Abstract
The two GATA transcription factors ELT-2 and ELT-7 function in the differentiation of the C. elegans intestine. ELT-2 loss causes lethality. ELT-7 loss causes no obvious phenotype but enhances the elt-2(−) intestinal phenotype. Thus, ELT-2 and ELT-7 appear partially redundant, with ELT-2 being more influential. To investigate the different regulatory roles of ELT-2 and ELT-7, we compared the transcriptional profiles of pure populations of wild-type, elt-2(−), elt-7(−), and elt-7(−); elt-2(−) double mutant L1-stage larvae. Consistent with the mutant phenotypes, loss of ELT-2 had a >25 fold greater influence on the number of significantly altered transcripts compared to the loss of ELT-7; nonetheless, the levels of numerous transcripts changed upon loss of ELT-7 in the elt-2(−) background. The quantitative responses of individual genes revealed a more complicated behaviour than simple redundancy/partial redundancy. In particular, genes expressed only in the intestine showed three distinguishable classes of response in the different mutant backgrounds. One class of genes responded as if ELT-2 is the major transcriptional activator and ELT-7 provides variable compensatory input. For a second class, transcript levels increased upon loss of ELT-2 but decreased upon further loss of ELT-7, suggesting that ELT-7 actually overcompensates for the loss of ELT-2. For a third class, transcript levels also increased upon loss of ELT-2 but remained elevated upon further loss of ELT-7, suggesting overcompensation by some other intestinal transcription factor(s). In spite of its minor loss-of-function phenotype and its limited sequence similarity to ELT-2, ELT-7 expressed under control of the elt-2 promoter is able to rescue elt-2(−) lethality. Indeed, appropriately expressed ELT-7, like appropriately expressed ELT-2, is able to replace all other core GATA factors in the C. elegans endodermal pathway. Overall, this study focuses attention on the quantitative intricacies behind apparent redundancy or partial redundancy of two related transcription factors.
1. Introduction
Multiple transcription factors belonging to the same transcription factor family are often expressed in the same set of cells during embryonic development, raising the question how they apportion their transcriptional responsibilities. For example,which factor influences which target genes, by how much and by what mechanism? What contribution does each factor make to the overall stability and robustness of the regulatory network?
A variety of models have been suggested to explain how multiple transcription factors work together in developmental networks, in which both the components and their importance can change with time (Davidson, 2001; Wagner, 2005; Davidson, 2006; Felix and Wagner, 2008; Peter and Davidson, 2016). In the extreme case of a simple linear hierarchy, a temporal cascade of successive transcription factors culminates in the activation of a “master regulator” that is then responsible for activating “all” downstream target genes in an organ (or tissue or cell lineage). In the opposite extreme of a distributed network, no single transcription factor dominates but rather a set of factors act within an egalitarian cross-regulating community and all participate in target gene control. Loss of function mutations can help to distinguish between these two extreme models. In the former case, loss of function of the master regulator would remove all of the differentiated markers that define the organ. In the latter case, loss of any one transcription factor would not produce an obvious phenotype because of the ability of redundant factors to compensate. Like most transcription factor networks in the real world, the network driving differentiation of the C. elegans endoderm appears to lie somewhere between these two extremes: some factors appear more important than others (McGhee et al., 2007; McGhee et al., 2009) but the factors also show partial redundancy (Sommermann et al., 2010).
The C. elegans intestine (endoderm or E-lineage) develops as a simple clone of cells under the control of a GATA-factor dominated transcriptional cascade (reviewed in (McGhee, 2007; Maduro, 2010; McGhee, 2013; Maduro, 2015, 2017). The E blastomere of the eight-cell embryo is the clonal progenitor of the intestine (Sulston et al., 1983) and is specified as endoderm by two largely redundant GATA-type transcription factors, END-1 and END-3 (Zhu et al., 1997; Maduro et al., 2005; Maduro et al., 2007; Owraghi et al., 2009; Boeck et al., 2011; Maduro et al., 2015). When the E lineage has two cells (2E cell stage), the combined action of END-1 and END-3 initiates expression of a third endoderm specific GATA factor, ELT-7 (Sommermann et al., 2010). At the beginning of the next cell cycle (the 4E cell stage), END-1, END-3, and ELT-7 combine to initiate the expression of a fourth endoderm-specific GATA factor, ELT-2 (Fukushige et al., 1998; Zhu et al., 1998; Sommermann et al., 2010; Du et al., 2016; Wiesenfahrt et al., 2016). ELT-2 and ELT-7 are then expressed throughout the remaining lifespan. Although END-1/END-3 are associated primarily with endoderm specification and ELT-2/ELT-7 are associated primarily with endoderm differentiation, each of these GATA factors by themselves appears capable of driving a significant fraction of the endodermal program. One striking example is the ability of ELT-7 to induce transdifferentiation or “transorganogenesis” of certain post-mitotic tissues into “intestine” (Riddle et al., 2013; Riddle et al., 2016). A second example is the ability of ELT-2, if expressed at the appropriate time and level, to replace all of the other core endoderm GATA factors and to drive both specification and differentiation of the C. elegans intestine (Wiesenfahrt et al., 2016).
In the present study, we focus on the relative roles of ELT-2 and ELT-7 in the differentiation of the C. elegans intestine. Most available evidence suggests that ELT-2 is more influential than ELT-7 (McGhee et al., 2007; McGhee et al., 2009). Null mutations in elt-2 are lethal; mutant animals hatch but arrest as L1 larvae with undersized distorted intestinal microvilli and a blocked intestinal lumen (Fukushige et al., 1998). Although null mutations in elt-7 show no obvious phenotype (McGhee et al., 2007; Sommermann et al., 2010), they significantly enhance the phenotype of the elt-2(−) mutant intestine (Sommermann et al., 2010). Thus, the wildtype elt-2 and elt-7 genes (and ELT-2 and ELT-7 proteins) act as if they are partially redundant.
In terms of gene regulation, the simplest model to explain ELT-2/ELT-7 partial redundancy is that ELT-7 contributes to the transcriptional control of a subset of the genes normally controlled by ELT-2 (McGhee et al., 2007). However, (Sommermann et al., 2010) showed that this partial redundancy can encompass a range of different quantitative responses. For two intestinal differentiation markers, ELT-2 and ELT-7 appeared completely redundant (a phenotype was detected only in the elt-7(−); elt-2(−) double mutant, not in either of the single mutants); for three other markers, ELT-2 and ELT-7 appeared partially redundant (loss of ELT-2 by itself produced a phenotype that was exacerbated by loss of ELT-7) and for two other markers, ELT-2 and ELT-7 appeared non-redundant (loss of ELT-2 produced a phenotype that was not exacerbated by further loss of ELT-7) (Sommermann et al., 2010).
To investigate the ELT-2/ELT-7 interplay in greater depth, we used automated flow sorting to isolate pure populations of arrested wild-type, elt-2(−), elt-7(−), and elt-7(−); elt-2(−) double mutant larvae. RNA-seq performed on these populations allowed us to define quantitative transcriptional responses of C. elegans genes to loss of ELT-2 and/or loss of ELT-7. Focusing on a set of genes expressed specifically in the intestine, we found that three equally sized classes of dynamically expressed genes could be distinguished. For genes in the first class, transcript levels show the expected response spectrum of ELT-2/ELT-7 redundancy/partial redundancy/non-redundancy. For genes in the second class, transcript levels increase in the elt-2(−) mutant but decrease in the elt-7(−); elt-2(−) double, suggesting that ELT-7 may actually overcompensate for loss of ELT-2. (Because transcript levels in the absence of ELT-2 actually increase above wildtype levels, we refer to this as overcompensation rather than compensation.) For genes in the third class, transcript levels also increase in the elt-2(−) mutant but remain at least partially elevated in the elt-7(−); elt-2(−) double, suggesting that some other intestinal transcription factor(s) can overcompensate for the absence of ELT-2. Overall, the results reveal unexpected diversity in the quantitative responses of individual ELT-2/ELT-7 target genes. Finally, in spite of the consistent evidence that ELT-2 is more influential than ELT-7, we show that ELT-7 can actually replace ELT-2 if expressed under control of the elt-2 promoter. Indeed, if ELT-7 is expressed under control of both the end-1 and elt-2 promoters, ELT-7 can substitute for END-1, END-3 and ELT-2, driving both specification and differentiation of the C. elegans intestine.
2. Results
2.1. Isolation of pure populations of mutant L1 larvae prior to RNA-seq
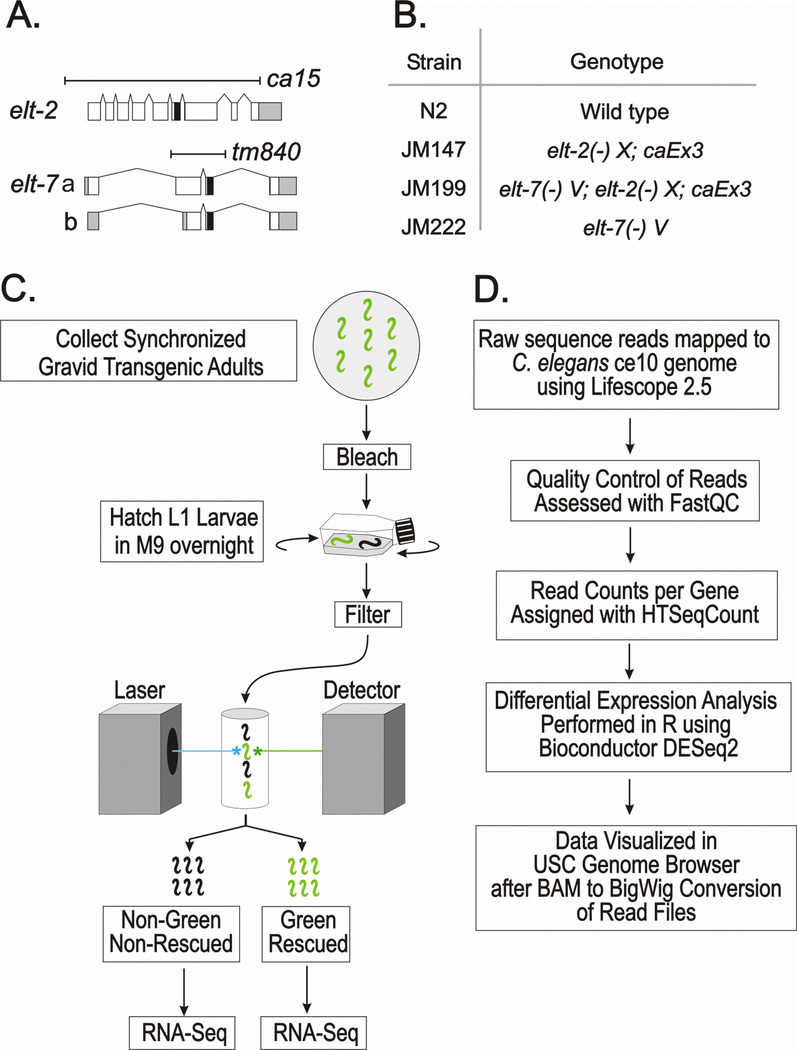
Null mutations are available for both elt-2 and elt-7 (Fig. 1A; alleles ca15 and tm840 respectively, hereafter referred to as elt-2(−) and elt-7(−)). To compare the transcriptomes of single mutants, double mutants and controls, four strains were constructed (Fig. 1B). The lethality associated with elt-2(−) and elt-7(−); elt-2(−) in strains JM147 and JM199 respectively was rescued by a wildtype copy of elt-2 on a non-integrated multicopy transgenic array (caEx3). The caEx3 array also contained a GFP-reporter, allowing array-positive rescued larvae to be separated from their array-negative non-rescued siblings by passage through the COPAS biosorter (see Methods for details). Wildtype N2 animals and animals from the viable strain JM222 elt-7(−) were also passed through the Biosorter in exact parallel. Sample preparation and bioinformatics analysis are summarized in Fig. 1C and 1D, respectively. Each sorting experiment resulted in ~100,000 animals, which were frozen and later processed to generate transcriptome profiles by RNA-seq (see Methods). Either three or four independent biological replicates of each sorted population were collected. Typical Biosorter profiles of the four strains (six populations) are shown in Supplementary Fig. 1. The purity of the rescued and non-rescued segregants from JM147 and JM199 populations was estimated from fluorescent microscopy to be ~99% (Supplementary Fig. 1).
