Abstract
Post-translational modifications on histone proteins play critical roles in the regulation of chromatin structure and all DNA-templated processes. Accumulating evidence suggests that these covalent modifications can directly alter chromatin structure, or they can modulate activities of chromatin modifying and remodeling factors. Studying these modifications in the context of the nucleosome, the basic subunit of chromatin, is thus of great interest, however, the generation of specifically modified nucleosomes remains challenging. This is especially problematic for most structural biology approaches in which a large amount of material is often needed. Here we discuss the strategies currently available for generation of these substrates. We put a particular focus on novel ideas and discuss challenges in the application to structural biology studies.
Keywords: Histones: The proteins involved in formation of chromatin.; Nucleosome: The basic subunit of chromatin consisting of a core of histones H2A, H2B, H3, and H4 wrapped by ~147 base-pairs of DNA.; Post-translational modification (PTM): Covalent modifications placed on amino acid side-chains after their translation.; Analogue: An engineered modified amino that is roughly (but not totally) equivalent to the native modified amino acid.; Reader domain: A protein domain capable of specifically recognizing the modification state of a histone.
Introduction
The eukaryotic genome is packaged into the cell nucleus in complex with histone proteins to form chromatin. This not only compacts a substantial amount of DNA into the small space of the nucleus, but also provides a mechanism for regulation of the genome. Chromatin structure is dynamically regulated throughout development and the life of the cell. One way in which chromatin structure is modulated is through post-translational modification (PTM) of the histone proteins1,2. These PTMs are thought to either directly alter chromatin structure through modulation of histone-histone or histone-DNA contacts, or to indirectly alter the structure through contributing to the occupancy of, or regulating the activity of chromatin modifying or remodeling factors. A large number of histone PTMs have now been identified, and genome wide are seen to be strongly correlated with particular DNA processes or elements3–5. Moreover, the dysregulation of histone PTMs is associated with a large number of diseases and disorders6,7. Thus, the study of histone PTMs and their regulatory effect is of great interest.
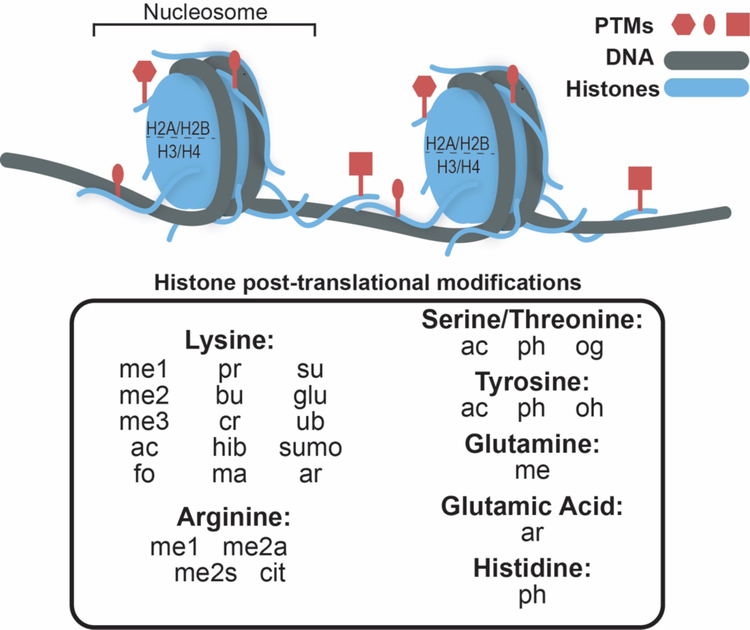
The basic subunit of chromatin is the nucleosome, consisting of an octamer of histone proteins (two each of H2A, H2B, H3, and H4) wrapped by ~147 base pairs of DNA. Protruding from the wrapped core are the N-termini of all of the histones as well as the C-termini of the H2A histones, which are collectively referred to as the histone tails. Modifications are found throughout the histone proteins, but are especially enriched in the histone tails (see Fig. 1). These PTMs are largely reversible, with their placement being catalyzed by enzymes often referred to as writers and their removal being catalyzed by enzymes of known as erasers8. Modified histone tails are a major site for binding of chromatin regulatory factors, and these interactions are mediated through subdomains referred to as reader domains9,10. Histone PTMs display distinctive correlations and anti-correlations genome wide, and are thought to function in combination, expanding their regulatory capacity. This includes the recent discovery that modifications can exist asymmetrically on single nucleosome11, e.g. differential modification of each histone copy. Notably, histone reader domains often exist in multiples within a given chromatin regulatory protein or protein complex. This provides the capacity to interact with chromatin in a multivalent manner, recognizing these patterns of modifications12. This can include multiple modifications on a single histone tail, on multiple tails within a single nucleosome, or on multiple tails spanning distinct nucleosomes.
Figure 1. Histone post-translation modification.
The nucleosome, the basic subunit of chromatin, consists of an octamer of histone proteins H2A, H2B, H3, and H4 (blue) wrapped by ~147 base pairs of DNA (gray). Histone post-translational modifications (red) are enriched in the tail domains. A list of identified histone modifications is shown below. Abbreviations are: monomethylation (me1), di-methylation (me2), tri-methylation (me3), acetylation (ac), formylation (fo), propionylation (pr), butyrylation (bu), crotonylation (cr), 2-hydroxylisobutyrylation (hib), malonylation (ma), succinylation (su), glutarylation (glu), ubiquitylation (ub), sumoylation (sumo), ADP ribosylation (ar), symmetric di-methylation (me2s), asymmetric di-methylation (me2a), citrullination (cit), phosphorylation (ph) and O-GlcNacylation (og), and hydroxylation (oh).
