Abstract
Pluripotency describes the developmental capacity to give rise to all cell types in the adult body. A comprehensive understanding of the molecular mechanisms that regulate pluripotency is important for both basic and translational research. While earlier studies mostly focused on signaling pathways, transcriptional regulation, and epigenetic modifications, recent investigations showed that RNA binding proteins, RNA processing machineries, and regulatory RNA molecules also play essential roles. Here, we provide a concise review on the latest findings and developments in post-transcriptional regulation of the pluripotent state.
Introduction
Pluripotency is defined as the developmental potential to give rise to all cell types formed by the three germ layers [1]. It is a unique property of the epiblast cells in blastocyst stage embryos, and it can also be captured in vitro in embryonic stem cells (ESCs), epiblast stem cells (EpiSCs), embryonic germ cells (EGCs), germline pluripotent stem cells (gPSCs), and induced pluripotent stem cells (iPSCs) [2–6]. Mouse ESCs cultured with MEK and GSK3 inhibitors (2i) show transcriptional and epigenetic similarity to the naïve pluripotent state in the pre-implantation epiblast [7], which can contribute to both blastocyst chimeras and the germline. In contrast, mouse EpiSCs derived from the post-implantation embryos represent the primed state. They are more primed for differentiation, and cannot integrate into the blastocyst or give rise to the germ cells The molecular mechanisms that regulate these pluripotent states have been extensively investigated, as they not only provide insights to early development but also facilitate the use of pluripotent stem cells in therapeutic applications.
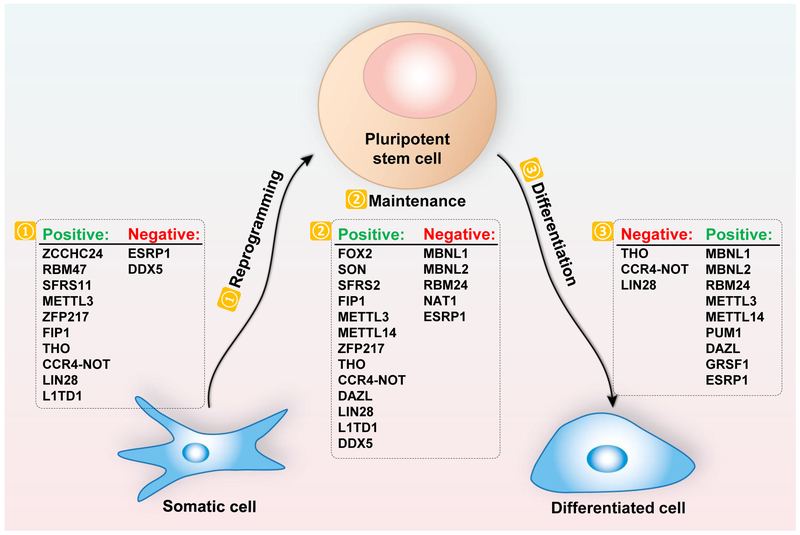
From studies in mouse ESCs and other systems, it has been shown that the pluripotent state is controlled by a combination of signal-transduction pathways, transcription factors, epigenetic modifiers, RNA binding proteins (RBPs), RNA processing machineries, and regulatory RNA molecules [8–11]. However, most of the early research focused on the transcriptional and epigenetic regulation. In comparison, the role of post-transcriptional regulation on pluripotency has only begun to be revealed in more recent years. One study using genomic and proteomic-approaches suggested that post-transcriptional regulation may be responsible for a large proportion of protein level changes during ESC fate transition [12]. Indeed, with emerging technologies such as high-throughput sequencing, large-scale screening, and systematic identification of protein-RNA interactions or RBPs, more and more post-transcriptional mechanisms have been uncovered in the regulation of the pluripotent state. Here, we provide an overview of the latest findings and developments in the post-transcriptional regulation of pluripotency, and we propose that post-transcriptional regulation adds important layers of controls to fine-tune the gene expression program in pluripotent stem cells (Figure 1, Table 1).
Figure 1.
Post-transcriptional Regulation of the Pluripotent State
Table 1.
