Abstract
Galectins, highly conserved β-galactoside-binding lectins, have diverse regulatory roles in development and immune homeostasis and can mediate protective functions during microbial infection. In recent years, the role of galectins in viral infection has generated considerable interest. Studies on highly pathogenic viruses have provided invaluable insight into the participation of galectins in various stages of viral infection, including attachment and entry. Detailed mechanistic and structural aspects of these processes remain undetermined. To address some of these gaps in knowledge, we used Zebrafish as a model system to examine the role of galectins in infection by infectious hematopoietic necrosis virus (IHNV), a rhabdovirus that is responsible for significant losses in both farmed and wild salmonid fish. Like other rhabdoviruses, IHNV is characterized by an envelope consisting of trimers of a glycoprotein that display multiple N-linked oligosaccharides and play an integral role in viral infection by mediating the virus attachment and fusion. Zebrafish’s proto-typical galectin Drgal1-L2 and the chimeric-type galectin Drgal3-L1 interact directly with the glycosylated envelope of IHNV, and significantly reduce viral attachment. In this study, we report the structure of the complex of Drgal1-L2 with N-acetyl-d-lactosamine at 2.0 Å resolution. To gain structural insight into the inhibitory effect of these galectins on IHNV attachment to the zebrafish epithelial cells, we modeled Drgal3-L1 based on human galectin-3, as well as, the ectodomain of the IHNV glycoprotein. These models suggest mechanisms for which the binding of these galectins to the IHNV glycoprotein hinders with different potencies the viral attachment required for infection.
Keywords: carbohydrate-recognition, galectins, IHNV, innate-immunity, viral attachment
Introduction
Galectins, a family of β-galactoside-binding lectins characterized by a unique sequence motif in their carbohydrate-recognition domains (CRDs) (Hirabayashi and Kasai 1993; Cummings et al. 2017), are evolutionary conserved and widely distributed in vertebrates, invertebrates, and some fungal taxa (Vasta 2009). Based on their domain organization, vertebrate galectins are classified into three major types: (a) proto-type, i.e., those possessing a single globular CRD; (b) chimera-type, i.e., those with one CRD and an N-terminal peptide; and (c) tandem-repeat-type, i.e., those with two CRDs joined by a linker peptide (Hirabayashi and Kasai 1993). Multiple galectin subtypes have been identified in vertebrates; all except the so-called galectin-related proteins fitting one of three types; they are numbered from 1 to 16 following the order of their discovery (Cummings et al. 2017). As proto- and chimera-type galectins can form oligomeric structures (dimers for the proto-type, and trimers or pentamers for the chimera-type), all three galectin types display multiple CRDs (from two to five) and therefore can establish cooperative interactions with multivalent carbohydrate ligands or cross-link glycan moieties that are present on the cell surface (Vasta 2012) enabling the formation of lattices (Rabinovich et al. 2007) or agglutinating cells (Camby et al. 2006). Galectins were initially shown to mediate cell adhesion in developmental processes and tissue organization in embryogenesis by binding to endogenous ligands (Ahmed et al. 1996; Rabinovich et al. 2007). However in the past two decades, their participation in processes such as cancer proliferation (Kaltner et al. 2017; Mendez-Huergo et al. 2017), immune homeostasis (Liu et al. 2012; Thiemann and Baum 2016; Sundblad et al. 2017), angiogenesis (Mendez-Huergo et al. 2017) and adipogenesis (Yang et al. 2011b) have become increasingly understood. Furthermore, it has been firmly established that mammalian galectins can mediate protective functions during microbial infections by, for example, recruiting neutrophils and activating lymphoid cells (Smith and Cummings 2008; Mendez-Huergo et al. 2017). In recent years, evidence has accumulated in support of the notion that galectins can directly recognize glycans on the surface of viruses, bacteria, pathogenic yeasts and parasites and function not only as pattern recognition receptors (PRRs) (Vasta 2009, 2012; Cummings et al. 2017), but also as effector factors by displaying bactericidal activity (Stowell et al. 2010). In addition, some pathogens have co-evolved with their hosts to take advantage of the recognition functions of the galectins to facilitate their entry and infection (Vasta 2009; Stowell et al. 2010; Thiemann and Baum 2016; Lujan et al. 2018).
The roles of mammalian galectins in viral infections have been examined for highly pathogenic viruses such as influenza (Yang et al. 2011a; Nita-Lazar et al. 2015a, 2015b), HIV (Mercier et al. 2008; Sato et al. 2012), Nipah (Levroney et al. 2005) and dengue (Toledo et al. 2014). These studies have provided invaluable insight into the participation of galectins in various stages of viral infection, including attachment, entry into the host cell, and proliferation (Levroney et al. 2005; Mercier et al. 2008; Yang et al. 2011a; Sato et al. 2012; Toledo et al. 2014; Nita-Lazar et al. 2015a, 2015b). However, key mechanistic and structural aspects of these processes remain to be fully elucidated.
We are using zebrafish (Danio rerio) as a genetically tractable model system to examine in detail the roles of galectins in infection by infectious hematopoietic necrosis virus; (IHNV) Rhabdoviridae, genus Novirhabdovirus, a pathogen responsible for significant losses in both farmed and wild salmon and trout populations (Batts et al. 1991; Crane and Hyatt 2011; Yang et al. 2011a). IHNV is a bullet-shaped negative-sense single-stranded RNA virus (Troyer and Kurath 2003). Its envelope consists of trimers of a glycoprotein that displays N-linked oligosaccharides and plays an integral role in the pathogenesis of viral infection. Like in other rhabdoviruses, rabies (RV), and vesicular stomatitis (VSV) the envelope glycoprotein (GP) of IHNV mediates viral attachment to the target cell surface receptors (Harmache et al. 2006; Nita-Lazar et al. 2016). While it is known that IHNV enters the host through the skin at the base of the fins and possibly the oral and gill epithelia (Harmache et al. 2006), viral attachment and entry mechanisms are not clearly understood.
Previous studies showed that zebrafish galectins Drgal1-L2 (proto-type) and Drgal3-L1 (chimera-type) expression in fish epithelial cells downregulates after exposure to INHV (Nita-Lazar et al. 2016). A preliminary glycomic study of the IHNV envelope glycoprotein identified oligosaccharides that may potentially constitute the galectin ligands responsible for the observed inhibition of viral attachment to the host (Nita-Lazar et al. 2016). The zebrafish Drgal1-L2 and Drgal3-L1 recognize in vitro non-reducing terminal LacNAc moieties of N-linked oligosaccharides in the IHNV GP (Nita-Lazar et al. 2016). These results suggest that the two zebrafish galectins Drgal1-L2 and Drgal3-L1 may inhibit IHNV attachment by binding to the virion envelope glycoprotein masking the recognition sites of the viral receptors, but the structural aspects of this interaction remain unknown.
The crystallization Drgal1-L2/Lactose (Galβ1,4Glc) complex in reducing conditions in a P212121 space group with a dimer in the asymmetric unit of the crystal but not the structure has been reported previously (Scott et al. 2010).
In this study, we successfully determined the structure of the Drgal1-L2 in complex with N-acetyl-d-lactosamine (LacNAc; Galβ1,4GlcNAc), at 2.0 Å resolution. This structure and the comparative models of Drgal3-L1 and the viral receptor were used to provide insight into the structural basis of the inhibitory effects of these galectins on IHNV attachment to the epithelial cells of host fish.
