Abstract
Human immunodeficiency virus type 1 (HIV-1) encodes a transactivator of transcription (Tat) protein, which has several functions that promote viral replication, pathogenesis, and disease. Amino acid variation within Tat has been observed to alter the functional properties of Tat and, depending on the HIV-1 subtype, may produce Tat phenotypes differing from viruses representative of each subtype and commonly used in in vivo and in vitro experimentation. The molecular properties of Tat allow for distinctive functional activities to be determined such as the subcellular localization and other intra- and extracellular functional aspects of this important viral protein influenced by variation within the Tat sequence. Once Tat has been transported into the nucleus and becomes engaged in transactivation of the long terminal repeat (LTR), various Tat variants may differ in their capacity to activate viral transcription. Post-translational modification patterns based on these amino acid variations may alter interactions between Tat and host factors, which may positively or negatively affect this process. Additionally, the ability of HIV-1 to utilize or not utilize the transactivation response (TAR) element within the LTR, based on genetic variation and cellular phenotype, adds a layer of complexity to the processes that govern Tat-mediated proviral DNA-driven transcription and replication. In contrast, cytoplasmic or extracellular localization of Tat may cause pathogenic effects in the form of altered cell activation, apoptosis, or neurotoxicity. Tat variants have been shown to differentially induce these processes, which may have implications for long-term HIV-1-infected patient care in the antiretroviral therapy era. Future studies concerning genetic variation of Tat with respect to function should focus on variants derived from HIV-1-infected individuals to efficiently guide Tat-targeted therapies and elucidate mechanisms of pathogenesis within the global patient population.
Keywords: HIV-1, Tat, genetic variation, transcription, pathogenesis
I. Introduction: Overview of Sequence Variation across Two Exons of Tat
Human immunodeficiency virus type 1 (HIV-1) encodes the transactivator of transcription (Tat), which is a small basic protein of about 14–16 kilodaltons (kDa), that enhances the elongation of the HIV-1 progeny viral mRNA during the viral LTR-directed transcription process [1,2]. This occurs through the recruitment of cellular positive transcription elongation factor (P-TEFb) to the transactivation response (TAR) element, an RNA secondary stem-loop structure encoded by the HIV-1 long terminal repeat (LTR), which consists of identical non-coding regions that flank the 5’ and 3’ ends of the proviral genome [3]. P-TEFb is comprised of two major subunits, cyclin-dependent kinase 9 (CDK9) and cyclin-T1 (CYCT1) [4]. The recruitment of P-TEFb to the LTR by Tat results in the hyperphosphorylation of RNA polymerase II (RNAPII), which increases the rate of transcriptional elongation and allows for the accumulation of a much higher level of full-length viral transcripts [5]. Tat is encoded by two exons that are separated by approximately 2,300 nucleotides and requires alternative splicing to become a full-length message and hence protein [6]. HIV-1 Tat was initially thought to span 86 amino acid residues, but it was later determined that the full-length protein consists of 101 residues [5]. The functionality of each domain within Tat often overlaps and there are relatively few conserved residues across the entire protein [2], allowing for extensive variation within the Tat protein to occur, ultimately causing a myriad of activating or inhibitory effects on viral and cellular gene expression.
Most of the functional domains of Tat reside in the first exon and include: the proline-rich, cysteine-rich, core, arginine-rich, and glutamine-rich domains [2]. The proline-rich domain, also known as the acidic N-terminal region, contains the first 21 amino acids, and is responsible for mediating LTR transactivation through interactions with CYCT1, in conjunction with the cysteine-rich and core domains. While much of the proline-rich domain is somewhat variable, residue 11 is a well conserved tryptophan and is required for efficient secretion of Tat [7]. The cysteine-rich domain spans residues 22 to 37 and is named for the abundance of highly conserved cysteine residues, which are located at positions 22, 25, 27, 30, 31, 34, and 37. These closely associated cysteines are responsible for the formation of intra-molecular disulfide bonds [8,9]. Notably, residue 31 may encode a cysteine-to-serine mutation that is prevalent in HIV-1 subtype C and is the subject of much debate because of its potential role in the reduction in neurocognitive impairment in patients infected with subtype C virus [10]. The core domain contains residues 38 to 48 and, in conjunction with the proline-rich and cysteine-rich domains, is responsible for interactions with CYCT1 [11]. In coordination with the cysteine-rich domain, the core domain has also been demonstrated to mediate cofactor binding, specifically with CREB-binding protein (CBP)/p300, histone acetyltransferase (HAT), and the Sp1 transcription factor [12,13]. The arginine-rich domain, also referred to as the basic domain, begins at residue 49 and ends at residue 57. It contains a well-conserved sequence, 49RKKRRQRRR57, that is crucial for the interaction with TAR as well as the secretion and uptake of Tat [7,14]. The glutamine-rich domain contains the remainder of the first exon, from residue 59 to 72. In conjunction with the arginine-rich domain, it is referred to as the basic region and is responsible for nuclear localization and mediates binding to CCATT enhancer binding protein (C/EBP) [15,16,14]. The second exon of Tat is less conserved compared to the first exon and is classically characterized as its own distinct domain [17], but has been demonstrated to be crucial for efficient replication of macrophage-tropic strains of HIV-1 and contributes to mechanisms of viral persistence [18–20]. However, the overall genetic diversity of exon II has been observed to vary in patient-derived sequences depending on the tropism of the virus; Tat from T-cell-tropic virus tended to exhibit less diversity than macrophage-tropic virus [21]. The second exon also contains a 73RGD75 motif which, in conjunction with the basic domain sequence, allows for interactions with molecules on the cell surface, such as integrins [7,22], and can trigger intracellular signaling cascades [23]. Additionally, there is a frequent mutation within the second exon at residue 87 that causes a premature stop codon, encoding for the truncated Tat86 variant that is frequently utilized in laboratory investigations [17], despite the observation that Tat101 has been shown to be much more prevalent in HIV-1-infected patients [24].
Within infected patients, HIV-1 is subject to selective pressures, such as from the immune system, antiretroviral therapy (ART), or the HIV-1-encoded error-prone reverse transcriptase [25,26]. These pressures cause accumulation or depletion of specific mutations across the viral genome, leading to the development of large numbers of genetic variants or quasispecies within the patient [27]. Tat, being encoded by the virus, is also susceptible to mutations, and the genetic variation within and between patients that can be observed in all HIV-1 subtypes globally [28,29]. Recently, cohorts of HIV-1 subtype B, such as the Bridging the Evolution and Epidemiology of HIV in Europe (BEEHIVE) and the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) as well as cohorts of other HIV-1 subtypes, have been used to initiate cross-sectional and/or longitudinal studies to examine sequence variation within Tat [17,27,30]. Using the CARES Cohort, it was determined that in well-suppressed patients on ART, the first exon of Tat had a mutational rate of 0.636 nucleotides/kilobase/year [27]. In the BEEHIVE Cohort, it has been demonstrated that Tat101 was the most prevalent in their patients but that no specific mutations appeared to be selected for or against over time [17].
The predominant and canonical function of Tat is to transactivate the HIV-1 LTR, which is mediated by the proline-rich, cysteine-rich, and core domains [31]. Single residue Tat genetic variants derived from HIV-1-infected individuals, such as P21A in subtype C virus, can affect LTR transactivation [32]. The cysteine residue at position 31 is another notable example of a specific residue variant that is often mutated to a serine in HIV-1-infected subtype C patients [10]. This positional variation inhibits various functions of Tat, such as a reduction in the rate of HIV-1 infection of CD4+ T cells, reduced neurotoxicity, and dysfunctional monocyte chemotactic activity [33,34,10], and is the subject of debate regarding its involvement in the reduction of neurocognitive impairment in patients infected with HIV-1 subtype C [35]. Likewise, variation within the HIV-1 LTR can influence LTR transactivation, and as such, impact interactions with factors that mediate transactivation and pathogenesis, including Tat and viral protein R (Vpr) [36–38]. HIV-associated neurocognitive disorders (HAND) has been in the past referred to neurocognitive impairment caused by HIV-1 infection, and while the complete mechanism of the pathogenesis associated with the etiology of HAND has yet to be elucidated, Tat has been determined to be a crucial contributor to this pathogenic process [39–41]. Genetic variation within Tat has been implicated as a factor for differential neuropathogenesis observed between patients [2,42]. Tat can also alter the expression of tight junctions, mimic chemokines, upregulate proinflammatory cytokines, induce oxidative stress [43–46], and modulate immune responses by upregulating IL-10, which has been argued to be crucial for viral persistence [47]. A single residue change can affect the function of Tat, and this review will examine the alterations observed in experimental and patient models of HIV-1 infection that are caused by Tat genetic variation within the context of HIV-1 subtype.
II. Subcellular Localization
Nucleus versus Nucleolus
HIV-1 Tat is expressed within infected cells soon after integration of the provirus into the host genome, and its production has been shown to initiate highly processive viral transcription [48–50]. Tat is encoded by two exons that are transcribed, spliced, and translated by host enzymes [51], following the dogmatic process of eukaryotic protein production. Because it is transcribed and translated by host cellular machinery, Tat must be able to effectively traffic between subcellular compartments during the course of the viral life cycle. HIV-1 Tat contains a unique and atypical nuclear localization signal (NLS) sequence, 49RKKRRQRRR57, within its arginine-rich domain [52], which is a crucial characteristic, as transcription of the integrated provirus must occur in the nucleus of the infected cell. This arginine-rich domain NLS, when isolated from the rest of Tat protein and produced as a fusion peptide, is capable of trafficking even large proteins into the nucleus [53]. The arginine-rich motif peptide can be turned into an even stronger NLS upon mutagenesis of the tri-arginine stretch within the domain peptide to encode 55GGG57, but this effect is unconfirmed in full-length Tat [54]. The Tat arginine-rich domain peptide containing the NLS has been shown to be capable of directly interacting with importin-A and importin-B nuclear import proteins [54,55], but these interactions have also not been confirmed for full-length Tat.
Wild-type Tat has been shown to localize densely in the nucleolus and is otherwise diffuse throughout the nucleus [56]. Both Tat86 and Tat101 length variants reside mainly within the nucleus, with Tat101 strictly in the nucleus and densely in the nucleolus, and Tat86 somewhat dispersed throughout the cytoplasm [57], though further study of each Tat length variant’s distinct localization pattern is needed. Given that the arginine-rich domain of Tat acts as an NLS, variation within this amino acid stretch could impact the efficiency of Tat nuclear translocation. Nucleolar localization of Tat is dependent on the conservation of the arginine-rich domain [57,58]. Deletion of this entire domain from Tat results in its exclusive accumulation in the cytoplasm and is also correlated with inhibited HIV-1 LTR transactivation [59], which may be a consequence of Tat’s exclusion from the nucleus.
Specific amino acid variation within the arginine-rich domain can help dictate the trafficking of Tat protein. Variation of Tat residues 50 and 51 has been shown to negatively affect nuclear import of Tat [56,60]. Lys50 and Lys51 are acetyl-accepting sites [61], so amino acid changes that prevent acetylation at these residues may affect the ability of Tat to traffic into the nucleus. Substitution of Lys50 or Lys51 with glutamine, chosen to neutralize the charge of lysine, resulted in the diffuse distribution of Tat throughout the cytoplasm and nucleus, as opposed to wild-type Tat, which mostly resided in the nucleus and nucleolus [56,60]. The K50Q Tat variant also showed a 4-day replication delay [56], possibly because of the diffuse subcellular distribution of Tat. The presence of a delay has indicated that the Tat variant was at least partly functional, but perhaps was not concentrated densely enough in the nucleus to promote transactivation and more intense replication. In contrast to K50Q, K50R assisted in the exclusive localization of Tat to the nucleus, as the positive charge of the additional arginine contributed to the localization potential of the NLS [56,60].
Variation outside of the arginine-rich domain can also dictate Tat subcellular localization. Mutation of Lys28 or Lys41 to generate K28Q, K28R, or K41A Tat variants disrupted the wild-type localization phenotype. Diffuse distribution within the cytoplasm in addition to the nucleus was observed in cells transfected with the K28Q Tat variant [56]. This variant, however, as well as the K28R variant and the K41A variant, exhibited a perinucleolar distribution within the nucleus [56], indicating that the conservation of Lys28 and Lys41 are required for Tat accumulation in the nucleolus. These three variants also showed significantly reduced potential for LTR transactivation, and the Lys28 mutants also displayed delayed replication kinetics as expected [56]. The exact mechanism that dictates alteration of subcellular localization of the Lys28 and Lys41 variants has remained unclear and will require more investigation, but may be related to the ability of Lys28 to act as an acetyl-acceptor [62]. Interestingly, K41R and K41Q variants have shown opposing activity in Tat-transfected HEK 293 cells, where K41R decreased nuclear localization and K41Q promoted nuclear localization [60]. The difference between these Lys41 variants may be due to the change in the charge of the substituted amino acid, where the positive charge is neutralized when replaced by glutamine. Additionally, exon II variation has also been shown to affect nuclear trafficking of Tat, specifically within the tri-lysine motif that spans positions 88 to 90. These effects were position-specific, as K89R increased Tat101 nuclear localization, yet K88R and K90R decreased its nuclear entry [60]. The variants that affect Tat localization and trafficking are summarized in Figure 1.
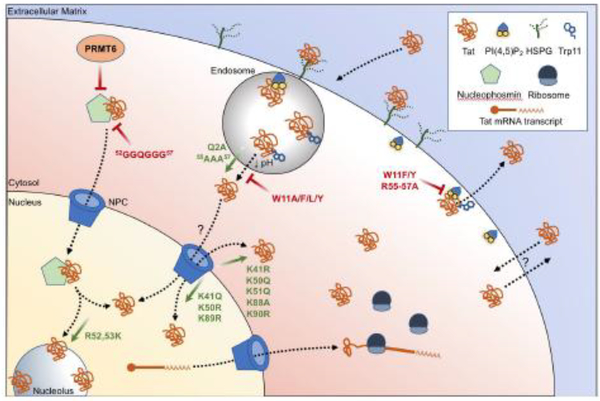
Fig. 1. Changes in the subcellular localization of HIV-1 Tat based on variation.
Variation within the Tat amino acid sequence dictate changes in the subcellular localization of HIV-1 Tat. Tat may be transcribed from integrated HIV-1 proviral DNA, processed, and translated by host machinery, or enter a cell from the extracellular matrix by way of interactions with surface heparan sulfate proteoglycans (HSPGs) and uptake via endocytic pathways. Translocation of Tat from endosomes has been shown to be mediated by the exposure of tryptophan residue 11 (Trp11) upon late endosome acidification. Cytosolic Tat may associate with nucleophosmin for nuclear import through the nuclear pore complex (NPC), or enter and exit otherwise by mechanisms that are not clearly defined. Tat egress from the cytoplasm is thought to be mediated by interactions with phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) and by the insertion of Trp11 into the plasma membrane. Amino acid variation within Tat that promotes (green arrows) or inhibits (red bars) certain localization patterns are shown.
Interactions between Tat and host factors may also contribute to its trafficking between subcellular compartments. Nucleophosmin is a ubiquitous nucleolar phosphoprotein that has been shown to be able to shuttle proteins between the nucleolus, nucleus, and cytoplasm [63]. HIV-1 Tat72 has been observed to directly interact with nucleophosmin via the involvement of a Tat nucleolar localization signal (NoLS), which was demonstrated in cell-free experiments using a Tat peptide derived from the arginine-rich domain [64], providing a potential mechanism for the shuttling of Tat in and out of the nucleus/nucleolus. Mutation of the arginine residues in the putative NoLS to glycine, which generated a sequence change from 52RRQRRR57 to 52GGQGGG57, resulted in the loss of association of Tat and nucleophosmin [64], although it is unclear which of the five substitutions conferred the greatest cost to the loss of this interaction. Tat and nucleophosmin were frequently spatially associated in the nucleolus of Jurkat T cells transfected with Tat constructs, as well as in the nucleus and cytoplasm [65]. This strong association throughout the cell may be essential for Tat to localize to the nucleolus [64,65], as mutations in nucleophosmin that prevented its trafficking to the nucleus and nucleolus also resulted in the retention of Tat in the cytoplasm [64]. Further study of this interaction revealed that acetylation of nucleophosmin was essential for Tat’s localization into the nucleus [66]. Absence of nucleophosmin acetylation completely prevented Tat trafficking into the nucleus, as well as Tat-mediated transactivation of the HIV-1 LTR [66], which may again be a consequence of defective nuclear trafficking.
Another host factor, protein arginine methyltransferase 6 (PRMT6), has been shown to potentially be important in Tat localization. PRMT6 has been shown to interact with and methylate HIV-1 Tat at residues 52 and 53 [67,68]. The methylation of Arg52 and Arg53 resulted in proportional decreases in Tat activity, based on the amount of PRMT6 present in the cell [68]. Overexpression of PRMT6 also correlated with the exclusion of Tat101 from the nucleolus, but did not disrupt Tat trafficking to the nucleus [57]. The replacement of Arg52 and Arg53 with lysines recovered nucleolar localization in the presence of PRMT6 [57], as the R52,53K variants were not methylated at those mutated residues due to the restriction of PRMT6 activity to arginine residues [69]. Interestingly, the analysis of nucleophosmin-Tat nucleolar co-trafficking in relation to PRMT6 showed that nucleophosmin still localized to the nucleolus without Tat upon PRMT6 overexpression [57]. Mutation of PRMT6, however, showed Tat colocalization with nucleophosmin in the nucleolus [57], which is intriguing, as it suggests that the co-trafficking of Tat and nucleophosmin is dependent on the conservation of Tat residues Arg52 and Arg53 and may act as interacting residues that are disrupted by PRMT6 methylation (Figure 1). Further elucidation of the interactions between nucleophosmin, Tat, and PRMT6 will assist in the understanding of Tat nucleolar import, as will host factor interactions that prevent it.
Phosphorylation of Tat serine and threonine residues has also been reported to effect nuclear localization of Tat86. Cellular double-stranded RNA-dependent protein kinase (PKR) has been shown to phosphorylate Tat86 at several residues, including Thr23, Thr40, Ser46, Ser62, and Ser68 [70]. HeLa cells incubated with either normal or fully-phosphorylated Tat86 recombinant protein show differential localization, where the result was the trapping of fully-phosphorylated Tat86 within the cytoplasm [70]. The substitution of serine or threonine with alanine at these positions did not affect the localization of Tat86 to the nucleus, while substitution with aspartate maintained localization to the cytoplasm [70]. Though aspartate substitution does not permit phosphorylation, it is possible that the replacement of Thr23, Thr40, Ser46, Ser62, and Ser68 with a negatively-charged amino acid may have altered the charge of Tat overall, altering its nuclear localization dynamics. Consequentially, LTR transactivation in HeLa cells co-transfected with FLAG-Tat86 aspartate mutants and LTR luciferase reporter plasmids showed decreased LTR transactivation [70], possibly due to the inability of aspartate-mutated Tat86 proteins to traffic into the nucleus to promote LTR activity. Alanine-substituted mutants were not significantly impaired in LTR transactivation capacity, with the exception of the T23A mutant, possibly because the nuclear localization of these mutants was not impaired [70].
The dense localization of Tat to the nucleolus begs the question of Tat’s function or involvement with cell processes once there. The nucleolus is the subcellular site of ribosomal RNA (rRNA) synthesis and processing, as well as rRNA assembly with ribosomal subunits [71]. Examination of Tat’s activities in the nucleolus using Drosophila-based methods have shown that Tat was capable of interfering with normal nucleolar activities. Indeed, the presence of Tat in the nucleolus corresponded with a reduction in the total amount of 80S ribosomes present in Drosophila cells [72]. This effect may be caused by the observed interference in the first steps of pre-rRNA processing pathways, and could account for the decreased rRNA biogenesis seen upon the introduction of Tat to the nucleolus [72]. In Jurkat T cells, however, transfection with a Tat86 construct has been shown to modulate the composition of nucleolar proteins to favor functions such as ribosomal biogenesis, glycolytic and amino acid metabolism, stress response, and T-cell signaling [73], which suggested a shift toward cell activation. Because the length of Tat used in the Drosophila experiments is unknown, the effect of Tat length between the Drosophila and Jurkat T cells studies cannot be compared. The contrasting data gathered from these studies should be further investigated in human cells and in the context of Tat length and variation to understand the downstream effects of these observations on HIV-1-infected cells. Other viruses are known to interfere with or completely shut down host cell translation [74–76], and this phenomenon may present a mechanism for host cell translational interference upon infection by HIV-1.
Cytosol versus Extracellular
HIV-1 Tat has also been observed to traffic between cells via endogenous cellular secretion and uptake pathways [77]. The interactions and mechanisms governing these processes are not fully understood, but variation of HIV-1 Tat at residue 11 has provided some insight into Tat cellular uptake and secretion. Tryptophan at position 11 (Trp11) has been identified as a conserved residue of HIV-1 Tat that is essential for both trafficking mechanisms. Studies have shown that Tat can enter cells via clathrin-mediated endocytosis, and subsequently translocate into the host cell cytoplasm upon endosome acidification [78]. The drop in pH results in a conformational change in Tat that has been attributed to the presence of an endogenous low pH sensor present in the protein [79]. This endogenous sensor involves interactions between Tat residue Gln2 and a tri-arginine stretch within the arginine-rich domain that spans residues 55 to 57 [79]. The involvement of residue 2 in the low pH sensor is consistent with a structural analysis of Tat, which demonstrated that residue 2 forms intramolecular bonds with residues within the arginine-rich domain [80] (Figure 2). The low pH-dependent conformational change of Tat that occurred upon endosome acidification exposed the Trp11 residue, which enabled its insertion into endosomal bilayers in vitro [79]. Substitution of Gln2 or the arginine-rich domain tri-arginine motif with alanine allows insertion of Trp11 at both low and neutral pH [79]. Prior investigation has also noted that Tat protein containing alanine substitutions at each arginine within the arginine-rich domain was unable to transactivate the HIV-1 LTR because it did not enter host cell cytoplasm when introduced extracellularly [81]. This suggests that the pH sensor dictates the ability of Tat to bind endosomal bilayers via Trp11 insertion and that the conservation of the amino acids that comprise it are essential for its function (Figure 1).
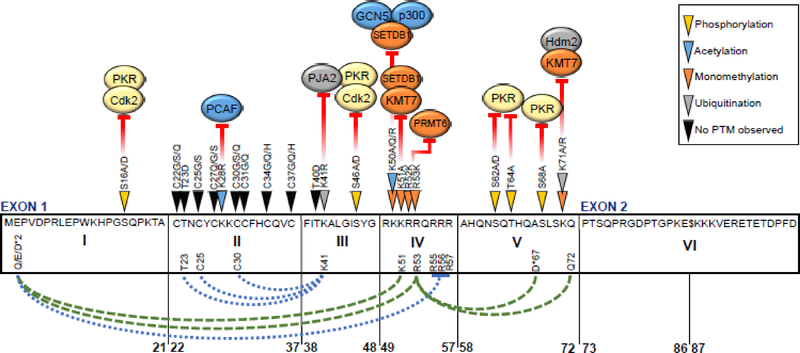
Fig. 2. Effect of HIV-1 Tat amino acid variation on TAR-dependent LTR transactivation, post-translational modifications, and intramolecular interactions.
Variation within the amino acid sequence of HIV-1 Tat contributes to altered LTR transactivation capacity, modeled on the subtype B HXB2 reference sequence. An alternative STOP codon at position 87 is represented by the symbol “$”. Filled arrows along the length of Tat indicate residues with variation that affects LTR transactivation directly or impairs interactions with, or addition of, any of a number of post-translational modifications (PTM) by corresponding host factors. Kinases CDK2 and PKR shown in yellow; histone acetyltransferases PCAF, GCN5, and p300 in blue; monomethyltransferases KMT7, PRMT6, and SETDB1 shown in orange; and E3 ubiquitin ligases PJA2 and Hdm2 shown in gray. Intramolecular interactions observed between Tat residues (bottom) are shown for subtype B (blue dotted) and subtype D (green dashed) Tat. An asterisk next to an amino acid indicates that the dominant amino acid observed differs from the HXB2 sequence.
The activity of the pH sensor at low pH is consistent with the ability of Trp11 to penetrate lipid monolayers in vitro [79]. At neutral pH, Trp11 did not insert into biological membranes, but shifting the conditions to an acidic pH enabled this activity [79]. Tat variants with Trp11 substitutions, such as W11A, W11F, W11L, and W11Y, were observed to have decreased LTR transactivation capacity when added to the extracellular environment of Jurkat cells [79], suggesting that they did not translocate into the cytosol or nucleus. None of these substitutions, however, affected the packaging of Tat into endosomes [79], which implies that the conservation of Trp11 mainly functions in Tat cytoplasmic translocation. Therefore, the mechanism of Tat uptake via clathrin-mediated endocytosis may also rely on the ability of Trp11 to insert into the lipid bilayer of the endosome. This mechanism is similar to that of the Pseudomonas exotoxin A cellular entry, where a Trp residue that is sequestered in a hydrophobic pocket at neutral pH is exposed upon pH acidification and is able to insert into endosomal membranes [82].