Post-transcriptional Regulators of the Pluripotent State
| Biological process | Regulators | Mechanisms | PMID |
|---|---|---|---|
| mRNA processing | FOX2 | Facilitates pluripotency-specific AS in human ESC | 19136955 |
| SON | 24013217 | ||
| SFRS2 | Regulates AS of Mbd2 to support human ESC and iPSC self-renewal | 24813856 | |
| ZCCHC24, RBM47 | Regulates phase-specific AS during reprogramming | 27050523 | |
| SFRS11 | Regulates AS for genes that are critical for human somatic cell reprogramming | 27292646 | |
| MBNL1, MBNL2 | Promotes differentiation-specific AS patterns in human ESC and reprogramming | 23739326 | |
| RBM24 | Regulates AS events that favor cardiac specification during mouse ESC differentiation | 26990106 | |
| FIP1 | Activates ESC-specific APAs for pluripotency-associated genes in mouse ESC and reprogramming | 24596251 | |
| mRNA modification | METTL3, METTL14 | Promotes degradation of target mRNAs via m6A in mouse ESC | 25456834, 25569111, 24394384 |
| ZFP217 | Promotes mouse somatic cell reprogramming via m6A deposition on target mRNAs | 26526723 | |
| mRNA export | THOC2, THOC5 | Facilitates pluripotency gene transcript nuclear export to support mouse ESC self-renewal and somatic cell reprogramming | 24315442 |
| poly(A) tail | CCR4-NOT | Promotes differentiation gene mRNA deadenylation and degradation to support mouse ESC self-renewal and epiblast maintenance; also supports somatic cell reprogramming and germ cell development | 22367759, 27746116, 27037025, 28297718 |
| Promotes planarian stem cell differentiation via deadenylation and degradation of stem cell gene mRNAs | 24367277 | ||
| mRNA translation | NAT1 | Promotes Map3k3 and Sos1 mRNA translation and mouse ESC differentiation | 11032820, 28003464 |
| DAZL | Promotes Tet1 mRNA translation and supports the naïve pluripotency state | 26077710 | |
| Suppresses Oct4 mRNA translation in both human and mouse ESCs | 27768780, 23298641 | ||
| Multifunctional | LIN28 | Promotes human somatic cell reprogramming and transition to the primed pluripotent state via let-7-dependent and independent pathways; couples MAPK/ERK signaling to post-transcriptional control | 27320042, 27992407 |
| Inhibits Hmga2 mRNA translation in mouse ESC | 27920151 | ||
| Interacts with RHA to promote Oct4 mRNA translation and human ESC self-renweal | 19966271 | ||
| L1TD1 | Interacts with post-transcriptional regulators and pluripotency factors in human ESC | 25702638 | |
| Interacts with LIN28 to regulate Oct4 mRNA translation in human ESC | 22162396 | ||
| ESRP1 | Fine-tunes pluripotency gene mRNA translation in mouse ESC | 24015231 | |
| Promotes pluripotency-specific AS events during mouse iPSC generation | 27050523 | ||
| DDX5 | Represses the expression and function of RYBP via miR-125b during mouse somatic cell reprogramming | 28111200 |
Post-transcriptional regulation of pluripotency
Gene expression can be regulated at every stage during the making of the gene product. While transcriptional regulations often function as on-off switches, post-transcriptional mechanisms can act as rheostats to refine the output of gene expression. After transcription initiation, the primary transcripts undergo a series of steps including processing, export, modification, translation, and degradation to complete their life cycles. Almost all of these steps are subjected to regulation to influence the final production of the protein. In the following sub-sections, we review the intricate post-transcriptional regulations in pluripotent stem cells in a temporal order, based on the sequence of events that happen to an RNA molecule after its synthesis.
mRNA processing
The primary transcripts generated by the RNA polymerase must first be processed into mature mRNAs. RNA processing includes 5’-capping, splicing, and 3’-end processing. Both alternative splicing (AS) and alternative polyadenylation (APA, during 3’-end processing) can lead to the production of multiple mRNA variants from the same transcript, which in turn greatly increases the complexity of gene expression and facilitate cell type-specific gene regulation without editing the genome [13].
AS was found to play an important role in both the maintenance of the pluripotent state as well as the re-establishment of pluripotency during somatic reprogramming in mouse and human cells [14–16]. Many pluripotency factors, such as Oct4, Nanog, Sall4, Tcf3, Foxp1, Mbd2, and Yy2 [16–22], have multiple isoforms that vary in expression, intracellular localization, stability, or function due to differences in their coding exons or untranslated regions from AS. Furthermore, AS regulators are differentially expressed in pluripotent stem cells and somatic cells [14]. They control the proper splicing of cell-state specific transcripts, and can rewire AS networks during cell fate transitions. Specifically, FOX2, SON, SFRS2, MYC, GCN5, ZCCHC24, and RBM47 facilitate pluripotency-specific AS of their target genes [14,21,23–25]. In contrast, MBNL1, MBNL2, RBM24, and SFRS11 promote differentiation-specific AS patterns for a large number of splicing events [16,26,27]. Together, these studies demonstrated an active role of AS in regulating both self-renewal and differentiation, and it will be important to further understand how specific AS signatures are established and maintained in different developmental states.
