab/brain_rnaseq.html) based on genes that were exclusively expressed in each cell type and had FPKM expression values at least five time higher than in the rest of the other cell types.
DEG = differentially expressed genes.
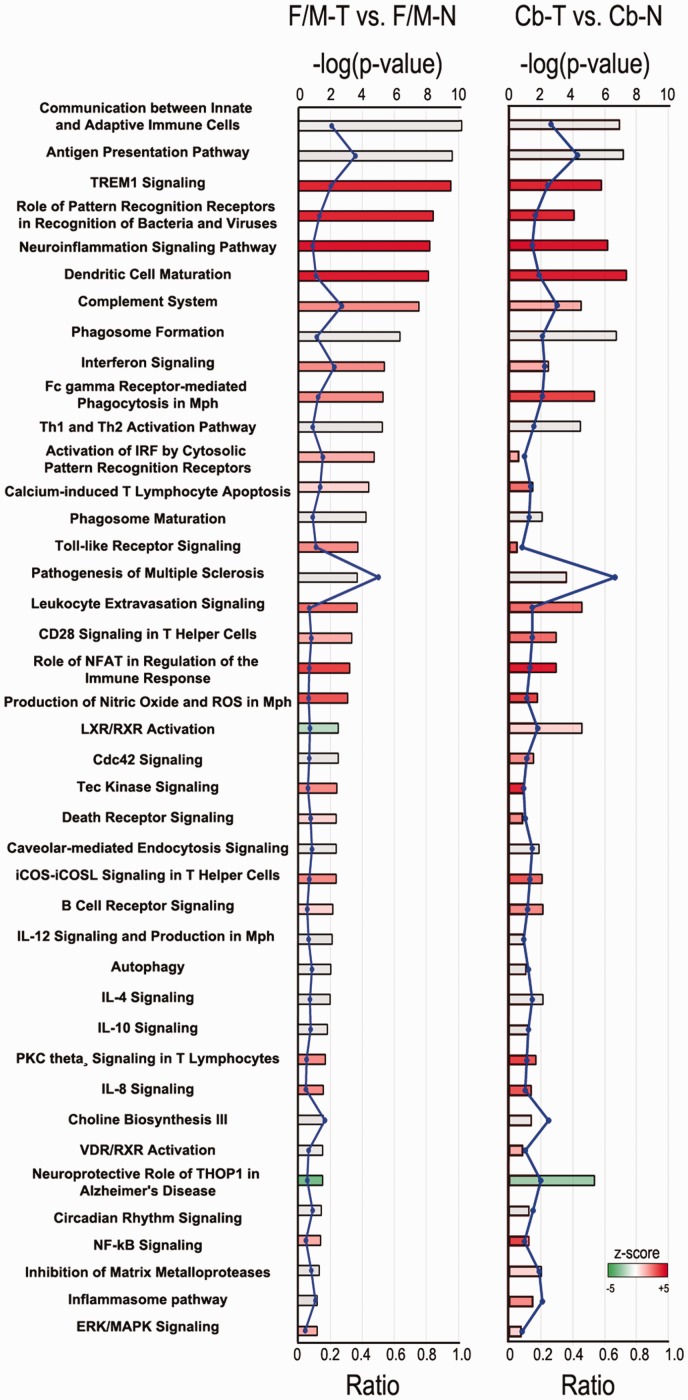
In addition, cellular processes and upstream regulators associated with the DEG found in our Tpp1–/– samples relative to controls were identified using QIAGEN’s IPA® (www.qiagen.com/ingenuity). A selected list of canonical pathways detected are represented in Figure 4 including a representation of the modified gene expression ratio (number of genes from the DEG list that map to the pathway divided by the total number of genes mapping to the same pathway) and a representation of the z score, where a positive z score (red) indicates a predicted activation, and a negative z score (green) indicates a predicted inactivation of the enriched pathway. Several of these canonical pathways, which can be considered relevant to this disease, are further highlighted in Table 1, with a detailed list of DEG found to be involved in each pathway. For a complete list of canonical pathways and involved DEG, see Table S2 for Cb and Table S3 for F/M areas.
Figure 4.
Ingenuity Pathway Analysis (IPA) from DEG in Tpp1–/– versus N mouse brain. Selected canonical pathways identified using IPA gene ontology algorithms for F/M and Cb areas scored as −log(p value) from Fisher’s exact test, set here to a threshold of 1.25. Bars are colored according to the z score (positive z score is red, negative z score is green, and no activity pattern available is gray). The ratio (blue dots connected by a line) indicates the ratio of genes from the dataset that map to the pathway divided by the total number of genes that map to the same pathway. For a complete list of canonical pathways and genes involved, see Table S2 and S3.
F/M-N = control forebrain/midbrain; F/M-T = Tpp1–/– forebrain/midbrain; Cb-N = control cerebellum; Cb-T = Tpp1–/– cerebellum; NFAT = nuclear factor of activated T-cells; ROS = reactive oxygen species; LXR = liver X receptor; RXR = retinoid X receptor; iCOS = inducible T-cell costimulator; iCOSL = inducible T-cell costimulator ligand; IL = interleukin; VDR = vitamin D receptor; PKC= protein kinase C; NFκB = nuclear factor kappa-light-chain-enhancer of activated B cells; ERK = extracellular signal-regulated kinases; MAPK = mitogen-activated protein kinases.
Table 1.
Selected Canonical Pathways and Associated Differentially Expressed Genes in Tpp1–/– Versus Wild-Type Cerebellum (Cb) and Forebrain/Midbrain (F/M).
| Tissue | Ingenuity canonical pathways | –log(p value) | Ratio | z score | DEG |
|---|---|---|---|---|---|
| Cb | TREM1 signaling | 5.82 | 0.246 | 3.638 | Icam1, Tyrobp, Nlrp10, Lat2, Itga5, Cd83, Fcgr2b, Tlr9, Tlr2, Nlrc5, Ccl2, Plcg2,Tlr7, Tlr13, Cd86, Tlr3, Itgax |
| F/M | 9.52 | 0.203 | 3.207 | Sigirr, Nlrp3, Icam1, Tyrobp, Lat2, Fcgr2b, Tlr2, Nlrc5, Ccl12, Tlr1, Tlr7, Tlr13, Cd86, Itgax | |
| Cb | Neuroinflammation signaling pathway | 6.2 | 0.147 | 5 | B2m, Gabra5, Tgfbr1, Icam1, Bdnf, Pycard, H2-Ab1, H2-Aa, Cx3cr1, Ccl5, H2-Q6, Cxcl10, Gabrg3, Hmox1, Ccl12, H2-DMa, Tgfb1, Pik3cg, Pla2g5, Cybb, Tlr7, Tlr3, Gabrq, Tnfrsf1a, Tyrobp, Pla2g4c, Pla2g3, Tlr9, Csf1r, Tlr2, Slc6a11, Irf7, Gabrr2, Trem2, Syk, Plcg2, Pla2g4b, Ncf2, Cd86, Tlr13, Mmp9, H2-Eb1 |
| F/M | 8.21 | 0.0877 | 4.491 | B2m, H2-Oa, Nlrp3,Icam1,Ager,Tyrobp,H2-Q6,H2-Ab1, Ccl5, Ngf, Tlr2, Cxcl10,Hmox1, Irf7,Ccl12,Tgfb1, Trem2, Ncf2,Tlr1, Cybb, Tlr7, Tlr13, Cd86, Stat1, H2-Eb1 | |
| Cb | Complement system | 4.5 | 0.303 | 1.633 | C4a/C4b, Itgb2, Itgam, C3, Masp2, C1qc, C1qa, C1qb, C3ar1, Itgax |
| F/M | 7.52 | 0.273 | 2.236 | C4a/C4b,Itgb2,Cd59,C3,C1qc,C1qa,C1qb, C3ar1, Itgax | |
| Cb | Role of NFAT in regulation of the immune response | 2.93 | 0.131 | 4.69 | Blnk, Gng4, Plcb2,H2-Q6,Fcgr2a,H2-Aa, H2-Ab1, Fcgr2b,Tlr9, Fcgr1a, Btk, Lck, Gna15,H2-DMa, Pik3cg, Plcg2,Syk, Zap70, Fcer1g,Cd86lcp2,H2-Eb1, Fcgr3a/Fcgr3b |
| F/M | 3.22 | 0.0682 | 3.464 | Btk, H2-Oa, Gna15, H2-Q6, Fcgr2a, H2-DAa, Fcer1g, Cd86, Fcgr2b, Fcgr1a, H2-Eb1, Fcgr3a/Fcgr3b | |
| Cb | Leukocyte extravasation signaling | 4.55 | 0.146 | 2.887 | Rac2, Icam1, Rock2, Timp1, Pik3cg, Cyba, Cybb, Mmp12, Mmp19, Timp2, Pxn, Cxcr4, Itga5, Mmp2, Rapgef3, Ncf4, Tlr9, Selplg, Btk, Itgb2, Ncf1, Itgam, Arhgap9, Was, Plcg2, Ncf2, Cd44, Vav1,Cldn14, Mmp9 |
| F/M | 3.67 | 0.0683 | 2.309 | Rac2, Icam1, Ncf4, Btk, Itgb2, Was, Timp1, Cyba, Prkcd, Ncf2, Cybb, Cldn14, Vav1, Mmp12 | |
| Cb | Phagosome formation | 6.71 | 0.208 | n.a.p.a | Mrc1, Plcb2, Fcgr2a, Itga5, Plcl2, Plch2, Fcgr2b, Tlr9, Fcgr1, Inpp5d, Tlr2, Itgb2, Fcrls, Rhov, Itgam, Syk, Pik3cg, Plcg2, Fcer1g, Tlr7, Tlr13, Tlr3, C3ar1, Fcgr3a/Fcgr3b, Itgax |
| F/M | 6.33 | 0.117 | n.a.p.a | Tlr2, Itgb2, Fcrls, Fcgr2a, Prkcd, Tlr1, Tlr7, Fcer1g, Tlr13, Fcgr2b, Fcgr1a, C3ar1, Fcgr3a/Fcgr3b, Itgax | |
| Cb | Phagosome maturation | 2.09 | 0.124 | n.a.p.a | B2m, Lpo, H2-Q6, Ctsw, Dynlt1, Tap1, Prdx6, Ctsz, Ctsd, Tubb6, Ctsh, Ctss, Ncf2, Rab7b, Cybb, Ctsc, H2-Eb1 |
| F/M | 4.24 | 0.0876 | n.a.p.a | B2m, Ctsz, Ctsd, Ctss, Ctsh, H2-Q6, Lpo, Ncf2, Cybb, Ctsc, Tap1, H2-Eb1 | |
