Abstract
The growing complexity and volume of clinical data and the associated decision-making processes in oncology promote the advent of precision medicine. Precision (or personalised) medicine describes preventive and/or treatment procedures that take individual patient variability into account when proscribing treatment, and has been hindered in the past by the strict requirements of accurate, robust, repeatable and preferably non-invasive biomarkers to stratify both the patient and the disease. In oncology, tumour subtypes are traditionally measured through repeated invasive biopsies, which are taxing for the patient and are cost and labour intensive. Quantitative analysis of routine clinical imaging provides an opportunity to capture tumour heterogeneity non-invasively, cost-effectively and on large scale. In current clinical practice radiological images are qualitatively analysed by expert radiologists whose interpretation is known to suffer from inter- and intra-operator variability. Radiomics, the high-throughput mining of image features from medical images, provides a quantitative and robust method to assess tumour heterogeneity, and radiomics-based signatures provide a powerful tool for precision medicine in cancer treatment. This study aims to provide an overview of the current state of radiomics as a precision medicine decision support tool. We first provide an overview of the requirements and challenges radiomics currently faces in being incorporated as a tool for precision medicine, followed by an outline of radiomics’ current applications in the treatment of various types of cancer. We finish with a discussion of possible future advances that can further develop radiomics as a precision medicine tool.
Introduction
Background
Technological advances have led to an abundance of novel diagnostic techniques and imaging modalities available to oncology.1 Additional complexity is added by genetic2 and micro environmental3 heterogeneity of tumours and between patients.4 Due to the large volumes and complexity of modern data,5 new methods to facilitate clinical decision-making are required.
Precision (or personalised) medicine describes preventive and treatment procedures that take into account an individual patient’s characteristics together with their specific disease(s).6 A common approach to precision medicine is data mining, i.e. discovering patterns in large databases of diversified cohorts using powerful computational tools such as machine learning. Patterns can be discovered within the variability of patient populations that allow for the stratification of patient groups and the identification of the ideal treatment for the individual patient,7 thus improving patient outcome.8–10 However, this requires large databases of patients in order to cover as much of the variations within a population as possible.
An important source of large-scale data that could be used are radiological images derived during routine oncological examinations. Tumours exhibit phenotypical differences which can be visualised through routine medical imaging,11 which in turn allows for visualisation of the entire tumour volume or subregions on a macroscopic level, at baseline and longitudinally. However, imaging in a clinical setting is primarily used qualitatively, and clinical decision-making is based on visual assessments of the tumour by radiologists. Radiomics offers a quantitative alternative to assess tumour heterogeneity quantitatively. Radiomics is an advanced image feature analysis methodology, which formats standard clinical images from CT, MRI and/or positron emission tomography (PET) into a multidimensional source for data mining.12 A large number of image features are extracted from imaging data using various mathematical algorithms. These features, together with gold standard information, are used by machine-learning algorithms, computational methods that “learn” correlations from data, creating models that automate and improve classification of tumour phenotype and genomic profile13–15 as imaging biomarkers.
Radiomics-based imaging biomarkers have shown to outperform common prognostic models based on clinical parameters such as the Tumor-Node-Metastasis staging system (TNM).13 However, radiomics does not intend to replace current clinical decision-making, but rather aims to provide a supplement to current measures such as clinical, treatment and genomic data, all incorporated into a decision support system.16 To do so, a robust, repeatable and cost-effective method to clinically implement radiomics is required.
Radiomics workflow
A typical radiomics analysis starts with data selection: choosing the image to analyse, the imaging protocol to use and the correlated outcome. The image typically contains the primary tumour volume, which is analysed and linked to a certain outcome, such as tumour type, overall survival, or tumour recurrence. Proper data selection is important to create useful models, as it needs to be reproducible and applicable across sizeable cohorts. Large heterogeneous datasets are required to provide enough data to validate the model on different subsamplings of the data (cross-validation).17 In addition, the quality of the data is dependent on the image acquisition protocols used in clinical centres, which can often vary extensively, as well as the imaged site, scanner properties, reconstruction methods and motion artefacts.18, 19 Guidelines for image acquisition and standardised protocols are therefore beneficial for producing large, high-quality datasets.20 In the case of non-standardised imaging protocols, sharing of imaging protocols should be encouraged.
After image acquisition and volume reconstruction, a region of interest is defined, usually, but not necessarily, through slice-by-slice delineation of the tumour in the case of three-dimensional images. This is a labour-intensive process, and the variance caused by inter- and intra-operator variability is an issue.21, 22 A (semi-) automatic segmentation method to reduce workload and uncertainty caused by human error is therefore preferred. Besides operator variability, image segmentation, protocol standardisation, slice interval, reconstruction method, time-point and respiratory motion have all been found to have effects on feature reproducibility.23–35 Methods to improve reproducibility include multiple segmentations by different clinicians and phantom studies to determine the effects caused by different scanners.