In order to understand the effect of histone modification and mechanisms of readout of PTMs, structural analysis of modified histones and histone binding is key. The most desirable approach to tackle this, is to study these modifications in the context of the nucleosome. This is obviously essential for understanding how modifications in the core of the nucleosome alter its structure, but recent studies have also shown that the nucleosome architecture strongly influences histone tail accessibility to modifications and binding. The binding dynamics and structural basis of interaction of reader domains with modified histone tails has been broadly studied using peptide fragments9,10,13. However, studies with nucleosomes, especially structural studies, has proven to be difficult due to challenges in obtaining modified nucleosomes containing the desired PTMs at high levels of homogeneity. Here we review developed methods for generating designer modified nucleosomes, and discuss the application of these approaches in studying the mechanism of function of histone modifications in the nucleosome context.
Generation of nucleosomes for in vitro studies
The most common strategy for generating nucleosomes for use in biochemical or structural studies is through reconstitution of individually purified fragments14,15. Recombinant histones are purified from E. Coli and refolded into the octamer, which is then reconstituted into the nucleosome using an engineered fragment of DNA. The most commonly utilized DNA sequences are the Widom 601 and a-satellite sequences as they yield strongly positioned nucleosomes. Flanking DNA of variable length can be added outside the core 601 sequence, and di-nucleosomes (or greater numbers) can be generated on a larger template with uniquely spaced positioning repeats, or through ligation of mono-nucleosomes. This reconstitution approach is advantageous as it provides the greatest control of the composition of the nucleosome substrates.
Notably, histones purified from E. Coli are free from any post-translational modification. This is advantageous as the installation of modifications can be carefully controlled. However, this also poses a challenge as the installation process is not trivial. Modification through treatment with enzymes has proven to be suboptimal as the full composition of the enzymatic complexes are often not known or not easily obtained. In addition, they can lead to heterogeneous modification and/or low levels of modified product. Thus, alternative approaches have been developed. These include genetic installation of modified amino acids, native chemical ligation or expressed protein ligation, and modified amino acid analogues. The details of these methods have recently been reviewed extensively16,17. Here we briefly discuss each approach, and then focus on the application in structural biology studies of the nucleosome.
Genetic installation of modified amino acids
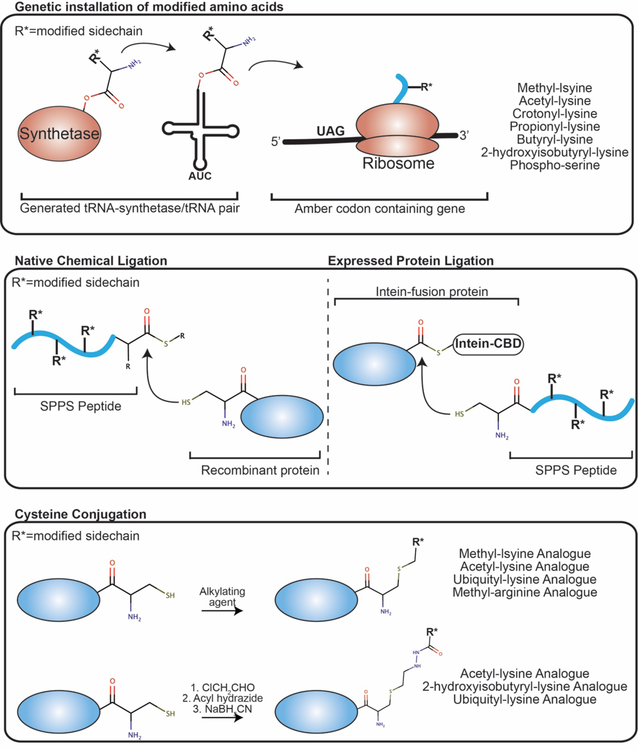
Modified amino acids can be installed in a genetic manner during the expression of a protein in E. Coli through the use of a generated orthogonal tRNA-synthetase/tRNA pair and/or an orthogonal ribosome that incorporates the modified amino acid in response to non-sense codons or four-base frame shift codons18 (see Fig. 2). In certain cases, these have been successfully coupled to incorporate two different non-natural amino acids on the same protein chain19,20. In the case of histones, tRNA-synthetase/tRNA pairs have now been engineered to install acetyl-lysine, crotonyl-lysine, propionyl-lysine, butyryl-lysine, 2-hydroxyisobutyryl-lysine, mono/di/trimethyl-lysine, and phospho-serine21–28. This method is advantageous in that it incorporates the true modified residue. However, the yield of the modified histone as compared to unmodified histones is often substantially lower, limiting its application.
Figure 2. Methods for installation of histone PTMs.
Shown are schematics for (top) genetic installation of modified amino acids, (middle) native chemical ligation and expressed protein ligation, (bottom) cysteine conjugation. Histone components are shown in blue, R* represents the modified side-chain.
Native chemical ligation and expressed protein ligation
Native chemical ligation (NCL) and expressed protein ligation (EPL) entails ligating two or more polypeptides together to generate a full-length protein (see Fig. 2)29–32. Ligation is achieved through fusion of a thioester at the C-terminus of one polypeptide to a cysteine at the N-terminus of another. Generation of fragments through solid phase peptide synthesis (SPPS) allows for incorporation of modifications of choice at any residue. The remaining fragment can be synthesized or generated recombinantly. Recombinant protein with a cysteine at the N-terminus is usually generated through cleavage from a tag using Factor Xa or TEV protease and then fused to the synthesized N-terminal fragment. Alternatively, recombinant protein with a thioester at the C-terminus can be generated through thiolysis from an intein fusion, which is subsequently fused with a synthesized C-terminal fragment30.
