Abstract
Heterogeneity in the metabolic properties of adipocytes in white adipose tissue has been well documented. We sought to investigate metabolic heterogeneity in adipocytes of brown adipose tissue (BAT), focusing on heterogeneity in nutrient uptake. To explore the possibility of metabolic heterogeneity in brown adipocytes, we used nanoscale secondary ion mass spectrometry (NanoSIMS) to quantify uptake of lipids in adipocytes interscapular BAT and perivascular adipose tissue (PVAT) after an intravenous injection of triglyceride-rich lipoproteins (TRLs) containing [2H]triglycerides (2H-TRLs). The uptake of deuterated lipids into brown adipocytes was quantified by NanoSIMS. We also examined 13C enrichment in brown adipocytes after administering [13C]glucose or 13C-labeled mixed fatty acids by gastric gavage. The uptake of 2H-TRLs-derived lipids into brown adipocytes was heterogeneous, with 2H enrichment in adjacent adipocytes varying by more than fourfold. We also observed substantial heterogeneity in 13C enrichment in adjacent brown adipocytes after administering [13C]glucose or [13C]fatty acids by gastric gavage. The uptake of nutrients by adjacent brown adipocytes within a single depot is variable, suggesting that there is heterogeneity in the metabolic properties of brown adipocytes.
Keywords: Brown adipose tissue, White adipose tissue, NanoSIMS imaging, Nutrient uptake, Fatty acids
1. Introduction
There is substantial heterogeneity in the gene expression, metabolism, and morphology of adipocytes in different white adipose tissue (WAT) depots [1, 2]. Even within a single WAT depot, the properties of adipocytes are heterogenous. For example, some adipocytes (beige adipocytes) are capable of responding to cAMP with high levels of UCP1 expression and high respiration rates [3]. Also, Boumelhem et al. [4] found heterogeneity in the expression of CD36, a fatty acid transporter, in WAT depots, and Valamov et al. [5] observed heterogeneous uptake of a fluorescently labeled fatty acid by adipocytes in WAT explants from rhesus monkeys. Heterogeneity in the uptake of the fluorescently labeled fatty acid was also observed in adipocytes generated from human stromal vascular cells [5]. These studies employing a fluorescently labeled fatty acid, while not quantitative, supported the idea that the metabolic properties of cultured adipocytes are variable, but they left open the question of whether the same sort of metabolic heterogeneity exists within WAT depots of live animals.
In the case of brown adipocytes, the regulation of metabolic activity has been investigated intensively [6]. However, whether heterogeneity exists in the metabolic properties of individual brown adipocytes has received little attention, and no one has tested whether nutrient uptake is uniform or heterogeneous. In the current studies, we tested whether the uptake of nutrients in brown adipocytes of mice is uniform or heterogeneous. Rather than using fluorescent fatty acids and adipose tissue explants, we injected mice with stable isotope–labeled nutrients and used NanoSIMS [7–11] to quantify uptake of the stable isotopes by brown adipocytes. An attractive feature of this approach is the ability to quantify the heterogeneity of nutrient uptake in brown adipocytes in living animals.
2. Materials and Methods
2.1. Mouse Strains
Wild-type mice and Gpihbp1–/– mice [12] (both strain C57BL/6J), 3–4 months of age, were housed in an accredited barrier vivarium with a 12-h light-dark cycle and fed a chow diet. All animal procedures were approved by UCLA’s institutional animal care and use committee (the Chancellor’s Animal Research Committee).
2.2. Metabolic studies
2H-labeled triglyceride-rich lipoproteins (2H-TRLs) were prepared as described [7]. Briefly, Gpihbp1–/– mice were given multiple doses of 2H-labeled mixed fatty acid over 4.5 days. Four hours after the final dose, blood was collected and 2H-TRLs were isolated from the plasma by ultracentrifugation. The 2H/1H ratio in 2H-TRLs was assessed by NanoSIMS; ~25–35% of the hydrogen atoms were 2H.
2H-TRLs (~40 μg triglycerides in a volume of 200 μl) were injected into anesthetized 3–4-month-old wild-type mice [7]. After 1, 2, 15, or 30 min, the mice were sacrificed, perfusion-fixed with 4% N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (carbodiimide) and 0.4% glutaraldehyde in a 0.1 M phosphate buffer [7]. 1-mm3 pieces of BAT (trimmed of all WAT) were placed in 4% carbodiimide and 2.5% glutaraldehyde overnight at 4°C. In other experiments, 3–4-month-old mice were given 80 μl of [13C]fatty acids (1 mg/μl; Cambridge Isotope Laboratories; two doses 12 h apart) or 75 μl of [13C]glucose (1 mg/μl, Cambridge Isotope Laboratories; three doses 12 h apart) by gastric gavage. BAT was harvested 22 h after the final dose of fatty acids or 4 h after the final dose of glucose, and sections were prepared for NanoSIMS as described [7].