In addition to AS, the majority of mammalian genes also generate alternatively polyadenylated mRNAs. In most cases, APA leads to the production mRNAs with different 3’-untranslated regions, which can impact mRNA stability, translation, or intracellular localization [28,29]. Global profiling showed that widespread APA occurs during early mouse development, mouse ESC differentiation, and somatic cell reprogramming, suggesting that APA is tightly regulated during cell fate transitions [30]. Consistently, genes involved in 3’-end processing were implicated in mouse ESC maintenance in several genetic screens [31,32]. Furthermore, the FIP1 subunit in the cleavage and polyadenylation specificity factor (CPSF) complex was shown to promote mouse ESC self-renewal and somatic cell reprogramming [33]. It activates an ESC-specific APA pattern on a group of pluripotency-associated genes to enhance their expression. Fip1 expression and the FIP1-dependent APA program change during ESC differentiation and are restored to an ESC-like state during somatic reprogramming. Thus, similar to AS, APA plays a significant role in regulating pluripotent stem cell fate specification. In this case, Fip1 expression level is at least partly responsible for the pluripotency-specific APA patterns. However, there likely exist other factors that regulates APA in development and diseases.
mRNA modification
In addition to RNA processing, RNA can be chemically modified and RNA modifications serve as another layer of post-transcriptional control in gene expression [34,35]. There are more than 100 distinct RNA modifications, such as N6-adenosine methylation (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), and pseudo-uridine (Ψ) [36]. Among them, m6A is the most abundant internal modification in mRNAs and has been extensively investigated in recent years. m6A is catalyzed by writers (METTL3, METTL14,), removed by erasers (ALKBH5 and FTO), and interpreted by readers (YTH domain family proteins, HNRNP proteins, and EIF3) [34,37]. In addition, m6A level can also be regulated by miRNAs and transcription factors. m6A is involved in multiple aspects of RNA metabolism, including degradation, splicing, transport, localization and translation, and its level is dynamically regulated in different cell types and during cell fate transitions.
It was initially shown that m6A modification destabilizes developmental regulators in in mouse ESCs [38]. However, further studies showed that in both mouse and human ESCs, m6A is enriched in pluripotency gene transcripts, and it promotes differentiation by facilitating pluripotency gene transcripts degradation [39,40]. In mouse EpiSCs, m6A is also found in differentiation gene transcripts and promotes EpiSC maintenance by driving differentiation gene transcript removal [40]. Therefore, it appears that m6A modification of mRNAs regulate pluripotent stem cell fate by acting on cell-type specific transcripts. It will be interesting to further dissect the underlying molecular mechanism to reconcile the results from different groups.
In mouse somatic cell reprogramming, the role of m6A is also complicated. One study showed that elevated m6A level resulted from human METTL3 overexpression enhances reprogramming efficiency of mouse embryonic fibroblasts [41]. However, a second study indicated that increased m6A level accompanying Zfp217 depletion impairs reprogramming [42]. This apparent discrepancy may be explained by how the m6A level was manipulated and which target mRNAs were involved. But these results suggested that m6A deposition is exquisitely regulated by different factors, and the resulting phenotype is highly dependent on specific m6A target mRNAs.
mRNA export
After processing, the mature mRNAs need to be exported from the nucleus to the cytoplasm for translation, and the mRNA export serves as the next step in post-transcriptional gene regulation. The main components of the mRNA export machinery have been well characterized, and a key player is the TRanscription and EXport (TREX) complex. Components of TREX are associated with other mRNA processing machineries, suggesting that mRNA export may act as an important interface in various processing steps to fine-tune gene expression [43]. The core of TREX is formed by the hexameric sub-complex THO. In mammals, THO is composed of THOC1, THOC2, THOC3, THOC5, THOC6, and THOC7, among which THOC2 functions as a scaffold for the complex and THOC5 acts as an adaptor for mRNA binding. THO plays a pivotal role in normal development and cellular differentiation, as its disruption leads to early embryonic lethality, as well as defects in hematopoietic progenitor survival, intestine stem cell homeostasis, and testis development [44,45].
THOC2 and THOC5 were identified as novel regulators of ESC maintenance in genetic screens [31]. It was further shown that THO preferentially interacts with pluripotency gene transcripts via THOC5, and regulates their export and expression [46]. During differentiation, THO loses its interaction with those transcripts due to reduced Thoc5 expression, facilitating the down-regulation of pluripotency gene expression and the exit from the pluripotent state. Finally, THO is also important for the establishment of pluripotency, as its depletion inhibits somatic cell reprogramming and blastocyst development. Thus, by regulating the export and expression of pluripotency genes at the post-transcription level, THO provides an extra layer of fast and potentially non-committal control to fine-tune the balance between self-renewal and differentiation. Importantly, the specificity of THO toward pluripotency gene mRNAs can be partly explained by the expression level of the Thoc5 subunit. This is reminiscent of the APA regulation, in which Fip1 expression also contributes to substrate selectivity. As will be seen in more examples below, the expression and composition of the RNA processing machinery may play a critical role in carrying out
mRNA poly(A)-tail length
Once the mRNAs are exported into the cytoplasm, they are subjected to additional regulations, such as by the enzymes that control the poly(A)-tail length. Most mRNAs are polyadenylated by poly(A) polymerases, and the poly(A)-tail length can be modulated to influence mRNA stability or translation. Indeed, global measurement of poly(A)-tail length suggested that it is highly involved in development and cell fate transition [47,48]. Among the regulatory mechanisms controlling poly(A)-tail length, cytoplasmic deadenylation plays a pivotal role. The best characterized deadenylases so far include the CCR4-NOT complex, the poly(A)-specific ribonuclease (PARN), and the poly(A) nuclease (PAN), with CCR4-NOT being the predominant deadenylase in all eukaryote cells [49].
