Abstract
We present the development, optimization, and application of constructs, cell lines, covalent crosslinking methods, and immunoprecipitation strategies that enable robust and accurate determination of collagen interactomes via mass spectrometry-based proteomics. Using collagen type-I as an example, protocols for working with large, repetitive, and GC-rich collagen genes are described, followed by strategies for engineering cells that stably and inducibly express antibody epitope-tagged collagen-I. Detailed steps to optimize collagen interactome crosslinking and perform immunoprecipitations are then presented. We conclude with a discussion of methods to elute collagen interactomes and prepare samples for mass spectrometry-mediated identification of interactors. Throughout, caveats and potential problems researchers may encounter when working with collagen are discussed. We note that the protocols presented herein may be readily adapted to define interactomes of other collagen types, as well as to determine comparative interactomes of normal and disease-causing collagen variants using quantitative isotopic labeling (SILAC)- or isobaric mass tags (iTRAQ or TMT)-based mass spectrometry analysis.
Keywords: Collagen proteostasis network, co-immunoprecipitation, proteomics, crosslinking, mass spectrometry
1. Introduction
Collagen is the molecular scaffold for animal life [1,2]. In addition to forming the primary proteinaceous component of bone, skin, cartilage, basement membranes, and other tissues, collagen also facilitates diverse phenomena such as cell–cell signaling, wound healing, and cell migration [3]. Proper execution of the folding, modification, and quality control processes required for biogenesis of this complex protein is, therefore, critical [4,5]. Collagen proteostasis, encompassing all of these factors, can be disrupted by mutations in collagen molecules themselves or by dysfunction of the proteostasis network [6–9], which is the integrated system of intracellular chaperones, quality control factors, and trafficking mechanisms that addresses cellular protein production challenges [10,11]. The hierarchical nature of collagenous matrices allows the resulting defects in individual collagen molecules to propagate to the level of tissues [12,13], engendering a broad category of disease known as the collagenopathies [7,14]. Current therapies remain inadequate for alleviating pathologic manifestations of these diseases, owing in part to an incomplete understanding of collagen biogenesis.
Developing a molecular-level understanding of intracellular collagen folding, processing, assembly, and quality control requires that we first identify the players. Unfortunately, the lack of an amenable collagen-I expressing cell model system, the absence of high quality immunoprecipitation (IP)-grade collagen antibodies, and the challenges associated with performing molecular biology on a lengthy, GC-rich, and highly repetitive gene had, prior to our work [15], precluded systematic study of the collagen proteostasis network. After addressing practical problems associated with handling collagen genes, we generated panels of fibrosarcoma and osteosarcoma cell lines that inducibly express collagen variants tagged with distinct antibody epitopes to overcome these roadblocks. Covalent crosslinking and selective immunoprecipitation of collagen-I from these cells, followed by mass spectrometry-based proteomic analysis (Figure 1), yielded the first unbiased map of the wild-type collagen-I proteostasis network (Table 1). Our strategy robustly identified virtually all previously known collagen-I interactors, as well as ~30 putative new players in collagen-I biogenesis. The novel interactors we identified encompass a wide range of proteostasis-relevant endoplasmic reticulum proteins, including cochaperones, oxidoreductases, and trafficking mechanisms. We validated a number of the newly identified players in collagen-I proteostasis by immunoprecipitations and genetic knockdowns in osteoblast-like cells that natively produce collagen-I. We also discovered a novel collagen-I post-translational modification, aspartyl hydroxylation, via these interactome studies.
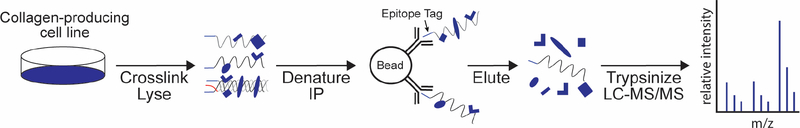
Figure 1:
Schematic representation of collagen interactomics workflow.
Table 1:
Selected wild-type collagen-I interactors reproducibly identified by mass spectrometry [15].
| Protein Name | Gene Name | Total Peptides | Unique Peptides |
|---|---|---|---|
| Collagen α1(I) chain | Col1A1 | 915 | 63 |
| Protein disulfide isomerase | P4HB | 129 | 20 |
| Prolyl 4-hydroxylase subunit α2 | P4HA2 | 69 | 15 |
| Prolyl 4-hydroxylase subunit α1 | P4HA1 | 68 | 15 |
| 78 kDa glucose-regulated protein (BiP/GRP78) | HSPA5 | 56 | 15 |
| Collagen α2(I) chain | Col1A2 | 42 | 13 |
| Endoplasmin (GRP94) | HSP90B1 | 35 | 12 |
| Prolyl 3-hydroxylase 1 | LEPRE1 | 25 | 9 |
| Peptidyl-prolyl cis-trans isomerase FKBP10 | FKBP10 | 19 | 8 |
| Prelamin-A/C | LMNA | 14 | 8 |
| Transitional endoplasmic reticulum ATPase | VCP | 13 | 8 |
| Protein disulfide isomerase A3 | PDIA3 | 16 | 7 |
| Fibronectin | FN1 | 11 | 7 |
| Cytoskeleton-associated protein 4 | CKAP4 | 10 | 7 |
| Peptidyl-prolyl cis-trans isomerase B | PPIB | 25 | 6 |
| Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | PLOD2 | 20 | 6 |
| Procollagen-lysine,2-oxoglutarate-5-dioxygenase 3 | PLOD3 | 17 | 6 |
| Annexin A2 | ANXA2 | 15 | 6 |
| C-type mannose receptor 2 | MRC2 | 12 | 6 |
| Procollagen-lysine,2-oxoglutarate-5-dioxygenase 1 | PLOD1 | 16 | 5 |
| Cartilage-associated protein | CRTAP | 14 | 5 |
| Protein disulfide isomerase A6 | PDIA6 | 12 | 5 |
| Serpin H1 (HSP47) | SERPINH1 | 10 | 5 |
| Peptidyl-prolyl cis-trans isomerase FKBP9 | FKBP9 | 12 | 4 |
| Protein disulfide isomerase A4 | PDIA4 | 8 | 4 |
| Galectin-3-binding protein | LGALS3BP | 7 | 4 |
| Peroxiredoxin-1 | PRDX1 | 5 | 3 |
| Peptidyl-prolyl cis-trans isomerase A | PPIA | 5 | 3 |
| Endoplasmic reticulum resident protein 44 | ERP44 | 5 | 3 |
| Calreticulin | CALR | 5 | 3 |
| Golgi apparatus protein-1 | GLG1 | 4 | 3 |
| Thioredoxin domain-containing protein 5 | TXNDC5 | 4 | 3 |
| Hypoxia up-regulated protein 1 | HϒOU1 | 3 | 3 |
| Calnexin | CANX | 5 | 2 |
| Polyubiquitin-C | UBC | 5 | 2 |
| Reticulocalbin-1 | RCN1 | 5 | 2 |
| Endoplasmic reticulum resident protein 29 | ERP29 | 4 | 2 |
| Golgi integral membrane protein 4 | GOLIM4 | 4 | 2 |
| Thioredoxin domain-containing protein 12 | TXNDC12 | 3 | 2 |
| Calumenin | CALU | 3 | 2 |
| DnaJ homolog subfamily B member 11 | DNAJB11 | 2 | 2 |
| Peptidyl-prolyl cis-trans isomerase FKBP4 | FKBP4 | 2 | 2 |
| Reticulocalbin-3 | RCN3 | 2 | 2 |
| Thioredoxin-related transmembrane protein 1 | TMX1 | 2 | 2 |
Using our work with wild-type collagen-I as a template for the description of a generalizable strategy, we present a detailed protocol to map collagen interactomes, including approaches for manipulating collagen genes, developing stable cell lines expressing antibody epitope-tagged collagens, optimizing and validating mass spectrometry-grade immunoprecipitations and covalent crosslinking reactions, preparing samples for proteomic analysis, and performing mass spectrometry experiments. The protocol should prove useful not just for studying wild-type collagen-I, but also for quantitatively evaluating how cells engage misfolding, disease-causing collagen-I variants. Moreover, the protocol is readily adaptable to studying the interactome of any of the other twenty-seven types of collagen [16].
2. Materials
2.1. Molecular biology and cell culture
Genes, vectors, and DNA oligomers: Human Col1A1 and Col1A2 genes (Origene), pENTR1A vector (Thermo Fisher Scientific), pTRE-Tight vector (Clontech), pLenti.CMV/TO.DEST vectors with assorted resistance cassettes (Addgene) [17], puromycin and hygromycin linear selection markers (Clontech), DNA primers (Sigma), and RRE, REV, and VSVG vectors for lentivirus production (Addgene).
Enzymes: Q5 Polymerase, BamHI, EcoRV, NotI, T4 ligase, and Antarctic phosphatase (New England BioLabs; NEB) and LR Clonase (Thermo Fisher Scientific).
Omega Gel Purification Kit (Omega BioTech).
Bacterial cell lines: Sure2 E. coli (Agilent) and DH5α electrocompetent and Stbl3 competent E. coli (Thermo Fisher Scientific).
Human cell lines: Human bone osteosarcoma Saos-2 cells (ATCC), human fibrosarcoma HT-1080 Tet-Off cells that constitutively express the tetracycline (Tet) transactivator (Clontech), and HEK293FT cells (Thermo Fisher Scientific).
Cell culture: Dulbecco’s Modified Eagle Medium (DMEM), Minimum Essential Medium (MEM), fetal bovine serum (FBS), and trypsin-EDTA (all from Corning Cellgro), tetracycline-free FBS (Clontech), and Opti-MEM media (Thermo Fisher Scientific),
Transfection reagents: XFect (Clontech) and Lipofectamine 2000 (Thermo Fisher Scientific).
Antibiotics: Penicillin and streptomycin (Thermo Fisher Scientific), puromycin (Corning Cellgro), hygromycin (Enzo Life Sciences), and blasticidin (Sigma).
Lenti-X GoStix (Takara).
Chemicals: Doxycycline hyclate, L-glutamine, poly-D-lysine, and polybrene (Sigma).
Antibodies: anti-HSP47 (ADI-SPA-470; Enzo Life Sciences), anti-HA (sc-7392; Santa Cruz), anti-PDI (sc-20132; Santa Cruz), anti-Colα1(I) (LF68; Kerafast), anti-Colα2(I) (SAB4500363; Sigma), and secondary antibodies 800CW goat anti-rabbit, 800CW goat anti-mouse, 680LT goat anti-rabbit, and 680LT goat anti-mouse (LiCor Biosciences).
2.2. Preparation of protein samples, immunoprecipitation, protein elution and precipitation
All chemicals and supplies used in these protocol steps must be mass spectrometry-grade.
RNase/DNase-free microcentrifuge tubes employed can be from a variety of vendors, but should not be autoclaved prior to use.
Radioimmunoprecipitation assay (RIPA) buffer: 50 mM Tris (VWR), 0.5% sodium deoxycholate (Alfa Aesar), 1% Triton (INTEGRA Chemical), 0.1% sodium dodecyl sulfate (SDS; Alfa Aesar), 50 mM NaF (Alfa Aesar), 5 mM β-glycerol phosphate (Sigma), 1 mM sodium metavanadate (Sigma), 5 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA; STREM Chemicals), 5 mM ethylenediaminetetraacetic acid (EDTA; Aqua Solutions), 100 μM sodium pyruvate (Sigma), 1 mM phenylmethylsulfonyl fluoride (PMSF; Thermo Fisher Scientific), and protease inhibitor mixture (Pierce) at pH 7.4 in dd-H2O.
Lysis buffer: 50 mM Tris, 1% Triton, 1 mM PMSF, 150 mM NaCl (VWR), 1.5 mM MgCl2 (Alfa Aesar), 1 mM EDTA, and protease inhibitor mixture (Biotool) or tablets (Pierce) at pH 7.4.
Phosphate-buffered saline (Corning).