This approach works well for generating modified histones, as many modifications of interest are in the N- or C-terminus and histones are cysteine depleted. It is also a very powerful approach because it allows for multiple types of modifications to be installed at multiple residues along the synthesized portion of the histone. Notably, some strides have also been made in total synthesis of histones, which also provides complete control of the modification state33. However, NCL, EPL, and total synthesis can become cost prohibitive when needing to produce these histones in large amounts.
Cysteine conjugation
Generation of analogues is another approach to incorporating modifications (see Fig. 2). There is naturally a dearth of cysteines in histone proteins, with only H3 containing a single cysteine at position 110, which is tolerant to mutation to alanine. Thus, several approaches have been explored for chemical modification of cysteine in order to generate modification analogues at specific positions in histones. Direct alkylation of cysteine has been successfully used to install mono-, di-, and tri-methyl lysine, acetyl-lysine, GlcNAc-serine, and methyl-arginine analogues34–38. More recently, a new cysteine modification strategy was employed leading to installation of hydrazide analogues to generate acetyl-lysine, 2-hydroxyisobutyryl-lysine, and ubiquityl-lysine analogues39. The analogue approach is advantageous over NCL/EPL and genetic installation as it is possible to generate large quantities of the modified histone relatively inexpensively, and is generally straightforward to carry out. However, it is limited in that only one type of modification can be installed per histone protein, and as these are analogues, they do not represent the true modification.
All of the analogues generated using cysteine contain a sulfur instead of carbon at the γ position, and the hydrazide mimics also contain an additional nitrogen between the ε-amino group and the carbonyl of the acyl group (see Fig. 2). A crystal structure of a tri-methyl-lysine analogue in complex with a PHD finger demonstrates that the sulfur results in a slightly increased side-chain length of ~0.3 Å due to the increased C-S bond length, as well as more compressed C-S-C bond angle by ~12°40. Notably, despite this, the analogues can be recognized by antibodies and are good substrates for modifying enzymes, as well as being broadly recognized by reader domains41,42. There are however differences in binding affinities and in the coordination of the side-chain40,42. These effects vary depending on the substrate and binding protein, from very minor to more severe. Thus, it is critical to assess how the analogue alters binding in the peptide form before using the analogue-harboring nucleosomes in experiments.
Functionalization of dehydroalanine
An alternative approach to cysteine conjugation first converts the cysteine to dehydroalanine, which can then be converted to a variety of modified side-chains. This approach has been used to generate mono-, di-, and tri-methyl lysine and acetyl-lysine, as well as phosphoserine, and GlcNAc-serine analogues43. Similar to direct conjugation of cysteine, these analogues have a C to S substitution in the side-chain. However, two reports have recently shown that using carbon free radical chemistry, dehydroalanine can be converted to a number of modified amino acids that contain the natural C-C linkage44,45. This approach was demonstrated to be successful for generation of phosphor-serine, and methylated lysine and arginine.
Methods for histone ubiquitylation
Ubiquitin (Ub) is unique in that it is a small protein as opposed to a small chemical group. It is natively attached by through conjugation of a lysine ε-amine with the C-terminus of ubiquitin. Several methods have now been developed for its installation, and largely applied to generation of ubiquitylated H2A and H2B. EPL approaches (discussed above) involve three-piece ligation of first a histone fragment to Ub, followed by attachment to the remaining histone46. NCL and total chemical synthesis (both discussed above) have also been utilized to generate ubiquitylated peptides and histones47,48. Though these lead to a native linkage, the approaches can be cumbersome and difficult to obtain in high yield. Alternatively, Ub can be ligated through disulfide linkage, in which the histone residue to be modified is mutated to a cysteine and an aminoethanethiol is generated through intein-mediated trans-thioesterification49. A nonhydrolyzable mimic has also been developed, which is also generated through di-sulfide linkage but is further stabilized by cross-linking with 1,3-dicholoroacetone50. Finally, though enzymatic approaches generally lead to extremely low yield, an H2A/H2B fusion was found to be extremely efficient for enzymatic installation of Ub at K13 or K15 of H2A51.
Requirements for structural biology studies
The three major techniques for studying the structure of nucleosomes and nucleosome complexes are X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and most recently cryo-electron microscopy (cryo-EM). Each has its advantages and limitations as well as unique sample requirements.
NMR spectroscopy and X-ray crystallography provide the ability to explore the nucleosome in near atomic level detail, providing a powerful approach to investigating the effects of histone modifications on nucleosome structure. Indeed, a large number of X-ray crystal structures of the nucleosome have been determined. However, challenges arise due to the apparent propensity of the nucleosome to be stabilized in a single conformation during crystallization52. In addition, the histone tails rarely resolve, and it has proven difficult to obtain structures of the nucleosome in complex with bound factors. NMR spectroscopy is advantageous in this regard in that it can be applied in the solution state, can resolve dynamic regions such as the histone tails, and can be used to monitor binding dynamics53. However, the nucleosome core pushes the size limit for NMR and thus specialized, expensive approaches to isotopic enrichment are sometimes necessary54.
Both X-ray crystallography and especially NMR spectroscopy require large amounts of nucleosome, sometimes up to ~15 mg per sample. In addition, it is critical that the nucleosome composition is as homogenous as possible. For this reason, NCL/EPL and genetic installation of modified amino acids can be cost prohibitive, and cysteine conjugation is the most common method for generating modified nucleosomes for these approaches. In contrast, single particle cryo-EM requires substantially smaller amounts of sample. This opens the door to utilization of NCL/EPL approaches for sample preparation, and provides the potential for looking at nucleosomes with more complicated patterns of modifications. In addition, the ability to pick and classify individual particles allows for heterogeneity in the sample. Technical advances have led to the ability to use cryo-EM to investigate particles as small as the nucleosome55. However, small conformational changes are often undetectable, and conformational dynamics can lead to averaging out of regions such as the histone tails and bound factors. It is most likely the combination of structural approaches that will yield the most insight into the mechanism of histone modification function in the nucleosome context.