| Cb | Production of nitric oxide and reactive oxygen species | 1.77 | 0.109 | 3.771 | Apoe, Ptpn6, Tnfrsf1a, Map3k13, Ncf4, Tlr9, Spi1, Irf1, Tlr2, Rhov, Lyz, Ncf1, Pik3cg, Plcg2, Cyba, Ncf2, Cybb, Irf8, Tnfrsf1b, Clu |
| F/M | 3.07 | 0.0656 | 2.53 | Tlr2, Lyz, Ptpn6, Prkcd, Ppm1j, Cyba, Ncf2, Cybb, Ncf4, Irf8, Stat1, Spi1 | |
| Cb | Toll-like receptor signaling | 0.48 | 0.0833 | 2.449 | Tlr2, Il33, Tlr7, Cd14, Tlr3, Tlr9 |
| F/M | 3.71 | 0.111 | 1.89 | Sigirr, Tlr2, Ly96, Il1a, Tlr1, Tlr7, Cd14, Eif2ak2 | |
| Cb | Autophagy | 1.05 | 0.119 | n.a.p.a | Ctsz, Ctsd, Ctss, Ctsh, Ctsw, Ctsc, Atg9b |
| F/M | 2 | 0.0847 | n.a.p.a | Ctsz, Ctsd, Ctss, Ctsh, Ctsc | |
| Cb | Circadian rhythm signaling | 1.21 | 0.152 | n.a.p.a | Per1, Vipr2, Adcyap1, Vip, Per2 |
| F/M | 1.44 | 0.0909 | n.a.p.a | Per1, Vipr2, Per2 | |
| Cb | Inflammasome pathway | 1.49 | 0.211 | 2 | Naip, Aim2, Pycard, Panx1 |
| F/M | 1.18 | 0.105 | n.a.p.a | Naip, Nlrp3 |
Note. DEG = differentially expressed genes; n.a.p.a. = no activity pattern available; NFAT = nuclear factor of activated T-cells.
Neuroinflammation Signaling Pathway
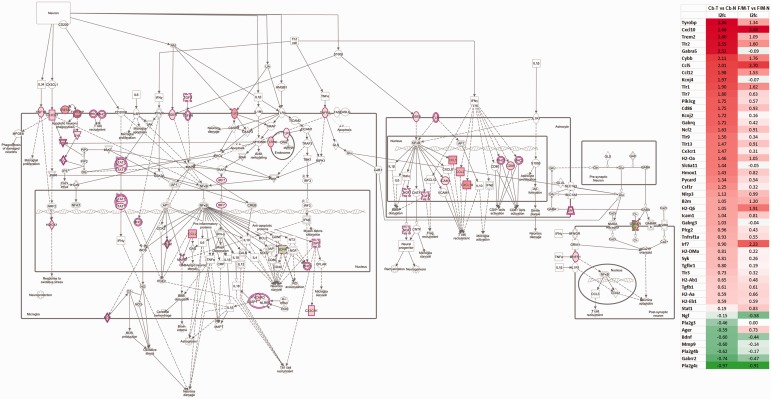
The list of DEG reveals a large number of genes with roles in the inflammatory process, in particular those reported to be expressed by microglia and astrocytes at their activated stage; thus, it is not surprising to see the neuroinflammation signaling pathway as one of those most significantly altered in the Tpp1–/– Cb and F/M in our IPA (Figures 4 and 5; Table 1). As indicated, 25 upregulated genes were found in the F/M area, and 42 altered expression genes were found in Cb (36 upregulated and 6 downregulated), all associated with the neuroinflammation pathway (Figure 5). In-depth analysis of the genes affected in this pathway highlights the possible regulatory process activated in microglia and astrocytes which may lead to altered postsynaptic behavior (Figure 5), in particular in Cb where gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 (Gabrr2) is strongly downregulated. More than 75% of the DEG associated with this pathway in F/M areas were also altered in Cb, indicating similar inflammatory processes are occurring in both areas, but with a higher positive z score, that is, stronger involvement, in the Cb where more gene expressions are altered, and levels of gene activation are higher (Figure 5).
Figure 5.
Neuroinflammation pathway. Canonical pathway analysis by IPA highlights the genes affected in the neuroinflammation pathways for Cb and F/M. Involved DEG detected by RNA-seq analysis and change ratio (l2fc=log2 of T. N ratio) are indicated to the right.
F/M-N = control forebrain/midbrain; F/M-T = Tpp1–/– forebrain/midbrain; Cb-N = control cerebellum; Cb-T = Tpp1–/– cerebellum.
IPA analysis also identifies, in both brain areas, several highly significant upstream regulators; that is, the transcription factors, Irf3 (F/M: p < 8.5E-36, z = 5.2; Cb: p < 6.6E-17, z = 4.7); Irf7 (F/M: p < 1.1E-28, z = 4.6; Cb: p < 5.7E-15, z = 4.1); the interleukin Ifng (interferon gamma; F/M: p < 9.9E-33, z = 6.3; Cb: p < 1.5E-26, z = 7.1); and interferon alpha/beta receptors (Ifnar1 and Ifnar2; F/M: p < 1.4E-44, z = 6.2; Cb: p < 1.5E-25, z = 6.2). Among the factors in this list, only Irf7 message is actually upregulated in both brain areas. Furthermore, Irf7, a critical transcription factor for the induction and positive feedback regulation of type I IFN signaling (Seth et al., 2006), has been found upregulated in experimental autoimmune encephalomyelitis (EAE; Salem et al., 2011) and is known to play a critical role with Tgfβ in microglia inflammatory response (Cohen et al., 2014).
Many of the highly significantly modified canonical pathways are associated with the immune system response and are consistent with findings for other neurodegenerative diseases such as multiple sclerosis (MS) and Alzheimer’s disease (AD; see Figure 4). For example, the use of single-cell RNA-seq gene expression profiling recently allowed the characterization of Alzheimer’s disease-associated microglia (DAM; Deczkowska et al., 2018) as having increased levels of Trem2 and Tyrobp expression, which was consistent with the increases observed in our data (Figure 5).
Histological and Quantitative Changes Associated With Astrocytes and Microglia
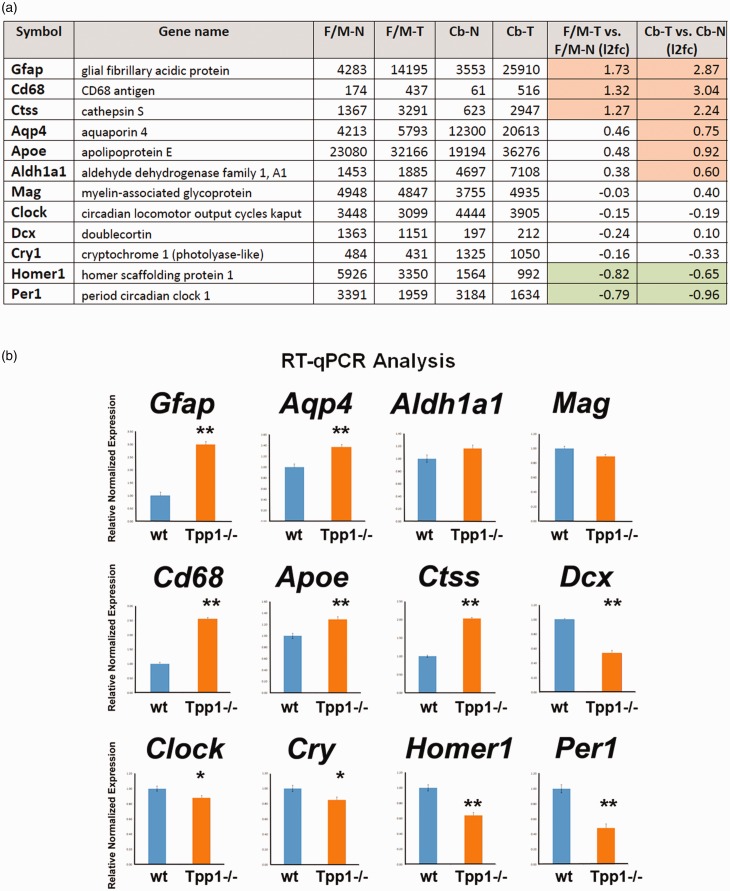
To confirm and validate the RNA-seq data, several genes were selected, and transcriptional differences were confirmed by RT-qPCR and mRNA ISH. The genes chosen encompass a broad range of expression levels and include significant (highlighted in color) and nonsignificant up- and downregulated genes (Figure 6(a)). Consistent results were obtained for Cb and F/M samples; thus, only F/M samples are represented (Figure 6(b)). The magnitude and directions of the differences obtained by qPCR are consistent with the RNA-seq data. Moreover, in F/M areas, tendencies in the RNA-seq that do not reach significance, such as Apoe and Aqp4, were confirmed to be significantly changed by qPCR (Figure 6(b)). These results may indicate that due to the high level of stringency in the statistical analysis of RNA-seq, the differences reported by this technique are underestimating the total numbers of differentially expressed transcripts and stress the need to extend the assessment of DEG to other techniques as well.