Dithiobis(succinimidyl propionate) (DSP; Sigma).
Ultrasound processor (Cole Parmer), tube rotator (VWR), refrigerated benchtop centrifuge (Thermo Scientific), vortex mixer (Corning).
Anti-HA agarose beads (A2095; Sigma).
Elution buffer: 6% SDS in 1 M aqueous Tris buffer at pH 6.8.
Solvents: MeOH and CHCl3 (Thermo Fisher Scientific).
Vacuum centrifuge dryer (Cole Parmer).
2.3. Preparation of tryptic peptides and mass spectrometry
All chemicals and supplies used in these protocol steps must be mass spectrometry-grade.
RNase/DNase-free microcentrifuge tubes employed can be from a variety of vendors, but should not be autoclaved prior to use.
Chemicals: Iodoacetamide (IAA; Sigma), 1,4-dithiothreitol (DTT; Sigma), and urea and formic acid (Fluka).
Buffers: 100 mM NH4HCO3(aq) at pH 8.0, 0.1% aqueous trifluoroacetic acid (TFA; Sigma), 0.1% aqueous formic acid (Sigma), 0.1% TFA in 9:1 MeCN:H2O (MeCN from Sigma), 0.1% formic acid in 4:1 MeCN:H2O.
Pierce C18 Peptide Desalting Spin Column (Thermo Fisher Scientific).
Aluminum foil (Reynolds).
Water bath at 56 °C.
EASY-nLC 1000 or similar HPLC with autosampler connected to a Thermo-Fisher Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer, or any similar high-resolution, accurate-mass mass spectrometer.
Proteome Discoverer 2.2. with Mascot search engine.
3. Methods
3.1. Creating cellular platforms for biochemical studies of collagen
Stable cell systems are valuable to enable robust and reproducible determination of collagen-I interactomes via an IP/MS-based proteomics approach. In the absence of MS-grade collagen-I antibodies, to permit the necessary IPs we use cloning strategies to introduce short, well-defined HA or FLAG antibody epitopes between an ER-targeting signal sequence and collagen-I’s N-propeptide. More recently, Cas9-based approaches may make it possible to introduce such epitopes directly in endogenous gene loci [18,19]. With necessary genes encoding antibody epitope-tagged collagens in hand, we next engineer cell lines that stably and inducibly express the tagged collagen proteins. As described below, in HT-1080 cells, we used a transfection-based strategy to deliver genes [15]. In Saos-2 cells, we used a lentivirus-based strategy [15,20]. Work in both cell types is described below. Either may be used to introduce genes for other collagens or collagen disease-causing variants into essentially any cell system of interest. An advantage of HT-1080 cells for collagen-I studies is that, although they are capable of synthesizing and secreting various collagen types, they do not express collagen-I and therefore the collagen-I interactome can be readily enriched. On the other hand, Saos-2 cells do endogenously express collagen-I. In such a system, it is more straightforward to mimic autosomal dominant collagen-I pathologies by simply introducing the disease-causing collagen-I variant in a background of endogenous wild-type collagen-I expression. Note that we have found that adenoviruses may also be used for robust transient expression of collagen (not described below, as the focus is on stable cell line development). An adenovirus-based strategy can be especially valuable for transient collagen variant expression in primary cells.
3.1.1. Transfection approach to prepare stable HT-1080 cells inducibly expressing collagen genes
Col1A1 and Col1A2 genes were obtained from Origene prior to transfer into an appropriate expression vector (Figure 2a), as described below (see Note 1).
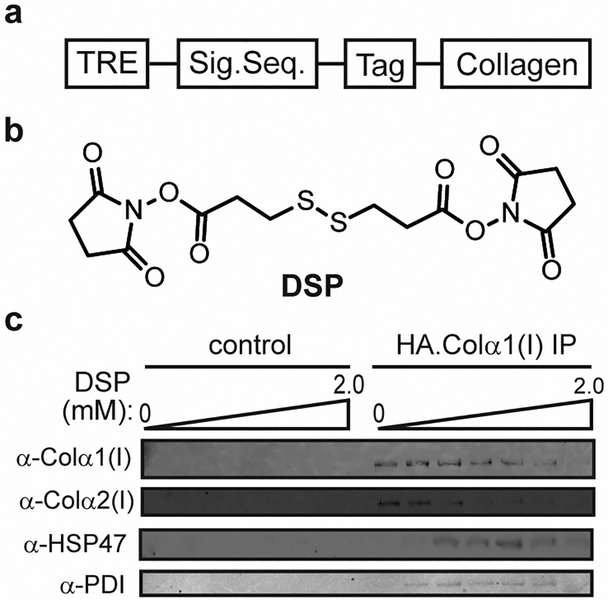
Figure 2:
(a) Generalized schematic for collagen-I expression constructs. Either the endogenous collagen signal sequence or the preprotrypsin signal sequence can be employed. The tag can be HA, FLAG, c-MYC, or any of a number of other short antibody epitope tags. (b) Structure of dithiobis(succinimidyl propionate) (DSP). (c) Optimizing DSP cross-linker concentration by evaluating the coimmunoprecipitation of known collagen-I interactors in the HT-1080 cell system. The control sample represents HT-1080 cells that do not express HA-tagged collagen-I upon induction. This figure was adapted from DiChiara et al [15].
Using appropriate restriction sites (e.g. BamHI and EcoRV), insert a DNA sequence (IDT) encoding the preprotrypsin signal sequence (PPT; to target collagen strands to the secretory pathway), the desired epitope tag (e.g., HA or FLAG), and a NotI restriction digestion site (i.e., PPT.HA.NotI or PPT.FLAG.NotI) into a pTRE-Tight vector (see Notes 2 and 3).
- PCR-amplify (using the primers shown below which remove the native signal sequence) collagen-I genes over 35–40 PCR cycles. For amplification of collagen-I genes, we recommend the use of Q5 Polymerase with the addition of the High GC Enhancer (protocol from NEB) and a 2 min extension cycle (see Note 4).