A similar mechanism has been described for Tat secretion, where the mutation of Trp11 to phenylalanine or tyrosine prevented Tat secretion from Jurkat cells by about 80%, when compared to wild-type [7]. In HIV-1-infected primary CD4+ T cells, Tat accumulated along the inner plasma membrane, and this effect has been corroborated in ex vivo studies using CD4+ T cells transfected with wild-type Tat [7]. Transfection with Tat Trp11 variants, however, resulted in Tat localization mainly throughout the cytoplasm, with no clear accumulation at the inner plasma membrane [7]. This effect may have been caused by a 300-fold decrease in the avidity of Tat for the known binding partner phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2, which was also required for Tat secretion in this system [7]. PtdIns(4,5)P2 is a phospholipid that is involved in various cellular processes, including trafficking, signaling, and endocytosis and exocytosis, and is primarily located in the inner leaflet of the plasma membrane [83,84]. As shown in Figure 1, the colocalization of Tat and PtdIns(4,5)P2 at the inner plasma membrane and the loss of avidity for PtdIns(4,5)P2 upon amino acid substitution of Trp11 has suggested that the interaction between these two components relies on the pH-dependent availability of Trp11 for insertion into the plasma membrane.
Variation within Tat that changes or abolishes its subcellular trafficking or localization may serve as a selective constraint for the strain of HIV-1 that contains the variation. Because Tat is essential for efficient LTR transactivation and HIV-1 transcription, the capacity for activity in Tat based on subcellular localization may be indicative of the fitness of each HIV-1 strain. Impaired trafficking of Tat because of amino acid variation at specific residues may impart a fitness deficit, as defective trafficking to the nucleus by Tat variants would result in a marked difference in LTR transactivation. In contrast, changes in subcellular localization with respect to Tat variation may serve as a functional switch; the determination of whether Tat will enter the nucleus or nucleolus versus exit the cell via secretion may depend on the amino acid sequence of the protein. For example, K50Q variants that localize to the cytoplasm cause viral replication deficits [56], plausibly because they do not concentrate densely enough in the nucleus for LTR transactivation. This variant could be predominantly secreted for uptake by bystander cells, though this hypothesis has not yet been tested. Another point of consideration is that of Tat accumulation in the nucleolus. Although there has been some effort to investigate nucleolus-specific Tat activity, it is unclear if Tat has a specific function within this subcellular compartment. Interference with rRNA assembly could impact global translation in the host cell, but additional nucleolus-specific functions of Tat have not been described. Certainly, it is possible that the observed dense accumulation of Tat in the nucleolus may confer some benefit to HIV-1 in the form of host cell translational alterations, but if so, that benefit has yet to be clearly defined.
III. LTR Transactivation and Activity
Variation within HIV-1 Tat has been well-studied with regard to LTR transactivation, and the molecular diversity seen within Tat has been demonstrated as a modulating factor of this function [2]. Analysis of Tat sequences isolated from the central nervous system (CNS) tissue of HIV-1-infected patients with HAND exhibited genetic heterogeneity and brain-derived Tat variants had differential LTR transactivation in a number of in vitro systems [85,86], indicating that Tat variability may be an important predictor of viral pathogenesis. These results are supported by the observation that subtype B, C, and E viruses possessed dissimilar LTR transactivation potentials in a Jurkat T-cell transfection model [87]. Cellular phenotype has also been implicated as a factor for differential LTR transactivation, and may be a contributing factor to the wide range of results observed in vitro, as well as in TAR-independent LTR transactivation studies [88,89]. Despite the fact that much of the primary literature has focused on and used experimental models to investigate downstream effects of amino acid substitutions, patient data has translationally corroborated and guided this research. This section will focus on genetic determinants within Tat that have been shown to alter LTR transactivation in a number of model systems and patient studies, as well as with regard to alternative transactivation mechanisms. These results have been summarized in Figures 2 and 3.
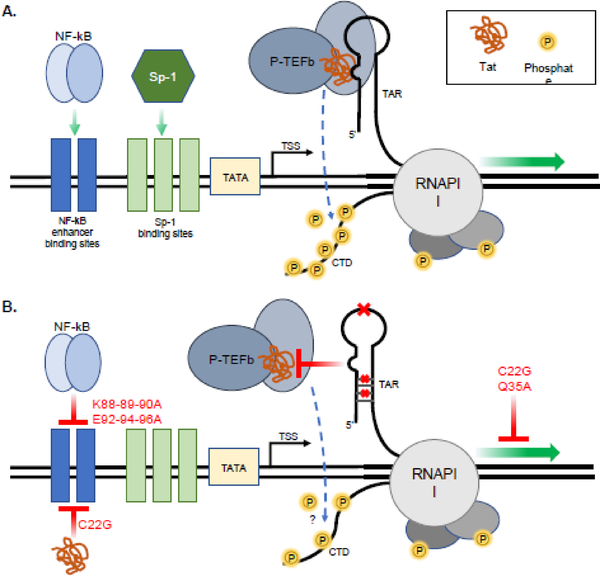
Fig. 3. TAR-independent functionality is limited by Tat variation.
(A) Overview of TAR-dependent LTR transactivation. The formation of the Tat-P-TEFb-TAR complex results in hyperphosphorylation of the RNA polymerase II (RNAPII) C-terminal domain (CTD) by P-TEFb, significantly increasing RNAPII processivity and transcriptional elongation. Transcription factor binding of enhancer sites upstream of the transcriptional start site (TSS) promote transcriptional initiation. (B) TAR-independent LTR transactivation is inhibited by Tat variation. TAR stem-loop point mutations or stem nucleotide mutations that destroy TAR secondary structure (red x’s) inhibit the formation of the Tat-P-TEFb-TAR complex, though the hyperphosphorylation status of the CTD in TAR-independent LTR transactivation is still unclear. Exon II Tat mutations are capable of inhibiting NF-κB association with upstream NF-κB enhancer binding sites. Cysteine-rich domain variation blocks both direct interactions of Tat with NF-κB enhancer binding sites and affects proviral DNA-directed transcription.
TAR-dependent Activity
Effects of Tat Variation on Phosphorylation of Tat
Because Tat is heavily post-translationally modified by host factors, multiple sites of phosphorylation are plausible and may have differing effects on Tat function [90]. The association of HIV-1 Tat and CDK2 has been shown to contribute to HIV-1 transcriptional elongation via enhanced RNAPII C-terminal domain (CTD) phosphorylation, a process dependent on the conservation of residue Cys22, certain residues within the core, basic, and glutamine-rich domains of Tat, and the phosphorylation of Tat itself [91]. CDK2 phosphorylated Tat residues Ser16 and Ser46 (Figure 2) in cells infected with Tat-containing adenovirus, an effect that could be significantly inhibited by CDK-specific siRNA [92]. S16A and S46A Tat mutant plasmids transfected into 293T cells each yielded Tat protein that had 2–3 times less total phosphorylation than wild-type Tat, with an S16,46A double mutant being even less phosphorylated than the single mutants [92]. Along with decreased total phosphorylation, total LTR transactivation was reduced to about 75%, 50%, or 33% of wild-type levels for S16A, S46A, and double mutants, respectively [92]. A separate study confirmed that Ser16 could be phosphorylated by CDK2. Using in vitro phosphorylation assays, Tat fragments containing potential phosphorylation sites, such as Ser16 and Ser46, were incubated with CDK2 and Cyclin E and the Tat fragment containing Ser16 was predominantly phosphorylated [93]. Likewise, when the same phosphorylation assay was conducted with Tat fragments and PKR, the Tat fragment containing Ser46 was predominantly phosphorylated [93]. When either S16A or S46A Tat101 mutants from this study were expressed in 293T cells with luciferase reporter genes for LTR transactivation, the total luciferase expression significantly decreased when compared to wild-type Tat101 [93], suggesting a crucial role for Ser16 and Ser46 phosphorylation in efficient LTR transactivation. The use of HLM-1 cells, which are CD4+ HeLa cells with one integrated copy of HIV-1 proviral genome per cell and a defective Tat sequence, allowed the observation of the S16A and S46A mutants’ effect on HIV-1 replication that arose because of decreased LTR transactivation. Transfection of mutant Tat vectors into the HLM-1 cell line resulted in replication deficits for each single mutant, and an additive replication deficit in the double mutant, when compared to wild-type Tat, as determined by p24 capsid protein readout [92]. This effect on replication could be due to the observed reduced LTR transactivation by Tat that contained S16,46A mutations [92], emphasizing the necessity of conserved Ser16 and Ser46 residues and dependence on Cdk2 and PKR for optimal function of Tat protein.
Analysis of 158 Tat sequences deposited in the PubMed database showed that though Ser16 and Ser46 were conserved in nearly 100% of the sequences, there was additional variation within the CDK2 consensus motif [92]. Because the CDK2 consensus motif dictates that the fourth position of the motif is either a lysine or an arginine as part of a S/T+0P+1X+2K/R+3 sequence [94,95], the conservation of downstream amino acids may also affect LTR transactivation and viral replication capacities. For example, when Tat residues 19 and 49 (corresponding to the fourth amino acid in the motifs containing S16 and S46, respectively) were analyzed, Arg49 was totally conserved across all sequences, while the amino acid composition of residue 19 in these sequences varied [92]. In the analyzed sequences, K/A19T/A/G substitutions present in 53% of HIV-1-infected patients correlated with a healthy, “non-progressor” phenotype, even when the patients were not being treated with antiretroviral therapies [92]. It is unclear if, experimentally, K/R19T/A/G Tat variants contribute to changes in the phosphorylation of Ser16, and if those changes are causative for differential health status. Prior research on the impact of viral replication capacity on HIV-1 disease severity has shown that the ability of virus quasispecies present at acute infection to replicate efficiently strongly associated with CD4+ T-cell decline [96]. Similarly, patients infected with virus that had less capacity for productive replication were projected to progress into clinically defined AIDS at a slower rate [96], suggesting that non-progressor patient phenotypes may arise from their infecting swarm of HIV-1 quasispecies. Because Tat is necessary for efficient viral replication, variation that affects its LTR transactivation may also dictate the rate at which an HIV-1-infected individual progresses to AIDS. Further analysis of this effect within larger patient groups is necessary to understand any downstream translational implications of this occurrence, though the variation within the CDK2 consensus motif in Tat and its relation to disease progression may represent an important mechanism that may affect the disease progression of HIV-1-infected individuals. This example not only demonstrates the significance of amino acid sequence variation on the disease progression of HIV-1-infected patients, but provides a bridge between the molecular mechanisms that govern the functional aspects derived from variation of HIV-1 Tat and the phenotypes that emerge as a result.
The DNA-dependent protein kinase (DNA-PK) has recently been shown to interact with Tat86, with the interaction resulting in phosphorylation of Tat Ser16, Ser62, and Ser75 [97]. In this study, HeLa-derived HL3T1 cells, which contain one copy of stably integrated HIV-1 LTR per genome linked to a CAT reporter construct, were treated with 50 to 200 nanograms of exogenous recombinant wild-type Tat86 protein or S16A or S62A variants. These alanine-substituted Tat86 variants had approximately a 10–20% decrease in CAT activity when compared to wild-type Tat86, with the largest decrease observed in HL3T1 cells treated with an S16,62A double mutant [97]. Similarly, when Tat86 or variants were endogenously expressed in Jurkat cells containing a luciferase gene under the control of HIV-1 LTR, luciferase expression was somewhat reduced, indicating a modest decrease in LTR transactivation by either the S16A or S62A variant when compared to wild-type Tat86 [97]. The S16,62A double mutant, however, was able to decrease the luciferase readout by about two-fold [97], suggesting that the S16A, S62A, and S16,62A variants were impaired in the LTR transactivation function of Tat86.
Tat86 and its truncated variant, Tat72, can both interact with and act as a substrate for PKR, an interferon-inducible Ser/Thr protein kinase [98,99]. Tat is known to compete with the translational regulator eIF2a as a substrate of PKR to promote viral mRNA translation in infected cells as previously reviewed [100]. The interaction between Tat86 and PKR is predicted to occur by the formation of several electrostatic, aromatic, and hydrogen bonds between amino acid interfaces of each protein [101]. Of these predicted bonds, the involvement of Tat residues Lys 19, Lys28, Lys29, Lys 51, and Lys71 in salt bridge formation and Lys41 in aromatic interactions with PKR are of interest, as the alteration of these residues could negatively affect LTR transactivation and affect the stability of the Tat-PKR interaction [101]. PKR has been shown to phosphorylate residues Ser46, Ser62, Thr64, and Ser68, all of which are located in either Tat’s core domain or glutamate-rich domain [99,58,102,93] (Figure 2). Ser62, Thr64, and Ser68 were also previously deemed necessary for optimal phosphorylation of Tat by PKR [103]. The substitution of each of these residues with alanine yielded decreased phosphorylation efficiency, with the least amount of total phosphorylation observed in a triple mutant [103]. Decreased phosphorylation of Tat by PKR corresponded to decreased HIV-1 LTR transactivation, with the triple Tat mutant conferring a four-fold decrease in LTR transactivation when compared to wild-type [103]. The mutation of Ser68 had the highest associated cost with regard to loss of amino acid phosphorylation, as other substitutions in combination with S68A generated more severe transactivation deficits when compared to combinations of substitutions without S68A [103]. Interestingly, although the T64A single mutation did result in decreased LTR transactivation by about 40% [103], it was observed at a 15% frequency in HIV-1 subtype B sequences [86]. It is possible that this Tat variant confers a fitness cost that is well-tolerated by HIV-1, as it still allowed for about 60% of total wild-type LTR transactivation. Even in the triple mutant, transactivation was not fully ablated by the loss of phosphorylated residues in Tat [103], so other mechanisms governing this activity could be involved. Although these mechanisms have not been resolved, they likely do not rely on the conservation of glutamine-rich region residues Ser62, Thr64, or Ser68.
Lysine Variants in Tat That Cause Loss of Acetylation and Effects on LTR Transactivation
Lysines 28, 50, and 51 of Tat can be acetylated by Tat-associated host histone acetyltransferases (HATs) in vitro and in vivo [104,105]. Tat acetylation at these residues has been shown to both positively and negatively direct interactions with host factors important for HIV-1 proviral DNA-directed transcription, such as histone acetyltransferases p300, hCGN5, and PCAF [104,106–108], summarized in Figure 2, and the p-TEFb complex [109]. The importance of Lys28 residue conservation for Tat activity was exemplified by the consequential deficits that occur when altered. K28R or K28Q substitutions in Tat resulted in significant viral replication deficits when introduced to an integrated HIV-1 293T cell model system [56]. Furthermore, K28R and K28Q Tat variants transfected into HeLa P4 luciferase reporter cells caused a 6.5- or 5-fold reduction in LTR transactivation, respectively [56]. These Lys28 variants formed less efficient interactions with CYCT1 [56], which may have impacted the formation of the Tat-TAR-p-TEFb ternary complex, and thus, total LTR transactivation. Additionally, K28R and K28Q Tat variants showed atypical subcellular localization with respect to wild-type Tat [56], which may have impaired the variants from ever reaching the site of activity and initiating LTR transactivation.
The histone acetyltransferase PCAF was previously observed to be able to interact with and acetylate HIV-1 Tat [56,106]. PCAF-mediated acetylation of Tat was demonstrated as important for optimal LTR transactivation [106,62], though the integrity of the interaction between PCAF and Tat is potentially as important as the acetylation itself. Tat-PCAF binding was dependent on interactions between the PCAF bromodomain and lysine 50-acetylated Tat [110]. K28R and K41A Tat variants interacted only weakly with PCAF, while K28Q, K50Q, and K50R variants interacted with and immunoprecipitated with PCAF [56]. HIV-1 Tat that was treated with PCAF had 5-fold enhanced affinity for CyclinT1, which in turn, increased ternary complex formation (Tat-TAR-p-TEFb) by over 110-fold compared to untreated Tat, and successfully stimulated LTR transactivation [62]. Variation at Lys28 may or may not affect acetylation, and thus, has varying effects on LTR transactivation. The K28R variant was not acetylated by PCAF and showed delayed viral replication kinetics in this system [62]. The K28Q variant, however, maintained similar biochemical properties as Lys28, and was still able to bind CyclinT1 when treated with PCAF, as well as increase ternary complex formation by about 65-fold when compared to untreated Tat K28Q [62]. Although K28Q bound CyclinT1 and induced ternary complex formation, it still did not transactivate the LTR [62], consistent with other reports [56]. PCAF has also been shown to acetylate Tat at lysine residues 50 and 51 [106], despite the proposed requirement for pre-acetylated Lys50 [110]. In this case, Tat K50R and K51R mutations conferred a 70% reduction in LTR transactivation when compared to wild-type Tat [106]. This was possibly due to the presence of Tat Y47A and R53A or R53E variants, which were observed as suppressing Tat-PCAF interactions in vivo and consequently displayed reduced Tat activity [106]. The largest decrease in transactivation activity was observed in the R53E variant, followed by the Y47A variant [106]. The R53E mutant exhibited a partial defect in TAR-RNA binding, but the Y47A mutant as well as the R53A mutant were still able to effectively bind TAR-RNA [106]. The apparent ability of the alanine-substituted variants to bind the TAR, but not PCAF, has suggested that Tat can bind TAR RNA even when it does not have acetylation patterns that enhance LTR transactivation.
There is conflicting evidence on whether Lys50 Tat variants confer a functional deficit to LTR transactivation. HAT hGCN5 has been shown to acetylate Tat Lys50 and Lys51, a process reliant on interactions between the cysteine-rich and core domains of HIV-1 Tat101 and hGCN5 [108]. Lys50 Tat variants with glutamine substitutions (K50Q) have failed to transactivate the HIV-1 LTR in HeLa P4 cell lines as well as wild-type Tat, and was accompanied by replication deficits of the mutant virus [56]. The K50A and K50R variants have also been shown to have decreased LTR transactivation when compared to wild-type Tat in HeLa cells co-transfected with LTR reporter and Tat constructs [61]. In this study, the transactivation deficit was correlated with the inability of p300/(C/EBP-binding protein) CBP to acetylate K50R Tat, an effect that prevented Tat from interacting with CYCT1 [61]. Furthermore, Tat double-mutants containing K50,51R substitutions had an additive decline in activity, which resulted in a more severe HIV-1 LTR transactivation deficit [61]. In contrast, the K50R Tat variant used in an alternate study did not have a transactivation deficit and achieved activity levels similar to wild-type Tat in the HeLa P4 cell line [56]. This variant, however, still showed a replication-deficient phenotype despite having wild-type levels of Tat activity [56]. Lys50 and Lys51 substitutions caused significant conformational changes in Tat that may have resulted in structural instability within the protein [80], likely inhibiting any conformation-dependent interactions. This may be a contributing factor for explaining the transactivation deficit observed in Lys50 Tat variants, but does not account for Lys50 variants that maintain wild-type levels of activity. The difference in LTR transactivation between transactivating and non-transactivating K50R Tat variants requires more investigation, as Tat-independent LTR transactivation in the presence of the K50R variant has not been observed experimentally.
Acetylation of Tat lysines 41, 50, and 51 has been observed to allow interactions with binding partner Brahma-related gene 1 (BRG1) [111], which is a component of the SWI/SNF chromatin remodeling complex [112]. Experimentally, acetylated Tat promoted LTR transactivation upon BRG1 binding, possibly due to the subsequent recruitment of BRG1 by Tat to the HIV-1 transcriptional start site-occupying nucleosome (nuc-1) [111,113]. K41R, K50R, or K51R Tat variants bound less efficiently to BRG1 and do not promote LTR transactivation [111], perhaps as a consequence of decreased recruitment of BRG1 to nuc-1. Moreover, the K50R and K51R variants do not recruit BRG1 to nuc-1 [111], highlighting the importance of Lys41, 50, and 51 acetylation for recruitment of chromatin remodeling complexes to the transcriptional start site of HIV-1. Through the binding of BRG1, Tat can direct entire host complexes to DNA loci of functional significance and potentially induce chromatin modifications to enhance viral transcription.
The Role of Tat Variation on Lysine Monomethylation
Recently, the role of Tat residue monomethylation has been investigated with respect to conferred stability or conformational preference for TAR-dependent activity, as monomethylation of Tat Lys51 has been shown to enhance Tat-TAR-p-TEFb complex interactions and positively affect HIV-1 transcription [114]. Monomethylation has been observed to occur on both Lys51 and Lys71 residues of HIV-1 Tat (Lys51me and Lys71me) [114,115]. This post-translational modification (PTM) was caused by the robust activity of human lysine monomethyltransferase KMT7, which monomethylated both Lys51 and Lys71 of Tat101 or the splice variant, Tat72, though kinetic assays showed K71 was the preferred methylation target [115,114]. Substitution of Lys71 to generate K71R or K71A variants had no visible methylation by KMT7 in Tat101-expressing HEK 293T cells when probed on western immunoblotting by K71me-specific antibody [115]. K71R Tat variants were shown to have about 50% decreased LTR transactivation activity compared to wild-type, and K51R mutants displayed a similar activity deficit [115]. A double mutant nearly lacked transactivation activity altogether [115], emphasizing the cumulative importance of these residues to Tat function. Knocking down KMT7 methyltransferase in TZMbl cell lines prior to transfection of wild-type Tat101 yielded a 4-fold decrease in LTR transactivation in Tat-producing cells [115], suggesting that the loss of monomethylation of Lys71 and the substitution of Lys71 with Arg had similar effects on HIV-1 transcription. These studies show that Lys71me is necessary for optimal LTR transactivation, though not essential to achieve partial LTR transactivation by Tat. Therefore, conservation of the Lys 51 and Lys71 residues likely have a role in maintaining monomethylation patterns important for interactions that stabilize the Tat-TAR-p-TEFb complex and enhance HIV-1 transcription as previously proposed [114]. Further investigation is required to determine if other interactions or host factors account for the remaining transcriptional activity when Lys51 and Lys71 monomethylation is absent.
In contrast, methylation of Tat by the SETDB1 or PRMT6 methyltransferases produced a transactivation-silencing effect [116,67,68]. Both SETDB1 and PRMT6 activity reduced the ability of Tat to transactivate the HIV-1 LTR and knockdown assays for both SETDB1 and PRMT6 increased LTR transactivation of wild-type Tat [116,68]. SETDB1 specifically interacts with and methylates Tat Lys50 and Lys51, so long as the two lysine residues have no other PTMs [116]. K50A and K51A Tat variants, therefore, caused a 2-fold or 10-fold drop in methylation activity by SETDB1, respectively, and a K50,51A double variant abolished the methylation of Tat by SETBD1 altogether [116]. The transactivation silencing activity of SETDB1 in the context of Lys50 and Lys51 methylation could be due to inhibition of HAT activity at these resides that have generally stimulated LTR transactivation. As such, SETDB1 may contribute to the restriction of viral replication, as it competed with transcription-activating post-transcriptional modifications to dampen viral transcription. Likewise, Tat has been shown to be a substrate for and interact with PRMT6 [67,68], which has been shown to methylate Tat residues Arg52 and Arg53 [68]. This methylation pattern interfered with Tat-CYCT1 binding efficiency, which in turn negatively affected the ability of the Tat-TAR-CYCT1 complex to form [68], accounting for the transactivation-silencing effect observed. Consequently, the R52K variant had 4-fold reduced LTR transactivation activity and the R52,53K double variant had even more severely reduced LTR transactivation activity [68]. The use of lysine in these variants maintained the positive charge of arginine at these positions, but because PRMT6 activity was restricted to arginine residues [69], it should have theoretically resulted in the loss of monomethylation. Additionally, an R49,53A double variant had nearly abolished transactivation activity, even though R49 was not identified as a methyl-accepting residue, possibly due to the loss of charge from substituting the arginines with alanines [68]. These variants each had reduced methylation [68], which should theoretically help to ameliorate the negative effects of PRMT6 activity by increasing transactivation capacity. Because the transactivation deficit was still observed in Tat variants in the absence of optimal methylation, it appeared that the transactivation deficit seen in the R52,53K Tat variants was independent of PRMT6 activity. Overall, monomethylation of Tat by host factors may enhance or inhibit Tat function and has seemed to be dependent on the position of the added methyl group. The pattern of PTMs of the arginine-rich domain and surrounding residues may represent an interesting interplay between activating and inhibitory protein states. Because several residues within the arginine-rich domain have been shown to be modified in experimental systems, often with overlapping locale, as shown in Figure 2, these modifications may be sterically unlikely, and the occurrence of one over the other may determine whether the HIV-1 LTR is transactivated with consequential provirus activation, or rather the occurrence of LTR silencing with the result of proviral DNA latency. This observation should be studied in depth, as recent efforts to eradicate HIV-1 from the infected patient have focused on viral latency as a tool to achieve additional clinical benefit [117–119] and a more thorough understanding of these intrinsic viral mechanisms through accurate analysis and evaluation will most likely yield important results with respect to more effective therapeutic intervention.