Figure 6.
Corroboration of RNA-seq results by RT-qPCR for selected genes. (a) Table counts per million (CPM) data from RNA-seq study and log2 of Tpp1–/– (T) versus control (N) ratio (l2fc) in Cb and F/M for 12 genes. Significant change ratios are highlighted in pink (upregulated) and green (downregulated genes). (b) Relative transcript expression of the same 12 genes quantified by qPCR in independent F/M RNA extractions, performed in triplicate. Relative normalized expression values were determined from the ΔΔCt for the indicated target genes relative to Actb (β–actin) expression in control brains. *p < .02; **p < .0001.
F/M-N = control forebrain/midbrain; F/M-T = Tpp1–/– forebrain/midbrain; Cb-N = control cerebellum; Cb-T = Tpp1–/– cerebellum; l2fc=log2 of T. N ratio; Tpp1 = tripeptidyl peptidase 1; RT-qPCR = quantitative real-time reverse transcription polymerase chain reaction; Gfap = glial fibrillary acidic protein; Aqp4 = aquaporin 4; Cd68 = CD68 antigen; Per1 = period1.
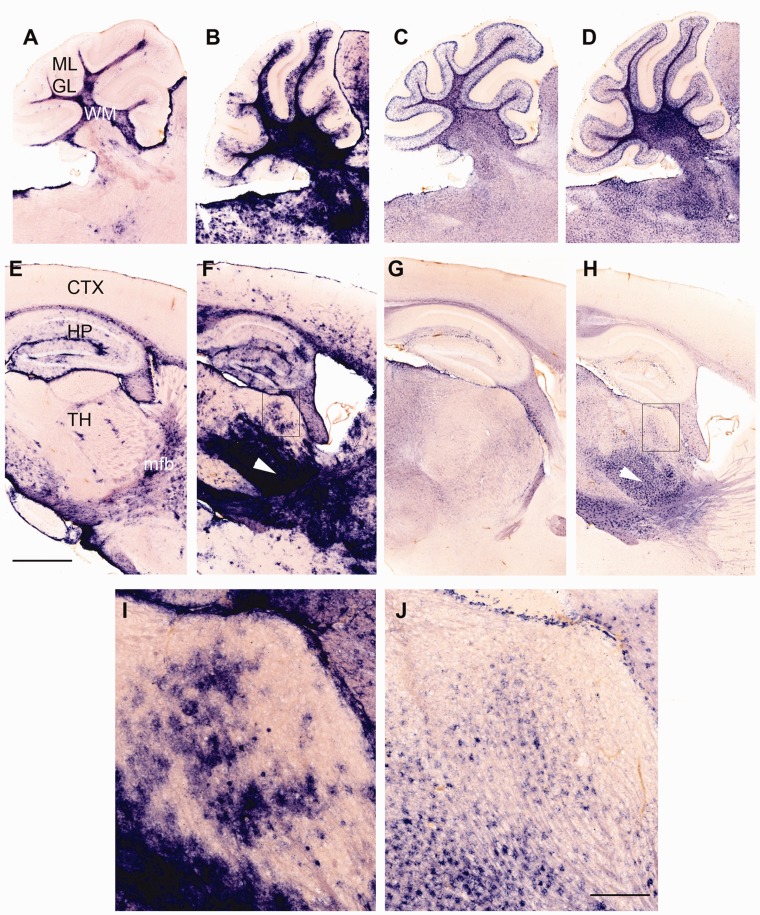
Transcripts from astrocytes and microglia were found to be strongly dysregulated in the Tpp1–/– mouse brain on the basis of our DEG cell-type enrichment profile (Figure 3); thus, we analyzed the histopathological changes in these two cell types using mRNA ISH. For astrocytes, ISHs in brain sagittal sections were performed using Gfap and Aqp4 probes (Figure 7). The Gfap transcript, which is well known to be upregulated in reactive astrocytes (Pekny et al., 2014), was found to be increased in all areas of the brain in a patchy or punctuated pattern (Figure 7(f) and (i)). Certain areas such as the ventral thalamus, medial forebrain bundle (Figure 7(f) and (i)), cerebellar white matter, and granular layer (Figure7(b)) were more distinctly affected, confirming the previously described distribution of reactive astrocytes in Tpp1–/– brains by immunohistochemistry (Sleat et al., 2004). The aquaporin 4 (Aqp4) transcript, which has also been reported to be expressed by astrocytes and increasingly so after brain injury (Vizuete et al., 1999; Saadoun and Papadopoulos, 2010; Domowicz et al., 2018), was also upregulated in the same areas as that of Gfap. The granular distribution of Aqp4 mRNA-expressing cells provides confirmation that astrocytes are being activated on-site in areas of neuronal damage (Figure 7(h) and (j)).
Figure 7.
Altered expression patterns of astrocytic markers in Tpp1–/– 4-month-old brains. Expression patterns of GFAP (a, b, e, f, i) and Aqp4 (c, d, g, h, j) transcripts in Tpp1–/– (b, f, i and d, h, j) and control (a, e and d, h) in 4-month-old brains by in situ hybridization (blue staining). Cerebellar (a to d) and forebrain areas (e to i) are shown. (i) and (j) depict close-ups of areas indicated in (f) and (h). White arrowhead in (f) and (h) indicates ventral thalamus. Scale bars: (a) to (g) = 1,000 μm; (i) and (j) = 200 μm.
ML = molecular layer; GL = granular layer; WM = molecular matter; CTX = cortex; HP = hippocampus; TH = thalamus; mfb = medial forebrain bundle.
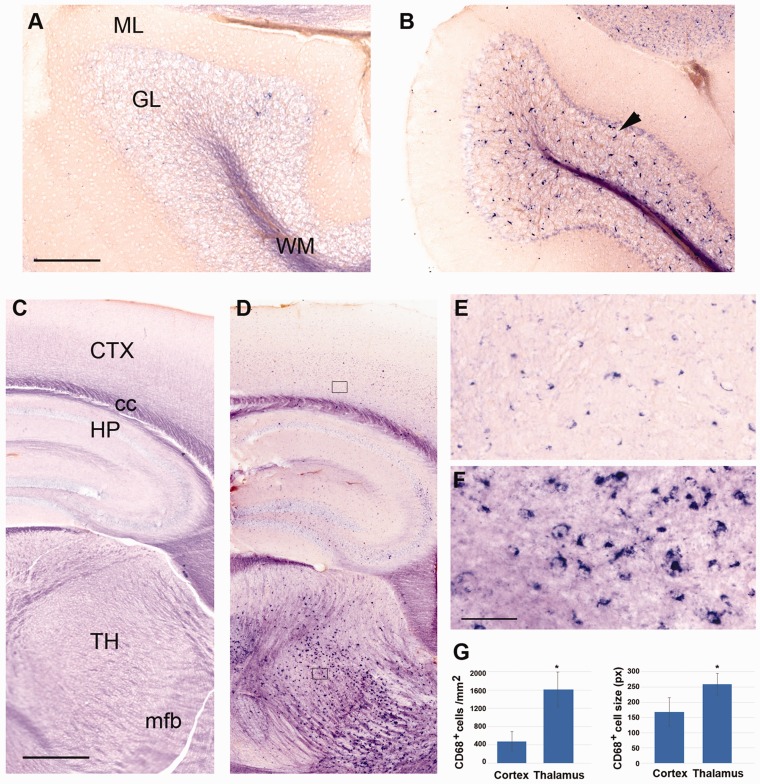
Neuronal damage is effectively sensed by microglia (Ransohoff and Perry, 2009), so it is not surprising that a large number of genes associated with microglia activation were found among the identified DEG (Figure 3). For instance, CD68 is a lysosomal protein that is upregulated in microglia in the brains of several neurodegeneration disorders (Bevan et al., 2018; Linnartz-Gerlach et al., 2019), as well as in human microglia from aged populations (Raj et al., 2017; Olah et al., 2018) and is frequently used as a marker for reactive microglia. Marked elevation of Cd68 expression was observed in the mRNA quantifications and by ISH in the Tpp1–/– brains (Figure 8). While CD68-expressing cells are rarely detected in normal brain (Figure 8(a) and (c)), we found elevated Cd68 expression in morphologically altered microglia throughout the entire 4-month-old Tpp1–/– brain (Figure 8(b), (d) to (f)). In thalamic and cerebellar areas, these changes were more marked with higher microglia density and clustered expression (Figure 8(b), (d) to (f)), indicating preferential areas of microglia activation. Significant increases in the number and size of CD68+ cells were also quantified in thalamus compared with cortex in Tpp1–/– brains (Figure 8(g)).
