Abstract
The hallmark function of αβ T cell antigen receptors (TCRs) involves the highly specific co-recognition of a major histocompatibility complex molecule and its carried peptide. However, the molecular basis of the interactions of TCRs with the lipid antigen–presenting molecule CD1c is unknown. We identified frequent staining of human T cells with CD1c tetramers across numerous subjects. Whereas TCRs typically show high specificity for antigen, both tetramer binding and autoreactivity occurred with CD1c in complex with numerous, chemically diverse self lipids. Such extreme polyspecificity was attributable to binding of the TCR over the closed surface of CD1c, with the TCR covering the portal where lipids normally protrude. The TCR essentially failed to contact lipids because they were fully seated within CD1c. These data demonstrate the sequestration of lipids within CD1c as a mechanism of autoreactivity and point to small lipid size as a determinant of autoreactive T cell responses.
The recognition of major histocompatibility (MHC)-peptide complexes by T cell antigen receptors (TCRs) is known as ‘co-recognition’ because the TCR makes simultaneous contact with the peptide and the MHC protein1. In humans, four types of CD1 proteins (CD1a, CD1b, CD1c and CD1d) function to display lipid antigens for recognition by T cells2–4. The structure of CD1 molecules is ideally suited for the capture of lipid antigens3. CD1 clefts derive from deep invaginations into the CD1 core structure and form two or four pockets5–9. In general, the pockets surround a large portion of the lipidic antigens so that their hydrocarbon moieties are sequestered from solvent and the hydrophilic headgroups protrude for T cell contact. However, each of the four types of human CD1 proteins has a cavity with unique architecture, which endows each CD1 isoform with the ability to present specific types of lipids. Whereas MHC proteins allow broad access to peptides that span the entire platform, CD1 proteins possess an A′-roof that blocks access of the TCR to the contents of the A′-pocket2 so that antigens are less exposed to solvent2.
Most evidence indicates that the recognition of CD1-lipid complexes by T cells follows the paradigm of MHC-peptide co-recognition1,2. Natural killer T cell receptors (NKT TCRs) show simultaneous contact with CD1d and protruding antigens10. Similarly, TCRs co-contact CD1b and the exposed polar moiety of glycolipid and phospholipid antigens11,12. However, each human CD1 isoform possesses a different platform structure, and the total number of solved TCR-lipid-CD1 structures remains limited. CD1a has been solved in complex with one autoreactive αβ TCR, which showed direct recognition of CD1a rather than of the lipid carried13. CD1c binds to αβ TCRs and γδ TCRs14,15, but any structural knowledge of TCR-CD1c contact is limited to mutational analyses16.
A role for self lipids in T cell autoreactivity is emerging17,18. For example, certain NKT TCRs show extremely high affinity for CD1d, which enables TCRs to bind CD1d carrying self-lipid phospholipids19–21. CD1a- and CD1c-autoreactive T cells can be detected at a high frequency in the blood of human subjects14,22. Moreover, CD1a-autoreactive T cells secrete interferon-γ (IFN-γ) and interleukin 22 (IL-22)23, both of which mediate autoimmune disease. CD1a mediates polyclonal responses to allergens24–26. CD1c can display cholesterol esters and tumor neo-antigens27,28.
CD1c appears on myeloid cells after exposure to bacterial products, the cytokine GM-CSF or IL-129,30. CD1c can be expressed on activated dendritic cells and marginal-zone B cells in lymph nodes or secondary follicles arising at the site of organ-specific autoimmune disease and in human leukemic cells30,31. However, the particular roles of T cells’ autoreactivity to CD1c remain undefined. We identified unexpectedly common CD1c tetramer staining on peripheral T cells in a large proportion of human subjects studied, which led to detailed studies of the formation of TCR-CD1c-lipid complexes through the use of tetramers, activation assays, lipid-elution assays and TCR-binding measurements32. On the basis of the determination of a TCR-CD1c-lipid ternary complex, we show how T cell–mediated autoreactivity to CD1c can operate outside the co-recognition paradigm and manifests as a polyspecific response to many types of CD1c-lipid complexes.
Results
CD1c tetramer staining of human T cells.
Using reported32 and newly designed expression systems, we produced CD1c monomers that were tetramerized with avidin linked to phycoerythrin (PE), allophycocyanin (APC) or Brilliant Violet 421 (BV421). Unexpectedly, we found that CD1c tetramers carrying endogenous lipids (CD1c-endo) consistently stained a large proportion of CD3+ cells from healthy donors (Fig. 1a). Conventional models of TCR specificity cannot explain how CD1c-endo tetramers could bind extensively to TCRs. Each arm of the tetramer would be expected to carry different ligands, so polyvalent binding to clonal TCRs would not be expected33. However, the staining phenomenon was robust. It was observed at moderate to high frequencies (0.06–3.0% of T cells) and was seen when CD1c tetramers were coupled to PE-, APC- or BV421-labeled streptavidin (Fig. 1a,b and Supplementary Fig. 1).
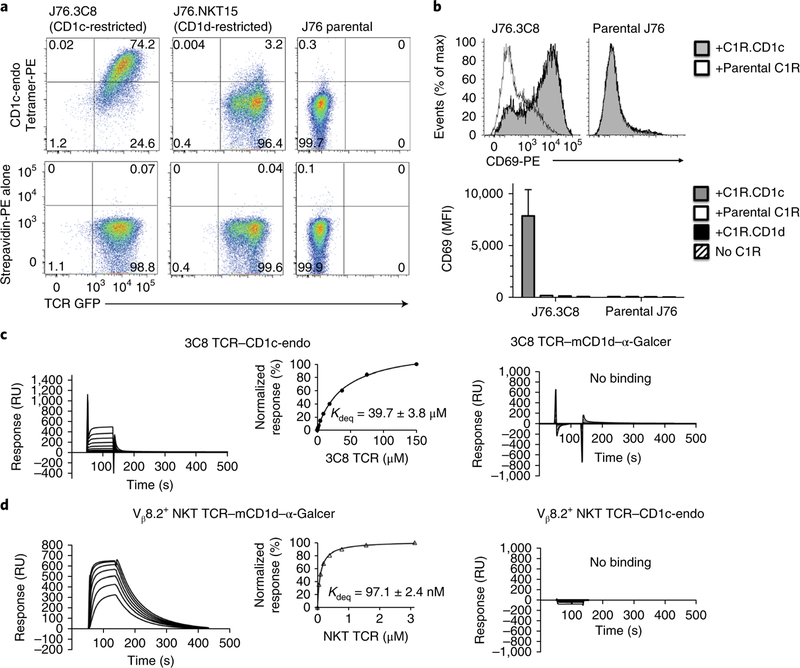
Fig. 1 |. CD1c tetramers stain human polyclonal T cells.
a, Flow cytometry of freshly isolated peripheral blood mononuclear cells (PBMCs) stained either with CD1c-endo tetramers conjugated separately to APC, PE or BV421 (top row and bottom left) or with a premixed combination of PE-conjugated and BV421-conjugated CD1c-endo tetramers (bottom right); cells were pre-gated as CD3+CD14–CD19–. Numbers adjacent to outlined areas indicate percent tetramer-positive cells. Data are representative of three independent experiments with similar results. b, Flow cytometry of PBMCs obtained from healthy donors (identified along left margin) and stimulated once ex vivo, then stained with CD1c-endo–APC or CD1b-endo–APC (above plots) produced in HEK293 human embryonic kidney cells; identical rectangular tetramer-positive gates were drawn for all subjects (numbers adjacent indicate percent tetramer-positive cells). Data are from three independent experiments, each with a single biological sample, representative of 28 experiments (one per subject). c, Frequency of cells positive for the CD1c-endo tetramer (CD1c-endo+) or CD1b-endo tetramer (CD1b-endo+) in the CD3+ population, assessed as in b, for all subjects (n = 28). Each symbol represents an individual subject; small horizontal lines indicate the median. The Wilcoxon signed rank test (two sided) with continuity correction and the Shapiro-Wilk test were used to establish non-normality. Data are from 28 experiments (one per subject).