Effect of Tat Lysine Variation on Ubiquitination Patterns and LTR Transactivation
Host E3 ubiquitin ligases have previously been studied for their role as restriction factors relative to HIV-1 infection [120,121], though ubiquitination of HIV-1 Tat has been demonstrated to promote efficient LTR transactivation. In GST-Tat pulldown assays of 35S-labeled, in vitro translated Tat, the RING finger-containing Hdm2 proto-oncoprotein interacted with Tat72 and Tat101 length variants that were incubated with purified, bacterially-translated Hdm2 [122]. Hdm2 has been shown to be the human ortholog of Mdm2, which ubiquinates p53 to target it for proteasomal degradation resulting in the maintenance of cell homeostasis [123,124], however, Tat was not targeted for degradation ubiquitination, suggesting an alternate function of ubiquitination [122]. The Tat-Hdm2 interaction was a direct interaction – the addition of CYCT1, a component of P-TEFb known to directly interact with Tat [125], to the assay still resulted in the exclusive pulldown of GST-Tat with Hdm2 [122]. Hdm2 was also found to ubiquitinate both Tat72 and Tat101 in vitro and in vivo [122], and the potential lysine residues of Tat that could accept ubiquitin (Ub) were substituted with arginines to probe for ubiquitination patterns, including Lys12, Lys19, Lys28, Lys29, Lys50, and Lys71. The only Tat variant that had decreased total ubiquitination upon substitution with arginine was Lys71 [122], indicating that Lys71 was the primary Hdm2-associated ubiquitin attachment site of HIV-1 Tat protein. Consideration, however, must be made for Lys71 with regard to PTMs, as it was also a position reported to be methylated (Figure 2) [115]. The determination of which PTM was added to this site has required further investigation, but may have represented redundant activating mechanisms, as both K71me and K71-Ub promoted LTR transactivation. In HeLa P4 LTR-LacZ reporter cells transfected with Tat K71R substitution variants, the transactivation potential of Tat decreased 4-fold when compared to wild-type Tat and could not be rescued by the presence of Hdm2 [122]. The fusion of ubiquitin to the C-terminus of the Tat K71R variant construct, to bypass the need for the addition of ubiquitin to Lys71, rescued transcriptional activation to levels near that of wild-type Tat, even when Hdm2 expression was transiently silenced with siRNA [122]. Although the Tat-Ub fusion protein had ubiquitin attached to an alternate physical location than Lys71, it was still able to transactivate the HIV-1 LTR. This was an interesting observation because it has placed into question the necessity for precise positioning of ubiquitin along the Tat protein, despite Lys71 being the preferred ubiquitin-acceptor for Hdm2. Even so, the interaction between Tat72 or Tat101 and Hdm2 and the addition of ubiquitin to Tat72 or Tat101 depended on the conservation of Tat residue Cys22, where its substitution to glycine (C22G) ablated both functions [122]. Further study of the Hdm2-Tat interaction will be required to determine the basis for the loss of the interaction upon the introduction of a glycine substitution at Cys22.
Another E3 ligase that was experimentally able to add ubiquitin to Tat is the PJA2 E3 ubiquitin ligase, which added ubiquitin to multiple residues of Tat, though Lys28 was the preferred target [126]. Lys41 has been determined to be absolutely essential for efficient LTR transactivation [127], but did not act as a ubiquitin acceptor in the case of PJA2 activity, despite mediating the interaction between PJA2 and Tat [126]. Indeed, when all lysines except for Lys28 in Tat were substituted with arginine, there was no PJA2-mediated ubiquitination, an effect that could be reversed by the reintroduction of just Lys41 [126]. Similar to ubiquitination via Hdm2, the C22A Tat variant was unable to immunoprecipitate with PJA2 [126], which may be due to the inability of the C22A to properly fold [128]. In terms of each variant’s effect on LTR transactivation, the K41R variant reduced Tat activity to 10% of wild-type activity and the K28R variant maintained only about 25% of wild-type activity [126], deeming the conservation of both lysine residues essential for efficient LTR transactivation in this system. When only Lys28 and Lys41 were conserved and all other lysines were substituted with arginine, transactivation capacity was about 80% of wild-type, and the reintroduction of just one more lysine – for a total of three conserved lysines in Tat – restored Tat activity to wild-type levels [126]. The location of the third lysine could vary between Lys12, Lys51, or Lys85, but was always able to restore activity [126], which indicated that only three lysines were required for efficient LTR transactivation and that the site of lysine ubiquitination was also flexible. The theme of flexibility in location of ubiquitin on Tat has been curious; it has suggested that ubiquitination of Tat doesn’t necessarily change its ability to interact with host factors that may rely on structural conformation or unmodified residues. Closer investigation of the nature of the ubiquitination state of Tat should assist in the understanding of the purpose and function of this PTM and how it affects HIV-1 pathogenesis.
Variation within the Cysteine-Rich Domain and Tat Transactivation Activity
The involvement of an intact and functional cysteine-rich domain in HIV-1 Tat has been shown to be essential for its function and optimal LTR transactivation. Early mutational studies of Tat revealed that single substitutions of Cys with either Glu or Gly at residues 22, 30, 31, 34, or 37 resulted in near abolition of LTR transactivation when transfected into HeLa or COS cell lines [58,129,130]. The cysteine residues at Tat positions 22, 25, 27, 30, 34, and 37 have been predicted to be able to support the coordination of two zinc ions [131]. Therefore, amino acid variation at these sites could disrupt amino acid interactions with zinc ions and account for the reduction in LTR transactivation observed in some of these Tat variants [58]. Further investigation has corroborated LTR transactivation activity depletion by Tat cysteine variants through the use of C22G, C30G, and C31G substitutions, where the C22G and C30G variants had little activity, and the C31G variant only maintained partial Tat activity [132]. Another study showed that in U-937 promonocytic cells, the C22G and C31G variants had essentially no transactivation capacity, even when the cells were activated with PMA [133]. Additionally, an H13L variant had greatly reduced LTR transactivation, which was only partly restored with PMA-induced activation [133]. PMA stimulation induced the formation of the Tat-P-TEFb complex, as shown by immunoprecipitation of P-TEFb components CyclinT1 and CDK9 with Tat [133]. The H13L, C22G, and C31G Tat variants were all unable to bind CDK9 [133], indicating that the conservation of these three positions were essential for Tat-P-TEFb complex formation. The lack of association between the Tat variants and CDK9 prevented LTR transactivation by inhibiting P-TEFb activity, as demonstrated by the total abolition of RNAPII CTD phosphorylation [133]. Despite the observation that Cys31 variants, depending on the substitution, may not associate with P-TEFb, subtype C Tat containing the C31S variation maintains its activity. Tat C31S transfected into HEK 293T cells was able to transactivate an LTR reporter construct, while the C30S subtype C variant had significantly decreased ability to transactivate the LTR [10]. This observation has been very interesting, as subtype C C31S Tat has been well-studied for its reduced capacity to induce monocyte chemotaxis [10], suggesting that the functions of LTR transactivation and chemokine mimicry are independent of one another.
The Tat C22S mutation, originally observed in the subtype C HIV-1OYI strain that was isolated from a Gabonese patient, conferred an LTR transactivation-defective phenotype [134]. When introduced to subtype B HIV-1BRU, a strain that does not normally encode a C22S variation, the same defective phenotype was observed [134]. Although the C22S TatOYI mutant appeared as a minor variant in the study, it was mainly found in healthy HIV-1-infected patients that did not quickly progress to AIDS or develop AIDS-like symptoms [134]. This correlation suggested that HIV-1 strains with defective or sub-optimally performing Tat may be unable to efficiently replicate in host cells. Furthermore, it has opened the question as to whether defective Tat found in HIV-1 strains was representative of the fitness of the entire proviral genome, or if a viral replication deficit was solely dependent on the integrity of Tat as previously discussed [134]. Other studies have suggested that Cys22 amino acid substitutions may contribute to the partial unfolding of Tat [128], which may impair the ability of Tat to interact with its targets. Regardless of the interactions in which Tat may participate, the conformation of the viral trans-activator protein has been thought to be critically important for its activity. An unfolded Tat protein may not allow efficient LTR transactivation due to a lack of intact structural domains and essential interactions with other viral and cellular proteins, though further structural analysis and experimental evidence will be required to elucidate these mechanisms.
Downstream of Cys22, the mutation of cysteine residues 34 and 37 to histidine significantly reduced Tat activity and LTR transactivation [130]. Because of the LTR transactivation deficit, there was a parallel reduction in viral protein synthesis with each of the C34H and C37H Tat variants [130]. In contrast, the C31H substitution variant had a less dramatic decrease in LTR transactivation and viral protein synthesis [130]. The disparity of Tat activity between the C31H and C34H and C37H variants has indicated that not all cysteines within the cysteine-rich domain contribute equally to Tat activity. Moreover, these cysteine substitutions did not affect nuclear or nucleolar localization of Tat within the cell [130], so the observed reduction in Tat activity was not likely due to Tat trafficking dysfunction. The conferred deficit may directly impact Tat’s ability to transactivate the HIV-1 LTR, since it reached the nucleus in this model, but still lacked the capacity for full transactivation. Additionally, the alteration of non-cysteine residues within and near the cysteine-rich domain, such as H33A, F38A, and K41A substitutions, also imparted a Tat activity deficit [132]. Similarly, the introduction of a K41T mutation into HIV-1 Tat101 has been observed to result in failure to activate HIV-1 LTR in vitro, as K41T HIV-1 Tat101 could bind TAR, but was transcriptionally inactive [127]. This has implied that the conservation of both cysteine and non-cysteine residues located in and near to the cysteine-rich region was equally important for maintenance of Tat function. The complete summary of cysteine-rich domain variation that contributes to effects on LTR transactivation has been shown in Figure 2.
TAR-dependent Tat Activity and Variation in Patients
Many of the residue substitutions that produce a functional change in the LTR transactivation capacity of Tat have been identified in the DNA of CNS, cerebral spinal fluid (CSF), peripheral blood mononuclear cells (PBMC), spleen, and lymph node samples from HIV-1-infected patients. In one study, 44 deceased human AIDS patients that had dementia and HIVE were analyzed, and 46 unique tat alleles were identified [86]. The substitutions of note within Tat from these samples included variation at amino acid positions 2, 11, 16, 22, 28, 31, 34, 41, 46, 47, 50, 51, 56, 57, 62, and 64 [86], many of which have been studied in the context of effects on LTR transactivation, as discussed in the above sections. Although the functional variants containing substitutions at amino acids 2 and 11 have not been observed to directly influence transactivation, residue Asp2 has been shown to participate in an intramolecular hydrogen bond with residues Lys51 and Arg53 that stabilized the tertiary structure of the protein [135], and the introduction of a stop codon that replaced Trp11 resulted in a severely truncated Tat protein that was unlikely to interact with transactivation components [86]. Therefore, even though variation at Tat residues 2 and 11 did not directly affect LTR transactivation, the indirect consequences of substitutions at these positions may have associated costs that restrict Tat’s function, such as Gln2 and Trp11 variants that impair the efficient trafficking of Tat [7,79,136]. This study, along with the study that isolated the HIV-1 subtype C TatOYI C22S variant that is associated with slow progression to AIDS [134] and the subtype C Tat dicysteine motif variants that correlate with reduced neurocognitive impairment [10,137], are examples of how functional variation in residues that confer LTR transactivation deficits may be useful with respect to HIV-1 translational research. Certainly, more investigation concerning the implications of these variants on HIV-1 disease severity is necessary, but these preliminary data provide the framework for these studies.
TAR-independent Activity
Implication of TAR-defective HIV-1 with respect to Tat Activity
In contrast to the TAR-dependent mechanism of LTR transactivation, Tat can also stimulate HIV-1 transcription in a TAR-independent manner. The first study investigating the phenomenon of TAR-independent LTR transactivation arose from the notion that TAR serves as a site of attachment for host proteins involved in transcriptional elongation of HIV-1 mRNA transcripts rather than directly participating in LTR transactivation itself (Figure 3A), as hypothesized by Berkhout, et al. [138]. Subsequent studies have attempted to address the host and viral factors that are required for LTR transactivation when TAR-RNA binding is inhibited. When the TAR sequence was mutated in HIV-1 LTR-CAT reporter constructs to prevent TAR-dependent host factor interactions and transfected into Jurkat T cells along with Tat constructs, there was an observed decrease in LTR transactivation that could be somewhat ameliorated upon T-cell activation with PMA [139]. These TAR mutations included a single point mutation in the loop sequence, a 4-base pair substitution of loop nucleotides, interruption of the stem sequence that preserved base-pairing and secondary structure, and interruption of the stem sequence that disrupted TAR stem secondary structure. LTR transactivation and viral replication were most severely affected when the stem secondary structure was disrupted, though loop base pair substitutions also greatly impacted these functions [139] (Figure 3B). In both the PMA-stimulated and nonstimulated Jurkat cell experiments, LTR transactivation was never entirely abolished [139], indicating that although the TAR was defective, some remaining LTR transactivation still occurred and the remaining transactivation under these conditions could be enhanced by T cell activation. Moreover, there was still a strict requirement for Tat, as well as the endogenous LTR Sp1 and TATA binding domains, for LTR transactivation [139], though it was unclear from this study whether Tat bound other factors or DNA to promote the residual levels of LTR transactivation in the absence of TAR.
The conservation of certain Tat residues in TAR-independent LTR transactivation appeared to be as important as residue conservation in its TAR-dependent counterpart. As in TAR-dependent transactivation [58,133], cysteine-rich region deletions in Tat conferred a transactivation-defective phenotype to the TAR-mutated in vitro model [139]. The inability of C22G Tat mutants to bind upstream promotors was observed in HeLa cells transfected with LTR TAR-deleted reporter and Tat constructs, as evident by reduced LTR transactivation [140]. H33A, F38A, and K41A Tat variants also negatively affected LTR transactivation in a TAR-independent manner, though all four Tat variants still had deleterious effects on TAR-dependent LTR transactivation when TAR-intact constructs were used [140]. Overall, it seemed that there was some redundancy in the function of these residues in Tat, which may serve as an evolutionary constraint to HIV-1. If Tat contains an amino acid substitution at one of the above residues that caused activity deficits in both TAR-dependent and TAR-independent manners, then HIV-1 transcription surely could not proceed. This may be a contributing factor behind the strict conservation of the TAR sequence. More information regarding TAR-independent transactivation will be required to truly understand what this redundancy means with respect to HIV-1 transcription. The further study of functional Tat variant redundancy present in LTR transactivation that are dysfunctional for both TAR-dependent and TAR-independent mechanisms will help clarify this relationship.
In an alternate TAR-deleted HIV-1 model using a luciferase reporter assay and 293T cells infected with a viral strain with the Y26A variant of HIV-1 Tat, selected based on its previously observed role in LTR transactivation deficits in TAR-dependent models [141], showed a 4-fold decrease in LTR transactivation when compared to wild-type Tat, but maintained wild-type levels of virus replication, despite decreased Tat activity at the LTR [142]. This result suggested that the Y26A amino acid substitution didn’t affect the ability of the virus to replicate under TAR-independent transactivation conditions, and that this mutation mainly affected TAR-dependent viral transcription in that system. Indeed, in a presumably TAR-dependent model, SupT1 cells transfected with LTR-CAT reporter and HIV-1 Tat constructs, the Y26A variant produced a 93% decrease in LTR transactivation activity and abolished the replication ability of HIV-1 in cells transfected with the subtype B LAI infectious HIV-1 clone construct [141]. In primary PBMCs, however, the requirement for wild-type Tat sequence conservation was less stringent, as the Y26A variant still had low levels of virus replication upon electroporation with the mutant construct [141]. Transactivation studies on Y26A Tat have demonstrated that there may be a different repertoire of variants that affect TAR-independent versus TAR-dependent LTR transactivation. This could be important for future investigations, as variants used in classical TAR-dependent LTR transactivation studies may not be useful for studies focused specifically on TAR-independent transactivation.
Direct Interactions between Tat and LTR DNA at Upstream Enhancer Sites
During TAR-dependent LTR transactivation, Tat was shown to interact with CYCT1, and indirectly with CDK9 to form P-TEFb, which then directly interacted with the TAR RNA (Figure 3A) [5]. In contrast, the manner in which Tat is required for TAR-independent LTR transactivation seems to rely somewhat on the proposed and hypothetical ability of Tat to bind to LTR DNA or to direct other cellular factors to bind LTR DNA, possibly with the use of canonical transcription factors. TAR-independent LTR transactivation may afford the opportunity for Tat to interact directly with LTR DNA transcription factor enhancing regions, as observed by structural studies. Wild-type Tat was shown to be able to interact with the NF-κB enhancer element in nuclear extracts of Jurkat (J6) cells, although this interaction depended on the conservation of Tat residue Cys22 [143]. As shown in Figure 3B, the C22G Tat variant lost its ability to interact with the NF-κB enhancer element and transactivate the LTR, as observed by electrophoretic mobility gel shift (EMS) assays [143]. NF-κB enhancer element oligonucleotides that interacted with Tat were deduced by screening an oligonucleotide library using SELEX software for use in circular dichroism (CD) experiments, and subsequent analysis of the oligonucleotide hits by MEME motif analysis software was used to identify the interacting regions of the NF-κB enhancer domain locus [143]. CD spectra of Tat86 protein incubated with oligonucleotides containing the NF-κB enhancer element showed a negative band of reduced intensity when compared to Tat86 protein alone [143], indicating an interaction between the two components. This association was sequence-dependent, as incubation of Tat86 with mutated NF-κB enhancer element oligonucleotide fragments resembled the CD spectrum of Tat86 alone [143]. This study demonstrated that the interaction between HIV-1 Tat protein and LTR DNA was plausible when observed at the molecular level. The presence of endogenous NF-κB and Sp1 enhancer binding sites located upstream of the TAR in the LTR [138] may enable the interaction of Tat or NF-κB and Sp1 transcription factors with LTR DNA to aid in LTR transactivation. Variation within these sites and Tat may complicate direct interactions for TAR-independent LTR transactivation [144]. It is still unclear if Tat can interact directly with DNA in vivo or if it requires recruitment of host cellular factors to achieve optimal activity in all cases. Further investigation, however, is necessary to ensure that these interactions occur with integrated HIV-1 provirus and in physiologically relevant conditions, such as in native or modified chromatin environments. Additionally, because of the apparent nucleotide sequence specificity, variation in the LTR sequence may affect the DNA-binding potential of Tat. The degree of variation tolerated for both the NF-κB enhancer sequence and Tat protein should be investigated to enhance knowledge of this interaction.
Tat Variation and Impact of Upstream Enhancer Elements on Transactivation
The observation made using CD that Tat may bind directly to the NF-κB enhancer sequence [143] has contributed some supporting evidence to the hypothesis that Tat can bind upstream genetic elements to stimulate LTR activity. Indeed, the ability of HIV-1 Tat to activate LTR-mediated transcription was inhibited in the absence of TATA domains, NF-κB-, or Sp1-binding sites in the HIV-1 LTR enhancer and promotor regions [127,140,138]. Likewise, in a TAR-mutated model, deletions of the Sp binding sites or TATA domains resulted in a transactivation-defective phenotype [139]. NF-κB and Sp enhancer binding sites were determined to be essential for optimal transactivation of the HIV-1 LTR, as shown in a study where all NF-κB and Sp enhancer binding sites were deleted from the HIV-1 LTR, but replaced with the SV40 promotor sequence to preserve functionality [138]. Although the SV40 promotor was a strongly enhancing, constitutive promotor [145], it failed to transactivate the LTR in response to Tat exposure in the absence of the endogenous NF-κB and Sp enhancer binding sites, even in the presence of an intact TAR sequence [138]. The TAR was, however, dispensable when Tat was guided to the LTR by a fused Jun domain [138], supporting the hypothesis that the TAR serves merely as a point of physical attachment for transactivation-associated factors.
The development of an in vitro, TAR-independent HIV-1 model system has helped define the mechanism by which Tat may transactivate the HIV-1 LTR in the absence of TAR. The HIV reverse tetracycline-controlled transactivator (HIV-rtTA) lacks a functional TAR sequence, but contains an incorporated Tet-on gene expression system within the 5’ and 3’ LTRs between the endogenous NF-κB and Sp binding sites to preserve transcriptional activation in the absence of TAR, as well as an optimized rtTA transcriptional activator protein gene sequence that replaces the nef gene [146–148]. The introduction of a Tat one-nucleotide frameshift mutation to generate a mutant containing only 19 N-terminus residues from Tat or the substitution of a STOP codon at the Tat START codon to prevent Tat production altogether into the HIV-rtTA model rendered Tat functionally defective and resulted in significantly reduced transactivation and reduced replication, even with the availability of the TAR-independent transcriptional activation pathway in the model [142]. This has suggested that even though HIV-rtTA tolerated TAR deletion in this system, it still required Tat activity for effective transactivation. The presence of Sp or TATA binding sites in the HIV-1 LTR in the HIV-rtTA model system dictated LTR responsiveness to Tat [142], as LTR deletion mutants lacking the Sp binding sites, NF-κB enhancer binding sites, TATA domains, or TAR were unresponsive to Tat, but could be rescued upon the addition of the Sp enhancer sites and the TATA domain back into the LTR [142].
Recent studies have revealed that the involvement of the NF-κB enhancer sequence may occur in physiologic circumstances where Tat concentrations were low. Transfection of the NF-κB reporter and 100 ng of wild-type Tat101-expressing plasmid DNA into Jurkat T cells resulted in significantly increased NF-κB activity, which was accompanied by LTR stimulation, and both effects were abrogated by the introduction of K88,89,90A or E92,94,96A variation [149] (Figure 3B). At transfection concentrations of 500 ng/mL or higher, however, LTR stimulation was mainly TAR-dependent, as observed by a drop in NF-κB activity at the higher Tat101 transfection concentration [149]. The altered functionality of these exon II mutants illustrated the importance of conservative amino acid motifs in the C-terminus of Tat for TAR-independent LTR transactivation. These Tat variants also presented the opportunity to study potential Tat-host protein interactions that involve Tat exon II and affect overall Tat activity. A remaining question centers on intracellular Tat concentrations; the use of physiological concentrations will assist in research relevant to the role of amino acid variation in Tat-mediated LTR transactivation. The complete role of Tat exon II in LTR transactivation is still unclear, but could help define the relationship between TAR-dependent and TAR-independent Tat activity.
Cell Type and Ability of Tat to Transactivate the LTR
The study of TAR-independent LTR transactivation has revealed a surprising detail about Tat activity with respect to cell type. An initial observation regarding the requirement of TAR for transactivation was that the deletion of TAR abolished LTR transactivation in cell types that were non-glial, such as unstimulated T cell lines or epithelial cell types, like HeLa cells, but proceeded in both the rodent C6 and human U138MG glial cell lines [89]. Furthermore, when the TAR was intact, transactivation in glial cells only reached about 30% of non-glial transactivation levels [89], suggesting that TAR-independent LTR transactivation may proceed preferentially in glial cells. This was an important observation, as it could mean that TAR-independent activity was restricted to certain tissues infected with HIV-1, specifically peripheral and central nervous system glial cells. The Tat residues that modulate TAR-dependent and TAR-independent transactivation also differ between glial and non-glial cells, particularly within the cysteine-rich domain. The introduction of C22G and Q35A substitutions to HIV-1 Tat significantly decreased LTR transactivation in U87MG glial cells with TAR-deleted LTRs (Figure 3B), while the presence of other Tat cysteine-rich or core domain variants T23A, N24A, Y26A, K28,29A, C31G, H33A, K41A, and Y47A only showed transactivation deficits with TAR-intact LTRs with intact TAR elements [150]. Both the C22G and Q35A mutations have, however, been shown to inhibit LTR transactivation in other LTR systems with intact TAR elements [132,122,133], so these should be verified in TAR-independent models with further experimentation. This effect may be astrocyte-dependent, as a subsequent study isolated an NF-κB isoform from astrocytic cells that differed from “prototypical” NF-κB, such as that observed in T cells [151] and prior data has shown that NF-κB enhancer domains in U87MG glial cells mediated Tat activity with the TAR-deleted mutant HIV-1 LTR [89]. These data lend plausibility to a mechanism exclusive to glial cells, as other studies have declared the necessity for the NF-κB enhancer binding sites for TAR-independent LTR transactivation [138,143]. Moreover, nuclear extract from PMA-stimulated U87MG glial cells was capable of initiating transactivation in LTR reporter HeLa cell line extracts that contained TAR deletions in the LTR [89]. The astrocyte-specific NF-κB isoform, which was presumably present in astrocyte nuclear extracts, may interact with upstream NF-κB enhancer elements to stimulate LTR transactivation in the absence of TAR. Overall, TAR-independent LTR transactivation may represent an alternative mechanism for viral transcription and may help explain the low levels of HIV-1 expression observed in infected astrocytic cell populations [152]. Additionally, curative therapeutic strategies, such as Tat inhibitors or CRISPR-Cas9 excision therapies aimed at TAR regions, may face obstacles in glial cell types, as TAR-independent mechanisms may impact the efficacy of both of these methodologies. Confirmation of this transactivation pathway as preferentially used in glial cell types would be an important finding relevant to the study of HIV-1 pathogenesis, as HAND still occurs in patients that adhere to ART and HIV-1-infected glial cells have been proposed to be an HIV-1 tissue reservoir prior to and after initiation of ART [153,154].
IV. Tat-mediated Apoptosis
The effects of HIV-1 Tat on both infected and uninfected cells are diverse, but one of the most severe for individual cells is the induction of apoptosis. Tat may cause apoptosis in a range of cells, including T cells and neurons [155–158], though the exact mechanisms governing the initiation of apoptosis in each cell type are not fully understood. Early studies concerning the apoptotic potential of Tat focused on the susceptibility of Tat-transfected cells to intrinsic cell death. These original observations included increased doubling time in Jurkat E6–1 T cells that were stably transfected with Tat or incubated overnight with exogenous Tat, which was attributed to the loss of Tat-transfected cells from culture via programmed cell death [155]. Further transfection experiments confirmed that Tat-transfected cells were, indeed, more susceptible to apoptosis than non-producing cells [159]. Much of the early research in this area focused on T cells and T cell lines because of the observation of CD4+ T-cell depletion in HIV-infected patients [160], though additional studies have identified specific HIV-1 Tat amino acid variants that have a role in promoting or preventing Tat-mediated apoptosis in T-cell lines, macrophage cell lines, and neuronal cell cultures [161,162,156].
