Abstract
Objectives:
To propose a reliable and practical method for automatically segmenting the mandible from CBCT images.
Methods:
The marker-based watershed transform is a region-growing approach that dilates or “floods” predefined markers onto a height map whose ridges denote object boundaries. We applied this method to segment the mandible from the rest of the CBCT image. The height map was generated to enhance the sharp decreases of intensity at the mandible/tissue border and suppress noise by computing the intensity gradient image of the CBCT itself. Two sets of markers, “mandible” and “background” were automatically placed inside and outside the mandible, respectively in a novel image using image registration. The watershed transform flooded the gradient image by dilating the markers simultaneously until colliding at watershed lines, estimating the mandible boundary. CBCT images of 20 adolescent subjects were chosen as test cases. Segmentation accuracy of the proposed method was evaluated by measuring overlap (Dice similarity coefficient) and boundary agreement against a well-accepted interactive segmentation method described in the literature.
Results:
The Dice similarity coefficient was 0.97 ± 0.01 (mean ± SD), indicating almost complete overlap between the automatically and the interactively segmented mandibles. Boundary deviations were predominantly under 1 mm for most of the mandibular surfaces. The errors were mostly from bones around partially erupted wisdom teeth, the condyles and the dental enamels, which had minimal impact on the overall morphology of the mandible.
Conclusions:
The marker-based watershed transform method produces segmentation accuracy comparable to the well-accepted interactive segmentation approach.
Keywords: CBCT, image segmentation, image registration, mandible, marker-based watershed transform
Introduction
Three-dimensional mandibular models are useful for planning maxillofacial surgery and orthodontic treatment.1,2 In studies of growth, mandibular models are important for assessing morphological changes over time.3,4 Such models are typically obtained from conventional computed tomography (CT, using high radiation dose to capture fine detail of the bony structure. Cone beam CT (CBCT) shows promise for oral and craniofacial imaging applications due to lower radiation dose, lower cost and shorter acquisition time compared to CT. However, CBCT images have lower contrast and higher levels of noise than conventional CT, making mandible segmentation a challenging task.5
Segmenting a 3D mandible is typically done ‘interactively’ in computer software on a case by case basis. Threshold-based algorithms or morphological operations are commonly used first for the separation of bony structures from soft tissues.6–8 Then manual work is needed to separate the mandible from the cranial base and the maxilla because the algorithms cannot distinguish between different facial bones with similar intensity values. We refer to this combination of computerized operations and manual editing as ‘interactive’ segmentation. Specific issues in mandible segmentation include intercuspation occlusion and low contrast of condyles relative to surrounding structures. Intercuspation leads to connection between upper and lower teeth while low contrast leads to difficult to define boundaries on the condyles. These require slice-by-slice based manual editing, which is tedious, time-consuming and operator-dependent as it produces slightly different outlines after continuous interventions.6
A robust automated mandible segmentation approach is thus desirable. In other applications, simple automated methods based on voxel intensity or edge intensity can be very effective.9,10 However, the mandible is not the only bone structure in CBCT images of the head, and the intensity of bone varies considerably. More sophisticated methods which incorporate prior information about the expected shapes and position of objects to be segmented are required. To date only five publications, using statistical shape models,11–13 multi atlas label registration14 or machine learning,15 have been proposed to automate the mandibular segmentation from CBCT images. All these methods required collection of large amounts of manually segmented mandibles as training data, which may be impractical in clinical situations. Three of them have not incorporated the teeth in their models because the number of teeth may vary between patients, which created more challenges in the segmentation task.11–13
The watershed method is a classic, computationally simple technique for object segmentation in images. The original grayscale image can be regarded as a topographic relief, with brightness/intensity corresponding to altitude, and thus identifying watershed lines provides a method for segmenting an image into separate spatial regions.16,17 The original grayscale image is transformed into a ‘height map’ which emphasizes discontinuities in image intensity (such as occur at object boundaries) and dampens continuous regions (such as homogeneous intensity region within the tissue). The marker-based watershed transform dilates, or floods, from markers that are provided to the algorithm. The number of markers determines the number of regions that will be created by the watershed transform.18 The watershed markers can be manually or automatically set. The marker-based watershed transform has been successfully used to segment breast lesions on ultrasound19 and lymphoma in sequential CT images.20 Use of watershed methods to segment teeth has been described.21–24 Naumovich et al suggested that the watershed method could be extended to segment ‘the jaws’ from CBCT, but did not report images of such segmentation.24 Each of these methods of dental segmentation requires manual placement of markers in each case.