Furthermore, CD1c-endo staining was seen consistently in blood from a larger cohort of healthy donors that lacked positive antigen-recall tests for tuberculosis. Although treatment of tuberculosis-naive donors with CD1c-endo tetramers was conceived as a negative control arm of the study, staining with CD1c-endo tetramer was higher in frequency and mean fluorescence intensity than was staining with CD1b-endo tetramer assembled with the same streptavidin-fluorophore (Fig. 1b). Overall, CD1c-endo tetramer staining was brighter and more frequent than was CD1b-endo tetramer staining for 26 of 28 subjects tested, and the mean fluorescence intensity for the whole group was about eightfold higher for CD1c-endo tetramer than for CD1b-endo tetramer (Fig. 1c).
Published studies have suggested high frequencies of CD1c-autoreactive T cells in activation assays14,34, but CD1c-autoreactive T cells have not been previously enumerated with tetramers ex vivo. One interpretation of our findings would be that the staining represented direct detection of a large autoreactive CD1c-restricted T cell pool. However, false-positive binding of tetramers to cells can occur on the basis of non-specific adherence or the presence of other CD1c-binding receptors, which we considered, since CD1c-endo tetramers detectably bound to CD3– cells, albeit at a lower frequency (Supplementary Fig. 2). Nevertheless, these unexpected results hinted at previously unknown mechanisms for the interaction of the TCR with CD1c.
Binding of the TCR to CD1c presenting endogenous self lipids.
We studied the 3C8 αβ TCR (TRAV29-TRBV7–2), as it represents the first report of a CD1c-autoreactive αβ TCR35. We transduced the 3C8 TCR into the Jurkat 76 (TCRα−TCRβ−) human T lymphocyte cell line to generate J76.3C8 cells. The CD1c-endo tetramers bound to J76.3C8 cells but not to untransfected J76 cells or J76 cells bearing an NKT TCR (Fig. 2a). Furthermore, J76.3C8 cells upregulated their expression of the activation marker CD69 specifically in the presence of CD1c+ antigen-presenting cells (Fig. 2b). In surface plasmon resonance experiments, purified 3C8 TCR bound CD1c-endo but did not bind CD1d–α-galactosylceramide (Fig. 2c,d). The 3C8 TCR bound to CD1c-endo with an equilibrium dissociation constant of 40 μM, which is a relatively high affinity for an autoreactive TCR1,36. While CD1c-endo was coupled to the chip to approximately 3,000 response units (RU), the maximal response was observed at about 400 response units (Fig. 2c). This suggested that 15–20% of the CD1c-endo proteins permitted the interaction with the 3C8 TCR, which raised questions about which lipids bound in CD1c-endo complexes might control binding of the TCR.
Fig. 2 |. Binding analysis of the 3C8 TCR by tetramer staining and surface plasmon resonance.
a, Flow cytometry analyzing the staining of J76.3C8 cells, Jurkat 76 cells bearing an NKT TCR (J76.NKT15) and Jurkat 76 cells (J76 parental) (above plots) with CD1c-endo tetramer (top row) or streptavidin-PE (bottom row). Numbers in quadrants indicate percent cells in each. GFP, green fluorescent protein. Data are representative of two independent experiments, each performed in technical duplicate, with similar results. b, Expression of CD69 on J76.3C8 cells and Jurkat 76 T cells in the presence of C1R.CD1c, C1R.CD1d or parental C1R antigen-presenting cells or no C1R cells (key). MFI, median fluorescent intensity. Data are representative of two independent experiments (top) or are from two independent experiments, each performed in technical duplicate (bottom; average and s.e.m.). c,d, Sensorgrams of concentration series for the 3C8 TCR (c) and the Vβ8.2+ NKT TCR (d) passed over CD1c-endo or mouse CD1d–α-galactosylceramide (mCD1d–α-GalCer) (above plots); inset (left), steady-state equilibrium versus concentration, with the equilibrium dissociation constant (Kdeq).RU, response units. Data are representative of two independent experiments, with each TCR in duplicate, with similar results.
Overview of the TCR-CD1c-lipid structure.
The binding of TCRs to antigen-presenting molecules usually requires a specific antigen; however, we determined the structure of 3C8 TCR–lipid–CD1c in the absence of a defined lipid ligand (Fig. 3 and Supplementary Tables 1 and 2). We took this unusual approach due to the lack of known antigens with corresponding clonal TCRs and because the feasibility of determining the TCR-CD1-lipid structure without added ligand was supported by the binding of untreated tetramer to T cells (Fig. 1) The 3C8 TCR docked at 66° across the long axis of the CD1c (Fig. 3a,b). The TCR bound the A′-roof of CD1c, which blocked contact of the TCR with the lipid(s) within the A′-pocket (Fig. 3a,b). The lipid(s) did not protrude through the F′-portal to the CD1c platform (Fig. 3c), which provided a straightforward structural explanation for staining of TCRs by CD1c tetramers carrying diverse lipids (Figs. 1 and 2).
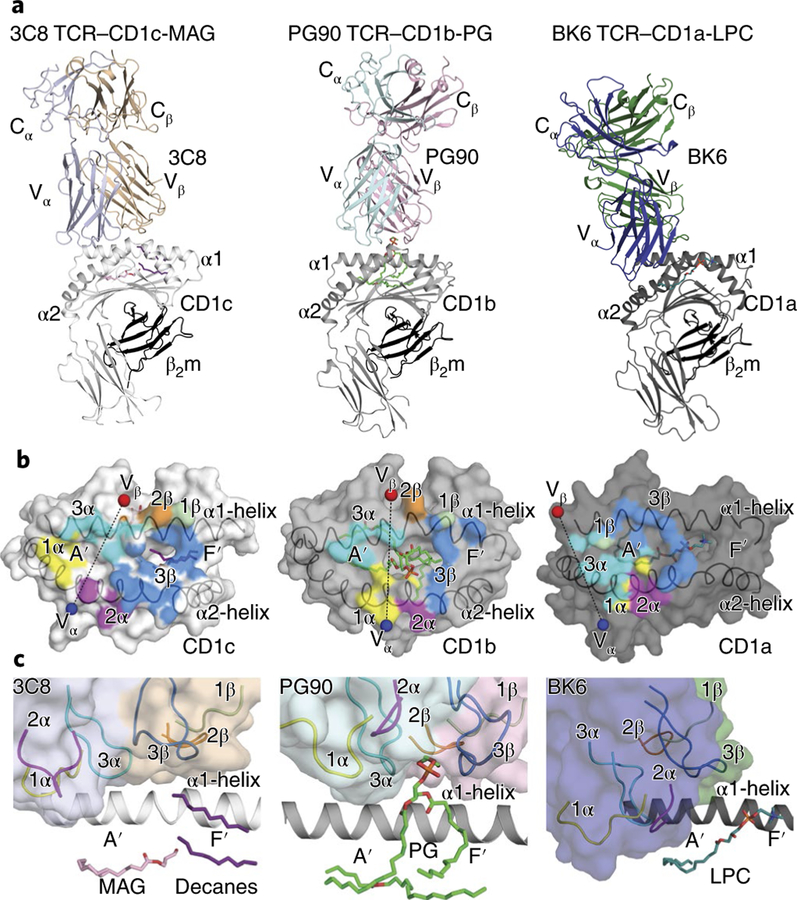
Fig. 3 |. Autoreactive T cell recognition of CD1a, CD1b and CD1c.
a, Ternary crystal structures of 3C8 TCR–CD1c-lipid, PG90 TCR–CD1b-PG and BK6 TCR–CD1a-LPC (above diagrams). b, Surface representation of the structure footprint of the autoreactive 3C8, PG90 and BK6 TCRs on CD1c, CD1b and CD1a, respectively12,13; colors of TCR docking footprints indicate the position of each CDR loop, and spheres indicate the center of mass of the Vα domain (blue) and Vβ domain (red) of the TCR. c, Enlarged view of the autoreactive TCR–CD1 binding interfaces as in a.