Caspase and Fas-mediated Apoptosis
Prior studies have observed the modulation or induction of apoptotic pathways in HIV-1-infected cells or bystander cells, which are uninfected, but are located near infected cells. These accounts include: the upregulation of Fas ligand that is further accelerated by the presence of Tat [163], increased TNF-related apoptosis-inducing ligand (TRAIL) expression on the surfaces of CD4+ T cells and PBMCs [164], and the presence of activated, immunoreactive caspase-3 enzyme in cerebrocortical brain tissue of HIV-1-infected patients [165]. The apoptotic pathways are composed of signaling and proteolytic cascades that culminate in physiological and morphological cell changes, leading to the death of the cell. Activated initiator caspases 8 and 9 are the enzymes responsible for the propagation of death signals via the cleavage of effector caspases 3, 6, and 7, and subsequently, apoptosis [166]. Subtype B Tat86 has been shown to induce 4-fold more apoptosis in both Jurkat T cells or CD4+ primary T cells in culture than the subtype B Tat101 isoform [167]. This contrasted with the ability of each length isoform to transactivate the LTR, as the Tat101 isoform had increased activity when compared to the Tat86 isoform in T cells [167]. Tat86 was significantly more capable of initiating apoptosis than Tat101 due to increased expression of procaspase-8, procaspase-9, and FADD (Fas-associated death domain) [167]. Variation in the length of the Tat C-terminus may contribute to its potential to induce apoptosis, though no specific residues in the C-terminal domain between residues 86 and 101 have been identified as responsible for this disparity.
Certain amino acid determinants within the Tat protein have been associated with the regulation or triggering of these pathways. Both Jurkat T cells and primary CD4+ T cells that were transfected with Tat or infected with HIV-1 were more sensitive to apoptosis than cells that were uninfected or did not express Tat [168]. In this study, this effect was not dependent on the presence of other HIV-1 accessory proteins, since cells infected with Vpu-, Vpr-, Rev-, Vif-, or Nef-deleted HIV-1 strains were still more susceptible to apoptosis than uninfected cells [168]. Increased caspase-8 expression in these cells was implicated as the causative basis for the increased susceptibility to apoptosis in this system, as the addition of a caspase-8 inhibitor prevented apoptosis of nearly all Tat-producing cells [168]. The C22G and K41A HIV-1 Tat variants, which have been reported as LTR transactivation-deficient, had no effect on the ability of Tat to induce apoptosis, and were able to cause apoptosis in a similar percentage of cells as wild-type Tat [168]. Though specific Tat residues have not been shown to alter caspase-8 specifically, the second exon of Tat may be essential for the induction of this pathway, as one-exon Tat (Tat72) did not cause apoptosis of HIV-1-infected Jurkat cells, despite its ability to transactivate the HIV-1 LTR [168].
Early research exploring the effects of HIV-1 Tat protein on neuronal cells demonstrated that exposure of primary hippocampal neurons in culture to exogenous Tat caused significant cell death [158]. This was accompanied by increased caspase-3 activity, significant influx of calcium into the cytoplasm and mitochondria, and the accumulation of reactive oxygen species (ROS) within mitochondria that was paired with membrane depolarization [158], all of which were characteristic of apoptosis [169]. The C22G Tat variant was significantly less neurotoxic upon extracellular incubation with primary hippocampal cell cultures when compared to incubation with wild-type Tat, manifesting as abrogation of apoptosis [156]. The underlying cause was revealed to be the inability of Tat C22G to activate intrinsic initiator caspase-9 and effector caspase-3/7, where wild-type HIV-1 subtype B Tat initiated the activity of both initiator and effector caspases [156]. The interactions leading to procaspase-9 cleavage in this case have not yet been elucidated, though the advent of caspase-mediated apoptosis in response to wild-type Tat exposure suggested a potential mechanism of neuronal damage that could contribute to HAND. Furthermore, it has suggested that HIV-1 strains containing nonsynonymous amino acid mutations at residue 22 caused less severe HAND, though historically, Cys22 is a strictly conserved residue in subtype B Tat [170].
Microtubule Polymerization
Studies have shown that Tat protein found in the HIV-1 subtype D proviral genome from rapidly-progressing Ugandan patients induced widespread apoptosis upon extracellular incubation with the Jurkat T-cell line [171]. In contrast, Tat protein generated from the sequences of subtype D infected long-term survivors prompted about 41% less apoptosis in Jurkat cells when compared to the rapidly-progressing group [171]. Structural studies that compared HIV-1 subtype D Tat from one long-term survivor and one rapid progressor against Tat Mal, a subtype D reference strain, showed defining variation within the glutamine-rich domain [171]. The presence of Q63H and T64A substitutions were present in the amino acid sequence from the long-term survivor and distorted the short alpha-helix secondary structure of this domain [171]. Whether this variation accounts for the difference in apoptosis induced between HIV-1-infected rapid progressors and long-term survivors remains to be investigated, but the mechanism by which apoptosis occurs in these cells may be due to previously observed phenomena.
Prior studies had shown that Tat could directly interact with αβ tubulin dimers and stabilize polymerized microtubules in Tat-exposed cells [162,172,173], an observation that was also observed with subtype D Tat derived from rapid progressor Ugandan patients [171]. Tat from long-term survivors did not bind free tubulin with the same affinity in vitro as Tat derived from a rapid progressor [171], suggesting a functional difference between Tat sequences of each patient group that affected the ability to bind free αβ tubulin dimers. This effect was also demonstrated across HIV-1 subtypes, where Tat from subtypes generally classified as being more toxic was more efficient at promoting microtubule polymerization [172]. These “toxic” strains included the subtype B HXB2 strain and subtype D Eli strain, which were isolated from patients that were rapid progressors [6,174]. In contrast, Tat from the subtype C strain HIV-1OYI – originally isolated from a highly exposed but persistently seronegative patient – did not efficiently promote microtubule polymerization in vitro [172,134].
A mutant form of Tat containing alanine substitutions from amino acid positions 36–39 was incapable of binding tubulin αβ dimers [162], suggesting that the peptide motif spanning residues 36–39 in Tat was necessary for association with free tubulin. The introduction of the alanine stretch did not affect Tat uptake or secretion, as these processes were unchanged by the mutations [162]. Moreover, this form of Tat could still transactivate the HIV-1 LTR [162], suggesting that Tat(36–39)A was biologically active in this model. The phenomenon of subtype B HIV-1 Tat binding to free tubulin in target cell cytoplasm was accompanied by the promotion of microtubule elongation and stabilization of formed microtubule filaments [162,172]. Eventual cell death in response to Tat exposure was triggered by the prevention of microtubule depolymerization, an effect also observed when cells are treated with paclitaxel, a chemotherapeutic that prevents disassembly of microtubules [162]. Perturbation of microtubule disassembly by paclitaxel or wild-type subtype B Tat in this study resulted in the cleavage of procaspase 9, initiating the downstream apoptotic cascade [162]. Tat(36–39)A was incapable of inducing procaspase-9 cleavage in Jurkat cells because of its inability to bind both αβ tubulin dimers and polymerized microtubules [162]. The lack of procaspase-9 cleavage in Tat(36–39)A-treated Jurkat cells prevented apoptosis in these cells and conveyed the importance of the tubulin-binding domain for Tat-meditated intrinsic cell death. In comparison, extrinsic, Fas-FasL-mediated cell death was not found to be a major pathway in Tat-induced apoptosis in this system [162].
HIV-1 subtype B Tat residue 37, which was within the mutated motif in the prior study, usually presents as a conserved cysteine and has been deemed important for the coordination of a zinc ion and is central to Tat’s structural stability [131]. Indeed, the zinc-coordinated form of Tat promoted the formation of Tat-tubulin complexes in both microtubule assembly and non-assembly cell conditions [175]. The replacement of cysteine at this residue position in Tat may account for the loss of tubulin-binding activity and induction of caspase-9-mediated apoptosis, though more specific mutational analysis is required to fully understand this effect. In contrast, extracellular treatment of Jurkat T cells with the C22G Tat variant has been reported to induce apoptotic cell death at levels similar to wild-type Tat in Jurkat T-cell culture, though this conflicted with data from other studies using the C22G Tat variant [162,156]. Cys22 is another of the seven cysteine residues within the cysteine-rich domain, and like Cys37, its conservation may also be central to zinc ion coordination by Tat [131]. Moreover, structural analyses have indicated that Cys22 and Cys37 could be involved in the coordination of the same zinc ion [131]. Unlike Tat(36–37)A, the Tat C22G variant had significantly reduced capacity for LTR transactivation [132,133,140,150]. This has prompted the question of the relative contributions of each cysteine to the functional integrity of Tat cysteine variants and whether each cysteine within the amino acid sequence of Tat has a single function, or is capable of functional redundancy. Additionally, it challenges the definition of biological activity for Tat. Is the capacity for HIV-1 LTR transactivation a firm requirement for biological activity, or are there other functions that qualify Tat as being biologically active in lieu of LTR transactivation?
RGD Motif in Subtypes B and C Alters Apoptosis Induction
The second exon of HIV-1 subtype B Tat contains an RGD motif that spans residue positions 78–80, while subtype C Tat was found to code for a QGD motif at these residues [161]. Subtype B Tat that contained the exon II RGD motif caused about 29% cell apoptosis in a Tat-producing PMA-stimulated THP-1 monocyte cell line culture, compared to a baseline 11% apoptosis in cells not exposed to Tat [161]. Interestingly, subtype C Tat-treated cells underwent apoptosis at a level comparable to the baseline, at 14% [161], indicating that the presence of the RGD motif in Tat exon II contributed to increased apoptotic cell death in this system. The mutation of residue 78 from Gln to Arg in subtype C Tat to introduce the RGD motif to the second exon induced additional apoptosis in PMA-stimulated THP-1 cells, even though the converse introduction of QGD into subtype B Tat exon II did not abrogate apoptosis induction, as one would expect [161]. This data demonstrated that the presence of the RGD motif in exon II is sufficient to trigger apoptosis, although its absence does not prevent apoptosis altogether.
Tat-mediated Upregulation of Bcl-2 – Promotes and Prevents Apoptosis
Tat has been shown to be a modulator of anti-apoptotic Bcl-2 protein expression and activity in HIV-1-infected cells. Prior research has investigated the role of a number of HIV-1 proteins in the regulation of Bcl-2 [176,177], but a bulk of the studies have focused on the effect of Tat on this important pathway. The role of Bcl-2 as a mediator of Tat-induced apoptosis has been controversial, as both the promotion and prevention of apoptosis have been observed upon analysis of the relationship between Bcl-2 and Tat [178–180]. The upregulation of Bcl-2 has been shown to protect Tat-transfected Jurkat T cells from apoptosis, even when cultured in conditions that promote apoptosis, such as serum-free culture, or treatment with cytotoxic agent TNF-α or anti-Fas antibody [181,182]. Similarly, MDMs (monocyte-derived macrophages) exposed to soluble Tat86 exhibited increased Bcl-2 mRNA and protein production [183]. It has been hypothesized that because Tat-producing or Tat-exposed cells may be protected from apoptosis, there is a window of time for viral production or viral spreading before apoptotic cascades overcome anti-apoptotic signals [182,183]. In this way, the infected cell would not be eliminated by host defenses prior to viral dissemination or integration of the HIV-1 genome into the host cell chromosome(s). In contrast, Jurkat cells transfected with the C22G variant of Tat showed levels of Bcl-2 mRNA and protein that were similar to control, untransfected cells [179]. In the same model, Tat C22G-transfected cells also exhibited a progressive increase in apoptosis over time [179], suggesting that conservation within the cysteine-rich domain is crucial for the protection of these cells from apoptosis, even though variation of Cys22 had the opposite effect of increasing the level apoptosis. Furthermore, Cys22 Tat variants have been reported as LTR transactivation-deficient [132,133,140,150], indicating a potential correlation between LTR transactivation and the upregulation of anti-apoptotic Bcl-2 expression. Contradictory evidence, however, has noted that Tat production by Tat-transfected Jurkat or H9 T cell lines can downregulate Bcl-2 expression and lead to increased apoptosis in serum-free conditions, as characterized by increased DNA fragmentation and expression of the pro-apoptotic protein, Bax [180]. This evidence agrees with reports on Tat-mediated apoptosis in T-cell lines [155,168], though contrasts with other major studies of Bcl-2 regulation by Tat [179]. Future investigation of the association between Tat and Bcl-2 activity will help elucidate the conditions that are responsible for the regulation of Bcl-2 in these systems. Additionally, in vivo studies of these pathways will add physiological significance to the mechanism.
V. Cell Activation
Endothelial Cells
Human endothelial tissues have represented an important population of bystander cells during the course of HIV disease; although they are not very susceptible to HIV-1 infection, they are very prone to the consequences of infection of other cell types and of secreted viral protein(s), including Tat. Tat has been shown to modulate the expression of endothelial cell (EC) surface proteins, tight junction proteins, and oxidative stress pathways, notably in brain microvascular endothelial cells, which are an important component of the blood brain barrier (BBB) [184,43,185] as well as populations of cardiac endothelial cells and other types of endothelial cell populations [186–188]. Therefore, changes in endothelial tissue behavior in response to Tat exposure in an HIV-1-infected patient may affect the integrity of the blood-brain barrier and provide an access point for viral entry into the CNS. Variation within Tat may affect disease severity by impairing this process.
Vascular endothelial growth factor receptor-2 (VEGFR-2) has been shown to be a receptor present on ECs that works to activate ECs mainly through interactions with ligand VEGF-A [189]. Activated ECs have been characterized by proliferative and migratory activity, as well as enhanced survival, and endothelial tissue permeability [189]. VEGFR-2 has been reported to act as a Tat receptor, where the engagement of VEGFR-2 by HIV-1 Tat activated vascular endothelial cells [190]. This was possible because of amino acid sequence homology between the arginine-rich domain of Tat and known human angiogenic, heparin-binding proteins, such as fibroblast growth factor, VEGF-A, and hepatocyte growth factor [191]. Likewise, Tat can bind heparin, as observed in sepharose column pulldown experiments, which most likely facilitates its uptake, as low concentrations of heparin have been observed to assist in Tat internalization into EaHy926 endothelial cells [191]. Low heparin concentrations can also enhance Tat-mediated EaHy926 endothelial cell and human umbilical vein endothelial cell (HUVEC) proliferation and migration [191], both of which are characteristic of VEGFR-2-mediated cellular activity [192]. Tat was also found capable to upregulate the expression of cell adhesion molecules on the surface of HUVEC monolayers in vitro, resulting in greater adherence of HL-60 monocytic cells to EC monolayers [193]. Interestingly, Tat peptides containing only the arginine-rich domain or part of the RGD motif present in exon II could elicit EaHy926 endothelial cell proliferation on their own [191], warranting a closer look at specific residue conservation within the known Tat domains that mediate endothelial cell behaviors.
Of the domains of HIV-1 Tat, the cysteine-rich domain, arginine-rich domain, and second exon RGD motif are all determined to have some effect on the ability of Tat to induce EC proliferation, migration, or adhesion to the extracellular matrix [194]. The cysteine-rich domain of Tat86 was found to be important for Tat-induced HUVEC proliferation and migration, but not adhesion to the extracellular matrix, as a variant with C(22,25,27)A substitutions was only competent at weakly activating migration and proliferation of these cells, and was still able to adhere to extracellular matrix at wild-type levels [194]. This effect may be due to the loss of zinc coordination by cysteines 22, 25, and 27, all of which are predicted to interact with a total of two zinc ions to assume the complete tertiary structure of Tat [131].
Mutations within the arginine-rich domain of Tat decreased the ability of Tat to activate EC proliferation, migration, and adhesion to the extracellular matrix. A Tat101 variant with alanine substitutions at each of the six arginines within the arginine-rich domain severely reduced these activities in HUVEC cultures [194], emphasizing the extreme importance of amino acid conservation within this region. Less severe reduction of HUVEC proliferation, migration, and extracellular matrix adhesion correlated with fewer substitutions in the arginine-rich domain of Tat86, as demonstrated upon the introduction of successive substitutions encoding either R49G/K50I/R52L/R53I or R49G/K50I, respectively [194]. The R49G/K50I Tat86 variant showed reduced migration and proliferation of HUVECs upon treatment, but did not affect the ability of the cells to adhere to extracellular matrix [194], indicating the number and charge of conserved arginine residues may be more important for EC adhesion than the maintenance of specific amino acids.
The RGD motif that is located in the second exon of HIV-1 subtype B Tat86 spans residues 78–80 and its conservation has been associated with increased apoptosis of Tat-producing PMA-stimulated THP-1 monocytic cells in vitro [161]. In the context of EC activation, mutation of residue 80 to produce a D80E variant prompted EC migration and proliferation, while treatment with a double variant containing an additional R78K substitution in the RGD motif resulted in reduced HUVEC proliferation, migration, and adhesion to extracellular matrix [194]. Though the effects of treatment with a single R78K variant was not studied in this investigation, it is unclear if these EC behaviors were developed by the disruption of Arg78 alone or if Asp80 substitutions also contributed to the outcome. ECs express VEGFR-2 receptors that have been observed to bind extracellular Tat with high affinity [190]. The R78K/D80E variation did not affect the adherence of Tat86 to EC monolayers [194], which has suggested that the ability of Tat to activate ECs via VEGFR-2 engagement likely did not depend on residue conservation within the second exon RGD motif in this model system. Alternatively, cysteine-rich and arginine-rich domain variants demonstrated lower affinities for EC monolayer cells than wild-type Tat [194].
Furthermore, the activation of ECs may depend on the ability of Tat variants to promote phosphorylation of VEGFR-2. VEGFR-2 phosphorylation in vitro may be indicative of dimerization that occurs upon receptor engagement by a ligand, leading to auto- or trans-phosphorylation of the dimerized VEGFR-2 units [189]. Tat binding to VEGFR-2 resulted in phosphorylation, which stimulated a variety of signal transduction pathways that characterize EC activation, including those involved in EC proliferation, migration, cell survival, transcriptional activation, and vascular permeability [189]. Arginine-rich mutants of Tat86 have been reported to have reduced ability to promote VEGFR-2 phosphorylation in EC monolayers when compared to wild-type Tat86 [194]. As with proliferation, migration, and adhesion to extracellular matrix, the severity of this effect was dependent on the number of substitutions within the arginine-rich domain, where total replacement of all arginines in the domain resulted in the greatest observed defect, and R49G/K50I/R52L/R53I and R49G/K50I Tat86 variants promoted lesser, but still significant, reduction of VEGFR-2 phosphorylation [194]. The C(22,25,27)A cysteine-rich domain Tat86 variant also demonstrated reduced phosphorylation of VEGFR-2 in EC monolayers upon treatment [194]. Therefore, reduced phosphorylation of VEGFR-2 in response to subtype B Tat variation has suggested that the ability of Tat to act as a strong VEGFR-2 ligand depends on amino acid conservation within the arginine-rich and cysteine-rich domains. Altogether, it has appeared that the ability of HIV-1 Tat variants to engage VEGFR-2 on ECs determines the degree of EC activation by Tat. This could be important in the context of BBB permeability, as it has been proposed to contribute to the development of HAND in HIV-1-infected patients [195]. Variation within Tat may abrogate the activation of ECs at the BBB, but this hypothesis will still require experimental validation.
Immune Cells - Chemokine Mimicry
The ability of HIV-1 Tat to dictate cell migration has been well-documented [196–198]. Tat has been shown to be capable of initiating chemotaxis of endothelial cells, monocytes, dendritic cells, PMNs, and B cells, all of which are hypothesized to respond accordingly to interactions between functional domains of Tat and chemokine receptors endogenous to each cell type [196]. Early observations concerning the chemotactic capacity of Tat relied on the potential of certain domains to prompt cell migration, where arginine-rich or RGD motif-containing Tat peptide fragments spanning positions 46 to 60 and 65 to 80, respectively, were found to be sufficient to elicit chemotaxis of primary monocytes or primary monocyte-derived dendritic cells through microchambers, though only achieving about 50% efficacy of Tat86 [197]. Tat peptides encompassing both the cysteine-rich and arginine-rich domains (spanning residues 24 to 51) were also able to potently induce primary monocyte migration and polarization to a degree similar to Tat86 [198], indicating a key role for these domains in Tat-mediated chemotaxis. While various Tat fragments may elicit chemotaxis of monocytes, studies have shown that Tat86 is more efficient in this process and shows higher affinity for surface receptor binding [197,198]. Therefore, the analysis of specific amino acid variants within Tat that affect its chemotactic properties are of interest.
The dicysteine motif present in the cysteine-rich domain of HIV-1 Tat has been studied for its chemotactic potential. The CCF motif sequence observed in HIV-1 subtype B Tat86 amino acid sequences spanning residues 30 to 32 has molecular homology to several known β-chemokines, including MCP-1, −2, and −3, as well as RANTES, MIP-1α, and MIP-1β [44]. Mutation of the cysteine residues in this motif to serines abolished cytoplasmic influx of Ca2+ in primary monocytes indicative of β-chemokine receptor engagement and was observed upon treatment with CCF-containing β-chemokines or Tat86 [44,199]. Analysis of the consensus sequence of the Tat cysteine-rich domain in HIV-1 subtype C isolates from several countries revealed that the dicysteine motif encodes a serine at position 31, resulting in a C30S31 motif [10]. Experimentally, the use of modified Boyden chemotactic chambers to observe primary PBMC migration in response to dicysteine motif subtype C Tat variants showed that the alteration of the motif affected the ability of PBMCs to migrate toward the Tat chemotactic gradient, where the C30C31 variant elicited the most monocyte migration and the C30S31 and S30C31 variants elicited markedly less [10]. This function of Tat has been identified as a potential mechanism for monocyte entry into the CNS, which has been thought to contribute to neuropathology observed in HAND [200–202].
Regulation of Gene Expression
HIV-1 has been shown to cause global gene expression changes in infected cells [203]. Research concerning the regulation of HIV-1-mediated host gene expression has revealed that HIV-1 and Tat were capable of inducing the production of inflammatory cytokines and chemokines in a number of cell types [204–206]. It has been documented that Tat can also act as a chemokine [10], and the regulation of these and other genes may represent redundant and independent functional characteristics of Tat. These regulatory effects have been demonstrated to occur both directly and indirectly.
Indirect mechanisms associated with the regulation of gene expression the by HIV-1 Tat protein include the modulation of NF-κB activity in Tat-producing cells. The acetylation status of the NF-κB p65 subunit determines its activity and ability to initiate the transcription of NF-κB-inducible genes, including those that control T-cell activation and cytokine regulation, and is part of a signaling pathway known to be appropriated during viral infection [207,208]. HIV-1 wild-type Tat101 has been shown to cause cell hyperactivation by inducing hyperacetylation of NF-κB, a downstream effect of inhibiting the activity of deacetylase SIRT1 [209]. In an in vitro cell system that utilized 293T cells transfected with expression vectors encoding Tat, SIRT1, HAT p300, and the p65 subunit of NF-κB, wild-type Tat was shown to interact with SIRT1 and prevent deacetylation of p65 at residue Ser310, thus maintaining the active form of NF-κB that can promote the expression of NF-κB-inducible genes [209]. Cysteine-rich and core domain Tat variants including C22G, F38A, or K41A substitutions decreased hyperacetylation of the p65 subunit of NF-κB in the same model system [209], indicating that these residues were essential for the interaction between Tat protein and SIRT1 leading to the inhibition of SIRT1 activity, though this still requires experimental validation.
Wild-type Tat101 strongly induced the expression of IL-2 in Jurkat T cells infected with LTR-Tat-GFP-expressing lentiviral vectors [209], thereby providing a system to facilitate the characterization of the downstream effects of SIRT1 inhibition. IL-2 is a cytokine that is expressed after viral infection, is a growth factor that supports T-cell survival and proliferation, and functions to stimulate immune responses [210]. The hyperactivation of T cells in response to HIV-1 infection has been previously attributed to Tat101 protein expression and occurred only in the presence of full-length Tat [211]. The K41A Tat variant lacked the ability to super-induce the expression of IL-2 mRNA in Jurkat T cells when compared to wild-type Tat or alternate lysine residue variants, K50A and K51A [209], suggesting that though Tat may normally induce the overexpression of IL-2 in T cells, the substitution of Lys41 affects this function. Therefore, Lys41 may have special importance in the process of IL-2 overexpression in Tat-producing T cells and may be essential for this function of Tat.
HIV-1 Tat may also be able to regulate gene expression of HIV-1-infected cells via direct interactions with promotor regions of specific genes. ChIP-seq analysis of an HIV-1 lentivirus-infected THP-1 monocyte cell line demonstrated that Tat could directly interact with promotor regions for 66 cellular genes [212]. This study focused on those genes that may be important for the development of HAND, such as brain-derived neurotrophic factor (BDNF), cytokine receptor-like factor 2 (CRFL2), complement component 5 (CR5), and amyloid beta precursor protein-binding family A, member 1 (APBA1) [212]. The presence of Tat increased the expression of each of these genes except CRLF2, for which expression was decreased [212]. As mentioned above, the K50A variation blocked acetylation of Tat by p300 HAT or methylation by SETDB1 and affected LTR transactivation [61,116], but did not affect binding to THP-1 monocytic DNA regions, despite changes in the expression patterns of the downstream genes [212]. The K50A substitution altered Tat-mediated gene expression, resulting in significant increases in the expression of CRLF2, BDNF, APBA1, and decreased expression of C5 by 48 hours post infection with HIV-1 K50A Tat-expressing lentivirus [212]. This has suggested that Lys50 plays a substantial role in the regulation of these genes, though the consequences of altered expression by Tat still requires investigation. Overall, it underscores the importance of posttranslational modifications of Tat residues, ranging from effects on LTR transactivation to modified host gene expression.