In this article, we propose an automatic approach for segmenting mandibles from CBCT using a marker-based watershed transform. We fully automate the segmentation by automated watershed marker placement using image registration. Segmentation accuracy of the proposed automated method is assessed by comparing outcomes with a well-accepted interactive segmentation method described in the literature.1,2
Methods and materials
Image data
CBCT images were obtained from 21 adolescent subjects with a mean age of 13.68 years (SD:1.27; nine males) from an orthodontic clinic, where images had previously been obtained for clinical indications. Images were taken using an i-Cat Flx machine (Imaging Sciences International, PA). The FOVs in the selected samples were 16 × 13 cm with a scan time of 8.9 s and exposure time of 3.7 s. Exposure parameters for CBCT image were 120 kVp and 5 mA. The original isotropic voxel size was 0.4 mm3. The DICOM files were then deidentified and downsized to an isotropic voxel size of 0.5 mm3 to decrease the computational power for later registration propose. Patients were instructed to bite into maximum intercuspation during scanning. Ethics approval to use the images was obtained from.
Overview of the marker-based watershed mandible segmentation
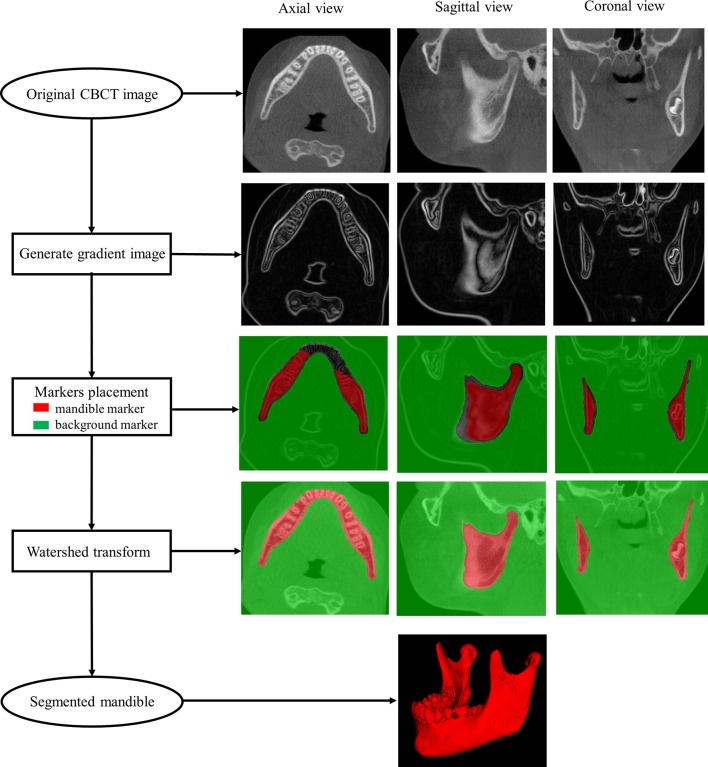
In a CBCT image, the mandible has typically high intensity and therefore appears brighter than its surrounding tissue (muscles or air) with a marked drop in intensity at the boundaries. The height map is constructed to enhance these boundaries and suppress homogeneous regions. Here the height map was generated by transforming the CBCT image into gradient image of itself using the Derivative of Gaussian (Full Width at Half Maximum 1 mm) kernel, which highlighted boundaries of sharply changing intensity in the original image. Initially, two sets of watershed markers were placed on the gradient image, one set within the mandible and the other set in the rest of the image. The watershed transform floods the gradient image by dilating the markers simultaneously until colliding at watershed lines, estimating the mandible boundary. Illustration of the marker-based watershed transform method was shown in Figure 1. This method was used in both generating template data and automating segmentation of the mandible from novel images.