Figure 8.
Altered expression patterns of Cd68 in Tpp1–/– 4-month-old brains. Expression pattern of CD68 mRNA in cerebellum (a and b) and forebrain (c to f) in Tpp1–/– (b, d, e, f) and control (a, c) 4-month-old brains by in situ hybridization (blue staining). (e) and (f) represent an enlargement of sections boxed in (d). Quantification of the number of CD68+ cell per mm2 and cell size (in pixels) for Tpp1–/– cortex and thalamus are compared in (g). Black arrowhead indicates CD68+ cells in granular cell layer of Tpp1–/– cerebellum. Scale bars: (a) and (b) = 200 μm; (c) and (d) = 500 μm; (e) and (f) = 50 μm.
ML = molecular layer; GL = granular layer; WM = white matter; CTX = cortex; cc = corpus callosum; HP = hippocampus; TH = thalamus; mfb = medial forebrain bundle.
Circadian Rhythm
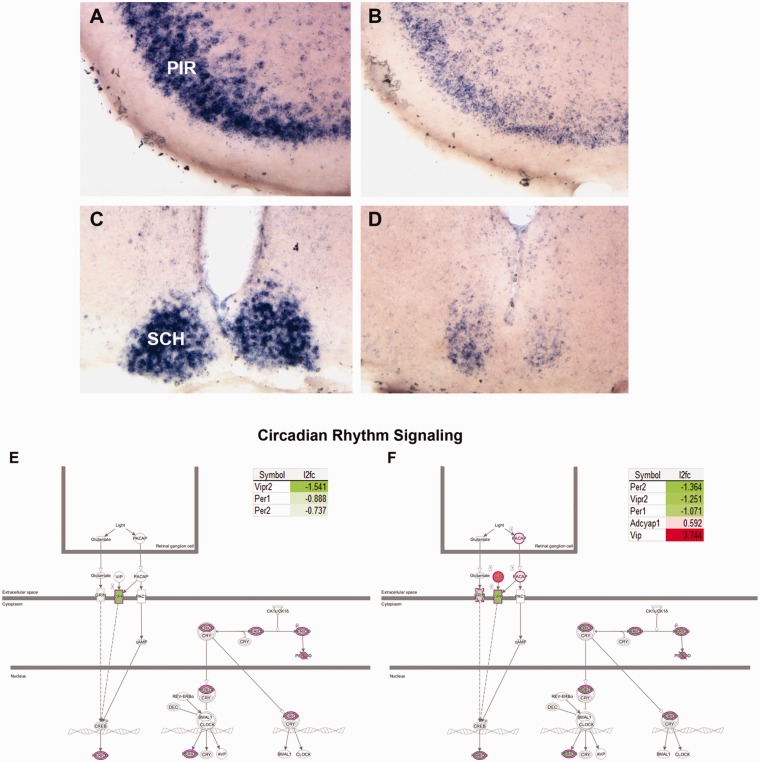
Levels of Per1 and Per2 were downregulated in theTpp1–/– mouse model, which is indicative of circadian rhythm abnormalities and correlates with the altered sleep patterns of Batten patients (Lehwald et al., 2016; Williams et al., 2017). In fact, IPA demonstrated significant alterations in the circadian rhythm pathway (Figure 9); these were further confirmed by the downregulation of Per1 found by qPCR (Figure 6) and mRNA ISH, which showed neuronal populations of the piriform cortex and the suprachiasmatic nuclei’s being more affected (Figure 9 (a) to (d)). The latter structure is known to have innervations from retinal ganglion cells and is key in coordinating the light cycle with sleep patterns (Abreu and Braganca, 2015; Ono et al., 2015; Roenneberg and Merrow, 2016). Alterations in the circadian rhythm signaling pathways for Cb and F/M are shown in Figure 9 (e and f) and include several genes besides Per1 and Per2. Interestingly, this study includes Clock and Cry (analyzed by qPCR, see Figure 6(b)) in the list of genes with altered expression in this pathway. In addition, Homer1, one of the rhythmic control transcripts (Maret et al., 2007), was also downregulated in the F/M transcriptome of the Tpp1–/– mouse brain.
Figure 9.
Loss of circadian rhythm gene expression in Tpp1–/– brains. Loss of Per1 mRNA expression in PIR (a and b) and the SCH (c and d) in Tpp1–/– (b, d) 4-month-old brains compared with control heterozygous (a, c). Canonical pathway analysis by IPA highlights the genes affected in the circadian rhythm signaling pathways for F/M (e) and Cb (f). Involved DEG detected by RNA-seq analysis and change ratio (l2fc=log2 of T vs. N ratio) are indicated.
PIR = piriform cortex; SCH = suprachiasmatic nuclei.
Discussion
A few reports have analyzed brain gene expression profiles in NCL animal models (Cln1, Cln5, and Cln3; Brooks et al., 2003; Elshatory et al., 2003; von Schantz et al., 2008) as well as in a cln3- Dictyostelium model (Mathavarajah et al., 2018). Because most of these reports used microarray methodology and the diseases have different times of onset, data comparison is difficult to perform, and only a few common gene expression changes can be confirmed in Tpp1–/– brains. Here, we report the first RNA-seq analysis in a Batten disease animal model. Our findings show that the transcriptional characterization of brain gene expression changes due to the lack of TPP1 during the end stages (at 4 months of age) of the disease not only defined the type of neuroinflammation process occurring but also identified a novel affected pathway (i.e., circadian rhythm), both of which contribute to a better understanding of disease pathophysiology in Batten disease (NCL).
In several NCL forms, studies on the involvement of microglia in the inflammation process have been reported. In the most aggressive form of NCL (infantile NCL due to CLN1 mutations), increased proinflammatory cytokine production and peripheral immune cell infiltration of the brain were described (Macauley et al., 2012). Furthermore, anti-inflammatory treatments were successful in increasing the survival rate (Macauley et al., 2012). In animal models of JNCL, characterized by mutations of CLN3, primary microglia were found to be in a proinflammatory state (Xiong and Kielian, 2013), and microglial and astrocytic activation were shown to precede neural cell death (Pontikis et al., 2004). Our study extends these observations to a mouse model of cLINCL in which upregulation of Cd68, visualized by ISH (Figure 8), and transcriptional changes associated with microglia (Figure 3 and Supplemental Table S1) confirm its activation through upregulation of cell surface receptors such as Csf1r, Cx3cr1, Trem2, Tyrobp, Tgfr1, and Tnfrsf1a. Similar to AD and in aging (Zhang et al., 2013; Soreq et al., 2017; Mostafavi et al., 2018), microglia constitute the predominant transcriptional footprint in Tpp1 mutant mouse brain. We also confirmed differential increases in the number of cells in particular brain areas as well as morphological changes (Figure 8) in the Tpp1–/– brain. Recently, a study using single-cell RNA-seq technology in Tg-AD models revealed a two-step pathway of DAM development; an initial TREM2-independent pathway (Stage 1) followed by a TREM2-dependent pathway (Stage 2). Signatures for both types of microglia stages were found in our analysis of Tpp1–/– brain (Stage 1 DAM: Tyrobp, Ctsd, Apoe, B2m, and Lyz2; and Stage 2: Axl, Trem2, Itgax, Csf1, Cst7, Lpl, Cd9, Ccl6, Lilrb4a, and Timp2; Keren-Shaul et al., 2017). Thus, it can be expected that microglia in both stages are present in the Tpp1–/– brain during the end stage of the disease. It remains to be studied whether the Stage-2 DAM that have increased phagocytic activities are functioning normally in the mutant brain, as lysosomal processes might be altered in the absence of TPP1 and defects in microglial function might contribute to the rate of progression of the disease.
