Abstract
The APOBEC3 enzymes can induce mutagenesis of HIV-1 proviral DNA through the deamination of cytosine. HIV-1 overcomes this restriction through the viral protein Vif that induces APOBEC3 proteasomal degradation. Within this dynamic host-pathogen relationship, the APOBEC3 enzymes have been found to be beneficial, neutral, or detrimental to HIV-1 biology. Here, we assessed the ability of co-expressed APOBEC3F and APOBEC3G to induce HIV-1 resistance to antiviral drugs. We found that co-expression of APOBEC3F and APOBEC3G enabled partial resistance of APOBEC3F to Vif-mediated degradation with a corresponding increase in APOBEC3F-induced deaminations in the presence of Vif, in addition to APOBEC3G-induced deaminations. We recovered HIV-1 drug resistant variants resulting from APOBEC3-induced mutagenesis, but these variants were less able to replicate than drug resistant viruses derived from RT-induced mutations alone. The data support a model in which APOBEC3 enzymes cooperate to restrict HIV-1, promoting viral inactivation over evolution to drug resistance.
Keywords: Immunology, Microbiology
1. Introduction
In order to infect host cells, the Human Immunodeficiency Virus (HIV) must overcome the activity of several host restriction factors [1, 2]. Due to the lifelong infection caused by HIV, there is extensive adaptation between the host restriction factors and their counteracting viral accessory proteins [3, 4, 5, 6, 7, 8, 9, 10, 11]. HIV restriction factors can prevent viral budding, decrease nuclear accumulation of viral cDNAs, interfere with reverse transcription, and induce mutagenesis of proviral DNA [1, 2]. While viral evolution to continually evade host restriction occurs constantly through selected mutations resulting from reverse transcriptase (RT) error there may also be a contribution from a group of host restriction factors [12, 13, 14, 15, 16]. The APOBEC3 (A3) restriction factors of HIV can induce mutagenesis of proviral DNA by deaminating cytosine to form promutagenic uracil in (−) DNA when it is single stranded [1, 2, 17]. Copying of these uracils during (+) DNA synthesis results in guanine to adenine mutations [1, 2, 17]. The intensity of these mutations is dependent on both the activity of the A3 enzymes and the ability of the HIV protein Vif to suppress A3 activity [18].
The HIV-1 protein Vif mediates the proteasomal degradation of several human A3 enzymes that are expressed in CD4 + T cells by interacting with several host factors to form a Cullin 5 E3 ubiquitin ligase assembly [18, 19, 20, 21, 22]. This results in the polyubiquitination and degradation of A3 enzymes [23, 24, 25, 26, 27, 28]. Without the action of Vif, higher numbers of A3 enzymes would bind HIV-1 genomic RNA or cellular RNA that is also bound by HIV-1 Gag and be encapsidated into virions [29, 30, 31]. Of the seven human A3 enzymes, A3C, A3D, A3F, A3G, and A3H interact with Vif, resulting in their proteasomal degradation [32, 33, 34, 35, 36, 37]. Even if A3 enzymes escape Vif-mediated degradation, Vif can block encapsidation of A3s into virions by a degradation-independent mechanism and Vif can inhibit the activity of A3G when co-encapsidated [38, 39, 40, 41]. Despite the multi-modal action of Vif, A3-induced mutagenesis of proviral genomes in HIV+ people occurs [42, 43]. This may be due to fortuitous escape from Vif or that the ability of Vif to induce A3 degradation can decrease over the course of an infection [44, 45].
Regardless of how A3 enzymes escape Vif-mediated inhibition, they remain within the capsid of the virus and exert their restriction activity in the next target cell that the virus infects. When the HIV-1 reverse transcriptase synthesizes the (−) DNA from the genomic RNA (gRNA) template, the RNase H domain becomes able to degrade the gRNA, exposing single-stranded regions of the (−) DNA [17, 46]. A3 enzymes can only deaminate single-stranded (ss) DNA and it has been demonstrated that more A3-induced mutations accumulate in proviral DNA in regions that are the last to become double-stranded. These vulnerable regions are on the 5′-side of both the central and 3′ polypurine tracts (PPTs) and result in two gradients of mutations [47, 48, 49]. The A3 enzymes combat this limited time of substrate availability by processively scanning ssDNA to enable multiple deaminations in a single enzyme-substrate encounter, which greatly increases the deamination efficiency of the enzymes [17, 18]. Of the five A3 enzymes that can encapsidate into HIV-1 virions and are targeted by Vif, each have a different activity that can be discerned based on their respective levels of processivity [17, 18]. In the absence of Vif, when A3 activity is at its highest, A3G can induce the most mutations and is the most processive [17, 18]. As processivity declines so do deamination levels in ΔVif HIV-1 virions where A3H, A3F, and A3C show a decreasing gradient of activity after A3G [17, 18, 50, 51, 52, 53]. There are also other factors that affect A3 activity, such as the polymorphisms present in the human population [54, 55, 56, 57, 58, 59, 60, 61, 62] and the preferred deamination motif [63]. A3G deaminates in 5′CC motifs on the (−) DNA for approximately 90% of the deamination events, with deaminations in 5′TC motifs contributing to the other 10% of deamination sites [17, 63, 64, 65, 66]. This results in predominantly 5′GG → 5′AG mutations and a lesser amount of 5′GA → 5′AA mutations induced by A3G in the (+) DNA. The 5′GG motif occurs in codons for the amino acids glycine, aspartate, and glutamate, where a 5′AG mutation results in a nonconservative missense mutation, and tryptophan, where a 5′AG mutation results in a nonsense mutation [17]. The other A3s all prefer to deaminate 5′TC motifs on the (−) DNA, resulting in 5′GA → 5′AA mutations in the (+) DNA [17]. For A3F, deaminations in 5′TC motifs occur for approximately 90% of the deamination events [65, 66]. These resulting 5′GA → 5′AA mutations cause a wide variety of missense mutations and can cause nonsense mutations, but nonsense mutations occur at least 2-fold less than from A3G deaminations, depending on the experimental system [17, 64].
Since viral restriction by A3s is based on functional inactivation of proviral DNA, the number and type of mutations that an A3 enzyme induces is an important factor. Low levels of mutations or missense mutations may result in either HIV-1 evolution or have a neutral effect, rather than HIV-1 inactivation [16, 42, 67, 68, 69, 70, 71]. Studies have demonstrated that due to the A3G preferred deamination motif, A3G-induced mutagenesis is likely to induce viral inactivation [50, 64, 66, 72, 73, 74]. However, the A3s that cause 5′GA → 5′AA mutations (A3F, A3H, and A3C) may be more likely to induce HIV-1 genetic diversity and evolution [17, 64, 74]. These data in combination with data demonstrating that during chronic HIV-1 infection Vif can acquire mutations that partially decrease its ability to induce A3 degradation suggest that HIV-1 can manipulate A3 activity like a mutational rheostat [44, 45, 68]. Evidence of this has come from studies that have assessed antiviral drug resistance, CTL epitopes, and co-receptor usage, some of which used less effective Vif variants isolated from HIV-1+ individuals [12, 15, 16, 68, 69, 70, 74, 75, 76, 77].
Of interest to the present work were the studies assessing whether A3-induced mutagenesis can cause drug resistance. In Mulder et al., the authors used a less active Vif variant (K22H) isolated from an HIV+ individual to create a hypermutated virus population that was largely unable to replicate, but the integrated proviral genomes contained the M184I mutation in pol that conferred resistance to the antiretroviral (ARV) drug 3TC (Lamivudine, 2′-deoxy-3′-thiacytidine) [68]. By co-infecting cells with a replication competent virus and the hypermutated ones, the authors were able to show that viral recombination could rescue replication in the presence of high amounts of 3TC [68]. In contrast, a combined experimental and in silico study demonstrated that super-infection of cells and viral recombination between inactivated and competent virus is unlikely to occur and suggested that hypermutated proviruses are a dead-end [67]. Nonetheless, the Vif K22H mutation correlated with increased virological treatment failure in HIV-1+ individuals, which may be due to increased levels of preexisting drug resistance mutations induced by A3 enzymes, even though many of the proviral genomes were nonfunctional [45, 78, 79]. However, none of these studies addressed if there could be drug resistance in a wild type (WT) HIV-1 infection before A3 hypermutation and if drug resistance was instead induced by A3 sublethal mutagenesis. Kim et al. addressed this by using CEM-SS cells that do not express A3s and created a CEM-SS cell line stably expressing A3G and showed with WT HIV-1 that resistance to 3TC only arose in the cells expressing A3G [71]. This study supports the hypothesis that Vif suppresses A3-induced hypermutation, but still allows for a small enough number of mutations to occur that provides a benefit to the HIV-1 genetic diversity [71]. However, clinical studies have not found a significant correlation between A3 enzymes and HIV-1 drug resistance [80, 81, 82, 83], which may be due to the principle of purifying selection and that drug resistance mutations induced by A3 enzymes may have too many other collateral mutations to produce replication competent virus or be rescued by viral recombination [67, 84].
Despite these previous cell-based and clinical studies there are several factors regarding A3-induced HIV-1drug resistance that have not been addressed. First, HIV-1 RT also has a preference to cause G → A mutations and it has not been established whether the A3 sequence contexts of 5′GG or 5′GA can properly discriminate between A3- and RT- induced mutations [85, 86, 87]. Second, several studies have used “hypermutated” viruses due to the absence of Vif, use of less active Vif variants, or use of proviral DNA sequences from databases [14, 67, 68], but the effect of A3s with a WT HIV-1 and Vif are not known; in particular, we still do not know the average number of deaminations an A3 can induce in a single round of replication with WT HIV-1. Third, several studies have tested only A3G [14, 68, 71] even though A3 enzymes are known to be coordinately expressed and A3F and A3G have been shown to interact as a hetero-oligomer that has enhanced HIV-1 restriction activity [66, 88, 89].
Here, we assessed if the A3 enzymes A3F and A3G could cause drug resistance to evolve in HIV-1. We chose to study A3F and A3G since they are most commonly the active A3s found in the human population [60]. We examined how the enzymes influenced drug resistance against each 3TC and AZT (Azidothymidine) alone. Since drug resistance can easily develop with monotherapies, we also tested a mixed ARV regimen of TDF (Tenofovir Disoproxil Fumarate), 3TC, and EFV (Efavirenz) (TLE regimen). We found that co-expression of A3F and A3G in HIV-1LAI producer cells resulted in deaminations that were above the RT-induced G → A mutation rate and drug resistant variants in the progeny virus. However, the drug resistant variants resulting from A3-induced mutagenesis were less able to replicate than drug resistant viruses derived from RT-induced mutations alone. Altogether, the data support a model in which A3 enzymes cooperate to restrict HIV-1, which promotes viral inactivation more than viral evolution.
2. Results
2.1. Co-expression of A3G and A3F results in partial protection of A3F from Vif-mediated degradation
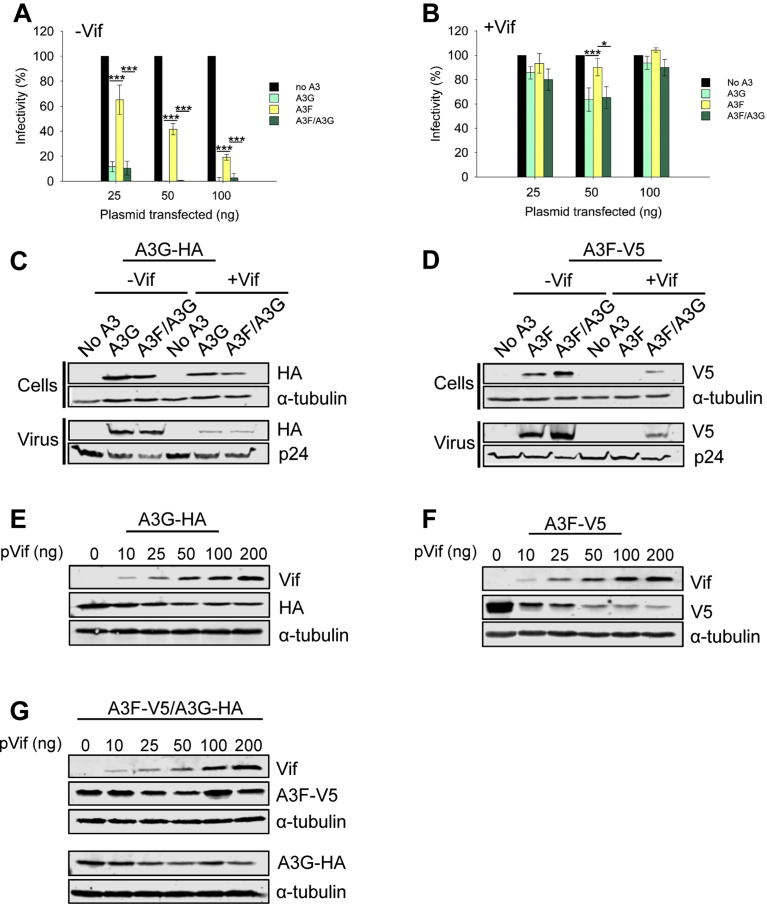
In the absence of Vif, A3G and A3F have different abilities to restrict HIV-1. In single-cycle replication assays, across a range of transfected A3 plasmid, A3F restricts ΔVif HIV-1 6- to 10- fold less than A3G (Fig. 1A). We used the A3F variant 108S/231I [60, 90]. However, both A3G and A3F are suppressed by Vif in a single-cycle replication assay with HIV-1LAI, as demonstrated by high HIV-1 infectivity levels (Fig. 1B). To more accurately assess the impact of A3G and A3F, which are coordinately expressed in CD4+ T cells and have been previously shown to hetero-oligomerize and synergistically restrict HIV-1 [66], we used a vector with two multiple cloning sites so that on a per cell basis each transfected cell would express both A3G and A3F (see Materials and Methods and Ref. [66]). Under these conditions, the ΔVif HIV-1 restriction by co-expressed A3F and A3G was equal to that of A3G alone (Fig. 1A) and the HIV-1LAI appeared to be able to suppress both A3G and A3F (Fig. 1B). The expression levels of the co-expressed or singly expressed A3F or A3G were directly comparable since the same MCS in the dual expression vector was used to express each A3 alone (see Materials and Methods and Ref. [66]).