Since the values of extracted features (mostly mathematical formulas using pixel/voxel intensities as input) depend on image reconstruction and pre-processing methods, proper reporting of methods such as filtering techniques, intensity discretisation and voxel resampling is critical for inter-operability of the radiomics features. Many of the extracted features are noise driven and need to be removed to improve model performance. The same applies to features that are highly correlated with other features or existing clinical parameters, as they do not provide any meaningful addition to the model. Test-retest studies which repeat the imaging processes after a short period of time are indispensable, as they measure the amount of variation inherent in the measurements. Stability and correlation tests can be used to make a selection based on the most robust, repeatable and non-redundant features.36, 37
The extracted features are fed into machine-learning methods together with clinical outcomes or pathology results to construct classification, predictive, or prognostic models. Prognostic models aim to predict a certain outcome regardless of therapy, while predictive models provide information about the effects of a certain therapeutic intervention. However, the number of extracted features is often larger than the number of patients included in a cohort, which risks overfitting the model. The best solution to prevent overfitting is to increase the number of samples used to train the model. While clinical data is abundant compared to research trial data, sharing between institutes has proven to be difficult due to various ethical, political and administrative issues.38 An alternative to large datasets is to reduce the number of features to a subset of the most relevant features. Various filtering-based techniques for feature selection can be used, such as the univariate Fisher score and Gini index tests, or multivariate algorithms such as mutual information or Conditional infomax feature extraction,39 which identify and select a subset of features based on predictive power. Valid predictive modelling requires separate independent datasets for training and validation.
Various different machine-learning models are available, such as neural networks, decision trees, support vector machines and multiple regression techniques. The modelling procedure has been shown to affect performance of prediction models based on radiomics features.39) Common measures of predictive performance of models are discrimination and calibration.40 Discrimination is a measure of the model to assign a higher risk-prediction to patients positive to a certain outcome, compared to patients without the outcome, which can be quantified using the sensitivity, specificity, or through the area under the curve (AUC) of the receiver operating characteristic. The AUC is equal to the probability that a positive event is correctly labelled as a positive event, and is given in the range of 0 to 1. Alternatively, the Concordance Index (CI), a measure of goodness of fit for classification models with binary outcome ranging from 0 to 1, can be used. Both AUC and CI show a perfect predictive performance at 1, while at 0.5 the predictive performance is completely random. Calibration is an internal measure of the model’s agreement between observed outcomes and predicted outcomes. The calibration is usually assessed through a calibration slope, where different resamplings of observed outcomes are plotted against predicted outcomes. If 100% agreement between these two is found at multiple samplings, then the calibration slope will be 1. Finally, a log-rank test is usually used to test the significance of the difference between survival curves of two patient groups. This is used when separating patients in low- and high-risk groups based on radiomics features.
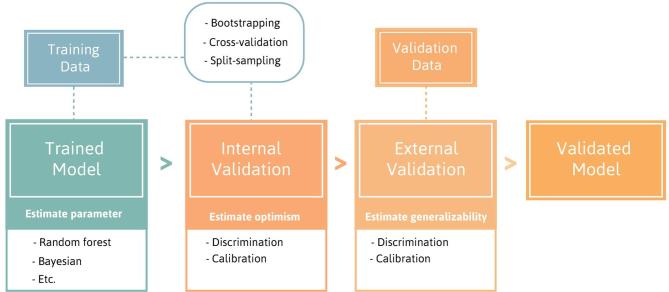
These measures of predictive performance are used to internally and externally validate the model. Internal validation is necessary to estimate and reduce the optimism in model performance, which is the degree a trained model fits worse on new data than it does on the data used to train the model. Internal validation uses the data used to train the model, and can be performed through methods such as bootstrap analysis or cross-validation.41 External validation uses an independent, external dataset to validate the accuracy of the predictive model, and to assess the generalisability of a predictive model.41 Figure 1 shows an overview of the steps involved to train and validate a predictive model.
Figure 1.
Overview of the steps involved to train and validate a predictive model.
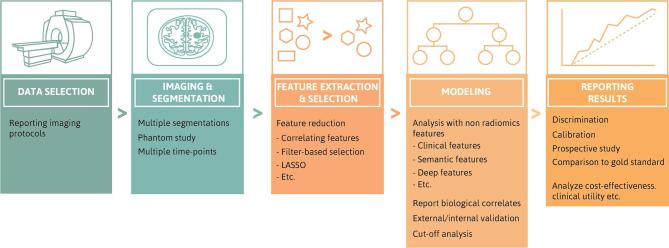
Effective and transparent radiomics studies require rigorous compliance with several guidelines, including effective validation. The Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) initiative is a set of guidelines made for studies creating and/or validating prediction models.42 There are guidelines for the source and specific information of data, the type of predictive model, procedures for building the model and the method for internal validation and measurements of model performance. Whereas the TRIPOD initiative covers prediction models in general, the Radiomics Quality Score (RQS)43 is being developed specifically for radiomics studies. The RQS assesses the quality of a study using a checklist and reports compliance as a percentage. Some of the guidelines include robust segmentations, test-retest stability of the determined features, the standardisation or thorough description of imaging protocols used, valid feature selection and internal/external validation.44 An overview of the different steps and the RQS criteria is shown in Figure 2.
Figure 2.
Overview of steps of a Radiomics analysis (top) with corresponding RQS score criteria for each step (bottom). RQS, Radiomics Quality Score.
