Abstract
Rapid advancements in next-generation sequencing (ngs) technology have created an unprecedented opportunity to decipher the molecular profile of tumours to more effectively prevent, diagnose, and treat cancer. Oncologists now have the option to order molecular tests that can guide treatment decisions. However, to date, most oncologists have received limited training in genomics, and they are now faced with the challenge of understanding how such tests and their interpretation align with patient management. Guidance on how to effectively use ngs technology is therefore needed to aid oncologists in applying the results of genomic tests. The Canadian guideline presented here describes best practices and unmet needs related to ngs-based testing for somatic variants in oncology, including clinical application, assay and sample selection, bioinformatics and interpretation of reports performed by laboratories, patient communication, and clinical trials.
Keywords: Next-generation sequencing, somatic variants, oncologists, sequencing, molecular genomics, pathology, guidelines
INTRODUCTION
Next-generation sequencing (ngs) is the sequencing of millions of small fragments of dna in a massively parallel fashion1. Computer algorithm–based bioinformatics analyses are then used to piece the fragments together by mapping individual digital reads to a reference genome sequence to identify potential variants. Application of ngs technology can provide high-throughput sequencing capacity as large as the whole genome, or deep coverage of small dna regions for many samples thereby improving the throughput of the assay. Using ngs, fragmented dna from formalin-fixed paraffin-embedded tissue (ffpe) can be sequenced without pre-existing knowledge of the individual’s genome2. The technique has the ability to detect low-frequency events—for example, resolving the existence of minor clones and identifying rare circulating tumour dna (ctdna) or cell-free total nucleic acid (cftna). It therefore provides an unparalleled opportunity to increase the understanding of the molecular profile of tumours to more effectively apply precision medicine to cancer diagnosis and treatment.
With the identification of an increasing number of genes affecting risk, prognosis, and response to radiochemotherapy and molecularly targeted therapy, it is essential that health care providers determine how ngs technology fits within the constructs of patient care. However, to date, oncologists in Canada have received limited training in genomics and might not be aware of the benefits, limitations, and differences between the many ngs-based tests that are available or in development.
Guidance about interpreting the results of genomic panels should aid oncologists in using such technologies effectively. The guideline that follows was developed by a steering committee of pathologists, geneticists, oncologists, and genetic counsellors from across Canada. It provides guidance for oncologists about the use of ngs for the identification of somatic variants in adult cancers. Best practices and unmet needs related to ngs-based testing in oncology—including clinical application, assay and sample selection, bioinformatics and interpretation of reports, patient communication, and clinical trials—identified from the best available evidence are discussed.
NGS WORKFLOW
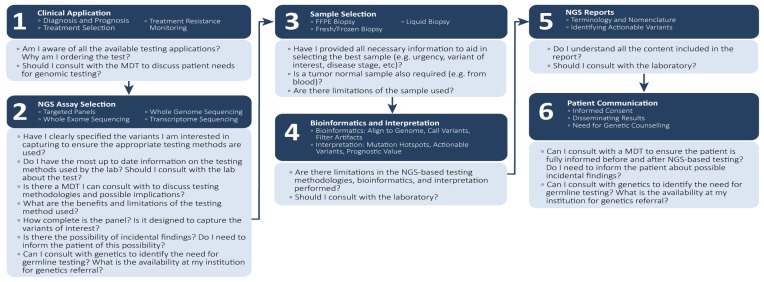
The proposed use of ngs to identify clinically actionable variants is described in Figure 1. At each step in the process, oncologists should be mindful of key questions that can inform decisions for optimal patient management. Each step in the process is discussed in detail in the subsections that follow.
FIGURE 1.
Next-generation sequencing (NGS): workflow considerations for oncologists. MDT = multidisciplinary team; FFPE = formalin-fixed, paraffin-embedded.
Clinical Application
For oncologists, ngs testing has many possible applications ranging from diagnosis and prognosis to treatment and monitoring for resistance. Given the complexity and multiple uses of genetic testing in oncology, it is advisable that clinicians discuss patient needs as part of a multidisciplinary team including oncologists, pathologists, medical geneticists, genetic counsellors, and laboratory science staff. Providing information about family history, phenotype, and results from other diagnostic investigations is important to aid in the clinical interpretation of variants3.
Diagnosis and Prognosis
Next-generation sequencing can aid in diagnosis by identifying alterations associated with individual cancers. When biopsy samples are too small to produce a reliable histology diagnosis (for example, fine-needle samples from thyroid nodules), ngs can be used to detect alterations frequently found in the particular cancer2. In addition, ngs can be used to identify pathognomonic molecular signatures associated with a diagnostic entity such as MYD88 L265P in Waldenström macroglobulinemia or BRAF V600E in hairy cell leukemia4. Next-generation sequencing is also able to identify biomarkers associated with poor prognosis such as KRAS variants in pancreatic cancer2 or TP53 alterations in myelodysplastic syndrome or acute myeloid leukemia4. Although other tests such as Sanger sequencing and quantitative polymerase chain reaction (pcr) can also be used for those purposes, ngs has the added advantage of interrogating samples for multiple variants to aid in making a diagnosis when both a pre-test knowledge of the diagnosis and the diagnostic material are limited2.
Treatment Decision Algorithms
Early testing is important to ensure prompt and effective treatment, especially for aggressive cancers. For example, reflex testing including multiple genes has been shown to save valuable biopsy tissue and to reduce time to treatment for patients with lung cancer5. In those patients, whose prognosis is typically poor, reflex testing (compared with on-demand testing) has been shown to provide more timely access to treatment5,6. The advantage of ngs compared with single-gene testing is that ngs has the ability to screen for a broader set of variants in one complete test, making the most efficient use of limited biopsy tissue7. In addition, when multiple genes are tested, greater cost-effectiveness has been shown for ngs compared with sequential single-gene testing modalities8. Given an increase in clinically validated biomarkers and a reduction in cost and testing time, the benefits of ngs are expected to grow. However, the use of ngs in place of companion diagnostic tests depends on the funding of ngs-based testing across Canada, for which no process is currently defined.
Systemic and Molecularly Targeted Therapies
A number of targeted therapies have been approved in Canada for use in patients harbouring specific gene variants9–28 (Table I). By correctly identifying patients who might respond to those agents, oncologists can both treat those who could derive benefit and spare those who are unlikely to benefit from unnecessary toxicities and unwarranted costs. Moreover, ngs can be used to identify patients who could be susceptible to drug toxicities. For example, patients with germline variants in the DYPD gene are at risk of toxicity from 5-fluorouracil, resulting in greater neutropenia, mucositis, and diarrhea29.
TABLE I.