Application of structural studies to modified nucleosomes
The ability to generate modified nucleosomes through the abovementioned approaches has enabled the study of the effect of modifications on nucleosome structure and dynamics, the effect of nucleosome composition on histone tail binding and modification, and the mechanisms by which larger chromatin regulator proteins or complexes recognize modified nucleosomes. The focus here is on the application of structural analyses to these questions, but it should be noted that substantial information has also been garnered through the use of other biophysical and biochemical approaches including fluorescence techniques, that are not discussed here. Below is a sampling of several recent structural studies. It is by no means a comprehensive listing of all reports, but highlights some key findings and demonstrates how important it is to investigate histone modifications in the context of the nucleosome.
Histone modification effects on nucleosome structure and dynamics
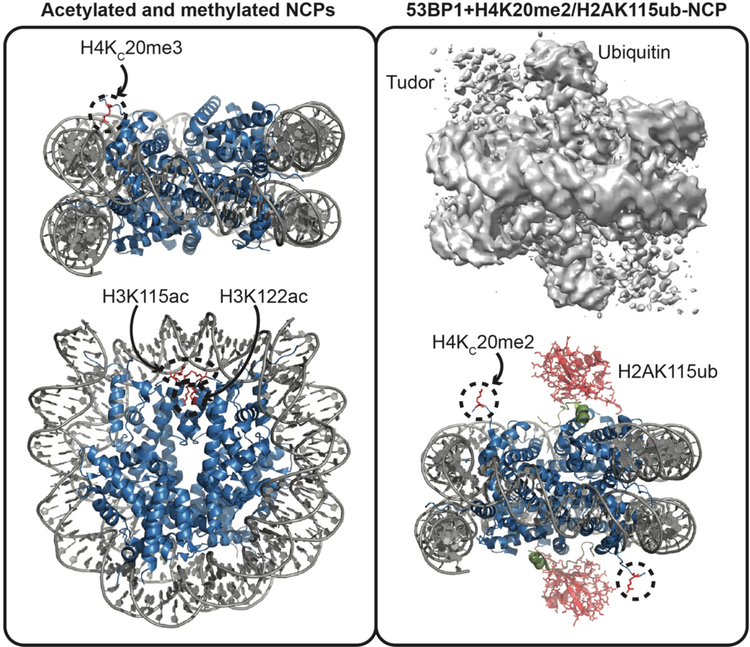
Several crystal structures of modified nucleosomes have been solved using the discussed approaches for installing modifications (see Fig. 3). One of the first applications of methyl-lysine analogues in structural studies was the crystallization of the nucleosome containing a tri-methyl analogue at lysine 20 of histone H4 (H4KC20me3)56. Comparison with the unmodified nucleosome revealed small changes in the conformation of K20me3 as well as adjacent residues H18 and R19 due to the methyl mark. Lu et al. further found that inclusion of H4KC20me3 in nucleosome arrays leads to increased compaction as compared to unmodified arrays. Importantly, the authors demonstrated that this was methylation dependent and not due to the analogue, as an unmethylated analogue did not produce this effect. Similar local effects were seen upon acetylation of H3. In a rare example of an EPL generated histone for use in structural studies, the crystal structure of the nucleosome containing H3K115ac and H3K122ac was solved57. The structure revealed decreased resolution of the acetyl-lysine sidechains, suggesting increased flexibility. Though the overall nucleosome conformation was not seen to change, increased nucleosome disassembly by the ATP-dependent remodeling complexes RSC and SWI/SNF was observed with the acetylated substrate compared to non-acetylated substrate.
Figure 3. Structural studies on modified nucleosomes.
Left, crystal structures of the nucleosome core particle (NCP) containing H4KC20me3 (top, PDB ID 3C1B) or H3K115ac/H3K122ac (bottom, PDB ID 4YS3). Histones are shown in blue, DNA in gray, and modified residues as red sticks. Right, the cryoEM reconstruction of the 53BP1 nucleosome binding region (tandem tudor domain plus ubiquitin dependent recruitment motif) in complex with the NCP containing H4KC20me2 and H2AK115ub. The cryoEM map is shown (top, EMDB ID 8246) and fit model (PDB ID 5KGF) is shown bottom. Histones are shown in blue, DNA in gray, 53BP1 in green, and modifications as red sticks.
As noted, the histone tails generally do not resolve in crystal structures of the nucleosome, consistent with biochemical data that indicate a high level of conformational flexibility58,59. However, NMR spectroscopy has recently proven incredibly powerful for investigating the structure and conformation of the histone tails in the nucleosome. Studies on the H3 tail in the context of the nucleosome revealed substantial conformational flexibility but also a robust interaction between the tails and the nucleosomal or linker DNA53,60–63. Complementary NMR and biochemical studies demonstrated that the intra-nucleosomal histone tail-DNA interactions reduce accessibility of unmodified H3 tails by up to a factor of ~10 in physiologically relevant conditions62. Furthermore, in low ionic strength (50 mM NaCl) less than 1% of H3 tails are in the DNA-unbound state within the nucleosome and therefore available for interactions, but this availability can be altered by PTMs62,64. A model was proposed in which the H3 tails are robustly but dynamically associated with DNA in the chromatin environment, sampling an ensemble of DNA bound states63. Installation of the H3KC4me3 analogue led to only modest perturbations in the association with DNA, however, mutation of additional histone residues led to more substantial release. Future incorporation of additional analogues should provide insight into the effect of various combinations of modifications on the H3 tail dynamics.