The aim of these guidelines is to provide key details of model development and validation, which in turn allows for better reproducibility and critical appraisal of predictive models. For future and past studies, authors should check the RQS score and TRIPOD initiative to determine the quality of their methodology and allow the field of radiomics to mature. The ultimate objective of precision medicine is to link the tumour phenotype to a certain clinical endpoint, with the goal of improving clinical decision-making. Therefore, the next section will describe the use of radiomics in various studies and their efficacy in determining clinical endpoints.
Role in precision medicine
Aerts et al13 performed a radiomics analysis on a large CT dataset (N = 1019) of lung- and head and neck (h-n) cancer patients. Using a feature selection algorithm to reduce the number of features from 440 to a prognostic signature of 4 features, they found that a model built using this signature was significantly more prognostic of overall survival (OS) than a measure of tumour volume, and combining the radiomics signature with tumour volume also provided a better prognostic ability. The model was validated on different patient groups and cancer types.13 The radiomics signature showed slightly higher prognostic performance when validated in an external lung dataset than TNM or tumour volume (CI of 0.65 vs 0.63 and 0.60 respectively). For two external h-n cohorts, the signature showed higher performance compared to volume or TNM in one case (CI of 0.69 vs 0.65 and 0.66 respectively), and similar performance in the other (CI = 0.69 vs 0.68 and 0.69 respectively). This radiomics signature was also externally validated in a study by Leijenaar et al.45 on a large set of oropharyngeal squamous cell carcinoma patients (N = 542).45 The signature showed good discrimination and calibration (CI = 0.628 and calibration slope of 0.855), and after the population were split in two groups using the median value of the signature score, significant differences in OS (long rank p-value = 2e-5) could be observed.
Furthermore, CT radiomics features have been shown to be prognostic of distant metastasis and 12-month survival in glioblastoma,46 and pathological response to treatment,47 local recurrence,48, 49 histology subtype,50, 51 OS36, 50 and prediction of radiation induced pneumonitis52, 53 for lung cancer. In h-n squamous cell carcinomas, radiomics has proven to improve the prediction overall and progression free survival, and determining Human papillomavirus (HPV) status.54
Delta-radiomics is an alternative analysis which measures the change of radiomics features longitudinally. Certain features have been proven to change during treatment, indicating that this may be an additional source of information.55 Delta-radiomics on CT has shown to be prognostic in non-small cell lung cancer (NSCLC) for OS, local recurrence and distant metastasis.56 For h-n cancer patients, delta-radiomics features have proven to be a predictive and prognostic biomarker, as well provide additional information of the presence of HPV for patient stratification.
An additional source of routine medical images for radiomics analysis are cone-beam CT (CBCT) images, often used in radiotherapy for daily positioning before treatment. Van Timmeren et al57 have used CBCT data of NSCLC patients to validate a previously constructed CT radiomics signature. The signature was found to be predictive of OS in three different independent CBCT datasets (CI = 0.59–0.66), indicating CBCT could potentially be a useful source of information for radiomics analysis.57
Fludeoxyglucose-PET (FDG-PET)-based quantitative image analysis shows promise in improving prognosis in pancreatic cancer. A study by Cui et al used quantitative imaging features to predict OS, and showed better prognostic compared to the use of conventional prognostic variables of tumour volume and maximum standardised uptake value (CI of 0.66 vs 0.48-0.64).60 FDG-PET-based radiomics features correlate to mortality, local failure and distant metastasis for pancreatic cancer,61 and have also shown to be predictive in oesophageal cancer,62 tumour response in cervical cancer63, 64 and local control65 and OS63 in h-n cancer.
MRI-based radiomics has shown promise in prostate cancer: a study by Shoshana et al. (2016) used T 2 weighted MRI radiomics features to differentiate between peripheral and transition zone prostate tumours (AUC = 0.61–0.71), in a patient dataset from three different institutions.66 Furthermore, a study by Vallières et al67 use a combination of FDG-PET and MRI texture features to predict the lung metastasis in soft-tissue sarcomas. They found that a multivariable model was highly predictive of lung metastasis in soft-tissue sarcomas (AUC = 0.98), validated through bootstrapping procedures. However, the study lacked external validation for a valid conclusion.67 In the context of glioblastoma, several studies using MRI data have shown that a radiomics model may accurately detail the molecular subtype of the tumour,68–70 OS69–71 and predict short v s long-term survival.72 Finally, for an imaging method outside of radiology, Zhang et al.73 proposed a radiomics approach to ultrasound elastography, to use the density of tumour tissue for classification as benign or malignant. A signature of seven features, out of a total of 364 extracted features, was able to accurately (AUC = 0.92) discriminate between benign and malignant tumour tissue.73
To reduce inter- and intra-observer delineation variability and to reduce workload, a number of (semi-) automatic segmentation methods have been proposed and tested in radiomics studies in recent years. Several studies have shown that (semi-) automatic segmentation methods reduce inter-observer delineation variability compared to manual segmentation of lung lesions.74–77 For example, a study by Parmar et al.77 compared the robustness of 56 radiomics features derived with manual segmentation of tumour volume by five experts to a semi-automatic method performed two times by three experts, and showed that semi-automatically derived features have significantly higher reproducibility compared to manually derived features.77 Full automatic segmentation of tumours is also a possibility, as shown by Li et al.78 This study used radiomics features in a random forest model to classify tumour tissue on a voxel level. The algorithm was trained and tested on publically available datasets, and showed promising accuracy in classifying tumour tissue, necrosis, normal tissue and oedema.78