Fig. 1. Overall experimental design: Genotypes, sorting protocol and outline of bioinformatic methods.
A. Diagram of the elt-2 and elt-7 genes; white, gray and black boxes represent coding regions, untranslated regions and the zinc finger DNA binding domains, respectively. Both deletion alleles remove the DNA binding domain and are presumed null mutations.
B. Genotypes of the four strains used in the present analysis. elt-2(−) refers to allele ca15; elt-7(−) refers to allele tm840. In the two strains JM147 and JM199, the lethality associated with the elt-2(−) mutation is rescued by the same multicopy extra-chromosomal array caEx3[elt-2(+); rol-6(su1006); sur-5::GFP], which is lost in ~50% of the progeny of a transgenic mother.
C. Summary of sample preparations prior to RNA-Seq. Arrested L1 larvae from all four strains (N2, JM147, JM199 and JM222) were passed through the COPAS Biosorter in exact parallel.
D. Summary of major steps in the bioinformatic analysis.
2.2. Loss of ELT-2 exerts a greater influence on the larval transcriptome than does loss of ELT-7
To determine the overall RNA-seq data structure, 11 independent loss-of function experimental samples (elt-2(−), elt-7(−) and the double mutant), together with 4 wild type control samples, were analyzed by a distance matrix approach (Fig. 2). As expected for reproducibly successful sorting, this unsupervised clustering method grouped replicates of the same genotype together. The transcriptomes of elt-7(−) and wild type larvae were closer to each other (median distance of 44 arbitrary units) than either was to the transcriptomes of elt-2(−) and elt-7(−); elt-2(−) larvae (median distances ~100 arbitrary units). Thus, as predicted from earlier phenotypic studies, loss of ELT-2 has a much greater influence on the L1 larval transcriptome than does loss of ELT-7 (whether in the presence or absence of ELT-2).
Fig. 2. Distance matrix illustrating Euclidean distances between RNA-seq samples.
Samples with lower distances between them (darker blue) have more closely correlated gene expression patterns. Distances were calculated from log-stabilized normalized read counts of all detected transcripts. “rep” denotes data from an independent replicate.
2.3. Levels of larval transcripts, both intestinal and non-intestinal, can be either decreased or increased in response to loss of ELT-2 and/or ELT-7
Fig. 3A shows pairwise comparisons of transcript levels for 16,708 genes between all four mutant backgrounds. The results show that loss of ELT-2 affected a larger number of genes, either directly or indirectly, than did loss of ELT-7. We identified 619 genes that were down-regulated upon loss of ELT-2 (i.e. elt-2(−) compared to wild type) but identified no genes significantly down-regulated upon loss of ELT-7 (i.e. elt-7(−) compared to wild type). Further, we identified 25 times as many genes up-regulated upon loss of ELT-2 as upon loss of ELT-7 (1152 genes v. 46 genes). (These various gene sets are listed in Supplementary Tables 1, 2, 3 and 4). The greater influence of ELT-2 was also seen when the analysis was restricted to genes enriched in the intestine (and expressed elsewhere as well) or to genes expressed specifically in the intestine and nowhere else in the worm (Supplementary Fig. 2.). Overall, the observed differences in transcriptomes of the single mutants are consistent with the more severe phenotype of the elt-2(−) mutant compared to the elt-7(−) mutant (Fukushige et al., 1998; McGhee et al., 2007; Sommermann et al., 2010). The elt-7(−) mutation enhanced the elt-2(−) intestinal phenotype at the morphological level (Sommermann et al., 2010) and, as shown in Fig. 3A, the elt-7(−) mutation also enhanced the elt-2(−) phenotype at the transcriptome level. In pairwise comparisons (Fig. 3A, Supplementary Tables 3 and 4), 647 genes were significantly down-regulated in the elt-7(−); elt-2(−) double mutant compared to the elt-2(−) mutant alone. Such behaviour is expected if ELT-7 contributes to the transcriptional control of a subset of the genes controlled by ELT-2.
Fig. 3. Pairwise comparisons of transcriptomes of the four strains: wild-type, elt-2(−), elt-7(−), and elt-7(−); elt-2(−).
A. Pairwise comparisons between transcriptome profiles (MA plots) of four strains of L1 worms: N2 (wild type), elt-2(−), elt-7(−), and elt-7(−); elt-2(−). The log2-fold change of gene expression (y-axis) between the two labeled samples is plotted against the mean intensity (logarithmic scale) for a given gene (x-axis). Differentially expressed genes for each comparison are shown in red (criteria for differential expression: total read counts > 10, BH-adjusted Wald Test p-value < 0.1, and log2-fold change > 0.8). The total number of genes in each plot is 16,708. Genes that were down-regulated in elt-7(−) compared to wild type would be identified within the green ellipse. The orange ellipse contains genes down-regulated in elt-2(−) compared to wild type. The blue ellipse contains genes down-regulated in the elt-7(−); elt-2(−) double mutant compared to the elt-2(−) single mutant. Up-regulated and down-regulated genes in key differentially expressed subsets were searched for over-represented categories using the WORMEXP tool (Yang et al., 2016).
B. Promoter sequences from genes categorized in Fig. 3A as “down-regulated in elt-2(−) versus wt” (n = 619, orange ellipse) or “down-regulated in elt-7(−);elt-2(−) versus elt-2(−)” (n = 647, blue ellipse) were searched for enriched binding motifs using HOMER, (Heinz et al., 2010). The top hit for each search is shown; the two top-scoring motifs are highly similar to each other and to the extended TGATAA motif previously found to be associated with intestine-specific genes (McGhee et al., 2007; McGhee et al., 2009).
C. The number of incidences of the extended TGATAA motif discovered in B within promoters of each gene set is given. Motifs occurred either once, twice, or three times per promoter as tabulated by the stacked barplot.
D. The locations of TGATAA motifs discovered in B were identified within promoters of genes “down-regulated in elt-2(−) versus wt” or “down-regulated in elt-7(−);elt-2(−) versus elt-2(−)”. A histogram (10 bp bins) of the motif locations in relation to the transcriptional start site (TSS) is shown.
We looked for sequence features that could distinguish the genes that were down-regulated upon loss of ELT-2 (Fig. 3A, orange ellipse) from the genes that were down-regulated upon loss of ELT-7 in the elt-2(−) mutant background (Fig. 3A, blue ellipse). Searches for enriched sequence motifs in promoters of these two gene sets returned essentially identical extended TGATAA motifs (Fig. 3B). Further, the number and the position of these TGATAA motifs in the two classes of promoters were indistinguishable (Fig. 3C,D). As will be discussed below, the types of genes enriched in each set were similar, providing no evidence that the two gene sets have different functions. Thus, we currently do not have a satisfactory explanation, at the molecular level, why one intestinal gene responds strongly when ELT-2 alone is lost but a different intestinal gene responds strongly only when both ELT-2 and ELT-7 are lost.
2.4. ELT-7 contributes to transcriptional overcompensation in the absence of ELT-2
Consideration of the types of genes that are up-regulated or down-regulated in the different mutant backgrounds revealed that the transcriptional response of many target genes was more complicated than could be explained by simple ELT-2/ELT-7 redundancy, either partial or complete.
Fig. 3A lists the most highly represented categories of C. elegans genes (Yang et al., 2016) that were up-regulated or down-regulated in the various mutant backgrounds. Genes down-regulated in the elt-2(−) mutant were enriched in categories associated with the intestine, as expected if ELT-2 acts as a transcriptional activator of intestinal differentiation. Genes up-regulated in the elt-2(−) mutant fell into two classes. One class had no obvious connection to the intestine and we suggest that these were indirectly up-regulated in response to starvation, stress or thwarted development. However, a second class of elt-2(−) up-regulated genes was associated with the intestine; such behaviour is not expected if ELT-2 is acting as a simple transcriptional activator. The class of intestine-associated genes up-regulated in the elt-2(−) mutant is no longer detected as up-regulated in the elt-7(−); elt-2(−) double mutant. However, intestine-associated genes now appear in the down-regulated genes in the double mutant, suggesting that ELT-7 contributes to the transcriptional overcompensation of these genes in the absence of ELT-2. Genes that remain up-regulated in the double mutant seem generally not to be associated with the intestine and we suggest these are non-intestinal genes that are responding indirectly to stress, etc.
One model to explain the involvement of ELT-7 in transcriptional overcompensation of intestine genes when ELT-2 is missing would propose that ELT-7 levels are normally repressed by ELT-2. Three pieces of evidence argue against this model. We measured GFP intensity from an elt-7promoter::GFP-lacZ reporter in an elt-2(−) mutant background, either rescued or non-rescued by a RFP-marked transgenic array (strain JM278): intestinal GFP levels in the elt-2(−) non-rescued larvae decreased by 37 +/− SD = 21 % (n=37) relative to their rescued siblings. Although levels of elt-7 transcripts do appear to increase ~1.5 fold in the presence of the elt-2(−) mutation (Supplementary Fig. 3), this increase is not statistically significant. Finally, (Sommermann et al., 2010) reported that ectopic expression of ELT-2 caused ectopic activation, not repression, of an elt-7 reporter. We will suggest alternative models for ELT-7 dependent overcompensation in the discussion section. Levels of elt-2 transcripts are unchanged in the elt-7(−) mutant (Supplementary Fig. 3).