In addition to direct interactions between Tat and host genomic DNA, HIV-1 Tat has been observed to associate with human cellular mRNA transcripts in primary CD4+ T cells [213]. The use of MEME-ChIP analysis revealed that Tat could bind mRNAs containing stem-loop structures resembling those of TAR [213]. This interaction was found to be mediated by Tat lysine residues 50 and 51, as K50S/K51G variants were unable to efficiently bind RNA, as evidenced by an 85% decrease in the immunoprecipitation of RNA with Tat variants in a CEM T-cell line infected with a Tat-encoding lentiviral vector [213]. Intriguingly, Gene Ontology analysis of the Tat-interacting mRNA showed many shared functional annotations of the transcripts, including nucleotide binding, transferase activity, and tRNA metabolic processes [213], suggesting that in addition to transcriptional regulation, HIV-1 Tat may be able to regulate RNA and protein expression. Although the downstream effects of Tat-mRNA interactions in this system have not yet been thoroughly detailed, increased expression of many of the genes identified in this analysis have been previously shown to be involved in HIV-1 pathogenesis [214,215,24,216], and Tat residues Lys50 and Lys51 may play a role in their positive regulation.
Experimental subtype B HIV-1 Tat variants that have the observed capacity to regulate gene expression are generally founded in amino acid residue substitutions that interfere with LTR transactivation, such as those at Cys22, Lys41, Lys50, and Lys51. Sequence analysis of HIV-1 isolates classified as subtype E, however, has been observed to code for a naturally-occurring tryptophan at Tat amino acid position 32, which possessed differential functionality with respect to TNF gene expression, although it still maintained the ability to transactivate HIV-1 LTR [217]. Where subtype E Tat contains tryptophan at position 32, the consensus sequences of subtype B and subtype C molecular clones encoded phenylalanine or tyrosine, respectively [217]. TNF gene expression and cytokine release from HIV-1-infected T cells has been implicated as a factor guiding nuclear translocation of NF-κB, as well as stimulating HIV-1 proviral genome expression [218–220]. When co-transfected with a TNF-α promotor-luciferase reporter construct into Jurkat T cells, the subtype E Trp32 variant exhibited suppressed TNF gene expression and subsequent decreased TNF-α protein production, as compared to Tat from subtypes B and C [217]. Mutation of the Trp32 to encode a subdominant W32G variant partially restored TNF-α transcription and protein production to about 30% of that found with subtype B, despite the replacement of a large ring-containing amino acid with glycine [217]. DNase I hypersensitivity assays, which were used to identify areas of decondensed chromatin within genomic DNA, demonstrated that the TNF-α gene was precluded in PMA-stimulated Jurkat cells transfected with subtype E Tat86 harboring the Trp32 sequence variant, but was accessible upon transfection with the subtype B LAI molecular clone [217]. Further investigation in this model cell system demonstrated that the Trp32 variant interfered with the ability of P/CAF and GCN5 HATs to remodel chromatin in the regions surrounding the TNF-α locus [217]. The reduced recruitment of P/CAF and GCN5 to the TNF-α locus could be partly restored with the introduction of the W32G mutation, though still did not fully recover function to levels near that of the subtype B LAI molecular clone [217]. Overall, this study demonstrated a disparity between HIV-1 subtypes regarding transcriptional functionality and the induction of cytokine gene expression. The differences in pathogenesis between HIV-1 Tat subtypes represents an area of research that is important for a deeper understanding of HIV-1 altogether, as modulation of gene expression by the Tat protein has the potential to affect HIV-1-mediated inflammatory responses.
VI. Tat-mediated Neurotoxicity
Tat-NMDAR Interactions
The study of Tat-mediated neurotoxicity began with the observation that certain peptides derived from subtype B Tat86 were able to induce excessive neuroexcitation in human fetal neurons in culture. This study identified Tat residues 31 to 61 as a neurotoxic domain within Tat [221]. Variation within this region is therefore of interest in the continued investigation of Tat, as it may provide insight to clinical examples of Tat-induced neurotoxicity. Further research identified a mechanism to explain the effects originally observed. HIV-1 Tat has been shown to elicit neurotoxicity via interactions with N-methyl-D-aspartate receptors (NMDARs), which occurred between Tat and NMDAR NR1 and NR2a subunits [222]. This interaction was between amino acid residue Cys744 of the NR1 subunit and conserved Tat residue Cys31 [222]. NR1 Cys744 normally forms a dicysteine bond with downstream Cys798,but was disrupted by the presence of Tat when Tat Cys31 paired with NR1 Cys744, forming an alternative disulfide bridge between the two proteins (Figure 4) [222]. To investigate the consequences of this effect, studies performed utilized the HEK 293T cell line, which was transfected with pertinent NMDAR subunits and, similar to cells that endogenously express NMDARs, there were decreases shown in cell viability upon exposure to excessive concentrations of NMDA [222]. Subtype B Tat, in which Cys31 was strongly conserved, induced neurotoxicity in the form of reduced NMDAR-expressing HEK 293T cell viability to a degree similar to NMDA-treatment [222]. Subtype C Tat, which encodes a serine at residue 31, showed attenuated neurotoxicity when compared to Tat or NMDA treatments [222]. As expected, variation of Cys31 to an amino acid that inhibits disulfide bond formation with the NR1 Cys744 residue blocked this interaction and reduced neurotoxicity by Tat. Indeed, subtype C patient samples that had a conserved Cys31 caused similar neurotoxicity to samples from patients infected with subtype B [222]. Similarly, mutation of NMDAR NR1 residue Cys744 from cysteine to alanine decreased toxicity to the NMDAR-expressing HEK cells [222]. The C31S variation present in subtype C Tat protected cells from toxicity, but did not prevent Tat binding to NMDARs, as the Tat-NMDAR interaction was only inhibited upon the deletion of the arginine-rich domain [222], suggesting that binding may occur at a site other than the NR1 Cys744 residue. These data reveal an intricate, two-step binding and activation mechanism for Tat-mediated neurotoxicity that requires residue conservation in both the arginine-rich and cysteine-rich domain, though clarification of the specific residues required for binding in the arginine-rich domain require further analysis. The downstream effects of NMDAR-mediated neurotoxicity include the production of reactive oxygen species and nitric oxide, which accumulate in neurons and may lead to cell death [223,224], representing an important pathway of damage involved in Tat-mediated neurotoxicity.
Fig. 4. Proposed model of the effect of cysteine-rich domain HIV-1 Tat variants on Tat-mediated neuronal neurotoxicity.
Overview of HIV-1 Tat variants that produce neurotoxic effects differing from wild-type (WT) Tat. (Top left) Interaction between Tat Cys31 and NMDAR NR1 Cys744. Excitotoxicity generated by the formation of a disulfide bond between WT Tat Cys31 and Cys744 of the NMDAR NR1 subunit is reduced with the replacement of Tat Cys31 with serine. This is presumably due to the inability of Tat C31S to form this disulfide bond. (Bottom left) Tat variants modulate F-actin puncta loss in neuronal dendrites. Although the mechanism of Tat-mediated synaptodendritic injury is not fully understood, treatment of primary hippocampal neurons with WT Tat results in decreased F-actin puncta, the presence of which normally indicates pre- and post-synaptic structural integrity. The Tat-mediated decrease in F-actin puncta is not seen upon treatment with C22G or C31S. (Right) Amyloid beta production is altered by exposure to Tat variants. Treatment of primary hippocampal neurons with WT Tat promoted beta amyloid 1–42 production and release into cell culture supernatant, where it may aggregate and form stiff fibrils in the presence of WT Tat. The introduction of the C22G or C31S variant restored beta amyloid 1–42 production to control levels. EC, extracellular; IC, intracellular.
Synaptodendritic Damage
Regarding neuronal damage at synapses, the three HIV-1 subtype B isoforms – including the 72, 86, and 101 amino acid length proteins – have all been shown to cause a similar degree of synaptodendritic damage when exposed to primary rat fetal hippocampal neurons in culture [225]. In this cell culture model, synaptodendritic injury was represented by decreased F-actin puncta accompanied by decreased dendritic branching and increased cell death after treatment with Tat (Figure 4) [225]. The number of F-actin puncta present within neuronal dendrites are indicative of the overall capability of a neuron to maintain synapse structure [226]. Therefore, decreased F-actin puncta in hippocampal neuron culture in response to Tat treatment conveys that Tat mediated the disruption of synaptic structure, and likely function, within affected cells. The cysteine-rich domain of Tat has been implicated as essential for the generation of synaptodendritic damage in hippocampal neurons in culture, as deletion of the entire cysteine-rich domain failed to cause the loss of F-actin puncta, decreased dendrite branching, or reduced cell viability that occurred upon exposure to wild-type Tat [225].
Similar to its complete removal, variation within the cysteine-rich domain of subtype B Tat has also been shown to attenuate synaptodendritic damage. The C22G Tat86 variant did not significantly reduce the number of F-actin puncta, affect cell viability, or alter the number of dendrite branches within primary rat fetal hippocampal neurons [225]. Rather, the use of this variant for treatment of neuronal cells in culture generated activity levels similar to controls [225], suggesting that Cys22 in subtype B Tat was essential to produce synaptodendritic injury in this model. Subtype C Tat101, which encodes the naturally-occurring C31S variation, also showed similar abrogation of synaptodendritic injury in cultured hippocampal neurons as the untreated control, as well as the subtype B C22S Tat86 [225]. These results have suggested that the conservation of the cysteine residues within the cysteine-rich domain are necessary for Tat to impart damage to the synaptic terminals of neurons, regardless of the exact location of the variation within the domain and the length of the Tat isoform.
Beta Amyloid Aggregation
Studies concerning the effect of beta amyloid protein (Aβ) aggregation in the CNS have supported its role in neurodegeneration and destabilization of neurons [227–229]. Increased Aβ deposition has been observed in the brains of HIV-1-infected patients when compared to age-matched HIV-1-negative patients, even when treated with ART [230]. Recently, the study of Aβ in the context of Tat-mediated neurotoxicity has revealed important mechanisms of neuronal damage. Beta amyloid concentrations were reported to be significantly increased in the cell bodies of neurons from HIV-1-infected patients in the absence or presence of HIV encephalopathies (HIVE) and were more likely to extend into the axons of neurons from patients with HIVE [231]. The aggregation of Aβ in HIV-1-infected patients is structurally different than that of Alzheimer’s Disease (AD) – while Aβ aggregates in AD form neuritic plaques or tangles, HIV-associated Aβ aggregates lacked these features [231]. In contrast, HIV-1-negative patients showed little to no beta amyloid accumulation in or around CNS neurons [231]. In vitro assays demonstrated that the normal drop in endolysosomal pH was disrupted by HIV-1 Tat72 upon entry into primary rat cortical neurons [232]. This observation preceded Tat72-induced production and accumulation of Aβ within the endolysosomes, leading to their enlargement and dysfunction [232], indicating a possible mechanism of neuronal damage upon Tat72 uptake. Lysosomal dysfunction in neurons has been closely linked to neurodegeneration in Aβ disorders [233,234] and the dysfunction observed upon Tat uptake may prevent the degradation of lysosome contents and promote Aβ aggregation.
Tat has been observed to bind amyloid precursor protein (APP), a large transmembrane protein that is proteolytically cleaved into shorter Aβ peptides within endosomes [235]. The ability of Tat to bind APP was dependent on the presence of the cysteine-rich domain, although cysteine-rich domain variants C22G, H33A, or a C34Q35 deletion mutant had no effect on the Tat-APP interaction [236]. Recent results have supported that Tat could also bind Aβ, an interaction that led to the formation of rigid, aggregating Tat-Aβ fibrils, which generated a synergistic neurotoxic effect in neuronal cell culture (Figure 4) [237]. The exact nature of the Tat-Aβ interaction is not fully understood, but may occur at the external surface of the Aβ fibrils, as predicted from crystal structure analysis [237]. Identification of the interacting residues in both Tat and Aβ is vital for the understanding of this pathogenesis, and if certain Tat variants may exhibit attenuated neurotoxicity for this reason.
Other studies have examined amino acid variation within the cysteine-rich domain of Tat to identify required residues for Tat-mediated beta amyloid production and aggregation. In vitro investigation of hippocampal neuron cultures exposed for three days to subtype B Tat86 showed significant increases in Aβ1–42 in conditioned culture medium as well as decreased cell viability [238]. The introduction of the transactivation-defective C22G variation in this system both restored Aβ1–42 production to control levels and rescued cell viability (Figure 4) [238]. Congo red staining of these cells exposed to subtype B Tat86 demonstrated that as a consequence of Tat treatment, over 20% of cells produced Aβ, while non-treated cells produced very little [238]. Subtype C Tat101 that contained the C31S variation but maintained Cys22, however, stimulated Aβ production in hippocampal neuron cultures that was similar, if not less than, control cultures that were not treated with Tat at all, and was also capable of rescuing cell viability [238]. An interesting takeaway from this study is apparent from the Tat variants used. The ability of subtype B Tat86 to induce Aβ1–42 production was lost after the introduction of the transactivation-defective C22G variation, suggesting that the ability to stimulate LTR transactivation may be linked to increased Aβ production. However, subtype C Tat101 was not able to induce significant concentrations of Aβ1–42 [238], despite prior reports that show the subtype C C31S protein retains LTR transactivation potential [10], but also because subtype C Tat has about 5-fold greater transactivation capacity than subtype B Tat [239]. Similarly, subtype B 101 amino acid Tat isoforms have also been reported to have significantly greater transactivation activity than the 86-amino acid protein [167]. Certainly, it is well-known that HIV-1 subtypes B and C differentially affect cells in culture and within hosts, including the incidence of neurotoxicity, strength of transactivation, and degree of induced cytokine expression [34,240,241]. Granted, the differences in amino acid length between the two forms of Tat could account for the Aβ disparity in this case, though evidence of this remains to be determined.
The presence of Tat in the brain may also modulate intrinsic host factors to prevent the clearance of Aβ from CNS tissues. Neprilysin is a ubiquitously-expressed endopeptidase that is a major host factor responsible for Aβ clearance in the CSF [242]. Inhibition of neprilysin has been shown to increase the accumulation of Aβ in the brain and reduced neprilysin activity may contribute to the generation of extracellular amyloid beta aggregates [242]. Through the investigation of neprilysin inhibition by HIV-1 Tat peptides, the conserved cysteine-rich domain KCCF motif that spans residues 29–32 was identified as a key amino acid stretch responsible for neprilysin-inhibited accumulation of beta amyloid [243]. This was confirmed by a subsequent study, though the same effect was not validated using full-length Tat [244].
Overall, the fact that Tat can interact with amyloidogenic proteins in the CNS is significant to the study of cognitive impairment associated with HIV-1 infection in era of cART. Aβ has been extensively studied for its role in the development of AD and neurodegeneration [229,245,246]. Moreover, if Aβ accumulation in the CNS was responsible for the development of neurocognitive impairment, genetic predispositions for developing AD [247] in the HIV-1-infected patient could potentially increase the risk of neurocognitive impairment. Further investigation of the variation within Tat that dictates the degree of Aβ accumulation and Tat-Aβ complex formation may assist in the diagnosis and treatment of HAND.
VII. Discussion
The study of HIV-1 Tat variants has provided perspective to the study of HIV-1 pathogenesis overall. Variation within Tat may occur at amino acid residues spanning the entire protein and is one component of the overall genetic variation across the HIV-1 genome that is observed during the development of the HIV-1 quasispecies during the course of HIV-1 infection and disease. The complete list of these variants and their effect on known functions is summarized in Table 1. While the properties of each Tat variant may not be fully understood, the range of functions that variation affects are as broad as the functions themselves. This review has focused on specific amino acid changes within Tat and how those alterations have been experimentally shown to shift the functional properties of the protein.
Table 1.
Overview of experimental HIV-1 Tat functional variants
| Tat Residue Variant | Length | Subtype | Functional Alteration(s) | Reference | Year |
|---|---|---|---|---|---|
| Q2A | 86 | B | Trp11 exposure at neutral pH | Yezid, et al | 2009 |
| W11A | 86 | B | Decrease in LTR transactivation | Yezid, et al | 2009 |
| W11L | Decrease in LTR transactivation | Yezid, et al | 2009 | ||
| W11Y | Decrease in LTR transactivation | Yezid, et al | 2009 | ||
| Unable to be secreted | Rayne, et al | 2010 | |||
| W11F | Decrease in LTR transactivation | Yezid, et al | 2009 | ||
| Unable to be secreted | Rayne, et al | 2010 | |||
| H13L | 72 | B | Decreased LTR transactivation; unable to bind p-TEFb | Reza, et al | 2003 |
| S16A | 86 | B | Reduction in posttranslational | Ammosova, et al | 2006 |
| phosphorylation; decrease in LTR transactivation; reduction in viral replication | |||||
| 86 | Not specified | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Tyagi, et al | 2011 | |
| 101 | Not specified | Resistant to phosphorylation by CDK2; decrease in LTR transactivation | Ivanov, et al | 2018 | |
| S16D | 101 | Not specified | Resistant to phosphorylation by CDK2; decrease in LTR transactivation | Ivanov, et al | 2018 |
| S16,46A | 86 | B | Reduction in posttranslational phosphorylation; decrease in LTR transactivation; reduction in viral replication | Ammosova, et al | 2006 |
| C22A | Not specified | Not specified | Ablation of binding to P-TEFb | D’Orso, et al | 2012 |
| 86 | B | Unable to bind PJA2 E3 ubiquitin ligase | Faust, et al | 2017 | |
| C22G | 72 | B | Decrease in LTR transactivation | Reza, et al | 2003 |
| 72, 101 | Not specified | Resistant to ubiquitination by Hdm2 | Bres, et al | 2003 | |
| 86 | B | Reduction in binding efficiency to NF-κB | Dandekar, et al | 2004 | |
| 86 | B | Reduction in neurotoxicity; inability to activate caspase-9, caspase-3, and caspase-7 | Aksenov, et al | 2009 | |
| 86 | B | Restored normal levels of | Aksenov, et | 2010 | |
| amyloid beta production; rescued cell viability | al | ||||
| 101 | Not specified | Reduction of hyperacetylation of the p65 subunit of NF-κB | Kwon, et al | 2008 | |
| 86 | B | Protection from reduction of F-actin puncta; protection from reduction in total dendrite branch number | Bertrand, et al | 2013 | |
| 86 | B | Decrease in LTR transactivation | Rice and Carlotti | 1990 | |
| 86 | B | Decrease in TAR-independent LTR transactivation | Yang, et al | 1997 | |
| Not specified | B | Unable to bind LTR promotor domains | Southgate and Green | 1991 | |
| Not specified | Not specified | Increased apoptosis of Jurkat T cells; decreased Bcl-2 levels | Zauli, et al | 1995 | |
| C22S | 72 | C | Decrease in LTR transactivation | Huet, et al | 1989 |
| 72 | B | Decrease in LTR transactivation | Garcia, et al | 1988 | |
| 86 | B | Reduced synaptodendritic injury | Bertrand, et al | 2013 | |
| C22,25,2 7A | 86 | Not specified | Reduction in HUVEC proliferation and migration; reduction in VEGFR-2 phosphorylation | Mitola, et al | 2000 |
| T23D | 86 | Not specified | Decrease in LTR transactivation | Yoon, et al | 2015 |
| Y26A | 86 | B | Decrease in LTR transactivation; reduction in viral replication | Verhoef, et al | 1997 |
| 72 | B | Decrease in TAR- | Das, et al | 2011 | |
| independent LTR transactivation | |||||
| C27S | 72 | B | Decrease in LTR transactivation | Garcia, et al | 1988 |
| K28Q | 72 | B | Decrease in nuclear import; decrease in LTR transactivation; delayed replication kinetics; reduction in binding efficiency to CycT1; atypical subcellular localization | Bres, et al | 2002 |
| Not specified | Not specified | Decrease in LTR transactivation | D’Orso and Frankel | 2009 | |
| K28R | 72 | B | Decrease in nuclear import; decrease in LTR transactivation; reduction in binding efficiency to CycT1; atypical subcellular localization; reduction in binding efficiency to PCAF | Bres, et al | 2002 |
| 86 | B | Decrease in LTR transactivation | Faust, et al | 2017 | |
| Not specified | Not specified | Reduction in acetylation by PCAF; delayed viral replication kinetics | D’Orso and Frankel | 2009 | |
| C30G | 86 | B | Decrease in LTR transactivation | Rice and Carlotti | 1990 |
| C30S | 72 | C | Reduction in primary PBMC migration; decrease in LTR transactivation | Ranga, et al | 2004 |
| C31G | 86 | B | Decrease in LTR transactivation | Rice and Carlotti | 1990 |
| 72 | B | Decrease in LTR transactivation | Reza, et al | 2003 | |
| C31S | 72 | C | Reduction in primary | Ranga, et al | 2004 |
| PBMC migration | |||||
| 72 | C | Protection from Tat-mediated neurotoxicity | Li, et al | 2008 | |
| 101 | C | Restored normal levels of amyloid beta production; restored cell viability | Aksenov, et al | 2010 | |
| 101 | C | Reduced synaptodendritic injury | Bertrand, et al | 2013 | |
| C31H | Not specified | B | Decrease in LTR transactivation | Sadaie, et al | 1990 |
| W32G | 101 | E | Restoration of P/CAF and GCN5 HAT chromatin remodeling; restored expression of TNF genes | Ranjbar, et al | 2006 |
| H33A | Not specified | B | Decrease in TAR-independent LTR transactivation | Southgate and Green | 1991 |
| 86 | B | Decrease in LTR transactivation | Rice and Carlotti | 1990 | |
| C34H | Not specified | B | Decrease in LTR transactivation | Sadaie, et al | 1990 |
| C34,37H | Not specified | B | Decrease in LTR transactivation | Sadaie, et al | 1990 |
| C34S | 72 | B | Decrease in LTR transactivation | Garcia, et al | 1988 |
| C34G | 67 | B | Decrease in LTR transactivation | Kuppuswam y, et al | 1989 |
| Not specified | B | Decreased protein stability | Sadaie, et al | 1990 | |
| Q35A | 86 | B | Decrease in TAR-independent LTR transactivation | Yang, et al | 1997 |
| 36,37,38, 39A | Not specified | B | Reduction in binding efficiency to tubulin; unable to activate | Chen, et al | 2002 |
| caspase-9 | |||||
| C37H | Not specified | B | Decrease in LTR transactivation | Sadaie, et al | 1990 |
| C37G | 67 | B | Decrease in LTR transactivation | Kuppuswam y, et al | 1989 |
| F38A | 101 | Not specified | Reduction of hyperacetylation of the p65 subunit of NF-κB | Kwon, et al | 2008 |
| Not specified | B | Decrease in TAR-independent LTR transactivation | Southgate and Green | 1991 | |
| 86 | B | Decrease in LTR transactivation | Rice and Carlotti | 1990 | |
| T40D | 86 | Not specified | Decrease in LTR transactivation | Yoon, et al | 2015 |
| K41A | 101 | Not specified | Reduction of hyperacetylation of the p65 subunit of NF-κB; increase the expression of IL-2 | Kwon, et al | 2008 |
| 72 | B | Decrease in nuclear import; decrease in LTR transactivation; reduction in binding to CycT1; reduction in binding efficiency to PCAF | Bres, et al | 2002 | |
| Not specified | Not specified | Decrease in efficiency of p-TEFb binding | D’Orso, et al | 2012 | |
| Not specified | B | Decrease in TAR-independent LTR transactivation | Southgate and Green | 1991 | |
| 86 | B | Decrease in LTR transactivation | Rice and Carlotti | 1990 | |
| K41Q | 101 | Not specified | Increase in nuclear localization | He, et al | 2013 |
| K41R | 86 | B | Reduction in binding efficiency to BRG1; decrease in LTR transactivation | Agbottah, et al | 2006 |
| 86 | B | Decrease in LTR transactivation; resistant to ubiquitination by PJA2 | Faust, et al | 2017 | |
| 101 | Not specified | Decreased nuclear localization | He, et al | 2013 | |
| K41T | 101 | Not specified | Decrease in LTR transactivation | El Kharroubi, et al | 1998 |
| S46A | 86 | B | Reduction in posttranslational phosphorylation; decrease in LTR transactivation; reduction in viral replication | Ammosova, et al | 2006 |
| 101 | Not specified | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Ivanov, et al | 2018 | |
| S46D | 86 | Not specified | Decrease in LTR transactivation | Yoon, et al | 2015 |
| 101 | Not specified | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Ivanov, et al | 2018 | |
| Y47A | 101 | Not specified | Reduction in binding efficiency to PCAF; decrease in LTR transactivation | Dorr, et al | 2002 |
| R49,53A | 72 | B | Decrease in LTR transactivation | Xie, et al | 2007 |
| R49G,K50I | 86 | Not specified | Reduction in HUVEC proliferation and migration; reduction in | Mitola, et al | 2000 |
| VEGFR-2 phosphorylation | |||||
| R49G,K5 0I,R52L, R53I | 86 | Not specified | Reduction in HUVEC proliferation and migration; reduction in VEGFR-2 phosphorylation | Mitola, et al | 2000 |
| K50A | 101 | Not specified | Increase in expression of CRLF2, BDNF, APBA1; decrease in expression of C5 | Carvallo, et al | 2017 |
| 101 | Not specified | Decrease in LTR transactivation | Ott, et al | 1999 | |
| 86 | Not specified | Resistant to methylation by SETDB1 | Van Duyne, et al | 2008 | |
| K50R | 101 | Not specified | Decrease in LTR transactivation | Dorr, et al | 2002 |
| 72 | B | Increase in nuclear localization; reduction in viral replication | Bres, et al | 2002 | |
| 101 | Not specified | Decrease in LTR transactivation; reduction in binding efficiency to CycT1; resistant to acetylation by p300/CBP | Ott, et al | 1999 | |
| 86 | B | Reduction in binding efficiency to BRG1; decrease in LTR transactivation | Agbottah, et al | 2006 | |
| K50Q | 72 | B | Decrease in nuclear import; decrease in LTR transactivation | Bres, et al | 2002 |
| K50,51A | 86 | Not specified | Resistant to methylation by SETDB1 | Van Duyne, et al | 2008 |
| K50,51R | 101 | Not | Decrease in LTR | Ott, et al | 1999 |
| specified | transactivation | ||||
| K50S,K5 1G | 101 | B | Reduced RNA binding efficiency | Bouwman, et al | 2014 |
| K51A | 86 | Not specified | Resistant to methylation by SETDB1 | Van Duyne, et al | 2008 |
| K51R | 72 | Not specified | Decrease in LTR transactivation | Ali, et al | 2016 |
| 86 | B | Reduction in binding efficiency to BRG1; decrease in LTR transactivation | Agbottah, et al | 2006 | |
| 101 | Not specified | Decrease in LTR transactivation | Dorr, et al | 2002 | |
| K51,71R | 72 | Not specified | Decrease in LTR transactivation | Ali, et al | 2016 |
| 52GGQGGG57 | 72 | Not specified | Inhibition of association with nucleophosmin; decreased nuclear localization | Li, et al | 1997 |
| R52K | 72 | B | Decrease in LTR transactivation | Xie, et al | 2007 |
| 101 | Not specified | Resistant to methylation by PRMT6 | Fulcher, et al | 2016 | |
| R52,53K | 72 | B | Decrease in LTR transactivation | Xie, et al | 2007 |
| R53A | 101 | Not specified | Reduction in binding efficiency to PCAF; decrease in LTR transactivation | Dorr, et al | 2002 |
| R53K | 101 | Not specified | Resistant to methylation by PRMT7 | Fulcher, et al | 2016 |
| R53E | 101 | Not | Reduction in binding | Dorr, et al | 2002 |
| specified | efficiency to PCAF | ||||
| 55AAA57 | 86 | B | Trp11 exposure at neutral pH | Yezid, et al | 2009 |
| S62A | 86 | B | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Endo-Munoz, et al | 2005 |
| 86 | Not specified | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Tyagi, et al | 2011 | |
| S62D | 86 | Not specified | Decrease in LTR transactivation | Yoon, et al | 2015 |
| S62,64,68 A | 86 | B | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Endo-Munoz, et al | 2005 |
| T64A | 86 | B | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Endo-Munoz, et al | 2005 |
| S68A | 86 | B | Resistant to phosphorylation by PKR; decrease in LTR transactivation | Endo-Munoz, et al | 2005 |
| S68D | 86 | Not specified | Decrease in LTR transactivation | Yoon, et al | 2015 |
| K71A | 72 | Not specified | Resistant to methylation by KMT7 | Ali, et al | 2016 |
| K71R | 72 | Not specified | Decrease in LTR transactivation; resistant to methylation by KMT7 | Ali, et al | 2016 |
| 72, 101 | Not specified | Decrease in LTR transactivation, resistant to ubiquitination by | Bres, et al | 2003 | |
| Hdm2 | |||||
| Q78R | Not specified | C | Increased apoptosis of THP-1 cells | Sood, et al | 2008 |
| R78K/D8 0E | 86 | B | Reduced HUVEC proliferation, migration, and adhesion to extracellular matrix | Mitola, et al | 2000 |
| K88R | 101 | Not specified | Decrease in nuclear localization | He, et al | 2013 |
| K89R | 101 | Not specified | Increase in nuclear localization | He, et al | 2013 |
| K88,89,9 0A | 101 | B | Decrease in TAR-independent LTR transactivation | Mahlknecht, et al | 2008 |
| K90R | 101 | Not specified | Decrease in nuclear localization | He, et al | 2013 |
| E92,94,9 6A | 101 | B | Decrease in TAR-independent LTR transactivation | Mahlknecht, et al | 2008 |
Tat trafficking, potential for LTR transactivation, and mediation of pathogenicity all provide context for the impact of genetic variation within the protein. Future studies of Tat-mediated pathogenesis should incorporate variants, as their use may provide some mechanistic understanding, since the domains of Tat have been well-annotated through the years. Yet despite the reinforcement that each piece of validating data may add to the growing body of literature on Tat’s functions, the full gamut of activities assigned to Tat will obviously continue to increase. The tendency of HIV-1 to develop into genetically unique quasispecies dictates that Tat should also change in genetic composition. The selective pressures for these changes are not totally resolved, but may include CD8+ T-cell epitope escape [248,249] and the gradual shift between HIV-1 co-receptors [250] as well as other pressures associated with therapy, the immune response, and others.