Semantic features, unique qualitative characteristics that provide information about the prognosis and (sub) type of lesions, are an alternative method to describe tumour (sub) type, and could be useful in improving prediction of certain endpoints. Some examples of semantic features are the presence of cavitation or calcification in the tumour, or features describing the roundness or spiculation of the tumours. In a study on NSCLC, Yip et al79 studied 9 semantic features, consisting of 3 binary features and 6 categorical classifiers, and 57 radiomics features describing NSCLC cancer phenotypes. To study the correlation between features they used Spearman’s Rank-Order Correlation, which is a measure of the strength and direction of association between two variables. Spearman’s rank ranges from −1 to 1, with both extremes signifying perfect correlation between two variables. The study found significant association between radiomics features and binary semantic features (AUC = 0.56–0.76), but no or weak correlation was found between classification semantic and radiomics features (Spearman’s correlation = 0.002–0.65). This indicates that radiomics and semantic features have complementary but distinct roles in outcome prediction, as they have both been proven to be able to significantly improve prediction outcomes.79
Lastly, deep learning tools, such as convolutional neural networks (CNNs), could be a method to augment radiomics analysis. Deep learning algorithms are able to learn features from imaging data without much manual input, provided that a large amount of data is available. Deep learning has been successfully implemented in a number of different studies using medical imaging data.80, 81 Orlando et al82 used a combination CNN-learned and hand-crafted discriminative features to detect red lesions (a collective term for micro aneurysms and haemorrhages), one of the earliest signs in diabetic retinopathy. The combination of features was used in a random forest classifier to discriminate between true and false red lesion candidates, and compared against either set of features separately. The combination achieved higher AUC values compared to the separate feature prediction models (AUC of 0.89 vs 0.79/0.73 for CNN and hand-crafted features respectively). Recently published articles have already shown that radiomics analysis may benefit from incorporating deep learning methods.(82-85) For example, Lao et al83 used a combination of hand-crafted and deep radiomics features to predict OS for Glioblastoma Multiforme patients on MRI images. After feature selection, a radiomics signature was created, using exclusively deep-learned features, that was able to accurately predict OS in an independent validation dataset (AUC = 0.71). Deep learning augmented radiomics analysis has also been reported to be effective in assessing treatment response in bladder cancer,84 where conversely a signature built solely on hand-crafted features was found to have better prognostic performance. These results indicate that deep learning will have an increasingly important role in predictive modelling,82 and have a complementary role with hand-crafted features in a radiomics analysis framework.
Discussion
Radiomics has been shown to be suitable for classification, prediction and prognosis of various clinical endpoints and tumour types. Many studies show a clear improvement over conventional measures predicting clinical endpoints, although variation in feature stability due to different scanners, imaging protocols and tumour motion still leaves a lot of room for improvement.13, 60 The segmentation of tumours also proves to be a small but persistent obstacle, as it is a labour- and time-intensive process and is heavily influenced by inter- and intra- segmentation variation.21, 22 However, numerous studies have reported methods to allow for a more automatic approach to segmentation,74–78 which in turn could lead to a more robust radiomics analysis.
Combining radiomics features with deep learning features or semantic features may be able to further improve prognostic performance. Several studies have proven the effectiveness of using these features independently in predictive modelling.80–87 In studies comparing the prognostic performance of these features to hand-crafted radiomics features, results were found to be mixed, indicating these methods may have distinct and complementary roles in improving prognosis.
A larger hurdle for radiomics is the transition to clinical implementation. While routine delineation is already in place in radiotherapy settings, a clinical platform to easily perform radiomics analysis during routine check-up/treatment is not. The main challenge of precision treatment is to correctly integrate various sources of data quantitatively and subsequently use this data to provide specific clinical predictions that accurately and robustly estimate outcomes as a function of the possible decisions. Numerous methods, besides radiomics, are currently in use that make use of novel biomarkers, as well as conventional clinical factors. However, many of these methods lack external validation of their legitimacy, reproducibility, or clinical validity.88 Radiomics offers a solution that integrates multiple measures into one prediction of outcome, with the added benefit of automation, which could save time and money in a clinical environment.
While many radiomics studies include external validation steps, sharing of clinical data is still an issue.89 The difficulty in sharing data may be overcome through a centralised database, or conversely through decentralised distributed learning platforms.90 To facilitate a centralised database, data has to be made available in accordance with the FAIR principles: Findability, Accessibility, Inter-operability and Reusability.89 An example of an effort to increase data shareability is through the development of ontologies to describe radiomics features.29
The distributed learning method instead aims to solve the problem of sparse data by avoiding the numerous ethical, legal and administrative issues involved in sharing data between institutes. Instead of the images being collected from numerous institutes in one central location, the model is sent and trained on site without any data leaving a particular institute. The trained models are then collected, analysed and integrated into a single model. Several proof-of-concept studies have proven that a distributed learning approach is feasible using clinical parameters,90–92 and the next step would be to integrate radiomics features, by sending a platform to extract radiomics features on-site in conjunction with the untrained predictive model. This way, a distributed learning method could provide the necessary volume and variety in data to achieve a machine-driven approach to medicine.