Targeted therapies approved by Health Canada that require testing for molecular biomarkers
| Gene | Nature of aberration | Cancer type | Targeted therapy |
|---|---|---|---|
| Solid tumours | |||
|
| |||
| ALK1 | Fusion | Lung cancer | Alectinib, brigatinib, ceritinib, crizotinib |
|
| |||
| BRAF | Variant | Melanoma | Cobimetinib–vemurafenib, dabrafenib with or without trametinib, trametinib monotherapy, vemurafenib monotherapy |
| Variant | Lung cancer | Dabrafenib with trametinib | |
|
| |||
| BRCA1/2 | Variant, copy number | Ovarian, fallopian tube, primary peritoneal cancer | Olaparib |
| Germline and HER2-negative | Variant, copy number | Breast cancer | Olaparib |
|
| |||
| EGFR | Variant, insertion or deletion | Lung cancer | Afatinib, erlotinib, gefitinib, osimertinib |
| KRAS wild type | Variant | Colon cancer | Cetuximab |
| RAS wild type | Variant | Colon cancer | Panitumumab |
|
| |||
| Kit (CD117) | Variant | Gastrointestinal stromal tumours | Imatinib |
|
| |||
| ROS1 | Fusion | Lung cancer | Crizotinib |
|
| |||
| HER2 | |||
| Overexpression | Copy number | Breast cancer | Lapatinib, pertuzumab, trastuzumab, trastuzumab emtansine |
| Negative | Copy number | Breast cancer | Everolimus |
|
| |||
| PD-L1 | Protein expression | Lung cancer | Pembrolizumab |
| Hematologic cancers | |||
|
| |||
| 17p deletion | Copy number | CLL | Venetoclax |
|
| |||
| BCR–ABL1 | Fusion | CML | Bosutinib, dasatinib, imatinib, nilotinib, ponatinib |
|
| |||
| FIP1L1–PDGFRA | Fusion | CEL | Imatinib |
|
| |||
| PDGFR | Variant | Myelodysplastic disease | Imatinib |
|
| |||
| PML/RAR | Fusion | APL | Arsenic trioxide |
CLL = chronic lymphocytic leukemia; CML = chronic myeloid leukemia; CEL = chronic eosinophilic leukemia; APL = acute promyelocytic leukemia.
Despite the success of targeted therapies, most patients will eventually experience disease progression7. Next-generation sequencing can then be useful in the detection of resistance variants that might cause treatment failure to guide selection of subsequent therapies. For example, KRAS, NRAS, and BRAF variants are associated with resistance to therapy targeting the epidermal growth factor receptor2.
Immunotherapies
Immune checkpoint inhibitors such as inhibitors of ctla4, PD-1, and PD-L1 have been shown to improve outcomes in melanoma and cancers of the lung, kidney, and bladder7. In non-small-lung cancer, immunotherapy is especially active in patients with high PD-L1 expression on tumour or immune cells. However, immunohistochemistry assays to measure PD-L1 expression lack sensitivity and specificity in identifying treatment-sensitive patients7,30. In other cancer types, PD-L1 expression also seems to be less important for predicting response to immunotherapy.
A number of studies of various tumour types—including lung cancer31,32, bladder cancer33, and melanoma32—have suggested that patients with a higher tumour mutational burden (tmb) experience increased clinical benefit from immune checkpoint inhibitors. Elevated somatic tmb is speculated to increase the quantity of tumour-associated neoantigens (which serve as targets of antitumour response) and is associated with improved clinical response to immunotherapy34,35. Thus, tmb is proving to be a useful biomarker for response to immunotherapies. Microsatellite instability (msi) and dna mismatch repair (mmr)—inherited or acquired cellular mechanisms that also lead to increased dna damage—have been identified as biomarkers for response to immune checkpoint inhibitors, agnostic of cancer histology36. The challenge with measuring tmb is how much of the genome must be explored to accurately measure the level of dna damage within the cell. It has been proposed that the mutational burden must be examined for at least hundreds of genes to accurately measure tmb37. To guide the selection of checkpoint inhibitors, ngs methods can provide information about tmb, as well as about msi and mmr35. Recently, the U.S. National Comprehensive Cancer Network’s guidelines for colon cancer38 indicated that msi or mmr can be tested using either traditional immunohistochemistry screening-based workflows or ngs methods. In addition, guidelines from both the National Comprehensive Cancer Network39 and the European Society for Medical Oncology40 have recommended tmb testing for non-small-lung cancer.
Best Practices
■ Use of multidisciplinary teams that meet regularly to discuss the needs of patients is critical to ensure access to the expertise required to select, perform, and interpret the results of genomic tests.
■ Compared with on-demand testing, reflex testing is time- and cost-effective.
■ When multiple genes must be tested, the use of ngs is especially justified to save valuable limited biopsy tissue.
Unmet Needs
■ Next-generation sequencing infrastructure, local expertise, methodologic validation, and implementation have to be funded and standardized across Canada.
■ Further development and harmonization of ngs methods to measure tmb, msi, and mmr could aid in the selection of immunotherapies.
NGS Assay Selection
When using ngs to identify alterations in the cancer genome, a number of methods can be used. Those methods vary in their specificity, sensitivity, specimen efficiency, and ability to capture genes of interest. For oncologists, working with geneticists and pathologists to gain an appreciation of the benefits and limitations of each assay is important to maximize the successful return of a result and to better interpret accuracy and completeness for detecting variants of interest (Table II). The subsections that follow describe the key types of ngs assays used in oncology.
TABLE II.
Comparison of next-generation sequencing assaysa
| Assay | Advantages | Disadvantages |
|---|---|---|
| Targeted panels |
|
|
| Whole-exome sequencing |
|
|
| Whole-genome sequencing |
|
|
| RNA sequencing |
|
|
Adapted from Horak et al., 201641.
Targeted Gene Panels
Targeted gene panels, which sequence a discrete number of target loci or genes of interest, are the method of choice for most laboratories, given their lower cost and simplicity for data generation and analysis2. Gene panels are either pcr amplicon–driven or they rely on a hybrid-capture enrichment approach7. The former type are designed for the identification of single nucleotide variants and small insertions or deletions; the latter type can also detect a number of fusion events and copy number changes. Amplicon sequencing is cheaper and faster, and typically requires less input dna; enrichment methods provide superior data quality, allow for the detection of fusions, and are better for fragmented samples42. Because of the small number of loci analyzed in gene panels compared with whole-exome (wes) or whole-genome sequencing (wgs), ngs reads are able to achieve the greatest depth of coverage, which represents the number of times a specific nucleotide has been compared against the reference genome, thereby increasing the sensitivity of detecting an alteration41. Depth of coverage is crucial in the consistent calling of variants and affects the efficiency, sensitivity, and specificity of the analysis. Targeted panels are also better able to identify tumour variants from less-than-ideal tissue specimens with low tumour content or low allelic variation2. However, they do not identify aberrations outside their regions of design. Communication between end users and experts, such as laboratory staff geneticists or pathologists, might be needed to ensure that the assay is compatible with the information being sought.
Targeted gene panels can vary significantly in size from a small number of actionable genes in a region of interest (“hotspot” panels) to hundreds of genes used to better characterize the genetic profile of cancers. Larger panels might be able to report alterations in a greater number of genes, including calculation of tmb, but at the expense of decreased throughput, increased cost, and greater bioinformatics support. A number of commercial gene panels are available that can easily be standardized and validated in laboratories without the need for extensive bioinformatics support41. Those panels can also be customized to better meet the needs of the treating physician, but would require ample time for development and validation. Alternatively, laboratories could build gene panels in-house to meet their testing needs. However, laboratories vary in achieving minimal technical standards—such as Clinical Laboratory Improvement Amendments or College of American Pathologists certification—for ngs testing. In instances in which current in-house options do not sufficiently cover variants of interest, a number of external services are available at an additional cost (for instance, Guardant Health and Foundation Medicine, among others). However, funding, availability, and completeness of gene panels available for testing vary significantly across Canada. It is therefore important that oncologists understand the completeness and features of the panel being used to ensure that it captures the variants of interest.