Fig. 1.
Co-expression of A3G and A3F results in protection of A3F from Vif mediated degradation. (A) In a ΔVif ΔEnv HIV-1LAI (−Vif) single-cycle replication assay, A3G and co-expressed A3F and A3G (A3F/A3G) restrict viral replication more than A3F alone. (B) In a ΔEnv HIV-1LAI (+Vif) single-cycle replication assay A3G, A3F, and A3F/A3G are suppressed by Vif. (A–B) The A3G was HA-tagged and the A3F was V5-tagged. Data were produced from three independent experiments and error bars represent the standard deviation from the mean. Designations for significant difference of values were a P value of ≤0.001 (***), ≤0.01(**), or ≤0.05 (*). (C–D) Immunoblots from the experiments in panels A and B using 50 ng transfected A3 plasmid. (C) Vif induces degradation of HA-tagged A3G when expressed alone or co-expressed with V5-tagged A3F. (D) Vif induces degradation of V5-tagged A3F, but A3F is partially rescued from degradation when expressed in the presence of HA-tagged A3G. (E–G) Further analysis of the A3G-mediated protection of A3F from Vif was carried out using increasing amounts of Vif expression plasmid that was cotransfected with a constant amount of (E) HA-tagged A3G, (F) V5-tagged A3F, and (G) co-expression of the HA-tagged A3G and V5-tagged A3F (A3F-V5/A3G-HA). The data show that A3F, but not A3G is protected from Vif mediated degradation when A3F/A3G are co-expressed. (C–G) Immunoblots were conducted for three independent experiments and a representative blot is shown. Full immunoblots from which representative images were obtained are in Supplementary Fig. S1.
To track the A3G and A3F we immunoblotted cell and virus lysates from the single-cycle replication assays (50 ng transfected A3 plasmid). The A3G-HA was sensitive to Vif-mediated degradation, resulting in less A3G encapsidation in the presence of Vif, although A3G was still present in cells infected with HIV-1 LAI (Fig. 1C). In contrast, the A3F-V5 was not detected in cells or viruses in the presence of Vif (Fig. 1D). Upon co-expression of A3G and A3F, we found no change in A3G sensitivity to Vif-mediated degradation or encapsidation (Fig. 1C), but there was a change in A3F protein levels (Fig. 1D). In ΔVif HIV-1 infected cells, the co-expression of A3G with A3F stabilized the steady state level of A3F in cells and resulted in more A3F being encapsidated (Fig. 1D). During a HIV-1LAI infection, the presence of co-expressed A3G and A3F resulted in incomplete Vif-mediated degradation of A3F and encapsidation of A3F (Fig. 1D). To determine the extent that A3F is protected from Vif-mediated degradation when co-expressed with A3G, we co-expressed the A3s with different amounts of Vif expressed from a plasmid, rather than the HIV-1 genome (50 ng transfected A3 plasmid). We determined that A3F was more sensitive to Vif-mediated degradation than A3G (Fig. 1E and F). In contrast, when co-expressed with A3G, the A3F was less sensitive to Vif-mediated degradation whereas A3G degradation was the same as when expressed alone (Fig. 1G). These results are consistent with previously published data that showed A3F 108S/231I can be partially protected from Vif-mediated degradation by A3G or A3F 108A/231V [90]. Altogether, these results demonstrate that there is not complete Vif-mediated degradation of A3F when it is co-expressed with A3G. These data prompted us to determine the A3-induced mutation frequency of HIV-1 proviral DNA in the presence of Vif, since it may have been previously underestimated due to the use of single A3s.
2.2. Level of A3G and A3F induced G → A mutations exceed those of RT in a single-cycle of HIV-1 replication
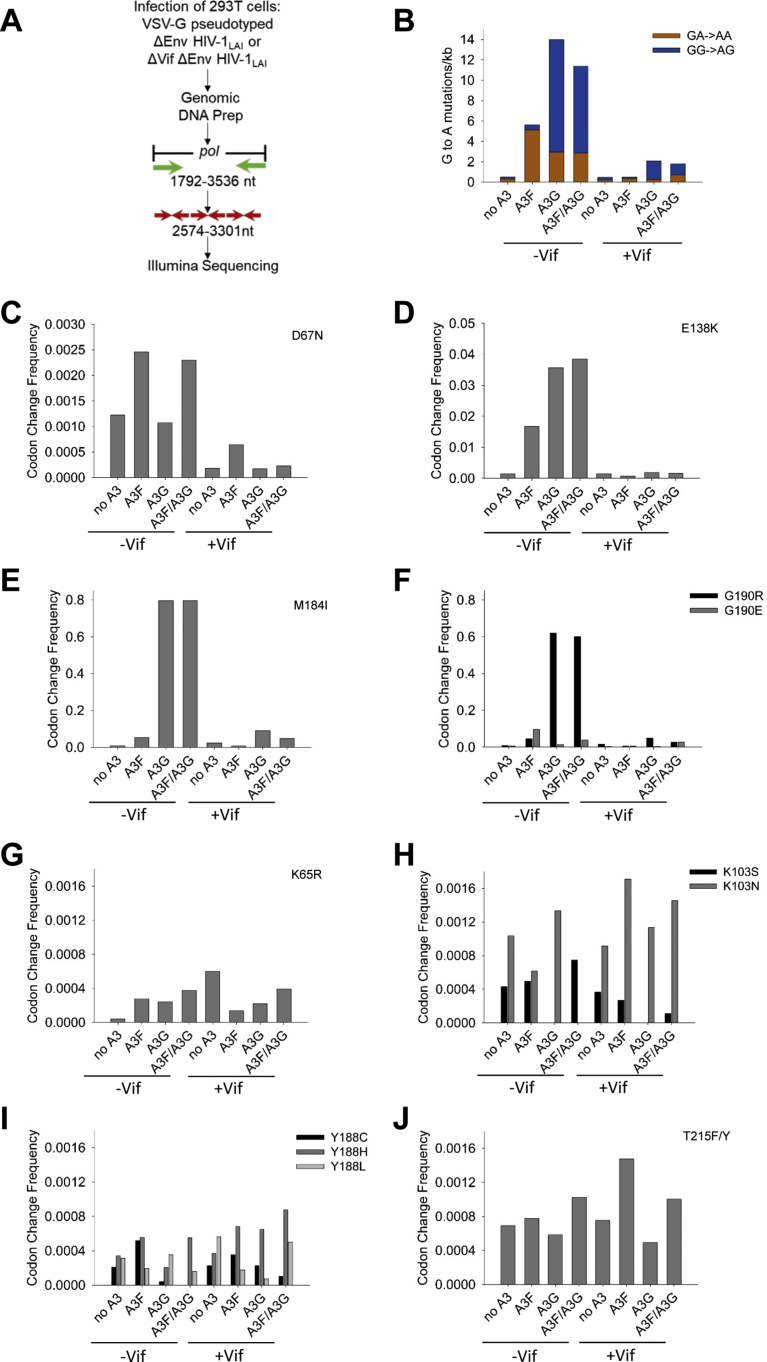
To determine the maximum numbers of A3-induced mutations during a HIV-1LAI infection we amplified part of pol from proviral DNA of single-cycle replication assays from the 25 ng transfected A3 plasmid condition (Fig. 1A and B, HIV-1:A3 plasmid molar ratio of 12:1) and sequenced the amplicons on an Illumina MiSeq platform (Fig. 2A). The sequences were processed through a customized pipeline to determine the flanking nucleotides of deaminations (see Materials and Methods). The proviral DNA was analyzed for the G → A mutation frequency in the presence and absence of Vif. In the absence of A3 enzymes and in the presence (ΔEnv HIV-1LAI) or absence (ΔVif ΔEnv HIV-1LAI) of Vif, the RT rate of G → A mutations stayed relatively the same and was ∼0.5 mutations/kb (Fig. 2B). In the absence of Vif, both A3F and A3G induced high numbers of mutations with A3F inducing 5.6 mutations/kb and A3G inducing 14 mutations/kb (Fig. 2B). From these values, we defined A3-induced G → A hypermutation as being at least 10-fold above the RT G → A mutation rate. In the presence of Vif, the A3F-induced mutations were not significantly different from the no A3 condition, consistent with the immunoblot that showed no detectable A3F encapsidation in the presence of Vif (Figs. 1B and 2B, 0.5 mutations/kb). In the presence of Vif, A3G was able to induce 2.1 mutations/kb, although this appeared not to have any significant effect on the infectivity level of the HIV-1 (Figs. 1B and 2B). When A3F and A3G were co-expressed in the absence and presence of Vif, the mutation frequency was similar to A3G alone and was 11 mutations/kb and 1.8 mutations/kb, respectively (Fig. 2B). These data indicated that in a HIV-1LAI infection with Vif, the A3s can induce a G → A mutation frequency in proviral DNA that is up to ∼4-fold above the RT G → A mutation frequency in a single-cycle of replication. Since this mutation frequency did not functionally effect viral infectivity (Fig. 1B), the data suggest that hypermutated proviral DNA isolated from HIV-1+ people (observed to have at least 10 mutations/kb) was exposed to A3s over multiple rounds of infection [67].
Fig. 2.
Drug resistant mutations can arise from A3- or RT- induced mutations in a single round of virus replication. (A) Proviral DNA resulting from single-cycle replication assays with ΔVif ΔEnv HIV-1LAI and ΔEnv HIV-1LAI from Fig. 1 (25 ng transfected A3 plasmid) was used as a template to PCR amplify 1744 nt of pol (1792–3536), which was further amplified with nested primers that contained Illumina adapters, resulting in a total 728 nt region of pol that was analyzed by next-generation sequencing. The 728 nt was amplified as three separate segments. (B) The G → A mutations in both the GA → AA (A3F context) and GG → AG (A3G context) were determined from ΔVif ΔEnv HIV-1LAI (Vif-) and ΔEnv HIV-1LAI (Vif+) infections. (C–F) Drug resistant mutations that can be induced by A3-mediated mutagenesis due to a deamination motif within the (−) DNA sequence. Note that not all y-axis scales are the same due to different mutation frequencies imposed by the timing of (+) DNA synthesis. (C) The D67N mutation was preferentially induced by A3F. (D) The E138K mutation was induced by A3F and A3G in Vif-, but not Vif+ infections. (E) The M184I mutation was preferentially induced by A3G. (F) The G190R mutation, but not the G190E mutation, was preferentially induced by A3G. (G–J) Drug resistance mutations that can only result from RT-induced error. The RT-induced errors resulted in mutations at (G) K65R, (H) K103S/N, (I) Y188C/H/L, and (J) T215F/Y. Drug resistant mutations relevant to A3-mediated mutagenesis and the drug resistance selection experiments are shown. Other drug resistance mutations are in Supplementary Table S1.
To determine which A3 was inducing the G → A mutations, we further analyzed the sequencing data to determine the GG → AG and GA → AA mutation frequencies. Since A3G preferentially deaminates within 5′CC contexts, the (+) DNA mutations are GG → AG. This is discernable from preferential A3F deaminations that occur within 5′TC contexts and result in (+) DNA mutations that are in the GA → AA context. Analysis of the ΔVif ΔEnv HIV-1LAI condition (−Vif) demonstrated that the random RT mutations that fall into an A3 context are equivalent to the total RT-induced G → A mutations, demonstrating that in context G → A mutations a priori cannot solely be attributed to A3 deamination activity (Fig. 2B). The A3F-induced mutations were 91% in the GA → AA context (Fig. 2B). The A3G-induced mutations were less stringent with the preferred GG → AG context being mutated 79% of the total (Fig. 2B). These values were consistent with their previously published deamination preferences that have been used to determine their respective deamination frequencies in HIV genomes [64, 65, 66]. In the ΔEnv HIV-1LAI condition (+Vif) when each A3 is expressed alone the A3F-induced mutations were 69% in the GA → AA context and the A3G-induced mutations were 87% in the GG → AG context (Fig. 2B). In the −Vif condition with combined A3F/A3G expression, the GA → AA mutations and GG → AG mutations were similar to A3G alone (Fig. 2B, A3F/A3G has 2.8 GA → AA and 8.5 GG → AG mutations/kb), likely due to high A3G activity that can obscure the cooperative effects of coencapsidated A3F/A3G [66]. In the +Vif condition the mutations induced by the combined expression of A3F/A3G resulted in a 60/40% split between GG → AG/GA → AA mutations with the total amount of GA → AA mutations being more than when A3F or A3G were expressed alone (Fig. 2B). This was consistent with Vif-mediated degradation data that showed A3F can be partially rescued from Vif-mediated degradation in the presence of A3G (Fig. 1D and G). Altogether, the data demonstrated that the combined expression of A3G and A3F resulted in a combined effect of both A3s on proviral mutagenesis in the presence of Vif, likely due to the previously characterized ability of A3G and A3F to form a hetero-oligomer and co-encapsidate into HIV-1 virions [65, 66].