Conclusion
In conclusion, radiomics provides a novel non-invasive method of assessing tumour subtype, using the mostly untapped source of data of routine clinical images. The technique is often hampered by studies with small sample sizes and lack of external validation. In addition, variability in features caused by differences in imaging modality, protocols and respiratory motion, and a lack of inter-operability, may decrease the generalisability of the created radiomics models. In the future, research should be informed by guidelines such as RQS and TRIPOD, which improve the validity of radiomics as a clinically accepted field. The clinical value of the technique has already been described for a wide range of tumours and a number of different clinical outcomes. The added fact that the analysis can be performed in an automated fashion makes the technique attractive for clinical implementation to reduce workload. Performing studies on different tumour sites/types in future research may prove the generalisability of the method, and consequently lead to radiomics becoming a standard method clinically. In the future, larger volumes of data will be available for use in Radiomics studies by means of centralised, publically accessible datasets and distributed learning. Combining radiomics with other parameters will lead to high-quality decision support systems, and deep learning and semantic feature approaches may be combined with radiomics analyses to increase predictive accuracies of these models even further. Radiomics has a way ahead before full implementation in clinic is a reality, but may prove to be invaluable in realising precision medicine in cancer treatment.
Footnotes
Acknowledgements: We thank Jean Coenen, from MAASTRO clinic, for providing us with the figures included in this paper.
The authors Arthur Jochems and Henry C Woodruff contributed equally to the work.
Contributor Information
Simon A Keek, Email: simon.keek@gmail.com; simon.keek@maastro.nl; s.keek@maastrichtuniversity.nl.
Ralph TH Leijenaar, Email: ralph.leijenaar@maastro.nl.
Arthur Jochems, Email: arthur.jochems@maastro.nl.
Henry C Woodruff, Email: henry.woodruff@maastro.nl.
REFERENCES
- 1. Burstein HJ, Krilov L, Aragon-Ching JB, Baxter NN, Chiorean EG, Chow WA, et al. Clinical cancer advances 2017: annual report on progress against cancer from the American society of clinical oncology. J Clin Oncol 2017; 35: 1341–67. doi: 10.1200/JCO.2016.71.5292 [DOI] [PubMed] [Google Scholar]
- 2. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–92. doi: 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Milosevic MF, Fyles AW, Wong R, Pintilie M, Kavanagh MC, Levin W, et al. Interstitial fluid pressure in cervical carcinoma: within tumor heterogeneity, and relation to oxygen tension. Cancer 1998; 82: 2418–26. [DOI] [PubMed] [Google Scholar]
- 4. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486: 346–52. doi: 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abernethy AP, Etheredge LM, Ganz PA, Wallace P, German RR, Neti C, et al. Rapid-learning system for cancer care. J Clin Oncol 2010; 28: 4268–74. doi: 10.1200/JCO.2010.28.5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol 2013; 31: 1803–5. doi: 10.1200/JCO.2013.49.4799 [DOI] [PubMed] [Google Scholar]
- 7. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015; 372: 793–5. doi: 10.1056/NEJMp1500523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aerts HJ, Bussink J, Oyen WJ, van Elmpt W, Folgering AM, Emans D, et al. Identification of residual metabolic-active areas within NSCLC tumours using a pre-radiotherapy FDG-PET-CT scan: a prospective validation. Lung Cancer 2012; 75: 73–6. doi: 10.1016/j.lungcan.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 9. Aerts HJ, van Baardwijk AA, Petit SF, Offermann C, Loon J, Houben R, et al. Identification of residual metabolic-active areas within individual NSCLC tumours using a pre-radiotherapy (18)Fluorodeoxyglucose-PET-CT scan. Radiother Oncol 2009; 91: 386–92. doi: 10.1016/j.radonc.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suit H, Skates S, Taghian A, Okunieff P, Efird JT. Clinical implications of heterogeneity of tumor response to radiation therapy. Radiother Oncol 1992; 25: 251–60. doi: 10.1016/0167-8140(92)90244-O [DOI] [PubMed] [Google Scholar]
- 11. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278: 563–77. doi: 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. doi: 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grossmann P, Stringfield O, El-Hachem N, Bui MM, Rios Velazquez E, Parmar C, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017; 6: e23421. doi: 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panth KM, Leijenaar RT, Carvalho S, Lieuwes NG, Yaromina A, Dubois L, et al. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol 2015; 116: 462–6. doi: 10.1016/j.radonc.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 16. Lambin P, Zindler J, Vanneste BG, De Voorde LV, Eekers D, Compter I, et al. Decision support systems for personalized and participative radiation oncology. Adv Drug Deliv Rev 2017; 109: 131–53. doi: 10.1016/j.addr.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 17. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012; 30: 1234–48. doi: 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nehmeh SA, Erdi YE. Respiratory motion in positron emission tomography/computed tomography: a review. Semin Nucl Med 2008; 38: 167–76. doi: 10.1053/j.semnuclmed.2008.01.002 [DOI] [PubMed] [Google Scholar]