-
Col1A1: For. 5’-ACATCAGCGGCCGCACAAGAGGAAGGCCAAGTCGAG-3’Rev. 5’- AAAAAAGTCGACTTACAGGAAGCAGACAGGG-3’
-
Col1A2: For. 5’-AAAAAAGCGGCCGCAACATGCCAATCTTTACAAGAGGAAAC-3’Rev. 5’-AAAAAAGATATCTTATTTGAAACAGACTGGGCCAATG-3’
-
Restriction digest amplicons using the NotI and EcoRV restriction enzymes (protocol from NEB).
Gel purify samples using Omega Gel Purification Kits.
Ligate into digested pTRE-Tight vectors prepared in Step 3.1.1.1 using T4 ligase (protocol from NEB) followed by transformation into competent E. coli (see Note 5).
Transfect antibody epitope-tagged, tet-regulated collagen-I expression vectors using XFect into HT-1080 Tet-Off cells that constitutively express the tetracycline transactivator along with either a puromycin or hygromycin linear selection marker (protocol from Clontech). Co-transfection with a linear selection marker at a fixed ratio of 25:1 (5 μg : 0.2 μg) reduces the probability of a cell being transfected with the linear selection marker alone, potentially conferring resistance without integration of the collagen gene.
Change the media 18–24 h post-transfection.
48 h post-transfection, treat cells with antibiotic (experimentally optimized to be 0.25 μg/mL puromycin or 150 μg/mL hygromycin) for 10–12 days (changing the media every 2 days) for stable selection.
Amplify heterostable cell lines, and then split to establish genetically homogenous single colonies to analyze for collagen-I expression and secretion (Figure 2a). Repeat above process to create cell lines that express both Col1A1 and Col1A2 or other collagen variants, as desired (see Note 6).
3.1.2. Lentiviral approach to prepare stable Saos-2 cells inducibly expressing collagen genes
Construct pENTR1A vectors encoding the Col1A1 and Col1A2 genes by ligating Col1A1 or Col1A2 extracted from pTRE.Tight vectors (described above) using the NotI and EcoRV restriction enzymes into a pENTR1A vector already encoding an ER-targeting PPT signal sequence and an HA antibody epitope tag (or other desired tag; see Note 7). All pENTR1A vectors must be sequence-confirmed before proceeding.
Introduce the resulting construct into the appropriate pLenti.CMV/TO.DEST Gateway destination vector via LR Clonase-mediated recombination (protocol from Thermo Fisher Scientific). The resulting pLenti vectors must be sequence-confirmed before proceeding to lentiviral production.
Produce lentivirus using the third-generation system by co-transfecting HEK293FT cells with the lentiviral plasmids and packaging vectors encoding RRE, REV, and VSVG using Lipofectamine 2000. Briefly, (1) incubate the plasmid mixture (pLenti vector, 15 μg; RRE, 15 μg; REV, 6 μg; and VSVG, 3 μg) with 60 μL of Lipofectamine 2000 in 3 mL of Opti-MEM media for 45 min at rt; (2) add the resulting mixture dropwise to a 10 cm dish of HEK293FT cells (~8 × 106 cells plated on poly-D-lysine-coated plates ~12 h pre-transfection) and incubate at 37 °C for 12 h; (3) remove and replace media with DMEM (6 mL) and incubate the plates for another 36 h, monitoring for successful production of lentivirus using Lenti-X GoStix (protocol from Takara); (4) collect viral supernatant 48 h post-transfection, and (5) use supernatant immediately for transductions of Saos-2-TREx cells.
Transduce Saos-2TREx (see Note 8) cells with a range of titers of crude viral supernatants of Tet-responsive HA-collagen-α1(I). Add polybrene to the viral supernatant (final concentration of 4 μg/mL during transductions) to increase transduction efficiency.
Select heterostable cell lines using 250 μg/mL hygromycin or other appropriate selection agent and isolate single colonies (Figure 2a; see Note 9).
Assay for HA-collagen-α1(I) expression in isolated single colonies using immunoblotting.
3.2. Immunoisolation of the collagen-I interactome for proteomic analysis
The protocol below uses Saos-2TREx cells inducibly expressing PPT.HA.Colα1(I) (Saos-2Colα1(I) cells) as an example, but can be adapted for essentially any cell type of interest. Note that a control IP should also always be performed on cells not expressing the antibody epitope-tagged collagen.
Plate Saos-2Colα1(I) cells (~6 × 106) in 10 cm dishes with 10 mL of DMEM supplemented with 15% heat-inactivated FBS, penicillin (100 IU/mL), streptomycin (100 μg/mL), hygromycin (150 μg/mL), and 2 mM L-glutamine. Maintain cells as a monolayer overnight at 37 °C in a humidified atmosphere of 5% CO2(g).
Induce HA-tagged wild-type collagen-I expression by overnight incubation with media containing 4 μg/mL doxycycline (see Note 10) in the presence of 50 μM ascorbate, a required co-factor for proper collagen hydroxylation and for induction of synthesis and secretion of the endogenous, untagged collagen-I (see Note 11).
After 48 h, remove media, wash with PBS, and add 1 mL of trypsin-EDTA for 5 min. Neutralize trypsin by adding 7 mL of complete DMEM. Pellet cells by spinning at 1000 × g for 5 min.
Remove media and wash cells twice with PBS at rt.
Resuspend cell pellet in a 15 mL Falcon tube containing 10 mL of PBS and an experimentally optimized concentration of DSP (75 μM for Saos-2Colα1(I) cells; Figure 2b). Rotate tubes for 30 min at rt to allow the crosslinking to proceed (see Note 12).
Quench excess DSP by adding 1 mL of 1 M aqueous Tris at pH 8 and rotating for 10–15 min at rt.
Pellet cells by centrifuging at 1000 × g for 5 min at 4 °C, followed by removal of the supernatant.
Lyse cells by resuspending them in 1 mL of RIPA buffer at 4 °C. Vortex vigorously for 30 s, then immediately sonicate (130 Watt, 20 kHz, 70% amplitude) for 6 cycles of one 15 s pulse each, returning tube to ice for 15 s between pulse cycles. Lysates should be clear and fluid after sonication (see Note 13).