2.3. A3G and A3F can induce drug resistant mutations in proviral DNA
We next determined the nature of the mutations in the proviral DNA to establish whether A3F and A3G could contribute to preexisting drug resistance mutations in proviral DNA. We examined several drug resistance changes along the pol gene and noted a significant increase in mutations from 5′ → 3′ in the (+) DNA, consistent with gradients of mutations forming in proximity to polypurine tract (PPT) regions (Fig. 2C, D, E, F, y-axis) [47, 48, 49]. The HIV-1 central PPT (cPPT) is within pol, with mutations peaking directly before the cPPT and within pol. We examined mutations that could result from A3 deaminase activity, which correspond to drug resistant mutations D67N, M184I, E138K, and G190R/E (Table 1). We used the ‘No A3’ transfection condition to represent the RT induced mutations. In the absence of Vif, A3F was able to induce the D67N mutation 2-fold, the M184I mutation 7-fold, the E138K mutation 11-fold, the G190R 6-fold, and the G190E 15-fold more than RT (Fig. 2C, D, E, F). In contrast, in the absence of Vif, A3G preferentially caused mutations more than RT at E138K (23-fold more than RT), M184I (108-fold more than RT), G190R (74-fold more than RT), and G190E (2-fold more than RT), but the D67N mutations were not significantly different from the no A3 condition (Fig. 2C, D, E, F). When the A3F and A3G were co-expressed in the absence of Vif, the mutation pattern matched both A3F and A3G alone consistent with previous data showing that the co-expression and co-encapsidation of A3F and A3G did not alter their deamination preferences (Fig. 2C, D, E, F) [65, 66]. For A3F in the presence of Vif, only the D67N mutation (3.5-fold) and G190E (2-fold) were significantly mutated in comparison to the No A3 condition (Fig. 2C, D, E, F). For A3G in the presence of Vif, there were more mutations induced than for the No A3 condition at the M184I site (3.8-fold) and G190R (2.8-fold) sites (Fig. 2C, D, E, F). When A3F and A3G were co-expressed, only the D67N site (1.2-fold more than RT), M184I site (2-fold more than RT), G190R site (1.6-fold more than RT) and G190E site (9.5-fold more than RT) were preferentially mutated (Fig. 2C, D, E, F). Notably, although the D67N mutation was a preferred site for A3F-induced mutations, as predicted by sequence context, in the absence of Vif, the E138K mutation was induced by both A3F and A3G, although the sequence context is predicted to be preferred by A3F (Fig. 2C and D and Table 1). Similarly, the G190 position contains a predicted A3G deamination motif in the (-)DNA, but in the absence of Vif can be deaminated by A3G and A3F (Fig. 2F and Table 1). These deviations from the predicted deamination motifs are likely due to the effects of secondary structure or larger sequence context that is known to effect A3 deamination [91, 92]. In particular, A3G can deaminate the canonical A3F context approximately 13–20% of the time in this region of RT (Fig. 2B, A3G and A3G/A3F conditions) and may be more able to deaminate 5′TC containing sites than A3F, such as E138K, due to the higher deamination activity (Fig. 1A). Nonetheless, in the presence of Vif when the encapsidated A3 levels are lower, the predicted deamination motifs are consistent with the observed mutations with A3F preferring a 5′TC deamination resulting in D67N and A3G preferring 5′CC deaminations resulting in M184I ((−) DNA context is 5′CCAT (codon is underlined)) and G190R (Table 1). Altogether, these data suggest that in a HIV-1LAI infection the mutation bias of A3G and A3F to cause drug resistant mutations are distinct.
Table 1.
HIV-1 nucleotide and non-nucleotide reverse transcriptase inhibitors and possible resistance mutations.
| Drug class | Drug | Primary drug resistance mutationsa | Possible A3-induced mutation | Codon in (+)/(−) DNA | Probable A3b |
|---|---|---|---|---|---|
| NRTI | 3TC | K65R | M184I | 5′ATG/3′CAT | A3F or A3G |
| Emtricitabine | M184I/V | ||||
| Abacavir | K65R | M184I | 5′ATG/3′CAT | A3F or A3G | |
| Didanosine | K70E | ||||
| L74V/I | |||||
| Y115F | |||||
| M184I/V | |||||
| TDF | M41L | D67N | 5′GAC/3′CTG | A3F | |
| Stavudine | K65R | ||||
| D67N | |||||
| K70E/R | |||||
| L210W | |||||
| T215Y/F | |||||
| AZT | M41L | D67N | 5′GAC/3′CTG | A3F | |
| D67N | |||||
| K70R | |||||
| L210W | |||||
| T215Y/F | |||||
| NNRTI | Etravirine | L100I | E138K | 5′GAA/3′TTC | A3F |
| Rilpivirine | K101E/P | G190E | 5′GGA/3′TCC | A3G | |
| E138A/G/K/Q | |||||
| Y181C/I/V | |||||
| Y188L | |||||
| G190E | |||||
| M230L | |||||
| Nevirapine | L100I | G190E | 5′GGA/3′TCC | A3G | |
| EFV | K101 E/P | ||||
| K103 N/S | |||||
| V106 A/M | |||||
| Y181C/I/V | |||||
| Y188L/C/H | |||||
| G190 A/S/E | |||||
| M230L |
Drug resistant mutations are listed in bold face type if they are associated with the highest levels of resistance, mutations in italics contribute to resistance, and mutations in plain type contribute to resistance in combination with other resistance mutations. Data from the Stanford University HIV Drug Resistance Database [123, 124].
The probable A3 is based on the preferred sequence context for deaminase activity in the viral (−) DNA which is 5′TC for A3F and 5′CC for A3G.
We also determined the level mutations that could only be induced by RT, such as K65R, T215Y/F, K103S/N, and Y118L/C/H to determine if the ability of A3s to interfere with RT polymerization also effects its insertion fidelity (Table 1). We found that RT could induce mutations at K65R, K103S/N, Y188C/H/L, and T215F/Y, among others (Fig. 2G, H, I, J and Supplementary Table S1). For the K103S/N, Y188C/H/L, and T215F/Y mutations, the No A3 condition in the absence or presence of Vif had similar levels of these mutations (Fig. 2G, H, I, J). However, for the K65R, the presence of Vif resulted in a 14-fold increase in the frequency, but overall this mutation occurred at a low level (Fig. 2F, 4 × 10−5, −Vif; 6 × 10−4, +Vif). For the K65R mutation in the absence of Vif, the presence of A3F, A3G or A3F/A3G increased the mutation 6- to 8- fold (Fig. 2G). This effect by A3F, A3G or A3F/A3G on the K65R mutation was not observed in the presence of Vif (Fig. 2G). For the K103S/N mutation, the presence of A3G, either in the absence or presence of Vif resulted in an absence of the K103S mutation, although the K103N mutation was not changed (Fig. 2H). For the Y188C/H/L mutation, the presence of A3G in the absence of Vif resulted in 5-fold less Y188C mutations (Fig. 2I). However, in the presence of Vif, A3F, A3G, and A3F/A3G all caused changes in the mutation frequencies. The presence of A3F or A3G decreased the amount of Y188L mutations 3-fold (+Vif, A3F) to 7.5-fold (+Vif, A3G) and increased the amount of Y188H mutations by 1.8-fold (+Vif, A3F and A3G) (Fig. 2I). The A3F/A3G condition resulted in 2-fold less Y188C mutations (Fig. 2H). In the presence of Vif, for the T215F/Y mutation, the presence of A3F resulted in a 2-fold increase in the mutation rate (Fig. 2J). Since the (−) DNA sequence for these codons have no deamination motifs, these data suggest that A3 enzymes can influence the RT insertion fidelity either by physical inhibition of RT or nearby A3-catalyzed uracils [93, 94].
2.4. Drug resistant HIV-1 can be selected for from spreading infections of peripheral blood mononuclear cells
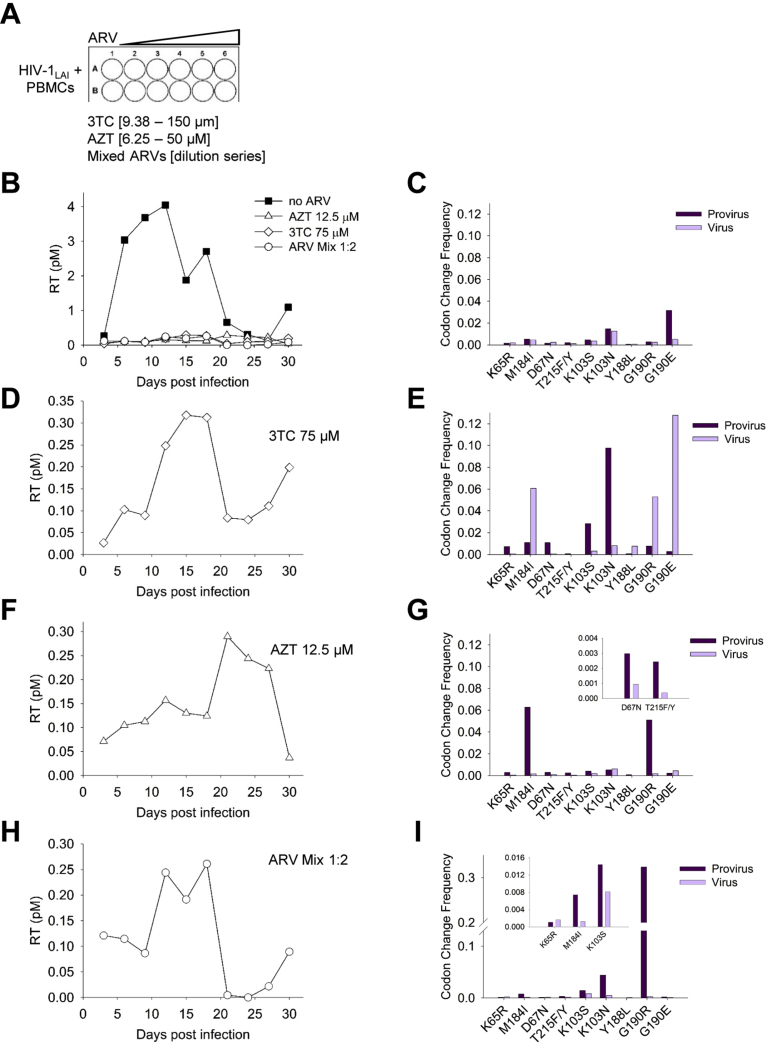
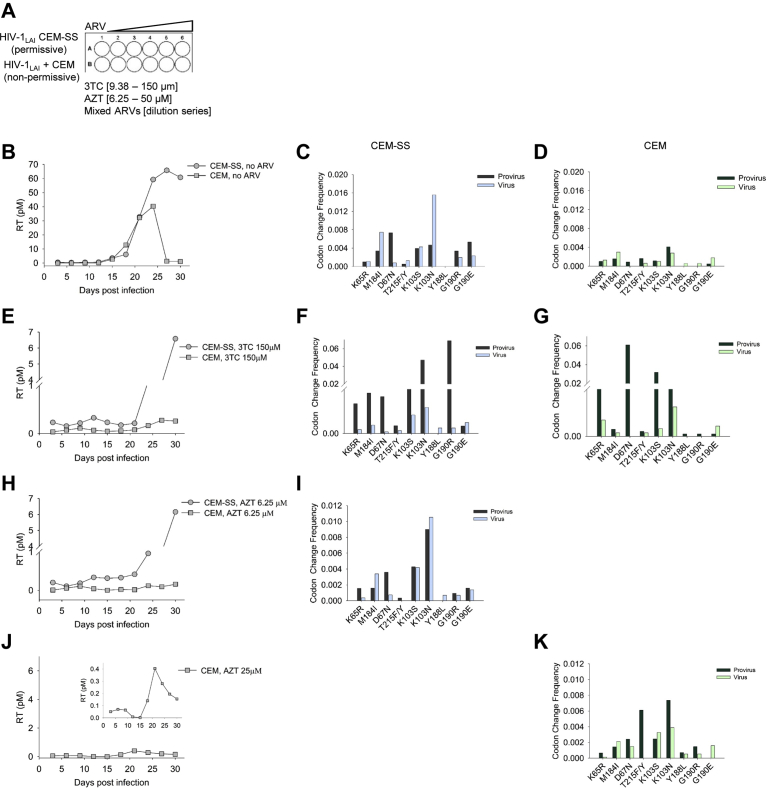
After determining the capability of A3F and A3G to induce drug resistant mutations in the presence of Vif, we sought to test if these mutations would result in functional drug resistant HIV-1LAI. We determined if the mutations we observed could cause viral growth in the presence of ARVs with endogenously expressed A3s from PBMCs. A3F commonly occurs as two alleles that result in 108S/A and 231I/V polymorphisms [60, 95]. Usually the polymorphisms are genetically linked as 108S/231I and 108A/231V [60, 95], but the PBMC donor genotype for A3F was 108S/231V, a less commonly linked genotype and different from our A3F lab clone which was 108S/231I (see Materials and Methods). We could not detect expression of A3F and A3G in the PBMCs due to the absence of sensitive native antibodies, but did confirm expression of A3F and A3G mRNA by qPCR under the experimental conditions (data not shown). The selection of drug resistant virus used a two-fold serial dilution of the ARV, either AZT (6.25–50 μM), 3TC (9.38–150 μM), or the Mixed ARVs (TDF, 3TC, and EFV; TLE regimen) (Fig. 3A). Mutations conferring resistance to AZT (D67N), 3TC (M184I), and EFV (G190E/R) can directly arise from A3-induced mutagenesis (Fig. 2C, E, F and Table 1). The EFV resistance mutations K103S/N and Y188L/C/H and TDF resistance mutation K65R can only arise from RT-induced mutagenesis (Fig. 2H, I and Table 1). After infection of PBMCs with WT HIV-1LAI, the RT activity in supernatants was measured every third day and used to detect if virus was present. The growth took place over 30 days with the total viral supernatant being added to fresh PBMCs from the same donor at day 21. The normal growth of HIV-1 in the PBMCs resulted in a peak at day 12 (Fig. 3B). Proviral DNA was isolated for sequencing at day 30 and genomic RNA was isolated for cDNA synthesis and sequencing at the peak of virus replication. The sequencing data was analyzed using the WT HIV-1LAI genome as the reference genome.
Fig. 3.
Drug resistant HIV-1 can be isolated from infected PBMC cultures. (A) PBMCs were cultured for 30 d with no drug or in the presence of increasing amounts of 3TC (9.38–150 μM), AZT (6.25–50 μM), or Mixed ARVs (dilution series). (B) Virus replication was measured using an RT assay and showed that the virus replication peaked in the PBMC culture at day 12. Drug resistant variants were also isolated, although the level of virus in the supernatant determined by the RT assay was approximately 10-fold less. (C) In the absence of ARVs, drug resistance mutations were present in both provirus DNA and virus gRNA. (D) Virus resistant to 75 μM 3TC shown in panel A, but on an expanded scale. (E) Virus resistant to 3TC had the corresponding drug resistance mutation at M184I in gRNA, along with drug resistance mutations for other ARVs. (F) Virus resistant to 12.5 μM AZT shown in panel A, but on an expanded scale. (G) Virus resistant to AZT had the corresponding drug resistance mutations at D67N and T215F/Y, along with drug resistance mutations for other ARVs. (H) Virus resistant to a 1:2 dilution of Mixed ARVs (TDF, 3TC, EFV) shown in panel A, but on an expanded scale. (I) Virus resistant to TDF, 3TC, and EFV had corresponding mutations in K65R, M184I, and K103S/N, respectively, along with drug resistance mutations for other ARVs. Other drug resistance mutations are in Supplementary Table S1.