Figure 1.
Illustration of the marker-based watershed transform method. The original image is transformed into the gradient image which highlights boundaries of sharply changing intensity in the original image. The mandible marker (in red) and the background marker (in green) are placed within the mandible and at the rest of the structures, separately. The watershed transform floods the gradient image by dilating the markers simultaneously until colliding at watershed lines, estimating the mandible boundary. The segmented mandible is reconstructed in below. The pipeline is demonstrated in axial (column 1) sagittal (column 2) and coronal (column 3) views.
Template construction
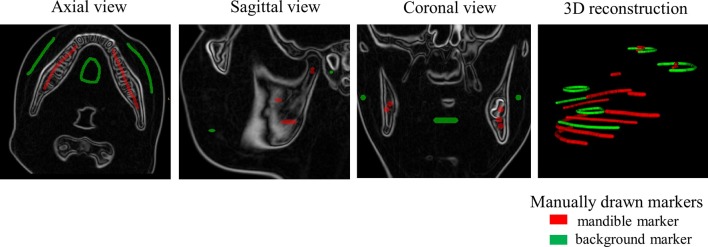
The template data comprised of a CBCT image and associated mandible and background markers that can later be propagated onto the novel image. An image of a 12.65-year-old male patient was selected to create this template data. The mandible and background markers were defined semi-automatically on the template image by applying the watershed method described above to manually drawn markers (lines or circles that were drawn unambiguously within or without the mandible as shown in Figure 2). The watershed transform segmented the mandible, and the remainder of the image was labelled as background. These segmentations were then eroded by one voxel to form the markers that have an unlabeled gap where the expected mandible boundary location resides. The dental crowns were manually removed from the mandible marker because the eruption stages and the number of the teeth could be different between the template image and the test images and the teeth were not of primary interest. Figure 1 shows a template image slice with the final mandible and background markers overlaid in red and green, respectively.
Figure 2.
Manually drawn markers on the template image.
Applying to a novel image
Markers were placed automatically inside and outside the mandible on a novel image from which the mandible was to be segmented. This was achieved by warping the template image onto each novel image using voxel-based image registration. Watershed markers, placed on the template, were warped along with this image, placing them into appropriate positions in each novel image.
In this study, the voxel-based image registration estimated a spatial transformation to be applied to each voxel of the novel image to corresponding voxels on the template image. The transformation involves both linear registration (translation and rotation) and non-linear registration (warp or stretch). The linear registration was used to coarsely align each test image to the template image using FLIRT registration in FSL open source tools (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL). The non-linear registration then deformed the voxels on the test image more precisely into the template image using the advanced normalization tools, or ANTS (http://stnava.github.io/ANTs/). Further details on these methods are provided in the supplementary materials. Once the markers were automatically placed by the transformation, watershed segmentation proceeded as described above.
Segmentation accuracy evaluation
Images of the 20 subjects not used for the template were used as test images in this study. The segmentation accuracy of the proposed method was assessed through the comparison to a well-accepted interactive segmentation method described in previous studies.1,2 This method was performed with open-source software ITK-SNAP (http://www.itksnap.org/pmwiki/pmwiki.php) by an experienced orthodontist (Hidden Content) and checked by a dentist (Hidden Content). Firstly, thresholding was used to grossly generate the main part of the mandible. The “region competition snake” method was used to generate the condyles. Slice-by-slice editing in all three orthogonal views was required for further trimming the condyles and the lower teeth.