CCR4-NOT is a highly conserved multi-protein complex from yeast to human. It has been implicated in gene regulation at many steps throughout the lifetime of mRNAs. The CNOT1, CNOT2, and CNOT3 subunits of the CCR4-NOT complex were identified as regulators of the pluripotent state in mouse ESCs, and were shown to prevent differentiation into extraembryonic lineages [50]. Further, CNOT3 was found to be required for epiblast cell maintenance during embryonic development [51]. Mechanistically, Cnot3 deletion results in increases in the poly(A)-tail lengths, half-lives, and steady-state levels of a subset of differentiation gene mRNAs. Consistently, the half-lives of these CNOT3 target mRNAs, but not those of housekeeping or pluripotency gene mRNAs, are shorter in ESCs and become longer during normal differentiation. Together, these results revealed that the CCR4-NOT complex maintains the pluripotent state by promoting differentiation gene mRNA deadenylation and degradation. More importantly, they strongly argued that poly(A) tail-length regulation is yet another critical post-transcriptional mechanism that controls pluripotency. In line with this notion, forced expression of CCR4-NOT components were shown to promote somatic cell reprogramming [52,53].
Intriguingly, other studies suggested that CCR4-NOT likely plays a much more complex role. First, the RNA binding protein PUM1 is required for the exit of the pluripotent state in ESCs [54]. It targets pluripotency gene mRNAs and accelerates their degradation at the onset of differentiation. As PUM1 is known to interact with CCR4-NOT, this study implied a potential involvement of CCR4-NOT in the regulation of pluripotency gene mRNAs. Second, in somatic cells the m6A reader YTHDF2 directly interacts with CNOT1 and recruits CCR4–NOT to m6A-containing mRNAs [55]. This recruitment is essential for the deadenylation and degradation of m6A-containing mRNAs. As m6A can mediate pluripotency gene mRNA degradation in ESCs, this study again suggested a possible connection between CCR4-NOT and pluripotency gene mRNAs. Integrating all the above findings, it is possible that different subunits in CCR4-NOT and/or different RBPs may facilitate the recognition of specific mRNA substrates by CCR4-NOT, allowing the complex to selectively target different functional groups of mRNAs for deadenylation.
mRNA translation
Finally, mRNA translation can profoundly regulate protein levels [56], and factors involved in translation control have been implicated in ESC biology. For example, the mechanistic target of rapamycin (mTOR) pathway is a master regulator of protein synthesis. It has been shown that protein synthesis and protein content significantly increases during mouse ESC differentiation. Such an increase can be partly attributed to the activation of the mTOR pathway and a hierarchy of translational regulators including 4EBP1, DAZL and GRSF1 [57]. Consistent with that, a recent study showed that mTOR inhibition can promote the maintenance of the pluripotent state both in vivo during embryonic diapause and in vitro in cultured ESCs [58]. Thus, mRNA translation clearly plays a critical role in pluripotent stem cells, and serves as a means for extracellular signals to influence cell fate. Intriguingly, mTOR inhibition was found to impair the long-term self-renewal of human ESCs [59], but in this case the function of mTOR was attributed to the transcriptional repression of developmental and growth-inhibitory genes.
Besides the mTOR pathway, additional translational regulators have also been implicated in ESC maintenance. The NAT1 protein binds to eukaryotic translation initiation factors and ribosomal proteins [60]. It promotes mouse ESC differentiation by enhancing the translation of genes involved in differentiation, mitochondrial oxidative respiration, and chromatin modification via a noncanonical, cap-independent mechanism [61]. The RNA-binding protein DAZL marks a sub-population of mouse ESCs [62]. It associates with Tet1 mRNA and enhances its translation, promoting global cytosine hydroxymethylation. The DAZL-mediated translational control promotes the conversion between the naïve and primed pluripotent state. However, how NAT1 and DAZL act on selected target mRNAs to promote pluripotency remains to be elucidated.
Multi-functional RBPs
In addition to the above regulators that act at specific steps during the life cycle of an RNA, some RBPs have been shown to regulate gene expression at multiple levels in pluripotent stem cells. We discuss their functions separately below.