Three autoreactive TCR modes for the recognition of self lipid.
Docking of the TCR onto CD1c (Fig. 3) was in contrast to that of two other determined CD1-autoreactive TCRs, PG90 and BK6, which bind to CD1b-phosphatidylglycerol (CD1b-PG) and CD1a-lysophosphatidylcholine (CD1a-LPC), respectively12,13 (Fig. 3). For CD1b-PG, the PG90 TCR sat similarly to the positioning of its counterpart in the 3C8 TCR–CD1c-endo complex. However, a key difference was that PG protruded through the F′ -portal so that its phosphoglycerol headgroup contacted the TCR12 (Fig. 3c). For CD1c and CD1a, the 3C8 and BK6 TCRs essentially failed to contact the lipid ligand but did so via two distinct mechanisms. For CD1a-LPC, phosphocholine exited through the F′ -portal but sat in an ectopic location on the right side of the platform, whereas the BK6 TCR bound toward the left, on the A′ -roof of CD1a13 (Fig. 3a,c). For CD1c, the TCR bound more centrally, with the F′ -portal fully plugged. The lack of lipid contact resulted from the lack of protrusion of the lipid ligands from CD1c. Thus, the three ternary structures for autoantigens presented by CD1a, CD1b and CD1c documented three distinct modes of lipid recognition (Fig. 3).
TCR-CD1c platform interactions.
The total buried surface area (BSA) of the 3C8 TCR–CD1c interaction was ≈ 2,000Å2. Equivalent contributions were made by the 3C8 TCR α -chain and its β –chain (Fig. 3b and Supplementary Table 2). Here, the CDR3β loop from the TCR β -chain (37% BSA) made the principal interaction with CD1c, relative to the interaction provided by CDR1β (4% BSA) and CDR2β (10% BSA). The CDR1α loop (23% BSA) and CDR3α loop (15% BSA) made larger contributions than did CDR2α (11% BSA) (Fig. 4a–d)
Fig. 4 |. Interaction between the 3C8 TCR and CD1c.
a–d, Enlarged view of the interactions between residues of the 3C8 TCR CDR3α loop (a), CDR1α loop (b), CDR2α loop (c) and CDR3β loop (d) and the CD1c α-helices. Spheres indicate glycine side chains; red dotted lines indicate salt-bridge interactions; black dotted lines indicate hydrogen-bond interactions. e, Expression of CD69 by J76.3C8 cells after co-culture with C1R cells expressing wild-type CD1c (CD1c WT) or various CD1c alanine-substitution mutants (horizontal axis), or parental C1R cells, C1R.CD1d cells or no C1R cells (right end); results were normalized to those of C1R cells expressing wild-type CD1c, set as 100%, and dashed horizontal lines indicate a 25% or 50% reduction (as marked), relative to that expression (top). Below, the CD1c surface, showing the side-chain residues; colors indicate effect on activation (red, > 50% decrease; orange, 25–50% decrease; blue, < 25% decrease). Data represent two independent experiments, each in duplicate (e).
The TCR α-chain was positioned on the A′-roof, where the CDR3α loop provided mostly hydrophobic interactions with the α1 helix of CD1c (Fig. 4a). Furthermore, Lys111α formed a salt bridge to Asp65, which made additional hydrogen-bond interactions with Asn27α of the CDR1α loop (Fig. 4a,b). Notably, five of the six residues from the CDR1α loop contacted both α-helices of CD1c (Fig. 4b). The CDR1α-loop interactions were enhanced by additional van der Waals interactions (Fig. 4b). Finally, the CDR2α loop, which contacted solely the α2 helix, was driven mainly by van der Waals interactions and was supplemented by a hydrogen-bond interaction between Ser58α and Asn161 of CD1c (Fig. 4c).
While the TCR α-chain bound to the left of the F′-portal, the TCR β-chain bound to the right margin of the F′-portal so that the overall TCR footprint plugged the site from which antigens would normally protrude. Only two residues from the CDR1β and CDR2β loops contacted CD1c (Supplementary Table 2). In contrast, the CDR3β loop had a major role in interacting with CD1c, covering the F′-portal (Figs. 3b and 4d and Supplementary Table 2). This network of van der Waals and polar interactions included a hydrogen bond between Tyr109β and Glu80, which in turn made additional salt-bridge interactions with Arg110β of the TCR. Furthermore, polar interactions were made between Gln151 and the main chain of Arg110β and Gly111β, with the latter forming an additional hydrogen bond with Gly154 (Fig. 4d). Overall, the CDR1α, CDR3α and CDR3β loops of the 3C8 TCR had a major role in contacting CD1c.
Critical residues that enable the CD1c-restricted response.
To identify CD1c residues needed for activation, we generated alanine-substitution mutants of residues exposed for possible TCR contact, as determined by a published CD1c-lipid crystal structure8. In analyses of parental C1R human lymphoid cells and C1R cells transduced with CD1c (C1R.CD1c cells) or CD1d (C1R.CD1d cells), the J76.3C8 reporter cell line responded only to C1R.CD1c cells (Fig. 4e). In the five alanine-substitution mutants with a negligible effect on activation, four of the residues replaced (Glu61, Asp83, Ser143 and Leu147) were not positioned within the 3C8 TCR footprint. Glu157 contacted three CDR loops, so the lack of effect after substitution was unexpected. Substitution of Arg79 or Asn161 abolished contact with the CDR1β loop or CDR2α loop, respectively, and reduced activation by 75%. Seven residues, scattered across the A′-roof and around the F′-portal, were essential for the 3C8 TCR–CD1c interaction (Fig. 4e). These included Glu62, Leu68, Phe72, Arg79, Glu80 and Tyr152 (Supplementary Table 2). The essential role of the last three residues, which were located on the right margin of the F′-portal, indicated not only that the 3C8 TCR footprint covered the F′-portal but also that key contact residues needed for activation fully surrounded that portal, consistent with the interpretation that the TCR acts as a plug to prevent egress of lipids to the surface.
The mutagenesis patterns noted above also showed commonalities with studies of CD1c-reactive TCRs that recognize mycobacterial phosphomycoketide16. Furthermore, when we compared the CD1c positions crucial for binding of the 3C8 TCR with those of other members of CD1 family, we found that Glu80 and Tyr152 were shared solely with CD1b and Asn161 was present in CD1a, while Leu68 and Phe72 were unique to CD1c (Supplementary Fig. 3). Therefore, the structural and energetic footprints also provided a basis for understanding the CD1c-specific nature of this autoreactive T cell response.
Mass spectrometry reveals exclusion of lipids after engagement of the TCR.
To identify lipids in CD1c-endo and TCR–CD1c-endo complexes, we treated them with chloroform and methanol. High-performance liquid chromatography (HPLC)–time-of-flight (TOF) electrospray ionization (ESI) mass spectrometry (MS) in negative mode detected many ions that were present in both complexes. Among the many ions detected, three matched the mass of [M-H]– of C16, C18 and C18:1 fatty acids (m/z = 255.233, 283.264 and 281.249, respectively). In the positive mode, we detected ions corresponding to the [M + H]+ mass of monoacylglycerol (MAG) (m/z = 331.284, 359.316 and 357.300), sphingomyelin (SM) (m/z = 703.575, 731.606, 811.669 and 813.685) and phosphatidylcholine (PC) (m/z = 732.554, 760.585, 786.601 and 788.617) (Fig. 5). Whereas SM and PC are known CD1 ligands13,18,23, the detection of MAG was unexpected. Further collisional MS experiments with an authentic standard confirmed the MAG structure (Supplementary Fig. 4).
Fig. 5 |. Isolation of lipids from CD1c and 3C8 TCR–CD1c complexes.
Ion chromatography of lipids in the CD1c-endo and 3C8 TCR–CD1c-endo (CD1c 3C8) proteins used for crystallography (left margin); these were treated with chloroform and methanol, which yielded lipid eluents separated by normal-phase HPLC-TOF-MS and analyzed in the positive-ion mode (left) and negative-ion mode (middle); labels indicate the most-abundant lipid in each named class. Right, quantification of the results at left, determined as individual molecular species within each class detected separately as ion chromatograms and compared with ion chromatograms of external standards, and presented as molar ratios. Data are representative of three independent experiments (normal phase), with similar results were obtained with reversed-phase chromatography (Supplementary Fig. 5).