The segmented mandibles of these two approaches were compared by computing a Dice similarity coefficient for the overlapping voxels. This index ranges from 0 (no overlap) to 1 (complete overlap). The outer surfaces of the mandible were generated with the marching cubes algorithm in MATLAB (https://au.mathworks.com/help/matlab/ref/isosurface.html). The boundary agreement between these two approaches were calculated as the surface distance between the two surfaces. This was quantified and visualized by a colormap.
Results
Timing
Automatic segmentation of each mandible executed in 12–14 min on an Ubuntu Linux machine, with an Intel i7-4770 @ 3.4 GHz CPU. The compared interactive method typically required 30 to 40 min.
Accuracy
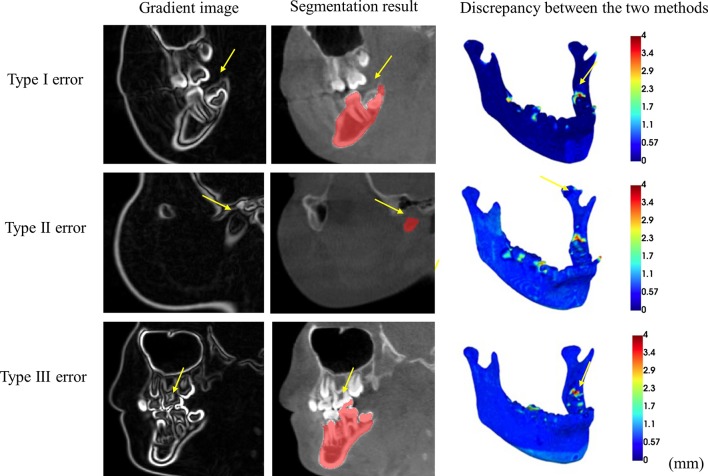
Mandibles segmented from the proposed automated method were compared against the interactive segmentation results. Dice similarity coefficients were 0.97 ± 0.01(mean ± SD), indicating almost complete overlap between the automatically segmented mandibles and the interactive segmented mandibles. Boundary deviations were predominantly under 1 mm over most of the mandibular surfaces (Figure 3). The errors were mostly from the bones around partially erupted wisdom teeth, the condyles and the dental enamels, which had minimal impact on the overall morphology of the mandible (Figure 4).
Figure 3.
The discrepancy between the proposed automatic method and the interactive method in 20 test cases.
Figure 4.
Segmentation errors for the proposed automatic approach. Type I error occurs at the partially erupted wisdom tooth. Type II error occurs at the ill-defined condyle. Type III error occurs at the dental enamel.
Discussion
The quality of the mandibular segmentation determines the accuracy of subsequent applications, such as orthognathic treatment planning or orthodontic treatment evaluation. To date, most software-based mandibular segmentation involves continuous manual intervention, which is tedious and time-consuming, making it impractical for dealing with large numbers of subjects. In this study, we propose and evaluate an automated mandibular segmentation method using the marker-based watershed transform. This approach demonstrates time efficiency and comparable segmentation accuracy with a well-accepted interactive segmentation method.
In CBCT images, speckles or noise are more prominent than conventional CT images, which reduces the contrast and makes it difficult to differentiate low-density tissue in the image.25 Segmentation accuracy of common provided algorithms in software is often limited by image related artifacts. For example, simple thresholding is effective in depicting the condyles from conventional CT images, where the image intensity histogram has a deep and sharp valley between two peaks representing the condyles and the soft tissue nearby. An adequate threshold can be chosen at the bottom of this valley to separate them. However, detecting the valley bottom precisely in CBCT images is difficult because the valley is flat and broad, imbued with noise. In this study, the Derivative of Gaussian filter is used to construct the height map, this not only enhances the intensity of the edges and dampen non-edges in the original image but also has noise suppression properties. Explicit placement of two markers, one inside and one outside the mandible ensures that the image is segmented into only two regions.