LIN28 is an RBP that can regulate both miRNA let-7 biogenesis and other mRNAs. It is highly expressed in mouse ESCs, further induced in EpiSCs, and down-regulated upon differentiation [63,64]. In human ESCs, LIN28 interacts with RNA helicase A and regulates Oct4 mRNA translation to support the pluripotent state [65]. Consistently, it promotes the reprogramming of human somatic cells [64], and the conversion from the naïve to the primed pluripotent state in mouse cells [66,67]. On the molecular level, LIN 28 acts in both let-7-dependent and independent manner. In the let7-independent axis, it binds to metabolic gene mRNAs and represses their expression, conferring the metabolism characteristic of primed state pluripotency [67]. The let-7-independent function of LIN28 may be regulated via MAPK/ERK-mediated phosphorylation [68].
L1td1 (LINE-1 type Transposase Domain-containing 1) was originally identified as an ESC-associated transcript. It is highly expressed in the inner cell mass of mouse blastocysts, and is also rapidly activated during somatic cell reprogramming. While it is dispensable for mouse early development and iPSC derivation [69,70], it is required for human ESC self-renewal [71]. It is an RBP that interacts with LIN 28 via RNA, and may regulate pluripotency gene expression post-transcriptionally [71]. Further analysis of L1TD1 interactome revealed that it indeed binds to many RNA processing factors, adding additional support for its role in RNA regulation in pluripotent stem cells [72].
ESRP1 was initially shown to be a negative regulator of pluripotency. Its silencing inhibits mouse ESC differentiation, and it binds to Oct4 and Sox2 mRNA 5’-UTR, and prevents their efficient loading into the polysomes [73]. However, a later study found that ESRP 1 is differentially expressed during mouse somatic cell reprogramming [14]. Its overexpression promotes iPSC generation and facilitates the establishment of the pluripotent-specific AS of an epithelial specific transcription factor Grhl1 transcript. These observations suggested that ESRP1 have multiple functions at different post-transcriptional steps, and it can influence pluripotent stem cell fate via different mechanisms.
Finally, two RNA interactome studies examined RBPs in mouse ESCs. One identified a list of novel RBPs that are selectively expressed in the pluripotent state [69], and the other provided a high-resolution mapping of RNA-binding regions of known and unknown RBPs [58]. Another study investigated the proteomic changes during somatic cell reprogramming, and uncovered many RNA processing factors that show stage-specific expression [74]. These systematic studies expanded the atlas of RBPs involved in pluripotency and provided a useful resource to study post-transcriptional gene regulation in pluripotent stem cells.
microRNAs
Beyond the protein factors, microRNAs (miRNAs) also play essential roles during post-transcriptional regulation in pluripotent cells. miRNAs are 20–21 base noncoding RNAs. They bind target mRNAs and regulate their stability or translation via the seed sequence in the 5’-region of the miRNAs. The involvement of miRNAs in ESCs maintenance and pluripotency was first revealed by the observations that the disruption of miRNA processing machineries led to impaired growth and differentiation [75,76]. In addition, the miRNA profile of pluripotent stem cells is well documented. In mouse ESCs, the polycistronic clusters miR-290–295 and miR-17–92b are dominantly expressed [77,78]. In mouse EpiSCs or human ESCs, the miR-302–367 cluster is highly expressed [79]. These pluripotency-specific miRNAs are activated by pluripotency transcription factors. Importantly, they all share the same seed sequence, and target cell cycle inhibitors p21, Lats2, and Rbl2 to maintain the distinct ESC cell cycle. In addition to these clusters of miRNAs, the let-7 family of miRNAs were shown to be important for ESC differentiation [80]. They target hundreds of pluripotency gene transcripts for degradation, and are required for the dismantlement of the pluripotent state. Consistent with results from ESCs, overexpression of many miRNAs, especially those pluripotency-specific ones, was shown to enhance somatic cell reprogramming in the presence of other reprogramming factors [81]. Furthermore, while still in debate, miRNA-only reprogramming has also been reported [82–84]. Finally, DDX5, a component of the Drosha miRNA processing complex, was recently reported to inhibit somatic cell reprogramming. DDX5 promotes miR-125b processing to repress the expression and function of the non-canonical polycomb complex 1 (PRC1) subunit RYBP, thereby impairing iPSC generation [85]. Together, these findings highlight the significance of miRNAs as a means for post-transcriptional regulation of pluripotency.