Results
Drgal1-L2 structure
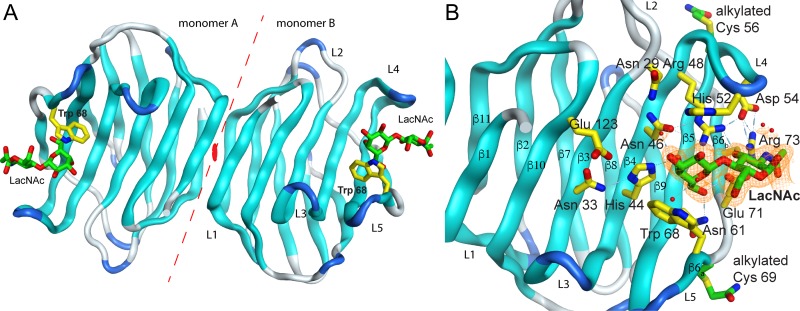
The Drgal1-L2 complex crystalizes as a dimer in the hexagonal space-group P 61. Each monomer has a bound LacNAc molecule in its lower energy conformation (Figure 1A). The electron density for the amino acids spanning residues 2 to 133 in the monomer A and 2 to 134 in the monomer B, including the electron densities of the alkyl groups (acetamide, -(C = O)NH2) modifying cysteine residues (Cys 56 of monomer A and Cys 56 and Cys 69 of monomer B), were observed in the crystal. Two magnesium metal ions from the buffer are also observed mediating crystal contacts. The final model was refined to a Rwork/Rfree of 0.16/0.22 with 94.57% of the residues in allowed regions of the Ramachandran plot (Table I). The 2mFo-DFc sigma weighted electron density map in the region around the ligand shows excellent agreement (Figure 1B). The Drgal1-L2 dimer buries 600 Å2 of accessible surface area (ASA), forming 10 hydrogen bonds and one salt bridge between Glu A84 and Lys B134. The root-mean-square deviation (rmsd) between the structures of Drgal1-L2 and the molecular replacement search model, toad (Bufo arenarum) galectin-1—Protein Data Bank identification code (PDB ID) 1GAN (Bianchet et al. 2000)—is 1.0 Å for 131 Cα-atoms aligned of the monomers and 1.56 Å for the 261 α-atom aligned of the dimers. The rmsd for 132/265 (monomer/dimer) Cα-atoms of Drgal1-L2 aligned with human—1GZW, 39.5% identity (Lopez-Lucendo et al. 2004)—and bovine—1SLA, 36.8% identity (Bourne et al. 1994)—galectin-1 are 1.09/1.92 Å and 0.85/2.42 Å, respectively (Supplementary Figure S2). Drgal1-L2 monomers overlay well with other structurally characterized galectins-1 monomers. Only loop L5 shows differences with other galectins-1 (Supplementary Figure S1). However, the zebrafish galectin dimer showed larger discrepancies (higher rmsds) between dimers caused by the smaller twist (2.7°) of the extended β-sheet formed by the Drgal1-L2 dimer compared to other structurally characterized galectins-1 that showed a better agreement between human, cow, rat, mouse, chicken and toad galectin-1 dimers (Supplementary Figure S1).
Fig. 1.
Drgal1-L2 complex with LacNAc. (A) Cartoon of the dimer in the asymmetric unit showing the bound disaccharide and Trp 68. The dashed line indicates the dimer interface and the orange oval the position of the dimer two-fold. (B) The panel shows the binding site in monomer B displaying the residues that participate in the LacNAc recognition and a portion of the refined electron density around the carbohydrate. Electron density of the final sigmaA-weighted 2mFo-DFc map (orange) is contoured at one σ. Sugar interacting residues are displayed with its carbon atoms colored yellow in the protein and green in LacNAc. The alkylated cysteine residues are shown in (B) with carbon atoms in green, and sulfur atoms are colored in yellow. All nitrogen atoms are colored in blue and oxygen atoms are colored in red.
Table I.
Data collection and refinement statistics
| Wavelength (Å) | CuKα (1.541 Å) |
|---|---|
| Resolution range (Å) | 35.0–2.0 (2.03–2.0) Å |
| Space group | P 61 |
| Unit cell | 40.4, 40.4, 303.6 Å, 90.0, 90.0, 120.0° |
| Total reflections | 342,227 |
| Unique reflections | 18,798 (905) |
| Multiplicity | 3.5 (2.7)* |
| Completeness (%) | 99.2 (97.3)* |
| Mean I/sigma(I) | 33.5 (5.7)* |
| Wilson B-factor (Å2) | 25.9 |
| Rmerge | 0.046 (0.22)* |
| Reflections used for R-free | 963 |
| R-work | 0.16 |
| R-free | 0.23 |
| CC(work) | 0.961 |
| CC(free) | 0.938 |
| Number of non-hydrogen atoms | |
| Total | 2372 |
| Macromolecules | 2147 |
| Ligands | 52 |
| Metal ions | 3 |
| Water | 213 |
| Protein residues | 268 |
| Rmsd (bonds) | 0.016 |
| Rmsd (angles) | 1.85 |
| Ramachandran favored (%) | 94.6 |
| Ramachandran allowed (%) | 5.04 |
| Ramachandran outliers (%) | 0.4 |
| Clashscore | 4 |
| Average B-factors (Å2) | |
| Macromolecules | 32.1 |
| Ligands | 24.2 |
| Solvent | 36.3 |
| Metal ions | 42.25 |
*Statistics for the highest-resolution shell are shown in parentheses.
Carbohydrate-binding site
Drgal1-L2 has an archetypical galectin binding site: a narrow cleft in the concave side at one of the ends of the molecule. As in other galectin-1/carbohydrate complexes, the galactose ring docks between two aromatic residues: His 52 and Trp 68 (Figure 1B). Tightly packed by a hydrogen bond network, the side-chains of hydrophilic residues recognize the hydroxyl groups of the disaccharide: His 44 and Asn 46 make hydrogen bonds with the axial 4-OH group and the Asn 61 and Glu 71 make a hydrogen bonds with the 6-OH group of the galactose moiety. Arg 48, Glu 71 and Arg 73 make hydrogen bonds with the 3-OH group of the N-acetyl glucosamine moiety. A network of water molecules coordinated mostly by main chain groups completes the N-acetyl glucosamine coordination. The 3- and 2-OH of the galactose are free to form a glycosidic bond with other carbohydrate moieties in extended backbones or with capping carbohydrates such as sialic acid or fucose.
Drgal3-L1 CRD model
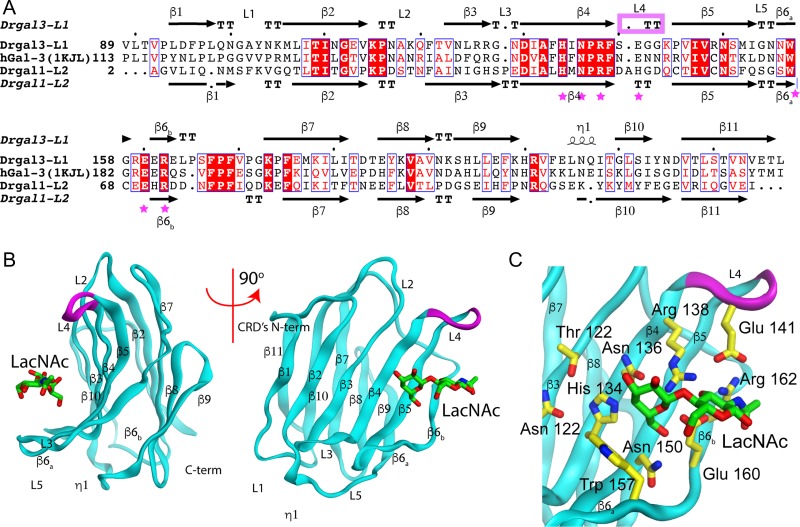
Drgal1-L2 and Drgal3-L1 recognize these hosts and viral glycans and this recognition is inhibited by LacNAc (Nita-Lazar et al. 2016). The differences in the recognition of the LacNAc moieties of the viral GP by these two secreted lectins may provide a rationale for their differential inhibition of viral attachment. To investigate this possibility, we modeled the structure of the Drgal3-L1’s CRD based on the hGal-3 CRD (Figure 2).