It is expected that microglia are initially activated by endogenous ligands generated as a result of neuronal damage, perhaps through toll-like receptors (TLRs; Fiebich et al., 2018) and the complement pathway. Upregulation of genes involved in the complement pathway (Figure 4) is probably linked to microglial and astrocytic response to neuronal damage. Genes from the classical (C4b, C1a, C1b, and C1c) and alternate pathways (C3) of complement activation as well as its receptor (C3ar1) are upregulated. It is possible that activated astrocytes increase the production and release of C3, which in turn interacts with neuronal and microglial C3aR, altering neuronal function and regulation of phagocytosis, respectively, as has been described for models of AD (Lian et al., 2015; Lian et al., 2016; Litvinchuk et al., 2018). TLRs have been shown to have an impact on noninfectious CNS disease and injuries because they can bind a number of endogenous molecules liberated from damaged tissues (Hanke and Kielian, 2011; Fiebich et al., 2018). Involvement of toll receptor signaling was also observed in Tpp1–/– brains; in particular, Tlr2 and Tlr7 were activated in both areas analyzed. In contrast with observations in AD, Parkinson’s disease, and amyotrophic lateral sclerosis patients (Letiembre et al., 2009; Casula et al., 2011), Tlr4 mRNA was not upregulated in Tpp1–/– brains, which argues for a unique pathway of microglia activation in this model. It is also important to point out that the three Tlrs—Tlr3, Tlr7, and Tlr9—known to be located in endosomes and lysosomes are elevated in Cb (Figure 5 inset) of Tpp1–/– animals.
Interestingly, two members of the CLN family are upregulated in the Tpp1–/– brain, cathepsin D (Ctsd), and granulin (Grn), but because these genes have been also found to be upregulated in microglia from other neurodegenerative diseases (i.e., amyotrophic lateral sclerosis), it is unclear if their upregulation in this cLINCL model is related to microglia activation or dysregulation of lysosomal function. Regarding the storage material that characterizes cLINCL, it is interesting that although the subunit C of mitochondrial ATP synthase is the major component of the storage material (Jalanko and Braulke, 2009), no transcriptional changes in the levels of this gene family (Atp5g1) were detected, confirming that the accumulation must be due to failure in catabolic protein processing. In contrast, changes related to metabolism of lipids and their transport were extensively represented in the enrichGO analysis (Table S4) in agreement with the multiple lipid alterations described previously for this family of diseases (Kollmann et al., 2013; Kim et al., 2017).
Extensive and severe loss of neurons has been described in cLINCL patients and the Tpp1-targeted mouse, and we identified the expression of a large number of neuronal genes affected, particularly in the Cb. Furthermore, the disease and functional networks found by IPA identified genes involved in inflammation of the CNS (Cb, 70 genes; F/M 41 genes). Seizure disorders (Cb, 46 genes), encephalitis (Cb, 65; F/M, 38), and EAE (Cb, 59; F/M, 34) are all known to be correlated with neuronal dysfunction (Table S5). Therefore, it is interesting that the Tpp1-targeted neuroinflammation process correlates more strongly with autoimmune types of neurodegenerative diseases (EAE and multiple sclerosis; see Figure 4) than with AD or Parkinson’s disease (Dendrou et al., 2016).
The entry of circulating immune cells into the brain has been described for several neurodegenerative diseases (Minagar et al., 2012; Dendrou et al., 2016; Fiebich et al., 2018), and our data suggest that this process is also active in Tpp1–/– brains as indicated by the activation of leukocyte extravasation signaling (Figure 4). Furthermore, a large number of endothelium-related changes are part of the set of DEG, suggesting disruption of the blood–brain barrier (Figure 3). Other genetic NCL models also showed compromised blood–brain barrier integrity (Okada et al., 2015; Costa-Silva et al., 2017). The involvement of the Th1 and Th2 activation pathways and Cd28 signaling in T-helper cells, in our model, may indicate T-cell extravasation, but confirmation of this possibility will require further experimental analysis. These pathways in EAE and MS usually involve a compromised blood–brain barrier (Kamimura et al., 2013; Dendrou et al., 2016; Lopes Pinheiro et al., 2016). Further studies will require confirmation of T-cell or other leukocyte infiltration of the brain and, more importantly, determining at which point in time this process is initiated.
Activation of the production of NO and ROS is another event that can lead to pathogenesis during chronic inflammation (Osorio et al., 2007; Urrutia et al., 2014; Gonzalez-Reyes et al., 2017; Bisht et al., 2018). Upregulation of neutrophil factors (Ncf) 1, 2, and 4, and cytochrome beta-245 alpha and beta chain genes (Cybb and Cyba) are indicative of the generation of ROS in Tpp1–/– brains. Thus, it is intriguing that even though inflammation in cLINCL may be triggered by accumulation of peptides or lipopeptides, similar to that observed in primary neurodegenerative diseases, it may lead to more intricate inflammatory processes that are accelerated by leukocyte infiltration, generation of NO and ROS, and activation of multiple interleukin receptors (listed here), all of which could add to the vicious cycle of neurotoxicity and neuronal death.
Interestingly, two population of reactive astrocytes have been identified, A1 and A2 (Zamanian et al., 2012; Liddelow et al., 2017). A1 upregulates classical components of complement cascade genes and was postulated to be harmful to synapses; A2 upregulates neurotrophic factors and is postulated to be protective (Zamanian et al., 2012). Signatures for both types of reactive astrocytes were found to be upregulated in our transcriptional analysis of Tpp1–/– brains at 4 months old (A1 type: Ggta1, Gbp2, Fbln5, Psmb8; A2 type: Tgm1, Ptx3, S100a10, Cd109, Ptgs2, Cd14). With regard to cross talk between astrocyte and microglia, it has been reported that the release of complement protein C3 activates expression of its receptor C3aR in microglia (Lian et al., 2016). Upregulation of mRNAs for both C3 and its receptor in the Tpp1–/– brain suggest that peptides and lipids accumulating during disease progression lead to microgliosis via the C3-C3aR pathway that usually engulfs particles through complement-mediated opsonization (Bodea et al., 2014). Conversely, during normal aging microglia may regulate the activation of astrocytes by upregulation of interleukin-1a and complement Cq1 (Clarke et al., 2018). Both of these components are also found to be upregulated in the Tpp1–/– brain and thus are possibly involved in sustaining astrocyte activation.
As in other neuroinflammatory diseases (Akiyama et al., 1994; Lue et al., 2001; Gowing et al., 2009), increases in Csf1 and Csf1R expression were also observed in the Tpp1–/– Cb. It has been suggested that the astrocytic CSF1 interacts with the microglial CSF1 receptor to promote microglial proliferation after injury (Tang et al., 2018), and the coincident localization of activated astrocytes and increased microglial number (Figure 7 and 8) in the Tpp1–/– brain support this notion. A common feature of neurodegenerative diseases is the presence of activated microglia in areas of neuronal death (Frank-Cannon et al., 2009), while in this cLINCL model, they appear to be in areas of axonal tracts (Figure 8), pointing to the development of an axonopathy as the disease progresses that should be a focus of future research.
Similar to several other neurodegenerative diseases (Heneka et al., 2018), evidence of inflammasome activation through Nlpr3 expression was detected in the Tpp1–/– mouse brain, including transcriptional upregulation of key components of that multimolecular signaling complex (i.e., Aim2, Pycard, Naip members, and Panx1). This complex serves as a platform to activate inflammatory caspases and the production of interleukin-1, as well as to recognize a broad range of aggregated substances, including perhaps the ones accumulated by lack of TPP1 (lipofuscins).
Furthermore, the dysregulation of phagosome formation and maturation (Table 1) may suggest functional implications related to protein clearance defects associated with the lack of Tpp1. Accumulation of phagosomes accompanying disturbed autophagy has been described in photoreceptors on a Cln3 null mouse model (Cao et al., 2006; Wavre-Shapton et al., 2015). Defective autophagy was also described in several other NCL mouse models (Seranova et al., 2017) as well as in fibroblasts from cLINCL patients (Vidal-Donet et al., 2013); thus, perhaps it is not surprising to find that autophagy pathways are also deregulated in Tpp1–/– brains. In this model, the implications of increased expression of Atg9b, an important component of the early autophagosome structures, will require further scrutiny.
A previous study on the pathological characteristics of the Tpp1-targeted mouse remarks that although retinal degeneration is a prominent feature in human cLINCL, in this model, no retinal pathology was observed, and only ascending visual pathways were mildly affected while extensive neurodegeneration was observed in the mouse auditory pathway (Sleat et al., 2004). Thus, our findings that there are global circadian rhythm abnormalities in this animal model may indicate that these effects are not necessarily linked to visual disturbances but rather to a more generalized effect, as we confirmed downregulation of Per1, not only in the suprachiasmatic nuclei but also in the piriform cortex (Figure 9) and Cb. In the Cb, clock gene rhythms have been detected, and the Cb circadian oscillators have been implicated in the response to food anticipation (Mendoza et al., 2010). Thus, it is feasible to suggest that the sleep abnormalities and food-intake disturbance seen in Batten’s patients may be linked to cerebral oscillator abnormalities. Furthermore, our findings in this model offer a molecular basis to investigate patients’ sleep disorders symptoms, which open a novel area of research for this disease.