2.3. NanoSIMS
Tissue sections were coated with 5 nm of platinum and analyzed with NanoSIMS 50L or NanoSIMS 50 instruments [7, 8]. A 16-KeV 133Cs+ beam was used to bombard the sample, and secondary electrons (SEs) and secondary ions (e.g., 12C14N–, 1H–, 2H–, 12C–, 13C–) were collected [7]. To assess nutrient uptake, regions-of-interest were selected and 2H/1H and 13C/12C ratios were measured with the OpenMIMS plugin in ImageJ and processed by GraphPad Prism 7.0.
3. Results
2H-TRLs were injected intravenously into wild-type mice. After 1, 15, or 30 min, interscapular BAT was harvested for NanoSIMS analyses (Fig. 1). 12C14N– NanoSIMS images were created to visualize tissue morphology, and 2H/1H ratio images were useful for defining 2H enrichment in adipocytes. The processing of 2H-TRLs and the subsequent uptake of 2H-labeled lipids by adipocytes was rapid; 2H enrichment in the cytosolic lipid droplets of brown adipocytes was easily detectable 1 min after the 2H-TRL injection. As expected [7], marginated 2H-TRLs were present along the capillary lumen at the 1-min time point but were much less common at the 15-and 30-min time points (Fig. 1). The 2H/1H ratio in adipocytes varied by more than fourfold at each of the three time points (n = 30–40 adipocytes/time point) (Fig. 1). Within individual adipocytes, the 2H content within different cytosolic lipid droplets was almost always homogeneous (Fig. 1). 2H/1H ratios were measured in cytosolic lipid droplets and in nitrogen-rich regions of the cytoplasm (primarily mitochondria), and those ratios were positively correlated (1 min, R2 = 0.56, P < 0.0001; 15 min, R2 = 0.58, P < 0.0001; 30 min, R2 = 0.33, P < 0.0001). The heterogeneity in 2H uptake in brown adipocytes exceeded that in left ventricular cardiomyocytes (n = 30–40), where 2H enrichment varied by only 1.5-fold.
Fig. 1.
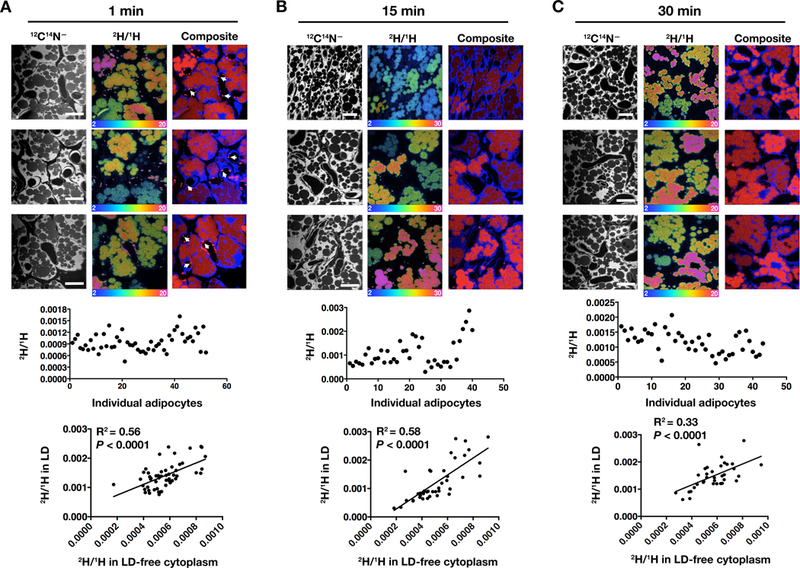
Heterogeneity in the uptake of deuterated fatty acids (derived from the lipolytic processing of 2H-TRLs) in the adipocytes of interscapular BAT. Four-month-old wild-type mice were fasted for 4 h and then injected with 200 μl of 2H-TRLs (see Methods). After 1 min (A), 15 min (B), or 30 min (C), the mice (1 mouse/time point) were perfusion-fixed with carbodiimide and glutaraldehyde, and tissue sections were prepared for NanoSIMS analysis. 12C14N– NanoSIMS images were generated to visualize tissue morphology; 2H/1H ratio images or composite 12C14N– (blue) and 2H/1H ratio (red) images revealed 2H enrichment in adipocyte lipid droplets and 2H-TRLs that had marginated along capillaries (arrows). The scale shows the 2H/1H ratio multiplied by 10,000. The 2H natural abundance, relative to 1H, is 0.000156. Scale bars, 10 μm. The 2H/1H ratios in all adipocytes (n = 30–40/time point) were quantified and plotted as a scatter plot. Also plotted is the correlation between 2H/1H ratios in cytoplasmic lipid droplets (LD) and nitrogen-rich LD-free regions of the cytoplasm (primarily mitochondria).