Fig. 2.
Drgal3-L1 carbohydrate recognition domain (CRD) model. (A) Sequence alignment of Drgal3-L1 aligned with the template hGal-3 and Drgal1-L2. Purple stars indicate residues involved in carbohydrate recognition. Secondary structure elements for Drgal3-L1 (top) and Drgal1-L2 (bottom) are also shown. The purple box indicates the short loop between β4 and β5. Boxes with red background and white letters indicate sequence identity and those with white background and red letters show similarity in size or properties (charge, hydropathy, etc.). (B) The panel shows the comparative model of Drgal3 L1(CRD)/LacNAc complex in two 90 degrees related views. The shorter loop is colored in purple. (C) Details of the binding site showing LacNAc bound to the Drgal3-L1 CRD. Residues interacting with the carbohydrate are shown in stick representation. Atoms are colored as in Figure 1B.
The sequence of the CRD in Drgal3-L1 (a.a. 89-227 of the 25 kDa protein) align with those of Drgal1-L2 and hGal-3 with 27.3% and 48% identity, respectively (Figure 2A). The final model of the Drgal3-L1’s CRD generated using hGal-3—PDB ID 1AKJ, (Sorme et al. 2005)—as a template and based on the Figure 2A alignment has 100% of its residues in the allowed region of the Ramachandran plot and an rmsd between model and template of 1.062 Å for 138 aligned Cα-atoms. Loop L4, the loop between β-strands 4 and 5, is shorter in Drgal3-L1 than in Drgal1-L2 by the deletion of a histidine residue that in the Drgal1-L2 complex stacks against the Gal moiety of the galactoside. This deletion in the loop makes room at the galactose non-reducing-end 2-OH group providing a better recognition platform for α-2-fucosyl group substitutions (Feng et al. 2013). Loops L3 and L5 of Drgal3-L3 showed small displacement compared to hGal-3 probably caused by Pro 165 insertion at the C-terminal of β6b. The model of Drgal3-L1’s CRD predicts that all the polar interactions with LacNAc observed in its complex with hGal-3 (Sorme et al. 2005) or Drgal1-L2 (this work) are conserved (Figure 2C).
Recognition of biantennary carbohydrates (BCs)
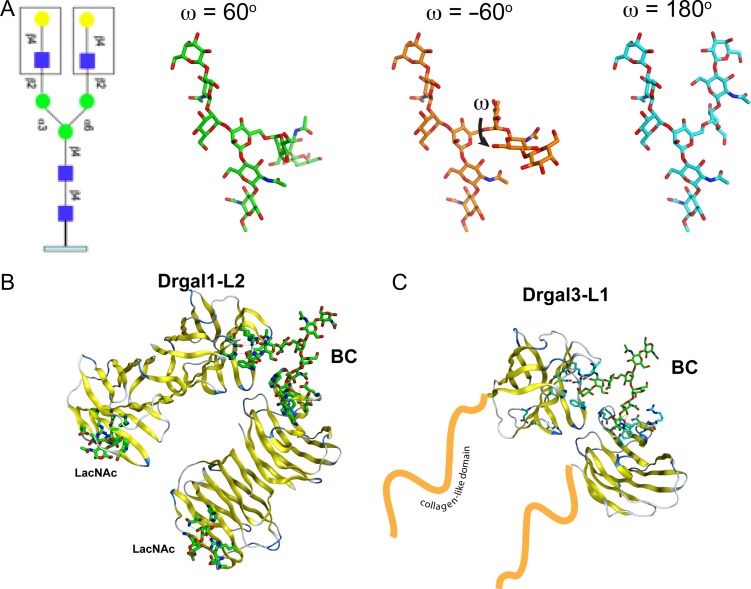
Biantennary N-glycans, such as the one shown in Figure 3, decorate host epithelial cells and the IHNV GP. Drgal1-L2 and Drgal3-L1 recognizes galactosides in these host viral glycans (Nita-Lazar et al. 2016). The several crystal structures of bovine galectin-1/BC show similar biantennary oligosaccharide linking bovine galectin-1 of different asymmetric units (Bourne et al. 1994; Yoshida et al. 2010). The BCs in the three crystal forms reported by Bourne et al. (1994) adopt conformations corresponding to low energy states (ω = −60, Figure 3A), reaching galectin binding sites 19.8 Å apart in the crystal arrangement. Modeling shows that a BC in an extended conformation (ω = 180) can be recognized by two closely packed Drgal1-L2 dimers or two Drgal3-L1 CRDs (Figure 3).
Fig. 3.
Model of the biantennary carbohydrate. (A) Common biantennary N-glycan observed in IHNV and fish epithelia. Monosaccharide symbols follow the symbol nomenclature for glycans system (PMID 26543186, Glycobiology 25:1323–1324, 2015). The three lowest energy conformations of the carbohydrate as calculated by the Glycam server (http://glycam.org). (B) Recognition of BC by two Drgal1-L2 dimers (left) and two Drgal3-L1 CRDs (right).
IHNV glycoprotein model
Upon pre-incubation of IHNV with either of the zebrafish galectins Drgal1-L2 (2.1 μM) or Drgal3-L1 (1.2 μM), viral attachment to the fish epithelial-cell membrane is inhibited by 40% and 65%, respectively, compared to the attachment of unexposed virus (Nita-Lazar et al. 2016). The lower inhibition with a molar excess of Drgal1-L2 is at odds with the expected similarity in the recognition of the GP carbohydrates by the two galectins. These prompted us to analyze how the differences in carbohydrate recognition and quaternary structure between Drgal1-L2 and Drgal3-L1 affects the inhibition of viral attachment. Further, to analyze in detail the inhibition of IHNV attachment by galectins, we modeled the IHNV GP ectodomain in complex with these galectins.
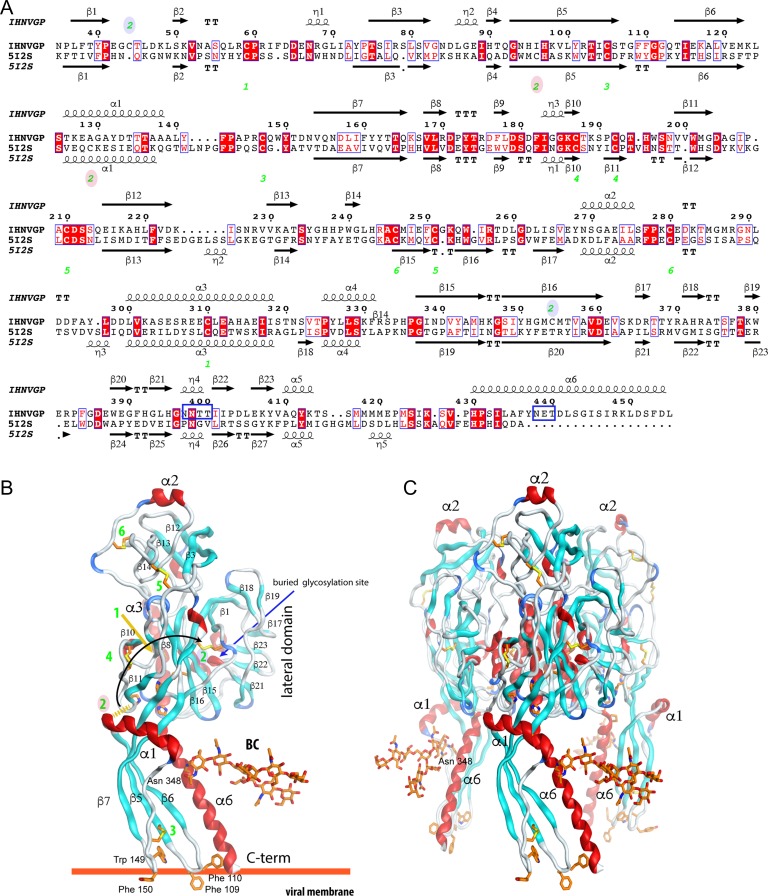
Figure 4 shows the sequence alignment used in the model of the envelope glycoprotein ectodomain of IHNV based on the crystal structure of its homologs in VSV (vesicular stomatitis virus) (Roche et al. 2008). Although the final sequence identity between target and template is low (18.9%), the conservation of most of the residues that make disulfide bridges, the aromatic residues at the tip of the fusion loops, the His-Pro sequence motifs that participates in a pH sensitive histidine cluster, and other key structural features (Albertini et al. 2012) reinforces the confidence in the modeling using VSV GP as a template (Figure 4). Cys 47 and 357 are only present in IHNV and in adjacent strands of the lateral domain—DI, using the nomenclature from Roche (Roche et al. 2008)—close enough to make a disulfide bridge to replace the non-conserved Cys 59-Cys 92 bridge of VSV.