2.5. Intestine-specific genes fall into three classes according to their response to loss of ELT-2 and/or ELT-7
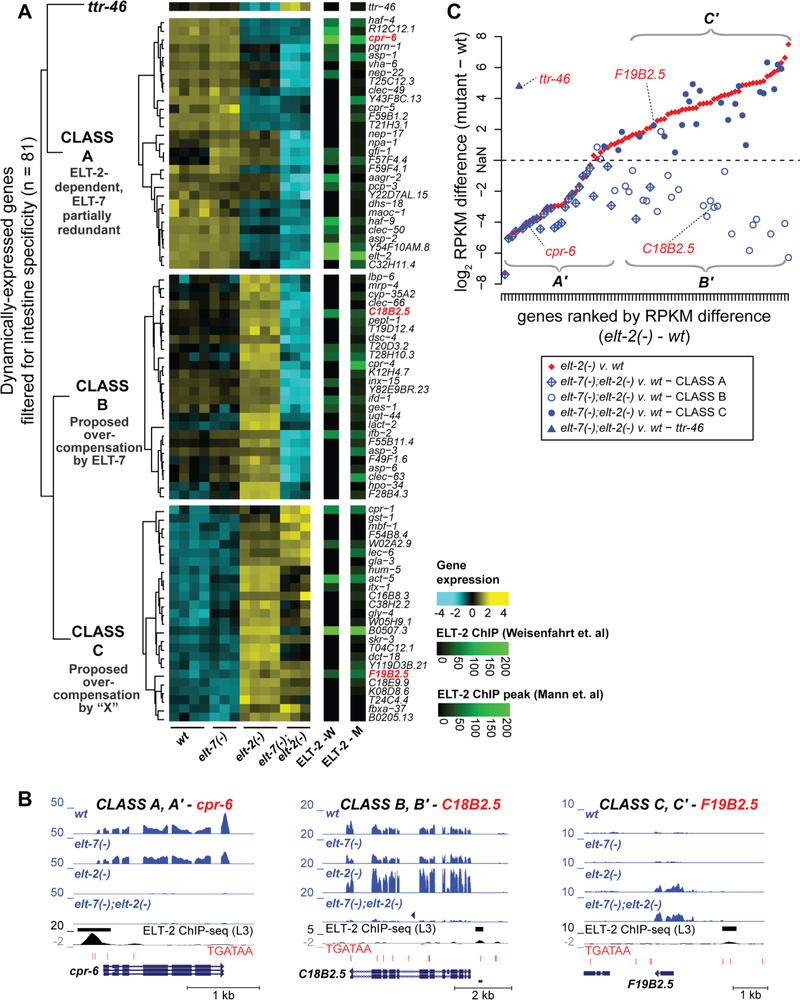
To track the response of individual intestine-specific genes to loss of ELT-2 and/or ELT-7 and to organize genes into clusters based on those responses, we performed hierarchical clustering of dynamically expressed genes across the different genotypes. Briefly, genes were filtered as dynamic by virtue of significant differential expression in any possible pair-wise comparison between genotypes (n = 3092), filtered further for genes annotated as “intestine-specific” (n = 81; Supplementary Table 5), row means normalized and finally clustered by Spearman correlation distances to emphasize the trends in expression patterns across genotypes as opposed to their expression level. The resulting heat map (Fig. 4A) clearly showed that for one-third of intestine-specific dynamic genes, expression was down-regulated by loss of elt-2; for the other two-thirds of such genes, expression was up-regulated. This latter class of intestine-specific genes whose expression was up-regulated by loss of elt-2(−) showed two distinctly different behaviors. For half of these genes, the up-regulation was elt-7-dependent; for the other half, the up-regulation was elt-7-independent. Therefore, we segregated the overall set of intestine-specific genes into three broad and roughly equal classes: A, B and C (plus one outlier). The main features of these three response classes are as follows:
For all classes, loss of ELT-7 showed little effect and transcription of intestinal-specific genes in elt-7(−) mutants appeared similar to wildtype. Two possible exceptions to this statement are the genes F57F4.4 and gfi-1 (Fig. 4A Class A), which were modestly up-regulated in the elt-7(−) mutant relative to wildtype. It is interesting that these genes are highly similar, lie adjacent to each other on the chromosome, and are expressed strongly in the anterior intestine (Cristina et al., 2009).
Class A genes were generally down-regulated in the elt-2(−) mutant and most were down-regulated further in the elt-7(−); elt-2(−) double mutant. For genes in this class, transcriptional activation is provided primarily by ELT-2, with a variable contribution provided by ELT-7. The cpr-6 gene is an example of a Class A gene. Transcript profiles in the different genetic backgrounds are shown below the heatmap in Fig. 4B.
Class B genes were up-regulated in the elt-2(−) mutant and then strongly downregulated in the elt-7(−); elt-2(−) double mutant, i.e., the overcompensation phenotype seen in the elt-2 single mutant was elt-7 dependent. The C18B2.5 gene provides an example of a Class B gene; transcript profiles are shown in Fig. 4B.
Class C genes were up-regulated in the elt-2(−) mutant but, unlike Class B genes, over-expression was largely elt-7-independent, i.e., expression remained elevated in the double mutant. We suggest that these are genes for which some other transcription factor or factors (“X”) overcompensates for loss of ELT-2. Expression of these genes would be lowered to the level found with Class B genes in the double mutant only if “X” were to be removed. F19B2.5 provides an example of a Class C gene; transcript profiles are shown in Fig. 4B.
Fig. 4A also summarizes ELT-2 peaks identified on intestinal specific gene promoters in two independent ELT-2 ChIP-Seq studies (Mann et al., 2016; Wiesenfahrt et al., 2016). ELT-2 was identified binding directly to genes belonging to each of the three major classes (A,B,C), supporting a direct link between ELT-2 and the resulting transcriptional output (Fig. 4A; see also Fig. 4B).
Fig. 4. Intestine-specific genes cluster into three main classes based on their response to loss of ELT-2 and subsequent loss of ELT-7.
A. The set of dynamically expressed genes (n = 3092) were filtered for those previously shown to be specifically expressed in the intestine and not in any other tissue (n = 81). Hierarchical clustering of their transcriptome dynamics was performed using Euclidean distances of row-means-normalized log-stabilized read counts. This effort identified three major classes of genes (Class A, Class B, and Class C) containing roughly equal numbers of members. ELT-2 ChIP-seq peak scores associated with these genes, as identified in two independent studies (Mann et al., 2016; Wiesenfahrt et al., 2016), are displayed to the right of the heat map.
B. Representative genome browser images are shown for the Class A gene cpr-6, the Class B gene C18B2.5, and the Class C gene F19B2.5. For each genome browser image, the normalized mapped read counts for all four conditions (wild type, elt-2(−), elt-7(−), and the double mutant elt-7(−);elt-2(−)) are shown. For each of the three representative genes, ELT-2 ChIP-seq peaks and peak regions (Wiesenfahrt et al., 2016) are shown, together with the incidence of TGATAA motifs and the gene models.
C. Deviations of transcript levels (RPKM) from wild-type is shown for elt-2(−) mutants and elt-2(−) elt-7(−) double mutant strains for the 81 intestine-specific dynamically changing genes (identified in Fig. 4A). Three classes of intestine-specific genes (A’, B’, C’) can be distinguished on the basis of their response to loss of ELT-2 and subsequent loss of ELT-7. RPKM differences between the elt-2 and wildtype are plotted in rank order (red points) beginning with the greatest decrease in transcript levels in the elt-2(−) mutant at rank # 1 and proceeding to the greatest increase in transcript levels in the elt-2(−) mutant at rank # 81. Keeping the same rank order as the x-axis, the difference in RPKM between the elt-7(−); elt-2(−) double mutant and the wildtype is then plotted for the same gene (blue points). The differences in RPKM are plotted as the signed log2(Absolute Value(RPKMfor mutant – RPKMfor wildtype control)), where the sign is negative for genes whose expression decreases upon loss of ELT-2 and positive for those genes whose expression increases. Because of this logarithmic transformation, the zero point is undefined. The different blue symbols map the different data points to membership of Class A,B or C genes on Fig. 4A.
A similar hierarchical clustering performed on all dynamically expressed genes in the data set (n = 3092), not just intestine-specific genes, resulted in six distinct gene sets (Supplementary Fig. 4A; Supplementary Table 6). We predicted that genes whose expression decreased in the elt-7(−);elt-2(−) double mutant (SET3, SET4, and SET6) would be more likely to show hallmarks of intestinal function and direct ELT-2 involvement in their expression. Indeed, genes in SET3, SET4, and SET6 exhibited higher percentages of ELT-2 ChIP-seq peaks (Mann et al., 2016; Wiesenfahrt et al., 2016) (S4A Figure), an over-representation of extended TGATAA motifs in their promoters (Suppelementary Fig. 4B), and an over-representation of GO ontology terms associated with intestinal tissue (Supplementary Fig. 4C). None of the gene sets showed the behaviour that would be expected if: (i) ELT-2 and ELT-7 were required to work together to activate transcription (i.e., single- and double mutant phenotypes would be expected to be equivalently strong), or; (ii) ELT-2 and ELT-7 functioned in an additive manner (i.e., equivalent loss of function phenotypes in the elt-2(−) and the elt-7(−) single mutants but a stronger phenotype in the double mutant.)
We also wished to analyze, in a more quantitative fashion, how the total transcript levels of intestine-specific genes changed in the elt-2(−) single mutant and in the elt-7(−);elt-2(−) double mutant relative to wildtype. The same 81 intestine-specific, dynamically changing genes used for Fig. 4A were first ranked with respect to the magnitude of the RPKM differences measured following loss of ELT-2 (RPKM = reads per kilobase gene model per million mapped reads). Because expression of an intestine-specific gene can either be decreased or increased upon removal of ELT-2 (Fig. 3A, 4A), differences in RPKM could be either negative or positive. A plot of the (arithmetic) magnitude of the differences in RPKM vs. gene rank was not informative because it was dominated by the few most strongly responding genes. Fig. 4C thus displays the differences in RPKM as the signed Log2(Absolute Value(RPKMfor mutant – RPKMfor wildtype control)), where the sign is negative for genes whose expression decreases upon loss of ELT-2 and positive for those genes whose expression increases; the zero-point on the Y-axis is undefined. (We recognize that RPKM transformations are not optimal for identifying differentially expressed genes because they can inappropriately skew count data. However, in the present analysis, our aim was not to compare the expression status of one gene across different conditions but to compare the absolute change in expression intensity of different genes; for this reason, normalizing for gene length was required.) The quantitative response of individual intestine-specific genes to loss of ELT-2 is thus captured on Fig. 4C as the smoothly varying closely-connected track of data points (red), trending from genes with the greatest decrease in gene expression on the lower left to genes with the greatest increase in gene expression on the upper right. Roughly one-third of the intestinal specific genes showed a decrease in RPKM upon loss of ELT-2 and two-thirds showed an increase, mirroring the proportions of genes in Class A vs. genes in Class B + C seen on Fig. 4A. Keeping the 81 intestine-specific genes in the same rank order on the X-axis, Fig. 4C adds a second set of data points (blue) corresponding, for each individual gene, to the differences in RPKM measured between the elt-7(−); elt-2(−) double mutant and wildtype. A vertical trajectory (i.e. maintaining the same rank) from red point to blue point thus describes how one particular intestine-specific gene responds to the loss of ELT-7 within a background that is already lacking in ELT-2.
The main features of Fig. 4C are as follows:
Fig. 4C clearly shows that there were three distinguishable ways by which the actual transcript levels (RPKM) of intestine-specific genes responded to loss of ELT-2 and to loss of both ELT-2 and ELT-7. By and large, these three response classes correspond to the A, B and C response classes identified by the clustering algorithm used to produce Fig. 4A and we designate the corresponding classes on Fig. 4C as A’, B’ and C’.
Transcript levels of Class A’ genes were lowered upon loss of ELT-2. Several genes in this class showed further reduction in transcript levels upon further loss of ELT-7 but most did not (at least on the logarithmic scale used for Fig. 4C). We suggest that ELT-2 acts as a transcriptional activator for Class A’ genes and ELT-7 can provide varying levels of transcriptional input in the absence of ELT-2.
Transcript levels of Class B’ genes increased upon loss of ELT-2 and underwent an ~equal and opposite decrease upon further loss of ELT-7. As noted above for Class B genes on Fig. 4A, we suggest that these are genes for which ELT-7 can transcriptionally overcompensate for loss of ELT-2. In the absence of ELT-2, ELT-7 is now the major transcriptional activator and the transcript levels of Class B’ genes drop when ELT-7 is also removed.
Transcript levels of Class C’ genes increased upon loss of ELT-2 but remained elevated above wildtype levels upon further loss of ELT-7. As noted above for Class A genes on Fig. 4A, we suggest that these are genes for which some as-yet-unidentified transcription factor or factors (“X”) can transcriptionally overcompensate for loss of ELT-2. Transcript levels remain elevated upon further loss of ELT-7 because “X” is now the main transcriptional activator. We note that, for several genes in Class C’, transcript levels did drop upon loss of ELT-7 although they remained above wildtype levels, suggesting that ELT-7 may also contribute to transcriptional activation of these genes in the absence of ELT-2.