WES
Whole-exome sequencing selects and sequences only the 1% of the genome involved in protein coding, which can detect up to 85% of disease-causing variants41. Depending on the application of wes, copy-number variations are more likely to be detected, although the method can still miss gene fusions. Given the greater amount of data captured than is captured in targeted gene panels, much more specimen material is required, and depth of coverage is less extensive, thereby affecting sensitivity and making data analysis more time-consuming. Moreover, differences between exome-capture methods, ngs platforms, and bioinformatics pipelines can create bias, leading to variations between testing centres. Given the large amount of data generated, greater bioinformatics support is needed, with a dedicated clinical infrastructure and workflow. Given the greater percentage of the genome being captured, a larger number of variants of unknown significance (vuss) can also be reported. Those vuss must be accurately interpreted and described to reduce confusion for clinicians and patients. However, wes can detect novel and unique variants that might be associated with cancer, but that are missed in the more restrictive targeted panels. Within Canada, wes is not yet routinely used as part of clinical practice in oncology; however, it is being used as part of a number of large-scale research efforts such as the Finding of Rare Disease Genes in Canada Consortium, which aimed to identify genes associated with rare pediatric single-gene disorders43. Given that the cost and turnaround time for wes is rapidly improving, its use in the clinical setting should become increasingly feasible.
WGS
Compared with other methods, wgs is the most expensive; however, it is able to identify the greatest number of changes to the genome, including germline and somatic single nucleotide variants, insertions or deletions, copy-number variations, and gene rearrangements7,41. Unlike wes, wgs does not require exon capture or enrichment, thereby reducing variation in sample preparation and sequencing. However, wgs has the lowest depth of coverage, and its identification of variants with low allelic frequency is less sensitive. Whole-genome sequencing has a greater chance of capturing vuss and represents the most complete and unbiased method for detecting changes in the cancer genome. In Canada, wgs is not currently being used in routine clinical practice, but is being used as part of a large-scale research effort by the Personalized Onco-Genomics program at BC Cancer in Vancouver (see NCT02155621 at http://ClinicalTrials.gov/)44. However, as sequencing and analytical costs decrease and a greater understanding of noncoding portions of the genome is reached, it is likely that the use of wgs will increase.
RNA Sequencing
Like dna sequencing, rna sequencing can be implemented using targeted panels to select specific transcripts of clinical interest or using non-targeted sequencing to achieve comprehensive profiling. The two key applications of rna sequencing include gene expression profiling and fusion gene detection41. The resulting information can be used to identify gene expression dysregulation caused by copy-number aberrations, to characterize molecular tumour subtypes by combining expression and genomic signatures, and to enhance detection of variants in low-purity tumour samples7. For example, measuring estrogen receptor gene expression can identify patients who are sensitive to endocrine therapy. Transcriptome sequencing is also used to detect fusions occurring at the rna level such as BCR-ABL1 in chronic myelogenous leukemia or in solid cancers such as sarcomas and cancers of the prostate and breast, among others45. However, rna sequencing is subject to variability stemming from low quantities of cancer cells within specimens, tumour heterogeneity, rna degradation in archived specimens, and inadequate references for baseline expression41.
Assay Considerations
Although the focus of this article is somatic variations in oncology, ngs analyses can also involve testing of normal samples, resulting in possible detections of germline variations. If germline normal testing will be done, it is important to understand which variants might be reported and to adequately inform patients before testing occurs. Communication with the laboratory and discussion within multidisciplinary team meetings can aid in ensuring that oncologists are up-to-date on the testing methods used at their institution. Even when only somatic variants are being assessed, potential germline variants that require further follow-up might, in some cases, be detected46. For example, BRCA variants in ovarian cancer could be either germline (13%–15% of cases) or somatic (3%–10% of cases), with parp1 inhibitors being effective in both types47. In such cases, oncologists might need to refer patients to a geneticist for germline testing. There is also a possibility that the results obtained during a retest at a later time point (for example, within clinical trials) might be different because of differences in the sensitivity and specificity of the testing methods. Because testing procedures can change over time, clinicians should ensure they have access to up-to-date information about the completeness of the gene panels used at their institution and the chance of incidental findings.
With the growing availability of larger gene panels and the increasing demand for wes and wgs, the complexity of genetic information will continue to rise. The need for expertise to advise institutions about optimal molecular testing, to provide bioinformatics support, and to aid in interpretation of results is increasing. Molecular tumour boards consisting of experts such as medical oncologists, pathologists, genome scientists, geneticists, genetic counsellors, computational biologists, and bioinformaticians can aid in informing clinical decisions48. Molecular tumour boards have been used successfully in large-scale research initiatives to aid in matching patients with appropriate therapies and could provide valuable expertise to institutions as the use of ngs expands.
Best Practices
■ To ensure that the correct testing method is used, oncologists should clearly specify the genes or genetic loci and the nature of the variants they are interested in capturing.
■ It is important to be aware of the completeness and availability of targeted panels. Oncologists should consult with the laboratory to ensure that they are up-to-date about the methods used at their institution.
■ Understanding the implication of vuss is important for the interpretation and dissemination of results. Appropriate tracking of vuss is key, because biologic and therapeutic significance might be discovered in the future and could affect patient management.
■ Physicians should be aware of the potential for results to be different when tissue is retested using a different method or sample. Differences can arise because of tumour heterogeneity and varying test methods (for example, different regions or different variant types detected by different panels).
■ To assess the risk of incidental findings, oncologists should consult with the laboratory and ensure that they have up-to-date information about whether germline normal samples will be tested. The risk should be communicated to patients before tests are ordered.
■ Where incidental findings from germline testing are reported or there is a need for germline testing, oncologists should consult with geneticists and be aware of the availability for genetics referral at their institution.
Unmet Needs
■ Minimally acceptable technical standards should be defined to ensure that all Canadian patients have access to high-quality ngs testing.
■ Canadian oncologists should have access to appropriate educational content on cancer genomics, testing, resources, and so on.
■ Canadian medical schools should emphasize appropriate content in the curriculum about cancer genomics, testing, resources, and so on.
■ There is a need for better access and resources for geneticists and genetic counselling services.
■ Molecular tumour boards will be increasingly important as the complexity of ngs assays, the need for bioinformatics support, and large-scale data interpretation increase.
Sample Selection
When ordering genomic testing, oncologists will find it helpful to consider the type of sample that will be used and its potential effect on results. Factors such as urgency, disease stage, previous treatments, and family history can affect the choice of sample and the testing method. It is also important to consider whether paired germline normal testing will be undertaken and what types of samples will be required for those tests. Communication between oncologists, surgeons, pathologists, and the laboratory should aid in determining the best sampling and testing methods.
FFPE Compared with Fresh or Frozen Biopsy Samples
The greatest proportion of samples assessed by Canadian pathologists are ffpe49. That sampling method has the advantage of reducing the logistic and ethics difficulties associated with obtaining fresh tissue biopsies41. However, ffpe preparation of samples can result in dna crosslinking or cytosine deamination, which can produce a large number of sequence artifacts. The artifacts can be removed with the use of biochemical or post-sequencing analytical methods and do not significantly affect the results of gene panel assays. However, when wes or wgs are being performed, age, fixation protocol, and storage method could affect the results. An understanding of the type and timeframe of the analysis to be undertaken is therefore necessary to determine whether ffpe or fresh or frozen biopsy is the best sampling method for research purposes. In clinical practice, ffpe remains the preferred tissue for panel-type assays.