- 19. Sonke JJ, Belderbos J. Adaptive radiotherapy for lung cancer. Semin Radiat Oncol 2010; 20: 94–106. doi: 10.1016/j.semradonc.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 20. de Jong EEC, van Elmpt W, Hoekstra OS, Groen HJM, Smit EF, Boellaard R, et al. Quality assessment of positron emission tomography scans: recommendations for future multicentre trials. Acta Oncol 2017; 56: 1459–64. doi: 10.1080/0284186X.2017.1346824 [DOI] [PubMed] [Google Scholar]
- 21. Erasmus JJ, Gladish GW, Broemeling L, Sabloff BS, Truong MT, Herbst RS, et al. Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol 2003; 21: 2574–82. doi: 10.1200/JCO.2003.01.144 [DOI] [PubMed] [Google Scholar]
- 22. Schwartz LH, Mazumdar M, Brown W, Smith A, Panicek DM. Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res 2003; 9: 4318–23. [PubMed] [Google Scholar]
- 23. Fried DV, Tucker SL, Zhou S, Liao Z, Mawlawi O, Ibbott G, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys 2014; 90: 834–42. doi: 10.1016/j.ijrobp.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, et al. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 2013; 52: 1391–7. doi: 10.3109/0284186X.2013.812798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med 2012; 53: 693–700. doi: 10.2967/jnumed.111.099127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016; 6: 23428. doi: 10.1038/srep23428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fave X, Mackin D, Yang J, Zhang J, Fried D, Balter P, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys 2015; 42: 6784–97. doi: 10.1118/1.4934826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol 2015; 8: 524–34. doi: 10.1016/j.tranon.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalpathy-Cramer J, Mamomov A, Zhao B, Lu L, Cherezov D, Napel S, et al. Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography 2016; 2: 430–7. doi: 10.18383/j.tom.2016.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beichel RR, Smith BJ, Bauer C, Ulrich EJ, Ahmadvand P, Budzevich MM, et al. Multi-site quality and variability analysis of 3D FDG PET segmentations based on phantom and clinical image data. Med Phys 2017; 44: 479–96. doi: 10.1002/mp.12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tan Y, Guo P, Mann H, Marley SE, Juanita Scott ML, Schwartz LH, et al. Assessing the effect of CT slice interval on unidimensional, bidimensional and volumetric measurements of solid tumours. Cancer Imaging 2012; 12: 497–505. doi: 10.1102/1470-7330.2012.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larue R, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol 2017; 56: 1544–53. doi: 10.1080/0284186X.2017.1351624 [DOI] [PubMed] [Google Scholar]
- 33. Van Timmeren J, Leijenaar R, van Elmpt W, Wang J, Zhang Z, Dekker A. Test–retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography 2016; 2: 361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 2017; 90: 20160665. doi: 10.1259/bjr.20160665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015; 5: 11075. doi: 10.1038/srep11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parmar C, Leijenaar RT, Grossmann P, Rios Velazquez E, Bussink J, Rietveld D, et al. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci Rep 2015; 5: 11044. doi: 10.1038/srep11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larue R, Van De Voorde L, van Timmeren JE, Leijenaar RTH, Berbée M, Sosef MN, et al. 4DCT imaging to assess radiomics feature stability: an investigation for thoracic cancers. Radiother Oncol 2017; 125: 147–53. doi: 10.1016/j.radonc.2017.07.023 [DOI] [PubMed] [Google Scholar]
- 38. Doshi P, Jefferson T, Del Mar C. The imperative to share clinical study reports: recommendations from the Tamiflu experience. PLoS Med 2012; 9: e1001201. doi: 10.1371/journal.pmed.1001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative radiomic biomarkers. Sci Rep 2015; 5: 13087. doi: 10.1038/srep13087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010; 21: 128–38. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steyerberg EW, Bleeker SE, Moll HA, Grobbee DE, Moons KG. Internal and external validation of predictive models: a simulation study of bias and precision in small samples. J Clin Epidemiol 2003; 56: 441–7. [DOI] [PubMed] [Google Scholar]
- 42. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med 2015; 13: 1. doi: 10.1186/s12916-014-0241-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radiomics world [Webpage]. Maastro clinic; c2014-2017. 2017. Available from: http://www.radiomics.world/. [updated 2017; cited 2017 25 August].