Fig. 4.
IHNV comparative model. (A) Sequence alignment of the GPs from IHNV and VSV used in the modeling. The green numbers indicate the associated disulfide bridges numbered in the alignment. The ones marked with a blue-shaded oval are new and the red shaded are those missed in IHNV GP with respect to VSV GP. The top (model) and bottom (template) secondary structure of the model (top) and template (bottom) are also represented. IHNV glycosylation sites are underlined in blue. (B-C) The final model of the IHNV GP ectodomain. Monomer (B) and trimer (C). Disulfide bridges are displayed and numbered as in (A); a black curved arrow connects the lost disulfide bridge with its replacement. The histidine residues associated with the fusion mechanism, hydrophobic residues at the tip of the fusion loops and BC attached to Asn 438 are displayed with orange carbon atoms. Helices are colored in red, β-strands in cyan, turns in blue, and loops in gray. Some of the structure secondary elements are labeled in both panels for orientation.
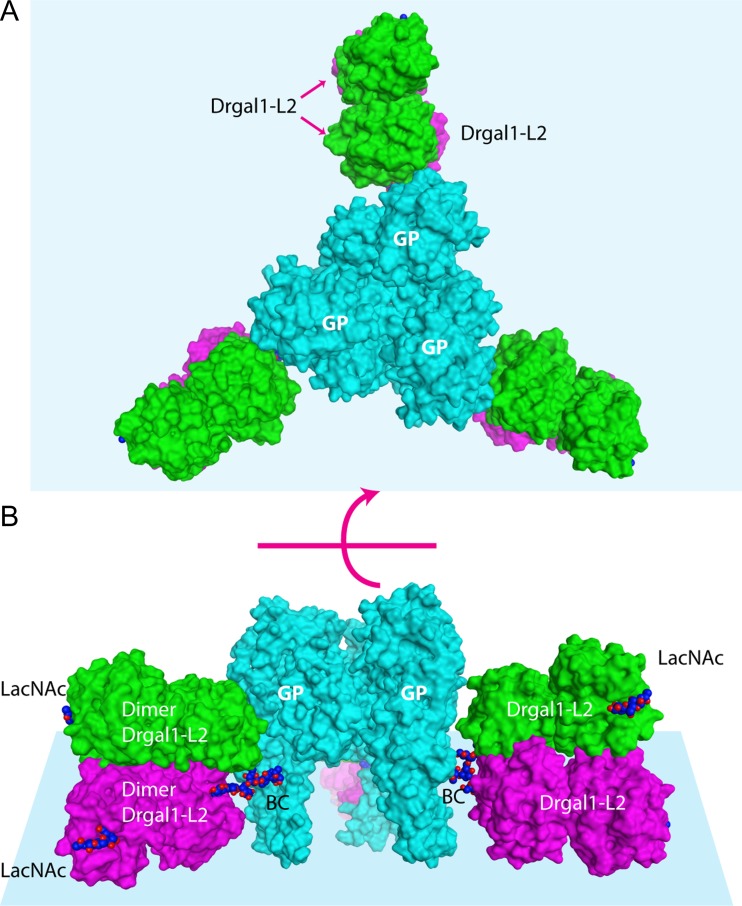
IHNV has three N-glycosylation sequence motifs (two overlapping) at positions not conserved with respect to those in the VSV GP. The model predicts that the overlapping N-glycosylation sites at Asn 400 and 401 will be buried in the lateral domain (a.a. 35–55, 331–411) and then expected not to be glycosylated (Pless and Lennarz 1977). The site at Asn 438 is the only one exposed on the trimer axis near the His-Pro motif. An O-GlcNAcylated serine residue observed in the recent structure of VSV GP is not conserved. The energy-minimized model of the IHNV GP shows 99.5% of the residues in the allowed region of the Ramachandran plot and 1.6 Å rmsd between model and template for 393 Cα-atoms aligned. In the model of the IHNV GP trimer fully decorated with Drgal1-L2 molecules bound to their BCs (Figure 5), two Drgal1-L2 galectin dimers can simultaneously recognize, without steric conflicts, the terminal LacNAc moieties on the two antennae of a biantennary oligosaccharide in an isolated pre-fusion GP trimer (Figure 5; coordinates included as Supplementary Materials).
Fig. 5.
IHNV GP trimer (peplomer) fully decorated with Drgal1-L2s. (A) Top view (viewed from the cell surface) of the IHNV GP model (cyan) with the BC in an extended conformation (ω = 180o) fully decorated by two Drgal1-L2 dimers (green and magenta). (B) Side-view. The BC and LacNAc in a CPK representation have their carbon-atoms colored blue. The light blue planes represent the viral surface.
Discussion
Despite the considerable phylogenetic distance between teleost fish and eutherian mammals, the determination of the structure of the zebrafish Drgal1-L2/LacNAc complex revealed a remarkable overall structural similarity between this molecule and the bovine (Bourne et al. 1994) and human (Lopez-Lucendo et al. 2004) galectins-1 (Supplementary Figure S1). This similarity extends to the topology of the CRD, including the position and identity of the residues that interact with the LacNAc ligand. This observation is in-line with the high evolutionary conservation of proto-type galectin sequences along the vertebrate lineages from bony fish to higher mammals—Supplementary Figure S2 (Stowell et al. 2010; Vasta 2012). Like human galectin-1, Drgal1-L2 dimerization made the same number of non-covalent bonds, but the dimer interface buries 5% more area than in human galectin-1 (571 Å2), suggesting a tighter dimer association than human galectin-1—Kd = 0.7 μM (Leppanen et al. 2005). In agreement with the fold conservation observed, the monomer superposition of Drgal1-L2 with other known galectin-1s showed a small rmsd. Small fold differences are concentrated in loops, e.g., loops L3, L4, and L5 (Figure 1 and Supplementary Figure S1). The differences in these loops accounts for the high pairwise rmsd of chicken galectin-1 with the others. The quaternary structure of characterized mammalian and amphibian galectins-1 shows a conserved twist between monomers in the dimer. However, Drgal1-L2 dimer has a significantly smaller twist than other known counterparts (Supplementary Figure S1). Interestingly, this difference in the galectin quaternary structure may be a response to evolutionary pressure for adaptation to a specific fish pathogen.
Zebrafish galectins Drgal1-L2 and Drgal3-L1 are secreted into the skin mucus in vivo, and it has been demonstrated to negatively modulate IHNV attachment to epithelial cells in vitro (Nita-Lazar et al. 2016). Glycomic studies of the IHNV GP and the fish epithelial cells reveal the presence of terminal LacNAcs in viral as well as host cell glycans (Nita-Lazar et al. 2016). Galectins binds to these glycotopes (Bourne et al. 1994; Yoshida et al. 2010) as it is safe to assume that both zebrafish galectins do as well. Drgal1-L2 and Drgal3-L1 bind to glycans on virions and glycosylated co-receptors inhibiting with different potency the virus attachment to the epithelial cells (Nita-Lazar et al. 2016). This inhibition could be the consequence of these galectins hindering the attachment sites of epithelial-cell receptors by steric conflict or by inducing changes in the GP distribution or conformation. The differences in such inhibition between Drgal1-L2 and Drgal3-L1 could be due to the differences in these galectins CRD’s affinity for the viral carbohydrates or because their different avidity for the multivalent display of glycotopes on the virion surface.