Heterogeneity in the uptake of 2H-TRL–derived lipids by individual brown adipocytes was very similar in perivascular adipose tissue (PVAT) (Fig. 2). To further explore nutrient uptake in brown adipocytes, mice were given 13C-labeled mixed fatty acids or [13C]glucose by gastric gavage. Interscapular BAT was harvested 22 h after the final dose of the mixed fatty acids or 4 h after the final dose of glucose, and NanoSIMS imaging was performed. We observed heterogeneity in 13C enrichment in brown adipocytes, varying by ~2.3-fold in both the fatty acid and glucose studies (Fig. 3).
Fig. 2.
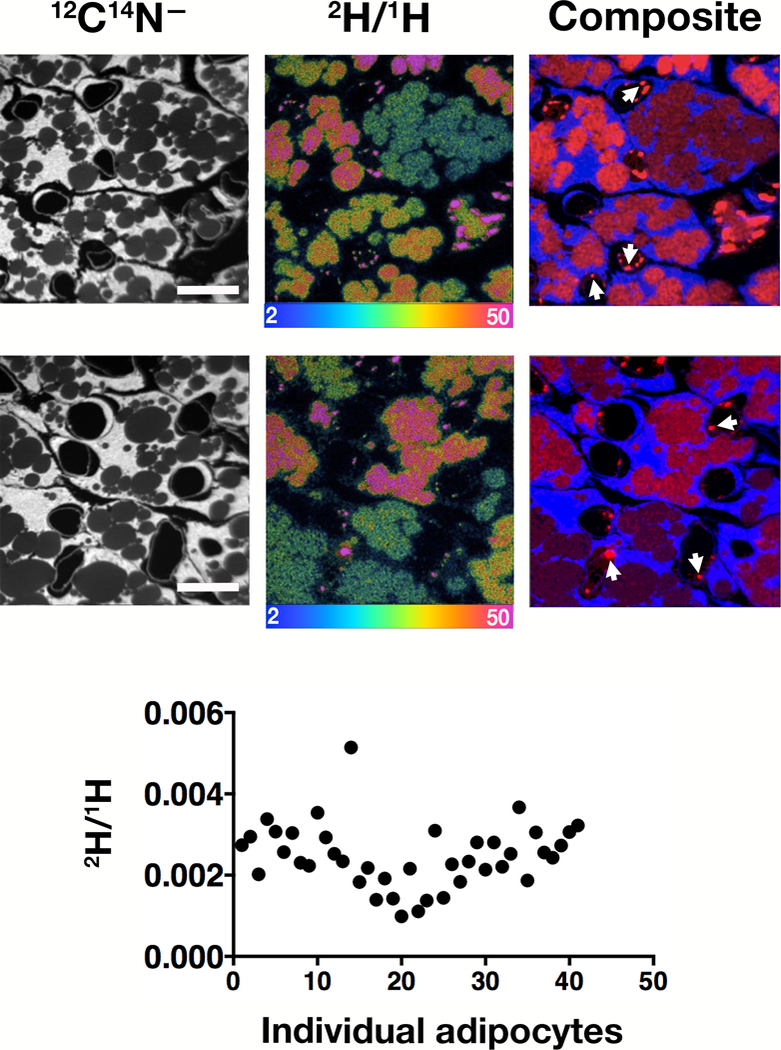
Heterogeneity in the uptake of deuterated fatty acids (derived from the lipolytic processing of 2H-TRLs) in the adipocytes of perivascular adipose tissue (PVAT). A four-month-old wild-type mouse was fasted for 4 h and then injected with 200 μl 2H-TRLs. After 1 min, the mouse was euthanized; perfusion-fixed with carbodiimide/glutaraldehyde; and sections of PVAT prepared to generate NanoSIMS images, exactly as in Fig. 1. Arrows point to marginated lipoproteins in capillaries.