In summary, this study highlights the prominent involvement of neuroinflammation and oxidative stress in the development of NCL due to lack of TPP1 activity. The initial lysosomal pathway defect associated with a lack of TPP1 leads to generation of lipid/protein aggregates and axonal toxicity that triggers the neuroinflammation process. This process is potentiated by astrocyte–microglia communication, leading to oxidative stress and, probably, disruption of the blood–brain barrier. Several neuroinflammation aspects were found to be unique to this model when compared with other neurodegenerative diseases, and this information should help tailor appropriate therapeutics for NCLs in the future. Considering the extensive involvement of microglia, astrocytes, and endothelial cells in the pathology, neuroinflammation, and neurodegenerative process, and that they are themselves compromised in their function due to the lack of TPP1, it is important to establish their potential as therapeutic targets for this disease. It will be also important to extend these findings and provide an in vivo timeline for understanding the developmental progression of the disease, in particular as it relates to the progression of the neuroinflammation state and the identification of initial triggers and responders to the dysfunctional lysosomal paradigm.
Summary
Brain transcriptome analysis in the Tpp1–/– mouse line identified specific brain inflammation pathways in which microglia and astrocyte activation potentiates neuronal dysfunction possibly by oxidative stress and damage of the blood–brain barrier integrity. Defects on the control of circadian rhythm were also observed.
Supplementary Material
Acknowledgment
We thank Drs. P. Lobel and D. E. Sleat for gifting Tpp1–/– mice line.
Author Contributions
M. S. D. and N. B. S. designed the study; M. S. D., P. C. V., J. G. H., and C. B. W. performed the experiments; W. C. and J. A. performed the informatic analysis; M. S. D., G. D., and N. B. S. contributed to the interpretation of the results. M. S. D. and N. B. S. wrote the article in consultation with G. D.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by The Children’s Brain Diseases Foundation.
References
- Abreu T., Braganca , M. (2015). The bipolarity of light and dark: A review on bipolar disorder and circadian cycles. J Affect Disord, 185, 219–229. [DOI] [PubMed] [Google Scholar]
- Akiyama H., Nishimura T., Kondo H.Ikeda, K., Hayashi, Y., & McGeer, P. L. (1994). Expression of the receptor for macrophage colony stimulating factor by brain microglia and its upregulation in brains of patients with Alzheimer’s disease and amyotrophic lateral sclerosis. Brain Res, 639, 171–174. [DOI] [PubMed] [Google Scholar]
- Anderson G. W., Goebel H. H., Simonati , A. (2013). Human pathology in NCL. Biochim Biophys Acta, 1832, 1807–1826. [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Bevan R. J., Evans R., Griffiths L.Watkins, L. M., Rees, M. I., Magliozzi, R., Allen, I., McDonnell, G., Kee, R., Naughton, M., Fitzgerald, D. C., Reynolds, R., Neal, J. W., & Howell, O. W. (2018). Meningeal inflammation and cortical demyelination in acute multiple sclerosis. Ann Neurol, 84, 829–842. [DOI] [PubMed] [Google Scholar]
- Bisht K., Sharma K., Tremblay , M. E. (2018). Chronic stress as a risk factor for Alzheimer’s disease: Roles of microglia-mediated synaptic remodeling, inflammation, and oxidative stress. Neurobiol Stress, 9, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodea L. G., Wang Y., Linnartz-Gerlach B.Kopatz, J., Sinkkonen, L., Musgrove, R., Kaoma, T., Muller, A., Vallar, L., Di Monte, D. A., Balling, R., & Neumann, H. (2014). Neurodegeneration by activation of the microglial complement-phagosome pathway. J Neurosci, 34, 8546–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert M. M., Erikson G. A., Shokhirev M. N.& Allen, N.J. (2018). The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep, 22, 269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks A. I., Chattopadhyay S., Mitchison H. M.Nussbaum, R. L., & Pearce, D. A. (2003). Functional categorization of gene expression changes in the cerebellum of a Cln3-knockout mouse model for Batten disease. Mol Genet Metab, 78, 17–30. [DOI] [PubMed] [Google Scholar]
- Burkovetskaya M., Karpuk N., Xiong J.Bosch, M., Boska, M. D., Takeuchi, H., Suzumura, A., & Kielian, T. (2014). Evidence for aberrant astrocyte hemichannel activity in juvenile neuronal ceroid lipofuscinosis (JNCL). PLoS One, 9, e95023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Espinola J. A., Fossale E.Massey, A. C., Cuervo, A. M., MacDonald, M. E., & Cotman, S. L. (2006). Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem, 281, 20483–20493. [DOI] [PubMed] [Google Scholar]
- Carcel-Trullols J., Kovacs A. D., Pearce , D. A. (2015). Cell biology of the NCL proteins: What they do and don’t do. Biochim Biophys Acta, 1852, 2242–2255. [DOI] [PubMed] [Google Scholar]
- Casula M., Iyer A. M., Spliet W. G.Anink, J. J., Steentjes, K., Sta, M., Troost, D., & Aronica E. (2011). Toll-like receptor signaling in amyotrophic lateral sclerosis spinal cord tissue. Neuroscience, 179, 233–243. [DOI] [PubMed] [Google Scholar]
- Chang M., Cooper J. D., Sleat D. E.Cheng, S. H., Dodge, J. C., Passini, M. A., Lobel, P., & Davidson, B. L. (2008). Intraventricular enzyme replacement improves disease phenotypes in a mouse model of late infantile neuronal ceroid lipofuscinosis. Mol Ther, 16, 649–656. [DOI] [PubMed] [Google Scholar]
- Clarke L. E., Liddelow S. A., Chakraborty C.Munch, A. E., Heiman, M., & Barres, B. A. (2018). Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A, 115, E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Matcovitch O., David E.Barnett-Itzhaki, Z., Keren-Shaul, H., Blecher-Gonen, R., Jaitin, D. A., Sica, A., Amit, I., & Schwartz, M. (2014). Chronic exposure to TGFbeta1 regulates myeloid cell inflammatory response in an IRF7-dependent manner. EMBO J, 33, 2906–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. D., Russell C., Mitchison , H. M. (2006). Progress towards understanding disease mechanisms in small vertebrate models of neuronal ceroid lipofuscinosis. Biochim Biophys Acta, 1762, 873–889. [DOI] [PubMed] [Google Scholar]
- Cooper J. D., Tarczyluk M. A., Nelvagal , H. R. (2015). Towards a new understanding of NCL pathogenesis. Biochim Biophys Acta, 1852, 2256–2261. [DOI] [PubMed] [Google Scholar]
- Costa-Silva J., Domingues D., Lopes , F. M. (2017). RNA-seq differential expression analysis: An extended review and a software tool. PLoS One, 12, e0190152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deczkowska A., Keren-Shaul H., Weiner A.Colonna, M., Schwartz, M., & Amit, I. (2018). Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell, 173, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Dendrou C. A., McVean G., Fugger , L. (2016). Neuroinflammation – Using big data to inform clinical practice. Nat Rev Neurol, 12, 685–698. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F.Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M., & Gingeras, T. R. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domowicz M., Wadlington N. L., Henry J. G.Diaz, K., Munoz, M. J., & Schwartz, N. B. (2018). Glial cell responses in a murine multifactorial perinatal brain injury model. Brain Res, 1681, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domowicz M. S., Henry J. G., Wadlington N.Navarro, A., Kraig, R. P., & Schwartz, N. B. (2011). Astrocyte precursor response to embryonic brain injury. Brain Res, 1389, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domowicz M. S., Sanders T. A., Ragsdale C. W.& Schwartz, N. B. (2008). Aggrecan is expressed by embryonic brain glia and regulates astrocyte development. Dev Biol, 315, 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y., Brooks A. I., Chattopadhyay S.Curran, T. M., Gupta, P., Ramalingam, V., Hofmann, S. L., & Pearce, D. A. (2003). Early changes in gene expression in two models of Batten disease. FEBS Lett, 538, 207–212. [DOI] [PubMed] [Google Scholar]
- Fiebich B. L., Batista C. R. A., Saliba S. W.Yousif, N. M., & de Oliveira, A. C. P. (2018). Role of microglia TLRs in neurodegeneration. Front Cell Neurosci, 12, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz M., AlSayed M., Burke D.Cohen-Pfeffer, J., Cooper, J. D., Dvorakova, L., Giugliani, R., Izzo, E., Jahnova, H., Lukacs, Z., Mole, S. E., Noher de Halac, I., Pearce, D. A., Poupetova, H., Schulz, A., Specchio, N., Xin, W., & Miller, N. (2016). Diagnosis of neuronal ceroid lipofuscinosis type 2 (CLN2 disease): Expert recommendations for early detection and laboratory diagnosis. Mol Genet Metab, 119, 160–167. [DOI] [PubMed] [Google Scholar]
