Abstract
The ribosomal synthesis of proteins in the eukaryotic cytosol has always been thought to start from the unformylated N-terminal (Nt) methionine (Met). In contrast, in virtually all nascent proteins in bacteria and eukaryotic organelles, such as mitochondria and chloroplasts, Nt-formyl-methionine (fMet) is the first building block of ribosomal synthesis. Through extensive approaches, including mass spectrometric analyses of the N-termini of proteins and molecular genetic techniques with an affinity-purified antibody for Nt-formylation, we investigated whether Nt-formylated proteins could also be produced and have their own metabolic fate in the cytosol of a eukaryote, such as yeast Saccharomyces cerevisiae. We discovered that Nt-formylated proteins could be generated in the cytosol by yeast mitochondrial formyltransferase (Fmt1). These Nt-formylated proteins were massively upregulated in the stationary phase or upon starvation for specific amino acids and were crucial for the adaptation to specific stresses. The stress-activated kinase Gcn2 was strictly required for the upregulation of Nt-formylated proteins by regulating the activity of Fmt1 and its retention in the cytosol. We also found that the Nt-fMet residues of Nt-formylated proteins could be distinct N-terminal degradation signals, termed fMet/N-degrons, and that Psh1 E3 ubiquitin ligase mediated the selective destruction of Nt-formylated proteins as the recognition component of a novel eukaryotic fMet/N-end rule pathway, termed fMet/N-recognin.
Keywords: Formyltransferase, Gcn2, Methionine, Proteolysis, Psh1, Ubiquitin
Nearly all proteins initially synthesized by ribosomes have methionine (Met) at their N-terminus. In eukaryotes, cytosolically synthesized proteins from nuclear DNA-encoded mRNA bear an unmodified N-terminal (Nt) Met residue. Co-translationally and irreversibly, this Nt-Met of eukaryotic proteins is often Nα-terminally acetylated by Nt-acetylases. Although Nt-acetylation is one of the most abundant modifications of eukaryotic proteins (about 60% and 80% of proteins in yeast Saccharomyces cerevisiae and human cells, respectively), a universal role for Nt-acetylation had been largely unknown until the Ac/N-degrons and the Ac/N-end rule pathway were found. The Ac/N-degrons are specific degradation signals created by the Nt-acetylation of eukaryotic proteins and are targeted by the Ac/N-end rule pathway, one branch of the N-end rule pathways (Hwang CS et al (2010) Science 327, 973–977).
The N-end rule pathways recognize, by N-recognins, proteins containing N-degrons, and causes their degradation by proteasomes or autophagy in eukaryotes. The N-degrons are N-terminal degradation signals determined by destabilizing Nt-residue of a protein. The N-recognins are recognition components of the N-end rule pathway, which are E3 ubiquitin ligases or other proteins which target N-degrons. Detailed investigations of N-degrons over 30 years have unveiled that nearly all species of amino acids can be destabilizing Nt-residues in analogous contexts of the sequences, indicating that many proteins can be short-lived N-end rule substrates, conditionally. Indeed, regulated determination of the in vivo half-life of proteins or natural protein fragments by the N-end rule pathway has been shown to control a variety of biological processes occurring in a cell. Currently, the identified N-end rule pathways in eukaryotes are the Arg/N-end rule pathway, the Ac/N-end rule pathway, and the Pro/N-end rule pathway.
In contrast to eukaryotes, the Nt-Met of nearly all nascent proteins is Nα-terminally formylated in bacteria and eukaryotic organelles, such as mitochondria and chloroplasts. This modification is a pre-translational process because the Met moiety of initiator Met-tRNAs is Nα-terminally formylated by formyltransferases (FMTs) using 10-formyltetrahydrofolate as a co-substrate. Although FMTs are universal among bacteria, the formylation of Nt-Met is not strictly essential for protein synthesis or even cell viability. Mitochondrial FMTs, which are encoded from nuclear DNA, synthesized by cytosolic ribosomes, and then translocated into the inner matrix of mitochondria, are highly conserved in the genomes of all investigated eukaryotes. This universality of FMTs might raise a fundamental question: Why has this metabolically expensive Nt-formylation been maintained across evolution?