Another advantage of the proposed automated method is that it allows segmentation of the teeth. Most previous mandibular segmentation studies either do not incorporate dental segmentation12,13 or use edentulous cases as test images.26 This limits the application of the segmentation results in dentistry. In clinical situations, CBCT scans are often acquired with the upper and lower teeth touching, making them hard to separate using methods such as thresholding. The watershed method, on the contrary, is particularly useful for splitting touching objects. For example, it has long been used to delineate touching cells or clustering nuclei from a microscopic image.27 In recent years, the watershed method has also been demonstrated in segmenting individual teeth from the jaw bones.21–24 This approach is better able to separate touching teeth because the boundaries of the upper and lower teeth are accentuated in the gradient image, reflecting the sharp intensity changes between the enamel and air.
We have been able to fully automate the segmentation by automated watershed marker placement. This is achieved by aligning each test image to the template image using a voxel-based registration algorithm. Registration using a single template yielded good results for all the adolescent test cases in this study. However, human mandibles change markedly from infancy through childhood to adolescence, and from early adulthood to old age.28 Age-appropriate templates may be necessary for accurate image registration at different ages.29 This will ensure that the regions with high inter age anatomical variability (such as the condyles and the coronoid processes) will be matched correctly and ensure the accuracy of the watershed marker placement.
Compared with the interactive segmentation, the proposed automatic approach will reduce the time investigators and clinicians spend at the console, which will be particularly beneficial for large-scale studies such as growth modelling or normal reference establishment. The proposed pipeline could be parallelized (on multiple machines) to run multiple segmentation jobs at the same time. Advances in 3D imaging technologies have generated large databases of 3D CBCT images which could be used for research. Few studies have made use of these large potential samples, the largest scale study to date using only 159 images to model the mandibular morphological change in 3D.30 This is likely due to the time required to segment each mandible. Using an automatic segmentation approach could open up large amounts of data to future studies.
The proposed method gives comparable accuracy as the interactive segmentation approach. We demonstrate almost complete overlap between the automatically segmented mandibles and the interactively segmented mandibles in our test cases. There were, however, some errors at certain anatomical regions. First, the watershed flooding stops at the dental enamel of the partially erupted wisdom tooth before it reaches the cortical bone above as the intensity drop at the dental enamel is sharp. Second, the watershed lines are unpredictable at ill-defined condyles because of poor image quality for the cartilage in CBCT modality. Third, errors occasionally occur due to over flooding to the enamels of the upper teeth. All these errors have minimal impact on the morphology of the mandible and can be easily fixed with a minimal amount of manual editing. However, further tests for images taken with different machine settings and from different age range patients are needed.
It should be noted that the interactive segmentation is less than a perfect gold standard. In this approach, a threshold was subjectively selected based on the intensity difference between the mandible and the rest of the structures. Differences in threshold selection due to blurring of the boundary or noise lead to slight changes in the final outline of the mandible. Slice-by-slice manual editing also results in jagged edges. The watershed method, on the other hand, implements a consistent definition of the boundary, corresponding to regions or rapid change in intensity. Therefore, some of the discrepancies between the interactively segmented mandible and the automated segmented mandible may be due to imperfections in the interactive segmentation.
Conclusions
In this study, we propose and evaluate a practical marker-based watershed algorithm for automatically segmenting the mandible from CBCT images. Compared with user-depended interactive segmentation, our approach showed promising time-efficiency and comparable accuracy.
Footnotes
Acknowledgment: We would like to thank Dr Paul Buchholz for sharing his CBCT images; Dr Arjun Atresh for providing the test cases and manually segmenting the mandibles. Dr Robert Wei for the instruction in using ITK-SNAP. We also thank Dr Wei-Yin Loh, Ms. Jian Chen and Ms. Claire Kelly for the valuable discussions.
Contributor Information
Yi Fan, Email: fan2@student.unimelb.edu.au.
Christopher Adamson, Email: chris.adamson@mcri.edu.au.