2.6. Assessing the contributions of the endoderm-specifying GATA factors END-1/3 to intestine differentiation in the absence of ELT-2 and/or ELT-7
A feature of the C. elegans endoderm is that all of the core regulators belong to the same transcription factor family, namely GATA factors, and thus they can all interact with the same DNA sequence motif, although the relative strengths of such interactions are not known (Du et al., 2016; Wiesenfahrt et al., 2016). In the normal course of endoderm development, ELT-7 is expected to be the immediate activator of elt-2 transcription (Sommermann et al., 2010) even though ELT-2 can be activated in the absence of ELT-7 (McGhee et al., 2007). To distinguish the relative contributions of END-1/3 and ELT-7 to the initial activation of elt-2 transcription, we measured the time at which an integrated elt-2-promoter::GFP transgenic reporter could first be detected in wildtype and elt-7(−) embryos (see Methods for details). As shown in Supplementary Fig. 5, elt-2 expression was delayed by 15–20 min by lack of ELT-7, indicating that ELT-7 does indeed contribute to the activation of elt-2. However, the effect is modest, suggesting that either END-1/3 are normally the major activators of elt-2 expression, END-1/3 are able to compensate in the absence of ELT-7, or there is some other activator of elt-2 yet to be discovered.
The short delay in elt-2 onset time in the elt-7(−) mutant does not lead to an overall retardation of C. elegans development, and elt-7(−) mutant embryos hatch at the same time as wildtype embryos (Supplementary Fig. 6). Indeed, even elt-2(−) mutant embryos, in spite of their imminent arrest, hatch at close to the same time as wildtype embryos. In contrast, hatching of the elt-7(−); elt-2(−) double mutant embryos is delayed by 3–7 hours (Supplementary Fig. 6).
As noted in the Introduction, the intestine of the elt-2(−) mutant is defective but is nonetheless clearly specified and highly differentiated (Fukushige et al., 1998; McGhee et al., 2007). The intestine of the elt-7(−); elt-2(−) double mutant is even more defective, especially evident at high resolution (Sommermann et al., 2010). However, at low resolution, the intestine of the elt-7(−); elt-2(−) double mutant appears both specified and differentiated; a typical example is shown in Supplementary Fig. 7 compared to a wildtype sibling. What transcription factor is responsible for activating the differentiation genes in the double mutant intestine? (Sommermann et al., 2010) had previously suggested that this residual differentiation could be the result of an unknown transcription factor (“X”) whose expression could be initiated by END-1/3. In the previous section, we provided evidence that, in the absence of ELT-2 (and independent of the presence of ELT-7), a subset of intestine-specific genes could be transcribed under control of an unknown factor(s) (also termed “X”) that may or may not be different from the Sommermann et al. (Sommermann et al., 2010) factor. An additional possibility is that END-1 and/or END-3 could activate intestinal differentiation genes early in embryogenesis and the gene products (either transcripts or proteins) could persist after END-1/END-3 decay at the 4E-to-8E-cell stage (see discussion in (Sommermann et al., 2010)). Thus, END-1/3 would contribute to activities of intestine-specific genes in the elt-7(−); elt-2(−) double mutant, as well as contributing to or influencing the double mutant phenotype. The behaviour of the ges-1 gene in elt-7(−); elt-2(−) mutant embryos provides an opportunity to assess whether END-1/END-3 contribute to at least this one example of intestinal differentiation. ges-1 encodes a non-specific esterase whose enzymatic activity can be detected starting at the late 4 E cell stage (Edgar and McGhee, 1986). Deletion or mutation of a tandem pair of GATA sites roughly 1.1 kb upstream of the ges-1 initiation codon essentially abolishes GES-1 reporter activity in the intestine (Egan et al., 1995). Thus, if either ges-1 transcripts or GES-1 enzymatic activity are not detected in the elt-7(−); elt-2(−) intestine, we can infer that a GATA factor is not likely to be involved in activating ges-1 transcription, i.e., this result would rule out the participation of END-1 and/or END-3 since both ELT-2 and ELT-7 are absent (and ignoring the non-functional ELT-4 (Fukushige et al., 2003)). First of all, the above RNA-seq data show that ges-1 transcripts can still be detected in the elt-7(−); elt-2(−) double mutant embryos (wildtype RPKM = 35 +/− 4; elt-2(−) RPKM = 42 +/− 3; elt-7(−) RPKM = 24 +/− 4; elt-7(−); elt-2(−) RPKM = 18 +/− 3). Secondly, GES-1 enzymatic activity can still be detected in the double mutant intestine (Supplementary Fig. 9), although activity is reduced (see also (Sommermann et al., 2010)) and now appears to be enriched in the intestine anterior. These results are consistent with a model in which the early activity of END-1 and/or END-3 contributes to GES-1 activity and perhaps other differentiated features found in the elt-7(−); elt-2(−) intestine. (ges-1 belongs to gene class B/B’ on Fig. 4A and 4C, suggesting that loss of ELT-2 does not lead to transcriptional overcompensation by factor “X”, which could then activate ges-1 by binding to a non-GATA site in the ges-1 promoter.)
2.7. Interchangeability of C. elegans endodermal GATA factors
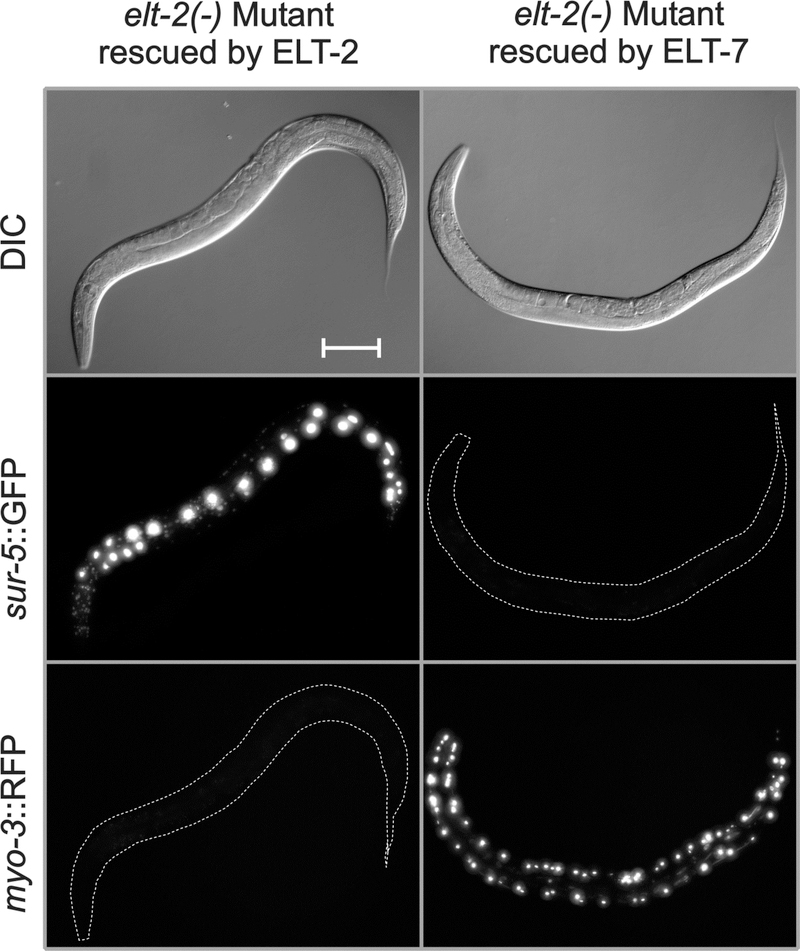
Loss of ELT-2 influences many more transcripts than does loss of ELT-7 but does this reflect greater molecular capabilities of the ELT-2 protein or different levels/patterns of elt-2 gene expression? To distinguish between these two possibilities, we placed elt-7 cDNA (with or without an epitope tag) under control of the elt-2 promoter and tested the ability of this construct to rescue the lethality of the elt-2(−) mutant. Several different injection markers and starting strains were used (see Methods for details) but the basic protocol was to introduce the elt-2-promoter::elt-7 cDNA construct as a new transgenic array into an elt-2(−) strain in which the elt-2(−) lethality was rescued by a pre-existing extrachromosomal transgenic array expressing the wildtype elt-2 gene. Progeny were then inspected for animals that had lost the original array but had retained the replacement array. Over the course of these experiments, some 100–150 elt-2(+)-rescued elt-2(−) hermaphrodites were injected and ten independent strains were eventually recovered in which transgenic arrays had been interchanged, i.e., in which ELT-7, expressed under control of the elt-2 promoter, was able to stably rescue the elt-2(−) mutation. One example is documented in Fig. 5. PCR assays confirmed the presence of the elt-2(−) deletion but failed to amplify the wildtype elt-2 gene or even the ELT-2 zinc finger domain (Supplementary Fig. 9). Although these strains were produced inefficiently, once produced or identified they appeared healthy. The brood size for one such strain (JM269 elt-2(−); caEx22 ) was measured as 208 +/− SD=20, compared to the N2 wildtype control of 270 +/− SD = 21 (four broods for each strain).
Fig. 5. ELT-7 expressed under control of the elt-2 promoter can rescue the elt-2(−) lethal arrest phenotype.
The three panels on the left are images of the starting strain JM147 in which the elt-2(−) mutation is rescued by an extrachromosomal transgenic array containing the genomic elt-2(+) gene and sur-5::GFP. As shown in the three panels on the right, the elt-2(+) rescuing array in JM147 can be replaced by a second extrachromosomal transgenic array containing the elt-2 promoter driving expression of an elt-7 cDNA as well as a myo-3::RFP construct expressed in bodywall muscle. Animals shown are one day past L4 stage. Images, from top to bottom, are Differential Interference Contrast (DIC), maximum point projection of a GFP stack, and maximum point projection of an RFP stack. Microscopic and image analysis parameters are identical between the two animals, except that the GFP exposure for the ELT-7 rescued strain was 4-fold higher than for the starting strain, in order to ensure that even faint GFP expression could have been detected. Scale bar = 100 microns.
We had previously shown that ELT-2, expressed under control of the end-1 promoter as well as its own endogenous promoter, was able to replace all other members of the core endoderm GATA factor network, i.e. END-1, END-3, ELT-7 (and ELT-4), and to direct both specification and differentiation of the C. elegans intestine (Wiesenfahrt et al., 2016). We showed that ELT-7 expressed under control of the end-1 promoter was also able to specify the C. elegans endoderm but full rescue required the endogenous elt-2 gene (Wiesenfahrt et al., 2016). Based on the results of the previous paragraph, we predicted that it should be possible to construct a viable strain in which “ELT-7 does it all”, i.e. in which elt-7 cDNA under control of both the end-1 and elt-2 promoters is able to rescue animals that are at least triply mutant (end-1(−) end-3(−); elt-2(−)). Three of the strains in which the elt-2-promoter::elt-7 cDNA rescued the elt-2(−) mutant also contained an end-1-promoter::elt-7 cDNA construct. As described in more detail in the Methods section, one of these strains (JM271) was crossed to a transgenically rescued elt-7(−) end-1(−) end-3(−) strain (JM243). An elt-7 rescued progeny strain (JM273) could indeed be identified. Supplementary Fig. 10 shows the results of PCR assays that verify that this strain is indeed deleted for all three of end-1, end-3 and elt-2 genes and, because of the nature of the cross, also deleted for elt-7.