Liquid Biopsies
The measurement of ctdna or cftna in peripheral blood samples might provide a viable alternative to invasive tumour biopsies7. Because ctdna or cftna can be measured repeatedly over time from a series of simple blood draws, it is possible to dynamically monitor tumour evolution, treatment response, and resistance mechanisms to more effectively tailor disease management. In addition, those methods can provide more representative samples, overcoming some of the limitations of tissue biopsies in regard to intra-tumour heterogeneity and differences between metastatic sites. However, low concentrations of ctdna or cftna can impair the sensitivity of the assay49. Further refinement and validation should aid in producing effective and noninvasive samples for ngs analyses. Currently, ctdna or cftna samples are used in clinical practice to screen for T790 resistance variants in patients with non-small-lung cancer progressing after treatment with tyrosine kinase inhibitors. If found to be negative for T790 variants, tumour tissue should still subsequently be analyzed to confirm findings50.
Sampling Considerations
Compared with biopsies for histology, samples for most ngs methods have to be relatively large, with a significant number of tumour cells51. Analytic failures in NGS stem, in greatest proportion, from the provision of insufficient tissue for dna extraction. Typically, those samples are less than 2 mm in their greatest dimension, the tissue contains less than 10% tumour, or the tumour is less than 10% viable. Larger samples also ensure that the heterogeneity of the tumour is well represented in test results. When a sample is insufficient to implement ngs, traditional low-throughput methods might be better able to obtain results. Moreover, some newer ngs methods that have now been developed can achieve results with smaller samples.
Best Practices
■ Before biopsy, communication between surgeons, pathologists, oncologists, and the laboratory should aid in determining the best sampling and testing methods.
■ Standard clinical assessment, including detailed phenotyping, should be undertaken before testing to facilitate interpretation of genome-wide variants. Details such as test urgency, variants of interest, disease stage, previous treatment, and family history will help in selecting the best sampling and testing methods.
■ It is important to consider whether paired germline normal testing will be undertaken and what types of samples will be required for those tests.
Unmet Needs
■ Further refinement and validation of ctdna or cftna analysis from liquid biopsies should aid in producing effective and noninvasive samples.
Bioinformatics and Interpretation
Although oncologists are not involved in the bioinformatics of ngs-based tests, it is important that they be aware of the limitations of testing methods. Despite rapid progression in the data-generating ability of ngs instruments, improvements in the software for analysis are needed—particularly improvements in clinical interpretation and report generation. Moreover, to extract clinically relevant information, the large volume of data generated from multigene panels requires experienced bioinformatics personnel and computational support.
Deciphering the large amount of data from ngs outputs poses a number of challenges. The platform used for ngs analysis can be based on hybrid capture or amplicon sequencing49. Hybrid capture methods hybridize dna sequences to complementary rna probes that are biotinylated, with the non-bound dna being washed away. That method maintains the relative proportion of dna fragments in the original cell, which aids in more accurately determining copy-number variations, but requires a higher depth of sequencing and more detailed bioinformatics. Amplicon sequencing uses pcr to enrich target regions for analysis. Compared with hybrid capture protocols, amplicon sequencing requires less dna and has a faster turnaround time. However, bias introduced by pcr can influence the observed allele fraction and reduce confidence in calling copy-number variations.
The larger the genomic territory explored, the greater the chance that, in mapping the reads to a location on the reference genome, the software will assign a percentage as being “unmappable,” which occurs when the read either cannot be aligned or could potentially align to multiple unique regions in the reference genome49. However, using additional analytic techniques, systematic biases of specific assays can often be successfully identified and removed during bioinformatics analysis. In addition, intra-tumour heterogeneity can make it difficult to determine whether a read of a low allele fraction is a true alteration or an artifact of the method that should be discarded. The latter factors can be improved with samples enriched for tumour relative to normal tissue and with greater depth of coverage during analysis.
In interpreting the data from ngs outputs, a number of available algorithms can identify clinically relevant alterations—for example, databases such as the Catalogue of Somatic Mutations in Cancer and My Cancer Genome (http://www.mycancergenome.org), among others49. Databases that show whether variants have been seen in published reports and whether they are of prognostic or therapeutic significance have also been created. Interpretation of clinically meaningful alterations therefore depends on the quality and completeness of the databases referenced. Ultimately, data-sharing and communication between centres is vital in improving access to good-quality data.
Best Practices
■ Oncologists should understand the limitations of ngs-based testing methods and the databases used to interpret findings.
Unmet Needs
■ Improved data interpretation will require access to appropriate repositories, data sharing, central bioinformatics, and appropriately scaled expertise.
NGS Reports
For oncologists, understanding the terminology and nomenclature used in describing variants is important in accurately interpreting and communicating the results of genomic tests. However, the structure of reports and the terminology used varies with the institution. To ensure that results are interpreted correctly, consulting with the laboratory should be considered. The term “mutation” is often used to describe a permanent alteration in the human genome52,53. However, some disciplines use the term to indicate all structural changes, and others limit its use to changes that are disease-causing. Similarly, “polymorphism” is used to denote a non-disease-causing change or a change found at a frequency of 1% or higher in the population. To standardize terms and to reduce confusion, guidelines from the Human Genome Variation Society (hgvs)53 and the American College of Medical Genetics and Genomics52 now recommend using the term “variant” to describe all genomic alterations.
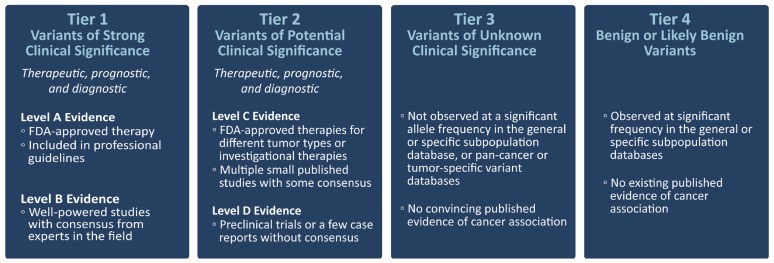
The American College of Medical Genetics and Genomics guideline. which has been endorsed by the Canadian College of Medical Geneticists46, recommends using the term “pathogenic” to identify possibly disease-causing variants and subdividing that designation into 5 categories of certainty52. In addition, a guideline jointly issued by the Association for Molecular Pathology, the American Society of Clinical Oncology, and the College of American Pathologists recommends categorizing somatic variants by their effect on clinical care, as related to therapeutic, prognostic, diagnostic, and preventive actionability (Figure 2). The guideline proposes 4 levels of clinical effect, with the highest level relating to biomarkers that predict response or resistance to approved therapies or that have been included in professional guidelines as one or more of therapeutic, diagnostic, or prognostic for specific tumour types.
FIGURE 2.
Evidence-based variant classification. Adapted from Li et al., 201754. FDA = U.S. Food and Drug Administration.
In reporting alterations, laboratory accreditation requires that standard nomenclature be followed, as described and maintained by the hgvs (http://varnomen.hgvs.org/). Nomenclature is continually updated by the hgvs as knowledge of the human genome progresses. Table III sets out definitions of common hgvs terms and variant reporting examples53.
TABLE III.