REFERENCES
- 1. Yatabe M, Garib D, Faco R, de Clerck H, Souki B, Janson G, et al. . Mandibular and glenoid fossa changes after bone-anchored maxillary protraction therapy in patients with UCLP: A 3-D preliminary assessment. Angle Orthod 2017; 87: 423–31. doi: 10.2319/052516-419.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cevidanes LH, Alhadidi A, Paniagua B, Styner M, Ludlow J, Mol A, et al. . Three-dimensional quantification of mandibular asymmetry through cone-beam computerized tomography. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2011; 111: 757–70. doi: 10.1016/j.tripleo.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kelly MP, Vorperian HK, Wang Y, Tillman KK, Werner HM, Chung MK, et al. . Characterizing mandibular growth using three-dimensional imaging techniques and anatomic landmarks. Arch Oral Biol 2017; 77: 27–38. doi: 10.1016/j.archoralbio.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andresen PR, Nielsen M, Kreiborg S. 4D shape-preserving modelling of bone growth. In: Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention Springer-Verlag 1998: 710–9. [Google Scholar]
- 5. Pauwels R, Araki K, Siewerdsen JH, Thongvigitmanee SS. Technical aspects of dental CBCT: state of the art 44. Dentomaxillofac Radiol 2015; 44: 20140224. doi: 10.1259/dmfr.20140224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wallner J, Hochegger K, Chen X, Mischak I, Reinbacher K, Pau M et al. . Clinical evaluation of semi-automatic open-source algorithmic software segmentation of the mandibular bone: Practical feasibility and assessment of a new course of action In: van Ooijen PMA. 13; 2018. pp e0196378. doi: 10.1371/journal.pone.0196378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. M. Nassef T. New segmentation approach to extract human mandible bones based on actual computed tomography data. Am J Biomed Eng 2012; 2: 197–201. doi: 10.5923/j.ajbe.20120205.01 [DOI] [Google Scholar]
- 8. Abdullah JY, Omar M, Pritam HMH, Husein A, Rajion ZA. Comparison of 3D reconstruction of mandible for pre-operative planning using commercial and open-source software. In 2016;: p. 020001.
- 9. Kiraly AP, Higgins WE, McLennan G, Hoffman EA, Reinhardt JM. Three-dimensional human airway segmentation methods for clinical virtual bronchoscopy. Acad Radiol 2002; 9: 1153–68. doi: 10.1016/S1076-6332(03)80517-2 [DOI] [PubMed] [Google Scholar]
- 10. Hu S, Hoffman EA, Reinhardt JM. Automatic lung segmentation for accurate quantitation of volumetric X-ray CT images. IEEE Trans Med Imaging 2001; 20: 490–8. doi: 10.1109/42.929615 [DOI] [PubMed] [Google Scholar]
- 11. Lamecker H, Zachow S, Wittmers A, Weber B, Hege HC, Elsholtz B. Automatic segmentation of mandibles in low dose CT-Data. Intenational J Comput Assist Radiol Surg 2006; 1: 393. [Google Scholar]
- 12. Kainmueller D, Lamecker H, Seim H, Zinser M, Zachow S. Automatic extraction of mandibular nerve and bone from cone-beam CT data. Med Image Comput Comput Assist Interv 2009; 12(Pt 2): 76–83. [DOI] [PubMed] [Google Scholar]
- 13. Gollmer ST, Buzug TM. Fully automatic shape constrained mandible segmentation from cone-beam CT data In: 9th IEEE International Symposium on Biomedical Imaging (ISBI) IEEE; 2012. pp 1272–5. [Google Scholar]
- 14. Wang L, Chen KC, Gao Y, Shi F, Liao S, Li G, et al. . Automated bone segmentation from dental CBCT images using patch-based sparse representation and convex optimization. Med Phys 2014; 41: 043503. doi: 10.1118/1.4868455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang L, Gao Y, Shi F, Li G, Chen K-C, Tang Z, et al. . Automated segmentation of dental CBCT image with prior-guided sequential random forests. Med Phys 2016; 43: 336–46. doi: 10.1118/1.4938267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beare R, Chen J, Adamson CL, Silk T, Thompson DK, Yang JY, et al. . Brain extraction using the watershed transform from markers. Front Neuroinform 2013; 7: 1063–74. doi: 10.3389/fninf.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beare R. A locally constrained watershed transform. IEEE Trans Pattern Anal Mach Intell 2006; 28: 1063–74. doi: 10.1109/TPAMI.2006.132 [DOI] [PubMed] [Google Scholar]