In the absence of any ARV drugs, the PBMCs accumulated drug resistance mutations. The highest levels of mutations in the proviral DNA were at K103N and G190E, conferring resistance to EFV (Fig. 3C). The gRNA had corresponding K103N and G190E mutations, although the G190E was significantly lower in gRNA consistent with the principle of purifying selection since G190E has been reported to create an aberrant protease cleavage site [96]. Mutations at other sites such as M184I and D67N that can be induced by A3G and A3F, respectively, were also present, consistent with the single-cycle replication data showing the preexistence of drug resistance mutations (Figs. 2C, E and 3C). When the HIV-1 was grown in the presence of ARVs, growth curves with delayed peaks were detected, but with approximately 10-fold less virus than in the absence of drug, consistent with low level variants emerging from the culture (Fig. 3B, D, F and H).
Each ARV selected for a unique combination of drug resistance mutations. The HIV-1 resistance to 3TC occurred at 75 μM 3TC and correlated with high levels of the M184I mutation in gRNA (Fig. 3D, E). In the gRNA, the G190E mutation was much higher, despite not being required for 3TC resistance, but suggests that G190 was hot spot for mutation either by RT or A3s. In proviral DNA, the K103N mutation was also higher than the M184I mutation, but was not in the gRNA, likely due to the lack of selection pressure for this mutation. The HIV-1 resistance to AZT occurred at 12.5 μM AZT and resulted in a similar growth curve, but with a more delayed peak of RT activity compared to growth with 3TC (Fig. 3F, day 21 compared to Fig. 3D, day 15). The high level of resistance to AZT conferred by the T215F/Y (RT induced mutation) and D67N (A3F induced mutation) was observed at low levels in culture likely due to the selection of drug resistant variants at a low concentration of AZT (Fig. 3G and inset). Consistent with the M184I mutation causing increased sensitivity to AZT, the mutation was found in the proviral DNA at a much greater extent than in the gRNA (Fig. 3G). The Mixed ARVs should select for resistance mutations at K65R, M184I, and any of the following K103S/N, Y188L/C/H, and G190E/R. At the 1:2 dilution, we recovered resistant virus that had mutations at K65R, M184I, and K103S (Fig. 3H, I). The G190R was present at high amounts in proviral DNA but was 3-fold less likely to be in gRNA compared to the K103S (Fig. 3I and inset). Consistent with M184I conferring increased sensitivity to TDF, there was a much higher amount of M184I mutations in the proviral DNA, than the gRNA (Fig. 3I and inset). Altogether, the data from virus growth on infected PBMCs with and without drug selection demonstrated that this experimental system could be used to isolate drug resistant viruses and recover drug resistant mutations through deep sequencing. Rather than analyze PBMCs from different donors that may introduce unknown variables into the analysis, we used different cell lines without or with endogenous A3 expression to test specific questions using the same experimental system.
2.5. Effect of exposure of HIV-1 to A3G and A3F in a single-cycle of replication on drug resistance
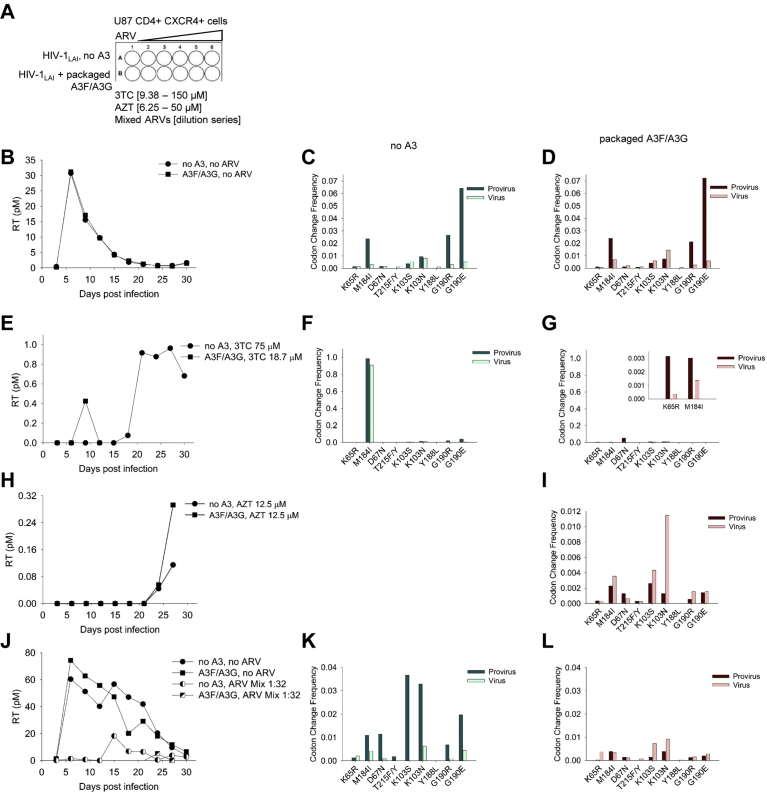
We next investigated if exposure to WT HIV-1LAI with encapsidated A3F and A3G could introduce drug resistant mutations that would be maintained in a spreading infection. For this experiment, we used U87 CD4+ CXCR4+ cells that do not express A3F or A3G and infected them with HIV-1LAI. Two stocks of HIV-1LAI were prepared, one stock was produced in the presence of A3F and A3G so that the A3s were encapsidated into the virions and the second stock was produced in the absence of A3F and A3G (see Materials and Methods). Importantly, the HIV-1LAI expressed Vif, which would induce degradation of A3F and A3G from virus producer cells resulting in low A3F and A3G encapsidation levels. Infection of U87 CD4+ CXCR4+ cells with the virus particles having encapsidated A3F/A3G would result in A3-induced mutagenesis occurring only in the first round of reverse transcription and proviral DNA synthesis. However, the cultures were kept for 30 days to determine the long-term fate of the A3 induced mutations on drug resistance to AZT, 3TC, and the Mixed ARV TLE regimen (Fig. 4A), in contrast to the single-cycle replication assay that only identified the presence of mutations (Fig. 2). In the absence of any ARVs, the RT activity of the U87 CD4+ CXCR4+ cells under both No A3 and A3F/A3G conditions had similar profiles and peaked at day 6 (Fig. 4B). The robust replication occurred, even with an MOI of 0.01 due to the high surface expression of the CD4+ and CXCR4+ [97]. Sequencing of the proviral DNA and cDNA produced from gRNA demonstrated the existence of drug resistant mutations, particularly M184I, K103S/N, and G190R/E (Fig. 4C, D). The mutation profiles of the No A3 and A3F/A3G condition were very similar, suggesting that the majority of mutations were made from RT-induced error or that RT and A3F/A3G have overlapping mutational hot spots (Fig. 4C, D).
Fig. 4.
Exposure of WT HIV-1LAI to A3G and A3F for a single round of replication can generate drug resistant viruses. (A) U87 CD4+ CXCR4+ cells were cultured for 30 d with no drug or in the presence of increasing amounts of 3TC (9.38–150 μM), AZT (6.25–50 μM), or Mixed ARVs. (B) Virus replication was measured using an RT assay and showed that the virus replication peaked in the U87 CD4+ CXCR4+ culture at day 6. (C–D) Drug resistance mutations were present in both provirus DNA and virus gRNA for the No A3 and co-expressed A3F and A3G (A3F/A3G) condition. (E) Virus resistant to 75 μM 3TC (No A3) and 18.7 μM 3TC (A3F/A3G) was isolated. (F–G) Virus resistant to 3TC had the corresponding drug resistance mutation at M184I in gRNA, along with drug resistance mutations for other ARVs. (G) The K65R mutation that provides additional resistance to 3TC and the M184I mutation were found in the A3F/A3G condition. (H) Virus resistant to 12.5 μM AZT was isolated from the A3F/A3G condition. (I) Virus resistant to AZT had the corresponding drug resistance mutations at D67N and T215F/Y, along with drug resistance mutations for other ARVs. (A–I) Data was obtained from the experiments listed in Table 2. Virus replication in the absence of drug was determined for each experiment in the No A3 and A3F/A3G condition, but only one representative curve is shown for each condition in panel B. (J) Virus resistant to a 1:32 dilution of Mixed ARVs (TDF, 3TC, EFV) was isolated, but similar to the other drug resistant viruses, the amount of RT activity in the supernatant was less than without the Mixed ARV. (K–L) Virus resistant to TDF, 3TC, and EFV had corresponding mutations in K65R, M184I, and K103S/N, respectively, along with drug resistance mutations for other ARVs. Other drug resistance mutations are in Supplementary Table S1.
To discern the impact of RT and A3F/A3G, we conducted 4 independent experiments for U87 CD4+ CXCR4+ cells grown in the presence of 3TC or AZT. The experiments were then scored for whether drug resistance viruses were obtained and if it occurred in the No A3 or A3F/A3G condition (Table 2). We found that for 3TC resistance, out of 4 experiments, resistance arose once in the No A3 condition and once in the A3F/A3G condition (Table 2). Although the drug resistant variants that grew from viruses that had packaged A3F/A3G arose early at day 9, they could not maintain growth in culture and by day 12 the RT activity in the supernatant was lost (Fig. 4E). In addition, the 3TC resistance from the A3F/A3G condition was at 18.7 μM, less than the 75 μM that arose in the PBMC culture (Figs. 3D and 4E). In contrast, the virus that had no A3 packaged resulted in robust drug resistant viruses emerging from the culture at day 18 in the presence of 75 μM 3TC (Fig. 4E). Accordingly, the drug resistant viruses from the No A3 condition had very high levels of the M184I mutation in proviral and cDNA isolated from gRNA (Fig. 4F). In contrast the A3F/A3G exposed viruses had lower amounts of the M184I, although the mutation was present in cDNA produced from gRNA (Fig. 4G and inset). The 3TC resistance is also strengthened by the K65R mutation, which can only be induced by RT and was also found in the A3F/A3G condition (Fig. 4G). Consistent with the D67N mutation being a hot spot for A3-induced mutagenesis in the presence of Vif, this mutation was found at the highest amount in the proviral DNA, but not in the viral cDNA since there was no AZT selection (Fig. 4G). When the experiments did use AZT, the only conditions that did produce drug resistant virus was the condition with packaged A3F/A3G where we isolated drug resistant virus for two of the four experiments (Table 2 and Fig. 4H, representative curve shown). Although we recovered virus in one experiment for the No A3 condition in the presence of AZT, the RT activity was 2.5-fold less than the A3F/A3G condition and we were unable to amplify the virus, which was required to isolate gRNA for cDNA synthesis and sequencing (Fig. 4H). The sequencing data from the A3F/A3G condition in the presence of 12.5 μM AZT showed that the main mutation mediating the resistance was D67N (Fig. 4I). Although the T215F/Y mutation that results in stronger AZT resistance was found in gRNA, the D67N mutation was 2.4-fold more frequent, consistent with A3F-induced mutagenesis (Figs. 2C and 4I). Altogether, these data suggest that 3TC resistance from the M184I mutation is more likely to occur from RT error than A3-induced mutagenesis, but AZT resistance from the D67N mutation is more likely to occur from A3-induced mutagenesis than RT.
Table 2.
Emergence of 3TC or AZT resistant viruses in the absence or presence of encapsidated A3F/A3G.
| Antiretroviral drug | Drug resistant virus |
|
|---|---|---|
| No A3 | Encapsidated A3F/A3G | |
| 3TC | ||
| Experiment 1 | Yes | |
| Experiment 2 | ||
| Experiment 3 | ||
| Experiment 4 | Yes | |
| AZT | ||
| Experiment 1 | ||
| Experiment 2 | Yes | |
| Experiment 3 | ||
| Experiment 4 | Yes | |
Experiment was carried out as detailed in Fig. 4A sketch for 3TC and AZT. Occurrence of drug resistant viruses was scored from four independent experiments. No notation means that drug resistant viruses were not isolated.
We also examined if exposure to A3F/A3G in a single round of replication could accelerate the development of viruses resistant to the Mixed ARV TLE regimen. We found that in the presence of the mixed ARVs the No A3 condition had a more robust RT signal at day 15 than the A3F/A3G exposed virus, which had an undetectable RT signal until day 24, and the peak was still 3.5-fold less signal than the no A3 condition (Fig. 4J). Proviral DNA and gRNA was isolated from each of these drug resistant virus populations and sequenced. For both the No A3 and A3F/A3G conditions, the same drug resistant mutations required for viral replication in the presence of the Mixed ARVs were found in the gRNA, which were K65R, M184I, K103S/N, and G190R/E. Although the mutation frequencies were higher in the proviral DNA for the No A3 condition, the virus from the No A3 and A3F/A3G conditions contained similar levels of mutations (Fig. 4K, L).
Although the A3s could contribute to drug resistant virus variants arising for 3TC and Mixed ARV treatments, the RT signal obtained was less than conditions with No A3 (Fig. 4E, J). Even with AZT, the RT signal was at least 3-fold less than virus isolated in the absence of A3s for the other single drug experiment with 3TC (Fig. 4E, H). These data suggest that A3 enzymes can contribute to drug resistance, but that there may be additional mutations occurring from A3-induced mutagenesis that compromises the replication capacity of the virus.
2.6. Drug resistant viruses derived from A3-induced mutagenesis have less replication capacity
To examine if there are consequences of A3-induced mutagenesis on virus replication capacity we tested for emergence of drug resistant variants using the genetically related CEM-SS and CEM cells that have nearly undetectable expression and normal A3F and A3G expression, respectively [88]. The CEM-SS/CEM cells were used to enable differentiation between RT- and A3- induced mutations, and the CEM cells enabled continuous encapsidation of endogenously expressed A3s during the experiment (Fig. 5A). The CEM-SS/CEM experimental system is in contrast to the U87 CD4+ CXCR4+ cells where A3 exposure only occurred in the first round of replication (Fig. 4) and provided the advantage of having a “no A3” cell line for comparison, in contrast to PBMCs (Fig. 3). We genotyped the A3F of the CEM cells and found that they have the commonly occurring heterozygous A3F genotype of 108S/231I and 108A/231V [60, 95]. In treatment naïve cohorts, the A3F 108A/231V genotype has been associated with lower set-point viral load and slower rate of progression to AIDS that appeared to be due to partial resistance to Vif [55]. Without any drug selection, the virus produced from CEM-SS and CEM cells both had peak RT activity after 24–27 days (Fig. 5B). The peak RT activity from the CEM cells was 1.6-fold less than from CEM-SS cells (Fig. 5B). Sequencing of proviral DNA and cDNA isolated from gRNA showed that although there were more drug resistance mutations in the CEM-SS conditions, the types of mutations between the no A3 (CEM-SS) and A3 (CEM) conditions were similar types (Fig. 5C, D).