To investigate the mechanism(s) by which Drgal1-L2 and Drgal3-L1 interfere with IHNV attachment and explain the observed differences in their inhibition potency, we developed models of (1) the CRD of Drgal3-L1 (Figure 2) and (2) the recognition by these zebrafish galectins of the IHNV GP pre-fusion trimer (Figures 4–5) and its arrangement on the viral particle surface (Figure 6). The Drgal3-L1 CRD model shows that all the polar interactions with the ligand are conserved with those of Drgal1-L2 (Figure 2). However, the shorter loop, L4 (loop between β4 and β5), in Drgal3-L1 losses a stacking interaction of the galactose C2-OH group with the aromatic ring of a histidine residue (His 52, Figure 1B). This also widens the type of glycans recognized by the Drgal3-L1 CRD (Feng et al. 2013) to include carbohydrates found in a small, probably coexisting, sub-populations of viral glycans (Nita-Lazar et al. 2016). Nevertheless, the opposite contributions of the loss of this stacking and the small increment in the number of targets recognized cannot explain the 62.5% of increase in inhibition (from 40% in Drgal1-L2 to 65% in Drgal3-L1).
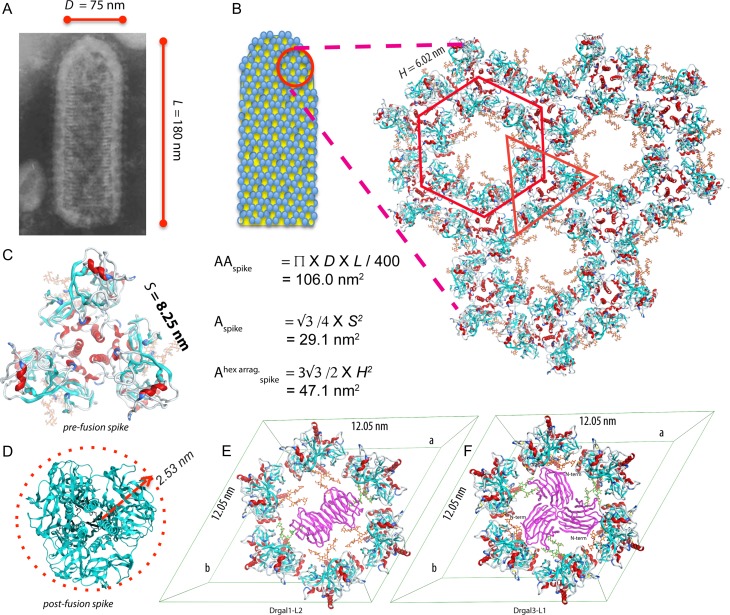
Fig. 6.
Recognition of IHNV GP by galectins on the viral surface. (A) The micrograph of a rabies rhabdovirus shows the dimensions of the virion. (This picture is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License.) The AAspike of an uncapped bullet-shape particle of diameter D (75 nm) and longitude L (180 nm) displaying around of 400 spikes is 106.0 nm2. A virion surface showing a hexagonal lattice of spikes with a dimension h (6.02 nm) formed by the equivalent of two spikes, has an area per spike (Ahexspike) of 47.1 nm2. The lattice shows a central void exposing six BCs (carbon atoms colored in orange). (C) The area per spike for the pre-fusion GP trimer has an area of 29.1 nm2, equivalent to the area of an equilateral triangle of side S (8.25 nm). (D) The area per spike for the post-fusion GP trimer is 19.6 nm2, corresponding to the area of a circle of radius r (r = 2.5 nm). (E) Drgal1-L2 dimer bound to the hexagonal arrangement of spikes, recognizing two BCs (orange carbon atoms) related by the two-fold axis of the center hexagonal arrangement. (F) Drgal3-L1 bound to the hexagonal arrangement of spikes. The full hexagonal arrangement is obtained by applying to the asymmetric unit formed by two adjacent GPs the three-fold axis at the center of the hexagon, one with the orange BC recognized by the galectin-3 and the other with the BC in green. The galectins in (E) and (F) are colored in purple.
Differences in these galectins avidity for the multivalent of ligands displayed on the virion surface may cause the pattern of inhibition observed. Drgal1-L2 is a proto-type galectin with a single CRD per subunit capable of forming dimers in a concentration-dependent manner. At the concentration range present in the fish mucus (2.1 μM), most of the Drgal1-L2 are associated as a homodimers. Dimers of Drgal1-L2 can simultaneously recognize carbohydrates 4.9 nm apart (Morris et al. 2004). In contrast, Drgal3-L1 belongs to the chimera-type galectin-3, a protein with two domains: a long and flexible collagen-like N-terminal domain and a C-terminal CRD. While galectin-3 is monomeric in solution (Morris et al. 2004), upon binding to multivalent ligands such as glycans displayed on the cell surface, galectin-3 can oligomerize via their N-terminal domains. Galectin-3 oligomers are capable of simultaneously recognize more glycans that are distributed further apart than those cross-linked by proto-type galectins (Morris et al. 2004; Fortuna-Costa et al. 2014). To explore differences in the avidity of these multivalent galectins for the arrangements of GP on the virion, we localized the GP glycosylation sites and modeled the attachment of these galectins (Figures 4 to 6). Rhabdoviruses are bullet-shaped with average dimensions of 75 nm of diameter and 180 nm in length (Figure 6A) (Hill et al. 1975; Pringle 2006). Currently, it is accepted that rabies virions display ~400 peplomers (spikes) attached to its external lipidic membrane; in an apparent honeycomb (hexagonal) arrangement (Hill et al. 1975). Hexagonal arrangements were observed in a rabies virus mutant briefly incubated under suboptimal fusion conditions (Gaudin et al. 1996) which suggests it plays a role after rhabdovirus attachment during the fusion of virion-host membranes (Roche et al. 2007). The hexagonal arrangement of GP trimers (Figure 6B), like one observed in the crystal structure of VSV GP, has been proposed in the rhabdavirus (Roche et al. 2007; 2008). Each spike is a trimer of envelope GPs that attaches to cell-membrane phospholipids and co-receptors such as the monomeric truncated form of fibronectin (Bearzotti et al. 1999; Liu and Collodi 2002). Pre-fusion and post-fusion states of GP are in a pH-dependent equilibrium making the need for a more significant number of spikes acting cooperatively during fusion (Roche and Gaudin 2002).
The area available per spike (AAspike) in an uniformly scattered arrangement of 400 spikes on a rabies virion—dimensions: 75 nm × 180 nm (Hill et al. 1975)—is 106.0 nm2 (Figure 6A). A similar AAspike is expected in IHNV, sufficient to accommodate either a pre-fusion spike (with a 29.5 nm2 footprint; Figure 6C) or the smaller footprint of a post-fusion spike (19.6 nm2; Figure 6D). Back-of-the-envelope calculations show that a pre-fusion spike fully decorated with Drgal1-L2 dimers (Figure 5) roughly requires to reach an area of 134.8 nm2, while with Drgal3-L1 CRDs only needs 91.0 nm2 (Supplementary Figure S3). A full decoration with Drgal1-L2 dimers is incompatible with the area available on the virion. Thus, partial decorations or a lower number of spikes fully decorated with Drgal1-L2 than with Drgal3-L1 are expected, correlating with their viral attachment inhibition potency.