3. Discussion
The ELT-2 and ELT-7 GATA factors provide an opportunity to explore the quantitative basis of transcription factor redundancy. Both factors are normally involved in regulating genes associated with differentiation of the C. elegans intestine but their loss causes quite different phenotypes. Loss of ELT-2 causes complete larval arrest and premature death, presumably because of the malformed intestine (Fukushige et al., 1998). Loss of ELT-7 causes no obvious phenotype (McGhee et al., 2007) but significantly exacerbates the phenotype caused by loss of ELT-2 (Sommermann et al., 2010). The customary view is thus that ELT-2 and ELT-7 are “partially redundant”. In the current study, we investigated the global transcriptional responses to loss of ELT-2 and/or ELT-7 as measured in first stage larvae. Results from the initial analysis were consistent with the different severities of the loss-of-function phenotypes. That is, loss of ELT-2 caused the downregulation of 25-fold more genes than did loss of ELT-7, loss of ELT-7 caused only minor transcriptional responses, and loss of both ELT-2 and ELT-7 caused mis-regulation of many genes in addition to those mis-regulated by loss of ELT-2 alone. It is not clear at the molecular level why some genes responded to loss of ELT-2 alone and other genes only responded strongly to loss of both ELT-2 and ELT-7. We searched for over-represented sequence motifs in the promoter regions from 619 genes significantly downregulated upon loss of ELT-2 and from 647 genes significantly downregulated upon loss of ELT-7 in the elt-2(−) background. Both analyses yielded essentially identical numbers and locations of the same extended TGATAA motif previously identified as associated with genes expressed in the intestine (Pauli et al., 2006; McGhee et al., 2007; McGhee et al., 2009).
When the behaviour of individual genes was tracked in the different mutant backgrounds, a richness of responses was revealed that was not apparent from consideration of either the entire data set (e.g. Fig. 2) or the numbers of significantly influenced genes (e.g. Fig. 3A). On the one hand, each individual gene seemed to have an essentially unique quantitative response to the different mutant backgrounds and these responses could not be captured by the usual binary representation of transcription factor redundancy using a Venn diagram. On the other hand, the different responses of genes expressed specifically in the intestine could be placed into three general classes, which can be briefly described as follows. Expression of genes in Class A/A’ (Fig. 4) largely depends on ELT-2, with ELT-7 providing a weak and variable degree of redundancy. Expression of genes in Class B/B’ (Fig. 4) increased upon loss of ELT-2 but decreased upon subsequent loss of ELT-7, suggesting that ELT-7 actually over-compensates when ELT-2 is missing. Finally, expression of genes in Class C/C’ (Fig. 4) increased upon loss of ELT-2 but remained elevated upon subsequent loss of ELT-7, suggesting that some other factor (“X”) can over-compensate in the absence of ELT-2. It is not yet clear what molecular mechanisms cause intestine-specific genes to fall into these three distinct classes. We note that examples from each gene class can be found on which prominent ELT-2 ChIP-seq peaks can be detected, implicating direct binding of ELT-2 in their control. Finally, we point out one class of intestine-specific genes that was not observed, namely genes whose transcript levels were influenced identically by loss of ELT-2, loss of ELT-7 or loss of both ELT-2 and ELT-7; the absence of such genes suggest that ELT-2 and ELT-7 do not “synergize” at the molecular level, for example by forming an obligatory hetero-dimer.
Three different models can be proposed to explain how ELT-7 could overcompensate for loss of ELT-2 in regulating genes of Class B/B’. In the first model, ELT-2 ordinarily activates Class B/B’ genes but represses the elt-7 gene, directly or indirectly; upon loss of ELT-2, ELT-7 protein levels would ultimately increase and lead to increased transcription of genes in Class B/B’. However, we could find no significant evidence that ELT-7 levels are elevated in the absence of ELT-2 (Supplementary Fig. 3). A second model is that ELT-2 directly represses genes of Class B/B’ and there are precedents for direct gene repression by GATA factors in other systems (Chlon and Crispino, 2012). In the absence of ELT-2 repression, ELT-7 activated transcript levels increase. In the third model, we suggest that ELT-2 and ELT-7 normally compete for binding to target TGATAA sites and consequent target gene activation; we further suggest that ELT-2 is a better competitor but a weaker transcriptional activator than is ELT-7. Thus, in the absence of ELT-2, the target TGATAA sites are now occupied by ELT-7 and levels of target gene transcripts increase. Evidence that ELT-7 is an intrinsically more powerful transcriptional activator than is ELT-2 is provided by the greater ability of ELT-7 to induce “transorganogenesis” when expressed ectopically (Riddle et al., 2013; Riddle et al., 2016). At the moment, we have no basis for favouring either of the last two models and perhaps different models apply to different members of Class B/B’ genes.
Before we performed the current study, we would have predicted that the majority of intestine-specific genes would belong to Class A/A’, in which ELT-2 acts as the predominant transcriptional activator. In the absence of ELT-2, the transcriptional contribution of ELT-7 could vary from none to complete depending on the individual gene (Sommermann et al., 2010). However, in the current analysis, only ~33% of dynamically-expressed, intestine-specific genes expressed in the embryo behaved as we would have predicted, in contrast to the ~60% estimated in our previous SAGE analysis (McGhee et al., 2009). It is possible that this discrepancy is primarily quantitative, resulting from the vastly different technologies and analysis methods used, or, our preferred explanation, resulting from the fact that the set of intestine-specific genes used in (McGhee et al., 2009) was biased towards genes whose expression patterns could be defined by in situ hybridization and that hence were highly expressed. These highly expressed genes (digestive enzymes, for example) might only require an on/off switch as a control mechanism and therefore might be the ones for which ELT-2 acts as a “simple” transcriptional activator. We further suggest that such genes will form a greater fraction of the expressed intestine-specific genes as the animal matures and emphasizes its nutrition over its development. Hence the quantitative transcriptional responses to loss of ELT-2/ELT-7 are likely to vary with stage of life and the transcriptional influence of ELT-7 might be greatest in embryos.
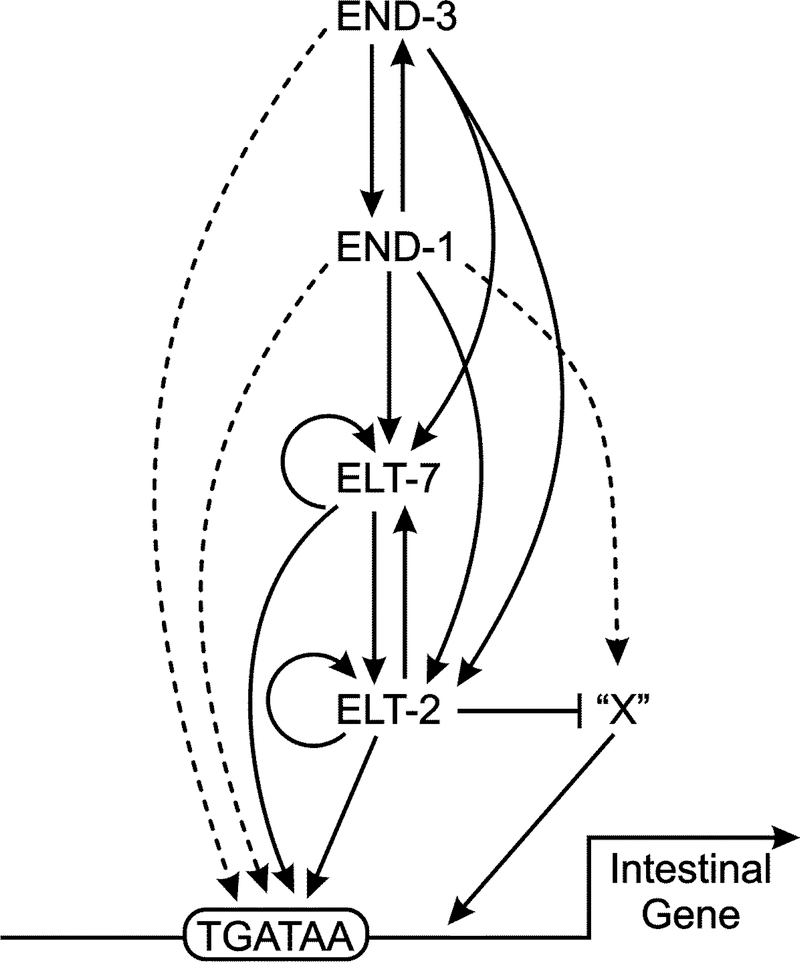
Our results suggest several minor adjustments to the current model of the core regulatory pathway driving C. elegans intestine development. The intestine of the elt-7(−); elt-2(−) double mutant still expresses differentiation markers in spite of its defects. GES-1 activity detected in the double mutant intestine is consistent with such residual intestinal differentiation deriving from the activity of END-1/END-3 in the early embryo and persisting either as transcripts or as proteins. To account for intestinal differentiation in the absence of ELT-2 and ELT-7, (Sommermann et al., 2010) had proposed an additional transcription factor “X”, initially under control of END-1/END-3 and auto-regulating thereafter (see their Fig. 6). As noted above, our present results suggest that the “X” model should be elaborated to include (or perhaps to solely consist of) one or more transcription factors that are induced by the loss of ELT-2 (Class C/C’ genes from Fig. 4)). We have previously identified several hundred transcription factors present in the embryonic endoderm that could be candidates for “X” (McGhee et al., 2009). The C. elegans core endoderm regulatory pathway incorporating these minor adjustments is shown on Figure 6.
Fig. 6.
Proposed current version of the core regulatory pathway driving differentiation of the C. elegans intestine
The last question we wish to consider is the ability of redundant transcription factors to replace each other. (Shoichet et al., 2000) had previously shown that the C. elegans END-1 protein was capable of inducing endoderm in embryos of the frog Xenopus laevis, presumably providing or augmenting the activity of an endogenous GATA factor. In the present study, we were able to demonstrate that ELT-7, if placed under control of the elt-2 promoter, is able to replace ELT-2 in the C. elegans intestine. Although the overall replacement/rescue/identification appeared inefficient, rescued strains were viable and appeared healthy. We find this interchangeability of ELT-2 and ELT-7 to be remarkable, considering that the transcriptional consequences of losing ELT-7 are relatively minor compared to the consequences of losing ELT-2, and also considering the low level of sequence conservation between ELT-2 and ELT-7. A sequence alignment between these two proteins is shown on Supplementary Fig. 11. Even the 25 residue zinc-finger DNA-binding domains do not appear to be particularly highly conserved. The zinc-finger domain shows 9 mismatches between ELT-2 and ELT-7 (64 % identical), whereas there are only 8 mismatches when either the ELT-2 or the ELT-7 zinc-fingers are compared with the similar domain of the hypodermal GATA factor ELT-3 (Gilleard et al., 1999). And yet, the ELT-2 zinc-finger is highly conserved in other nematodes (e.g. 2/25 mismatches with Haemochus contortus), suggesting that most residues are functionally important.
Finally, we showed that a strain of worms could be produced in which “ELT-7 does it all”, that is, ELT-7 both specifies and differentiates the intestine in the absence of END-1, END-3, ELT-2 and even endogenous ELT-7. Although the possibility of producing this strain was predicted by the results of the previous paragraph and by our previous demonstration that ELT-7 under the end-1promoter could specify the endoderm (Wiesenfahrt et al., 2016), we nonetheless find this result also remarkable. The limited sequence alignments (Supplementary Fig. 11) suggest that any “endoderm-specifying” or “endoderm-differentiating” domains held in common between the core endoderm GATA factors might be surprisingly short. Future work will focus on quantitative analysis of the growth and development of these various replacement strains, in order to define the molecular features of transcription factor redundancy (e.g., protein-DNA interactions, protein-protein interactions, expression levels and regulatory network structure) that provide the ability to specify endoderm fate and then to produce a robust functioning intestine.