Given that the locations of the formyl and acetyl groups are identical in Nα-terminally modified proteins, and that they are chemically similar to each other, it might be assumed that Nt-fMet has a role in the degradation of bacterial proteins. Indeed, similar to the Ac/N-degrons in eukaryotes, the Nt-fMet of nascent proteins can act as bacterial N-degrons, termed fMet/N-degrons. In addition, a previous study reported that the expression of bacteria Escherichia coli FMT in yeast S. cerevisiae converted about 70% of initiator Met-tRNA to fMet-tRNA and inhibited cell growth, suggesting the production of Nt-formylated proteins in the cytosol of the yeast.
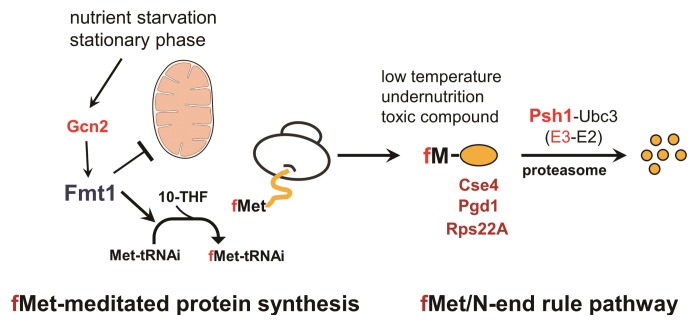
Through the mass analysis of enriched N-terminal peptides and, independent of mass spectrometry, the detection of Nt-fMet in proteins by an affinity-purified antibody, we found direct evidence that Nt-formylated proteins could be generated in the yeast cytosol and that Nt-formylation was mediated by Fmt1, the mitochondrial FMT of the yeast. As had been previously assumed, translocation of Fmt1 into the inner mitochondrial matrix was not sufficiently efficacious and consequently, mitochondrial Fmt1 transiently resided in the cytosol, even under non-stressed condition. We also detected greatly upregulated Nt-formylated proteins in the stationary phase or upon starvation for specific amino acids, such as histidine or lysine. The Gcn2 kinase, a multifunctional and stress-activated protein kinase in the yeast, was strictly required for the massive increase of Nt-formylated proteins and was found to regulate the enzymatic activity of Fmt1 and to mediate its retention in the cytosol (Diagram 1).
Diagram 1.
The fMet-mediated protein synthesis and fMet/N-end rule pathway in eukaryotes. In the stationary phase or upon starvation for nutrients, stress-activated kinase Gcn2 increases the cytosolic retention of mitochondrial formyltransferase (Fmt1) and also partly, its activity. As a result, Fmt1 augments the fMet-tRNAi in the cytosol and subsequently upregulates Nt-formylated proteins. These Nt-formylated proteins are important for the survival of cells against specific stresses and are destroyed in a proteasome-dependent manner through polyubiquitylation by Psh1 E3 ubiquitin ligase targeting the N-terminal fMet residues of Nt-formylated proteins.
We also found that the Nt-formylation of the proteins synthesized in the yeast cytosol created the previously unknown eukaryotic fMet/N-degrons and that Psh1 E3 ubiquitin ligase was the fMet/N-recognin, the recognition component of the eukaryotic fMet/N-end rule pathway. Downregulation of this Nt-formylation in the cytosol could lead cells to become hypersensitive to stress caused by undernutrition, prolonged cold, or toxic chemicals (Diagram 1).
The Nt-formylation of proteins had long been thought to be strictly limited to bacteria and bacteria-derived eukaryotic organelles. However, this pre-translational modification was found to also occur at the eukaryotic Nt-Met of proteins synthesized by cytosolic ribosomes. The Nt-formylated proteins in eukaryotes might be crucial for cell survival against, or adaptation to, natural but specific stresses because the eukaryotic Nt-formylation of proteins were massively upregulated by these stressors. The eukaryotic fMet/N-end rule pathway selectively destroys these Nt-formylated proteins in the eukaryotic cytosol. Given that FMTs are encoded by all examined eukaryotic genomes and the mechanism of N-degron recognition is strongly conserved, this previously unknown system might be universal among eukaryotes.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2016R1A6A3A11934059).
Abbreviations
- Met
methionine
- Nt
N-terminal
- tRNA
transfer RNA
- FMT
formyltransferase
- fMet
formylmethionine
- Fmt1
Formyl-Methionyl-tRNA Transformylase 1
- Gcn2
General Control Nonderepressible 2