Fig. 3.
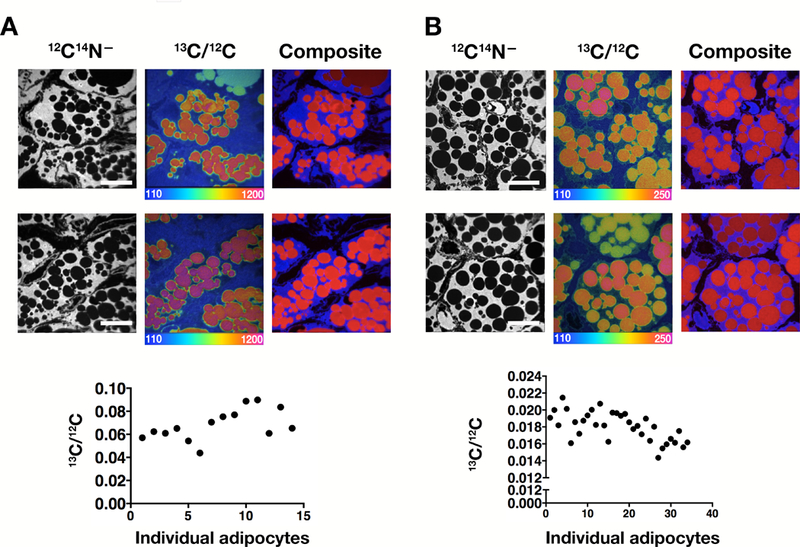
Heterogeneity of 13C enrichment in interscapular brown adipocytes following the administration of [13C]fatty acids or [13C]glucose. (A) 13C-labeled mixed fatty acids were administered to a 4-month-old wild-type mouse by gastric gavage (see Methods). 22 h after the final dose, BAT was harvested and prepared for NanoSIMS imaging. (B) [13C]glucose was administered to a 4-month-old wild-type mouse by gastric gavage (see Methods). 4 h after the final dose, BAT was harvested and prepared for NanoSIMS imaging. For panels B and C, 12C14N– images were generated to visualize tissue morphology, and the 13C/12C ratio images or composite 12C14N– (blue) and 13C/12C (red) images revealed heterogeneity of nutrient uptake. The scale shows the 2H/1H and 13C/12C ratio multiplied by 10,000. Scale bars, 10 μm. The average 2H/1H and 13C/12C ratio in each adipocyte (n = 15–40) was quantified and plotted as a scatter plot.
4. Discussion
We investigated heterogeneity in fatty acid uptake in brown adipocytes after an intravenous injection of 2H–TRLs. At each of the three time points examined, we observed substantial heterogeneity in 2H enrichment in brown adipocytes—even within adjacent adipocytes. Our findings are consistent with earlier studies showing heterogenous uptake of fatty acids by adipocytes of rhesus monkey WAT explants [5]. Heterogeneity in 13C enrichment in different brown adipocytes was also observed after administering 13C-labeled mixed fatty acids or [13C]glucose by gastric gavage, although the extent of the heterogeneity was somewhat lower. However, in considering the results of the latter experiments, it is important to keep in mind that the studies were performed nearly two days after administering the first doses of 13C-labeled nutrients, allowing ample time for 13C-labeled nutrients to be converted into other metabolites (e.g., nonessential amino acids) and taken up by cells [7].
The analytical method that we used to assess nutrient uptake, NanoSIMS imaging, is attractive because it allows one to examine and quantify nutrient uptake in vivo without relying on lipids containing a fluorescent tag. Also, because of the high resolution of NanoSIMS images, it is possible to assess and quantify stable isotope enrichment in both cytosolic lipid droplets and the lipid droplet–free nitrogen-rich regions of the cytoplasm (primarily mitochondria). A downside of NanoSIMS imaging is that it is very expensive and time-consuming, making it unrealistic to examine many different tissues or a host of genetically modified mouse models. In our experience, NanoSIMS imaging is particularly useful for examining metabolic questions with an anatomical angle—such as in the current study where we examined heterogeneity in nutrient uptake by individual brown adipocytes.
Metabolic heterogeneity in white adipocytes has been documented extensively [1–3], but heterogeneity in brown adipocytes has received much less attention. In future studies, it will be important to define the biochemical basis for the inhomogenous uptake of nutrients. A potential clue is that there is substantial heterogeneity in UCP1 expression in individual brown adipocytes in Sprague-Dawley rats [13]. It would be interesting to determine if similar heterogeneity in UCP1 expression exists in individual adipocytes of mouse BAT, and if so, whether the UCP1 expression correlates inversely with 13C or 2H enrichment after administering nutrients enriched in those isotopes.