4. Materials and methods
4.1. C. elegans strains.
Standard nematode handling conditions were used (Brenner, 1974). Animals were grown at 20°C unless otherwise indicated. Strains used were wild-type N2, JM69 dpy-5 unc-13/szT1[lon-2] I; elt-2(ca15)/szT1 X, JM147 elt-2(ca15) X; caEx3, JM199 elt-7(tm840) V; elt-2(ca15) X; caEx3, JM222 elt-7(tm840) V (retrieved as a segregant from an out-cross of JM199), JM149 caIs71, JM243 elt-7(tm840) end-1(ok558) end-3(ok1448) V; elt-4(ca16); caEx7, JM259 elt-7(tm840); caIs71, JM267 elt-2(ca15) X; caEx20, JM269 elt-2(ca15) X; caEx22, JM271 elt-2(ca15) X; caEx24, JM273 elt-7(tm840) end-1(ok558) end-3(ok1448) V; elt-2(ca15) X; caEx24, JM278 elt-2(ca15) X; wIs126(elt-7promoter::GFP-lacZ); caEx20, and JR2132 wIs126 (elt-7promoter::GFP-lacZ). The multi-copy extrachromosomal array caEx3 contains [pJM276 (full rescuing elt-2 genomic region), pTG96 (sur-5::GFP (Gu et al., 1998)) and pRF4 (rol-6(su1006) (Mello et al., 1991)]). The multi-copy extrachromosomal array caEx7 contains [pJM581 (end-1promoter::elt-7 cDNA) and pJM473 (myo-3promoter::tdTomato)]. The multi-copy integrated array caIs71 contains [pJM370 (5.2 kb elt-2promoter::nuclear-localized-GFP) and pRF4]. The multi-copy extrachromosomal array caEx20 contains [pJM276 (full rescuing elt-2 genomic region), pJM473 (myo-3promoter::tdTomato) and pMA122 (heat shock promoter::peel-1 (Frokjaer-Jensen et al., 2012))]. The multi-copy extrachromosomal array caEx22 contains [pJM773 (elt-2promoter::elt-7 cDNA), pJM762 (ifb-2promoter::GFP and pRF4]. The multi-copy extrachromosomal array caEx24 contains [pJM773 (elt-2promoter::elt-7 cDNA), pJM581 (end-1promoter::elt-7 cDNA), pJM762 (ifb-2promoter::GFP) and pRF4]. Strains JM269 and JM271 were produced by injections into JM267 hermaphrodites and identifying Rolling progeny that were Green Not-Red and that were able to survive heating for two hours at 34°C. Strain JM273 (in which ELT-7 replaces all other core endodermal GATA factors) was constructed by crossing JM243 males with JM271 hermaphrodites and identifying F2 progeny that were Rol Green not-Red and subsequently verified by PCR (Supplementary Fig. 10).
4.2. COPAS Biosorting and RNA Extraction.
Worms from starved NGM plates were transferred as chunks to fresh 150 mm diameter NGM plates (seeded with E. coli OP50) and allowed to develop at 20°C until the majority were gravid adults. Animals were collected in 15 ml of M9 buffer per 150 mm plate, concentrated by centrifugation (~200g, 5 min, room temperature) and washed once with 50 ml of M9 buffer. The final pellet was suspended in 5 ml of dH20, mixed with 17.5 ml of alkaline bleach solution (5ml 6% hypochlorite with 12.5 ml 1M NaOH), incubated 4–6 minutes (until ~50% of the gravid adults had burst), mixed with an equal volume of alkaline bleach stop solution (0.95 M Tris-HCl with 0.05 M Tris Base), vortexed briefly and centrifuged as above. Collected embryos were suspended in 50 ml of 0.45 μm filtered M9 buffer, re-centrifuged, re-suspended in 50 ml of filtered M9 buffer, transferred to a 75 cm2 tissue culture flask and allowed to hatch overnight on a shaker at room temperature. The next morning (12–14 hours after bleaching), the starved L1 larvae were passed twice through a 27 μm nylon mesh and placed on a shaker in 50 ml tubes. Filtered worms were then passed as continual batches through a Union Biometrica Complex Object Parametric Analyzer and Sorter (COPAS) platform with Biosort Device (488 nm excitation) capable of sorting green larvae from non-green. For each sort of strains JM147 and JM199, a total of 50,000 GFP positive and 50,000 GFP negative worms were isolated and two consecutive sorts of each type were usually pooled into one sample. For strains N2 and JM222 elt-7(−), 50,000 worms were collected from the sorter and an unknown number (>100,000) of worms collected from the filtered but not sorted populations. Passage of wildtype N2 control animals through the COPAS Biosorter changes their transcript profile (Supplementary Fig. 13). Hence, both wildtype N2 animals and animals from the viable strain JM222 elt-7(−) were passed through the COPAS Biosorter in exact parallel to both rescued and non-rescued segregants from strains JM147 and JM199. Larvae were stored in 50 ml tubes in M9 buffer on the shaker until the entire sample had been collected.
Pooled larvae from each sort were collected by centrifugation (~200g, 5 min, RT), the pellet transferred to an RNase free tube and re-centrifuged (~500g, 3 min, RT). The supernatant was removed, 1 ml of TRizol and 10 μl of β-mercaptoethanol were added to the pellet, and the sample was mixed by inversion, flash frozen in liquid nitrogen and stored at −80°C. The time between larval hatching and sample freezing is roughly 6–8 hours, by which time transcriptional responses to starvation have stabilized (Baugh et al., 2009). Total RNA extractions were performed using the Ambion PureLink RNA Mini Kit. Quality and concentration of RNA samples were assayed using an Agilent Tapestation. The qualities of RNA samples were uniformly high (average RIN = 9.5).
4.3. Library Preparation and RNAseq (U of Calgary Core DNA Services).
Poly-A+ mRNA was isolated using the Life Technologies Dynabeads mRNA Direct Kit (part #61021; standard method). Whole transcriptome libraries were prepared with the Life Technologies Solid Total RNA-Seq kit (part #4445374; standard method) using chemical fragmentation and Array Script reverse transcriptase. Sequencing was performed on a Life Technologies 5500xl genome analyzer on two separate runs generating 50 bp paired-end reads. The mean number of reads per sample was in the range of 15–20 million.
4.4. RNA-seq Alignment.
Raw sequence reads were mapped by Dr. Paul Gordon, (Genomics and Bioinformatics, University of Calgary) to the WS220 ce10 version of the C. elegans genome using Lifescope ver. 2.5. Quality control of sequence reads was assessed using FastQC version 0.52. The resulting alignments were tabulated for read counts associated with each gene model using HTseq 0.6.1 (Anders et al., 2015), Samtools 1.3.1 (Li, 2011), and the WS235 genome feature file (WS235, Ensemble 78, Caenorhabditis_elegans.WBcel235.78.gtf.gz). The genome feature file was filtered for protein coding sequences, transferred to the ce10 genome and renamed C_elegans_WS235_lifeover_ce10.gtf.gz. HTseq’s HTseq.scripts.count f unction was performed with the following options: --mode=union --stranded=no --minaqual=20. elt-2(−) samples that did not show sufficient reduction in elt-2 gene expression were filtered from analysis. This removed one sample from the study. The raw and processed RNA-seq data generated in this study as well as the genome feature file used for alignment have been deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE107175 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107175)
4.5. Differential Expression Analysis.
RNA-seq count data (Supplementary Table 1) were filtered for detected genes (> 10 counts across all samples) and analyzed using DESeq2 (Love et al., 2014) with Cook’s cutoff filtering set to FALSE. This procedure performed read depth normalization, corrected for gene-wise dispersion estimates, and performed negative binomial Wald Tests without outlier filtering. Count data was normalized, variance-stabilized, and log transformed using the rlog utility (Supplementary Table 2). To illustrate correlations between samples, sample-to-sample Euclidian distances were calculated using the Complete cluster method on normalized, variance-stabilized, log transformed count data (Fig. 2). Genes were counted as differentially expressed if they registered a Benjamini-Hochberg corrected p-value less than 0.05 and a log2-fold change greater than 1.2 between any pair-wise conditional comparison (Supplementary Tables 3 and 4).
ELT-2 ChIP-seq data from (Wiesenfahrt et al., 2016) and (Mann et al., 2016) were imported and ChIP-seq peaks were identified as being associated with a given gene promoter if they fell within 3kb upstream or 1kb downstream of the mean transcriptional start site reported for all transcripts arising from a given gene. If multiple ChIP-seq peaks fell within this region, their peak scores were summed and the number of total peaks was tabulated.
4.6. Clustering.
Differentially expressed, intestine-specific genes (n=81; Supplementary Table 5) were hierarchically clustered into classes using the complete method on the Spearmen correlation distances calculated on row-means-normalized, variance-stabilized, log transformed count data from the DESeq2 analysis, and a heat map was drawn depicting relative expression across all samples. The three main classes were identified as Class A, Class B, and Class C genes. The ELT-2 combined ChIP-seq peak scores were annotated onto this heat map (Supplementary Fig. 4A; Fig. 4A). A similar analysis was performed on all differentially expressed genes (n=3092; Supplementary Table 6).
The differential expression and clustering analyses employed R_3.2.4, DESeq2_1.10.1, pheatmap_1.0.8, RColorBrewer_1.1–2, circlize_0.3.7, ComplexHeatmap_1.6.0 (Gu et al., 2016), gplots_3.0.1, dplyr_0.5.0, GenomicRanges_1.22.4 (Lawrence et al., 2013), and IRanges_2.4.8).
4.7. Genome browser.
RNA-seq alignment files in .bam format were normalized for read depth, averaged, and converted into BigWig format using automated scripts that employed bedtools (bamtobed, sort, and genomecov) v2.26.0 (Quinlan and Hall, 2010), java-genomics-toolkit wigmath.Average, and the UCSC Kent utility wigToBigWig v4. Generated files have been deposited in the NCBI GEO database (GSE107175).
4.8. Motif Finding.
Over-represnted motifs were identified in gene promoter regions on sets of gene identifiers using homer v4.8 (Heinz et al., 2010) (promoter library ce10, v5.5) by searching for motifs of 8, 10, and 12 mer lengths. The occurences of top hitting motifs and the histograms of their occurrences across a meta-promoter region were calculated using homer and illustrated graphically in R.
4.9. Bifurcation Plot.
Differentially expressed, intestine-specific genes (n=81; Supplementary Tables 1 and 4) were filtered and calculated for RPKM (Reads Per Kilobase gene model per Million mapped reads) to allow for improved comparisons between the estimated changes in expression levels across genes. These genes were ranked by absolute differences in RPKM between elt-2(−) and wild-type strains. Logarithmic transformations of the absolute value of differences between the elt-2 and wild-type strains were calculated and plotted against the rank order calculated above (red, Fig. 4C), taking negative values into account. Similar logarithmic transformations of the absolute differences between the double mutant and wild-type strains were also calculated and plotted (blue, Fig. 4C) with shapes depicting the Class of each gene as identified on Fig. 4A.
4.10. Miscellaneous.
To measure the onset time of ELT-2 expression (Supplementary Fig. 5), embryos were picked at the two-cell stage in Boyd’s buffer (Murray et al., 2008), transferred to agarose pads on a microscope slide in a wet chamber, incubated at room temperature for 120–140 mins, covered with a cover slip, sealed with molten paraffin and GFP fluorescence recorded at timed intervals using low light settings and a five slice Z-stack. Image collection was stopped as soon as GFP could be unambiguously detected at high contrast. In this way, embryos were exposed to minimum light levels and the large majority of observed embryos hatched; the few embryos that did not hatch reached the pretzel stage. Embryo staining for GES-1 esterase activity (Supplementary Fig. 8) was performed basically as described by (Edgar and McGhee, 1986), except that the concentration of substrate α-naphthyl acetate was decreased two-fold; colour images were recorded using a Zeiss AxioCam HRc. For the two strains with rescued embryonic lethality (JM147 and JM199), non-Green (i.e. Non-rescued) embryos were selected using a fluorescent dissecting microscope prior to GES-1 staining.