Concluding remarks and perspectives
Pluripotent cells have a sophisticated gene expression program that controls the delicate balance between self-renewal and differentiation. In addition to the transcriptional regulations, post-transcriptional mechanisms bring additional layers to fine-tune gene expression and cell fate. Significant advances have been made in understanding the fundamental roles of post-transcriptional regulation in governing the pluripotent state. However, many questions remain to be answered. How are specific mRNAs being recognized and targeted at each of the regulatory steps? How can the regulatory machineries regulate different groups of mRNAs in different or sometimes even the same cellular context? Can functionally related mRNAs be co-regulated? Is there any coordination across the different regulatory factors or processes? Is there any crosstalk between transcriptional and post-transcriptional regulations? Along these lines, it has been proposed that functionally related mRNAs may be coordinately regulated as post-transcriptional RNA regulons by RBPs or RNA processing machineries [86]. Pluripotent stem cells appear to be an appealing system to further test this RNA regulon hypothesis. In addition to the conceptual challenges, technical improvements, such as those to identify protein-RNA interactions more convincingly, accurately measure poly(A)-tail length, and determine translation efficiency from limited materials, are also needed to move the research forward. Progress along these lines will provide a more comprehensive view of post-transcriptional regulation in the establishment, maintenance, and destabilization of the pluripotent state, as well as in other developmental and disease processes.
Finally, it may be interesting to look beyond the pluripotent stem cells to have a broader view on the post-transcriptional regulation of pluripotency. In particular, we propose that germ cells can provide a novel perspective. Germ cells harbor latent pluripotent potential, as they can re-acquire pluripotency via fertilization, teratocarcinogenesis, or spontaneous conversion during culture [87]. Furthermore, they express and require many of the same key post-transcriptional regulators, and may share similar regulatory mechanisms with pluripotent stem cells [47,48,51,54,62,78,88]. Therefore, a systematic investigation of post-transcriptional gene regulation in the pluripotency cycle between germ cells and pluripotent stem cells [87] will uncover new mechanistic and evolutionary insights to answer the conceptual questions listed above.
Acknowledgements
We apologize to those whose work were not cited due to space constraints. This work is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences Z01ES102745.
References and Recommended Readings
Papers of particular interest are highlighted below: ● of special interest, ●● of outstanding interest
- 1.Posfai E, Tam OH, Rossant J: Mechanisms of pluripotency in vivo and in vitro. Curr Top Dev Biol 2014, 107:1–37. [DOI] [PubMed] [Google Scholar]
- 2.Boroviak T, Nichols J: The birth of embryonic pluripotency. Philos Trans R Soc Lond B Biol Sci 2014, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols J, Smith A: Naive and primed pluripotent states. Cell Stem Cell 2009, 4:487–492. [DOI] [PubMed] [Google Scholar]
- 4.De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, et al. : Hallmarks of pluripotency. Nature 2015, 525:469–478. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Yamauchi T, Izpisua Belmonte JC: An overview of mammalian pluripotency. Development 2016, 143:1644–1648. [DOI] [PubMed] [Google Scholar]
- 6.Kalkan T, Smith A: Mapping the route from naive pluripotency to lineage specification. Philos Trans R Soc Lond B Biol Sci 2014, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martello G, Smith A: The nature of embryonic stem cells. Annu Rev Cell Dev Biol 2014, 30:647–675. [DOI] [PubMed] [Google Scholar]
- 8.Dejosez M, Zwaka TP: Pluripotency and nuclear reprogramming. Annu Rev Biochem 2012, 81:737–765. [DOI] [PubMed] [Google Scholar]
- 9.Hackett JA, Surani MA: Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 2014, 15:416–430. [DOI] [PubMed] [Google Scholar]
- 10.Ye J, Blelloch R: Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 2014, 15:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guallar D, Wang J: RNA-binding proteins in pluripotency, differentiation, and reprogramming. Front Biol (Beijing) 2014, 9:389–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. : Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 2009, 462:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licatalosi DD, Darnell RB: RNA processing and its regulation: global insights into biological networks. Nat Rev Genet 2010, 11:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ● 14.Cieply B, Park JW, Nakauka-Ddamba A, Bebee TW, Guo Y, Shang X, Lengner CJ, Xing Y, Carstens RP: Multiphasic and Dynamic Changes in Alternative Splicing during Induction of Pluripotency Are Coordinated by Numerous RNA-Binding Proteins. Cell reports 2016, 15:247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 15.Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. : Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A 2010, 107:10514–10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 16.Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. : MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 2013, 498:241–245.These studies revealed an integral role of alternative splicing in pluripotency regulation.
- 17.Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW: OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells 2008, 26:3068–3074. [DOI] [PubMed] [Google Scholar]
- 18.Das S, Jena S, Levasseur DN: Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. J Biol Chem 2011, 286:42690–42703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH: Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol 2010, 30:5364–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ● 20.Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. : An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 2011, 147:132–146. [DOI] [PubMed] [Google Scholar]
- ● 21.Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, et al. : Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell 2014, 15:92–101.These studies illustrated the role of alternative splicing in human ESC and reprogramming.