The area under the curves of ion chromatograms for the most abundant molecular species in each class (Fig. 5) and all molecular variants (Supplementary Fig. 5) showed differential trapping of lipids in CD1c-endo relative to that in TCR–CD1c-endo. In CD1c-endo we observed more-intense ion chromatograms for two polar lipids with large headgroups, SM and PC, which were relatively excluded from TCR–CD1c-endo. Conversely, fatty acids (FAs) and MAGs were present at trace amounts in CD1c-endo, and TCR–CD1c-endo complexes showed enrichment for these. Similar patterns were seen in reversed-phase analysis, which separated individual lipids more extensively; this provided further details about chain length and saturation variants in each class (Supplementary Fig. 5). These patterns of lipid exclusion held true after quantification of individual lipid species with authentic external standards (Fig. 5 and Supplementary Fig. 6). Thus, whereas CD1c bound lipids with small headgroups (FAs and MAGs) and large headgroups (PC and SM), the addition of the 3C8 TCR created ternary complexes that selectively trapped lipids with smaller hydrophilic headgroups.
Permissive and non-permissive lipids within CD1c.
The observed densities within the CD1c cleft were derived from many types of bound molecules, so our analysis focused mainly on the general size and position of densities in CD1c-lipid versus that in TCR–CD1c-lipid (Fig. 6a,b, Supplementary Table 1 and Supplementary Fig. 7), interpreted in conjunction with the associated MS analyses (Fig. 5). In the CD1c-endo binary structure, electron density within the antigen-binding cleft adopted the shape of a headgroup protruding from the cleft with two bifurcating tubes of electron density that extended into the A′- and F′-pockets (Fig. 6a). Given the strength of the SM and PC ion chromatograms in CD1c-endo (Fig. 5), we modeled SM (Fig. 6a) and PC (Supplementary Fig. 7d) into the density, which was generally consistent with the observed electron density within the CD1c-endo binary structure (Fig. 6a and Supplementary Fig. 7d). There was no distinct electron density representative of any phospholipid headgroup positioned outside the CD1c cleft within the 3C8 TCR–CD1c-endo ternary complex (Fig. 6b). Indeed, modeling SM was clearly non-permissive for the binding of 3C8 TCR (Fig. 6c), as predicted by portal-covering footprint and the selective elution of ‘headless’ lipids from TCR-CD1c (Figs. 3, 4e and 5).
Fig. 6 |. Models of the CD1c ligands in the X-ray crystal structures.
a,b, Electron-density maps of CD1c-SM (2Fo-Fc = 0.7 σ, Fo-Fc = 1.5 σ) (a) and 3C8 TCR–CD1c-MAG C16:0 (2Fo-Fc = 1.0 σ, Fo-Fc = 2.2 σ) (b), showing the 2Fo-Fc of the refined map (blue) and Fo-Fc of the omit map (green). c, Model of CD1c presenting antigens with a headgroup, showing of steric clashes with the CDR loops of the 3C8 TCR (loops and surface): gray, CD1c; dark blue, 3C8 TCR Vα; beige, 3C8 TCR Vβ; pink sticks, MAG; green sticks, SM; orange sticks, MPM; red sticks, GD3; and purple sticks, decanes.
In the TCR-CD1c ternary complex, three disconnected tubes of electron density were observed within the cleft (Fig. 6b) that matched those observed in a previously determined CD1c binary structure with unnamed ‘spacer lipids’ (CD1c-SL)27 (Supplementary Fig. 7a). Given the diversity of ligands present, these densities probably represented areas of the cleft that position lipids in a more uniform way, rather than the outlines of individual named molecules. For example, two tubes of electron density matched the size of C10–C12 spacer lipids (Fig. 6b, Supplementary Fig. 7a). C10–C12 fatty acids were not detected in MS experiments, but densities of that size were also seen in the same position within the F′-pocket of the CD1c-SL complex and thus might represent constrained portions of larger molecules27 (Supplementary Fig. 7a). The C16 MAG and the C18 stearic acid eluted from CD1c did match the size of the observed electron density within the A′-pocket (Fig. 6b, Supplementary Fig. 7b,c). We modeled MAG such that the glycerol moiety made a series of polar contacts at the extreme end of the A′-pocket, which could not occur in the reverse orientation (Supplementary Fig. 7e).
CD1c has been reported to exist in open or closed conformations8,27. In contrast to the open F′-roof structure seen in the published CD1c-phosphomycoketide (CD1c-PM) and CD1c–mannosyl-β1-phosphomycoketide (CD1c-MPM) structures8,16 (Supplementary Fig. 7g, right), we observed a closed F′-roof platform, which more closely mimicked the CD1c-SL structure27 (Supplementary Fig. 7f,g). Thus, both published work and our current structure are consistent with a role for lipids in stabilizing the closed conformation of CD1c. Overall, the structural data were consistent with a mechanism in which CD1c forms complexes with lipids possessing exposed headgroups, but the TCR selectively binds those CD1c complexes formed with smaller lipids that remain buried within the cleft and participate in generating a compact complex (Fig. 6c).
Recognition of CD1c-endo by the TCR generates functional autoreactivity.
We returned to our study of polyclonal T cells from tuberculosis-naive donors to identify the TCRs and lipid ligands that mediate CD1c-endo tetramer staining. Similar to mean values seen in the group of patients, approximately 1 in 1,000 T cells from a healthy donor (HD1) showed staining with CD1c-endo tetramers. After two rounds of sorting for CD1c tetramer–positive cells, we selected cells with particularly bright CD1c tetramer staining whose frequency increased ~500-fold to 58% of T cells and created a polyclonal T cell line from HD1 (Fig. 7a). We observed release of IFN-γ in response to each of two types of antigen-presenting cell, C1R cells and K562 human myelogenous leukemia cells transfected to express CD1c, but not in response to their control counterparts transfected to express CD1a or CD1b (Fig. 7a). Thus, tetramer binding translated into functional CD1c-directed autoreactivity. Furthermore, the pattern of response observed here in which CD1c, but not any specific lipid ligand, was required for activation matched the proposed mechanism by which CD1c-endo tetramers might bind polyspecific TCR(s) expressed in HD1.
Fig. 7 |. CD1c-endo tetramer staining involves TCR binding and T cells with functional autoreactivity to CD1c proteins.
a, Flow cytometry (top) of PBMCs freshly obtained from subject HD1 (left), sorted for binding to anti-CD3 and CD1c tetramers treated with MAG and fatty acid (First expansion; middle) then resorted a second time to generate the HD1 T cell line (right). ELISpot assay (below) of IFN-γ (assessing functional autoreactivity) in the HD1 T cell line after stimulation with K562 or C1R cells (above plots) transfected to express CD1a, CD1b or CD1c (horizontal axis). Numbers adjacent to outlined areas (top) indicate percent tetramer-positive CD3+ cells. Data are from two independent experiments representative of four experiments with similar results (bottom; error bars, s.e.m. of three technical replicates). b, Flow cytometry of HD1 T cells stained with a panel of monoclonal antibodies that specifically bind TCRs with various β-chain variable regions (vertical axes). Numbers in quadrants indicate percent cells in each. c, Flow cytometry of HD1 T cells sorted by binding to an antibody specific for Vβ5.1 and to CD1c tetramers (top left), followed by further sorting of Vβ5.1+ cells by expression of CD4 (top middle), and then further culture so that > 98% of cells stained with CD1c tetramer (Supplementary Fig. 8a). ELISpot assay of IFN-γ (assessing functional CD1c autoreactivity) by the Vβ5.1− cells (HD1TRBV5.1−; bottom left) and the CD4+ Vβ5.1+ cells (HD1CD4+; top right) and CD4− Vβ5.1+ cells (HD1CD4–; bottom right) after stimulation with K562 or C1R antigen-presenting cells (below plots) transfected to express CD1c ( + ) or not (–) (horizontal axis). Data are representative of one experiment (HD1TRBV5.1–) or two experiments (HD1CD4– and HD1CD4+) with each antigen-presenting cell line (error bars (bar graphs), s.e.m. of three technical replicates). d, Flow cytometry of CD4+ Vβ5.1+ (HD1CD4+) and CD4− Vβ5.1+ (HD1CD4–) HD1 T cell lines (left margin) stained first with CD1c-endo tetramer and then with anti-Vβ5.1 (Tetramer, antibody), first with anti-Vβ5.1 and then with CD1c-endo tetramers (Antibody, tetramer) or with anti-Vβ5.1 alone (Antibody only) (above plots). Numbers in quadrants indicate percent cells in each. Data are representative of four experiments with similar results.