Post-sonicating, place the lysate solution on ice for 30 min to ensure compete lysis.
Transfer cell lysates to 1.7 mL microcentrifuge tubes and spin for 15 min (21,100 × g) at 4 °C.
Transfer to new 1.7 mL tubes. Lysates may be stored at −80 °C at this stage, if desired.
Add 50 μL of HA bead slurry (25 μL of dried beads) to 1 mL of lysate solution (see Note 14) and incubate the sample overnight on a tube roller at 4 °C (see Note 15).
Pellet the beads at 2000 × g for 5 min at 4 °C.
Wash beads 4× with 1 mL RIPA buffer to remove non-specific binders, rotating 5 min at 4 °C during each wash.
Pellet beads at 2000 × g for 5 min at 4 °C after the last wash and remove the supernatant (see Note 16).
3.3. Collagen interactome elution and precipitation
To enhance the sensitivity of proteomic detection by minimizing the presence of any eluted IgG, we elute the proteins from beads using a non-reducing, denaturing buffer.
Elute pelleted beads using 50 μL of 6% SDS in 1 M aqueous Tris buffer at pH 6.8 by heating to 100 °C for 15 min (mix by shaking every 5 min during heating). Do not include a reducing agent during the elution, to minimize release of any antibody fragments via disulfide reduction (see Note 17).
Spin down at 2,000 × g for 5 min, carefully collecting the supernatant and avoiding uptake of any beads.
Repeat steps 1 and 2 above to elute a second time. Combine all supernatants.
Add 450 μL MeOH to microcentrifuge tubes containing 90% of the supernatant (see Note 18). Vortex vigorously for 30 s.
Add 150 μL CHCl3. Vortex vigorously for 30 s.
Add 450 μL H2O. Vortex vigorously 3 × 30 s. Spin down at 10,000 × g for 2 min, rotate the tube 180°, and spin down again at 10,000 × g for 2 min (see Note 19).
Discard all but ~100 μL of the upper layer of supernatant, being careful not to disturb the thin, white, protein-containing film at the interface between the H2O/MeOH upper layer and the CHCl3 lower layer.
Add 500 μL MeOH. Vortex vigorously for 30 s. White protein particles should be observable. Spin down at 16,300 × g for 2 min, followed by rotating the tube 180° and spin down again at 16,300 × g for 2 min.
Repeat step 3.3.8 3× to remove all residual SDS, which would otherwise interfere with MS analysis.
Use a speed-vacuum to dry the pellet. Samples can then be maintained at −20 °C until use.
3.4. Protein digestion and mass spectrometry-based proteomic studies
3.4.1. Trypsin digestion of protein pellets and preparation of desalted peptides for mass spectrometry analysis
All procedures should be carried out at rt unless otherwise specified.
Add 22.5 μL of 8 M aqueous urea to the precipitated protein pellet from Step 3.3.10. Vortex vigorously 3 × 30 s. Centrifuge at 3000 × g for 1 min.
Add 2.5 μL of 100 mM DTT dissolved in 100 mM NH4HCO3(aq) at pH 8 to yield a final concentration of 10 mM DTT.
Incubate at 56 °C for 1 h.
Cool samples to rt and centrifuge at 3000 × g for 1 min to collect any H2O condensed on the internal surface of the tube.
Add 2.75 μL of 550 mM iodoacetamide to yield a final concentration of 55 mM iodoacetamide. Wrap tubes in foil to protect from light and mix end-over-end for 45 min (see Note 20).
Add 169.75 μL of 100 mM NH4HCO3(aq) at pH 8. Vortex briefly and then add 2.5 μL of sequencing grade modified trypsin (0.4 μg/μL) to yield a final volume of 200 μL (see Note 21).
Mix end-over-end at rt for 12 h or for 3 h at 37 °C with agitation on a VWR thermomixer with hot plate.
Acidify the sample by adding 11 μL of 98% formic acid to achieve a final concentration of 5% formic acid.
Remove white tip from the bottom of the peptide desalting spin column. Place column into a 2 mL microcentrifuge tube. Centrifuge at 3000 × g for 1 min and discard flow-through.
Wash the column with 300 μL of 0.1% TFA in 90:10 MeCN:H2O, centrifuge at 3000 × g for 1 min, and discard flow through (see Note 22). Repeat once.
Wash the column with 300 μL 0.1% aqueous TFA, centrifuge at 3000 × g for 1 min, and discard flow through. Repeat once.
Add sample to the column, centrifuge at 3000 × g for 3 min, and discard flow through. Repeat centrifugation if not all of the sample passed through the column (see Note 23).
Wash the sample on the column by adding 50 μL of 0.1% aqueous formic acid. Centrifuge at 500 × g for 3 min. Repeat twice.
Place column into a fresh microcentrifuge tube and elute the peptides from the column by adding 50 μL of 0.1% formic acid in 80:20 MeCN:H2O. Centrifuge at 3000 × g for 3 min. Repeat this elution step once.
Evaporate solvent in a vacuum centrifuge until near dryness. Store at −20 °C until mass spectrometry analysis.
3.4.2. Mass spectrometry and data analysis
Solubilize digested peptide mixture into a working volume (we typically use 30 μL) of HPLC-grade 0.1% aqueous formic acid by vortexing 3× for 30 s each.
Inject samples (typically 1–5 μL volumes are reasonable) into a C18 reversed-phase resin (3 μm resin; 120 Å pore size) column (15 cm in length; 50 μm in diameter) on an EASY-nLC 1000 nanopump system connected to a Thermo Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer. Elute peptides into the mass spectrometer for analysis over a 140-min gradient of 0.1% aqueous formic acid solution ranging to a 95:5 MeCN:H2O in 0.1% formic acid solution (see Notes 24 and 25).
Operate the mass spectrometer in data-dependent mode with a full scan MS spectrum followed by MS/MS for the top 10 precursor ions in each cycle. Depending on instrumentation, the number of precursor ions selected in each cycle may be increased.