The effect of nucleosome nature on histone tail binding
A large number of reader domains have now been identified including methyl-lysine binding chromodomains, PHD fingers, Tudor and PWWP domains, and acetyl-lysine binding bromodomains9,10,13,65. The structural basis of these interactions have been extensively characterized using correspondingly modified histone peptides, but are only recently being characterized in the context of the nucleosome61–64,66–69. Interestingly, several histone reader domains have now been found to associate not only with histone tails but also with nucleic acids70–72. The use of nucleosomes as substrates in NMR experiments has been instrumental in advancing our understanding of the binding mechanisms. It allowed for detailed analysis of the multivalent engagement of the PSIP1 PWWP domain and the PHF1 Tudor domain with the nucleosome containing H3KC36me367,68. Similarly, it helped establish the mechanism of the BRDT bromodomain binding to the NCL generated H4K5acK8ac nucleosomes and enabled characterization of how paired readers, the tandem PHD fingers of CHD4 and CHD3, bind to unmodified nucleosomes62,64,69.
There is also recent evidence that histone tail binding is altered in the context of the nucleosome even in the absence of additional nucleosome contacts. The PHD finger of BPTF is known to readily associate with the H3K4me3 tail peptide, and the structure of this reader has been solved in complex with a histone peptide73,74. However, recent NMR studies revealed that binding of the BPTF PHD finger to the H3KC4me3 nucleosome is strongly inhibited due to the interaction of the histone tail with DNA, and therefore tail inaccessibility63. Mutation of additional residues in the H3 tail modulated this effect indicating that PTM cross-talk can be mediated by nucleosome conformation itself.
Mechanisms by which chromatin regulators recognize modified nucleosomes
The capability to install histone modifications on nucleosomes has also allowed for characterization of the mechanisms by which larger chromatin regulator constructs or even full complexes associate with modified nucleosomes (as opposed to just the histone reader domains). These complexes have been notoriously difficult to crystallize, but a few structures have now been determined. In addition, NMR spectroscopy has proven powerful for studying the more dynamic complexes, and advances in cryo-EM have made it possible to visualize some of these complexes by microscopy.
NMR spectroscopy was utilized to investigate the association of full length HP1b with the nucleosome66. HP1b contains an N-terminal H3K9me3 binding chromodomain, a chromoshadow domain, and a hinge region between the two. Using an analogue approach, complex formation between HP1b and the H3KC9me3 nucleosome was examined66. The chromodomain associated with the methylated histone tail of the nucleosome in a manner structurally similar to how it associated with a methylated peptide. However, two major differences were uncovered, namely that the hinge region was found to make additional contacts with the nucleosomal DNA and while binding of the chromodomain to an unmodified H3 tail peptide was observed, there was no binding observed to the unmodified H3 tail within the nucleosome. This likely has to do with the lack of histone tail accessibility as discussed above.
Recently the cryo-EM structure of the minimal nucleosome binding region of 53BP1 in complex with a modified nucleosome was obtained (see Fig. 3)75. 53BP1 interacts with H4K20me2 through its tandem tudor domain and with H2AK115ub through the ubiquitin dependent recruitment motif. Association with these modifications occurs during DNA damage response and notably 53BP1 is selective for K115ub over K113ub. To solve the structure, an analogue approach was used to install H4KC20me2 and H2AK115ub was installed enzymatically. The structure revealed an extensive set of contacts between 53BP1 and the nucleosome, including contacts between the H2A tail and the nucleosome, and ubiquitin and the nucleosome that together drive complex formation. Notably, the structure revealed that interaction of two arginine residues of the H2A tail with the nucleosomal DNA is necessary to position the ubiquitin properly for 53BP1 binding and discriminate against binding to H2AK113ub.
The mechanism underlying engagement of the histone methyltransferase complex PRC2 with a di-nucleosome was defined by Cryo-EM76. PRC2 methylates H3K27 through the enzymatic subunit, EZH2, and binds to its product (H3K27me3) through the EED subunit, which stimulates the catalytic activity and is thought to be important in spreading of this modification. The structure of PRC2 was determined with several di-nucleosomes separated by variable length linkers, with one nucleosome being unmodified and another containing H3K27me3. An analogue approach was used to generate the H3KC27me3 nucleosome. Remarkably, one cryo-EM structure showed that the PRC2 complex is capable of bridging the two nucleosomes. Extensive contacts with DNA position EZH2 towards the unmodified substrate, whereas EED engages the H3KC27me3 nucleosome in a more flexible manner, allowing for adaptation to variable linker lengths.
A similar bridging mechanism was observed for the DNA methyltransferase ZMET77. ZMET contains two H3K9me3 binding domains, a chromodomain and a BAH domain. To investigate how association with H3K9me3 in the nucleosome context may contribute to ZMET activity, mono- and di-nucleosomes containing H3KC9me3 were generated. A combination of biochemical, enzymatic, and negative stain EM studies revealed that the chromodomain and BAH domain each associate with one H3KC9me3-nucleosome, together spanning a di-nucleosome. The binding of both of these domains positions the methyltransferase domain near the substrate linker DNA, and in addition the BAH domain binding allosterically stimulates activity.