- 22.Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos-Pournatzis T, Matta-Camacho E, Tsukumo Y, Yanagiya A, Li WC, Atlasi Y, Caron M, et al. : Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proceedings of the National Academy of Sciences of the United States of America 2016, 113:12360–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH: An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 2009, 16:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Goke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH: SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol 2013, 15:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, et al. : Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev 2015, 29:803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, Lin Y, Liu J, Zhang ZG, Fu W, Guo LY, Pan L, Kong X, Zhang MK, Lu YH, et al. : Rbm24 Regulates Alternative Splicing Switch in Embryonic Stem Cell Cardiac Lineage Differentiation. Stem cells 2016, 34:1776–1789. [DOI] [PubMed] [Google Scholar]
- 27.Toh CX, Chan JW, Chong ZS, Wang HF, Guo HC, Satapathy S, Ma D, Goh GY, Khattar E, Yang L, et al. : RNAi Reveals Phase-Specific Global Regulators of Human Somatic Cell Reprogramming. Cell Rep 2016, 15:2597–2607. [DOI] [PubMed] [Google Scholar]
- 28.Elkon R, Ugalde AP, Agami R: Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 2013, 14:496–506. [DOI] [PubMed] [Google Scholar]
- 29.Tian B, Manley JL: Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 2013, 38:312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Z, Tian B: Reprogramming of 3’ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 2009, 4:e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ: A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes & development 2009, 23:837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. : A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 2009, 4:403–415. [DOI] [PubMed] [Google Scholar]
- ●● 33.Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, Xie X, Wan J, Xing Y, Freudenberg JM, et al. : Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J 2014, 33:878–889.This study showed that Fip1 promotes an ESC-specific APA pattern to fine-tune pluripotency gene expression, thereby illustrating the critical role of APA in pluripotency regulation.
- 34.Zhao BS, Roundtree IA, He C: Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017, 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T: RNA modifications: what have we learned and where are we headed? Nat Rev Genet 2016, 17:365–372. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert WV, Bell TA, Schaening C: Messenger RNA modifications: Form, distribution, and function. Science 2016, 352:1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR: 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 38.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC: N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 2014, 16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 39.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. : m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 40.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. : Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015, 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- ● 41.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. : m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16:289–301. [DOI] [PubMed] [Google Scholar]
- ● 42.Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee DF, Chen CH, Rengasamy M, Andino B, et al. : Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell 2015, 17:689–704.These studies investigated the role of mRNA m6A modification in ESC and reprogramming, and showed that different factors can regulate m6A deposition to influence cell fate.
- 43.Heath CG, Viphakone N, Wilson SA: The role of TREX in gene expression and disease. Biochem J 2016, 473:2911–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Chinnam M, Wang J, Wang Y, Zhang X, Marcon E, Moens P, Goodrich DW: Thoc1 deficiency compromises gene expression necessary for normal testis development in the mouse. Mol Cell Biol 2009, 29:2794–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mancini A, Niemann-Seyde SC, Pankow R, El Bounkari O, Klebba-Farber S, Koch A, Jaworska E, Spooncer E, Gruber AD, Whetton AD, et al. : THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol 2010, 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 46.Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I, et al. : The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell stem cell 2013, 13:676–690.This study showed that THO-dependent mRNA export promotes pluripotency gene expression in ESC maintenance, differentiation, somatic cell reprogramming, and blastocyst development.
- 47.Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP: mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim J, Lee M, Son A, Chang H, Kim VN: mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes & development 2016, 30:1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstrohm AC, Wickens M: Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008, 9:337–344. [DOI] [PubMed] [Google Scholar]
- ● 50.Zheng X, Dumitru R, Lackford BL, Freudenberg JM, Singh AP, Archer TK, Jothi R, Hu G: Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem cells 2012, 30:910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 51.Zheng X, Yang P, Lackford B, Bennett BD, Wang L, Li H, Wang Y, Miao Y, Foley JF, Fargo DC, et al. : CNOT3-Dependent mRNA Deadenylation Safeguards the Pluripotent State. Stem cell reports 2016, 7:897–910.These studies revealed a critical role of poly(A)-tail length regulation by the CCR4-NOT complex in the maintenance of the pluripotent state both in ESCs and during mouse early development.
- 52.Zukeran A, Takahashi A, Takaoka S, Mohamed HM, Suzuki T, Ikematsu S, Yamamoto T: The CCR4-NOT deadenylase activity contributes to generation of induced pluripotent stem cells. Biochemical and biophysical research communications 2016, 474:233–239. [DOI] [PubMed] [Google Scholar]
- 53.Kamon M, Katano M, Hiraki-Kamon K, Hishida T, Nakachi Y, Mizuno Y, Okazaki Y, Suzuki A, Hirasaki M, Ueda A, et al. : Identification of Ccr4-not complex components as regulators of transition from partial to genuine induced pluripotent stem cells. Stem Cells Dev 2014, 23:2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leeb M, Dietmann S, Paramor M, Niwa H, Smith A: Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 2014, 14:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L: YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 2016, 7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larsson O, Tian B, Sonenberg N: Toward a genome-wide landscape of translational control. Cold Spring Harb Perspect Biol 2013, 5:a012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 57.Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE: A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2008, 2:448–460. [DOI] [PubMed] [Google Scholar]
- ●● 58.Bulut-Karslioglu A, Biechele S, Jin H, Macrae TA, Hejna M, Gertsenstein M, Song JS, Ramalho-Santos M: Inhibition of mTOR induces a paused pluripotent state. Nature 2016, 540:119–123.These studies illustrated the role of mTOR and mRNA translation in pluripotency regulation.