Analyze raw data using Proteome Discoverer 2.2, performing a database search using Mascot for protein identification [21]. Depending on instrumentation and study objectives, search parameters and data analysis used may vary. For qualitative mapping of the collagen-I interactome in these example cellular systems, Mascot search parameters typically used were as follows: mass tolerance for precursor ions = 10 ppm; fragment ion mass tolerance = 15 mmu; missed cleavages of trypsin = 2; fixed modifications were carbamidomethylation of cysteine; variable modifications were methionine oxidation and hydroxylation of proline, lysine, aspartate, and/or asparagine. Peptides with unambiguous peptide spectrum match, Mascot scores ≥ 25, and isolation inference ≤ 30 were considered identified, resulting in an average false discovery rate of 0.0077 (see Notes 26 and 27).
4. Notes
Sequencing of numerous collagen genes received from commercial sources and other investigators revealed a number of missense mutations as well as, in some cases, large or small deletions of gene segments. We strongly recommend full sequencing of any collagen vector obtained from any source, and correction of defects using site-directed mutagenesis or other approaches. We also recommend complete sequencing of collagen genes after any manipulation, including even simple cut-and-paste reactions, not just PCR amplifications.
We incorporated an HA- or FLAG-epitope tag downstream of PPT but otherwise at the N-terminus of collagen strands to enable straightforward, orthogonal immunoblotting and IPs of collagen-I polypeptides. Other antibody epitopes such as cMyc, 3×FLAG, cleavable tags, etc. can also be used depending on the purpose of the study. The proteomic studies presented here use the HA epitope as an example. We note that, because collagen folding begins at the C-terminus, placement of these small tags at the N-terminus is unlikely to disrupt folding and assembly, which we confirmed for collagen-I via a variety of biochemical assays [15]. Note also that the N-propeptide and thus the epitope-tag will be cleaved extracellularly by N-propeptidases, depending on the cell system used.
Tet-inducible vectors were chosen to inducibly control the expression of collagen in engineered stable cell lines. Inducible expression minimizes potential negative selection pressure placed on cells forced to continuously express ectopic collagen that might render continuous propagation of stable cell lines challenging.
Each collagen-I gene is highly repetitive in the triple-helical domain and has about 65% GC-content overall, with local regions containing up to 85% GC content in a 50 base-pair span. These factors render PCR amplification of collagen genes problematic, resulting in low yields and many small PCR fragments, likely owing to incomplete gene synthesis. We found that the yield of full-length collagen-I genes was slightly boosted by increasing the recommended final extension time by about 1.25-fold.
Note that because each collagen-I gene is over 4000 base pairs in length, more than double the size of the pTRE-Tight expression vector, an inverted molar ratio is required when ligating the two linear pieces of DNA (with the insert more abundant than the vector). Even with inverted ratios, the most common ligation product observed was the pTRE-Tight vector ends being ligated together, despite the use of incompatible NotI and EcoRV restriction enzymes to digest the vector. Sequencing confirmed that, in a rare event, the sticky end of the NotI site was destroyed, leaving a blunt end of DNA that ligated to the EcoRV blunt end. This rare intramolecular ligation was more favorable and occurred more frequently than insertion of the collagen-I genes. To avoid this problem, dephosphorylation of the vector backbone with Antarctic phosphatase prior to ligating the two pieces of DNA proved useful in some cases for successful insertion of collagen genes. We ultimately succeeded in ligating pTRE-Tight and the collagen-I PCR amplicons, confirmed by sequencing the resulting clones with a primer that binds to the vector, providing evidence for the presence of both the 5’-and the 3’-ends of the collagen-I genes. However, sequencing of the full gene revealed that an apparent recombination event had removed > 1000 base pairs of the gene. Thereafter, we transformed collagen gene-containing vectors into recombination-deficient Sure2 E. coli cells. When grown at 30 °C, recombination is effectively shut down in these cells, allowing for the synthesis and propagation of intact collagen-I genes. We recommend the use of these recombination-deficient cells at 30 °C for work with collagen genes whenever possible.
Expression of the tetracycline transactivator allows doxycycline (dox)-inducible expression of the collagen-I genes of interest. We optimized HT-1080 culture conditions by varying the concentration of dox for 48 h, and then split each concentration into two conditions: continued culture in the same concentration of dox or no dox in Tet-free FBS containing media. We found that continuous culture in 1 ng/mL dox, 1000-fold lower than the manufacturer’s recommended concentration, efficiently suppresses both collagen-I genes. Removal of the dox-containing media and replacement with dox-free media induces both genes and the collagen proteins can be detected by immunoblotting in both cell lysates and media. Continuous culture in higher dox concentrations made it difficult to induce collagen-I expression via a media change, and so we recommend the use of 1 ng/mL dox during continuous culture.
Because we found the ligation of collagen genes into pENTR1A vectors to be extremely slow and inefficient, we used electroporation instead of heat shock to enhance transformation efficiency of the ligated vectors.
Expression of the tetracycline repressor in Saos-2-TREx cells allows doxycycline (dox)-inducible expression of the collagen-I gene under control of the tet-operon (in contrast to dox-repressed expression in Tet-Off cells like the HT-1080s employed above). To create Saos-2TREx cells, Saos-2 cells were stably transduced with a lentivirus encoding a constitutively expressed tetracycline repressor protein and a blasticidin resistance cassette. Stable cells were selected using 2 μg/mL blasticidin, and single colonies were isolated and assayed for the colony most responsive to dox treatment (using Tet-responsive CFP to visualize inducible control of the tetracycline repressor protein).
The production of single colony cell lines from stably transduced Saos-2 cells is challenging. Heterogeneous cells must be plated at a very low density (~1:10,000) in 10 cm dishes for 2–3 weeks prior to picking and expanding colonies.
The dox concentration for collagen induction should be separately optimized for each cell system. Note that high concentrations (≥ 10 μg/mL) can be toxic.
Depending on the time course used for collagen induction, ascorbate should be refreshed every 24 h owing to its poor stability.