- 44. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–62. doi: 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 45. Leijenaar RT, Carvalho S, Hoebers FJ, Aerts HJ, van Elmpt WJ, Huang SH, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol 2015; 54: 1423–9. doi: 10.3109/0284186X.2015.1061214 [DOI] [PubMed] [Google Scholar]
- 46. Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015; 114: 345–50. doi: 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SW, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016; 119: 480–6. doi: 10.1016/j.radonc.2016.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mattonen SA, Palma DA, Johnson C, Louie AV, Landis M, Rodrigues G, et al. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys 2016; 94: 1121–8. doi: 10.1016/j.ijrobp.2015.12.369 [DOI] [PubMed] [Google Scholar]
- 49. Huynh E, Coroller TP, Narayan V, Agrawal V, Romano J, Franco I, et al. Associations of radiomic data extracted from static and respiratory-gated CT scans with disease recurrence in lung cancer patients treated with SBRT. PLoS One 2017; 12: e0169172. doi: 10.1371/journal.pone.0169172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song J, Liu Z, Zhong W, Huang Y, Ma Z, Dong D, et al. Non-small cell lung cancer: quantitative phenotypic analysis of CT images as a potential marker of prognosis. Sci Rep 2016; 6: 38282. doi: 10.1038/srep38282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol 2016; 6: 71. doi: 10.3389/fonc.2016.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cunliffe A, Armato SG, Castillo R, Pham N, Guerrero T, Al-Hallaq HA. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol Biol Phys 2015; 91: 1048–56. doi: 10.1016/j.ijrobp.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anthony GJ, Cunliffe A, Castillo R, Pham N, Guerrero T, Armato SG, et al. Incorporation of pre-therapy 18 F-FDG uptake data with CT texture features into a radiomics model for radiation pneumonitis diagnosis. Med Phys 2017; 44: 3686–94. doi: 10.1002/mp.12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ou D, Blanchard P, Rosellini S, Levy A, Nguyen F, Leijenaar RTH, et al. Predictive and prognostic value of CT based radiomics signature in locally advanced head and neck cancers patients treated with concurrent chemoradiotherapy or bioradiotherapy and its added value to Human Papillomavirus status. Oral Oncol 2017; 71: 150–5. doi: 10.1016/j.oraloncology.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 55. van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Lambin P. Feature selection methodology for longitudinal cone-beam CT radiomics. Acta Oncol 2017; 56: 1537–43. doi: 10.1080/0284186X.2017.1350285 [DOI] [PubMed] [Google Scholar]
- 56. Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep 2017; 7: 588. doi: 10.1038/s41598-017-00665-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Oberije C, Monshouwer R, et al. Survival prediction of non-small cell lung cancer patients using radiomics analyses of cone-beam CT images. Radiother Oncol 2017; 123: 363–9. doi: 10.1016/j.radonc.2017.04.016 [DOI] [PubMed] [Google Scholar]
- 58. Leijenaar R, Nesteruk M, Feliciani G, Hoebers F, Van Timmeren J, Van Elmpt W, et al. EP-1608: Deriving HPV status from standard CT imaging: a radiomic approach with independent validation. Radiotherapy and Oncology 2017; 123: S868–S9. [Google Scholar]
- 59. Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol 2015; 5: 272. doi: 10.3389/fonc.2015.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cui Y, Song J, Pollom E, Alagappan M, Shirato H, Chang DT, et al. Quantitative analysis of (18)F-fluorodeoxyglucose positron emission tomography identifies novel prognostic imaging biomarkers in locally advanced pancreatic cancer patients treated with stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys 2016; 96: 102–9. doi: 10.1016/j.ijrobp.2016.04.034 [DOI] [PubMed] [Google Scholar]
- 61. Folkert MR, Setton J, Apte AP, Grkovski M, Young RJ, Schöder H, et al. Predictive modeling of outcomes following definitive chemoradiotherapy for oropharyngeal cancer based on FDG-PET image characteristics. Phys Med Biol 2017; 62: 5327–43. doi: 10.1088/1361-6560/aa73cc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Desbordes P, Ruan S, Modzelewski R, Pineau P, Vauclin S, Gouel P, et al. Predictive value of initial FDG-PET features for treatment response and survival in esophageal cancer patients treated with chemo-radiation therapy using a random forest classifier. PLoS One 2017; 12: e0173208. doi: 10.1371/journal.pone.0173208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit 2009; 42: 1162–71. doi: 10.1016/j.patcog.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang F, Thomas MA, Dehdashti F, Grigsby PW. Temporal analysis of intratumoral metabolic heterogeneity characterized by textural features in cervical cancer. Eur J Nucl Med Mol Imaging 2013; 40: 716–27. doi: 10.1007/s00259-012-2332-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bogowicz M, Leijenaar RTH, Tanadini-Lang S, Riesterer O, Pruschy M, Studer G, et al. Post-radiochemotherapy PET radiomics in head and neck cancer - The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother Oncol 2017; 125: 385-391. doi: 10.1016/j.radonc.2017.10.023 [DOI] [PubMed] [Google Scholar]
- 66. Ginsburg SB, Algohary A, Pahwa S, Gulani V, Ponsky L, Aronen HJ, et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study. J Magn Reson Imaging 2017; 46: 184–93. doi: 10.1002/jmri.25562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vallières M, Freeman CR, Skamene SR, El Naqa I, Naqa E I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015; 60: 5471–96. doi: 10.1088/0031-9155/60/14/5471 [DOI] [PubMed] [Google Scholar]