Human Genome Variation Society definitions of common terminology and nomenclaturea
| Term (reporting code) | Definition | Reporting example |
|---|---|---|
| Complementary DNA (cDNA) | DNA copy of a single-stranded RNA molecule synthesized using the enzyme reverse transcriptase | Not applicable |
| Deletion (del) | A sequence change in which, compared with a reference sequence, one or more nucleotides are not present | Breast BRCA2 c.7210_7211delAA |
| Duplication (dup) | A sequence change in which, compared with a reference sequence, a copy of one or more nucleotides is inserted directly 3′ of the original copy of that sequence | Breast BRCA1 c.5266dupC |
| Exon | Any nucleotide sequence within a gene that, during maturation of the RNA transcript, is not removed by RNA splicing | Not applicable |
| Intron | Any nucleotide sequence within a gene that, during maturation of the RNA transcript, is removed by RNA splicing | Not applicable |
| Insertion (ins) | A sequence change in which, compared with the reference sequence, one or more nucleotides are inserted and the insertion is not a copy of a sequence immediately upstream | Hematopoietic TP53 c.209_210insNN |
| Inversion (inv) | A sequence change in which, compared with the reference sequence, more than one nucleotide replacing the original sequence are the reverse complement of the original sequence | Lung ALK inv(2)(p23q35) |
| Missense | A variant in a protein sequence in which, compared with the reference sequence, an amino acid is replaced by another amino acid | Lung KRAS p.G12V |
| Nonsense | A variant in a protein sequence in which, compared with the reference sequence, an amino acid is replaced with a translational stop codon (termination codon) | Breast TP53 p.W146* |
| Nucleotide | A letter from the DNA code (A, C, G, or T) | Not applicable |
| Splicing | The process removing specific segments (the introns) of a precursor messenger RNA (pre-mRNA) transcript; when an intron is removed, the flanking RNA segments (the exons) are joined together (ligated) | Where a change is expected to affect splicing on the RNA level: r.spl? |
| Substitution (>) | A sequence change in which, compared with a reference sequence, one residue is replaced by one other residue | Melanoma NRAS c.181C>A |
| Translocation | A sequence change in which, compared with a reference sequence, all nucleotides upstream and downstream from a specific nucleotide position (the breakpoint) derive from different chromosomes | Ewing sarcoma EWSR1–FLI1 fusion t(11;22)(q24;q12) |
Adapted from Human Genome Variation Society recommendations (http://varnomen.hgvs.org/), the COSMIC (Catalog of Somatic Mutations in Cancer) database (https://cancer.sanger.ac.uk/cosmic), the NCBI (National Center for Biotechnology Information) database (https://www.ncbi.nlm.nih.gov/snp) and the Atlas of Genetics and Cytogenetics in Oncology and Haematology (http://atlasgeneticsoncology.org/).
Best Practices
■ Categorization by level of actionability is valuable in providing a uniform approach to determining the clinical effect of somatic variants.
■ Awareness of key terminology and nomenclature and any updates made by the hgvs over time is important in accurately interpreting the results of genomic tests.
■ To effectively interpret and disseminate results, oncologists should be familiar with the details of molecular reports, while considering limitations of the analyses.
■ Clinicians should consult with laboratory staff geneticists and pathologists to aid in the accurate interpretation of reports.
Unmet Needs
■ There is a need for standardized reporting within and between institutions to aid in the access to genomic information and interpretation of results. The laws and practices relevant to data-sharing, privacy, and ownership also have to be well defined.
Patient Communication
Oncologists face a number of challenges in communicating the results of ngs analyses to patients. When ngs testing is performed, incidental findings are a possibility that has to be communicated to patients. It is therefore important that processes be implemented to ensure that patients fully understand the tests undertaken and their potential consequences if they are to give adequate informed consent.
Counselling before tumour testing should include a discussion of the limitations of testing, the likelihood and implications of diagnosis and incidental findings, and the potential need for further analysis to facilitate clinical interpretation, including studies performed in a research setting. Further, processes should be put in place to initiate referrals for genetic counselling when pathogenic variants are identified, providing patients an opportunity to discuss the implications for themselves and their at-risk family members.
Occasionally, germline testing could also be required when testing identifies a variant that might have a germline origin. In those cases, oncologists should understand the policies at their institution for genetics referral. In addition, when analytic methods examine a greater number of genes, the likelihood of identifying vuss increases, making it difficult to counsel patients about the relationship between the disease and the variant. Regular consultations with multidisciplinary teams should aid oncologists in effectively disseminating ngs results.
Best Practices
■ Multidisciplinary teams including pathologists, surgeons, genetic counsellors, and oncologists should meet regularly to discuss the needs of patients before and after testing.
■ Before testing, patients should be counselled to ensure that they are informed about the limitations, implications, potential incidental findings, and possible need for further analysis of ngs-based test results.
■ In interpreting reports, oncologists should consult with genetics and be aware of the availability at their institution for genetics referral when subsequent germline testing is needed.
■ When disseminating results, referral to genetic counsellors might be necessary to effectively communicate results to patients and their families.
Unmet Needs
■ There is a need for improved communication within health care teams and between institutions to ensure the best in individualized patient care.
Clinical Research
Clinical Trials
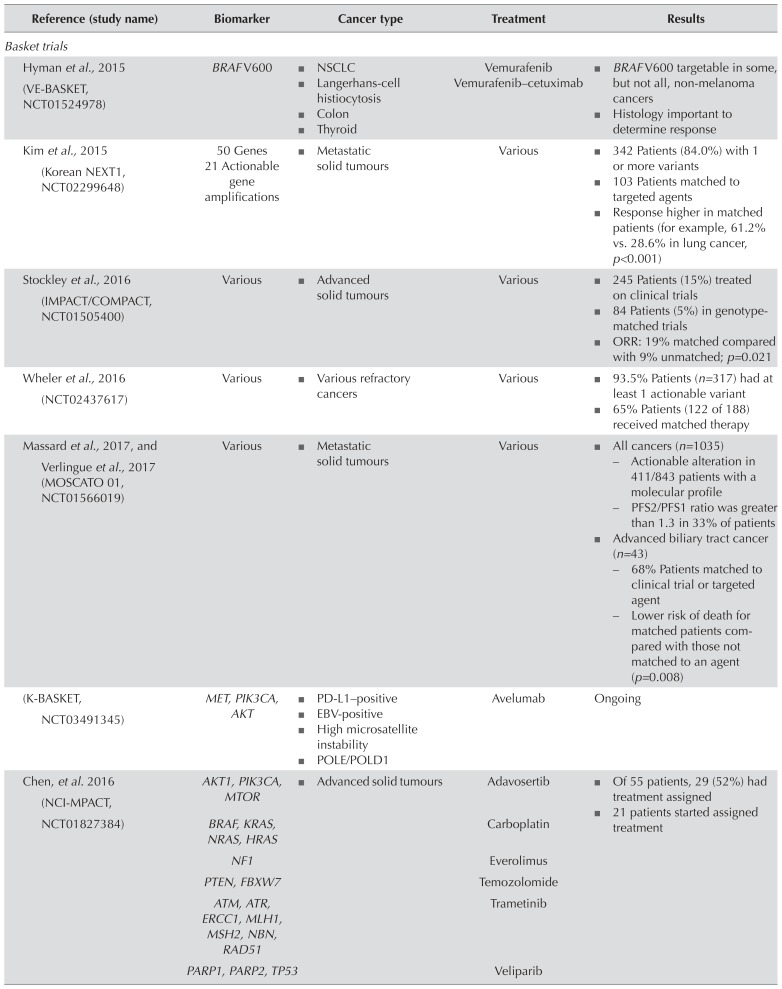
When patients are not eligible for available targeted therapies, clinical trials that include ngs analyses can match patients with experimental therapies. The identification of driver alterations that can guide treatment decisions is a key focus of ngs-based clinical trials in oncology. Given the rarity of many genomic alterations, two approaches are being used to develop effective ngs-based clinical trial designs. “Basket trials” include patients with 1 genomic biomarker associated with a large number of disease indications; “umbrella trials” focus on 1 disease indication, but include multiple biomarkers under investigation. Table IV presents examples of clinical trials using those approaches44,55–64.