To identify the TCRs, we first screened the HD1 T cell line with antibodies specific for the following TCR β-chain variable regions: Vβ13.1, Vβ8, Vβ2, Vβ4, Vβ9, Vβ21.3, Vβ7.1, Vβ13.6 and Vβ5.1. Each antibody stained a small population of tetramer-negative T cells, which provided positive controls, but only antibody to Vβ5.1 (anti-Vβ5.1) stained tetramer-positive cells (Fig. 7b). This pattern of staining effectively divided the CD1c tetramer–positive events into three populations with no staining, high staining or very high staining with anti-Vβ5.1 (Fig. 7b). Further sorting showed that the two partially overlapping CD1c tetramer–positive Vβ5.1+ populations represented distinct CD4+ and CD4– populations (Fig. 7c). After further cultivation to > 98% tetramer-positive cells (Supplementary Fig. 8a), we identified a single TCR β-chain sequence from each line. Sequencing confirmed the presence of TRBV5–1 gene segments in the CD4– HD1 T cell line (TRBV5–1 joined to TRBJ1–2 with the CDR3 sequence CASSAGQALYGYTFGSG) (called ‘HD1CD4–cells’ here) and the CD4+ HD1 T cell line (TRBV5–1 joined to TRBJ1–1 with the CDR3 sequence CASSLDGTGATDTEAFFGQG) (called ‘HD1CD4+ cells’ here). Initially we considered whether this finding might represent bias toward expression of TRBV5–1, but this TCR variable region is common in human T cells37 and is not overrepresented in the CD1c-reactive TCRs identified so far16,32. Instead, the key point was that all three CD1c tetramer–positive clones were selectively activated by CD1c+ target T cells in IFN-γ-release assays (Fig. 7c), which demonstrated functional autoreactivity to CD1c. Two different antigen-presenting cell lines were used (K562 and C1R); the results in each case led to the conclusion that CD1c expression was essential for T cell activation. Notably, monoclonal antibodies to the Vβ5–1 TCR and CD1c tetramers showed cross-blocking (Fig. 7d), which ruled in the possibility that TCRs were the binding partners of CD1c tetramers.
CD1c tetramers carry many self lipids.
The CD1c-restricted clones noted above provided an experimental system with which to determine the number and nature of lipids that can support binding of TCR-CD1c. In contrast to results obtained for HLA-DR4, the elution of lipids from the CD1c-endo monomers that were used to make tetramers produced a rich nanoelectrospray ionization mass spectrum (Fig. 8a), which suggested that many different lipids were bound in the CD1c cleft. Normal-phase HPLC-TOF-ESI-MS separates compounds according to their polarity and detects the number and mass of lipids eluted from CD1c. This system detected 388 distinct molecular events associated with CD1c (Fig. 8b), which demonstrated that CD1c-endo carried a large number of bound lipids. Furthermore, events were detected along the full spectrum of lipid polarity (Fig. 8b), such that CD1c-eluted lipids co-migrated with extreme hydrophobes, glycolipids, phospholipids and other charged lipids.
Fig. 8 |. CD1c monomers bind highly diverse self lipids, and tetramers derived therefrom bind to CD1c-autoreactive T cell clones.
a, Positive-mode nanoelectrospray ionization MS of eluents from CD1c monomers used to make CD1c-endo tetramers (left) or from a normalized amount of HLA-DR4 protein prepared in the same expression system (right). Data are from one experiment representative of two independent experiments with similar results. b, Normal-phase HPLC-TOF-ESI-MS profiling of CD1c eluents extracted (in triplicate) from 20 μM input protein, with signals generating nearly Gaussian ion chromatograms detected within narrow mass and retention-time windows in two or more replicates presented as molecular features (dots, top left); only events present at an intensity of over twofold higher in CD1c eluents than in HLA-DR4 eluents and with significantly higher intensity (corrected P value, < 0.05 (derived with Welch’s t-test) are shown (for less false-positive detection of ions in the medium or loosely bound to proteins), and events corresponding to isotopes, alternate adducts and events with m/z > 1,000, which are typically lipid dimers, were censored (for fewer false-positive results from the detection of one molecule as two ions). Below plot, retention times corresponding to fractions previously found to be enriched for hydrophobic lipids, glycolipids and charged lipids; the lipids noted (bottom) were solved on the basis of matches of mass to known compounds and co-elution with authentic standards (colors match those in plot). For the 388 total events detected (top left), unnamed compounds of known mass (gray) and named compounds (colors) are listed with mass, retention time, chain length, lipid unsaturation and adduct formation in Supplementary Table 3. Chain length and saturation variants within the same lipid class typically have similar retention times and m/z values and thus appear as clusters (top left). Right, delineation of sphingomyelin (pink) as 29 molecular species, presented as ‘CX:Y’, where ‘X’ is the total number of carbon atoms (C) in the combined fatty acyl and sphingosine units and ‘Y’ is the total number of unsaturations. Data are representative of two experiments. c, Ion chromatography of named lipids eluted from CD1c (top) and authentic lipid standards (bottom), presented as extracted chromatograms. Data are from one experiment representative of two independent experiments with similar results. d, Flow cytometry of the HD1CD4+ and HD1CD4– T cell lines (above plots) without tetramer staining (No tetramer) or stained with mock-treated CD1c tetramers (CD1c-endo) or CD1c tetramers treated with PC, SM, PM or MAG (key). Data are representative of two experiments.
In the m/z–versus–time plot, the 388 events formed approximately 40 clusters. Each such cluster typically represents one type of lipid that appears as a family of molecular variants with differing chain length and unsaturation. For example, we identified one cluster as SM, on the basis of collision-induced dissociation–MS, and then mapped 29 variants on the basis of lipid length and saturation variations (Fig. 8b). We report all events according to their m/z value (Supplementary Table 3). Next we selected for identification seven clusters with high signal intensity and diverse retention times. This identified 93 of the events as triacylglcyerides, diacylglycerides, hexosyl ceramides, PC, SM, phosphatidylinositol or LPC on the basis of m/z matches to databases and co-elution with authentic standards (Fig. 8c).
TCR-CD1c binding is blocked by lipids with large headgroups.
To determine if ligands could influence the binding of TCR-CD1c, we obtained synthetic lipids matching the identity of cellular lipids eluted from CD1c and treated them with CD1c-endo tetramers. In addition, we treated tetramers with bacterial PM as a positive control known to protrude from the CD1c cleft8. Whereas CD1c-endo tetramers stained both CD1c-reactive T cell lines brightly, treatment of CD1c with MAG further enhanced staining. Ligands with larger or charged headgroups (PM, PC, SM and MPM) all blocked tetramer staining of HD1CD4+ cells and HD1CD4– cells (Fig. 8d). As a control experiment to determine if SM somehow denatured or inactivated tetramers, we found that staining of HD1CD4+ cells and HD1CD4– cells was restored when CD1c-SM tetramers were subsequently treated with MAG and used to stain T cells (Supplementary Fig. 8b). Thus, SM probably functioned to bind and block the formation of a TCR epitope rather than to globally inactivate CD1c. Overall, our findings indicated that CD1c carries a mixture of lipids, including those that modulate binding to the TCR. The spectrum of lipids captured from human cells was sufficient to mediate tetramer binding and T cell activation. Skewing the spectrum toward smaller permissive or larger non-permissive ligands bound to CD1c moved the middle set point toward higher avidity or lower avidity for the TCR.