Fig. 5.
Drug resistant viruses produced from cells expressing A3G and A3F have less replication capacity than drug resistant viruses that arise from RT-induced error. (A) CEM-SS (permissive) or CEM (non-permissive) cells were cultured for 30 d with no drug or in the presence of increasing amounts of 3TC (9.38–150 μM) or AZT (6.25–50 μM). (B) Virus replication was measured using an RT assay and showed that the virus replication peaked in the CEM-SS culture at day 27 and the CEM culture at day 24. (C–D) Drug resistance mutations were present in both provirus DNA and virus gRNA for the No A3 and co-expressed A3F and A3G (A3F/A3G) condition. (E) Virus resistant to 150 μM 3TC was isolated from both CEM-SS and CEM cultures, but the CEM cells had 24-fold less peak RT activity. (F–G) Virus resistant to 3TC had the corresponding drug resistance mutation at M184I in gRNA, along with drug resistance mutations for other ARVs for (F) CEM-SS and (G) CEM cells. (H) Virus resistant to 6.25 μM AZT was isolated from cultures of CEM-SS and CEM cells. (I) Virus resistant to 6.25 μM AZT from the CEM-SS culture was sequenced and had the corresponding drug resistance mutations at D67N and T215F/Y, along with drug resistance mutations for other ARVs. (J) Virus resistant to 25 μM AZT was isolated from CEM cell culture and is shown on same scale as AZT resistant virus from CEM-SS cells in Panel H. Inset shows expanded scale. (K) Virus resistant to AZT from the CEM culture was sequenced and had the corresponding drug resistance mutations at D67N and T215F/Y, along with drug resistance mutations for other ARVs. Other drug resistance mutations are in Supplementary Table S1.
To examine specifically the function of A3s in emergence of drug resistance, we grew CEM-SS and CEM cells in either 3TC or AZT. Data from infections in U87 CD4+ CXCR4+ cells suggested that the M184I mutation in replicating viruses was induced more efficiently by RT error than A3-induced mutagenesis (Fig. 4). In support of this line of reasoning, we isolated viruses that had high RT activity from CEM-SS cells in the presence of 150 μM 3TC (Fig. 5E). Although we could isolate viruses from CEM cells at 150 μM 3TC, the level of RT activity was 24-fold lower than viruses from CEM-SS cells (Fig. 5E). The M184I was present in gRNA for both CEM-SS and CEM cells, but the frequency was 2.5-fold higher in virus from CEM-SS cells (Fig. 5F, G). There were also multiple other drug resistance mutations present, such as K103S/N in CEM-SS cells, consistent with RT-induced mutations, and D67N in CEM cells, consistent with A3-induced mutagenesis (Figs. 2C, H and 5F, G).
For selection of drug resistant variants in the presence of AZT, consistent with the results from U87 CD4+ CXCR4+ cells, the CEM cells resulted in AZT resistance at 25 μM AZT, but the CEM-SS cells resulted in resistance only at 6.25 μM AZT (Figs. 4H, I and 5H, J). We also obtained virus from CEM cells resistant to 6.25 μM AZT, but the RT activity was ∼40-fold less than drug resistant virus from CEM-SS cells (Fig. 5H). Even the CEM derived drug resistant virus isolated at 25 μM AZT had 15-fold less RT activity than CEM-SS derived virus resistant to 6.25 μM AZT (Fig. 5H, J). All the AZT resistance was from the D67N mutation in the gRNA, since we did not recover any T215F/Y resistance mutations in gRNA, although it was found in the proviral DNA of viruses isolated from both CEM-SS and CEM cells (Fig. 5I, K). These data collectively show that although A3s can induce drug resistance, the resulting viruses had less replication capacity than drug resistant viruses derived from RT-induced mutations alone.
3. Discussion
Some key limitations from previous studies for assessing if A3-induced mutagenesis can contribute to the development of drug resistance were that only A3G instead of multiple A3s was assessed, the bias of RT to induce G → A mutations was not considered, only one antiviral drug was used, and only viral recombination with ΔVif HIV-1 or HIV-1 with partially inactive Vif was considered. To overcome these past limitations, we used WT VifLAI and assessed the contribution of A3F and A3G on drug resistance to multiple drugs, alone and in combination, in comparison to conditions with no A3 enzymes. We determined that all the RT-induced mutations were in a G → A context that matched the A3F or A3G contexts, confounding the interpretation of many previous studies. Importantly, we found that expression of both A3F and A3G in the HIV-1 producer cells resulted in a protection of A3F from complete Vif-mediated degradation and resulted in a proviral mutational load from approximately equal contributions of A3F and A3G, consistent with a previous report [90], and despite A3F previously being found less active in ΔVif HIV-1 infections [50, 72, 73, 95, 98]. Altogether, in separate cell lines we found the consistent result that although A3 enzymes could induce drug resistant mutations that resulted in viral replication under drug selection pressure, the viruses had less replication capacity than drug resistant viruses induced by RT alone. Although A3F and A3G can induce nonsense mutations when codons for tryptophan are deaminated in (−) DNA [17], these would completely inactivate the virus and were not likely to contribute to replication competent drug resistant variants. The more common mutation induced by both A3G and A3F are missense mutations that may or may not inactivate the virus [17]. Due to the stochastic nature of A3-induced mutations and the possibility of several types of mutations causing less replication capacity, we could not identify the specific mutations responsible. Nonetheless, these data support a model in which A3 enzymes cooperate to safeguard against viral evolution and to promote viral suppression.
Consistent with previous studies, we also found in our mutation analysis that A3G could induce the M184I mutation in proviral DNA [68, 71]. We found in a single-cycle replication assay with WT HIV-1LAI that A3G could induce this mutation in the absence of 3 TC at a higher level than A3F and RT alone (Fig. 2). However, despite this ability we found that A3G was less able to produce drug resistant virus that could replicate as robustly as drug resistant viruses that emerged from the RT-induced mutations alone (Figs. 4 and 5). Further, in the U87 CD4+ CXCR4+ cells, the virus isolated from day 6 had approximately equal levels of the M184I mutation between the No A3 and A3F/A3G condition, in contrast to the single-cycle replication assay, and suggesting that RT over several replication cycles can also induce the M184I mutation (Figs. 2E and 4C, D). Nonetheless, that we did isolate replication competent 3TC resistant virus in the presence of A3F and A3G underscores that viral inactivation after A3-induced mutagenesis during a WT HIV-1LAI infection is not guaranteed (Figs. 4 and 5). There are several missense mutations that can be induced by A3 enzymes that do not inactivate key enzymes such as protease [17, 50, 99]. However, we observed that sublethally mutated and drug resistant proviral genomes that were exposed to A3F and A3G were unlikely to replicate as well in the presence of drug selection as WT HIV-1LAI that was not exposed to A3F and A3G. As a result, the differentiation between sublethal mutagenesis and viral suppression appears to be the number of mutations, in addition to the nature of the mutations. A3G has been shown to be very efficient at inactivation of ΔVif HIV-1 with some reports suggesting that even a single encapsidated A3G molecule can induce lethal mutations that inactivate the virus [72, 73, 100]. Even though A3G is more efficient at inducing mutations than A3F in a ΔVif HIV-1 infection, the co-expression of A3F and A3G resulted in partial protection of only A3F from Vif-mediated degradation (Fig. 1E, F, G). In our study, A3G was always co-expressed with A3F. This may have resulted in additional A3F-induced mutations that led to drug resistant viruses with less replication capacity [65, 66, 101, 102, 103].
Although A3F has been implicated more than A3G for inducing genetic diversity, no studies have directly tested the ability of A3F to promote HIV-1 drug resistance [50, 72, 74]. Here we showed that A3F can preferentially induce the D67N mutation that results in AZT resistance and this occurred in the presence of Vif (Figs. 2, 4, and 5). In U87 CD4+ CXCR4+ cells with virus exposed to A3F and A3G for only one round of replication, the A3F/A3G condition resulted in AZT resistant virus with more replication capacity than the No A3 condition for two of the four experimental replicates (Table 2, Fig. 4). Despite this induced resistance, we also found that in CEM cells where there is continual encapsidation of A3F and A3G, the resulting viruses still became resistant to AZT, but had less replication capacity than the AZT resistant viruses that emerged from CEM-SS cells in the absence of any A3s (Fig. 5). Our data demonstrated that this is because the coordinate expression of A3G with A3F results in more encapsidation of A3F due to protection from Vif-mediated degradation, resulting in almost equal contribution of mutations from A3F and A3G in a WT HIV-1LAI infection (Figs. 1D and 2B). This suggests that the combination of GG → AG and GA → AA mutations could have a higher inactivation potential than each alone in a WT HIV-1 infection [65, 104]. This may also be due to differences in enzyme processivity that result in A3F inducing mutations that are widely spaced, potentially inactivating or mutating more target proteins, and A3G inducing clusters of mutations that have high inactivation potential if they fall within conserved regions or enzyme active sites [50].
Our study also addressed the contribution of RT-induced G → A mutations and whether the number of A3-induced mutations could exceed the RT error rate in a single round of replication. Several studies have assessed levels of A3-induced mutagenesis using proviruses recovered after unknown numbers of multiple rounds of virus replication and sometimes from infections with naturally occurring Vif defective viruses [14, 44, 67, 68, 103]. If the proviral DNA is “hypermutated”, then recombination with a WT or sublethally mutated virus would be required to recover drug resistant mutations. While this may efficiently occur in cell culture, it appears less likely in a clinical infection, suggesting that this is not a main route of A3-induced drug resistance, if it occurs [67, 68]. Rather, the sublethal mutagenesis that would occur in the first round of replication would be the most likely mechanism for A3 enzymes to contribute to drug resistance. In this situation, we found evidence of potential sublethal mutagenesis since A3-induced mutations decreased from ∼10-20-fold above RT error in the absence of Vif to ∼4-fold above RT error in the presence of Vif (Fig. 2B). These mutations did not cause any decrease in viral replication in the absence of drug selection (Fig. 1B). However, in the presence of drug selection where only select viruses could replicate, the effect of the A3-induced mutations was visible because the virus replication, as measured by an RT assay, resulted in less signal in the presence of drugs than the no RT condition (Fig. 4, 3TC, combined ARVs; Fig. 5, 3TC, AZT). Importantly, in a spreading infection in the absence of drugs we found that the chance for RT to induce the M184I mutation in comparison to viruses exposed to A3G and A3F for a single round of replication was equal, but under drug selection pressure, the no A3 condition produced a much stronger pool of 3TC resistant virus (Figs. 4 and 5). Consistent with analysis of the mutational bias of HIV-1 proviral DNA from infected individuals [82] and studies characterizing RT fidelity [85, 86, 87], all of our quantified RT induced G → A mutations were within a relevant A3 mutation context (Fig. 2B). These data suggest that previous studies have underestimated the ability of RT alone to induce this mutation. Only the resistance to AZT was regularly induced by A3 enzymes, likely A3F, but AZT resistant viruses continually exposed to A3G and A3F had lower replication capacity than viruses not exposed to A3G and A3F (Fig. 5). With a combined ARV treatment, the single round of exposure to A3s enabled resistance, but it was less robust than RT alone (Fig. 4).
Results from our study differ from two previous studies that examined A3-induced drug resistance. Namely, both studies found that A3G induced 3TC drug resistance either through hypermutation and subsequent recombination [68] or through sublethal mutagenesis [71]. The key difference between our studies is that we also had A3F present, which may result in higher levels of mutagenesis and ensure viral inactivation. Despite different results from some cell-based experiments, our data are in agreement with in silico studies regarding the effects of collateral A3-induced mutations. An early study by Jern et al. found through simulation analysis that A3G deaminations have been a factor in HIV-1 evolution, but only a small proportion of the deaminations could be responsible for drug resistance mutations, indicating that there is not always a direct connection between resistance and A3-induced mutagenesis [14]. This was supported by a study by Delvis-Frankenberry et al. that suggested hypermutated proviruses occur, but are rarely rescued by recombination and that sublethal mutagenesis for A3G and A3F is negligible since it occurs at a rate less than the RT mutations [67]. While our single-cycle replication assays found a larger number of mutations due to A3G and A3F in the presence of Vif (Fig. 2), the overall conclusions were the same; that the effect of A3-induced mutagenesis appeared to be either be very low and insignificant or high enough to inactivate or partially suppress viral replication [67].
In particular, several studies have found stop codons almost concomitant with A3-induced mutagenesis, even with WT HIV-1 infections and in clinical studies [67, 79, 83, 84]. Notably, the M184I mutation causes RT to be less processive, although this can be compensated for with an E138K mutation [105, 106, 107, 108, 109, 110]. Nonetheless, the M184I mutation slows the synthesis of DNA and enables more time for A3-induced mutagenesis, resulting in more mutations and making viral inactivation more likely and the contribution of A3 enzymes to drug resistance less likely [81]. The level of A3-induced mutations differs for various anatomical compartments, but consistently A3 mutated proviral genomes contain a high proportion of inactivating mutations [80]. Further, during a WT HIV-1 infection, even the E138K mutation is more likely to be induced by RT, due to its G → A error bias [82], consistent with our sequencing data (Fig. 2D).
Collectively the data from our study and others suggests that there is a large dependence on the environmental selection pressures for whether A3-induced mutations result in replication competent drug resistant virus. Indeed, there is evidence of less fit HIV-1 persisting in a host since the lower replication rate would diminish the infectious burden and immune response [111, 112]. Depending on the selection pressures, a replication compromised proviral clone could emerge as a major component of the virus population [111, 112]. Nonetheless, the data from our work and others suggest that A3 enzymes are not predominantly responsible for inducing HIV-1 drug resistance despite having deamination hotspots that can induce drug resistance mutations in proviral DNA [14, 67]. Due to the chance of these A3-mutated viruses being replication incompetent, it is appears more likely that HIV-1 RT induced errors primarily drive HIV-1 evolution to drug resistance.