In a honeycomb-like (hexagonal) arrangement of spikes (Figure 6B), Drgal1-L2 dimers can crosslink BCs from two spikes related by the two-fold at the center of the hexagonal void (Figure 6F). On the other hand, the single C-terminal CRD of Drgal3-L1 allows decoration of three BCs from three-fold related spikes with enough room to accommodate the exit of the N-terminal domain to oligomerize (Figure 6G). These models suggest that Drgal1-L2 could occlude two of the six co-receptor attachment sites on the surface of the virus, while Drgal3-L1 could do the same with three sites. Also, the viral surface coverage by Drgal3-L1 attached GP is greater than that for Drgal1-L2 by a factor of 3/2. The ratio of the inhibitory efficiency estimated using this 3/2 model is around the observed ratio of 65%/40%.
These galectins are predicted to attach to a region of the INHV GP ectodomain that remains relatively unchanged between pre and post fusion conformations. Nevertheless, an equilibrium shift toward post-fusion states of the GP (more compact), becoming more favorable to the galectin conjugation, is probable. On the other hand, a spike decoration of a hexagonal lattice, as revealed by the aforementioned models (Figure 6A and B), may not only hinder virion-receptor interaction but also impair or even inhibit conformational changes of the spikes during viral fusion.
The roles of galectins in viral attachment and infection have only been investigated in recent years (Batts et al. 1991; Levroney et al. 2005; Crane and Hyatt 2011; Yang et al. 2011a; Sato et al. 2012; Toledo et al. 2014; Nita-Lazar et al. 2016). It is worth noting that for several viruses, recognition of the envelope glycoprotein by selected host galectins promotes viral attachment and infection. This suggests that close co-evolution of the viruses with their hosts has led to the functional hijacking of the recognition properties of the highly conserved and pleiotropic galectin family for host entry and infection (Vasta 2009). In some virus/host systems, however, galectins can function as successful innate immune recognition and effector factors, preventing viral attachment and infection, (Levroney et al. 2005; Yang et al. 2011a; Sato et al. 2012; Toledo et al. 2014; Nita-Lazar et al. 2016). The structural study reported here reveals the potential interactions of these galectins with the IHNV GP that may be responsible for their protective properties.
Materials and methods
Protein expression and purification
Recombinant Drgal1-L2 was expressed and purified as described previously (Nita-Lazar et al. 2016). Briefly, Rosetta (DE3)pLysS competent cells (Novagen) were transformed with a pET-28b (+) based plasmid expressing a 6xHis-tagged full-length Drgal1-L2 gene (134 amino acids, 15.4 kDa, Genbank accession no. AY421704)(Ahmed et al. 2004). Cultures of plasmid carrying bacteria were grown in Luria–Bertani broth with antibiotics (chloramphenicol, 34 μg/mL; kanamycin, 15 μg/mL), induced with 0.1 isopropyl d-thiogalactoside when the optical density reached 0.6, and grown for additional 16 h at 23°C. Overexpressed protein was purified from a clarified lysate of these cells by binding to an affinity chromatography column packed with 4 mL of divinyl sulfone-conjugated lactosyl-sepharose and eluted with 100 mM lactose (Nita-Lazar et al. 2016). Alkylation with iodoacetamide was used to prevent aggregation due to the exposure of two cysteine residues, Cys 56 and Cys 69.
Crystallization, data collection, structure determination and refinement of the zebrafish galectin Drgal1-L2
Alkylated Drgal1-L2 at a concentration of 10 mg/mL (in 50 mM Tris-HCl, 150 mM NaCl, pH7.5) was incubated with 20 mM of LacNAc on ice for 30 min before crystallization. Crystals of the Drgal1-L2/LacNAc complex were obtained by the hanging drop vapor diffusion method. One μL of Drgal1-L2/LacNAc was mixed with 1 μL of reservoir solution and equilibrated against 0.5 mL of reservoir solution (0.1 M HEPES/NaOH pH 7.5, 0.2 M MgCl2, 30% (v/v) PEG 400). Diffraction quality crystals grew in 12 h.
Data were collected at a home source, using an FRE+-high-brilliance X-ray generator equipped with a CCD detector Saturn-977 (Rigaku Inc., Texas), from a crystal flash-frozen in a drop held by a nylon loop containing reservoir solution with 15% (v/v) glycerol added as cryoprotectant; 720 frames were collected using a 0.25 degrees oscillation with 15 s of exposure. Diffraction data was reduced with HKL2000 and scaled using the program Scalepack (Otwinowski and Minor 1997). The crystal has two molecules in the asymmetric unit. The structure of Drgal1-L2/LacNAc complex was determined by molecular replacement with the program MOLREP (Vagin and Teplyakov 1997) using the coordinates of the amphibian Bufo arenarum galectin-1 structure (PDB ID 1GAN) previously determined by our group (Bianchet et al. 2000). The initial structure was manually rebuilt with the graphical program Coot (Emsley et al. 2010), and refined using the program REFMAC5 (Vagin et al. 2004). The crystal shows additional electron density in the ligand pocket associated with the disaccharides. After an initial rigid body refinement of each separated monomer, the carbohydrate was placed into the observed positive difference-Fourier density. The resulting model was subjected to cycles of maximum likelihood, real space, isotropic B-factor, and TLS refinement followed by manual rebuilding. The interface’s characterization was carried out with the program qtPISA (Krissinel and Henrick 2007). MOLREP, REFMAC, Coot, and qtPISA are included in the CCP4 program package v. 7.0.060 (Winn et al. 2011). To calculate the two-fold symmetry axis of the dimer, the monomers superposition matrix was diagonalized and the arc cosine of scalar product of the eigenvectors divided by their norms were used to calculate the differences between twist angles. Figures, rmsd calculations, sequence, and structural alignments were carried out with the programs MOE (v 2018.1, Molecular Operating Environment; Chemical Computing Group Inc., Canada). Sequence alignment figures were prepared with ESPrit 3 web server at http://espript.ibcp.fr (Robert and Gouet 2014). Coordinates and structure factors were deposited in the Protein-Data Bank (PDB ID 6E20).
Comparative modeling
Drgal3-L1 Model. A model of the zebrafish galectin Drgal3-L1 carbohydrate recognition domain (CRD, a.a. 89 to 227) in complex with LacNAc was built by comparative modeling using the crystal structure of human galectin-3—hGal-3; PDB accession code (PDB ID) 1KJL (Sorme et al. 2005)—as the template. The sequence of Drgal3-L1 (NCBI: NP_999858) was aligned to the sequences of hGal-3 and Drgal1-L2 structures (Figure 2A). The molecular modeling program MOE (Molecular Operating Environment, 2018.01; Chemical Computing Group ULC) was used for the comparative modeling of the galectin, side-chains conformation sampling, loop building, and energy minimization. Structural stress in the models was relaxed by several cycles of conjugate gradient minimization using an AMBER14HT force-field until the change in the gradient was below of 10−4 Kcal Mol−1Å−1.
Coordinates for the BC N-glycan in the three lowest energy conformation were obtained from the Glycam server (http://glycam.org). In the models of the complexes of the galectins with the BC (Figure 3), the galectin bound LacNAc guided the placement of the LacNAc terminals of the BC. The LacNAc disaccharide terminal of an extended biantennary oligosaccharide in its lower energy conformation was docked in the galectin binding-site using the structure of the bound LacNAc disaccharide as a target with the pair fitting routine of PyMol (v.1.7.1.3, ©2009-2014 Schödinger, LLC). Briefly, this routine minimizes the distance between pairs of equivalent atoms by moving one of the molecules as a rigid unit.