Transiently interacting chaperones, collagen modifying enzymes, and related biogenesis factors are often not observable via traditional IP workflows. Indeed, we find that covalent crosslinking is essential to co-immunoprecipitate even most of the well-established collagen-I interactors, such as protein disulfide isomerase and HSP47. DSP has dual NHS esters (see DSP structure in Figure 2b), thus targeting nearby primary amines from lysine residues in protein complexes and forming a conjugated network of closely interacting proteins [22,23]. DSP also has a disulfide bond that can be reduced to release cross-linked proteins. Generally, the choice and length of chemical cross-linkers used will impact the quality and the relevance of data produced. The optimal choice of reactive functional groups and the length of cross-linkers varies based on the objectives of a given study [24,25]. The optimal DSP concentration is cell line- and collagen expression-level dependent and therefore must be experimentally optimized to maximally immunoprecipitate genuine interactors while minimizing non-specific interactions. For HT-1080 cells, we used 150 μM DSP. For Saos-2 cells we used 75 μM DSP. To optimize the cross-linker concentration for immunoprecipitations (Figure 2c), we recommend using different concentrations ranging from 0–2 mM, and then choosing the lowest DSP concentration that yields excellent signal for HA-collagen and also allows detection of known, well-established collagen-I proteostasis network components such as PDI and HSP47. The optimal concentration is typically also the lowest DSP concentration at which the cell pellet becomes slightly viscous in the Falcon tube, providing a rough visual parameter to assess crosslinking success. If co-immunoprecipitations without covalent crosslinking are performed, a different protocol from that described in this protocol is required, employing much milder, non-denaturing buffers and gentler treatments of cell lysates – both of which can lead to high background owing to non-specific protein binding.
Likely owing to a small amount of residual DSP activity even after Tris quenching, the cell pellet must be lysed immediately with RIPA buffer to avoid the formation of large cell aggregates. Another option to avoid formation of large cell aggregates is to wash the cells repeatedly with PBS post-crosslinking.
Because RIPA buffer contains SDS, which interferes with standard protein concentration assays, we typically do not measure the protein concentration and instead assume that the protein concentration in the same volume of lysate from the same number of cells is the same.
Optional: Lysates can be precleared using 50 μL of packed sepharose or agarose beads for 1 h at 4 °C. The preclear step can help to minimize non-specific binding of proteins to beads.
If the immunoprecipitated samples will not be used immediately, we have found that dried beads can be stored at −20 °C for at least a few weeks without compromising sample integrity. However, we find that it is essential to first fully remove any residual RIPA buffer, which can be achieved using an additional wash step with a standard lysis buffer (e.g., 50 mM Tris, 1% Triton, 1 mM PMSF, and protease inhibitor mixture at pH 7.4).
Instead of SDS-elution, which is our recommended approach, proteins can alternatively be directly digested on the beads after the immunoprecipitation to prepare for proteomic analysis. This approach is less time-consuming and, in addition, because it skips elution and protein precipitation steps, the results can be somewhat less variable. However, a significant issue is that the HA antibody, which is chemically conjugated to the bead, will also be digested to some extent and the resulting abundant IgG tryptic peptides in the final peptide mixture will substantially reduce sensitivity of the MS-based proteomics. Therefore, we recommend the described SDS elution procedure.
To verify successful co-immunoprecipitation of collagen-I and known components of the collagen-I interactome, at this stage and prior to proceeding with any proteomic studies we typically use 10% of the supernatant volume to run a 10% SDS-PAGE gel, followed by transfer to a nitrocellulose membrane and probing with anti-HA, anti-PDI, and anti-HSP47 (see Figure 2c).
Small white droplet inclusions should form and be clearly observed by eye. After spinning and re-equilibration, two layers of solvent should be observed in the tube. Precipitated proteins will form a thin white layer on the surface between the two solvent layers. The thin white layer may stick to the wall of the tube, in which case the spinning should be repeated.
Iodoacetamide is unstable and light-sensitive.
Sequencing grade modified trypsin has methylated lysine residues to minimize autoproteolysis during the digestion step.
We recommend switching to a fresh microcentrifuge tube after every wash.
Optional: Reload flow through onto the column and centrifuge again to maximize the binding of peptides to the column.
Other instruments may be used depending on availability. Note that the solvent gradient and method setup were optimized for the instrument listed.
We have observed that in a typical experiment the proportion of tryptic peptides derived from collagen is > 50% of the total detected peptides. In some cases, to ensure deeper coverage of the interactome, we remove high levels of collagen-I proteins after elution from the beads via the following protocol: Treat eluted samples with 100 μM DTT to reduce the DSP crosslinker and release the interactome from the collagen-I bait protein. Pass each sample over a 100 kDa molecular weight cut off filter (Millipore) to remove collagen-I (and other large proteins). Precipitate the filtrates as in Section 3.3.1 and proceed. Another approach to ensure deeper coverage of the interactome is to (1) run the protein mixture that was eluted from bead on an SDS-PAGE gel, (2) silver stain the gel, (3) cut out the collagen band in the gel, and (4) digest the other bands using trypsin. A disadvantage of both these approaches is that some high molecular weight protein interactors will also be eliminated.
To exclude false positives and non-specific proteins binding to antibody-conjugated beads, control cells expressing HA-tagged CFP in the cytoplasm (or other appropriate control cells) should be used as a control. Proteins identified with high abundance in the control sample should be excluded from the list of collagen interactors. Proteins with only one unique peptide identified should also be excluded.
Tryptic peptides can be labelled with iTRAQ or TMT tags [26,27] before mixing together and injecting into the mass spectrometer if quantitative comparisons of collagen interactomes are desired (for example between wild-type versus disease-causing collagen variants). SILAC proteomics [28] can alternatively be also performed using cells incubated with media containing stable isotope labeling amino acids. See our recent paper for details on using the quantitative SILAC technique in our collagen system [15].
Acknowledgements
We thank Prof. Joseph Genereux (University of California–Riverside) for technical advice. This work was supported by NIH/NIAMS Grants R03AR067503 and R01AR071443 (to M.D.S.). N.-D.D. was supported by Fonds de Recherche du Québec – Santé (FRQS) and Canadian Institutes of Health Research postdoctoral fellowships. A.S.D. was supported by a NIH Ruth L. Kirschstein predoctoral fellowship (F31AR067615). This work was also supported in part by the NIH/NIEHS under award P30-ES002109 and by a Cancer Center Support (core) Grant P30-CA14051 from the NIH/NCI.