Discussion
The co-recognition model emphasizes precise discrimination, such that T cells scan many MHC or CD1 complexes on antigen-presenting cells but remain ‘off ‘ until they encounter a rare antigen that turns them ‘on’. The mechanism of human T cells’ autoreactivity to CD1c identified here showed extreme polyspecificity for many self lipids, which created a situation in which autoreactive T cells were able to respond to any CD1c-expressing cell tested. This mode of autoreactivity is explained by a straightforward mechanism involving contact of the TCR with CD1c rather than with the lipids carried. If antigens are defined as the target of TCR contact, then CD1c itself is the antigen. Certain ligands can ‘dial up’ or ‘dial down’ the response on the basis of their overall headgroup size, with antigenic function being inversely correlated with headgroup size.
The two distinct mechanisms by which TCRs contact CD1a or CD1c, while avoiding contact with the ligand, make different predictions about the nature of inhibitory and activating ligands. For CD1a, the autoreactive BK6 TCR involves an extreme left-shifted footprint that leaves the F′-portal uncovered13. While this allows polyspecific response to lipids, it makes no predictions about the ligands that can be recognized or block contact because the ligands can protrude and extend to the right side of the platform. In contrast, the sequestration mechanism for CD1c involved a centrally located TCR plugging the F′-portal and the seating of lipids fully within the CD1c cleft. Here, lipid sequestration allows general predictions about the types of lipids involved: activating self lipids must be small so that they can reside within the CD1c cleft. Larger lipid ligands with bulky headgroups will protrude and interfere with TCR binding.
The lipid polyspecificity, which manifested as autoreactivity of CD1c to untreated cells and ‘endo tetramer’ staining of T cells, indicates that any TCR with the properties found in the 3C8 TCR or the HD1-derived lines would be activated in the presence of CD1c-expressing cells. This invites consideration of the mechanisms that limit CD1c-mediated autoimmunity. First, although the spectrum of lipids normally bound to CD1c in cells allows T cell activation and tetramer binding, certain lipids, such as PC and SM, blocked TCR autoreactivity. Second, ligand-induced structural flexibility of CD1c could disrupt the ‘landing pad’ for autoreactive TCRs8,27. A third mechanism is simply the restricted expression of CD1c in the periphery, which limits contact with CD1c-autoreactive T cells38. Finally, autoreactivity does not always generate autoimmunity. For example, when CD1c-expressing B cells and dendritic cells do contact CD1c-autoreactive T cells, the responses might represent immunoregulatory cross-talk, as has been demonstrated for CD1d39.
Conventional models emphasize high antigen specificity and require an atomized T cell repertoire consisting of millions of non-crossreactive TCRs with many rare clones at low precursor frequency. Published studies have shown that CD1c- and CD1a-autoreactive T cells have a high precursor frequency in human blood14,22. Here we have provided a detailed exposition of how CD1 autoreactivity can show extreme polyspecificity for lipids. Because the response is controlled by CD1c, not by the carried ligand, it diverges from the co-recognition model that is entrenched in the roots of MHC biology. Given the low CD1c polymorphism in humans and the extreme nature of TCR crossreactivity for lipids shown here, the CD1c TCR repertoire might be less complex than that of the MHC system.
Methods
Lipid standards.
Triacylglycerol (TAG, # T5141), diacylglycerol (DAG, #D0138), monoacylglycerol (MAG, #M2015) and fatty acid (FA, #M3128) were purchased from Sigma-Aldrich. Glucosylceramide (#860539), phosphatidylcholine (PC, #850475), sphingomyelin (SM, 860584), phosphatidylinositol (PI, 840042) and lyso-PC (#845875) were purchased from Avanti polar lipids.
Lipid elution from proteins.
The lipid elution from human CD1c and HLA-DR4 proteins was performed in 15-ml glass tubes using chloroform, methanol and water, based on a published method40. The organic phase was separated from the aqueous phase and dried under a nitrogen stream. The eluent residue was redissolved in chloroform/methanol (1:2) and was stored at −20 °C for further mass analysis. For lipidomic analysis, CD1c and HLA-DR4 were extracted three times for triplicate runs via HPLC-MS.
Nanospray and HPLC-MS analysis of lipid eluents.
Extracted lipids from CD1c-endo and HLA-DR4 (~5 μl) were loaded onto a nanospray tip for nanoelectrospray ionization mass spectrometry (ESI-MS) using a linear ion-trap mass spectrometer (LXQ, Thermo Scientific). For the Q-TOF HPLC-MS analysis, the eluents from CD1c-endo and HLA-DR4 were normalized to 20 µM based on the input proteins, and 20 µl were injected (Agilent 6520 Accurate-Mass Q-TOF and 1200 series HPLC system using a normal phase Inertsil Diol column (150 mm × 2 mm, GL Sciences), running at 0.15 ml/min according to published methods41–43 with minor modifications. The lipid analysis for the eluents from 3C8 TCR–CD1c-lipid crystals was similar except that the injection quantity was calculated to normalized ~50 μg of input protein. For the reversed-phase HPLC-MS, an Eclipse Plus-C18 column (3.5 µM, 2.1 mm × 30 mm, Agilent Technologies) was used based on published methods44. The mobile phases were (A) 2 mM ammonium formate in methanol/water (90/10; v/v) and (B) 2 mM ammonium formate in 1-propanol/cyclohexane/water (90/10/0.1; v/v/v). The solvent gradient was: 0–4 min, 100% A; 4–13 min, from 100% A to 100% B; 13–18 min, 100% B; 18–20 min, from 100% B to 100% A; and 20–25 min, 100% A. To quantify the lipids, the peak areas of the time-intensity ion chromatograms were compared to the external standard curves.
Human subjects.
PBMCs were isolated from venous blood drawn from healthy volunteers, after informed consent was obtained on an individual basis and with ethical approval and oversight from the ethics committee of Cardiff University School of Medicine. Additional healthy donors from Lima, Peru were recruited under oversight from the Institutional Committee of Ethics in Research (CIEI) of the Peruvian Institutes of Health, the Institutional Review Board (IRB) of the Harvard Faculty of Medicine, and the Partners Healthcare IRB. Peruvian patients provided oral and written informed consent in Spanish and met study criteria for lack of prior tuberculosis infection, as follows: a negative Quantiferon test result and no clinical evidence of active tuberculosis. Separately, PBMCs were obtained from leukoreduction collars provided by the Brigham and Women’s Hospital Specimen Bank.
Generation of 3C8 TCR–transduced T cell line.
The Jurkat 76 (J76) T cell line was generated as previously described45. In brief, a pMIG construct containing 3C8 TCR sequence was co-transfected into HEK293T cells in the presence of the retroviral packaging vectors pPAM-E and pVSV-g. Supernatants from transfected HEK293T cells were harvested and used to transduce J76 cells. Following a 5-day transduction period, the J76.3C8 cells that had the highest surface expression, as determined by their GFP and CD3 expression, were enriched and sorted using a BD Aria III instrument (BD Biosciences).
Tetramer staining.