IHNV glycoprotein model. A structural model of the IHNV glycoprotein (GP) ectodomain (a.a. 38-455, GenBank ABA41533) pre-fusion form was generated using the crystal structure of the high pH form of VSV glycoprotein—PDB ID 5I2S, (Roche et al. 2007)—as the template (Figure 4). The sequence alignment between IHNV and VSV GPs used in the modeling (Figure 4A) was obtained from an initial automatic alignment by careful adjustments of insertions and deletions to maintain features observed in these types of viruses. The homology modeling program Modeller v. 9.20 (Sali and Blundell 1993) was used with this alignment to build the IHNV GP structural model (Figure 4B). An alpha-helical secondary structure constrain was used in the C-terminal region of the ectodomain (a.a. 430–455), because of the lack of a template. The putative new disulfide bridge between Cys 44 and Cys 354 was generated with a disulfide patch in the Modeller script. The structural model was optimized in Modeller using a energy minimization protocol consisting of a conjugate gradients, molecular dynamic, and final conjugate gradients minimization. The main-chain of the resulting model was improved by local optimization of Ramachandran outlier regions using an Amber 14HT (MOE v 2018.1) force field. The final Ramachandran plot (Supplementary Figure 4), the Modeller python script, and Modeller input are included as Supplementary Materials.
To model the glycosylation by BC, an N-glucosamine was built as a postranslational modification to the solvent-exposed Asn 438 with the program MOE. This glycosylation was used to fit the N-glucosamine of the first moiety of BC using PyMol as described above. For each galectin type, galectin molecules were docked to each antenna of the modeled biantennary glycans attached to Asn 438 using the LacNAc bound to Drgal1-L2 in the experimental structure as a guide, followed by a least squares fitting of equivalent atoms of the disaccharide terminal as described above.
The GP trimer and the hexagonal lattice were generated by MOE by applying the point-group P3 and P6 symmetry, respectively, to the GP model superposed with the template, using the hexagonal cell dimensions of the latter (a = b = 120.51 Å). PDB file of the asymmetric unit is included as Supplementary Materials.
The Drgal1-L2 dimer crosslinking two GPs across the void in Figure 6F was modeled by making the dimer two-fold axis to coincide with the two-fold axis of the hexagonal void by a multi-step process. (1) A pair of two-fold atoms one from each galectin bound LacNAc and the other from the BC terminal moieties were selected to guide the fitting. With the galectin/ligand as a rigid moving molecule, the distances between paired atoms were minimized by least squares fitting. (2) The BC glycoside-angles were manually adjusted to match their terminal disaccharide orientation with that of the galectin-bound disaccharide, and a final pair fitting was performed. (3) Finally, each BC terminal LacNAc was replaced by the corresponding LacNAc in the galectin-bound, generating a hybrid molecule by forming a distorted glycosyl bond between them. These hybrid BC were minimized under restrains with the program MOE using an Amber 14HT force field. Proteins and terminal LacNAc were restrained to their position allowing only the rest of BC to adjust.
A rigid-body pair fitting procedure was used to generate the model of Drgal3-L1/GP (Figure 6G). One Drgal3-L1 molecule was docked to one of the antennas (the most extended) of a pair of adjacent GPs in the hexameric arrangement. The model was generated by applying a three-fold axis located at the center of the hexagonal void to this dimer. The conformation of the unbound BC was manually adjusted to eliminate clashes with the galectins.
Supplementary Material
Abbreviations
- IHNV
infectious hematopoietic necrosis virus
- GP
glycoprotein
- CRD
carbohydrate-recognition domain
- VSV
vesicular stomatitis virus
- HIV
human immunodeficiency virus
- BC
bi-antennary carbohydrate
- LacNAc
N-acetyl-lactosamine, Galβ1,4GlcNAc
- hGal-3
human galectin-3
Supplementary data
Funding
This work was supported by Pilot Award component of the R25 Translational Research in NeuroAIDS and Mental Health (TRNAMH) Program [R25MH080661] and JHU CFAR NIH/NIAID fund [P30AI094189] awards to M.A.B., and National Institutes of Health [R01GM070589] and National Science Foundation [IOS-1656720 and IOS-1050518] grants to G.R.V.
Conflict of interest statement
The authors declare no competing interests.
Authors' contributions
A.B. carried out expression and purification of Drgal1-L2 in the GRV lab. A.G. performed the crystallization, diffraction data collection and crystal structure determination. M.A.B. contributed to modeling experiments, analysis and interpretation of the results. G.R.V., L.M.A. and M.A.B. wrote the manuscript with input from the other authors. M.A.B. was responsible for the overall direction of the project.
References
- Ahmed H, Du SJ, O’Leary N, Vasta GR. 2004. Biochemical and molecular characterization of galectins from zebrafish (Danio rerio): Notochord-specific expression of a prototype galectin during early embryogenesis. Glycobiology. 14:219–232. [DOI] [PubMed] [Google Scholar]
- Ahmed H, Fink NE, Pohl J, Vasta GR. 1996. Galectin-1 from bovine spleen: Biochemical characterization, carbohydrate specificity and tissue-specific isoform profiles. J Biochem. 120:1007–1019. [DOI] [PubMed] [Google Scholar]
- Albertini AAV, Baquero E, Ferlin A, Gaudin Y. 2012. Molecular and cellular aspects of rhabdovirus entry. Viruses. 4:117–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batts WN, Landolt ML, Winton JR. 1991. Inactivation of infectious hematopoietic necrosis virus by low levels of iodine. Appl Environ Microbiol. 57:1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearzotti M, Delmas B, Lamoureux A, Loustau AM, Chilmonczyk S, Bremont M. 1999. Fish rhabdovirus cell entry is mediated by fibronectin. J Virol. 73:7703–7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchet MA, Ahmed H, Vasta GR, Amzel LM. 2000. Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: Crystallographic studies of two protein-sugar complexes. Proteins. 40:378–388. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Bolgiano B, Liao DI, Strecker G, Cantau P, Herzberg O, Feizi T, Cambillau C. 1994. Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat Struct Biol. 1:863–870. [DOI] [PubMed] [Google Scholar]
- Camby I, Le Mercier M, Lefranc F, Kiss R. 2006. Galectin-1: A small protein with major functions. Glycobiology. 16:137R–157R. [DOI] [PubMed] [Google Scholar]
- Crane M, Hyatt A. 2011. Viruses of fish: An overview of significant pathogens. Viruses. 3:2025–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings RD, Liu FT, Vasta GR. 2017. Galectins In: Varki A, Cummings RD, Esko J, Freeze H, Hart G, Marth J, editors. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; p. 469–480. [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Ghosh A, Amin MN, Giomarelli B, Shridhar S, Banerjee A, Fernandez-Robledo JA, Bianchet MA, Wang LX, Wilson IB et al. 2013. The galectin CvGal1 from the eastern oyster (Crassostrea virginica) binds to blood group A oligosaccharides on the hemocyte surface. J Biol Chem. 288:24394–24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuna-Costa A, Gomes AM, Kozlowski EO, Stelling MP, Pavao MS. 2014. Extracellular galectin-3 in tumor progression and metastasis. Front Oncol. 4:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y, Raux H, Flamand A, Ruigrok RW. 1996. Identification of amino acids controlling the low-pH-induced conformational change of rabies virus glycoprotein. J Virol. 70:7371–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmache A, LeBerre M, Droineau S, Giovannini M, Bremont M. 2006. Bioluminescence imaging of live infected salmonids reveals that the fin bases are the major portal of entry for Novirhabdovirus. J Virol. 80:3655–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill BJ, Underwood BO, Smale CJ, Brown F. 1975. Physico-chemical and serological characterization of five rhabdoviruses infecting fish. J Gen Virol. 27:369–378. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J, Kasai K. 1993. The family of metazoan metal-independent beta-galactoside-binding lectins: structure, function and molecular evolution. Glycobiology. 3:297–304. [DOI] [PubMed] [Google Scholar]