- 68. Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 2014; 273: 168–74. doi: 10.1148/radiol.14131731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou H, Vallières M, Bai HX, Su C, Tang H, Oldridge D, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 2017; 19: 862–70. doi: 10.1093/neuonc/now256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys 2015; 42: 6725–35. doi: 10.1118/1.4934373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McGarry SD, Hurrell SL, Kaczmarowski AL, Cochran EJ, Connelly J, Rand SD, et al. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography 2016; 2: 223–8. doi: 10.18383/j.tom.2016.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol 2017; 27: 4188-4197. doi: 10.1007/s00330-016-4637-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Q, Xiao Y, Suo J, Shi J, Yu J, Guo Y, et al. Sonoelastomics for breast tumor classification: a radiomics approach with clustering-based feature selection on sonoelastography. Ultrasound Med Biol 2017; 43: 1058–69. doi: 10.1016/j.ultrasmedbio.2016.12.016 [DOI] [PubMed] [Google Scholar]
- 74. Rios Velazquez E, Aerts HJ, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists' delineations and with the surgical specimen. Radiother Oncol 2012; 105: 167–73. doi: 10.1016/j.radonc.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Heye T, Merkle EM, Reiner CS, Davenport MS, Horvath JJ, Feuerlein S, et al. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra- and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology 2013; 266: 812–21. doi: 10.1148/radiol.12120255 [DOI] [PubMed] [Google Scholar]
- 76. Velazquez ER, Parmar C, Jermoumi M, Mak RH, van Baardwijk A, Fennessy FM, et al. Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci Rep 2013; 3: 3529. doi: 10.1038/srep03529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014; 9: e102107. doi: 10.1371/journal.pone.0102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, et al. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep 2017; 7: 14331. doi: 10.1038/s41598-017-14753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yip SSF, Liu Y, Parmar C, Li Q, Liu S, Qu F, et al. Associations between radiologist-defined semantic and automatically computed radiomic features in non-small cell lung cancer. Sci Rep 2017; 7: 3519. doi: 10.1038/s41598-017-02425-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zheng Y, Liu D, Georgescu B, Nguyen H, Comaniciu D. 3D deep learning for efficient and robust landmark detection in volumetric data : Navab N, Hornegger J, Wells W. M, Frangi A, Medical image computing and computer-assisted intervention - MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part I. Cham: The British Institute of Radiology.; 2015. 565–72. [Google Scholar]
- 81. Ravishankar H, Sudhakar P, Venkataramani R, Thiruvenkadam S, Annangi P, Babu N. Understanding the mechanisms of deep transfer learning for medical images : Carneiro G, Mateus D, Peter L, Bradley A, Tavares J, Belagiannis V, Deep learning and data labeling for medical applications: first international workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings. Cham: The British Institute of Radiology.; 2016. 188–96. [Google Scholar]
- 82. Orlando JC, Prokofyeva E, del Fresno M, Blaschko MB. Learning to detect red lesions in fundus photographs: an ensemble approach based on deep learning. Medical Image Analysis 2018; 153: 115–27. [DOI] [PubMed] [Google Scholar]
- 83. Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, et al. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep 2017; 7: 10353. doi: 10.1038/s41598-017-10649-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cha KH, Hadjiiski L, Chan HP, Weizer AZ, Alva A, Cohan RH, et al. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep 2017; 7: 8738. doi: 10.1038/s41598-017-09315-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep learning based radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep 2017; 7: 5467. doi: 10.1038/s41598-017-05848-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kumar D, Chung AG, Shaifee MJ, Khalvati F, Haider MA, Wong A. Discovery radiomics for pathologically-proven computed tomography lung cancer prediction : Karray F, Campilho A, Cheriet F, Image analysis and recognition: 14th International Conference, ICIAR 2017, Montreal, QC, Canada, July 5–7, 2017, Proceedings. Cham: The British Institute of Radiology.; 2017. 54–62. [Google Scholar]
- 87. Jochems A, Hoebers F, De Ruysscher D, Leijenaar R, Walsh S, O'Sullivan B, et al. EP-1605: Deep learning of radiomics features for survival prediction in NSCLC and Head and Neck carcinoma. Radiotherapy and Oncology 123: S866. [Google Scholar]
- 88. Vickers AJ. Prediction models: revolutionary in principle, but do they do more good than harm? J Clin Oncol 2011; 29: 2951–2. doi: 10.1200/JCO.2011.36.1329 [DOI] [PubMed] [Google Scholar]
- 89. Lustberg T, van Soest J, Jochems A, Deist T, van Wijk Y, Walsh S, et al. Big Data in radiation therapy: challenges and opportunities. Br J Radiol 2017; 90: 20160689. doi: 10.1259/bjr.20160689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Jochems A, Deist TM, van Soest J, Eble M, Bulens P, Coucke P, et al. Distributed learning: developing a predictive model based on data from multiple hospitals without data leaving the hospital - a real life proof of concept. Radiother Oncol 2016; 121: 459–67. doi: 10.1016/j.radonc.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 91. Jochems A, Deist TM, Naqa E I, Kessler M, Mayo C, Reeves J, et al. Developing and validating a survival prediction model for NSCLC patients through distributed learning across three countries. International Journal of Radiation Oncology*Biology*Physics. 2017;. [DOI] [PMC free article] [PubMed]
- 92. Deist TM, Jochems A, van Soest J, Nalbantov G, Oberije C, Walsh S, et al. Infrastructure and distributed learning methodology for privacy-preserving multi-centric rapid learning health care: euroCAT. Clin Transl Radiat Oncol 2017; 4: 24–31. doi: 10.1016/j.ctro.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]