4. Materials and Methods
4.1. Single-cycle replication assay
HIV-1LAI ΔVif ΔEnv or HIV-1LAI ΔEnv (500 ng) were cotransfected into 1 × 105 293T cells with 25, 50, or 100 ng of A3G in MCS-II of pVIVO2 (Invivogen) vector with a C-terminal HA tag, A3F in MCS-I of pVIVO2 vector with a C-terminal V5 tag, or A3F/A3G where A3F was cloned into MCS-I of pVIVO2 with a C-terminal V5 tag and A3G was cloned into the MCS-II of the pVIVO2 vector with a C-terminal HA tag. Empty pVIVO2 vector was used for the no A3 condition. Construction of these vectors has been previously described [66]. The pVIVO2 vector contains two separate transcription units (SV40 or CMV) that independently result in strong, constitutive expression without transcriptional inference. The A3F clone is the 108S/231I polymorphism [60]. GeneJuice (Novagen) transfection reagent was used as per manufacturer's protocol and cells were maintained in DMEM supplemented with 10% FBS. After 20 h the media was changed and 48 h post transfection the virus-containing supernatants were harvested and filtered through a 0.45 μm polyvinylidene difluoride (PVDF) syringe filter. The cells were washed with PBS, lysed in Laemmli buffer and used for immunoblotting. For the infectivity assay, 1 × 104 of HeLa CD4+ CCR5+ LTR lacZ cells per well of a 96-well plate were infected with a 15-fold dilution of HIVLAI ΔVif ΔEnv or HIVLAI Δenv virus in the presence of 8 μg/mL of polybrene. At 48 h post infection infectivity was measured through a colorimetric detection using a β-galactosidase assay reagent (Pierce). Infectivity of each virus was compared by using the infectivity of the No A3 condition as 100%. Statistical significance of results was determined using one-way ANOVA. For sequencing of proviral DNA, 1 × 105 293T cells per well of a 24-well plate were infected in the presence of 8 μg/mL polybrene with 40 μL of supernatant containing virus from the 25 ng transfected A3 plasmid conditions. The plates were spinoculated at 800 × g for 1 h. Cells were harvested after 48 h by removing the media, washing with PBS, and lysing the cells and extracting DNA with DNAzol (Invitrogen). The proviral DNA was then PCR amplified and sequenced as described for “Deep sequencing”.
4.2. Immunoblotting
Tagged proteins (50 ng transfected A3 plasmid) in cells or virions were detected by rabbit anti-HA (1:1000, Sigma) or rabbit anti-V5 (1:500, Sigma). Mouse anti-α tubulin (1:1000, Sigma) and anti-p24 (1:1000, Cat# 3537, NIH AIDS Reagent Program) were used to detect the cell lysate loading control (α-tubulin) and the virus lysate loading control (p24) [113,114]. Secondary detection was performed using Licor IRDye antibodies produced in goat (IRDye 680-labeled anti-rabbit and IRDye 800-labeled anti mouse). For cell lysates, 40 μg total protein was used. For virions, a portion of the filtered supernatant was concentrated using Retro-X concentrator (Clonetech) according to manufacturer's instructions and 20 μL was used.
4.3. Vif degradation assay
To compare efficiency of Vif in mediating degradation of A3F, A3G and A3F/A3G, 1 × 105 293T cells per well of a 12-well plate were co-transfected with 50 ng of A3F-V5, A3G-HA, A3F-V5/A3G-HA or empty pVIVO2 (no A3) and a titration of VifLAI expression plasmid (0,10, 25, 50, 100, and 200 ng). The VifLAI expression plasmid has been previously described [115]. GeneJuice (Novagen) used as transfection reagent as described by the manufacturer. DMEM and 10% FBS was used to maintain the cells. Twenty hours post transfection the media was changed and 48 h post transfection cells were washed in PBS and lysed with Laemmli buffer. Vif was detected with HIV-1 Vif monoclonal antibody (1:1000; Cat# 6459, NIH AIDS Reagent Program) [116, 117, 118] and immunoblotting was carried out as described for cell lysates.
4.4. Deep sequencing
HIV genomic RNA was purified from the supernatant of infected cells using QIAamp Viral RNA Kit as per manufacturer's protocol. The cDNA was synthesized using AccuScript High Fidelity Reverse Transcriptase (Agilent) using the Half RT reverse primer (5′ TATTTCTGCTATTAA GTCTTTTGATGGGTCA). From the virus cDNA, pol nucleotides 1792–3536 were amplified by PCR with Pfu polymerase (Agilent) using Ext F p7.9 forward primer (5′ AAAGCATTGGGACCAGGAG CGACACTAGAAGARATGATGACAGCATGYCA) and Half RT reverse primer. The product of this reaction was then amplified (1807–3463 nt) in a nested PCR reaction with Pfu using Ext F p7.10 forward primer (5′ GGAGCGACACTAGAAGAAATGATGACAGCATGYCARGGAGTRG GRGGRCY) and Half Pol reverse primer (5′ TCTGCCAGTTCTAGCTCTGCTT). Proviral DNA was obtained by using DNAzol (Invitrogen) treatment, as described in the manufacturer's protocol. The extracted DNA was then treated with DpnI (NEB) for 1 hour at 37 °C to digest any contaminating transfected plasmid.
The region to be sequenced for both cDNA and proviral DNA was pol nucleotides 2573–3301 nt and was divided into three 360 nt overlapping segments. All segment PCR reactions were performed with Q5 High Fidelity polymerase (New England Biolabs). Segment 1 (2574–2933 nt) was amplified using primers D67N_F forward (5′ CCAGTAAAATTAAAGCCAGGAATG) and D67N_R reverse (5′ AGTATACTTCCTGAAGTCT TCATCT) primers. Segment 2 (2790–3149 nt) was amplified using primer E138K_F forward (5′ AATAAGAGAACTCAAGACTTCTGG) and E138K_R reverse (5′ TGTTCTATGCTGCCCTATTTCTA) primers. Segment 3 (2942–3301 nt) was amplified using primer M184I_F forward (5′ CATACCTAGTATAAACAATGAGACAC) and M184I_R reverse (5′ CTGTCTTTTTCTGGCAGCACTAT) primers. For use in the Illumina MiSeq we performed another PCR with the segments cognate MiSeq primer to add the necessary overhang adapter sequences (D67N_F_MS: 5′ TCGTCGGCAGCG TCAGATGTGTATAAGAGACAGCCAGTAAAATTAAAGCCAGGAATG; D67N_R_MS: 5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGAGTATACTTCCTGAAGTCTTCATCT; E138K_F_MS: 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAATAAGAGAACTC AAGACTTCTGG; E138K_R_MS: 5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAG ACAGTGTTCTATGCTGCCCTATTTCTA; M184I_F_MS: 5′ TCGTCGGCAGAGTCAGA TGTGTATAAGAGACAGCATACCTAGTATAAACAATGAGACAC; M184I_R_MS; 5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCTGTCTTTTTCTGGCAGCACTAT). The amplicons were then sequenced following the 16S Metagenomic Sequencing Library Preparation manual (Illumina) using the reagents described. Prepared samples were sequenced using MiSeq Nano V2 chemistry in a 250 × 250 paired-end reaction.
4.5. Deep sequencing analysis
The raw outputs from the Illumina MiSeq platform were demultiplexed using the manufacturer software and the resulting FASTQ files were processed through a customized version of the MiCall pipeline (http://github.com/cfe-lab/MiCall) [119]. In brief, the pipeline first identifies and censors problematic tile-cycle combinations (with run-specific empirical ϕX174 error rates above 7.5%) in the FASTQ data. Next, it performs a preliminary map of reads from a given sample to a curated set of reference sequences to determine the closest reference using bowtie2 (version 2.2.8) with local alignment, and subsequently uses all mapped reads to iteratively adapt the reference to the sample to maximize the mapped number of reads [120]. Finally, MiCall merges paired-end reads (using base quality scores to resolve discordant base calls in overlapping regions) and outputs the aligned nucleotide and amino acid frequencies relative to a user-specified coordinate reference. For this study, the MiCall pipeline was extended to extract both single- and double-nucleotide frequencies from each sample alignment. The pipeline outputs were next analyzed with Microsoft Excel functions to determine the level of deamination in an A3 relevant dinucleotide context. Overall deamination frequencies were derived by adding all the in-context deaminations for a sample across all three segments and dividing the sum by the total number of nucleotides sequenced (across all three segments). The FASTQ files will be made available upon request.
4.6. Production of viral stocks
HIV-1 for spreading infections was produced by transfection of HIV-1LAI plasmid into 1 × 106 293T cells cultured in DMEM with 10% FBS in a T75 flask. Virus particles were produced without or with A3 enzymes. For conditions with A3 enzymes the pVIVO2 plasmid (Invivogen) that has two MCS in the same vector was used. Untagged A3F (Accession BC038808) was cloned into MCS-I using XbaI and A3G (Accession NM_021822) was cloned into MCS-II using EcoRI. The transfection used 22.8 μg of HIV-1LAI plasmid and 1.2 μg of empty pVIVO2 plasmid or 1.2 μg of pVIVO2 plasmid with A3F/A3G. This resulted in a plasmid molar ratio of 12:1 for HIV-1:A3, to avoid overexpression artefacts [121]. Transfections used FuGene 6 transfection reagent according to manufacturer's protocol. Twenty hours after the transfection, the media was changed and 48 h after the transfection virus was harvested, cleared by centrifugation at 500 × g for 10 min, and then frozen at −80 °C. Virus was titered using an RT assay and the Reed and Muench method to determine the TCID50.
4.7. RT assay
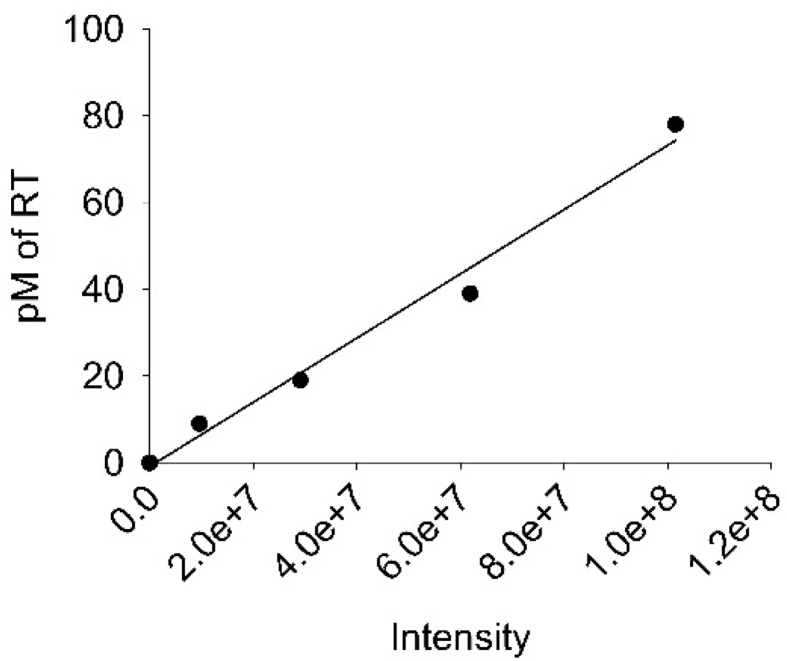
To titer and measure the viability of the virus an RT assay was conducted. The assay used 10 μL of supernatant in duplicate. RT buffer (25 mM Tris pH 7.8, 37.5 mM KCl, 2.5 mM MgCl2, 0.25% v/v NP-40, 0.025 U Poly rA p(dT) 12–18, and 1 mM DTT) was used to lyse virions and provide the DNA template. The reaction was incubated for 20 min at room temperature for lysis. Then, α-32P-TTP (800 Ci/mmol), diluted in RT buffer, was added to each sample and incubated for 2 h at 37 °C. Afterwards, the mixture was spotted onto a Whatman DE81 paper (Perkin Elmer) and dried for 10 minutes. This was followed by washing in saline-sodium citrate (SSC) buffer and 85% ethanol. The paper was then dried for 10 minutes at 65 °C and exposed overnight to a phosphorimaging screen. The screen was scanned on a Typhoon Trio Imager (GE Healthcare) and the file was analyzed with Image Quant Software (GE Healthcare). Purified RT of known concentration was used in the assay to construct a standard curve, which was used to determine the RT concentration in the culture supernatants. The RT (p66/p51) was purified as previously described [66]. The standard curve is shown in Fig. 6.
Fig. 6.
Standard curve used to determine RT levels for RT assays. Purified RT (p66/p51) was used in the assay at 10 pM, 20 pM, 40 pM, and 80 pM in order correlate 32P intensity measured by phosphorimaging with an RT concentration.
4.8. Spreading infection in PBMCs
Healthy donor PMBCs were isolated from whole blood samples using Ficoll-Paque (GE-Healthcare) density gradient centrifugation and SepMate-50 tubes (STEMCELL Technologies) as per manufacturer's protocol. Cells were cultured in RPMI 1640 media (Hyclone) with 10% heat inactivated FBS (Gibco), and 1% PenStrep (Hyclone). Cells were stimulated for 3 days at 37 °C with 10 ng/mL recombinant human IL-2 (Sigma) and 10 μg/mL Phytohemagglutinin-L (PHA-L) (Roche). Cells were then harvested and 8 × 105 cells/mL were dispensed to tubes and infected with 0.05 MOI of HIV-1LAI. Cells were gently vortexed and incubated with the virus at 37 °C for 16 h. Next, the cells were pelleted at 300 × g for 2 minutes at room temperature. The supernatant was removed and the cells were washed twice with PBS. Cell pellets were resuspended in complete RPMI with IL-2 alone or containing a titration of either: AZT (6.25–50 μM), 3TC (9.38–150 μM) or a 1:2 to 1:32 dilution of the Mixed ARVs: TDF, 3TC, and EFV with initial concentration of 100 μM, 300 μM, and 1 μM, respectively. The cells were then dispensed into a well of a 24-well plate. This constituted day 0. Three days later 180 μL of supernatant was harvested and saved at −80 °C and 180 μL was replaced to the well with its respective drug-containing media. Samples were taken every three days for the duration of twenty days. On day 21, the supernatant was harvested and applied to freshly stimulated PMBCs, prepared as described. This cycle continued until day 30, when the final supernatant was removed and the cellular genomic DNA was harvested by DNAzol (Invitrogen) treatment, following manufacturer protocol, for provirus sequencing. The frozen virus aliquots were subsequently used for RT assay and viral RNA extraction for cDNA synthesis.