- Kaltner H, Toegel S, Caballero GG, Manning JC, Ledeen RW, Gabius HJ. 2017. Galectins: Their network and roles in immunity/tumor growth control. Histochem Cell Biol. 147:239–256. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 372:774–797. [DOI] [PubMed] [Google Scholar]
- Leppanen A, Stowell S, Blixt O, Cummings RD. 2005. Dimeric galectin-1 binds with high affinity to alpha2,3-sialylated and non-sialylated terminal N-acetyllactosamine units on surface-bound extended glycans. J Biol Chem. 280:5549–5562. [DOI] [PubMed] [Google Scholar]
- Levroney EL, Aguilar HC, Fulcher JA, Kohatsu L, Pace KE, Pang M, Gurney KB, Baum LG, Lee B. 2005. Novel innate immune functions for galectin-1: Galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol. 175:413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FT, Yang RY, Hsu DK. 2012. Galectins in acute and chronic inflammation. Ann N Y Acad Sci. 1253:80–91. [DOI] [PubMed] [Google Scholar]
- Liu X, Collodi P. 2002. Novel form of fibronectin from zebrafish mediates infectious hematopoietic necrosis virus infection. J Virol. 76:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lucendo MF, Solis D, Andre S, Hirabayashi J, Kasai K, Kaltner H, Gabius HJ, Romero A. 2004. Growth-regulatory human galectin-1: Crystallographic characterisation of the structural changes induced by single-site mutations and their impact on the thermodynamics of ligand binding. J Mol Biol. 343:957–970. [DOI] [PubMed] [Google Scholar]
- Lujan AL, Croci DO, Gambarte Tudela JA, Losinno AD, Cagnoni AJ, Marino KV, Damiani MT, Rabinovich GA. 2018. Glycosylation-dependent galectin-receptor interactions promote Chlamydia trachomatis infection. Proc Natl Acad Sci USA. 115:E6000–E6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Huergo SP, Blidner AG, Rabinovich GA. 2017. Galectins: Emerging regulatory checkpoints linking tumor immunity and angiogenesis. Curr Opin Immunol. 45:8–15. [DOI] [PubMed] [Google Scholar]
- Mercier S, St-Pierre C, Pelletier I, Ouellet M, Tremblay MJ, Sato S. 2008. Galectin-1 promotes HIV-1 infectivity in macrophages through stabilization of viral adsorption. Virology. 371:121–129. [DOI] [PubMed] [Google Scholar]
- Morris S, Ahmad N, Andre S, Kaltner H, Gabius HJ, Brenowitz M, Brewer F. 2004. Quaternary solution structures of galectins-1, -3, and -7. Glycobiology. 14:293–300. [DOI] [PubMed] [Google Scholar]
- Nita-Lazar M, Banerjee A, Feng C, Amin MN, Frieman MB, Chen WH, Cross AS, Wang LX, Vasta GR. 2015. a. Desialylation of airway epithelial cells during influenza virus infection enhances pneumococcal adhesion via galectin binding. Mol Immunol. 65:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar M, Banerjee A, Feng C, Vasta GR. 2015. b. Galectins regulate the inflammatory response in airway epithelial cells exposed to microbial neuraminidase by modulating the expression of SOCS1 and RIG1. Mol Immunol. 68:194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar M, Mancini J, Feng C, Gonzalez-Montalban N, Ravindran C, Jackson S, de Las Heras-Sanchez A, Giomarelli B, Ahmed H, Haslam SM et al. 2016. The zebrafish galectins Drgal1-L2 and Drgal3-L1 bind in vitro to the infectious hematopoietic necrosis virus (IHNV) glycoprotein and reduce viral adhesion to fish epithelial cells. Dev Comp Immunol. 55:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326, Academic Press. [DOI] [PubMed] [Google Scholar]
- Pless DD, Lennarz WJ. 1977. Enzymatic conversion of proteins to glycoproteins. Proc Natl Acad Sci USA. 74:134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle CR. 2006. Rhabdoviruses. eLS.
- Rabinovich GA, Toscano MA, Jackson SS, Vasta GR. 2007. Functions of cell surface galectin-glycoprotein lattices. Curr Opin Struct Biol. 17:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Albertini AA, Lepault J, Bressanelli S, Gaudin Y. 2008. Structures of vesicular stomatitis virus glycoprotein: Membrane fusion revisited. Cell Mol Life Sci. 65:1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Gaudin Y. 2002. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 297:128–135. [DOI] [PubMed] [Google Scholar]
- Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 315:843–848. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- Sato S, Ouellet M, St-Pierre C, Tremblay MJ. 2012. Glycans, galectins, and HIV-1 infection. Ann N Y Acad Sci. 1253:133–148. [DOI] [PubMed] [Google Scholar]
- Scott SA, Cozier MO, Dubar PD, Ramakrishna M, Scott K, Blanchard H. 2010. Crystallization and preliminary X-ray crystallographic analysis of zebrafish prototype galectin Drgal1-L2. Acta Crystallogr Sect F Struct Biol Cryst Commun. 66:1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Cummings RD. 2008. Deciphering lectin ligands through glycan arrays In: Vasta GR, Ahmed H, editors. Animal Lectins: A functional View. Boca Raton, FL: Taylor & Francis; p. 49–62, 33487-2742. [Google Scholar]
- Sorme P, Arnoux P, Kahl-Knutsson B, Leffler H, Rini JM, Nilsson UJ. 2005. Structural and thermodynamic studies on cation-Pi interactions in lectin-ligand complexes: High-affinity galectin-3 inhibitors through fine-tuning of an arginine-arene interaction. J Am Chem Soc. 127:1737–1743. [DOI] [PubMed] [Google Scholar]
- Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B et al. 2010. Innate immune lectins kill bacteria expressing blood group antigen. Nat Med. 16:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundblad V, Morosi LG, Geffner JR, Rabinovich GA. 2017. Galectin-1: A Jack-of-all-trades in the resolution of acute and chronic inflammation. J Immunol. 199:3721–3730. [DOI] [PubMed] [Google Scholar]
- Thiemann S, Baum LG. 2016. Galectins and immune responses-just how do they do those things they do? Annu Rev Immunol. 34:243–264. [DOI] [PubMed] [Google Scholar]
- Toledo KA, Fermino ML, Andrade Cdel C, Riul TB, Alves RT, Muller VD, Russo RR, Stowell SR, Cummings RD, Aquino VH et al. 2014. Galectin-1 exerts inhibitory effects during DENV-1 infection. PLoS One. 9:e112474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Kurath G. 2003. Molecular epidemiology of infectious hematopoietic necrosis virus reveals complex virus traffic and evolution within southern Idaho aquaculture. Dis Aquat Organ. 55:175–185. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. 1997. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 30:1022–1025. [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 60:2184–2195. [DOI] [PubMed] [Google Scholar]
- Vasta GR. 2009. Roles of galectins in infection. Nat Rev Microbiol. 7:424–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta GR. 2012. Galectins as pattern recognition receptors: Structure, function, and evolution. Adv Exp Med Biol. 946:21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ML, Chen YH, Wang SW, Huang YJ, Leu CH, Yeh NC, Chu CY, Lin CC, Shieh GS, Chen YL et al. 2011. a. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J Virol. 85:10010–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RY, Yu L, Graham JL, Hsu DK, Lloyd KC, Havel PJ, Liu FT. 2011. b. Ablation of a galectin preferentially expressed in adipocytes increases lipolysis, reduces adiposity, and improves insulin sensitivity in mice. Proc Natl Acad Sci USA. 108:18696–18701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Teraoka M, Nishi N, Nakakita S, Nakamura T, Hirashima M, Kamitori S. 2010. X-ray structures of human galectin-9 C-terminal domain in complexes with a biantennary oligosaccharide and sialyllactose. J Biol Chem. 285:36969–36976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.