- Frank-Cannon T. C., Alto L. T., McAlpine F. E.& Tansey, M. G. (2009). Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener, 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraets R. D., Langin L. M., Cain J. T.Parker, C. M., Beraldi, R., Kovacs, A. D., Weimer, J. M., & Pearce, D. A. (2017). A tailored mouse model of CLN2 disease: A nonsense mutant for testing personalized therapies. PLoS One, 12, e0176526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes R. E., Nava-Mesa M. O., Vargas-Sanchez K.Ariza-Salamanca, D., & Mora-Munoz, L. (2017). Involvement of astrocytes in Alzheimer’s disease from a neuroinflammatory and oxidative stress perspective. Front Mol Neurosci, 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Skola D., Coufal N. G.Holtman, I. R., Schlachetzki, J. C. M., Sajti, E., Jaeger, B. N., O'Connor, C., Fitzpatrick, C., Pasillas, M. P., Pena, M., Adair, A., Gonda, D. D., Levy, M. L., Ransohoff, R. M., Gage, F. H., & Glass, C. K. (2017). An environment-dependent transcriptional network specifies human microglia identity. Science, 356, eaal322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowing G., Lalancette-Hebert M., Audet J. N.Dequen, F., & Julien, J. P. (2009). Macrophage colony stimulating factor (M-CSF) exacerbates ALS disease in a mouse model through altered responses of microglia expressing mutant superoxide dismutase. Exp Neurol, 220, 267–275. [DOI] [PubMed] [Google Scholar]
- Haimon Z., Volaski A., Orthgiess J.Boura-Halfon, S., Varol, D., Shemer, A., Yona, S., Zuckerman, B., David, E., Chappell-Maor, L., Bechmann, I., Gericke, M., Ulitsky, I., & Jung, S. (2018). Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat Immunol, 19, 636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltia M., Herva R., Suopanki J.Baumann, M., & Tyynela, J. (2001). Hippocampal lesions in the neuronal ceroid lipofuscinoses. Eur J Paediatr Neurol, 5(Suppl A), 209–211. [DOI] [PubMed] [Google Scholar]
- Hanke M. L., Kielian T. (2011). Toll-like receptors in health and disease in the brain: Mechanisms and therapeutic potential. Clin Sci (Lond), 121, 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M. T., McManus R. M., Latz E. (2018). Inflammasome signalling in brain function and neurodegenerative disease. Nat Rev Neurosci, 19, 610–621. [DOI] [PubMed] [Google Scholar]
- Jalanko A., Braulke T. (2009). Neuronal ceroid lipofuscinoses. Biochim Biophys Acta, 1793, 697–709. [DOI] [PubMed] [Google Scholar]
- Kamimura D., Yamada M., Harada M.Sabharwal, L., Meng, J., Bando, H., Ogura, H., Atsumi, T., Arima, Y., & Murakami, M. (2013). The gateway theory: Bridging neural and immune interactions in the CNS. Front Neurosci, 7, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A.Matcovitch-Natan, O., Dvir-Szternfeld, R., Ulland, T. K., David, E., Baruch, K., Lara-Astaiso, D., Toth, B., Itzkovitz, S., Colonna, M., Schwartz, M., & Amit, I. (2017). A unique microglia type associated with restricting development of Alzheimer’s disease. Cell, 169, 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- Kim K., Kleinman H. K., Lee H. J.& Pahan, K. (2017). Safety and potential efficacy of gemfibrozil as a supportive treatment for children with late infantile neuronal ceroid lipofuscinosis and other lipid storage disorders. Orphanet J Rare Dis, 12, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann K., Uusi-Rauva K., Scifo E.Tyynela J., Jalanko, A., & Braulke, T. (2013). Cell biology and function of neuronal ceroid lipofuscinosis-related proteins. Biochim Biophys Acta, 1832, 1866–1881. [DOI] [PubMed] [Google Scholar]
- Lehwald L. M., Pappa R., Steward S.& de Los Reyes, E. (2016) Neuronal ceroid lipofuscinosis and associated sleep abnormalities. Pediatr Neurol, 59, 30--35. [DOI] [PubMed] [Google Scholar]
- Letiembre M., Liu Y., Walter S.Hao, W., Pfander, T., Wrede, A., Schulz-Schaeffer, W., & Fassbender, K. (2009). Screening of innate immune receptors in neurodegenerative diseases: A similar pattern. Neurobiol Aging, 30, 759–768. [DOI] [PubMed] [Google Scholar]
- Lian H., Litvinchuk A., Chiang A. C.Aithmitti, N., Jankowsky, J. L., & Zheng, H. (2016). Astrocyte-microglia cross talk through complement activation modulates amyloid pathology in mouse models of Alzheimer's disease. J Neurosci, 36, 577--589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Yang L., Cole A.Sun, L., Chiang, A. C., Fowler, S. W., Shim, D. J., Rodriguez-Rivera, J., Taglialatela, G., Jankowsky, J. L., Lu, H. C., & Zheng, H. (2015). NFkappaB-activated astroglial release of complement C3 compromises neuronal morphology and function associated with Alzheimer’s disease. Neuron, 85, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liddelow S. A., Guttenplan K. A., Clarke L. E., et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz-Gerlach B., Bodea L. G., Klaus C.Ginolhac, A., Halder, R., Sinkkonen, L., Walter, J., Colonna, M., & Neumann, H. (2019). TREM2 triggers microglial density and age-related neuronal loss. Glia, 67(3), 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinchuk A., Wan Y. W., Swartzlander D. B.Chen, F., Cole, A., Propson, N. E., Wang, Q., Zhang, B., Liu, Z., & Zheng, H. (2018). Complement C3aR inactivation attenuates tau pathology and reverses an immune network deregulated in tauopathy models and Alzheimer’s disease. Neuron, 100, 1337–1353.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes Pinheiro M. A., Kooij G., Mizee M. R.Kamermans, A., Enzmann, G., Lyck, R., Schwaninger, M., Engelhardt, B., & de Vries, H. E. (2016). Immune cell trafficking across the barriers of the central nervous system in multiple sclerosis and stroke. Biochim Biophys Acta, 1862, 461–471. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue L. F., Rydel R., Brigham E. F.Yang, L. B., Hampel, H., Murphy, G. M. Jr., Brachova, L., Yan, S. D., Walker, D. G., Shen, Y., & Rogers, J. (2001). Inflammatory repertoire of Alzheimer’s disease and nondemented elderly microglia in vitro. Glia, 35, 72–79. [DOI] [PubMed] [Google Scholar]
- Lun M. P., Johnson M. B., Broadbelt K. G.Watanabe, M., Kang, Y. J., Chau, K. F., Springel, M. W., Malesz, A., Sousa, A. M., Pletikos, M., Adelita, T., Calicchio, M. L., Zhang, Y., Holtzman, M. J., Lidov, H. G., Sestan, N., Steen, H., Monuki, E. S., & Lehtinen, M. K. (2015). Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J Neurosci, 35, 4903–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley S. L., Roberts M. S., Wong A. M.McSloy, F., Reddy, A. S., Cooper, J. D., & Sands, M. S. (2012). Synergistic effects of central nervous system-directed gene therapy and bone marrow transplantation in the murine model of infantile neuronal ceroid lipofuscinosis. Ann Neurol, 71, 797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret S., Dorsaz S., Gurcel L. Pradervand, S., Petit, B., Pfister, C., Hagenbuchle, O., O'Hara, B. F., Franken, P., & Tafti, M. (2007). Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A, 104, 20090–20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta D., Tinelli E., Mole S. E. (2017). NCLs and ER: A stressful relationship. Biochim Biophys Acta, 1863, 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavarajah S., McLaren M. D., Huber R. J. (2018). Cln3 function is linked to osmoregulation in a Dictyostelium model of Batten disease. Biochim Biophys Acta Mol Basis Dis, 1864, 3559–3573. [DOI] [PubMed] [Google Scholar]
- Mendoza J., Pevet P., Felder-Schmittbuhl M. P.Bailly, Y., & Challet, E. (2010). The cerebellum harbors a circadian oscillator involved in food anticipation. J Neurosci, 30, 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagar A., Maghzi A. H., McGee J. C.& Alexander, J. S. (2012). Emerging roles of endothelial cells in multiple sclerosis pathophysiology and therapy. Neurol Res, 34, 738–745. [DOI] [PubMed] [Google Scholar]
- Mink J. W., Augustine E. F., Adams H. R.Marshall, F. J., &, Kwon, J. M. (2013). Classification and natural history of the neuronal ceroid lipofuscinoses. J Child Neurol, 28, 1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole S. E., Cotman S. L. (2015). Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim Biophys Acta, 1852, 2237–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S., Gaiteri C., Sullivan S. E., et al. (2018). A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat Neurosci, 21, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R., Wu Z., Zhu A.Ni, J., Zhang, J., Yoshimine, Y., Peters, C., Saftig, P., & Nakanishi, H. (2015). Cathepsin D deficiency induces oxidative damage in brain pericytes and impairs the blood-brain barrier. Mol Cell Neurosci, 64, 51–60. [DOI] [PubMed] [Google Scholar]