TABLE IV.
Key next-generation sequencing–based basket and umbrella trials assessing molecular biomarkers for adult cancer treatment
| Reference (study name) | Biomarker | Cancer type | Treatment | Results |
|---|---|---|---|---|
| Basket trials | ||||
|
| ||||
| Hyman et al., 2015 (VE-BASKET, NCT01524978) | BRAF V600 |
|
Vemurafenib Vemurafenib–cetuximab |
|
|
| ||||
| Kim et al., 2015 (Korean NEXT1, NCT02299648) | 50 Genes 21 Actionable gene amplifications |
|
Various |
|
|
| ||||
| Stockley et al., 2016 (IMPACT/COMPACT, NCT01505400) | Various |
|
Various |
|
|
| ||||
| Wheler et al., 2016 (NCT02437617) | Various |
|
Various |
|
|
| ||||
| Massard et al., 2017, and Verlingue et al., 2017 (MOSCATO 01, NCT01566019) | Various |
|
Various |
|
|
| ||||
| (K-BASKET, NCT03491345) | MET, PIK3CA, AKT |
|
Avelumab | Ongoing |
|
| ||||
| Chen, et al. 2016 (NCI-MPACT, NCT01827384) | AKT1, PIK3CA, MTOR |
|
Adavosertib |
|
| BRAF, KRAS, NRAS, HRAS | Carboplatin | |||
| NF1 | Everolimus | |||
| PTEN, FBXW7 | Temozolomide | |||
| ATM, ATR, ERCC1, MLH1, MSH2, NBN, RAD51 | Trametinib | |||
| PARP1, PARP2, TP53 | Veliparib | |||
|
| ||||
| (NCI-MATCH, NCT02465060) | >4000 Variants |
|
35 Treatment arms | Ongoing |
|
| ||||
| Toulmonde et al., 2015 (MOST, NCT02029001) | Various |
|
Nilotinib Everolimus Sorafenib Lapatinib Pazopanib Olaparib Durvalumab– tremelimumab |
Ongoing |
|
| ||||
| (CAPTUR, NCT03297606) | Various |
|
Approved targeted and immuno-oncology agents | Ongoing |
|
| ||||
| Umbrella trials | ||||
|
| ||||
| (LUNG-MAP, NCT02154490) | CDK4, PIK3CA, MET, FGFR |
|
Nivolumab vs. targeted therapy | Ongoing |
|
| ||||
| Gerber et al., 2015 (ALCHEMIST— screening: NCT02194738; EGFR: NCT02193282; ALK: NCT02201992) | EGFR, ALK |
|
Screening Erlotinib vs. placebo Crizotinib vs. placebo |
Ongoing |
|
| ||||
| Adams et al., 2018 (FOCUS4, ISRCTN 90061546) | RAS, BRAF, KRAS, PIK3CA |
|
Novel agent vs. placebo |
|
|
| ||||
| (ROCHE-BFAST, NCT03178552) | ALK, RET, bTMB-positive |
|
Alectinib Atezolizumab vs. standard treatment |
Ongoing |
NSCLC = non-small-cell lung cancer; ORR = overall response rate; EBV = Epstein–Barr virus; PFS = progression-free survival; MAMS = multi-arm, multi-stage; bTMB = tumour mutational burden in blood.
Unfortunately, no easy method to identify molecular-based clinical trials recruiting patients in Canada is currently available. Although Web sites such as ClinicalTrials. gov are useful for obtaining some study details, they do not include up-to-date information about recruitment and Canadian sites. Databases and resources that can identify available trials, with patient eligibility criteria, would be useful in ensuring that patients have access to all available and novel treatment options. Moreover, laboratories need time to modify and validate targeted panels that are used to assess patients for clinical trials. Involving laboratories long before patient recruitment begins to ensure that they can be ready in time is therefore important.
Canadian Research Initiatives
Canada is quickly becoming a global leader in ngs-based research, with a number of national and provincial initiatives underway. Exactis Innovation (https://www.exactis.ca/about) is a pan-Canadian nonprofit organization that provides accelerated access to clinical trials for precision cancer therapies in patients whose cancer has been molecularly profiled. Its Personalize My Treatment program aims to match patients to suitable clinical trials based on sequencing studies that will collect data from solid tumours, liquid biopsies, and immune-oncology assays.
In Ontario, the Cancer Targeted Nucleic Acid Evaluation study (NCT02906943 at http://ClinicalTrials.gov/) is an initiative that is using large targeted panels to provide ngs-based molecular profiling at multiple Ontario cancer centres to patients with advanced solid tumours65. Results will facilitate targeted therapy and immunotherapy clinical trials and identify patient subsets for additional research studies. The initiative aims to establish a province-wide repository of samples for future research, as well as a large database of genomically-characterized and clinically-annotated sequencing results. The Personalized Onco-Genomics program at BC Cancer is using whole-genome and transcriptome analysis to identify cancer drivers and corresponding drugs that might inhibit the relevant pathways in patients with advanced cancers44. From 2012 to 2014, 78 patients with incurable cancers underwent wgs, with results being categorized as actionable in 55 cases, and 23 patients receiving treatments based on wgs results. Based on the results of the foregoing initiatives, the Canadian Profiling and Targeted Agent Utilization Trial (a basket trial, NCT03297606 at http://ClinicalTrials.gov/) is taking on patients who have potentially actionable variants identified with an assay approved by the research group and providing them with access to approved targeted therapies.
Best Practices
■ When patients are not eligible for approved targeted therapies, ngs might be used to detect variants meeting eligibility criteria for clinical trials.
■ To develop and validate any required gene panels, laboratories have to be informed and to work with the study sponsor well in advance of patient recruitment into a clinical trial.
Unmet Needs
■ There is a need for easy access to information about available clinical trials and eligibility criteria for patient enrolment in Canada.
■ Clinical trials could aid in determining the benefit of multigene panels to guide treatment across cancer types.
SUMMARY
Rapid advancements and adoption of precision medicine allow for an unprecedented use of ngs technologies in clinical practice. Oncologists now have the ability to provide the highest level of personalized care to cancer patients to optimize treatment choices and outcomes. Next-generation sequencing allows for the identification of high-risk genes associated with hereditary cancers, more precise diagnosis and prognosis prediction, selection of patients for optimal targeted therapies and clinical trials, and detection of resistance alterations to guide subsequent treatment. Despite the rapid advances in ngs technology, clinicians must be aware of challenges and limitations in analyses, bioinformatics, data interpretation, and patient communication. Those issues can be mitigated through appropriate sampling, careful and detailed analysis, improved reporting and communication within the health care team, and data-sharing between institutions. With continued improvement in ngs assays and bioinformatics, oncologists will have powerful and precise tools to achieve the highest level of care for patients with cancer.