CD1c tetramers were made from CD1c monomers from the NIH Tetramer Facility as described32 and using a mammalian expression system and construct described below. The previously validated monomers were used for patient cohort studies (Figs. 1b,c, 7 and 8) and elution (Fig. 5), and the new design was coupled to multiple fluorophores and used for study of individuals (Fig. 1a) and staining of TCR-transfected cell lines (Fig. 2a,b). The J76.3C8, J76.NKT15 and parental J76 T cell lines were stained with PE-labeled tetramers of CD1c at 10 μg/ml in PBS containing 2% FCS (flow cytometry buffer). The cells and tetramers were incubated at 4 °C for 1 h and were washed twice in that flow cytometry buffer. Human PBMCs and T cells were stained with tetramers at 2 μg/ml in PBS containing 1% bovine serum albumin and 0.01% sodium azide for 20 min at 20 °C, followed by optimally titered antibodies for 20 min at 4 °C. In other experiments, human PBMCs were washed twice in lipid-free medium and were dually stained with PE- and BV421-conjugated tetramers for 20 min at 4 °C, followed by LIVE/ DEAD fixable Aqua (Life Technologies) for 10 min at 20 °C and the following optimally titered antibodies for 20 min at 4 °C: anti-CD14-V500 (clone M5E2, BD Horizon) and anti-CD19-V500 (clone HIB19, BD Horizon); anti-CD3-APC- Fire750 (clone SK7, BioLegend), anti-CD8-BV711 (clone RPA-T8, BioLegend) and anti-CCR7-Cy7-PE (clone 3D12, BD Pharmingen); anti-CD4-Cy5.5-PE (clone S3.5, Thermo Fisher Scientific); and anti-CD45RA-ECD (clone 2H4LDH11LDB9, Beckman Coulter) and anti-pan-γδ-Cy5-PE (clone IMMU510, Beckman Coulter).
For loading of CD1c monomers with defined ligands, 16 μg of PC or SM, or a combination of 5 μg C18:0 MAG and 5 μg C16 fatty acid, was sonicated for 30 min at 37 °C in 50 μl of 0.5% CHAPS and 50 mM sodium citrate buffer, pH 7.4. CD1c monomers (10 μg) were added to the tubes and incubated overnight at 37 °C. For loading of CD1c monomers with PM, 5 μg of PM was sonicated for 30 min at 37 °C in 28 μl of PBS. CD1c monomers (10 μg) were added to the tubes and incubated overnight at 37 °C. The next day, 17 μl PBS was added to reach a final concentration of 0.2 mg/ml loaded CD1c monomers. For reloading experiments, CD1c monomers (4 μg) were treated overnight with 0.4 μg SM or not. Subsequently, 12 μg of ceramide, MAG or diacylglycerol (DAG) was added, and the CD1c–ligand mixtures were incubated overnight for a second time. Monomers were tetramerized using streptavidin-APC (Molecular Probes), streptavidin-PE (Molecular Probes or Sigma-Aldrich) or streptavidin-BV421 (BioLegend). Cells were acquired using an LSRFortessa or a custom-modified FACSAria II flow cytometer (BD Biosciences). Flow cytometry data were analyzed with FlowJo software (Tree Star).
Generation and testing of HD1 T cells and subsets.
PBMCs from subject HD1 were sorted based on their binding to anti-CD3 (clone SK7, Becton Dickinson) and CD1c tetramers treated with a mixture of MAG and fatty acid. Population expansion of sorted cells was performed using anti-CD3 (clone OKT3, produced in-house), irradiated feeder cells and IL-2. After 2 weeks, the sorting and expansion procedure was repeated, and the resulting cell line was named ‘HD1’. IFN-γ ELISpot assays were performed using anti-1D1K and GB-11-biotin according to the manufacturer’s instructions (Mabtech). After an initial screen using Vβ-specific antibodies (Vβ2: IM1484; Vβ4: IM3602; Vβ5.1: IM1552; Vβ9: IM2003; Vβ13.6: IM1330 and Vβ7.1: IM2287 (Beckman Coulter), Vβ13.1 (clone H31, Ebioscience), Vβ8 (clone JR2, Biolegend), Vβ21.3 (Catalog: 1483, Immunotech), three subsets of HD1 were sorted using anti-Vβ5.1 and cultured in vitro. The TCR β-chain sequences of HD1CD4+ and HD1CD4– were determined from RNA isolated with an RNeasy Kit (Qiagen), with cDNA synthesized using a Quantitect Reverse Transcription Kit (Qiagen). V-segment usage was determined by PCR using primer set IPS000030, as described online (https://www.imgt.org/) and via a multiplex approach46.
CD1c-mutagenesis assays.
DNA constructs encoding single-residue CD1c mutants were synthesized (Life Technologies) and cloned into pMIG247, and retroviruses were transduced into C1R cells. Surface expression of each mutant was confirmed by flow cytometry using anti-CD1c (clone L161, BioLegend), and cell lines were purified via cell sorting based on co-expression of eGFP and CD1c. C1R.CD1d cells were generated previously via similar means48. C1R cells were co-cultured at a 1:1 ratio with J76.3C8 cells overnight, and CD69 expression on eGFP+ CD3+ cells was assessed using an LSRFortessa flow cytometer (BD Biosciences). CD69 median fluorescence intensity (MFI) is presented relative to activation levels induced by C1R.CD1c wild-type antigen-presenting cells.
Expression and purification of soluble 3C8 TCR and CD1c.
Genes encoding the extracellular domains of CD1c and β2-microglobulin were cloned into pHLsec vectors49 with the addition of C termini thrombin cleavage sites and leucine zippers (Fos, β2-microglobulin; Jun, CD1c; where CD1c contains a further BiRA-hexaHis tag). Soluble fusion proteins were expressed by cotransfection of HEK293S GnTI– cells with the plasmids pHLsec-β2-microglobulin-Fos and pHLsec-CD1c-Jun-BirA-hexaHis. Purified CD1c-β2-microglobulin heterodimers were obtained by nickel affinity followed by size-exclusion chromatography. The HLA-DR4 molecules were expressed in HEK293S cells, with the CLIP peptide, and purified using a similar protocol to that of CD1c.
Genes encoding the extracellular region of the 3C8 TCR (α-chain: TRAV29/DV5*01-TRAJ30*01, CDR3α 104CAASVGDKIIF114; β-chain: TRBV7–2*01- TRBD2*01-TRBJ2–1*01, CDR3β 104CASSSYRGPRMNEQFF119) were cloned into pET30a vectors and were expressed as insoluble inclusion bodies in Escherichia coli BL21 strain50. Inclusion bodies were purified from the bacterial cells and solublized in 8 M urea, 0.5 mM EDTA, 1 mM DTT and 20 mM Tris-HCl, pH 8.0. The inclusion bodies were used in the oxidative refold of the 3C8 TCR, and soluble TCRs were purified via a series of anion-exchange, size-exclusion and hydrophobic interaction chromatography methods.
Surface plasmon resonance measurements and analysis.
Steady-state equilibrium affinity of the 3C8 TCR–CD1c-endo interaction was assessed at 25 °C using a BIAcore 3000 instrument with 10 mM HEPES, pH 7.4, and150 mM NaCl as the running buffer. The Vβ8.2 NKT TCR and mouse CD1d-endo material were produced as described previously51. Approximately 3,000 response units of biotinylated CD1c-endo, mouse CD1d-endo and mouse CD1d–α -galactosylceramide were coupled to a SA sensor chip and analyzed against a twofold serial dilution of the 3C8 TCR (150 µM to 146 nM) or the Vβ8.2 NKT TCR (3.125 µM to 48.8 nM). The analyte was passed over the sensor chip at a flow rate of 5 µl/min for a period of 80 s, and the final response was subtracted from that of mouse CD1d-endo. The affinity value and sensorgram plots were generated using BIAevaluation and GraphPad Prism software.
Crystallization and structure determination.
The leucine zipper of the CD1c–β2-microglobulin protein was removed using soluble thrombin. The zipperless CD1c–β2-microglobulin protein (CD1c-endo) was further purified by size-exclusion chromatography. The 3C8 TCR and CD1c-endo complexes were mixed at a ratio of 1:5 and were concentrated to 8 mg/ml in 10 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Crystallization experiments were performed at 20 °C using the sitting-drop vapor-diffusion method. Crystals of the 3C8 TCR–CD1c-endo ternary and CD1c-endo binary complexes were obtained from the 3C8 TCR–CD1c-endo sample. Crystals of the 3C8 TCR–CD1c-endo ternary complex grew in precipitant containing 200 mM potassium thiocyanate and 20% PEG 3350 after 2 weeks. Crystals of the CD1c-endo binary complex were obtained in 10 mM MES, pH 6.5, 10% dioxane and 1.6 M ammonium sulfate after 3 months. Prior to X-ray diffraction data collection, crystals were cryoprotected in mother liquor (crystallization solution) containing 20–30% ethylene glycol and then flash frozen in liquid nitrogen. Data collection was conducted at 100 °K on the MX2 beamline at the Australian Synchrotron. The 3C8 TCR–CD1c-endo and CD1c-endo crystals diffracted in the P3121 and P6522 space groups, respectively.