4.9. Spreading infection in U87 CD4+ CXCR4+ cells
The U87 CD4+ CXCR4+ cells were cultured in DMEM (Hyclone) with 10% FBS (Gibco) [97]. For spreading infections, 1 × 105 cells in 1 mL were plated in each well of a 12 -well plate and infected with 0.01 MOI of HIV-1LAI with or without encapsidated A3F/A3G. Then, 16 h post infection, the media was replaced with 1.5 mL of DMEM with 10% FBS and 1% PenStrep (Hyclone) alone or containing a dilution of: AZT (6.25–50 μM), 3TC (9.38–150 μM) or 1:2 to 1:32 dilution of the Mixed ARVs: TDF, 3TC, and EFV with initial concentration of 100 μM, 300 μM, and 1 μM, respectively. Three days later 180 μL of supernatant was harvested and saved at −80 °C and 180 μL of appropriate media was replaced to the well. Samples were taken every three days for the duration of thirty days. The cellular genomic DNA was harvested by DNAzol (Invitrogen) treatment, following manufacturer protocol, for provirus sequencing. The frozen virus aliquots were subsequently used for RT assay and viral RNA extraction for cDNA synthesis.
4.10. Spreading infection in CEM/CEM-SS
The CEM and CEM-SS cells were cultured in RPMI 1640 media (Hyclone) plus 10% heat inactivated FBS (Gibco) medium containing 1% PenStrep (Hyclone) [122]. For stimulation, 10 ng/mL human IL-2 (Sigma) and 10 μg/mL PHA-L (Roche) was incubated with cells for 3 days. Cells were then harvested and 8 × 105 cells/mL of cells were dispensed to tubes and infected with 0.25 MOI of HIV-1LAI. The spreading infection then progressed as described for PBMCs. Samples were taken every three days for the duration of thirty days. The final supernatant was removed and the cellular genomic DNA was harvested by DNAzol (Invitrogen) treatment, following manufacturer protocol, for provirus sequencing. The frozen virus aliquots were subsequently used for RT assay and viral RNA extraction for cDNA synthesis.
4.11. Genotyping PBMCs and CEM cells
The presence of the A3F single nucleotide polymorphism (SNP) that results in the S108A variant was determined using the following primers: (forward) 5′ GTGATATCCCAGCCTGAGCA and (reverse) 5′ CTTCATCGTCCATAATCTTCACG. The presence of the A3F SNP that results in the I231V variant was determined using the following primers (forward) 5′ AGCCTATGGTCGGAACGAAA and (reverse) 5′ CTGGTTTCGGAAGACGCC. The PCR amplification used Q5 High Fidelity polymerase (New England Biolabs) and cycling conditions were 98 °C for 30 sec followed by 30 cycles of 98 °C for 10 sec, 64–67 °C for 20 sec, 72 °C for 20 sec and 2 minutes at 72 °C. Amplicons were cloned using the CloneJET PCR cloning kit (ThermoScientific) as outlined in the manufacturer's protocol. Analysis for A3F S108A and A3F I231V polymorphisms included 6–12 clones for each position from PBMCs and CEM cells to determine zygosity.
4.12. Ethics statement
This study has been reviewed and approved by the Research and Ethics Board at the University of Saskatchewan (Bio# 14-62). PBMCs were specifically isolated for this study from de-identified HIV- individuals after written consent was obtained. In accordance with OCAP® principles, no samples of Indigenous peoples were included.
Declarations
Author contribution statement
Nazanin Mohammadzadeh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Richard Gibson, Eric J. Arts: Conceived and designed the experiments.
Linda Chelico: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Robin P. Love: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Art F. Y. Poon: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
Linda Chelico was supported by the Canadian Institutes of Health Research grant (MOP137090). Art F. Y. Poon was supported by the Canadian Institutes of Health Research grant (PJT-155990).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We acknowledge Dr. Janet Hill and Champika Fernando (Western College of Veterinary Medicine, University of Saskatchewan) for assistance with Illumina sequencing. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Anti-HIV-1 p24 Monoclonal (183-H12-5C) (Cat# 3537) from Dr. Bruce Chesebro and Kathy Wehrly, HIV-1 Vif Monoclonal Antibody (#319) from Dr. Michael H. Malim, U87.CD4.CXCR4 from Dr. HongKui Deng and Dr. Dan R. Littman, CEM-SS Cells from Dr. Peter L. Nara, and CEM CD4+ Cells from Dr. J.P. Jacobs (cat# 117).
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Harris R.S., Hultquist J.F., Evans D.T. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon V., Bloch N., Landau N.R. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compton A.A., Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003135. PPATHOGENS-D-12-02303 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Compton A.A., Hirsch V.M., Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe. 2012;11:91–98. doi: 10.1016/j.chom.2011.11.010. S1931-3128(11)00402-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compton A.A., Malik H.S., Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120496. doi: 10.1098/rstb.2012.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P.S., Young J.M., Emerman M., Malik H.S. Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fregoso O.I., Ahn J., Wang C., Mehrens J., Skowronski J., Emerman M. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etienne L., Hahn B.H., Sharp P.M., Matsen F.A., Emerman M. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe. 2013;14:85–92. doi: 10.1016/j.chom.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris R.S., Anderson B.D. Evolutionary paradigms from ancient and ongoing conflicts between the lentiviral vif protein and mammalian APOBEC3 enzymes. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakano Y., Aso H., Soper A., Yamada E., Moriwaki M., Juarez-Fernandez G., Koyanagi Y., Sato K. A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology. 2017;14:31. doi: 10.1186/s12977-017-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauter D., Kirchhoff F. Multilayered and versatile inhibition of cellular antiviral factors by HIV and SIV accessory proteins. Cytokine Growth Factor Rev. 2018;40:3–12. doi: 10.1016/j.cytogfr.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Alteri C., Surdo M., Bellocchi M.C., Saccomandi P., Continenza F., Armenia D., Parrotta L., Carioti L., Costa G., Fourati S., Di Santo F., Scutari R., Barbaliscia S., Fedele V., Carta S., Balestra E., Alcaro S., Marcelin A.G., Calvez V., Ceccherini-Silberstein F., Artese A., Perno C.F., Svicher V. Incomplete APOBEC3G/F neutralization by HIV-1 Vif mutants facilitates the genetic evolution from CCR5 to CXCR4 usage. Antimicrob. Agents Chemother. 2015;59:4870–4881. doi: 10.1128/AAC.00137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duggal N.K., Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jern P., Russell R.A., Pathak V.K., Coffin J.M. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casartelli N., Guivel-Benhassine F., Bouziat R., Brandler S., Schwartz O., Moris A. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. J. Exp. Med. 2010;207:39–49. doi: 10.1084/jem.20091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant M., Larijani M. Evasion of adaptive immunity by HIV through the action of host APOBEC3G/F enzymes. AIDS Res. Ther. 2017;14:44. doi: 10.1186/s12981-017-0173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adolph M.B., Love R.P., Chelico L. Biochemical basis of APOBEC3 deoxycytidine deaminase activity on diverse DNA substrates. ACS Infect. Dis. 2018;4:224–238. doi: 10.1021/acsinfecdis.7b00221. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y., Baig T.T., Love R.P., Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front. Microbiol. 2014;5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desimmie B.A., Delviks-Frankenberrry K.A., Burdick R.C., Qi D., Izumi T., Pathak V.K. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager S., Kim D.Y., Hultquist J.F., Shindo K., LaRue R.S., Kwon E., Li M., Anderson B.D., Yen L., Stanley D., Mahon C., Kane J., Franks-Skiba K., Cimermancic P., Burlingame A., Sali A., Craik C.S., Harris R.S., Gross J.D., Krogan N.J. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., Evans S.L., Han X., Liu Y., Yu X.F. Characterization of the interaction of full-length HIV-1 Vif protein with its key regulator CBFbeta and CRL5 E3 ubiquitin ligase components. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W., Du J., Evans S.L., Yu Y., Yu X.F. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 23.Stanley B.J., Ehrlich E.S., Short L., Yu Y., Xiao Z., Yu X.F., Xiong Y. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 2008;82:8656–8663. doi: 10.1128/JVI.00767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y., Xiao Z., Ehrlich E.S., Yu X., Yu X.F. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergeron J.R., Huthoff H., Veselkov D.A., Beavil R.L., Simpson P.J., Matthews S.J., Malim M.H., Sanderson M.R. The SOCS-box of HIV-1 Vif interacts with Elongin BC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y., Dong L., Qiu X., Wang Y., Zhang B., Liu H., Yu Y., Zang Y., Yang M., Huang Z. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 27.Iwatani Y., Chan D.S., Liu L., Yoshii H., Shibata J., Yamamoto N., Levin J.G., Gronenborn A.M., Sugiura W. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19539–19544. doi: 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albin J.S., Anderson J.S., Johnson J.R., Harjes E., Matsuo H., Krogan N.J., Harris R.S. Dispersed sites of HIV Vif-dependent polyubiquitination in the DNA deaminase APOBEC3F. J. Mol. Biol. 2013;425:1172–1182. doi: 10.1016/j.jmb.2013.01.010. S0022-2836(13)00013-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York A., Kutluay S.B., Errando M., Bieniasz P.D. The RNA binding specificity of human APOBEC3 proteins resembles that of HIV-1 nucleocapsid. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apolonia L., Schulz R., Curk T., Rocha P., Swanson C.M., Schaller T., Ule J., Malim M.H. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., Suzuki A., Sugiura W., Iwatani Y. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 33.Nakashima M., Ode H., Kawamura T., Kitamura S., Naganawa Y., Awazu H., Tsuzuki S., Matsuoka K., Nemoto M., Hachiya A., Sugiura W., Yokomaku Y., Watanabe N., Iwatani Y. Structural insights into HIV-1 vif-APOBEC3F interaction. J. Virol. 2016;90:1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima M., Tsuzuki S., Awazu H., Hamano A., Okada A., Ode H., Maejima M., Hachiya A., Yokomaku Y., Watanabe N., Akari H., Iwatani Y. Mapping region of human restriction factor APOBEC3H critical for interaction with HIV-1 vif. J. Mol. Biol. 2017;429:1262–1276. doi: 10.1016/j.jmb.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Ooms M., Letko M., Simon V. The structural interface between HIV-1 vif and human APOBEC3H. J. Virol. 2017;91 doi: 10.1128/JVI.02289-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letko M., Booiman T., Kootstra N., Simon V., Ooms M. Identification of the HIV-1 vif and human APOBEC3G protein interface. Cell Rep. 2015;13:1789–1799. doi: 10.1016/j.celrep.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards C., Albin J.S., Demir O., Shaban N.M., Luengas E.M., Land A.M., Anderson B.D., Holten J.R., Anderson J.S., Harki D.A., Amaro R.E., Harris R.S. The binding interface between human APOBEC3F and HIV-1 vif elucidated by genetic and computational approaches. Cell Rep. 2015;13:1781–1788. doi: 10.1016/j.celrep.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binning J.M., Smith A.M., Hultquist J.F., Craik C.S., Caretta Cartozo N., Campbell M.G., Burton L., La Greca F., McGregor M.J., Ta H.M., Bartholomeeusen K., Peterlin B.M., Krogan N.J., Sevillano N., Cheng Y., Gross J.D. Fab-based inhibitors reveal ubiquitin independent functions for HIV Vif neutralization of APOBEC3 restriction factors. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opi S., Kao S., Goila-Gaur R., Khan M.A., Miyagi E., Takeuchi H., Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Britan-Rosich E., Nowarski R., Kotler M. Multifaceted counter-APOBEC3G mechanisms employed by HIV-1 Vif. J. Mol. Biol. 2011;410:1065–1076. doi: 10.1016/j.jmb.2011.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Y., Love R.P., Chelico L. HIV-1 viral infectivity factor (Vif) alters processive single-stranded DNA scanning of the retroviral restriction factor APOBEC3G. J. Biol. Chem. 2013;288:6083–6094. doi: 10.1074/jbc.M112.421875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruner K.M., Murray A.J., Pollack R.A., Soliman M.G., Laskey S.B., Capoferri A.A., Lai J., Strain M.C., Lada S.M., Hoh R., Ho Y.C., Richman D.D., Deeks S.G., Siliciano J.D., Siliciano R.F. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016;22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albin J.S., Harris R.S. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon V., Zennou V., Murray D., Huang Y., Ho D.D., Bieniasz P.D. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fourati S., Malet I., Binka M., Boukobza S., Wirden M., Sayon S., Simon A., Katlama C., Simon V., Calvez V., Marcelin A.G. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. Aids. 2010;24:2313–2321. doi: 10.1097/QAD.0b013e32833e515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris R.S., Dudley J.P. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suspene R., Rusniok C., Vartanian J.P., Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu C., Saenz D.T., Fadel H.J., Walker W., Peretz M., Poeschla E.M. The HIV-1 central polypurine tract functions as a second line of defense against APOBEC3G/F. J. Virol. 2010;84:11981–11993. doi: 10.1128/JVI.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wurtzer S., Goubard A., Mammano F., Saragosti S., Lecossier D., Hance A.J., Clavel F. Functional central polypurine tract provides downstream protection of the human immunodeficiency virus type 1 genome from editing by APOBEC3G and APOBEC3B. J. Virol. 2006;80:3679–3683. doi: 10.1128/JVI.80.7.3679-3683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ara A., Love R.P., Chelico L. Different mutagenic potential of HIV-1 restriction factors APOBEC3G and APOBEC3F is determined by distinct single-stranded DNA scanning mechanisms. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng Y., Love R.P., Ara A., Baig T.T., Adolph M.B., Chelico L. Natural polymorphisms and oligomerization of human APOBEC3H contribute to single-stranded DNA scanning ability. J. Biol. Chem. 2015;290:27188–27203. doi: 10.1074/jbc.M115.666065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adolph M.B., Ara A., Feng Y., Wittkopp C.J., Emerman M., Fraser J.S., Chelico L. Cytidine deaminase efficiency of the lentiviral viral restriction factor APOBEC3C correlates with dimerization. Nucleic Acids Res. 2017;45:3378–3394. doi: 10.1093/nar/gkx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hultquist J.F., Lengyel J.A., Refsland E.W., LaRue R.S., Lackey L., Brown W.L., Harris R.S. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An P., Bleiber G., Duggal P., Nelson G., May M., Mangeat B., Alobwede I., Trono D., Vlahov D., Donfield S., Goedert J.J., Phair J., Buchbinder S., O'Brien S.J., Telenti A., Winkler C.A. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An P., Penugonda S., Thorball C.W., Bartha I., Goedert J.J., Donfield S., Buchbinder S., Binns-Roemer E., Kirk G.D., Zhang W., Fellay J., Yu X.F., Winkler C.A. Role of APOBEC3F gene variation in HIV-1 disease progression and pneumocystis pneumonia. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harari A., Ooms M., Mulder L.C., Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ooms M., Brayton B., Letko M., Maio S.M., Pilcher C.D., Hecht F.M., Barbour J.D., Simon V. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 58.Reddy K., Ooms M., Letko M., Garrett N., Simon V., Ndung'u T. Functional characterization of Vif proteins from HIV-1 infected patients with different APOBEC3G haplotypes. AIDS. 2016;30:1723–1729. doi: 10.1097/QAD.0000000000001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.OhAinle M., Kerns J.A., Li M.M., Malik H.S., Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duggal N.K., Fu W., Akey J.M., Emerman M. Identification and antiviral activity of common polymorphisms in the APOBEC3 locus in human populations. Virology. 2013;443:329–337. doi: 10.1016/j.virol.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X., Abudu A., Son S., Dang Y., Venta P.J., Zheng Y.H. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittkopp C.J., Adolph M.B., Wu L.I., Chelico L., Emerman M. A single nucleotide polymorphism in human APOBEC3C enhances restriction of lentiviruses. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 64.Belanger K., Langlois M.A. Comparative analysis of the gene-inactivating potential of retroviral restriction factors APOBEC3F and APOBEC3G. J. Gen. Virol. 2015;96:2878–2887. doi: 10.1099/vir.0.000214. [DOI] [PubMed] [Google Scholar]