Supplementary Material
Supplementary Fig. 1.
Properties of the six L1 larval populations used for RNA-seq.
For each of the four strains (N2, JM147, JM199 and JM222; see Fig. 1 for genotypes), the COPAS Biosorter profile is shown (x-axis = L1 larval length (time of flight); y-axis = GFP intensity; collected area indicated by box in each profile). Representative sample of sorted populations shown as Differential Interference Contrast (DIC) image, and by GFP fluorescence; scale bar = 100 microns. Larvae from the two non-transgenic strains N2 and JM222 were “mock-sorted” by passage through the Biosorter. Larvae from the two transgenic strains JM147 and JM199 were sorted into “Non-Green Non-Rescued” and “Green Rescued” populations, depending on whether larvae had lost or had retained the caEx3 extrachromosomal transgenic array. An example of a rare mis-sorted (non-fluorescent) worm is shown by the white arrow in the JM199 GFP image.
Supplementary Fig. 10.
Gel images showing unsuccesful attempts to detect end-1, end-3, elt-7 and elt-2 coding region in the elt-7(−) end-1(−) end-3(−); elt-2(−) strains rescued by transgenic [elt-2promoter::elt-7 cDNA + end-1promoter::elt-7 cDNA]
Expected PCR-amplified fragment sizes are indicated; red “X” signifies that no band is expected if there is no coding region.
Supplementary Fig. 11.
Alignment of ELT-2, ELT-7, END-1 and END-3 amino acid sequences
Clustal-Omega algorithm with default settings, and displayed using MVIEW. The red box highlights the conserved zinc finger binding domains.
Supplementary Fig. 12.
Expression differences between COPAS sorted vs. filtered wildtype L1 worms
Fold change of gene expression (y-axis) between the filtered and sorted samples is plotted against the mean intensity for a given gene (x-axis). Differentially expressed genes for each comparison are shown in red (differential expression: total read counts > 10, and BH-adjusted Wald Test p-value < 0.1, and log2-fold change > 0.8). Of the 15,217 genes present, 108 were up-regulated in COPAS sorted conditions and 47 were down-regulated
Supplementary Table 1. Raw RNA-seq read counts tabulated for each sample.
Supplementary Table 2. Log-transformed, variance-stabilized RNA-seq read counts tabulated for each sample.
Supplementary Table 3. Results (res) tables from DESeq2 differential expression analyses generated from pair-wise comparisons between conditions.
Supplementary Table S4. Lists of genes significantly differentially expressed between key pairwise comparisons of conditions.
Supplementary Table S5. Hierarchical clustering output from 81 significantly changing genes that have been annotated as intestine-specific (Figure 4). The data table includes the gene’s assigned “class”, whether the gene contains an ELT-2 ChIP-seq peak, and whether the gene has been ascribed as intestine-enriched or intestine-specific.
Supplementary Table S6. Hierarchical clustering output from all 3092 significantly changing genes (Supplementary Figure 4). The data table includes the gene’s assigned “set”, whether the gene contains an ELT-2 ChIP-seq peak, and whether the gene has been ascribed intestine-enriched or intestine-specific status.
Supplementary Fig. 2.
Pairwise comparisons of RNA-seq data between wild-type and mutant strains focusing on all-genes, intestine-enriched genes, and intestine-specific genes.
Fold change of gene expression (y-axis) between the two labeled samples is plotted against the mean intensity for a given gene (x-axis). Differentially expressed genes for each comparison are shown in red (criteria for differential expression: total read counts > 10, BH-adjusted Wald Test p-value < 0.1, and log2-fold change > 0.8). The list of intestinal expressed genes was produced by querying WormBase (www.wormbase.org) for genes annotated to be expressed in the intestine/gut and verifying from WormBase that expression was primarily or exclusively in the intestine. This 2073 long gene list was supplemented with genes from intestinal SAGE libraries (McGhee et al., 2007; McGhee et al., 2009) and from (Sommermann et al., 2010). The final list was composed of 2210 genes.
Supplementary Fig. 3.
Levels of elt-2 and elt-7 transcripts in elt-7(−) and elt-2(−) mutants, respectively.
Transcriptional cross-regulation between elt-2 and elt-7 is shown as transcript levels calculated as RPKM (reads mapped per kilo-base gene model per million mapped reads) measured in wild type or single-mutant samples. Values above the comparisons are Benjamini-Hochberg corrected p-values. Note that the normalized read counts of the elt-7 transcript are not statistically different between wild type and elt-7(−) mutant because only the catalytically active DNA binding domain is deleted in the mutant. Much of the transcript still remains but the protein is not functional.
Supplementary Fig. 4.
Dynamically expressed genes cluster into six sets based on expression patterns.
A. We performed hierarchical clustering on dynamically expressed genes (n = 3092) and illustrate the results in a heat map. Clustering was performed using Spearman correlation distances of row-means-normalized log-stabilized read counts. This effort identified six sets of genes. The percentage of genes within each set that contain a significant ELT-2 Chip-seq peak (as determined by either of two published studies; (Mann et al., 2016; Wiesenfahrt et al., 2016)) is tabulated to the right of the heatmap.
B. The top over-represented motifs identified within the promoter regions of each gene set are illustrated as are the P-values associated with those motifs. Motifs and their P-values were determined using HOMER (Heinz et al., 2010).
C. The top gene categories associated with each gene set are tabulated along with their associated P values as determined using WORMEXP (Yang et al., 2016).
D. Depictions of potential types of epistatic relationships, their predicted expression patterns, and their representations in the heatmap above are shown.
Supplementary Fig. 5.
Loss of ELT-7 delays the onset time of elt-2promoter::GFP by 15–20 minutes.
The two strains JM149 and JM259 contain the same integrated transgenic array expressing elt-2promoter::GFP but are wildtype or null at the elt-7 locus, respectively. Embryos were picked at the 2-cell stage and the time at which GFP could first be detected was determined. Red vertical lines represent mean onset time for each strain. Number of embryos observed = 22 and 20 for strains JM149 and JM259, respectively. t-test probability that the onset times are equal < 10−5
Supplementary Fig. 6.
Time (20°C) between the 2-cell stage and embryo hatching for the four mutant strains.
From top to bottom, the panels correspond to strains N2, JM222 elt-7(−), JM147 elt-2(−); caEx3, and JM199 elt-7(−); elt-2(−); caEx3. In each panel, different symbols represent independent replicate experiments. For JM147 and JM199, filled symbols represent embryonic segregants that have lost the rescuing caEx3 array; open symbols represent segregants that have retained caEx3. The wide grey line corresponds to the range of data seen with independent N2 replicates (top panel) and is included for comparison.
Supplementary Fig. 7.
Representative images of JM199 elt-7(−); elt-2(−); caEx3 embryos squeezed from egg-shells at 15 hours after 2-cell stage (20°C).
The embryo/larva on the left is elt-7(−); elt-2(−) because it shows no GFP flurorescence marking the rescuing extrachromosomal transgenic array. The embryo/larva on the right is GFP-positive and therefore rescued by the extrachromsomal transgenic array. However, both embryos/larvae exhibit birefringence measuring intestinal differentiation (bottom panel, shown with inverted contrast). Ten micron diameter glass beads (visible in background) were placed around embryos to protect from over-squeezing. Scale bar = 20 microns.
Supplementary Fig. 8.
Staining for GES-1 esterase activity in ~1.5 fold stage embryos.
GES-1 esterase activity can be detected in the intestines of embryos with the following genotypes (from left to right) elt-7(−); elt-2(−), wild type strain N2, elt-2(−) and elt-7(−). Strain JM1041 contains two activity-lowering point mutations introduced into the ges-1 structural gene (McGhee et al., 1990); the low level of “background” staining represents the activity produced by other intestinal esterases and possibly from residual enzymatic activity in the mutated GES-1 protein.
Supplementary Fig. 9.
Gel images showing unsuccesful attempts to detect the ELT-2 coding region in the elt-2(−) strains rescued by elt-2promoter::elt-7 cDNA
Expected fragment sizes are indicated; red “X” signifies that no band is expected if there is no elt-2 coding region. Two pairs of primers are indicated for a particular feature (e.g. overall ca15 deletion) when nested PCR was performed.
Highlights:
The two GATA-type transcription factors ELT-2 and ELT-7 normally function in differentiation of the C. elegans intestine
ELT-2 is much more influential than is ELT-7 as judged by the transcriptional consequences of ELT-2 and/or ELT-7 loss of function in the embryo or early larva
ELT-2 activates roughly one-third of intestine-specific genes with ELT-7 only providing a modest contribution
For a second third of intestine-specific genes, ELT-7 appears to over-compensate for loss of ELT-2
For the final third of intestine-specific genes, some other factor besides ELT-7 appears to over-compensate for the absence of ELT-2
ELT-7 expressed under the elt-2 promoter can rescue the lethality caused by ELT-2 loss; indeed ELT-7 can replace all the other core endoderm GATA factors and both specify and differentiate the C. elegans intestine.