Retrospective analysis of the effects of Tat variants identified in the literature, however, is often complicated by the omission of Tat protein length. Protein length disclosure, in these cases, have been an important aspect of data interpretation, as the use of one-exon Tat versus the two-exon protein may mean a difference of 14 to 29 amino acids in Tat86 and Tat101, respectively. Viral subtype utilized also adds a layer of difficulty to the study of Tat variability, due to the presence of amino acid determinants that are unique to each subtype. Much of HIV-1 research occurs in the western hemisphere where subtype B is most common, leaving other subtypes underexplored despite higher incidence globally. Because these studies have shown that Tat from different HIV-1 subtypes displays different functional characteristics, results from studies on single subtypes may not be applicable to the understanding of the remaining viral subtypes.
Additionally, the half-life of functional Tat variants should be considered in future studies of Tat genetic variation. Though the pool of knowledge of Tat variant half-life has yet to develop, it could provide the means for important reevaluations of functional discrepancies between wild-type and variant functionality. For example, a study that analyzed differences in half-life between Tat of certain HIV-1 subtypes found that in [35S]Met/Cys pulse-chase experiments of HEK 293 cells transfected with subtype B, C, or E Tat constructs, subtype B and C Tat was shown to have a half-life of about 3 hours, while subtype E Tat had a half-life of over 6 hours [251]. This result was concurrent with subtype E Tat’s increased LTR transactivation capacity [251], suggesting that there could be genetic variability in subtype E Tat that differentiates it from subtype B or C Tat, affecting its half-life, and subsequently altering its functional efficiency. Though no specific amino acid variants were implicated in the half-life disparity observed in this study, it presents an interesting avenue for future research.
Another factor that needs to be considered in the interpretation of studies using Tat variants is the model system, especially in in vitro studies. Because in vitro studies often utilize cell lines to investigate the effects of Tat genetic variation on Tat function, it is important that the model system is able to simulate the likely conditions of in vivo conditions. For example, several HeLa-based cell lines, such as HLM-1, HL3T1, and HeLa P4 reporter cells, have been used to study Tat functional variation, mainly with respect to subcellular localization or LTR transactivation [56,70,92,97,122]. The use of HeLa-based cells, however, does come with caveats, as HeLa cells have been shown to exhibit chromosomal aberrations, abnormal cell biology, and dysregulated protein expression [252,253]. These cellular characteristics may not allow the accurate recapitulation of Tat expression in primary cell models or in vivo, potentially biasing the results of some studies. The Jurkat T-cell line has also been frequently used in Tat genetic variation studies [65,79,87,139,149,162,209], though a thorough analysis of changes in gene expression between the Jurkat T-cell line and primary CD4+ T cells with respect to HIV-1 Tat expression has not been done. More recently, primary cell culture models have been used to study the effects of Tat genetic variation [7,141,156,198,225,232]. Primary cell culture models provide the advantage of cell physiology more closely related to that of in vivo systems, though many of the effects seen in cell lines have not been corroborated in primary cell experiments, especially with regard to LTR transactivation data, levels of Tat produced in transfected cells, and post-translational modification patterns. Overall, much of the literature on functional Tat variants has utilized cell lines, likely because of their ease of use in culture and in transfection experiments. Although cell line-based studies have provided valuable information about Tat function, many of these functions have yet to be verified in primary cell culture, in in vivo models, such as rodent models, or in patient populations.
Control of HIV-1 replication in infected patients has been dramatically improved since the advent of ART, although a functional cure of HIV-1 infection may be possible through the treatments directed against Tat itself. Recently, the discovery and exploration of Tat inhibitor, didehydro-cortistatin A (dCA) has stimulated the discussion regarding Tat as a drug target. dCA has, most notably, been proclaimed as preventing the reactivation of HIV-1 from viral latency [117]. As Tat could be involved in low-level viral transcription in patients on ART (particularly in viral reservoirs), dCA inhibition of Tat activity to inhibit low level viral production could be an important component to the next generation of therapies for the treatment of HIV-1-infected individuals. Genetic variation of Tat, however, may present a unique obstacle, as dCA binds to the arginine-rich domain to prevent Tat-TAR interactions [254]. Although specific natural Tat variants that prevent dCA binding have not yet been encountered, it is possible that the overall quasispecies present in HIV-1-infected individuals may be pools for the expansion of pre-existing or newly generated dCA resistant quasispecies during the course of dCA therapy.
As Tat is essential for the productive replication of the HIV-1 proviral genome, it is produced early on in infection, and acts aggressively to recruit host factors to enhance viral transcription. In a system where viral transcription has been suppressed by ART, however, Tat functional activity still persists and is apparent in the development of HIV-1-associated neurocognitive dysfunction. Tat-associated toxicity is an important area of research in neuroscience, immunology, and viral pathogenesis that will benefit greatly from the continued genetic, structural, and functional analysis of patient-derived Tat sequences. However, basic discovery, translational, and clinical studies are not always geared toward analyzing genetic variation of quasispecies and HIV proteins within each patient. The BEEHIVE, Drexel CARES cohort, and several other studies, however, have focused on longitudinal analyses of HIV-1 genetic variation and evolution within individuals [17,27]. Results from these studies have provided valuable insight into viral genome structure, function, pathogenesis, and evolution/adaptation and should serve as important resources for future studies of variation within HIV-1 proteins as well as in the development of new vaccine and therapeutic strategies.
Vpr is another HIV-1 accessory protein that has been implicated as a causative factor for the development of HAND [255,256]. Just as Tat is subject to mutation during replication and selection, the Vpr coding sequence may vary as HIV-1 infection progresses. As such, Vpr derived from patient HIV-1 sequences have also been studied for genetic heterogeneity that affects its functionality. Specific genetic determinants in blood- and brain-derived Vpr, such as Ile37 and Ser41, have been correlated with more severe neurocognitive deficits in patient populations [257], which emphasizes the detrimental impact of certain genetic variants throughout the HIV-1 genome on CNS function. Vpr has been determined to be a cis-acting element produced by HIV-1 [258] and its likeness to Tat in pathogenic function suggest that variation within either protein may enhance or deplete neuropathogenesis in an additive manner. Studies concerning the effect of co-linear variation in both Tat and Vpr function have not yet been published, but may provide genetic signatures to help explain differential neurocognitive comorbidity in HIV-1-infected patients.
Overall, HIV-1 Tat represents a significant hurdle in the pursuit of a functional cure for HIV-1, as well as for the study of HAND disease progression. Recent evidence of Tat interactions with amyloid beta in the CNS represents an exciting area of investigation, as research on amyloid beta in the context of AD has generated several therapeutic approaches that may be useful in the treatment of HAND that is characterized by Aβ accumulation [259,260]. Genetic heterogeneity of Tat, however, may impact the potential efficacy of AD treatments on HIV-associated neurocognitive impairment, as several of them are monoclonal antibody therapies that target Aβ and may be blocked by the interaction of Tat with Aβ fibrils. Broadening our knowledge of Tat genetic architecture and function will be one of many critical areas of research in our quest to develop tomorrow’s diagnostics, vaccines, and therapeutic agents.
VIII. Acknowledgements
Funding: The authors were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke (NINDS) R01 NS089435 (PI, Michael R. Nonnemacher), the NIMH Comprehensive NeuroAIDS Center (CNAC) P30 MH092177 (Kamel Khalili, PI; Brian Wigdahl, PI of the Drexel subcontract involving the Clinical and Translational Research Support Core) and under the Ruth L. Kirschstein National Research Service Award T32 MH079785 (PI, Jay Rappaport; with Brian Wigdahl serving as the PI of the Drexel University College of Medicine component and Olimpia Meucci as Co-Director). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Disclosure of Conflict of Interest: The authors declare that they have no conflict of interest.
Literature Cited
- 1.Rana TM, Jeang KT (1999) Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch Biochem Biophys 365 (2):175–185. doi: 10.1006/abbi.1999.1206 [DOI] [PubMed] [Google Scholar]
- 2.Li L, Dahiya S, Kortagere S, Aiamkitsumrit B, Cunningham D, Pirrone V, Nonnemacher MR, Wigdahl B (2012) Impact of Tat Genetic Variation on HIV-1 Disease. Adv Virol 2012:123605. doi: 10.1155/2012/123605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu J, Babayeva ND, Suwa Y, Baranovskiy AG, Price DH, Tahirov TH (2014) Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4. Cell Cycle 13 (11):1788–1797. doi: 10.4161/cc.28756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price DH (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol 20 (8):2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeang KT, Xiao H, Rich EA (1999) Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem 274 (41):28837–28840 [DOI] [PubMed] [Google Scholar]
- 6.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, et al. (1985) Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313 (6000):277–284 [DOI] [PubMed] [Google Scholar]
- 7.Rayne F, Debaisieux S, Yezid H, Lin YL, Mettling C, Konate K, Chazal N, Arold ST, Pugniere M, Sanchez F, Bonhoure A, Briant L, Loret E, Roy C, Beaumelle B (2010) Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J 29 (8):1348–1362. doi: 10.1038/emboj.2010.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koken SE, Greijer AE, Verhoef K, van Wamel J, Bukrinskaya AG, Berkhout B (1994) Intracellular analysis of in vitro modified HIV Tat protein. J Biol Chem 269 (11):8366–8375 [PubMed] [Google Scholar]
- 9.Pierleoni R, Menotta M, Antonelli A, Sfara C, Serafini G, Dominici S, Laguardia ME, Salis A, Damonte G, Banci L, Porcu M, Monini P, Ensoli B, Magnani M (2010) Effect of the redox state on HIV-1 tat protein multimerization and cell internalization and trafficking. Mol Cell Biochem 345 (1–2):105–118. doi: 10.1007/s11010-010-0564-9 [DOI] [PubMed] [Google Scholar]
- 10.Ranga U, Shankarappa R, Siddappa NB, Ramakrishna L, Nagendran R, Mahalingam M, Mahadevan A, Jayasuryan N, Satishchandra P, Shankar SK, Prasad VR (2004) Tat protein of human immunodeficiency virus type 1 subtype C strains is a defective chemokine. J Virol 78 (5):2586–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92 (4):451–462 [DOI] [PubMed] [Google Scholar]
- 12.Marzio G, Tyagi M, Gutierrez MI, Giacca M (1998) HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A 95 (23):13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeang KT, Chun R, Lin NH, Gatignol A, Glabe CG, Fan H (1993) In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol 67 (10):6224–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauber J, Malim MH, Cullen BR (1989) Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol 63 (3):1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukerjee R, Sawaya BE, Khalili K, Amini S (2007) Association of p65 and C/EBPbeta with HIV-1 LTR modulates transcription of the viral promoter. J Cell Biochem 100 (5):1210–1216. doi: 10.1002/jcb.21109 [DOI] [PubMed] [Google Scholar]
- 16.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R, Haseltine WA, Rosen CA (1989) Structural and functional characterization of human immunodeficiency virus tat protein. J Virol 63 (1):1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Kuyl AC, Vink M, Zorgdrager F, Bakker M, Wymant C, Hall M, Gall A, Blanquart F, Berkhout B, Fraser C, Cornelissen M, Collaboration B (2018) The evolution of subtype B HIV-1 tat in the Netherlands during 1985–2012. Virus Res 250:51–64. doi: 10.1016/j.virusres.2018.04.008 [DOI] [PubMed] [Google Scholar]
- 18.Neuveut C, Scoggins RM, Camerini D, Markham RB, Jeang KT (2003) Requirement for the second coding exon of Tat in the optimal replication of macrophage-tropic HIV-1. J Biomed Sci 10 (6 Pt 1):651–660. doi: 10.1159/000073531 [DOI] [PubMed] [Google Scholar]
- 19.Kukkonen S, Martinez-Viedma Mdel P, Kim N, Manrique M, Aldovini A (2014) HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology 11:30. doi: 10.1186/1742-4690-11-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Huertas MR, Mateos E, Sanchez Del Cojo M, Gomez-Esquer F, Diaz-Gil G, Rodriguez-Mora S, Lopez JA, Calvo E, Lopez-Campos G, Alcami J, Coiras M (2013) The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4+ T lymphocytes: a potential mechanism for persistent viral production. J Biol Chem 288 (11):7626–7644. doi: 10.1074/jbc.M112.408294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aiamkitsumrit B, Dampier W, Martin-Garcia J, Nonnemacher MR, Pirrone V, Ivanova T, Zhong W, Kilareski E, Aldigun H, Frantz B, Rimbey M, Wojno A, Passic S, Williams JW, Shah S, Blakey B, Parikh N, Jacobson JM, Moldover B, Wigdahl B (2014) Defining differential genetic signatures in CXCR4- and the CCR5-utilizing HIV-1 co-linear sequences. PLoS One 9 (9):e107389. doi: 10.1371/journal.pone.0107389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiodelli P, Urbinati C, Mitola S, Tanghetti E, Rusnati M (2012) Sialic acid associated with alphavbeta3 integrin mediates HIV-1 Tat protein interaction and endothelial cell proangiogenic activation. J Biol Chem 287 (24):20456–20466. doi: 10.1074/jbc.M111.337139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urbinati C, Mitola S, Tanghetti E, Kumar C, Waltenberger J, Ribatti D, Presta M, Rusnati M (2005) Integrin alphavbeta3 as a target for blocking HIV-1 Tat-induced endothelial cell activation in vitro and angiogenesis in vivo. Arterioscler Thromb Vasc Biol 25 (11):2315–2320. doi: 10.1161/01.ATV.0000186182.14908.7b [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Huertas MR, Callejas S, Abia D, Mateos E, Dopazo A, Alcami J, Coiras M (2010) Modifications in host cell cytoskeleton structure and function mediated by intracellular HIV-1 Tat protein are greatly dependent on the second coding exon. Nucleic Acids Res 38 (10):3287–3307. doi: 10.1093/nar/gkq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canducci F, Marinozzi MC, Sampaolo M, Berre S, Bagnarelli P, Degano M, Gallotta G, Mazzi B, Lemey P, Burioni R, Clementi M (2009) Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects. Retrovirology 6:4. doi: 10.1186/1742-4690-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee SY, Fessel WJ, Zolopa AR, Hurley L, Liu T, Taylor J, Nguyen DP, Slome S, Klein D, Horberg M, Flamm J, Follansbee S, Schapiro JM, Shafer RW (2005) HIV-1 Protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J Infect Dis 192 (3):456–465. doi: 10.1086/431601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dampier W, Nonnemacher MR, Mell J, Earl J, Ehrlich GD, Pirrone V, Aiamkitsumrit B, Zhong W, Kercher K, Passic S, Williams JW, Jacobson JM, Wigdahl B (2016) HIV-1 Genetic Variation Resulting in the Development of New Quasispecies Continues to Be Encountered in the Peripheral Blood of Well-Suppressed Patients. PLoS One 11 (5):e0155382. doi: 10.1371/journal.pone.0155382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy CN, Khandaker I, Oshitani H (2015) Intersubtype Genetic Variation of HIV-1 Tat Exon 1. AIDS Res Hum Retroviruses 31 (6):641–648. doi: 10.1089/AID.2014.0346 [DOI] [PubMed] [Google Scholar]
- 29.Roy CN, Khandaker I, Oshitani H (2015) Evolutionary Dynamics of Tat in HIV-1 Subtypes B and C. PLoS One 10 (6):e0129896. doi: 10.1371/journal.pone.0129896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Aiamkitsumrit B, Pirrone V, Nonnemacher MR, Wojno A, Passic S, Flaig K, Kilareski E, Blakey B, Ku J, Parikh N, Shah R, Martin-Garcia J, Moldover B, Servance L, Downie D, Lewis S, Jacobson JM, Kolson D, Wigdahl B (2011) Development of co-selected single nucleotide polymorphisms in the viral promoter precedes the onset of human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol 17 (1):92–109. doi: 10.1007/s13365-010-0014-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Li J, Kim BO, Pace BS, He JJ (2002) HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J Biol Chem 277 (26):23854–23863. doi: 10.1074/jbc.M200773200 [DOI] [PubMed] [Google Scholar]
- 32.Rossenkhan R, MacLeod IJ, Sebunya TK, Castro-Nallar E, McLane MF, Musonda R, Gashe BA, Novitsky V, Essex M (2013) tat Exon 1 exhibits functional diversity during HIV-1 subtype C primary infection. J Virol 87 (10):5732–5745. doi: 10.1128/JVI.03297-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell GR, Loret EP, Spector SA (2010) HIV-1 clade B Tat, but not clade C Tat, increases X4 HIV-1 entry into resting but not activated CD4+ T cells. J Biol Chem 285 (3):1681–1691. doi: 10.1074/jbc.M109.049957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra M, Vetrivel S, Siddappa NB, Ranga U, Seth P (2008) Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: significance of dicysteine C30C31 motif. Ann Neurol 63 (3):366–376. doi: 10.1002/ana.21292 [DOI] [PubMed] [Google Scholar]
- 35.Tyor W, Fritz-French C, Nath A (2013) Effect of HIV clade differences on the onset and severity of HIV-associated neurocognitive disorders. J Neurovirol 19 (6):515–522. doi: 10.1007/s13365-013-0206-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah S, Alexaki A, Pirrone V, Dahiya S, Nonnemacher MR, Wigdahl B (2014) Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I. Virol J 11:92. doi: 10.1186/1743-422X-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B (2009) Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology 6:118. doi: 10.1186/1742-4690-6-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burdo TH, Nonnemacher M, Irish BP, Choi CH, Krebs FC, Gartner S, Wigdahl B (2004) High-affinity interaction between HIV-1 Vpr and specific sequences that span the C/EBP and adjacent NF-kappaB sites within the HIV-1 LTR correlate with HIV-1-associated dementia. DNA Cell Biol 23 (4):261–269. doi: 10.1089/104454904773819842 [DOI] [PubMed] [Google Scholar]
- 39.Maubert ME, Pirrone V, Rivera NT, Wigdahl B, Nonnemacher MR (2015) Interaction between Tat and Drugs of Abuse during HIV-1 Infection and Central Nervous System Disease. Front Microbiol 6:1512. doi: 10.3389/fmicb.2015.01512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clifford DB (2017) HIV-associated neurocognitive disorder. Curr Opin Infect Dis 30 (1):117–122. doi: 10.1097/QCO.0000000000000328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaul M, Garden GA, Lipton SA (2001) Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410 (6831):988–994. doi: 10.1038/35073667 [DOI] [PubMed] [Google Scholar]
- 42.Dahiya S, Irish BP, Nonnemacher MR, Wigdahl B (2013) Genetic variation and HIV-associated neurologic disease. Adv Virus Res 87:183–240. doi: 10.1016/B978-0-12-407698-3.00006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andras IE, Pu H, Deli MA, Nath A, Hennig B, Toborek M (2003) HIV-1 Tat protein alters tight junction protein expression and distribution in cultured brain endothelial cells. J Neurosci Res 74 (2):255–265. doi: 10.1002/jnr.10762 [DOI] [PubMed] [Google Scholar]
- 44.Albini A, Ferrini S, Benelli R, Sforzini S, Giunciuglio D, Aluigi MG, Proudfoot AE, Alouani S, Wells TN, Mariani G, Rabin RL, Farber JM, Noonan DM (1998) HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci U S A 95 (22):13153–13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pu H, Tian J, Flora G, Lee YW, Nath A, Hennig B, Toborek M (2003) HIV-1 Tat protein upregulates inflammatory mediators and induces monocyte invasion into the brain. Mol Cell Neurosci 24 (1):224–237 [DOI] [PubMed] [Google Scholar]
- 46.Agrawal L, Louboutin JP, Reyes BA, Van Bockstaele EJ, Strayer DS (2012) HIV-1 Tat neurotoxicity: a model of acute and chronic exposure, and neuroprotection by gene delivery of antioxidant enzymes. Neurobiol Dis 45 (2):657–670. doi: 10.1016/j.nbd.2011.10.005 [DOI] [PubMed] [Google Scholar]
- 47.Badou A, Bennasser Y, Moreau M, Leclerc C, Benkirane M, Bahraoui E (2000) Tat protein of human immunodeficiency virus type 1 induces interleukin-10 in human peripheral blood monocytes: implication of protein kinase C-dependent pathway. J Virol 74 (22):10551–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brady J, Kashanchi F (2005) Tat gets the “green” light on transcription initiation. Retrovirology 2:69. doi: 10.1186/1742-4690-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cullen BR (1991) Regulation of HIV-1 gene expression. FASEB J 5 (10):2361–2368 [DOI] [PubMed] [Google Scholar]
- 50.Feinberg MB, Baltimore D, Frankel AD (1991) The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A 88 (9):4045–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell DF, Martin MA (1993) Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67 (11):6365–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truant R, Cullen BR (1999) The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol 19 (2):1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efthymiadis A, Briggs LJ, Jans DA (1998) The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J Biol Chem 273 (3):1623–1628 [DOI] [PubMed] [Google Scholar]
- 54.Cardarelli F, Serresi M, Bizzarri R, Beltram F (2008) Tuning the transport properties of HIV-1 Tat arginine-rich motif in living cells. Traffic 9 (4):528–539. doi: 10.1111/j.1600-0854.2007.00696.x [DOI] [PubMed] [Google Scholar]
- 55.Smith KM, Himiari Z, Tsimbalyuk S, Forwood JK (2017) Structural Basis for Importin-alpha Binding of the Human Immunodeficiency Virus Tat. Sci Rep 7 (1):1650. doi: 10.1038/s41598-017-01853-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bres V, Kiernan R, Emiliani S, Benkirane M (2002) Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J Biol Chem 277 (25):22215–22221. doi: 10.1074/jbc.M201895200 [DOI] [PubMed] [Google Scholar]
- 57.Fulcher AJ, Sivakumaran H, Jin H, Rawle DJ, Harrich D, Jans DA (2016) The protein arginine methyltransferase PRMT6 inhibits HIV-1 Tat nucleolar retention. Biochim Biophys Acta 1863 (2):254–262. doi: 10.1016/j.bbamcr.2015.11.019 [DOI] [PubMed] [Google Scholar]