Data sets were processed using the CCP4 software suite52. Diffraction images were integrated using iMosflm, and intensities were scaled using SCALA. Ellipsoidal Truncation and Anisotropic Scaling were implemented using the UCLA Diffraction Anisotropy server53. The structure was determined via molecular replacement, using the CD1c-MPM binary complex (PDB accession code 3OV6) and the B7 TCR (PDB accession code 1BD2), with the CDR loops truncated, as search models in the maximum likelihood-based molecular replacement program PHASER54. Densities indicative of the presence of ligands in the CD1c binding pockets (A′, F′ and G′) became visible following initial rounds of refinements using the structure refinement program autoBUSTER55. Throughout the structure refinement process, COOT was used to build structure models56. Various ligand libraries were generated using the program JLigand from the CCP4 Suite and used as models to fit the ligand densities. Structure factors and models were validated using wwPDB.
Statistics.
The statistical significance of CD1b versus CD1c staining of PBMCs from 28 subjects was tested using the Wilcoxon signed rank test with continuity correction, after non-normality was established using the Shapiro-Wilk test.
Life Sciences Reporting Summary.
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Data Availability.
The data that support the findings of this study are available from the corresponding authors upon request. Structural information has been deposited in the Protein Data Bank under accession codes 6C09 (3C8 TCR–CD1c- lipid) and 6C15 (CD1c-lipid).
Supplementary Material
Acknowledgements
We thank S. A. Porcelli (Albert Einstein College of Medicine, New York) for the 3C8 T cell clone; the Monash Macromolecular crystallization Facility staff for assistance with crystallization and the staff at the Australian synchrotron for assistance with data collection; D. Ly for advice on tetramer staining patterns; and L. L. Tan, H. Halim and N. Williams for technical assistance. Supported by the US National Institutes of Health (R01 AR048632, R01 AI049313 and AI U19111224), the Australian Research Council (Laureate Fellowship to J.R.), the National Health and Medical Research Council (Early Career Fellowship to K.S.W.) and the Wellcome Trust (Senior Investigator Award to D.A.P.).
Footnotes
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41590–018-0065–7.
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41590–018-0065–7.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rossjohn J et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Van Rhijn I, Godfrey DI, Rossjohn J & Moody DB Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol 15, 643–654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams EJ & Luoma AM The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol 31, 529–561 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J & Moody DB The burgeoning family of unconventional T cells. Nat. Immunol 16, 1114–1123 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Gadola SD et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat. Immunol 3, 721–726 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Zajonc DM et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat. Immunol 6, 810–818 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zajonc DM, Elsliger MA, Teyton L & Wilson IA Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat. Immunol 4, 808–815 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Scharf L et al. The 2.5 Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 33, 853–862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch M et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol 6, 819–826 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Rossjohn J, Pellicci DG, Patel O, Gapin L & Godfrey DI Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol 12, 845–857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gras S et al. T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat. Commun 7, 13257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shahine A et al. A molecular basis of human T cell receptor autoreactivity toward self-phospholipids. Sci. Immunol 2, eaao1384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birkinshaw RW et al. αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat. Immunol 16, 258–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lalla C et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. Eur. J. Immunol 41, 602–610 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Roy S et al. Molecular analysis of lipid-reactive Vδ1 γδ T cells identified by CD1c tetramers. J. Immunol 196, 1933–1942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy S et al. Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by αβ T cells. Proc. Natl. Acad. Sci. USA 111, E4648–E4657 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagchi S et al. CD1b-autoreactive T cells contribute to hyperlipidemia-induced skin inflammation in mice. J. Clin. Invest 127, 2339–2352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Rhijn I et al. Human autoreactive T cells recognize CD1b and phospholipids. Proc. Natl. Acad. Sci. USA 113, 380–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matulis G et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol 8, e1000402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallevaey T et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 34, 315–326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumperz JE et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity 12, 211–221 (2000). [DOI] [PubMed] [Google Scholar]
- 22.de Jong A et al. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat. Immunol 11, 1102–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jong A et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat. Immunol 15, 177–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourgeois EA et al. Bee venom processes human skin lipids for presentation by CD1a. J. Exp. Med 212, 149–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH et al. CD1a on Langerhans cells controls inflammatory skin disease. Nat. Immunol 17, 1159–1166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramaniam S et al. Elevated and cross-responsive CD1a-reactive T cells in bee and wasp venom allergic individuals. Eur. J. Immunol 46, 242–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour S et al. Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells. Proc. Natl. Acad. Sci. USA 113, E1266–E1275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepore M et al. Targeting leukemia by CD1c-restricted T cells specific for a novel lipid antigen. OncoImmunology 4, e970463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakimchuk K et al. Borrelia burgdorferi infection regulates CD1 expression in human cells and tissues via IL1-β. Eur. J. Immunol 41, 694–705 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roura-Mir C et al. CD1a and CD1c activate intrathyroidal T cells during Graves’ disease and Hashimoto’s thyroiditis. J. Immunol 174, 3773–3780 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Lepore M et al. A novel self-lipid antigen targets human T cells against CD1c+ leukemias. J. Exp. Med 211, 1363–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ly D et al. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J. Exp. Med 210, 729–741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altman JD et al. henotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996). [PubMed] [Google Scholar]
- 34.Guo T et al. A subset of human autoreactive CD1c-restricted T cells preferentially expresses TRBV4–1+ TCRs. J. Immunol 200, 500–511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porcelli S, Morita CT & Brenner MB CD1b restricts the response of human CD4–8– T lymphocytes to a microbial antigen. Nature 360, 593–597 (1992). [DOI] [PubMed] [Google Scholar]
- 36.Yin Y, Li Y & Mariuzza RA Structural basis for self-recognition by autoimmune T-cell receptors. Immunol. Rev 250, 32–48 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Melenhorst JJ et al. Contribution of TCR-β locus and HLA to the shape of the mature human Vβ repertoire. J. Immunol 180, 6484–6489 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Dougan SK, Kaser A & Blumberg RS CD1 expression on antigen-presenting cells. Curr. Top. Microbiol. Immunol 314, 113–141 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Vincent MS et al. CD1-dependent dendritic cell instruction. Nat. Immunol 3, 1163–1168 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Bligh EG & Dyer WJ A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 41.Layre E et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol 18, 1537–1549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc. Natl Acad. Sci. USA 108, 19335–19340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madigan CA et al. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 109, 1257–1262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young DC et al. In vivo biosynthesis of terpene nucleosides provides unique chemical markers of Mycobacterium tuberculosis infection. Chem. Biol 22, 516–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gras S et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J. Exp. Med 207, 1555–1567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang GC, Dash P, McCullers JA, Doherty PC & Thomas PG T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci. Transl. Med 4, 128ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holst J, Vignali KM, Burton AR & Vignali DAA Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat. Methods 3, 191–197 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Uldrich AP et al. CD1d-lipid antigen recognition by the γδ TCR. Nat. Immunol 14, 1137–1145 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Aricescu AR, Lu W & Jones EY A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr 62, 1243–1250 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Kjer-Nielsen L et al. A structural basis for the selection of dominant αβ T cell receptors in antiviral immunity. Immunity 18, 53–64 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Pellicci DG et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the Vβ8.2 and Vβ7 semi-invariant NKT T cell receptors. Immunity 31, 47–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strong M et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103, 8060–8065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bricogne G et al. autoBUSTER, Version 1.6.0 Global Phasing Ltd, Cambridge, United Kingdom: (2011). [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request. Structural information has been deposited in the Protein Data Bank under accession codes 6C09 (3C8 TCR–CD1c- lipid) and 6C15 (CD1c-lipid).