Acknowledgements
The authors gratefully acknowledge Dr. Paul Gordon, Dr. Richard Pon and the University of Calgary Bioinformatics Core for their assistance in SOLiD Sequencing and bioinformatics. This work was supported by an operating grant provided by the Canadian Institutes of Health Research (CIHR) to JDM. JDM received salary support from the Canada Research Chairs and Alberta Innovates- Health Solutions. EON is an Investigator of the Boettcher Foundation. JHR was supported in part by grants from the NIH (#1R01HD082347 and NIH 1R01HD081266).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Anders S, Pyl PT, Huber W, 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW, 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324, 92–94. [DOI] [PubMed] [Google Scholar]
- Boeck ME, Boyle T, Bao Z, Murray J, Mericle B, Waterston R, 2011. Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development. Dev Biol 358, 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, 1974. The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlon TM, Crispino JD, 2012. Combinatorial regulation of tissue specification by GATA and FOG factors. Development 139, 3905–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C, 2009. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet 5, e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, 2001. Genomic Regulatory Systems: Development and Evolution. Academic Press, San Diego. [Google Scholar]
- Davidson EH, 2006. The Regulatory Genome. Academic, San Diego. [Google Scholar]
- Du L, Tracy S, Rifkin SA, 2016. Mutagenesis of GATA motifs controlling the endoderm regulator elt-2 reveals distinct dominant and secondary cis-regulatory elements. Dev Biol 412, 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar LG, McGhee JD, 1986. Embryonic expression of a gut-specific esterase in Caenorhabditis elegans. Developmental Biology 114, 109–118. [DOI] [PubMed] [Google Scholar]
- Egan CR, Chung MA, Allen FL, Heschl MF, Van Buskirk CL, McGhee JD, 1995. A gut-to-pharynx/tail switch in embryonic expression of the Caenorhabditis elegans ges-1 gene centers on two GATA sequences. Developmental Biology 170, 397–419. [DOI] [PubMed] [Google Scholar]
- Felix MA, Wagner A, 2008. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity 100, 132–140. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM, 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods 9, 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Goszczynski B, Tian H, McGhee JD, 2003. The evolutionary duplication and probable demise of an endodermal GATA factor in Caenorhabditis elegans. Genetics 165, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD, 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol 198, 286–302. [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD, 1999. ELT-3: A Caenorhabditis elegans GATA Factor Expressed in the Embryonic Epidermis during Morphogenesis. Dev Biol 208, 265–280. [DOI] [PubMed] [Google Scholar]
- Gu T, Orita S, Han M, 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol 18, 4556–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M, Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ, 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data Software for computing and annotating genomic ranges. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ, 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, 2010. Cell fate specification in the C. elegans embryo. Dev Dyn. [DOI] [PubMed] [Google Scholar]
- Maduro MF, 2015. Developmental robustness in the Caenorhabditis elegans embryo. Molecular reproduction and development 82, 918–931. [DOI] [PubMed] [Google Scholar]
- Maduro MF, 2017. Gut development in C. elegans. Semin Cell Dev Biol. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Choi H, Carranza F, Wu AC, Rifkin SA, 2015. MED GATA factors promote robust development of the C. elegans endoderm. Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH, 2007. Maternal deployment of the embryonic SKN-1-->MED-1,2 cell specification pathway in C. elegans. Dev Biol 301, 590–601. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess JR, Rothman JH, 2005. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev Biol 284, 509–522. [DOI] [PubMed] [Google Scholar]
- Mann FG, Van Nostrand EL, Friedland AE, Liu X, Kim SK, 2016. Deactivation of the GATA Transcription Factor ELT-2 Is a Major Driver of Normal Aging in C. elegans. PLoS Genet 12, e1005956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, 2007. The C. elegans intestine (March 27, 2007) WormBook, eg. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.133.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, 2013. The Caenorhabditis elegans intestine. Wiley Interdisciplinary Reviews: Developmental Biology 2, 347–367. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, Gaudet J, Kohara Y, Bossinger O, Zhao Y, Khattra J, Hirst M, Jones SJ, Marra MA, Ruzanov P, Warner A, Zapf R, Moerman DG, Kalb JM, 2009. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol 327, 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG, 2007. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol 302, 627–645. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO Journal 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH, 2008. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods 5, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF, 2009. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK, 2006. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 133, 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH, 2016. Implications of Developmental Gene Regulatory Networks Inside and Outside Developmental Biology. Current topics in developmental biology 117, 237–251. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM, 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle MR, Spickard EA, Jevince A, Nguyen KC, Hall DH, Joshi PM, Rothman JH, 2016. Transorganogenesis and transdifferentiation in C. elegans are dependent on differentiated cell identity. Dev Biol 420, 136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle MR, Weintraub A, Nguyen KC, Hall DH, Rothman JH, 2013. Transdifferentiation and remodeling of post-embryonic C. elegans cells by a single transcription factor. Development 140, 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet SA, Malik TH, Rothman JH, Shivdasani RA, 2000. Action of the Caenorhabditis elegans GATA factor END-1 in Xenopus suggests that similar mechanisms initiate endoderm development in ecdysozoa and vertebrates. Proc Natl Acad Sci U S A 97, 4076–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermann EM, Strohmaier KR, Maduro MF, Rothman JH, 2010. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and −7 mediates the specification-->differentiation transition. Dev Biol 347, 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Wagner A, 2005. Robustness and Evolvability in Living Systems. Princeton University Press. [Google Scholar]
- Wiesenfahrt T, Berg JY, Osborne Nishimura E, Robinson AG, Goszczynski B, Lieb JD, McGhee JD, 2016. The function and regulation of the GATA factor ELT-2 in the C. elegans endoderm. Development 143, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Dierking K, Schulenburg H, 2016. WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics 32, 943–945. [DOI] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH, 1998. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev 12, 3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH, 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev 11, 2883–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1.
Properties of the six L1 larval populations used for RNA-seq.
For each of the four strains (N2, JM147, JM199 and JM222; see Fig. 1 for genotypes), the COPAS Biosorter profile is shown (x-axis = L1 larval length (time of flight); y-axis = GFP intensity; collected area indicated by box in each profile). Representative sample of sorted populations shown as Differential Interference Contrast (DIC) image, and by GFP fluorescence; scale bar = 100 microns. Larvae from the two non-transgenic strains N2 and JM222 were “mock-sorted” by passage through the Biosorter. Larvae from the two transgenic strains JM147 and JM199 were sorted into “Non-Green Non-Rescued” and “Green Rescued” populations, depending on whether larvae had lost or had retained the caEx3 extrachromosomal transgenic array. An example of a rare mis-sorted (non-fluorescent) worm is shown by the white arrow in the JM199 GFP image.
Supplementary Fig. 10.
Gel images showing unsuccesful attempts to detect end-1, end-3, elt-7 and elt-2 coding region in the elt-7(−) end-1(−) end-3(−); elt-2(−) strains rescued by transgenic [elt-2promoter::elt-7 cDNA + end-1promoter::elt-7 cDNA]
Expected PCR-amplified fragment sizes are indicated; red “X” signifies that no band is expected if there is no coding region.
Supplementary Fig. 11.
Alignment of ELT-2, ELT-7, END-1 and END-3 amino acid sequences
Clustal-Omega algorithm with default settings, and displayed using MVIEW. The red box highlights the conserved zinc finger binding domains.
Supplementary Fig. 12.
Expression differences between COPAS sorted vs. filtered wildtype L1 worms
Fold change of gene expression (y-axis) between the filtered and sorted samples is plotted against the mean intensity for a given gene (x-axis). Differentially expressed genes for each comparison are shown in red (differential expression: total read counts > 10, and BH-adjusted Wald Test p-value < 0.1, and log2-fold change > 0.8). Of the 15,217 genes present, 108 were up-regulated in COPAS sorted conditions and 47 were down-regulated
Supplementary Table 1. Raw RNA-seq read counts tabulated for each sample.
Supplementary Table 2. Log-transformed, variance-stabilized RNA-seq read counts tabulated for each sample.
Supplementary Table 3. Results (res) tables from DESeq2 differential expression analyses generated from pair-wise comparisons between conditions.
Supplementary Table S4. Lists of genes significantly differentially expressed between key pairwise comparisons of conditions.
Supplementary Table S5. Hierarchical clustering output from 81 significantly changing genes that have been annotated as intestine-specific (Figure 4). The data table includes the gene’s assigned “class”, whether the gene contains an ELT-2 ChIP-seq peak, and whether the gene has been ascribed as intestine-enriched or intestine-specific.
Supplementary Table S6. Hierarchical clustering output from all 3092 significantly changing genes (Supplementary Figure 4). The data table includes the gene’s assigned “set”, whether the gene contains an ELT-2 ChIP-seq peak, and whether the gene has been ascribed intestine-enriched or intestine-specific status.
Supplementary Fig. 2.
Pairwise comparisons of RNA-seq data between wild-type and mutant strains focusing on all-genes, intestine-enriched genes, and intestine-specific genes.
Fold change of gene expression (y-axis) between the two labeled samples is plotted against the mean intensity for a given gene (x-axis). Differentially expressed genes for each comparison are shown in red (criteria for differential expression: total read counts > 10, BH-adjusted Wald Test p-value < 0.1, and log2-fold change > 0.8). The list of intestinal expressed genes was produced by querying WormBase (www.wormbase.org) for genes annotated to be expressed in the intestine/gut and verifying from WormBase that expression was primarily or exclusively in the intestine. This 2073 long gene list was supplemented with genes from intestinal SAGE libraries (McGhee et al., 2007; McGhee et al., 2009) and from (Sommermann et al., 2010). The final list was composed of 2210 genes.
Supplementary Fig. 3.
Levels of elt-2 and elt-7 transcripts in elt-7(−) and elt-2(−) mutants, respectively.
Transcriptional cross-regulation between elt-2 and elt-7 is shown as transcript levels calculated as RPKM (reads mapped per kilo-base gene model per million mapped reads) measured in wild type or single-mutant samples. Values above the comparisons are Benjamini-Hochberg corrected p-values. Note that the normalized read counts of the elt-7 transcript are not statistically different between wild type and elt-7(−) mutant because only the catalytically active DNA binding domain is deleted in the mutant. Much of the transcript still remains but the protein is not functional.
Supplementary Fig. 4.
Dynamically expressed genes cluster into six sets based on expression patterns.
A. We performed hierarchical clustering on dynamically expressed genes (n = 3092) and illustrate the results in a heat map. Clustering was performed using Spearman correlation distances of row-means-normalized log-stabilized read counts. This effort identified six sets of genes. The percentage of genes within each set that contain a significant ELT-2 Chip-seq peak (as determined by either of two published studies; (Mann et al., 2016; Wiesenfahrt et al., 2016)) is tabulated to the right of the heatmap.
B. The top over-represented motifs identified within the promoter regions of each gene set are illustrated as are the P-values associated with those motifs. Motifs and their P-values were determined using HOMER (Heinz et al., 2010).
C. The top gene categories associated with each gene set are tabulated along with their associated P values as determined using WORMEXP (Yang et al., 2016).
D. Depictions of potential types of epistatic relationships, their predicted expression patterns, and their representations in the heatmap above are shown.
Supplementary Fig. 5.
Loss of ELT-7 delays the onset time of elt-2promoter::GFP by 15–20 minutes.
The two strains JM149 and JM259 contain the same integrated transgenic array expressing elt-2promoter::GFP but are wildtype or null at the elt-7 locus, respectively. Embryos were picked at the 2-cell stage and the time at which GFP could first be detected was determined. Red vertical lines represent mean onset time for each strain. Number of embryos observed = 22 and 20 for strains JM149 and JM259, respectively. t-test probability that the onset times are equal < 10−5
Supplementary Fig. 6.
Time (20°C) between the 2-cell stage and embryo hatching for the four mutant strains.
From top to bottom, the panels correspond to strains N2, JM222 elt-7(−), JM147 elt-2(−); caEx3, and JM199 elt-7(−); elt-2(−); caEx3. In each panel, different symbols represent independent replicate experiments. For JM147 and JM199, filled symbols represent embryonic segregants that have lost the rescuing caEx3 array; open symbols represent segregants that have retained caEx3. The wide grey line corresponds to the range of data seen with independent N2 replicates (top panel) and is included for comparison.
Supplementary Fig. 7.
Representative images of JM199 elt-7(−); elt-2(−); caEx3 embryos squeezed from egg-shells at 15 hours after 2-cell stage (20°C).
The embryo/larva on the left is elt-7(−); elt-2(−) because it shows no GFP flurorescence marking the rescuing extrachromosomal transgenic array. The embryo/larva on the right is GFP-positive and therefore rescued by the extrachromsomal transgenic array. However, both embryos/larvae exhibit birefringence measuring intestinal differentiation (bottom panel, shown with inverted contrast). Ten micron diameter glass beads (visible in background) were placed around embryos to protect from over-squeezing. Scale bar = 20 microns.
Supplementary Fig. 8.
Staining for GES-1 esterase activity in ~1.5 fold stage embryos.
GES-1 esterase activity can be detected in the intestines of embryos with the following genotypes (from left to right) elt-7(−); elt-2(−), wild type strain N2, elt-2(−) and elt-7(−). Strain JM1041 contains two activity-lowering point mutations introduced into the ges-1 structural gene (McGhee et al., 1990); the low level of “background” staining represents the activity produced by other intestinal esterases and possibly from residual enzymatic activity in the mutated GES-1 protein.
Supplementary Fig. 9.
Gel images showing unsuccesful attempts to detect the ELT-2 coding region in the elt-2(−) strains rescued by elt-2promoter::elt-7 cDNA
Expected fragment sizes are indicated; red “X” signifies that no band is expected if there is no elt-2 coding region. Two pairs of primers are indicated for a particular feature (e.g. overall ca15 deletion) when nested PCR was performed.