- 18. Gao H, Xue P, Lin W. A new marker-based watershed algorithm. Proceedings of the IEEE International Symposium on Circuits and Systems IEEE 2004: 81–4. [Google Scholar]
- 19. Gómez W, Leija L, Alvarenga AV, Infantosi AF, Pereira WC. Computerized lesion segmentation of breast ultrasound based on marker-controlled watershed transformation. Med Phys 2010; 37: 82–95. doi: 10.1118/1.3265959 [DOI] [PubMed] [Google Scholar]
- 20. Yan J, Zhao B, Wang L, Zelenetz A, Schwartz LH. Marker-controlled watershed for lymphoma segmentation in sequential CT images. Med Phys 2006; 33: 2452–60. doi: 10.1118/1.2207133 [DOI] [PubMed] [Google Scholar]
- 21. Kakehbaraei S, Seyedarabi H, Zenouz AT. Dental segmentation in cone-beam computed tomography images using watershed and morphology operators. J Med Signals Sens 2017; 8: 119–24. [PMC free article] [PubMed] [Google Scholar]
- 22. Galibourg A, Dumoncel J, Telmon N, Calvet A, Michetti J, Maret D. Assessment of automatic segmentation of teeth using a watershed-based method 47. Dentomaxillofac Radiol 2018; 47: 20170220. doi: 10.1259/dmfr.20170220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia Z, Gan Y, Chang L, Xiong J, Zhao Q. Individual tooth segmentation from CT images scanned with contacts of maxillary and mandible teeth. Comput Methods Programs Biomed 2017; 138: 1–12. doi: 10.1016/j.cmpb.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 24. Naumovich SS, Naumovich SA, Goncharenko VG. Three-dimensional reconstruction of teeth and jaws based on segmentation of CT images using watershed transformation. Dentomaxillofac Radiol 2015; 44: 20140313. doi: 10.1259/dmfr.20140313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagarajappa AK, Dwivedi N, Tiwari R. Artifacts: The downturn of CBCT image. J Int Soc Prev Community Dent 2015; 5: 440–5. doi: 10.4103/2231-0762.170523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wallner J, Hochegger K, Chen X, Mischak I, Reinbacher K, Pau M, et al. . Clinical evaluation of semi-automatic open-source algorithmic software segmentation of the mandibular bone: Practical feasibility and assessment of a new course of action. PLoS One 2018; 13: e0196378–26. doi: 10.1371/journal.pone.0196378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang K, Liao Q, Dai S. A novel white blood cell segmentation scheme using scale-space filtering and watershed clustering. Proceedings of the International Conference on Machine Learning and Cybernetics IEEE 2003: 2820–5. [Google Scholar]
- 28. Enlow DH, Harris DB. A study of the postnatal growth of the human mandible. Am J Orthod 1964; 50: 25–50. doi: 10.1016/S0002-9416(64)80016-6 [DOI] [Google Scholar]
- 29. Chuang YJ, Doherty BM, Adluru N, Chung MK, Vorperian HK. A novel registration-based semiautomatic mandible segmentation pipeline using computed tomography images to study mandibular development. J Comput Assist Tomogr 2017; 42: 306–16. doi: 10.1097/RCT.0000000000000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coquerelle M, Bookstein FL, Braga J, Halazonetis DJ, Weber GW, Mitteroecker P. Sexual dimorphism of the human mandible and its association with dental development. Am J Phys Anthropol 2011; 145: 192–202. doi: 10.1002/ajpa.21485 [DOI] [PubMed] [Google Scholar]