ACKNOWLEDGMENTS
The authors acknowledge financial support from Hoffmann–La Roche, AstraZeneca, Pfizer, Illumina, and Thermo Fisher Scientific for medical writing services provided by Anna Christofides of impact Medicom Inc.
Footnotes
This guideline has been endorsed by the Canadian Association of Genetic Counsellors and the Canadian Association of Pathologists/Association canadienne des pathologistes.
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare the following interests: SY has received compensation from Bayer, Hoffmann–La Roche, and Pfizer for participating in advisory boards; AC has received funding from Hoffmann–La Roche, AstraZeneca, Pfizer, Illumina, and Thermo Fisher Scientific for medical writing services related to this paper; MRD has received compensation from Hoffman–La Roche and AstraZeneca for participating in advisory boards, and honoraria from AstraZeneca; II has received honoraria from Pfizer, AstraZeneca, Novartis, Hoffmann–La Roche, and Bayer for participating in advisory boards; JM has received honoraria from AstraZeneca; TS has received compensation from AstraZeneca, Bristol–Myers Squibb, and Janssen for participating in advisory boards, and has received research funding from AstraZeneca and Janssen; AS has participated in advisory boards for Merck, AstraZeneca, Janssen, Pfizer, Novartis, Roche, Bristol–Myers Squibb, and Myriad; he has also received research funding from Merck, AstraZeneca, Bristol–Myers Squibb, and Pfizer. The remaining authors have no conflicts to disclose.
REFERENCES
- 1.Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013;98:236–8. doi: 10.1136/archdischild-2013-304340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamps R, Brandao RD, Bosch BJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18020308. pii:E308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canadian College of Medical Geneticists (ccmg) CCMG Statement on Germline Variant Classification. Kingston, ON: ccmg; 2017. [Available online at: https://www.ccmg-ccgm.org/images/Guidelines/CCMG_Statement_on_Germline_Variant_Classification_FINAL_April_2017.pdf; cited 30 July 2018] [Google Scholar]
- 4.Yang F, Press RD. Next-generation sequencing multi-gene mutation panels in myeloid malignancies. Hematologist. 2016;13:6. [Google Scholar]
- 5.Melosky B, Blais N, Cheema P, et al. Standardizing biomarker testing for Canadian patients with advanced lung cancer. Curr Oncol. 2018;25:73–82. doi: 10.3747/co.25.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehrad M, Roy S, Bittar HT, Dacic S. Next-generation sequencing approach to non–small cell lung carcinoma yields more actionable alterations. Arch Pathol Lab Med. 2018;142:353–7. doi: 10.5858/arpa.2017-0046-OA. [DOI] [PubMed] [Google Scholar]
- 7.Cummings CA, Peters E, Lacroix L, Andre F, Lackner MR. The role of next-generation sequencing in enabling personalized oncology therapy. Clin Transl Sci. 2016;9:283–92. doi: 10.1111/cts.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennell NA, Mutebi A, Zhou ZY, et al. Economic impact of next generation sequencing vs. sequential single-gene testing modalities to detect genomic alterations in metastatic non–small cell lung cancer using a decision analytic model [abstract 9031] J Clin Oncol. 2018;36 doi: 10.1200/JCO.2018.36.15_suppl.9031. [Available online at: http://ascopubs.org/doi/abs/10.1200/JCO.2018.36.15_suppl.9031; cited 16 February 2019] [DOI] [PubMed] [Google Scholar]
- 9.AstraZeneca Canada Inc. Lynparza: Olaparib Capsules [product monograph] Mississauga, ON: AstraZeneca Canada Inc; 2018. [Google Scholar]
- 10.Eli Lilly Canada Inc. Erbitux: (Cetuximab) Intravenous Injection [product monograph] Toronto, ON: Eli Lilly Canada Inc; 2018. [Google Scholar]
- 11.Pfizer Canada Inc. Xalkori: Crizotinib Capsules [product monograph] Kirkland, QC: Pfizer Canada Inc; 2017. [Google Scholar]
- 12.Novartis Pharmaceuticals Canada Inc. Mekinist: Trametinib Tablets [product monograph] Dorval, QC: Novartis Pharmaceuticals Canada Inc; 2018. [Google Scholar]
- 13.Hoffmann–La Roche Limited. Zelboraf: Vemurafenib Film-Coated Tablet [product monograph] Mississauga, ON: Hoffmann–La Roche Limited; 2018. [Google Scholar]
- 14.Novartis Pharmaceuticals Canada Inc. Tafinlar: Dabrafenib (as Dabrafenib Mesylate) Capsules [product monograph] Dorval, QC: Novartis Pharmaceuticals Canada Inc; 2018. [Google Scholar]
- 15.Apotex Inc. Apo-Erlotinib: Erlotinib Hydrochloride Tablets [product monograph] Toronto, ON: Apotex Inc; 2017. [Google Scholar]
- 16.Boehringer Ingelheim (Canada) Ltd. Giotrif: Afatinib Tablets [product monograph] Burlington, ON: Boehringer Ingelheim (Canada) Ltd; 2018. [Google Scholar]
- 17.Ariad Pharmaceuticals Inc. Iclusig: Ponatinib: Tablets (as Ponatinib Hydrochloride) [product monograph] Cambridge, MA: Ariad Pharmaceuticals Inc; 2018. [Google Scholar]
- 18.Hoffmann–La Roche Limited. Alecensaro Alectinib (as Alectinib Hydrochloride) Capsule [product monograph] Mississauga, ON: Hoffmann–La Roche Limited; 2018. [Google Scholar]
- 19.Novartis Pharmaceuticals Canada Inc. Zykadia: Ceritinib Capsules [product monograph] Dorval, QC: Novartis Pharmaceuticals Canada Inc; 2018. [Google Scholar]
- 20.AstraZeneca Canada Inc. Tagrisso: Osimertinib Tablets [product monograph] Mississauga: ON: AstraZeneca Canada Inc; 2018. [Google Scholar]
- 21.Amgen Canada Inc. Vectibix (Panitumumab): Sterile Solution for Infusion [product monograph] Mississauga: ON: Amgen Canada Inc; 2017. [Google Scholar]
- 22.Hoffmann–La Roche Limited. Cotellic: Cobimetinib Tablets [product monograph] Mississauga: ON: Hoffmann–La Roche Limited; 2018. [Google Scholar]
- 23.Novartis Pharmaceuticals Canada Inc. Afinitor Disperz (Everolimus Tablets for Oral Suspension) [product monograph] Dorval, QC: Novartis Pharmaceuticals Canada Inc; 2017. [Google Scholar]
- 24.Takeda Canada Inc. Alunbrig: Brigatinib Tablets [product monograph] Oakville, ON: Takeda Canada Inc; 2018. [Google Scholar]
- 25.Novartis Pharmaceuticals Canada Inc. Tasigna (Nilotinib Capsules) [product monograph] Dorval, QC: Novartis Pharmaceuticals Canada Inc; 2015. [Google Scholar]
- 26.Bristol–Myers Squibb Canada. Sprycel: Dasatinib Tablets [product monograph] Montreal, QC: Bristol–Myers Squibb Canada; 2017. [Google Scholar]
- 27.Wyeth LLC. Bosulif: Bosutinib Tablets [product monograph] Kirkland, QC: Wyeth LLC; 2017. [Google Scholar]
- 28.Merck Canada Inc. Keytruda: Pembrolizumab Powder for Solution for Infusion [product monograph] Kirkland, QC: Merck Canada Inc; 2017. [Google Scholar]
- 29.Longley J, Naheed S, Copson E. Medical Oncology and Genomics. Cambridge, U.K.: phg Foundation; 2016. [Available online at: http://www.phgfoundation.org/documents/543_1456831319.pdf; cited 1 July 2018] [Google Scholar]