- 58.Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G (1989) Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res 17 (9):3551–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orsini MJ, Debouck CM (1996) Inhibition of human immunodeficiency virus type 1 and type 2 Tat function by transdominant Tat protein localized to both the nucleus and cytoplasm. J Virol 70 (11):8055–8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He M, Zhang L, Wang X, Huo L, Sun L, Feng C, Jing X, Du D, Liang H, Liu M, Hong Z, Zhou J (2013) Systematic Analysis of the Functions of Lysine Acetylation in the Regulation of Tat Activity. PLoS One 8 (6):e67186. doi: 10.1371/journal.pone.0067186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ott M, Schnolzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr Biol 9 (24):1489–1492 [DOI] [PubMed] [Google Scholar]
- 62.D’Orso I, Frankel AD (2009) Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc Natl Acad Sci U S A 106 (9):3101–3106. doi: 10.1073/pnas.0900012106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grisendi S, Mecucci C, Falini B, Pandolfi PP (2006) Nucleophosmin and cancer. Nat Rev Cancer 6 (7):493–505. doi: 10.1038/nrc1885 [DOI] [PubMed] [Google Scholar]
- 64.Li YP (1997) Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J Virol 71 (5):4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marasco WA, Szilvay AM, Kalland KH, Helland DG, Reyes HM, Walter RJ (1994) Spatial association of HIV-1 tat protein and the nucleolar transport protein B23 in stably transfected Jurkat T-cells. Arch Virol 139 (1–2):133–154 [DOI] [PubMed] [Google Scholar]
- 66.Gadad SS, Rajan RE, Senapati P, Chatterjee S, Shandilya J, Dash PK, Ranga U, Kundu TK (2011) HIV-1 infection induces acetylation of NPM1 that facilitates Tat localization and enhances viral transactivation. J Mol Biol 410 (5):997–1007. doi: 10.1016/j.jmb.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 67.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, Richard S (2005) Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol 79 (1):124–131. doi: 10.1128/JVI.79.1.124-131.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie B, Invernizzi CF, Richard S, Wainberg MA (2007) Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat Interactions with both cyclin T1 and the Tat transactivation region. J Virol 81 (8):4226–4234. doi: 10.1128/JVI.01888-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT (2002) The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem 277 (5):3537–3543. doi: 10.1074/jbc.M108786200 [DOI] [PubMed] [Google Scholar]
- 70.Yoon CH, Kim SY, Byeon SE, Jeong Y, Lee J, Kim KP, Park J, Bae YS (2015) p53-derived host restriction of HIV-1 replication by protein kinase R-mediated Tat phosphorylation and inactivation. J Virol 89 (8):4262–4280. doi: 10.1128/JVI.03087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL (2010) The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA 1 (3):415–431. doi: 10.1002/wrna.39 [DOI] [PubMed] [Google Scholar]
- 72.Ponti D, Troiano M, Bellenchi GC, Battaglia PA, Gigliani F (2008) The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol 9:32. doi: 10.1186/1471-2121-9-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jarboui MA, Bidoia C, Woods E, Roe B, Wynne K, Elia G, Hall WW, Gautier VW (2012) Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus. PLoS One 7 (11):e48702. doi: 10.1371/journal.pone.0048702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136 (4):731–745. doi: 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider RJ, Shenk T (1987) Impact of virus infection on host cell protein synthesis. Annu Rev Biochem 56:317–332. doi: 10.1146/annurev.bi.56.070187.001533 [DOI] [PubMed] [Google Scholar]
- 76.Bushell M, Sarnow P (2002) Hijacking the translation apparatus by RNA viruses. J Cell Biol 158 (3):395–399. doi: 10.1083/jcb.200205044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Debaisieux S, Rayne F, Yezid H, Beaumelle B (2012) The ins and outs of HIV-1 Tat. Traffic 13 (3):355–363. doi: 10.1111/j.1600-0854.2011.01286.x [DOI] [PubMed] [Google Scholar]
- 78.Vendeville A, Rayne F, Bonhoure A, Bettache N, Montcourrier P, Beaumelle B (2004) HIV-1 Tat enters T cells using coated pits before translocating from acidified endosomes and eliciting biological responses. Mol Biol Cell 15 (5):2347–2360. doi: 10.1091/mbc.E03-12-0921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yezid H, Konate K, Debaisieux S, Bonhoure A, Beaumelle B (2009) Mechanism for HIV-1 Tat insertion into the endosome membrane. J Biol Chem 284 (34):22736–22746. doi: 10.1074/jbc.M109.023705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pantano S, Tyagi M, Giacca M, Carloni P (2002) Amino acid modification in the HIV-1 Tat basic domain: insights from molecular dynamics and in vivo functional studies. J Mol Biol 318 (5):1331–1339 [DOI] [PubMed] [Google Scholar]
- 81.Tyagi M, Rusnati M, Presta M, Giacca M (2001) Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J Biol Chem 276 (5):3254–3261. doi: 10.1074/jbc.M006701200 [DOI] [PubMed] [Google Scholar]
- 82.Mere J, Morlon-Guyot J, Bonhoure A, Chiche L, Beaumelle B (2005) Acid-triggered membrane insertion of Pseudomonas exotoxin A involves an original mechanism based on pH-regulated tryptophan exposure. J Biol Chem 280 (22):21194–21201. doi: 10.1074/jbc.M412656200 [DOI] [PubMed] [Google Scholar]
- 83.De Matteis MA, Godi A (2004) PI-loting membrane traffic. Nat Cell Biol 6 (6):487–492. doi: 10.1038/ncb0604-487 [DOI] [PubMed] [Google Scholar]
- 84.Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443 (7112):651–657. doi: 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 85.Boven LA, Noorbakhsh F, Bouma G, van der Zee R, Vargas DL, Pardo C, McArthur JC, Nottet HS, Power C (2007) Brain-derived human immunodeficiency virus-1 Tat exerts differential effects on LTR transactivation and neuroimmune activation. J Neurovirol 13 (2):173–184. doi: 10.1080/13550280701258399 [DOI] [PubMed] [Google Scholar]
- 86.Cowley D, Gray LR, Wesselingh SL, Gorry PR, Churchill MJ (2011) Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia. J Neurovirol 17 (1):70–81. doi: 10.1007/s13365-010-0002-5 [DOI] [PubMed] [Google Scholar]
- 87.Roof P, Ricci M, Genin P, Montano MA, Essex M, Wainberg MA, Gatignol A, Hiscott J (2002) Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-kappaB and the Tat transactivator. Virology 296 (1):77–83. doi: 10.1006/viro.2001.1397 [DOI] [PubMed] [Google Scholar]
- 88.Dahiya S, Nonnemacher MR, Wigdahl B (2012) Deployment of the human immunodeficiency virus type 1 protein arsenal: combating the host to enhance viral transcription and providing targets for therapeutic development. J Gen Virol 93 (Pt 6):1151–1172. doi: 10.1099/vir.0.041186-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taylor JP, Pomerantz R, Bagasra O, Chowdhury M, Rappaport J, Khalili K, Amini S (1992) TAR-independent transactivation by Tat in cells derived from the CNS: a novel mechanism of HIV-1 gene regulation. EMBO J 11 (9):3395–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hetzer C, Dormeyer W, Schnolzer M, Ott M (2005) Decoding Tat: the biology of HIV Tat posttranslational modifications. Microbes Infect 7 (13):1364–1369. doi: 10.1016/j.micinf.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 91.Deng L, Ammosova T, Pumfery A, Kashanchi F, Nekhai S (2002) HIV-1 Tat interaction with RNA polymerase II C-terminal domain (CTD) and a dynamic association with CDK2 induce CTD phosphorylation and transcription from HIV-1 promoter. J Biol Chem 277 (37):33922–33929. doi: 10.1074/jbc.M111349200 [DOI] [PubMed] [Google Scholar]
- 92.Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S (2006) Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology 3:78. doi: 10.1186/1742-4690-3-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ivanov A, Lin X, Ammosova T, Ilatovskiy AV, Kumari N, Lassiter H, Afangbedji N, Niu X, Petukhov MG, Nekhai S (2018) HIV-1 Tat phosphorylation on Ser-16 residue modulates HIV-1 transcription. Retrovirology 15 (1):39. doi: 10.1186/s12977-018-0422-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stevenson-Lindert LM, Fowler P, Lew J (2003) Substrate specificity of CDK2-cyclin A. What is optimal? J Biol Chem 278 (51):50956–50960. doi: 10.1074/jbc.M306546200 [DOI] [PubMed] [Google Scholar]
- 95.Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y (1996) The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J 15 (24):7060–7069 [PMC free article] [PubMed] [Google Scholar]
- 96.Selhorst P, Combrinck C, Ndabambi N, Ismail SD, Abrahams MR, Lacerda M, Samsunder N, Garrett N, Abdool Karim Q, Abdool Karim SS, Williamson C (2017) Replication Capacity of Viruses from Acute Infection Drives HIV-1 Disease Progression. J Virol 91 (8). doi: 10.1128/JVI.01806-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tyagi S, Ochem A, Tyagi M (2011) DNA-dependent protein kinase interacts functionally with the RNA polymerase II complex recruited at the human immunodeficiency virus (HIV) long terminal repeat and plays an important role in HIV gene expression. J Gen Virol 92 (Pt 7):1710–1720. doi: 10.1099/vir.0.029587-0 [DOI] [PubMed] [Google Scholar]
- 98.McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR (1995) HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213 (2):413–424. doi: 10.1006/viro.1995.0014 [DOI] [PubMed] [Google Scholar]
- 99.Brand SR, Kobayashi R, Mathews MB (1997) The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem 272 (13):8388–8395 [DOI] [PubMed] [Google Scholar]
- 100.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70 (4):1032–1060. doi: 10.1128/MMBR.00027-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krishna KH, Vadlamudi Y, Kumar MS (2016) Viral Evolved Inhibition Mechanism of the RNA Dependent Protein Kinase PKR’s Kinase Domain, a Structural Perspective. PLoS One 11 (4):e0153680. doi: 10.1371/journal.pone.0153680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Campbell GR, Loret EP (2009) What does the structure-function relationship of the HIV-1 Tat protein teach us about developing an AIDS vaccine? Retrovirology 6:50. doi: 10.1186/1742-4690-6-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Endo-Munoz L, Warby T, Harrich D, McMillan NA (2005) Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol J 2:17. doi: 10.1186/1743-422X-2-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deng L, de la Fuente C, Fu P, Wang L, Donnelly R, Wade JD, Lambert P, Li H, Lee CG, Kashanchi F (2000) Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology 277 (2):278–295. doi: 10.1006/viro.2000.0593 [DOI] [PubMed] [Google Scholar]
- 105.Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18 (21):6106–6118. doi: 10.1093/emboj/18.21.6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dorr A, Kiermer V, Pedal A, Rackwitz HR, Henklein P, Schubert U, Zhou MM, Verdin E, Ott M (2002) Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J 21 (11):2715–2723. doi: 10.1093/emboj/21.11.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bres V, Tagami H, Peloponese JM, Loret E, Jeang KT, Nakatani Y, Emiliani S, Benkirane M, Kiernan RE (2002) Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J 21 (24):6811–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Col E, Caron C, Seigneurin-Berny D, Gracia J, Favier A, Khochbin S (2001) The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J Biol Chem 276 (30):28179–28184. doi: 10.1074/jbc.M101385200 [DOI] [PubMed] [Google Scholar]
- 109.Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M, Loret E, Cole PA, Verdin E, Ott M (2003) Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell 12 (1):167–176 [DOI] [PubMed] [Google Scholar]
- 110.Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM (2002) Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol Cell 9 (3):575–586 [DOI] [PubMed] [Google Scholar]
- 111.Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F (2006) Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 3:48. doi: 10.1186/1742-4690-3-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang L, Nogales E, Ciferri C (2010) Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog Biophys Mol Biol 102 (2–3):122–128. doi: 10.1016/j.pbiomolbio.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verdin E, Paras P Jr., Van Lint C (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J 12 (8):3249–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pagans S, Kauder SE, Kaehlcke K, Sakane N, Schroeder S, Dormeyer W, Trievel RC, Verdin E, Schnolzer M, Ott M (2010) The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription. Cell Host Microbe 7 (3):234–244. doi: 10.1016/j.chom.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ali I, Ramage H, Boehm D, Dirk LM, Sakane N, Hanada K, Pagans S, Kaehlcke K, Aull K, Weinberger L, Trievel R, Schnoelzer M, Kamada M, Houtz R, Ott M (2016) The HIV-1 Tat Protein Is Monomethylated at Lysine 71 by the Lysine Methyltransferase KMT7. J Biol Chem 291 (31):16240–16248. doi: 10.1074/jbc.M116.735415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Duyne R, Easley R, Wu W, Berro R, Pedati C, Klase Z, Kehn-Hall K, Flynn EK, Symer DE, Kashanchi F (2008) Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology 5:40. doi: 10.1186/1742-4690-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mousseau G, Kessing CF, Fromentin R, Trautmann L, Chomont N, Valente ST (2015) The Tat Inhibitor Didehydro-Cortistatin A Prevents HIV-1 Reactivation from Latency. MBio 6 (4):e00465. doi: 10.1128/mBio.00465-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF (2014) New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20 (4):425–429. doi: 10.1038/nm.3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rasmussen TA, Lewin SR (2016) Shocking HIV out of hiding: where are we with clinical trials of latency reversing agents? Curr Opin HIV AIDS 11 (4):394–401. doi: 10.1097/COH.0000000000000279 [DOI] [PubMed] [Google Scholar]
- 120.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W (2008) TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog 4 (2):e16. doi: 10.1371/journal.ppat.0040016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT (2012) SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18 (11):1682–1687. doi: 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M (2003) A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol 5 (8):754–761. doi: 10.1038/ncb1023 [DOI] [PubMed] [Google Scholar]
- 123.Moll UM, Petrenko O (2003) The MDM2-p53 interaction. Mol Cancer Res 1 (14):1001–1008 [PubMed] [Google Scholar]
- 124.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, Weissman AM, Vousden KH (2005) Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 7 (6):547–559. doi: 10.1016/j.ccr.2005.04.029 [DOI] [PubMed] [Google Scholar]
- 125.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA (1998) The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 12 (22):3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Faust TB, Li Y, Jang GM, Johnson JR, Yang S, Weiss A, Krogan NJ, Frankel AD (2017) PJA2 ubiquitinates the HIV-1 Tat protein with atypical chain linkages to activate viral transcription. Sci Rep 7:45394. doi: 10.1038/srep45394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.El Kharroubi A, Piras G, Zensen R, Martin MA (1998) Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol 18 (5):2535–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.D’Orso I, Jang GM, Pastuszak AW, Faust TB, Quezada E, Booth DS, Frankel AD (2012) Transition step during assembly of HIV Tat:P-TEFb transcription complexes and transfer to TAR RNA. Mol Cell Biol 32 (23):4780–4793. doi: 10.1128/MCB.00206-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garcia JA, Harrich D, Pearson L, Mitsuyasu R, Gaynor RB (1988) Functional domains required for tat-induced transcriptional activation of the HIV-1 long terminal repeat. EMBO J 7 (10):3143–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sadaie MR, Mukhopadhyaya R, Benaissa ZN, Pavlakis GN, Wong-Staal F (1990) Conservative mutations in the putative metal-binding region of human immunodeficiency virus tat disrupt virus replication. AIDS Res Hum Retroviruses 6 (11):1257–1263. doi: 10.1089/aid.1990.6.1257 [DOI] [PubMed] [Google Scholar]
- 131.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH (2010) Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 465 (7299):747–751. doi: 10.1038/nature09131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rice AP, Carlotti F (1990) Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J Virol 64 (4):1864–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reza SM, Rosetti M, Mathews MB, Pe’ery T (2003) Differential activation of Tat variants in mitogen-stimulated cells: implications for HIV-1 postintegration latency. Virology 310 (1):141–156 [DOI] [PubMed] [Google Scholar]
- 134.Huet T, Dazza MC, Brun-Vezinet F, Roelants GE, Wain-Hobson S (1989) A highly defective HIV-1 strain isolated from a healthy Gabonese individual presenting an atypical western blot. AIDS 3 (11):707–715 [DOI] [PubMed] [Google Scholar]
- 135.Pantano S, Tyagi M, Giacca M, Carloni P (2004) Molecular dynamics simulations on HIV-1 Tat. Eur Biophys J 33 (4):344–351. doi: 10.1007/s00249-003-0358-z [DOI] [PubMed] [Google Scholar]
- 136.Mele AR, Marino J, Chen K, Pirrone V, Janetopoulos C, Wigdahl B, Klase Z, Nonnemacher MR (2018) Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter. Traffic. doi: 10.1111/tra.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Paul RH, Joska JA, Woods C, Seedat S, Engelbrecht S, Hoare J, Heaps J, Valcour V, Ances B, Baker LM, Salminen LE, Stein DJ (2014) Impact of the HIV Tat C30C31S dicysteine substitution on neuropsychological function in patients with clade C disease. J Neurovirol 20 (6):627–635. doi: 10.1007/s13365-014-0293-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Berkhout B, Gatignol A, Rabson AB, Jeang KT (1990) TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell 62 (4):757–767 [DOI] [PubMed] [Google Scholar]
- 139.Harrich D, Garcia J, Mitsuyasu R, Gaynor R (1990) TAR independent activation of the human immunodeficiency virus in phorbol ester stimulated T lymphocytes. EMBO J 9 (13):4417–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Southgate CD, Green MR (1991) The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev 5 (12B):2496–2507 [DOI] [PubMed] [Google Scholar]
- 141.Verhoef K, Koper M, Berkhout B (1997) Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology 237 (2):228–236. doi: 10.1006/viro.1997.8786 [DOI] [PubMed] [Google Scholar]
- 142.Das AT, Harwig A, Berkhout B (2011) The HIV-1 Tat protein has a versatile role in activating viral transcription. J Virol 85 (18):9506–9516. doi: 10.1128/JVI.00650-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dandekar DH, Ganesh KN, Mitra D (2004) HIV-1 Tat directly binds to NFkappaB enhancer sequence: role in viral and cellular gene expression. Nucleic Acids Res 32 (4):1270–1278. doi: 10.1093/nar/gkh289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Antell GC, Dampier W, Aiamkitsumrit B, Nonnemacher MR, Jacobson JM, Pirrone V, Zhong W, Kercher K, Passic S, Williams JW, Schwartz G, Hershberg U, Krebs FC, Wigdahl B (2016) Utilization of HIV-1 envelope V3 to identify X4- and R5-specific Tat and LTR sequence signatures. Retrovirology 13 (1):32. doi: 10.1186/s12977-016-0266-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT (2010) Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One 5 (5):e10611. doi: 10.1371/journal.pone.0010611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Das AT, Harwig A, Vrolijk MM, Berkhout B (2007) The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol 81 (14):7742–7748. doi: 10.1128/JVI.00392-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marzio G, Verhoef K, Vink M, Berkhout B (2001) In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc Natl Acad Sci U S A 98 (11):6342–6347. doi: 10.1073/pnas.111031498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Das AT, Verhoef K, Berkhout B (2004) A conditionally replicating virus as a novel approach toward an HIV vaccine. Methods Enzymol 388:359–379. doi: 10.1016/S0076-6879(04)88028-5 [DOI] [PubMed] [Google Scholar]
- 149.Mahlknecht U, Dichamp I, Varin A, Van Lint C, Herbein G (2008) NF-kappaB-dependent control of HIV-1 transcription by the second coding exon of Tat in T cells. J Leukoc Biol 83 (3):718–727. doi: 10.1189/jlb.0607405 [DOI] [PubMed] [Google Scholar]
- 150.Yang L, Morris GF, Lockyer JM, Lu M, Wang Z, Morris CB (1997) Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat. Virology 235 (1):48–64. doi: 10.1006/viro.1997.8672 [DOI] [PubMed] [Google Scholar]
- 151.Taylor JP, Pomerantz RJ, Raj GV, Kashanchi F, Brady JN, Amini S, Khalili K (1994) Central nervous system-derived cells express a kappa B-binding activity that enhances human immunodeficiency virus type 1 transcription in vitro and facilitates TAR-independent transactivation by Tat. J Virol 68 (6):3971–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gendelman HE, Lipton SA, Tardieu M, Bukrinsky MI, Nottet HS (1994) The neuropathogenesis of HIV-1 infection. J Leukoc Biol 56 (3):389–398 [DOI] [PubMed] [Google Scholar]
- 153.Zhou L, Saksena NK (2013) HIV Associated Neurocognitive Disorders. Infect Dis Rep 5 (Suppl 1):e8. doi: 10.4081/idr.2013.s1.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simioni S, Cavassini M, Annoni JM, Rimbault Abraham A, Bourquin I, Schiffer V, Calmy A, Chave JP, Giacobini E, Hirschel B, Du Pasquier RA (2010) Cognitive dysfunction in HIV patients despite long-standing suppression of viremia. AIDS 24 (9):1243–1250. doi: 10.1097/QAD.0b013e3283354a7b [DOI] [PubMed] [Google Scholar]
- 155.Purvis SF, Jacobberger JW, Sramkoski RM, Patki AH, Lederman MM (1995) HIV type 1 Tat protein induces apoptosis and death in Jurkat cells. AIDS Res Hum Retroviruses 11 (4):443–450. doi: 10.1089/aid.1995.11.443 [DOI] [PubMed] [Google Scholar]
- 156.Aksenov MY, Aksenova MV, Mactutus CF, Booze RM (2009) Attenuated neurotoxicity of the transactivation-defective HIV-1 Tat protein in hippocampal cell cultures. Exp Neurol 219 (2):586–590. doi: 10.1016/j.expneurol.2009.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McCloskey TW, Ott M, Tribble E, Khan SA, Teichberg S, Paul MO, Pahwa S, Verdin E, Chirmule N (1997) Dual role of HIV Tat in regulation of apoptosis in T cells. J Immunol 158 (2):1014–1019 [PubMed] [Google Scholar]
- 158.Kruman II, Nath A, Mattson MP (1998) HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol 154 (2):276–288. doi: 10.1006/exnr.1998.6958 [DOI] [PubMed] [Google Scholar]
- 159.Li CJ, Friedman DJ, Wang C, Metelev V, Pardee AB (1995) Induction of apoptosis in uninfected lymphocytes by HIV-1 Tat protein. Science 268 (5209):429–431 [DOI] [PubMed] [Google Scholar]
- 160.Pantaleo G, Fauci AS (1995) Apoptosis in HIV infection. Nat Med 1 (2):118–120 [DOI] [PubMed] [Google Scholar]
- 161.Sood V, Ranjan R, Banerjea AC (2008) Functional analysis of HIV-1 subtypes B and C HIV-1 Tat exons and RGD/QGD motifs with respect to Tat-mediated transactivation and apoptosis. AIDS 22 (13):1683–1685. doi: 10.1097/QAD.0b013e3282f56114 [DOI] [PubMed] [Google Scholar]
- 162.Chen D, Wang M, Zhou S, Zhou Q (2002) HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J 21 (24):6801–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Peter ME, Ehret A, Berndt C, Krammer PH (1997) AIDS and the death receptors. Br Med Bull 53 (3):604–616 [DOI] [PubMed] [Google Scholar]