Outlook
Recent developments in the design of modified nucleosomes has expanded their utility in a range of studies, making high resolution structural characterization possible. Each approach has its benefits and drawbacks. Nucleosomes generated with NCL/EPL and total synthesis have the benefit of containing the native modified amino acid, and allow for multiple modifications to be added on the same protein. However, they can be cost prohibitive especially if large amounts are needed. Genetic installation of modifications also leads to the native amino acid, but is limited in the number that can be installed, and often suffers in yield. Alternative to these approaches is the use of analogues. The drawbacks to analogues are the deviation from the native modified amino acid, as well as difficulty in placing different modifications on the same protein, as currently all rely on cysteine chemistry. However, the analogue approach is extremely cost effective and thus can be used to generate large amounts of modified nucleosomes. As such, this is the most common approach being utilized to generate nucleosomes, especially in the application towards structural biology. Though still somewhat limited in the variety of modifications possible, recent developments of additional modification analogues is promising.
It is becoming exceedingly clear that to understand the biological roles of histone modifications, it is essential to use nucleosomes as substrates in experiments. Recent reports convincingly show that even for a single reader domain, there are substantial differences in binding to a histone peptide versus the intact nucleosome. Moreover, with the realization that histone PTM crosstalk can be mediated by nucleosome conformation even on a single histone tail, it is clear that nucleosome studies are crucial for uncovering how modifications function in combination. Newly developed commercially available designer nucleosomes provide an exciting opportunity to test a variety of combinations of modifications, and will hopefully be developed further such that a routine screen can be carried out.
The largest challenge is the continued development of methods for generating an expanded library of modifications that can be created at high sensitivity levels for reasonable cost. This includes additional modifications, as well as the ability to place multiple modifications on the same nucleosome. In parallel, methods development that allows for decreased amounts of sample, such as is underway in cryoEM, will push these studies forward even further.
Funding:
Work in the Musselman laboratory is funded by the National Science Foundation (CAREER-1452411) and the National Institutes of Health (GM128705). Work in the Kutateladze laboratory is funded by the National Institutes of Health (GM106416, GM125195, GM100907).
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Contributor Information
Catherine A. Musselman, Department of Biochemistry, University of Iowa Carver College of Medicine, Iowa City, IA 52246
Tatiana G. Kutateladze, Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO 80045
References
- (1).Bannister AJ, and Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zentner GE, and Henikoff S (2013) Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol 20, 259–266. [DOI] [PubMed] [Google Scholar]
- (3).Zhou VW, Goren A, and Bernstein BE (2010) Charting histone modifications and the functional organization of mammalian genomes : Abstract : Nature Reviews Genetics. Nat Rev Genet [DOI] [PubMed] [Google Scholar]
- (4).Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, and Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rivera CM, and Ren B (2013) Mapping Human Epigenomes. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Maunakea AK, Chepelev I, and Zhao K (2010) Epigenome mapping in normal and disease States. Circulation Research 107, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Portela A, and Esteller M (2010) Epigenetic modifications and human disease. Nature Biotechnology 28, 1057–1068. [DOI] [PubMed] [Google Scholar]
- (8).DesJarlais R, and Tummino PJ (2016) The Role of Histone-modifying Enzymes and their Complexes in Regulation of Chromatin Biology. Biochemistry 55, 1584–1599. [DOI] [PubMed] [Google Scholar]
- (9).Rothbart SB, and Strahl BD (2014) Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 1839, 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Andrews FH, Strahl BD, and Kutateladze TG (2016) Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol 12, 662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Voigt P, LeRoy G, Drury WJ III, Zee BM, Son J, Beck DB, Young NL, Garcia BA, and Reinberg D (2012) Asymmetrically Modified Nucleosomes. Cell 151, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ruthenburg AJ, Li H, Patel DJ, and David Allis C (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Musselman CA, Lalonde M-E, Côté J, and Kutateladze TG (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol 19, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lee KM, and Narlikar G (2001) Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol Chapter 21, Unit 21.6. [DOI] [PubMed] [Google Scholar]
- (15).Bao Y, Chakravarthy S, Muthurajan UM, and Luger K (2003) Reconstitution of Nucleosome Core Particles from Recombinant Histones and DNA. Methods in … [DOI] [PubMed] [Google Scholar]
- (16).Müller MM, and Muir TW (2014) Histones: At the Crossroads of Peptide and Protein Chemistry. Chem. Rev 115, 2296–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nadal S, Raj R, Mohammed S, and Davis BG (2018) Synthetic post-translational modification of histones. Curr Opin Chem Biol 45, 35–47. [DOI] [PubMed] [Google Scholar]
- (18).Kim CH, Axup JY, and Schultz PG (2013) Protein conjugation with genetically encoded unnatural amino acids. Curr Opin Chem Biol 17, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, and Liu WR (2010) A Facile System for Genetic Incorporation of Two Different Noncanonical Amino Acids into One Protein in Escherichia coli. Angewandte Chemie 122, 3279–3282. [DOI] [PubMed] [Google Scholar]