- Olah M., Patrick E., Villani A. C.Xu, J., White, C. C., Ryan, K. J., Piehowski, P., Kapasi, A., Nejad, P., Cimpean, M., Connor, S., Yung, C. J., Frangieh, M., McHenry, A., Elyaman, W., Petyuk, V., Schneider, J. A., Bennett, D. A., De Jager, P. L., & Bradshaw, E. M. (2018). A transcriptomic atlas of aged human microglia. Nat Commun, 9, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D., Honma K., Honma S. (2015). Circadian and ultradian rhythms of clock gene expression in the suprachiasmatic nucleus of freely moving mice. Sci Rep, 5, 12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio N. S., Carvalho A., Almeida A. J.Padilla-Lopez, S., Leao, C., Laranjinha, J., Ludovico, P., Pearce, D. A., & Rodrigues, F. (2007). Nitric oxide signaling is disrupted in the yeast model for Batten disease. Mol Biol Cell, 18, 2755–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. N., Barry L. A., Tyynela J.& Cooper, J. D. (2013). NCL disease mechanisms. Biochim Biophys Acta, 1832, 1882–1893. [DOI] [PubMed] [Google Scholar]
- Palmer D. N., Oswald M. J., Westlake V. J. & Kay, G. W. (2002). The origin of fluorescence in the neuronal ceroid lipofuscinoses (Batten disease) and neuron cultures from affected sheep for studies of neurodegeneration. Arch Gerontol Geriatr, 34, 343–357. [DOI] [PubMed] [Google Scholar]
- Parviainen L., Dihanich S., Anderson G. W., Wong, A. M., Brooks, H. R., Abeti, R., Rezaie, P., Lalli, G., Pope, S., Heales, S. J., Mitchison, H. M., Williams, B. P., & Cooper, J. D. (2017). Glial cells are functionally impaired in juvenile neuronal ceroid lipofuscinosis and detrimental to neurons. Acta Neuropathol Commun, 5, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini M. A., Dodge J. C., Bu J.Yang, W., Zhao, Q., Sondhi, D., Hackett, N. R., Kaminsky, S. M., Mao, Q., Shihabuddin, L. S., Cheng, S. H., Sleat, D. E., Stewart, G. R., Davidson, B. L., Lobel, P., & Crystal, R. G. (2006). Intracranial delivery of CLN2 reduces brain pathology in a mouse model of classical late infantile neuronal ceroid lipofuscinosis. J Neurosci, 26, 1334–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M., Wilhelmsson U., Pekna M. (2014). The dual role of astrocyte activation and reactive gliosis. Neurosci Lett, 565, 30–38. [DOI] [PubMed] [Google Scholar]
- Pontikis C. C., Cella C. V., Parihar N.Lim, M. J., Chakrabarti, S., Mitchison, H. M., Mobley, W. C., Rezaie, P., Pearce, D. A., & Cooper, J. D. (2004). Late onset neurodegeneration in the Cln3-/- mouse model of juvenile neuronal ceroid lipofuscinosis is preceded by low level glial activation. Brain Res, 1023, 231–242. [DOI] [PubMed] [Google Scholar]
- Pontikis C. C., Cotman S. L., MacDonald M. E.& Cooper, J.D. (2005). Thalamocortical neuron loss and localized astrocytosis in the Cln3Deltaex7/8 knock-in mouse model of Batten disease. Neurobiol Dis, 20, 823–836. [DOI] [PubMed] [Google Scholar]
- Radke J., Stenzel W., Goebel H. H. (2015). Human NCL neuropathology. Biochim Biophys Acta, 1852, 2262–2266. [DOI] [PubMed] [Google Scholar]
- Raj D., Yin Z., Breur M.Doorduin, J., Holtman, I. R., Olah, M., Mantingh-Otter, I. J., Van Dam, D., De Deyn, P. P., den Dunnen, W., Eggen, B. J. L., Amor, S., & Boddeke, E. (2017). Increased white matter inflammation in aging- and Alzheimer’s disease brain. Front Mol Neurosci, 10, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R. M., Perry V. H. (2009). Microglial physiology: Unique stimuli, specialized responses. Annu Rev Immunol, 27, 119–145. [DOI] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D.Hu, Y., Law, C. W., Shi, W., & Smyth, G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T., Merrow M. (2016). The circadian clock and human health. Curr Biol, 26, R432–R443. [DOI] [PubMed] [Google Scholar]
- Saadoun S., Papadopoulos M. C. (2010). Aquaporin-4 in brain and spinal cord oedema. Neuroscience, 168, 1036–1046. [DOI] [PubMed] [Google Scholar]
- Salem M., Mony J. T., Lobner M.Khorooshi, R., & Owens, T. (2011). Interferon regulatory factor-7 modulates experimental autoimmune encephalomyelitis in mice. J Neuroinflammation, 8, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A., Ajayi T., Specchio N.de Los Reyes, E., Gissen, P., Ballon, D., Dyke, J. P., Cahan, H., Slasor, P., Jacoby, D., Kohlschutter, A., & Group CLNS. (2018). Study of intraventricular cerliponase alfa for CLN2 disease. N Engl J Med, 378, 1898–1907. [DOI] [PubMed] [Google Scholar]
- Schulz A., Kohlschutter A., Mink J.Simonati, A., & Williams, R. (2013). NCL diseases – Clinical perspectives. Biochim Biophys Acta, 1832, 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seranova E., Connolly K. J., Zatyka M.Rosenstock, T. R., Barrett, T., Tuxworth, R. I., & Sarkar, S. (2017). Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem, 61, 733–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R. B., Sun L., Chen Z. J. (2006). Antiviral innate immunity pathways. Cell Res, 16, 141–147. [DOI] [PubMed] [Google Scholar]
- Sleat D. E., Donnelly R. J., Lackland H.Liu, C. G., Sohar, I., Pullarkat, R. K., & Lobel, P. (1997). Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science, 277, 1802–1805. [DOI] [PubMed] [Google Scholar]
- Sleat D. E., Wiseman J. A., El-Banna M.Kim, K. H., Mao, Q., Price, S., Macauley, S. L., Sidman, R. L., Shen, M. M., Zhao, Q., Passini, M. A., Davidson, B. L., Stewart, G. R., & Lobel, P. (2004). A mouse model of classical late-infantile neuronal ceroid lipofuscinosis based on targeted disruption of the CLN2 gene results in a loss of tripeptidyl-peptidase I activity and progressive neurodegeneration. J Neurosci, 24, 9117–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq L., UK Brain Expression Consortium, North American Brain Expression ConsortiumRose, J., Soreq, E., Hardy, J., Trabzuni, D., Cookson, M. R., Smith, C., Ryten, M., Patani, R., & Ule, J. (2017). Major shifts in glial regional identity are a transcriptional hallmark of human brain aging. Cell Rep, 18, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Liu L., Xu D.Zhang, W., Zhang, Y., Zhou, J., & Huang, W. (2018). Interaction between astrocytic colony stimulating factor and its receptor on microglia mediates central sensitization and behavioral hypersensitivity in chronic post ischemic pain model. Brain Behav Immun, 68, 248–260. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi, A., Kwan, G., van Baren, M. J., Salzberg, S. L., Wold, B. J., & Pachter, L. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol, 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynela J., Cooper J. D., Khan M. N.Shemilts, S. J. & Haltia, M. (2004). Hippocampal pathology in the human neuronal ceroid-lipofuscinoses: Distinct patterns of storage deposition, neurodegeneration and glial activation. Brain Pathol, 14, 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia P. J., Mena N. P., Nunez M. T. (2014). The interplay between iron accumulation, mitochondrial dysfunction, and inflammation during the execution step of neurodegenerative disorders. Front Pharmacol, 5, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Donet J. M., Carcel-Trullols J., Casanova B.Aguado, C., & Knecht, E. (2013). Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PLoS One, 8, e55526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizuete M. L., Venero J. L., Vargas C.Ilundain, A. A., Echevarria, M., Machado, A., & Cano, J. (1999). Differential upregulation of aquaporin-4 mRNA expression in reactive astrocytes after brain injury: Potential role in brain edema. Neurobiol Dis, 6, 245–258. [DOI] [PubMed] [Google Scholar]
- von Schantz C., Saharinen J., Kopra O.Cooper, J. D., Gentile, M., Hovatta, I., Peltonen, L., & Jalanko, A. (2008). Brain gene expression profiles of Cln1 and Cln5 deficient mice unravels common molecular pathways underlying neuronal degeneration in NCL diseases. BMC Genomics, 9, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wavre-Shapton S. T., Calvi A. A., Turmaine M.Seabra, M. C., Cutler, D. F., Futter, C. E., & Mitchison, H. M. (2015). Photoreceptor phagosome processing defects and disturbed autophagy in retinal pigment epithelium of Cln3Deltaex1-6 mice modelling juvenile neuronal ceroid lipofuscinosis (Batten disease). Hum Mol Genet, 24, 7060–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. E., Adams H. R., Blohm M.Nouri, N., Zhou, L., Giffard, R. G., & Barres, B. A. (2017). Management strategies for CLN2 disease. Pediatr Neurol, 69, 102–112. [DOI] [PubMed] [Google Scholar]
- Xiong J., Kielian T. (2013). Microglia in juvenile neuronal ceroid lipofuscinosis are primed toward a pro-inflammatory phenotype. J Neurochem, 127, 245–258. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L. G., Han Y. & He, Q. Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian J. L., Xu L., Foo L. C.Nouri, N., Zhou, L., Giffard, R. G., & Barres, B. A. (2012). Genomic analysis of reactive astrogliosis. J Neurosci, 32, 6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Gaiteri C., Bodea L. G., et al. (2013). Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell, 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S. A.Bennett, M. L, Scholze, A. R., O'Keeffe, S., Phatnani, H. P., Guarnieri, P., Caneda, C., Ruderisch, N., Deng, S., Liddelow, S. A., Zhang, C., Daneman, R., Maniatis, T., Barres, B. A., & Wu, J. Q. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci, 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.