- 164.Herbeuval JP, Grivel JC, Boasso A, Hardy AW, Chougnet C, Dolan MJ, Yagita H, Lifson JD, Shearer GM (2005) CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood 106 (10):3524–3531. doi: 10.1182/blood-2005-03-1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Garden GA, Budd SL, Tsai E, Hanson L, Kaul M, D’Emilia DM, Friedlander RM, Yuan J, Masliah E, Lipton SA (2002) Caspase cascades in human immunodeficiency virus-associated neurodegeneration. J Neurosci 22 (10):4015–4024. doi:20026351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Taylor RC, Cullen SP, Martin SJ (2008) Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 9 (3):231–241. doi: 10.1038/nrm2312 [DOI] [PubMed] [Google Scholar]
- 167.Campbell GR, Watkins JD, Esquieu D, Pasquier E, Loret EP, Spector SA (2005) The C terminus of HIV-1 Tat modulates the extent of CD178-mediated apoptosis of T cells. J Biol Chem 280 (46):38376–38382. doi: 10.1074/jbc.M506630200 [DOI] [PubMed] [Google Scholar]
- 168.Bartz SR, Emerman M (1999) Human immunodeficiency virus type 1 Tat induces apoptosis and increases sensitivity to apoptotic signals by up-regulating FLICE/caspase-8. J Virol 73 (3):1956–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35 (4):495–516. doi: 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kamori D, Ueno T (2017) HIV-1 Tat and Viral Latency: What We Can Learn from Naturally Occurring Sequence Variations. Front Microbiol 8:80. doi: 10.3389/fmicb.2017.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Campbell GR, Pasquier E, Watkins J, Bourgarel-Rey V, Peyrot V, Esquieu D, Barbier P, de Mareuil J, Braguer D, Kaleebu P, Yirrell DL, Loret EP (2004) The glutamine-rich region of the HIV-1 Tat protein is involved in T-cell apoptosis. J Biol Chem 279 (46):48197–48204. doi: 10.1074/jbc.M406195200 [DOI] [PubMed] [Google Scholar]
- 172.de Mareuil J, Carre M, Barbier P, Campbell GR, Lancelot S, Opi S, Esquieu D, Watkins JD, Prevot C, Braguer D, Peyrot V, Loret EP (2005) HIV-1 Tat protein enhances microtubule polymerization. Retrovirology 2:5. doi: 10.1186/1742-4690-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Battaglia PA, Zito S, Macchini A, Gigliani F (2001) A Drosophila model of HIV-Tat-related pathogenicity. J Cell Sci 114 (Pt 15):2787–2794 [DOI] [PubMed] [Google Scholar]
- 174.Alizon M, Wain-Hobson S, Montagnier L, Sonigo P (1986) Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell 46 (1):63–74 [DOI] [PubMed] [Google Scholar]
- 175.Egele C, Barbier P, Didier P, Piemont E, Allegro D, Chaloin O, Muller S, Peyrot V, Mely Y (2008) Modulation of microtubule assembly by the HIV-1 Tat protein is strongly dependent on zinc binding to Tat. Retrovirology 5:62. doi: 10.1186/1742-4690-5-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Strack PR, Frey MW, Rizzo CJ, Cordova B, George HJ, Meade R, Ho SP, Corman J, Tritch R, Korant BD (1996) Apoptosis mediated by HIV protease is preceded by cleavage of Bcl-2. Proc Natl Acad Sci U S A 93 (18):9571–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jacotot E, Ferri KF, El Hamel C, Brenner C, Druillennec S, Hoebeke J, Rustin P, Metivier D, Lenoir C, Geuskens M, Vieira HL, Loeffler M, Belzacq AS, Briand JP, Zamzami N, Edelman L, Xie ZH, Reed JC, Roques BP, Kroemer G (2001) Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein rR and Bcl-2. J Exp Med 193 (4):509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zauli G, Gibellini D (1996) The human immunodeficiency virus type-1 (HIV-1) Tat protein and Bcl-2 gene expression. Leuk Lymphoma 23 (5–6):551–560. doi: 10.3109/10428199609054864 [DOI] [PubMed] [Google Scholar]
- 179.Zauli G, Gibellini D, Caputo A, Bassini A, Negrini M, Monne M, Mazzoni M, Capitani S (1995) The human immunodeficiency virus type-1 Tat protein upregulates Bcl-2 gene expression in Jurkat T-cell lines and primary peripheral blood mononuclear cells. Blood 86 (10):3823–3834 [PubMed] [Google Scholar]
- 180.Sastry KJ, Marin MC, Nehete PN, McConnell K, el-Naggar AK, McDonnell TJ (1996) Expression of human immunodeficiency virus type I tat results in down-regulation of bcl-2 and induction of apoptosis in hematopoietic cells. Oncogene 13 (3):487–493 [PubMed] [Google Scholar]
- 181.Zauli G, Gibellini D, Milani D, Mazzoni M, Borgatti P, La Placa M, Capitani S (1993) Human immunodeficiency virus type 1 Tat protein protects lymphoid, epithelial, and neuronal cell lines from death by apoptosis. Cancer Res 53 (19):4481–4485 [PubMed] [Google Scholar]
- 182.Gibellini D, Caputo A, Celeghini C, Bassini A, La Placa M, Capitani S, Zauli G (1995) Tat-expressing Jurkat cells show an increased resistance to different apoptotic stimuli, including acute human immunodeficiency virus-type 1 (HIV-1) infection. Br J Haematol 89 (1):24–33 [DOI] [PubMed] [Google Scholar]
- 183.Zhang M, Li X, Pang X, Ding L, Wood O, Clouse KA, Hewlett I, Dayton AI (2002) Bcl-2 upregulation by HIV-1 Tat during infection of primary human macrophages in culture. J Biomed Sci 9 (2):133–139. doi: 10.1159/000048209 [DOI] [PubMed] [Google Scholar]
- 184.Lafrenie RM, Wahl LM, Epstein JS, Hewlett IK, Yamada KM, Dhawan S (1996) HIV-1-Tat modulates the function of monocytes and alters their interactions with microvessel endothelial cells. A mechanism of HIV pathogenesis. J Immunol 156 (4):1638–1645 [PubMed] [Google Scholar]
- 185.Toborek M, Lee YW, Pu H, Malecki A, Flora G, Garrido R, Hennig B, Bauer HC, Nath A (2003) HIV-Tat protein induces oxidative and inflammatory pathways in brain endothelium. J Neurochem 84 (1):169–179 [DOI] [PubMed] [Google Scholar]
- 186.Raidel SM, Haase C, Jansen NR, Russ RB, Sutliff RL, Velsor LW, Day BJ, Hoit BD, Samarel AM, Lewis W (2002) Targeted myocardial transgenic expression of HIV Tat causes cardiomyopathy and mitochondrial damage. Am J Physiol Heart Circ Physiol 282 (5):H1672–1678. doi: 10.1152/ajpheart.00955.2001 [DOI] [PubMed] [Google Scholar]
- 187.Paladugu R, Fu W, Conklin BS, Lin PH, Lumsden AB, Yao Q, Chen C (2003) Hiv Tat protein causes endothelial dysfunction in porcine coronary arteries. J Vasc Surg 38 (3):549–555; discussion 555–546 [DOI] [PubMed] [Google Scholar]
- 188.Rusnati M, Presta M (2002) HIV-1 Tat protein and endothelium: from protein/cell interaction to AIDS-associated pathologies. Angiogenesis 5 (3):141–151 [DOI] [PubMed] [Google Scholar]
- 189.Koch S, Claesson-Welsh L (2012) Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb Perspect Med 2 (7):a006502. doi: 10.1101/cshperspect.a006502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Albini A, Soldi R, Giunciuglio D, Giraudo E, Benelli R, Primo L, Noonan D, Salio M, Camussi G, Rockl W, Bussolino F (1996) The angiogenesis induced by HIV-1 tat protein is mediated by the Flk-1/KDR receptor on vascular endothelial cells. Nat Med 2 (12):1371–1375 [DOI] [PubMed] [Google Scholar]
- 191.Albini A, Benelli R, Presta M, Rusnati M, Ziche M, Rubartelli A, Paglialunga G, Bussolino F, Noonan D (1996) HIV-tat protein is a heparin-binding angiogenic growth factor. Oncogene 12 (2):289–297 [PubMed] [Google Scholar]
- 192.Ferrara N, Gerber HP, LeCouter J (2003) The biology of VEGF and its receptors. Nat Med 9 (6):669–676. doi: 10.1038/nm0603-669 [DOI] [PubMed] [Google Scholar]
- 193.Dhawan S, Puri RK, Kumar A, Duplan H, Masson JM, Aggarwal BB (1997) Human immunodeficiency virus-1-tat protein induces the cell surface expression of endothelial leukocyte adhesion molecule-1, vascular cell adhesion molecule-1, and intercellular adhesion molecule-1 in human endothelial cells. Blood 90 (4):1535–1544 [PubMed] [Google Scholar]
- 194.Mitola S, Soldi R, Zanon I, Barra L, Gutierrez MI, Berkhout B, Giacca M, Bussolino F (2000) Identification of specific molecular structures of human immunodeficiency virus type 1 Tat relevant for its biological effects on vascular endothelial cells. J Virol 74 (1):344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Toborek M, Lee YW, Flora G, Pu H, Andras IE, Wylegala E, Hennig B, Nath A (2005) Mechanisms of the blood-brain barrier disruption in HIV-1 infection. Cell Mol Neurobiol 25 (1):181–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vene R, Benelli R, Noonan DM, Albini A (2000) HIV-Tat dependent chemotaxis and invasion, key aspects of tat mediated pathogenesis. Clin Exp Metastasis 18 (7):533–538 [DOI] [PubMed] [Google Scholar]
- 197.Benelli R, Mortarini R, Anichini A, Giunciuglio D, Noonan DM, Montalti S, Tacchetti C, Albini A (1998) Monocyte-derived dendritic cells and monocytes migrate to HIV-Tat RGD and basic peptides. AIDS 12 (3):261–268 [DOI] [PubMed] [Google Scholar]
- 198.Albini A, Benelli R, Giunciuglio D, Cai T, Mariani G, Ferrini S, Noonan DM (1998) Identification of a novel domain of HIV tat involved in monocyte chemotaxis. J Biol Chem 273 (26):15895–15900 [DOI] [PubMed] [Google Scholar]
- 199.Premack BA, Schall TJ (1996) Chemokine receptors: gateways to inflammation and infection. Nat Med 2 (11):1174–1178 [DOI] [PubMed] [Google Scholar]
- 200.Conant K, Garzino-Demo A, Nath A, McArthur JC, Halliday W, Power C, Gallo RC, Major EO (1998) Induction of monocyte chemoattractant protein-1 in HIV-1 Tat-stimulated astrocytes and elevation in AIDS dementia. Proc Natl Acad Sci U S A 95 (6):3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bonwetsch R, Croul S, Richardson MW, Lorenzana C, Del Valle L, Sverstiuk AE, Amini S, Morgello S, Khalili K, Rappaport J (1999) Role of HIV-1 Tat and CC chemokine MIP-1alpha in the pathogenesis of HIV associated central nervous system disorders. J Neurovirol 5 (6):685–694 [DOI] [PubMed] [Google Scholar]
- 202.Weiss JM, Nath A, Major EO, Berman JW (1999) HIV-1 Tat induces monocyte chemoattractant protein-1-mediated monocyte transmigration across a model of the human blood-brain barrier and up-regulates CCR5 expression on human monocytes. J Immunol 163 (5):2953–2959 [PubMed] [Google Scholar]
- 203.Geiss GK, Bumgarner RE, An MC, Agy MB, van ‘t Wout AB, Hammersmark E, Carter VS, Upchurch D, Mullins JI, Katze MG (2000) Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266 (1):8–16. doi: 10.1006/viro.1999.0044 [DOI] [PubMed] [Google Scholar]
- 204.Fan J, Bass HZ, Fahey JL (1993) Elevated IFN-gamma and decreased IL-2 gene expression are associated with HIV infection. J Immunol 151 (9):5031–5040 [PubMed] [Google Scholar]
- 205.Lafrenie RM, Wahl LM, Epstein JS, Yamada KM, Dhawan S (1997) Activation of monocytes by HIV-Tat treatment is mediated by cytokine expression. J Immunol 159 (8):4077–4083 [PubMed] [Google Scholar]
- 206.Nath A, Conant K, Chen P, Scott C, Major EO (1999) Transient exposure to HIV-1 Tat protein results in cytokine production in macrophages and astrocytes. A hit and run phenomenon. J Biol Chem 274 (24):17098–17102 [DOI] [PubMed] [Google Scholar]
- 207.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M (2003) Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem 278 (4):2758–2766. doi: 10.1074/jbc.M209572200 [DOI] [PubMed] [Google Scholar]
- 208.Lenardo MJ, Baltimore D (1989) NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell 58 (2):227–229 [DOI] [PubMed] [Google Scholar]
- 209.Kwon HS, Brent MM, Getachew R, Jayakumar P, Chen LF, Schnolzer M, McBurney MW, Marmorstein R, Greene WC, Ott M (2008) Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell Host Microbe 3 (3):158–167. doi: 10.1016/j.chom.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bachmann MF, Oxenius A (2007) Interleukin 2: from immunostimulation to immunoregulation and back again. EMBO Rep 8 (12):1142–1148. doi: 10.1038/sj.embor.7401099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ott M, Emiliani S, Van Lint C, Herbein G, Lovett J, Chirmule N, McCloskey T, Pahwa S, Verdin E (1997) Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 275 (5305):1481–1485 [DOI] [PubMed] [Google Scholar]
- 212.Carvallo L, Lopez L, Fajardo JE, Jaureguiberry-Bravo M, Fiser A, Berman JW (2017) HIV-Tat regulates macrophage gene expression in the context of neuroAIDS. PLoS One 12 (6):e0179882. doi: 10.1371/journal.pone.0179882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bouwman RD, Palser A, Parry CM, Coulter E, Rasaiyaah J, Kellam P, Jenner RG (2014) Human immunodeficiency virus Tat associates with a specific set of cellular RNAs. Retrovirology 11:53. doi: 10.1186/1742-4690-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sharma V, Knobloch TJ, Benjamin D (1995) Differential expression of cytokine genes in HIV-1 tat transfected T and B cell lines. Biochem Biophys Res Commun 208 (2):704–713. doi: 10.1006/bbrc.1995.1395 [DOI] [PubMed] [Google Scholar]
- 215.Dabrowska A, Kim N, Aldovini A (2008) Tat-induced FOXO3a is a key mediator of apoptosis in HIV-1-infected human CD4+ T lymphocytes. J Immunol 181 (12):8460–8477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Izmailova E, Bertley FM, Huang Q, Makori N, Miller CJ, Young RA, Aldovini A (2003) HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat Med 9 (2):191–197. doi: 10.1038/nm822 [DOI] [PubMed] [Google Scholar]
- 217.Ranjbar S, Rajsbaum R, Goldfeld AE (2006) Transactivator of transcription from HIV type 1 subtype E selectively inhibits TNF gene expression via interference with chromatin remodeling of the TNF locus. J Immunol 176 (7):4182–4190 [DOI] [PubMed] [Google Scholar]
- 218.Karin M, Lin A (2002) NF-kappaB at the crossroads of life and death. Nat Immunol 3 (3):221–227. doi: 10.1038/ni0302-221 [DOI] [PubMed] [Google Scholar]
- 219.Israel N, Hazan U, Alcami J, Munier A, Arenzana-Seisdedos F, Bachelerie F, Israel A, Virelizier JL (1989) Tumor necrosis factor stimulates transcription of HIV-1 in human T lymphocytes, independently and synergistically with mitogens. J Immunol 143 (12):3956–3960 [PubMed] [Google Scholar]
- 220.Hiscott J, Kwon H, Genin P (2001) Hostile takeovers: viral appropriation of the NF-kappaB pathway. J Clin Invest 107 (2):143–151. doi: 10.1172/JCI11918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nath A, Psooy K, Martin C, Knudsen B, Magnuson DS, Haughey N, Geiger JD (1996) Identification of a human immunodeficiency virus type 1 Tat epitope that is neuroexcitatory and neurotoxic. J Virol 70 (3):1475–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li W, Huang Y, Reid R, Steiner J, Malpica-Llanos T, Darden TA, Shankar SK, Mahadevan A, Satishchandra P, Nath A (2008) NMDA receptor activation by HIV-Tat protein is clade dependent. J Neurosci 28 (47):12190–12198. doi: 10.1523/JNEUROSCI.3019-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dawson VL, Dawson TM, Bartley DA, Uhl GR, Snyder SH (1993) Mechanisms of nitric oxide-mediated neurotoxicity in primary brain cultures. J Neurosci 13 (6):2651–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sengpiel B, Preis E, Krieglstein J, Prehn JH (1998) NMDA-induced superoxide production and neurotoxicity in cultured rat hippocampal neurons: role of mitochondria. Eur J Neurosci 10 (5):1903–1910 [DOI] [PubMed] [Google Scholar]
- 225.Bertrand SJ, Aksenova MV, Mactutus CF, Booze RM (2013) HIV-1 Tat protein variants: critical role for the cysteine region in synaptodendritic injury. Exp Neurol 248:228–235. doi: 10.1016/j.expneurol.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang W, Benson DL (2001) Stages of synapse development defined by dependence on F-actin. J Neurosci 21 (14):5169–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mattson MP, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel RE (1992) beta-Amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J Neurosci 12 (2):376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Loo DT, Copani A, Pike CJ, Whittemore ER, Walencewicz AJ, Cotman CW (1993) Apoptosis is induced by beta-amyloid in cultured central nervous system neurons. Proc Natl Acad Sci U S A 90 (17):7951–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Holscher C (2005) Development of beta-amyloid-induced neurodegeneration in Alzheimer’s disease and novel neuroprotective strategies. Rev Neurosci 16 (3):181–212 [DOI] [PubMed] [Google Scholar]
- 230.Green DA, Masliah E, Vinters HV, Beizai P, Moore DJ, Achim CL (2005) Brain deposition of beta-amyloid is a common pathologic feature in HIV positive patients. AIDS 19 (4):407–411 [DOI] [PubMed] [Google Scholar]
- 231.Achim CL, Adame A, Dumaop W, Everall IP, Masliah E, Neurobehavioral Research C (2009) Increased accumulation of intraneuronal amyloid beta in HIV-infected patients. J Neuroimmune Pharmacol 4 (2):190–199. doi: 10.1007/s11481-009-9152-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Chen X, Hui L, Geiger NH, Haughey NJ, Geiger JD (2013) Endolysosome involvement in HIV-1 transactivator protein-induced neuronal amyloid beta production. Neurobiol Aging 34 (10):2370–2378. doi: 10.1016/j.neurobiolaging.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Nixon RA, Cataldo AM (1995) The endosomal-lysosomal system of neurons: new roles. Trends Neurosci 18 (11):489–496 [DOI] [PubMed] [Google Scholar]
- 234.Bahr BA, Bendiske J (2002) The neuropathogenic contributions of lysosomal dysfunction. J Neurochem 83 (3):481–489 [DOI] [PubMed] [Google Scholar]
- 235.Annaert W, De Strooper B (2002) A cell biological perspective on Alzheimer’s disease. Annu Rev Cell Dev Biol 18:25–51. doi: 10.1146/annurev.cellbio.18.020402.142302 [DOI] [PubMed] [Google Scholar]
- 236.Kim J, Yoon JH, Kim YS (2013) HIV-1 Tat interacts with and regulates the localization and processing of amyloid precursor protein. PLoS One 8 (11):e77972. doi: 10.1371/journal.pone.0077972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hategan A, Bianchet MA, Steiner J, Karnaukhova E, Masliah E, Fields A, Lee MH, Dickens AM, Haughey N, Dimitriadis EK, Nath A (2017) HIV Tat protein and amyloid-beta peptide form multifibrillar structures that cause neurotoxicity. Nat Struct Mol Biol 24 (4):379–386. doi: 10.1038/nsmb.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Aksenov MY, Aksenova MV, Mactutus CF, Booze RM (2010) HIV-1 protein-mediated amyloidogenesis in rat hippocampal cell cultures. Neurosci Lett 475 (3):174–178. doi: 10.1016/j.neulet.2010.03.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Johri MK, Sharma N, Singh SK (2015) HIV Tat protein: Is Tat-C much trickier than Tat-B? J Med Virol 87 (8):1334–1343. doi: 10.1002/jmv.24182 [DOI] [PubMed] [Google Scholar]
- 240.Kurosu T, Mukai T, Komoto S, Ibrahim MS, Li YG, Kobayashi T, Tsuji S, Ikuta K (2002) Human immunodeficiency virus type 1 subtype C exhibits higher transactivation activity of Tat than subtypes B and E. Microbiol Immunol 46 (11):787–799 [DOI] [PubMed] [Google Scholar]
- 241.Gandhi N, Saiyed Z, Thangavel S, Rodriguez J, Rao KV, Nair MP (2009) Differential effects of HIV type 1 clade B and clade C Tat protein on expression of proinflammatory and antiinflammatory cytokines by primary monocytes. AIDS Res Hum Retroviruses 25 (7):691–699. doi: 10.1089/aid.2008.0299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Bayes-Genis A, Barallat J, Richards AM (2016) A Test in Context: Neprilysin: Function, Inhibition, and Biomarker. J Am Coll Cardiol 68 (6):639–653. doi: 10.1016/j.jacc.2016.04.060 [DOI] [PubMed] [Google Scholar]
- 243.Rempel HC, Pulliam L (2005) HIV-1 Tat inhibits neprilysin and elevates amyloid beta. AIDS 19 (2):127–135 [DOI] [PubMed] [Google Scholar]
- 244.Daily A, Nath A, Hersh LB (2006) Tat peptides inhibit neprilysin. J Neurovirol 12 (3):153–160. doi: 10.1080/13550280600760677 [DOI] [PubMed] [Google Scholar]
- 245.Hamley IW (2012) The amyloid beta peptide: a chemist’s perspective. Role in Alzheimer’s and fibrillization. Chem Rev 112 (10):5147–5192. doi: 10.1021/cr3000994 [DOI] [PubMed] [Google Scholar]
- 246.Butterfield DA (2002) Amyloid beta-peptide (1–42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer’s disease brain. A review. Free Radic Res 36 (12):1307–1313 [DOI] [PubMed] [Google Scholar]
- 247.Tanzi RE, Bertram L (2005) Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 120 (4):545–555. doi: 10.1016/j.cell.2005.02.008 [DOI] [PubMed] [Google Scholar]
- 248.Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O’Sullivan K M, Desouza I, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD (2005) Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol 79 (21):13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Allen TM, O’Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI (2000) Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407 (6802):386–390. doi: 10.1038/35030124 [DOI] [PubMed] [Google Scholar]
- 250.Xiao H, Neuveut C, Tiffany HL, Benkirane M, Rich EA, Murphy PM, Jeang KT (2000) Selective CXCR4 antagonism by Tat: implications for in vivo expansion of coreceptor use by HIV-1. Proc Natl Acad Sci U S A 97 (21):11466–11471. doi: 10.1073/pnas.97.21.11466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Desfosses Y, Solis M, Sun Q, Grandvaux N, Van Lint C, Burny A, Gatignol A, Wainberg MA, Lin R, Hiscott J (2005) Regulation of human immunodeficiency virus type 1 gene expression by clade-specific Tat proteins. J Virol 79 (14):9180–9191. doi: 10.1128/JVI.79.14.9180-9191.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stutz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, Gagneur J, Korbel JO, Huber W, Steinmetz LM (2013) The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3 (8):1213–1224. doi: 10.1534/g3.113.005777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Frattini A, Fabbri M, Valli R, De Paoli E, Montalbano G, Gribaldo L, Pasquali F, Maserati E (2015) High variability of genomic instability and gene expression profiling in different HeLa clones. Sci Rep 5:15377. doi: 10.1038/srep15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mousseau G, Clementz MA, Bakeman WN, Nagarsheth N, Cameron M, Shi J, Baran P, Fromentin R, Chomont N, Valente ST (2012) An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe 12 (1):97–108. doi: 10.1016/j.chom.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ferrucci A, Nonnemacher MR, Wigdahl B (2011) Human immunodeficiency virus viral protein R as an extracellular protein in neuropathogenesis. Adv Virus Res 81:165–199. doi: 10.1016/B978-0-12-385885-6.00010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.James T, Nonnemacher MR, Wigdahl B, Krebs FC (2016) Defining the roles for Vpr in HIV-1-associated neuropathogenesis. J Neurovirol 22 (4):403–415. doi: 10.1007/s13365-016-0436-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Dampier W, Antell GC, Aiamkitsumrit B, Nonnemacher MR, Jacobson JM, Pirrone V, Zhong W, Kercher K, Passic S, Williams JW, James T, Devlin KN, Giovannetti T, Libon DJ, Szep Z, Ehrlich GD, Wigdahl B, Krebs FC (2017) Specific amino acids in HIV-1 Vpr are significantly associated with differences in patient neurocognitive status. J Neurovirol 23 (1):113–124. doi: 10.1007/s13365-016-0462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hogan TH, Nonnemacher MR, Krebs FC, Henderson A, Wigdahl B (2003) HIV-1 Vpr binding to HIV-1 LTR C/EBP cis-acting elements and adjacent regions is sequence-specific. Biomed Pharmacother 57 (1):41–48 [DOI] [PubMed] [Google Scholar]
- 259.Yiannopoulou KG, Papageorgiou SG (2013) Current and future treatments for Alzheimer’s disease. Ther Adv Neurol Disord 6 (1):19–33. doi: 10.1177/1756285612461679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rygiel K (2016) Novel strategies for Alzheimer’s disease treatment: An overview of anti-amyloid beta monoclonal antibodies. Indian J Pharmacol 48 (6):629–636. doi: 10.4103/0253-7613.194867 [DOI] [PMC free article] [PubMed] [Google Scholar]