Supplementary Material
Acknowledgements
This work was supported by the Fondation Leducq Transatlantic Network Grant [12CVD04 (S.G.Y.)], the National Institutes of Health [HL090553 (S.G.Y.), HL087228 (S.G.Y.), HL125335 (S.G.Y.)], and a Ruth L. Kirschstein National Research Service Award [F32 HL132471 (C.H.)]. H.J. is supported by the Australian Research Council DECRA [DE180100080] and the Healy Research Collaboration Award. We are grateful for the NanoSIMS facilities at Caltech and the University of Western Australia.
Abbreviations
- NanoSIMS
Nanoscale secondary ion mass spectrometry
- TRLs
triglyceride-rich lipoproteins
- WAT
white adipose tissue
- BAT
brown adipose tissue
- cAMP
cyclic adenosine monophosphate
- UCP1
uncoupling protein 1
References
- [1].Sackmann-Sala L, Berryman DE, Munn RD, Lubbers ER, Kopchick JJ, Heterogeneity among white adipose tissue depots in male C57BL/6J mice, Obesity (Silver Spring), 20 (2012) 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kwok KH, Lam KS, Xu A, Heterogeneity of white adipose tissue: molecular basis and clinical implications, Exp Mol Med, 48 (2016) e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerback S, Schrauwen P, Spiegelman BM, Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human, Cell, 150 (2012) 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Boumelhem BB, Assinder SJ, Bell-Anderson KS, Fraser ST, Flow cytometric single cell analysis reveals heterogeneity between adipose depots, Adipocyte, 6 (2017) 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Varlamov O, Chu M, Cornea A, Sampath H, Roberts CT Jr., Cell-autonomous heterogeneity of nutrient uptake in white adipose tissue of rhesus macaques, Endocrinology, 156 (2015) 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hao Q, Yadav R, Basse AL, Petersen S, Sonne SB, Rasmussen S, Zhu Q, Lu Z, Wang J, Audouze K, Gupta R, Madsen L, Kristiansen K, Hansen JB, Transcriptome profiling of brown adipose tissue during cold exposure reveals extensive regulation of glucose metabolism, American Journal of Physiology. Endocrinology and Metabolism, 308 (2015) E380–392. [DOI] [PubMed] [Google Scholar]
- [7].He C, Weston TA, Jung RS, Heizer P, Larsson M, Hu X, Allan CM, Tontonoz P, Reue K, Beigneux AP, Ploug M, Holme A, Kilburn M, Guagliardo P, Ford DA, Fong LG, Young SG, Jiang H, NanoSIMS Analysis of intravascular lipolysis and lipid movement across capillaries and into cardiomyocytes, Cell Metab, 27 (2018) 1055–1066 e1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].He C, Hu X, Jung RS, Weston TA, Sandoval NP, Tontonoz P, Kilburn MR, Fong LG, Young SG, Jiang H, High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS, Proc Natl Acad Sci U S A, 114 (2017) 2000–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He C, Fong LG, Young SG, Jiang H, NanoSIMS imaging: an approach for visualizing and quantifying lipids in cells and tissues, J Investig Med, 65 (2017) 669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jiang H, Goulbourne CN, Tatar A, Turlo K, Wu D, Beigneux AP, Grovenor CR, Fong LG, Young SG, High-resolution imaging of dietary lipids in cells and tissues by NanoSIMS analysis, J Lipid Res, 55 (2014) 2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jiang H, Favaro E, Goulbourne CN, Rakowska PD, Hughes GM, Ryadnov MG, Fong LG, Young SG, Ferguson DJ, Harris AL, Grovenor CR, Stable isotope imaging of biological samples with high resolution secondary ion mass spectrometry and complementary techniques, Methods, 68 (2014) 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Davies BSJ, Beigneux AP, Barnes II RH, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyrén R, Goldberg IJ, Olivecrona G, Bensadoun A, Young SG, Fong LG, GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries, Cell Metab, 12 (2010) 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cinti S, Cancello R, Zingaretti MC, Ceresi E, De Matteis R, Giordano A, Himms-Hagen J, Ricquier D, CL316,243 and cold stress induce heterogeneous expression of UCP1 mRNA and protein in rodent brown adipocytes, The Journal of Histochemistry and Cytochemistry, 50 (2002) 21–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