References
- 1.Eyre DR (1980) Collagen: Molecular diversity in the body’s protein scaffold. Science 207:1315–1322. doi: 10.1126/science.7355290 [DOI] [PubMed] [Google Scholar]
- 2.Brinckmann J (2005) Collagens at a glance. Top Curr Chem 247:1–6. doi: 10.1007/b103817 [DOI] [Google Scholar]
- 3.Ricard-Blum S (2011) The collagen family. CSHL Perspect Biol 3:a004978. doi: 10.1101/cshperspect.a004978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoulders MD, Raines RT (2009) Collagen structure and stability. Annu Rev Biochem 78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa Y, Bachinger HP (2013) A molecular ensemble in the rER for procollagen maturation. Biochim Biophys Acta 1833:2479–2491. doi: 10.1016/j.bbamcr.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 6.Morello R, Bertin TK, Chen YQ, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, Bachinger HP, Pace JM, Schwarze U, Byers PH, Weis M, Fernandes RJ, Eyre DR, Yao ZQ, Boyce BF, Lee B (2006) CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127:291–304. doi: 10.1016/j.cell.2006.08.039 [DOI] [PubMed] [Google Scholar]
- 7.Myllyharju J, Kivirikko KI (2001) Collagens and collagen-related diseases. Ann Med 33:7–21. doi: 10.3109/07853890109002055 [DOI] [PubMed] [Google Scholar]
- 8.Barnes AM, Cabral WA, Weis M, Makareeva E, Mertz EL, Leikin S, Eyre D, Trujillo C, Marini JC (2012) Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum Mutat 33:1589–1598. doi:Doi 10.1002/Humu.22139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marini JC, Reich A, Smith SM (2014) Osteogenesis imperfecta due to mutations in non-collagenous genes: lessons in the biology of bone formation. Curr Opin Pediatr 26:500–507. doi:Doi 10.1097/Mop.0000000000000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labbadia J, Morimoto RI (2015) The biology of proteostasis in aging and disease. Annu Rev Biochem 84:435–464. doi: 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475:324–332. doi: 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- 12.Ottani V, Martini D, Franchi M, Ruggeri A, Raspanti M (2002) Hierarchical structures in fibrillar collagens. Micron 33:587–596. doi: 10.1016/S0968-4328(02)00033-1 [DOI] [PubMed] [Google Scholar]
- 13.Han L, Grodzinsky AJ, Ortiz C (2011) Nanomechanics of the cartilage extracellular matrix. Annu Rev Mater Res 41:133–168. doi: 10.1146/annurev-matsci-062910-100431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jobling R, D’Souza R, Baker N, Lara-Corrales I, Mendoza-Londono R, Dupuis L, Savarirayan R, Ala-Kokko L, Kannu P (2014) The collagenopathies: Review of clinical phenotypes and molecular correlations. Curr Rheumatol Rep 16:394. doi: 10.1007/S11926-013-0394-3 [DOI] [PubMed] [Google Scholar]
- 15.DiChiara AS, Taylor RJ, Wong MY, Doan N-D, Del Rosario AM, Shoulders MD (2016) Mapping and exploring the collagen-I proteostasis network. ACS Chem Biol 11:1408–1421. doi: 10.1021/acschembio.5b01083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuttner V, Mack C, Gretzmeier C, Bruckner-Tuderman L, Dengjel J (2014) Loss of collagen VII is associated with reduced transglutaminase 2 abundance and activity. J Invest Dermatol 134:2381–2389. doi: 10.1038/jid.2014.185 [DOI] [PubMed] [Google Scholar]
- 17.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufmann PD (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4:e0006529. doi: 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bressan RB, Dewari PS, Kalantzaki M, Gangoso E, Matjusaitis M, Garcia-Diaz C, Blin C, Grant V, Bulstrode H, Gogolok S, Skarnes WC, Pollard SM (2017) Efficient CRISPR/Cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Development 144:635–648. doi: 10.1242/dev.140855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savic D, Partridge EC, Newberry KM, Smith SB, Meadows SK, Roberts BS, Mackiewicz M, Mendenhall EM, Myers RM (2015) CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res 25:1581–1589. doi: 10.1101/gr.193540.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong MY, Doan N-D, DiChiara AS, Papa LJ 3rd, Cheah JH, Soulen CK, Watson N, Hulleman JD, Shoulders MD (2018) A high-throughput assay for collagen secretion suggests an unanticipated role for Hsp90 in collagen production. Biochemistry 57:2814–2827. doi: 10.1021/acs.biochem.8b00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:Doi [DOI] [PubMed] [Google Scholar]
- 22.Kim KM, Yi EC, Kim Y (2012) Mapping protein receptor-ligand interactions via in vivo chemical crosslinking, affinity purification, and differential mass spectrometry. Methods 56:161–165. doi: 10.1016/j.ymeth.2011.10.013 [DOI] [PubMed] [Google Scholar]
- 23.Corgiat BA, Nordman JC, Kabbani N (2014) Chemical crosslinkers enhance detection of receptor interactomes. Front Pharmacol 4:171. doi: 10.3389/fphar.2013.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitner A (2016) Cross-linking and other structural proteomics techniques: how chemistry is enabling mass spectrometry applications in structural biology. Chem Sci 7:4792–4803. doi: 10.1039/c5sc04196a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holding AN (2015) XL-MS: Protein cross-linking coupled with mass spectrometry. Methods 89:54–63. doi: 10.1016/j.ymeth.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 26.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C (2003) Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75:1895–1904. doi: 10.1021/ac0262560 [DOI] [PubMed] [Google Scholar]
- 27.Unwin RD, Pierce A, Watson RB, Sternberg DW, Whetton AD (2005) Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol Cell Proteomics 4:924–935. doi: 10.1074/mcp.M400193-MCP200 [DOI] [PubMed] [Google Scholar]
- 28.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1:376–386. doi: 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]