- 59.Zhou J, Su P, Wang L, Chen J, Zimmermann M, Genbacev O, Afonja O, Horne MC, Tanaka T, Duan E, et al. : mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proc Natl Acad Sci U S A 2009, 106:7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV Jr., Iwao H, Innerarity TL: Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J 2000, 19:5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugiyama H, Takahashi K, Yamamoto T, Iwasaki M, Narita M, Nakamura M, Rand TA, Nakagawa M, Watanabe A, Yamanaka S: Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc Natl Acad Sci U S A 2017, 114:340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welling M, Chen HH, Munoz J, Musheev MU, Kester L, Junker JP, Mischerikow N, Arbab M, Kuijk E, Silberstein L, et al. : DAZL regulates Tet1 translation in murine embryonic stem cells. EMBO reports 2015, 16:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moss EG, Tang L: Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Developmental biology 2003, 258:432–442. [DOI] [PubMed] [Google Scholar]
- 64.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. : Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318:1917–1920. [DOI] [PubMed] [Google Scholar]
- 65.Qiu C, Ma Y, Wang J, Peng S, Huang Y: Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res 2010, 38:1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parisi S, Passaro F, Russo L, Musto A, Navarra A, Romano S, Petrosino G, Russo T: Lin28 is induced in primed embryonic stem cells and regulates let-7-independent events. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 2016. [DOI] [PubMed] [Google Scholar]
- 67.Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, et al. : LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell stem cell 2016, 19:66–80. [DOI] [PubMed] [Google Scholar]
- 68.Tsanov KM, Pearson DS, Wu Z, Han A, Triboulet R, Seligson MT, Powers JT, Osborne JK, Kane S, Gygi SP, et al. : LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency. Nature cell biology 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN: The RNA-binding protein repertoire of embryonic stem cells. Nature structural & molecular biology 2013, 20:1122–1130. [DOI] [PubMed] [Google Scholar]
- 70.Iwabuchi KA, Yamakawa T, Sato Y, Ichisaka T, Takahashi K, Okita K, Yamanaka S: ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency. PloS one 2011, 6:e20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Narva E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nasti J, Autio R, Rasool O, Denessiouk K, Lahdesmaki H, et al. : RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem cells 2012, 30:452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emani MR, Narva E, Stubb A, Chakroborty D, Viitala M, Rokka A, Rahkonen N, Moulder R, Denessiouk K, Trokovic R, et al. : The L1TD1 protein interactome reveals the importance of post-transcriptional regulation in human pluripotency. Stem cell reports 2015, 4:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, Tolosano E, Provero P, Pandolfi PP, Silengo L, et al. : The RNA binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS One 2013, 8:e72300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J: Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell reports 2012, 2:1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K: Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005, 19:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 76.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R: DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007, 39:380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ● 77.Judson RL, Babiarz JE, Venere M, Blelloch R: Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 2009, 27:459–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ● 78.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R: Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 2008, 40:1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ● 79.Zhang Z, Hong Y, Xiang D, Zhu P, Wu E, Li W, Mosenson J, Wu WS: MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Reports 2015, 4:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●● 80.Melton C, Judson RL, Blelloch R: Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463:621–626.These studies demonstrated the essential roles of miRNAs in ESC and reprogramming.
- 81.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R: Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 2011, 29:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. : Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8:376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY: Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14:2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, et al. : Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011, 8:633–638. [DOI] [PubMed] [Google Scholar]
- 85.Li H, Lai P, Jia J, Song Y, Xia Q, Huang K, He N, Ping W, Chen J, Yang Z, et al. : RNA Helicase DDX5 Inhibits Reprogramming to Pluripotency by miRNA-Based Repression of RYBP and its PRC1-Dependent and -Independent Functions. Cell Stem Cell 2017, 20:571. [DOI] [PubMed] [Google Scholar]
- 86.Keene JD: RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007, 8:533–543. [DOI] [PubMed] [Google Scholar]
- 87.Leitch HG, Smith A: The mammalian germline as a pluripotency cycle. Development 2013, 140:2495–2501. [DOI] [PubMed] [Google Scholar]
- 88.Tong MH, Mitchell DA, McGowan SD, Evanoff R, Griswold MD: Two miRNA clusters, Mir-17–92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol Reprod 2012, 86:72. [DOI] [PMC free article] [PubMed] [Google Scholar]