- (20).Neumann H, Wang K, Davis L, Garcia-Alai M, and Chin JW (2010) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444. [DOI] [PubMed] [Google Scholar]
- (21).Neumann H, Peak-Chew SY, and Chin JW (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol 4, 232–234. [DOI] [PubMed] [Google Scholar]
- (22).Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, and Chin JW (2009) Genetically Encoding Nϵ-Methyl-l-lysine in Recombinant Histones. J. Am. Chem. Soc 131, 14194–14195. [DOI] [PubMed] [Google Scholar]
- (23).Nguyen DP, Alai M, Virdee S, and Chin JW (2010) Genetically Directing ɛ-N, NDimethyl-l-Lysine in Recombinant Histones. Chem. Biol [DOI] [PubMed] [Google Scholar]
- (24).Kim CH, Kang M, Kim HJ, Chatterjee A, and Schultz PG (2012) Site-Specific Incorporation of ε-N-Crotonyllysine into Histones. Angew. Chem. Int. Ed. Engl 51, 7246–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gattner MJ, Vrabel M, and Carell T (2013) Synthesis of ε- N -propionyl-, ε- N -butyryl-, and ε- N -crotonyl-lysine containing histone H3 using the pyrrolysine system. Chemical Communications 49, 379–381. [DOI] [PubMed] [Google Scholar]
- (26).Wilkins BJ, Hahn LE, Heitmüller S, Frauendorf H, Valerius O, Braus GH, and Neumann H (2015) Genetically Encoding Lysine Modifications on Histone H4 ACS Publications. [DOI] [PubMed] [Google Scholar]
- (27).Lee S, Oh S, Yang A, Kim J, Söll D, Lee D, and Park H-S (2013) A Facile Strategy for Selective Incorporation of Phosphoserine into Histones. Angew. Chem. Int. Ed. Engl 52, 5771–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Xiao H, Xuan W, Shao S, Liu T, and Schultz PG (2015) Genetic Incorporation of ε-N-2-Hydroxyisobutyryl-lysine into Recombinant Histones. ACS Chem. Biol 10, 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dawson PE, Muir TW, Clark-Lewis I, and Kent SB (1994) Synthesis of proteins by native chemical ligation. Science 266, 776–779. [DOI] [PubMed] [Google Scholar]
- (30).Muir TW, Sondhi D, and Cole PA (1998) Expressed protein ligation: A general method for protein engineering. Proceedings of the National … [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Berrade L, and Camarero JA Expressed protein ligation: a resourceful tool to study protein structure and function. Cell. Mol. Life Sci 66, 3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Holt M, and Muir T (2015) Application of the Protein Semisynthesis Strategy to the Generation of Modified Chromatin http://dx.doi.org.proxy.lib.uiowa.edu/10.1146/annurev-biochem-060614-034429. [DOI] [PMC free article] [PubMed]
- (33).Qi Y-K, Ai H-S, Li Y-M, and Yan B (2018) Total Chemical Synthesis of Modified Histones. Front Chem 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Simon MD (2001) Installation of Site-Specific Methylation into Histones Using Methyl Lysine Analogs Current Protocols in Molecular Biology, pp Unit 21.18.1–10 John Wiley & Sons, Inc., Hoboken, NJ, USA. [DOI] [PubMed] [Google Scholar]
- (35).Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, Marmorstein R, Cole RN, Khochbin S, and Cole PA (2010) Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation. J. Am. Chem. Soc 132, 9986–9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Dhall A, Weller CE, and Chatterjee C (2016) Chapter Seven - Rapid Semisynthesis of Acetylated and Sumoylated Histone Analogs. Meth. Enzymol [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Raj R, Lercher L, Mohammed S, and Davis BG (2016) Synthetic Nucleosomes Reveal that GlcNAcylation Modulates Direct Interaction with the FACT Complex. Angew. Chem. Int. Ed. Engl [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Le DD, Cortesi AT, Myers SA, Burlingame AL, and Fujimori DG (2013) Site-Specific and Regiospecific Installation of Methylarginine Analogues into Recombinant Histones and Insights into Effector Protein Binding. J. Am. Chem. Soc 135, 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Bhat S, Hwang Y, Gibson MD, Morgan MT, Taverna SD, Zhao Y, Wolberger C, Poirier MG, and Cole PA (2018) Hydrazide Mimics for Protein Lysine Acylation To Assess Nucleosome Dynamics and Deubiquitinase Action. J. Am. Chem. Soc 140, 9478–9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Chen Z, Notti RQ, Ueberheide B, and Ruthenburg AJ (2017) Quantitative and Structural Assessment of Histone Methyllysine Analogue Engagement by Cognate Binding Proteins Reveals Affinity Decrements Relative to Those of Native Counterparts. Biochemistry [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Jia G, Wang W, Li H, Mao Z, Cai G, Sun J, Wu H, Xu M, Yang P, Yuan W, Chen S, and Zhu B (2009) A systematic evaluation of the compatibility of histones containing methyllysine analogues with biochemical reactions. Cell Res 19, 1217–1220. [DOI] [PubMed] [Google Scholar]
- (42).Seeliger D, Soeroes S, Klingberg R, Schwarzer D, Grubmüller H, and Fischle W (2012) Quantitative assessment of protein interaction with methyl-lysine analogues by hybrid computational and experimental approaches. ACS Chem. Biol 7, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Chalker JM, Lercher L, Rose NR, Schofield CJ, and Davis BG (2012) Conversion of Cysteine into Dehydroalanine Enables Access to Synthetic Histones Bearing Diverse Post-Translational Modifications. Angewandte Chemie 124, 1871–1875. [DOI] [PubMed] [Google Scholar]
- (44).Yang A, Ha S, Ahn J, Kim R, Kim S, Lee Y, Kim J, Söll D, Lee H-Y, and Park H-S (2016) A chemical biology route to site-specific authentic protein modifications. Science 354, 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wright TH, Bower BJ, Chalker JM, Bernardes GJL, Wiewiora R, Ng W-L, Raj R, Faulkner S, Vallée MRJ, Phanumartwiwath A, Coleman OD, Thézénas M-L, Khan M, Galan SRG, Lercher L, Schombs MW, Gerstberger S, Palm-Espling ME, Baldwin AJ, Kessler BM, Claridge TDW, Mohammed S, and Davis BG (2016) Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 354, aag1465. [DOI] [PubMed] [Google Scholar]
- (46).McGinty RK, Kim J, Chatterjee C, Roeder RG, and Muir TW (2008) Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Kumar KSA, Spasser L, Ohayon S, Erlich LA, and Brik A (2011) Expeditious chemical synthesis of ubiquitinated peptides employing orthogonal protection and native chemical ligation. Bioconjug. Chem 22, 137–143. [DOI] [PubMed] [Google Scholar]