- 65.Desimmie B.A., Burdick R.C., Izumi T., Doi H., Shao W., Alvord W.G., Sato K., Koyanagi Y., Jones S., Wilson E., Hill S., Maldarelli F., Hu W.S., Pathak V.K. APOBEC3 proteins can copackage and comutate HIV-1 genomes. Nucleic Acids Res. 2016;44:7848–7865. doi: 10.1093/nar/gkw653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ara A., Love R.P., Follack T.B., Ahmed K.A., Adolph M.B., Chelico L. Mechanism of enhanced HIV restriction by virion coencapsidated cytidine deaminases APOBEC3F and APOBEC3G. J. Virol. 2017:91. doi: 10.1128/JVI.02230-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delviks-Frankenberry K.A., Nikolaitchik O.A., Burdick R.C., Gorelick R.J., Keele B.F., Hu W.S., Pathak V.K. Minimal contribution of APOBEC3-induced G-to-A hypermutation to HIV-1 recombination and genetic variation. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mulder L.C., Harari A., Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pollack R.A., Jones R.B., Pertea M., Bruner K.M., Martin A.R., Thomas A.S., Capoferri A.A., Beg S.A., Huang S.H., Karandish S., Hao H., Halper-Stromberg E., Yong P.C., Kovacs C., Benko E., Siliciano R.F., Ho Y.C. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. 2017;21:494–506.e4. doi: 10.1016/j.chom.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim E.Y., Lorenzo-Redondo R., Little S.J., Chung Y.S., Phalora P.K., Maljkovic Berry I., Archer J., Penugonda S., Fischer W., Richman D.D., Bhattacharya T., Malim M.H., Wolinsky S.M. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim E.Y., Bhattacharya T., Kunstman K., Swantek P., Koning F.A., Malim M.H., Wolinsky S.M. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J. Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zennou V., Bieniasz P.D. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 73.Chaipan C., Smith J.L., Hu W.S., Pathak V.K. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J. Virol. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato K., Takeuchi J.S., Misawa N., Izumi T., Kobayashi T., Kimura Y., Iwami S., Takaori-Kondo A., Hu W.S., Aihara K., Ito M., An D.S., Pathak V.K., Koyanagi Y. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monajemi M., Woodworth C.F., Benkaroun J., Grant M., Larijani M. Emerging complexities of APOBEC3G action on immunity and viral fitness during HIV infection and treatment. Retrovirology. 2012;9:35. doi: 10.1186/1742-4690-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monajemi M., Woodworth C.F., Zipperlen K., Gallant M., Grant M.D., Larijani M. Positioning of APOBEC3G/F mutational hotspots in the human immunodeficiency virus genome favors reduced recognition by CD8+ T cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Squires K.D., Monajemi M., Woodworth C.F., Grant M.D., Larijani M. Impact of APOBEC mutations on CD8+ T cell recognition of HIV epitopes varies depending on the restricting HLA. J. Acquir. Immune Defic. Syndr. 2015;70:172–178. doi: 10.1097/QAI.0000000000000689. [DOI] [PubMed] [Google Scholar]
- 78.Fourati S., Malet I., Lambert S., Soulie C., Wirden M., Flandre P., Fofana D.B., Sayon S., Simon A., Katlama C., Calvez V., Marcelin A.G. E138K and M184I mutations in HIV-1 reverse transcriptase coemerge as a result of APOBEC3 editing in the absence of drug exposure. AIDS. 2012;26:1619–1624. doi: 10.1097/QAD.0b013e3283560703. [DOI] [PubMed] [Google Scholar]
- 79.Fourati S., Lambert-Niclot S., Soulie C., Malet I., Valantin M.A., Descours B., Ait-Arkoub Z., Mory B., Carcelain G., Katlama C., Calvez V., Marcelin A.G. HIV-1 genome is often defective in PBMCs and rectal tissues after long-term HAART as a result of APOBEC3 editing and correlates with the size of reservoirs. J. Antimicrob. Chemother. 2012;67:2323–2326. doi: 10.1093/jac/dks219. [DOI] [PubMed] [Google Scholar]
- 80.Fourati S., Lambert-Niclot S., Soulie C., Wirden M., Malet I., Valantin M.A., Tubiana R., Simon A., Katlama C., Carcelain G., Calvez V., Marcelin A.G. Differential impact of APOBEC3-driven mutagenesis on HIV evolution in diverse anatomical compartments. Aids. 2014;28:487–491. doi: 10.1097/QAD.0000000000000182. [DOI] [PubMed] [Google Scholar]
- 81.Knoepfel S.A., Salisch N.C., Huelsmann P.M., Rauch P., Walter H., Metzner K.J. Comparison of G-to-A mutation frequencies induced by APOBEC3 proteins in H9 cells and peripheral blood mononuclear cells in the context of impaired processivities of drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants. J. Virol. 2008;82:6536–6545. doi: 10.1128/JVI.00554-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCallum M., Oliveira M., Ibanescu R.I., Kramer V.G., Moisi D., Asahchop E.L., Brenner B.G., Harrigan P.R., Xu H., Wainberg M.A. Basis for early and preferential selection of the E138K mutation in HIV-1 reverse transcriptase. Antimicrob. Agents Chemother. 2013;57:4681–4688. doi: 10.1128/AAC.01029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noguera-Julian M., Cozzi-Lepri A., Di Giallonardo F., Schuurman R., Daumer M., Aitken S., Ceccherini-Silberstein F., D'Arminio Monforte A., Geretti A.M., Booth C.L., Kaiser R., Michalik C., Jansen K., Masquelier B., Bellecave P., Kouyos R.D., Castro E., Furrer H., Schultze A., Gunthard H.F., Brun-Vezinet F., Metzner K.J., Paredes R. Contribution of APOBEC3G/F activity to the development of low-abundance drug-resistant human immunodeficiency virus type 1 variants. Clin. Microbiol. Infect. 2016;22:191–200. doi: 10.1016/j.cmi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 84.Russell R.A., Moore M.D., Hu W.S., Pathak V.K. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ji J.P., Loeb L.A. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 86.Abram M.E., Ferris A.L., Shao W., Alvord W.G., Hughes S.H. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 2010;84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mansky L.M., Temin H.M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Refsland E.W., Stenglein M.D., Shindo K., Albin J.S., Brown W.L., Harris R.S. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koning F.A., Newman E.N., Kim E.Y., Kunstman K.J., Wolinsky S.M., Malim M.H. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009;83:9474–9478. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mohammadzadeh N., Follack T.B., Love R.P., Stewart K., Sanche S., Chelico L. Polymorphisms of the cytidine deaminase APOBEC3F have different HIV-1 restriction efficiencies. Virology. 2018;527:21–31. doi: 10.1016/j.virol.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 91.Holtz C.M., Sadler H.A., Mansky L.M. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013;41:6139–6148. doi: 10.1093/nar/gkt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rausch J.W., Chelico L., Goodman M.F., Le Grice S.F. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J. Biol. Chem. 2009;284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pollpeter D., Parsons M., Sobala A.E., Coxhead S., Lang R.D., Bruns A.M., Papaioannou S., McDonnell J.M., Apolonia L., Chowdhury J.A., Horvath C.M., Malim M.H. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat. Microbiol. 2018;3:220–233. doi: 10.1038/s41564-017-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iwatani Y., Chan D.S., Wang F., Maynard K.S., Sugiura W., Gronenborn A.M., Rouzina I., Williams M.C., Musier-Forsyth K., Levin J.G. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7098. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mulder L.C., Ooms M., Majdak S., Smedresman J., Linscheid C., Harari A., Kunz A., Simon V. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. J. Virol. 2010;84:9613–9617. doi: 10.1128/JVI.02630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dunn L.L., Boyer P.L., McWilliams M.J., Smith S.J., Hughes S.H. Mutations in human immunodeficiency virus type 1 reverse transcriptase that make it sensitive to degradation by the viral protease in virions are selected against in patients. Virology. 2015;484:127–135. doi: 10.1016/j.virol.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bjorndal A., Deng H., Jansson M., Fiore J.R., Colognesi C., Karlsson A., Albert J., Scarlatti G., Littman D.R., Fenyo E.M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miyagi E., Brown C.R., Opi S., Khan M., Goila-Gaur R., Kao S., Walker R.C., Jr., Hirsch V., Strebel K. Stably expressed APOBEC3F has negligible antiviral activity. J. Virol. 2010;84:11067–11075. doi: 10.1128/JVI.01249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng Y., Chelico L. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J. Biol. Chem. 2011;286:11415–11426. doi: 10.1074/jbc.M110.199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Browne E.P., Allers C., Landau N.R. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. S0042-6822(09)00133-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krisko J.F., Martinez-Torres F., Foster J.L., Garcia J.V. HIV restriction by APOBEC3 in humanized mice. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liddament M.T., Brown W.L., Schumacher A.J., Harris R.S. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 103.Sato K., Izumi T., Misawa N., Kobayashi T., Yamashita Y., Ohmichi M., Ito M., Takaori-Kondo A., Koyanagi Y. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J. Virol. 2010;84:9546–9556. doi: 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Refsland E.W., Hultquist J.F., Harris R.S. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao L., Hanson M.N., Balakrishnan M., Boyer P.L., Roques B.P., Hughes S.H., Kim B., Bambara R.A. Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. J. Biol. Chem. 2008;283:9196–9205. doi: 10.1074/jbc.M710148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu H.T., Asahchop E.L., Oliveira M., Quashie P.K., Quan Y., Brenner B.G., Wainberg M.A. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J. Virol. 2011;85:11300–11308. doi: 10.1128/JVI.05584-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sharma P.L., Crumpacker C.S. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 1999;73:8448–8456. doi: 10.1128/jvi.73.10.8448-8456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao H.Q., Boyer P.L., Arnold E., Hughes S.H. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 109.Back N.K., Nijhuis M., Keulen W., Boucher C.A., Oude Essink B.O., van Kuilenburg A.B., van Gennip A.H., Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 110.Boyer P.L., Hughes S.H. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 1995;39:1624–1628. doi: 10.1128/aac.39.7.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lobritz M.A., Lassen K.G., Arts E.J. HIV-1 replicative fitness in elite controllers. Curr. Opin. HIV AIDS. 2011;6:214–220. doi: 10.1097/COH.0b013e3283454cf5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Henry K.R., Weber J., Quinones-Mateu M.E., Arts E.J. The impact of viral and host elements on HIV fitness and disease progression. Curr. HIV AIDS Rep. 2007;4:36–41. doi: 10.1007/s11904-007-0006-9. [DOI] [PubMed] [Google Scholar]
- 113.Toohey K., Wehrly K., Nishio J., Perryman S., Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 114.Wehrly K., Chesebro B. p24 Antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods. 1997;12:288–293. doi: 10.1006/meth.1997.0481. [DOI] [PubMed] [Google Scholar]
- 115.Baig T.T., Feng Y., Chelico L. Determinants of efficient degradation of APOBEC3 restriction factors by HIV-1 Vif. J. Virol. 2014;88:14380–14395. doi: 10.1128/JVI.02484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Simon J.H., Fouchier R.A., Southerling T.E., Guerra C.B., Grant C.K., Malim M.H. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 1997;71:5259–5267. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Simon J.H., Southerling T.E., Peterson J.C., Meyer B.E., Malim M.H. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 1995;69:4166–4172. doi: 10.1128/jvi.69.7.4166-4172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fouchier R.A., Simon J.H., Jaffe A.B., Malim M.H. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J. Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brumme C.J., Poon A.F.Y. Promises and pitfalls of Illumina sequencing for HIV resistance genotyping. Virus Res. 2017;239:97–105. doi: 10.1016/j.virusres.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 120.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soros V.B., Yonemoto W., Greene W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. 06-PLPA-RA-0322R2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Foley G.E., Lazarus H., Farber S., Uzman B.G., Boone B.A., McCarthy R.E. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer. 1965;18:522–529. doi: 10.1002/1097-0142(196504)18:4<522::aid-cncr2820180418>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 123.Rhee S.Y., Gonzales M.J., Kantor R., Betts B.J., Ravela J., Shafer R.W. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shafer R.W. Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 2006;194(Suppl. 1):S51–S58. doi: 10.1086/505356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.