- 30.Ionescu DN, Downes MR, Christofides A, Tsao MS. Harmonization of PD-L1 testing in oncology: a Canadian pathology perspective. Curr Oncol. 2018;25:e209–16. doi: 10.3747/co.25.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36:633–41. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–20. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 35.Nowak JA, Yurgelun MB, Bruce JL, et al. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn. 2017;19:84–91. doi: 10.1016/j.jmoldx.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang L, Chang M, Chang HM, Chang F. Microsatellite instability: a predictive biomarker for cancer immunotherapy. Appl Immunohistochem Mol Morphol. 2018;26:e15–21. doi: 10.1097/PAI.0000000000000575. [DOI] [PubMed] [Google Scholar]
- 37.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Comprehensive Cancer Network (nccn) NCCN Clinical Practice Guidelines in Oncology: Colon Cancer. Ver. 4.2018. Fort Washington, PA: nccn; 2018. [Current version available online at: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf (free registration required); cited 1 October 2018] [DOI] [PubMed] [Google Scholar]
- 39.National Comprehensive Cancer Network (nccn) NCCN Clinical Practice Guidelines in Oncology: Non–Small Cell Lung Cancer. Ver. 3.2019. Fort Washington, PA: nccn; 2019. [Current version available online at: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (free registration required); cited 1 October 2018] [Google Scholar]
- 40.Planchard D, Popat S, Kerr K, et al. on behalf of the esmo Guidelines Committee. Metastatic non–small cell lung cancer: esmo clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(suppl 4):iv192–237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 41.Horak P, Frohling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1:e000094. doi: 10.1136/esmoopen-2016-000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture–based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21:3631–9. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beaulieu CL, Majewski J, Schwartzentruber J, et al. forge Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am J Hum Genet. 2014;94:809–17. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskin J, Jones S, Aparicio S, et al. Lessons learned from the application of whole-genome analysis to the treatment of patients with advanced cancers. Cold Spring Harb Mol Case Stud. 2015;1:a000570. doi: 10.1101/mcs.a000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Razzaq SK, Vo AD, Gautam M, Li H. Identifying fusion transcripts using next generation sequencing. Wiley Interdiscip Rev RNA. 2016;7:811–23. doi: 10.1002/wrna.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canadian College of Medical Geneticists (ccmg), Ad Hoc Working Group on Next Generation Sequencing. CCMG Practice Guideline: Laboratory Guidelines for Massively Parallel Sequencing. Kingston, ON: ccmg; 2018. [Available online at: https://www.ccmg-ccgm.org/documents/1-CCMG-Guidelines-for-MPS_FINAL-June-11-2018.pdf; cited 27 July 2018] [Google Scholar]
- 47.Ratajska M, Koczkowska M, Zuk M, et al. Detection of BRCA1/2 mutations in circulating tumor dna from patients with ovarian cancer. Oncotarget. 2017;8:101325–32. doi: 10.18632/oncotarget.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Velden DL, van Herpen CML, van Laarhoven HWM, et al. Molecular tumor boards: current practice and future needs. Ann Oncol. 2017;28:3070–5. doi: 10.1093/annonc/mdx528. [DOI] [PubMed] [Google Scholar]
- 49.Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80. doi: 10.1186/s13073-015-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stockley T, Souza CA, Cheema PK, et al. Evidence-based best practices for EGFR T790M testing in lung cancer in Canada. Curr Oncol. 2018;25:163–9. doi: 10.3747/co.25.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Kateb H, Nguyen TT, Steger-May K, Pfeifer JD. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (ngs) Mol Oncol. 2015;9:1737–43. doi: 10.1016/j.molonc.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards S, Aziz N, Bale S, et al. on behalf of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Human Genome Variation Society (hgvs) Recommendations for the Description of Sequence Variants. Melbourne, Australia: hgvs; 2016. [Available online at: https://www.hgvs.org/mutnomen/recs.html; cited 25 June 2018] [DOI] [PubMed] [Google Scholar]
- 54.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373:726–36. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verlingue L, Malka D, Allorant A, et al. Precision medicine for patients with advanced biliary tract cancers: an effective strategy within the prospective moscato-01 trial. Eur J Cancer. 2017;87:122–30. doi: 10.1016/j.ejca.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 57.Kim ST, Lee J, Hong M, et al. The next-1 (Next Generation Personalized Tx with Multi-omics and Preclinical Model) trial: prospective molecular screening trial of metastatic solid cancer patients, a feasibility analysis. Oncotarget. 2015;6:33358–68. doi: 10.18632/oncotarget.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stockley TL, Oza AM, Berman HK, et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret impact/compact trial. Genome Med. 2016;8:109. doi: 10.1186/s13073-016-0364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Massard C, Michiels S, Ferte C, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the moscato 01 trial. Cancer Discov. 2017;7:586–95. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 60.Wheler JJ, Janku F, Naing A, et al. Cancer therapy directed by comprehensive genomic profiling: a single center study. Cancer Res. 2016;76:3690–701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 61.Chen AP, Williams M, Kummar S, et al. Feasibility of molecular profiling based assignment of cancer treatment (mpact): a randomized nci precision medicine study [abstract 2539] J Clin Oncol. 2016;34 [Available online at: http://ascopubs.org/doi/abs/10.1200/JCO.2016.34.15_suppl.2539; cited 16 February 2019] [Google Scholar]
- 62.Toulmonde M, Le Tourneau C, Italiano A, et al. A randomized, open-label, phase ii trial evaluating the clinical benefit of a maintenance treatment targeting tumor molecular alterations in patients with advanced solid tumors [abstract TPS2622] J Clin Oncol. 2015;33 doi: 10.1200/jco.2015.33.15_suppl.tps2622. [Available online at: http://ascopubs.org/doi/abs/10.1200/jco.2015.33.15_suppl.tps2622; cited 18 February 2019] [DOI] [Google Scholar]
- 63.Gerber DE, Oxnard GR, Govindan R. alchemist: bringing genomic discovery and targeted therapies to early-stage lung cancer. Clin Pharmacol Ther. 2015;97:447–50. doi: 10.1002/cpt.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams R, Brown E, Brown L, et al. on behalf of the focus4 trial investigators. Inhibition of egfr, her2, and her3 signalling in patients with colorectal cancer wild-type for BRAF, PIK3CA, KRAS, and NRAS (focus4-d): a phase 2–3 randomised trial. Lancet Gastroenterol Hepatol. 2018;3:162–71. doi: 10.1016/S2468-1253(17)30394-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ontario Institute for Cancer Research (oicr) Ontario-Wide Cancer Targeted Nucleic Acid Evaluation (OCTANE) Toronto, ON: oicr; 2019. [Available online at: https://oicr.on.ca/research-portfolio/octane; cited 7 July 2018] [Google Scholar]