- (48).Maity SK, Jbara M, and Brik A (2016) Chemical and semisynthesis of modified histones. Journal of Peptide Science [DOI] [PubMed] [Google Scholar]
- (49).Chatterjee C, McGinty RK, Fierz B, and Muir TW (2010) Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol 6, 267–269. [DOI] [PubMed] [Google Scholar]
- (50).Long L, Furgason M, and Yao T (2014) Generation of nonhydrolyzable ubiquitin-histone mimics. Methods 70, 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Hu Q, Botuyan MV, Cui G, Zhao D, and Mer G (2017) Mechanisms of Ubiquitin-Nucleosome Recognition and Regulation of 53BP1 Chromatin Recruitment by RNF168/169 and RAD18. Molecular Cell 66, 473–487.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Andrews AJ, and Luger K (2011) Nucleosome Structure(s) and Stability: Variations on a Theme. Annu Rev Biophys 40, 99–117. [DOI] [PubMed] [Google Scholar]
- (53).Zhou B-R, Feng H, Ghirlando R, Kato H, Gruschus J, and Bai Y (2012) Histone H4 K16Q mutation, an acetylation mimic, causes structural disorder of its N-terminal basic patch in the nucleosome. J. Mol. Biol 421, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kato H, van Ingen H, Zhou B-R, Feng H, Bustin M, Kay LE, and Bai Y (2011) Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proceedings of the … [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Cheng Y (2018) Single-particle cryo-EM-How did it get here and where will it go. Science 361, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, and Luger K (2008) The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol 15, 1122–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Chatterjee N, North JA, Dechassa ML, Manohar M, Prasad R, Luger K, Ottesen JJ, Poirier MG, and Bartholomew B (2015) Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF. Mol. Cell. Biol 35, 4083–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Luger K, Mäder AW, Richmond RK, Sargent DF, and Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- (59).Cutter AR, and Hayes JJ (2015) A brief review of nucleosome structure. FEBS Letters [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Gao M, Nadaud PS, Bernier MW, North JA, Hammel PC, Poirier MG, and Jaroniec CP (2013) Histone H3 and H4 N-Terminal Tails in Nucleosome Arrays at Cellular Concentrations Probed by Magic Angle Spinning NMR Spectroscopy. J. Am. Chem. Soc [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Stützer A, Liokatis S, Kiesel A, Schwarzer D, Sprangers R, Söding J, Selenko P, and Fischle W (2016) Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Molecular Cell 61, 247–259. [DOI] [PubMed] [Google Scholar]
- (62).Gatchalian J, Wang X, Ikebe J, Cox KL, Tencer AH, Zhang Y, Burge NL, Di L, Gibson MD, Musselman CA, Poirier MG, Kono H, Hayes JJ, and Kutateladze TG (2017) Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 8, 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, and Musselman CA (2018) The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 7, e31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Tencer AH, Cox KL, Di L, Bridgers JB, Lyu J, Wang X, Sims JK, Weaver TM, Allen HF, Zhang Y, Gatchalian J, Darcy MA, Gibson MD, Ikebe J, Li W, Wade PA, Hayes JJ, Strahl BD, Kono H, Poirier MG, Musselman CA, and Kutateladze TG (2017) Covalent Modifications of Histone H3K9 Promote Binding of CHD3. Cell Rep 21, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Taverna SD, Li H, Ruthenburg AJ, and Allis CD (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers : Article : Nature Structural & Molecular Biology. Nature structural & … [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Munari F, Soeroes S, Zenn HM, Schomburg A, Kost N, Schröder S, Klingberg R, Rezaei-Ghaleh N, Stützer A, Gelato KA, Walla PJ, Becker S, Schwarzer D, Zimmermann B, Fischle W, and Zweckstetter M (2012) Methylation of Lysine 9 in Histone H3 Directs Alternative Modes of Highly Dynamic Interaction of Heterochromatin Protein hHP1β with the Nucleosome. Journal of Biological … [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Musselman CA, Gibson MD, Hartwick EW, North JA, Gatchalian J, Poirier MG, and Kutateladze TG (2013) Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat Commun 4, 2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, and van Ingen H (2013) Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Miller TCR, Simon B, Rybin V, Grötsch H, Curtet S, Khochbin S, Carlomagno T, and Müller CW (2016) A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun 7, 13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Klein BJ, Muthurajan UM, Lalonde M-E, Gibson MD, Andrews FH, Hepler M, Machida S, Yan K, Kurumizaka H, Poirier MG, Côté J, Luger K, and Kutateladze TG (2015) Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Weaver T, Morrison E, and Musselman C (2018) Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 23, 2614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Klein BJ, Vann KR, Andrews FH, Wang WW, Zhang J, Zhang Y, Beloglazkina AA, Mi W, Li Y, Li H, Shi X, Kutateladze AG, Strahl BD, Liu WR, and Kutateladze TG (2018) Structural insights into the π-π-π stacking mechanism and DNA-binding activity of the YEATS domain. Nat Commun 9, 4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, and Patel DJ (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, and Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature [DOI] [PubMed] [Google Scholar]
- (75).Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien J-P, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, and Durocher D (2016) The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103. [DOI] [PubMed] [Google Scholar]
- (76).Poepsel S, Kasinath V, and Nogales E (2018) Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Stoddard CI, Feng S, Campbell MG, Liu W, Wang H, Zhong X, Bernatavichute Y, Cheng Y, Jacobsen SE, and Narlikar GJ (2018) A Nucleosome Bridging Mechanism for Activation of a Maintenance DNA Methyltransferase. Molecular Cell [DOI] [PMC free article] [PubMed] [Google Scholar]