Abstract
Mutations in cardiac myosin binding protein C (MYBPC3) represent the most frequent cause of familial hypertrophic cardiomyopathy (HCM), making up approximately 50% of identified HCM mutations[4]. MYBPC3 is distinct among other sarcomere genes associated with HCM in that truncating mutations make up the vast majority, whereas non-truncating mutations predominant in other sarcomere genes[4,84]. Several studies using myocardial tissue from HCM patients have found reduced abundance of wild-type MYBPC3 compared to control hearts[34,43,79,80,47,28], suggesting haploinsufficiency of full-length MYBPC3. Further, decreased mutant versus wild-type mRNA and lack of truncated mutant MYBPC3 protein has been demonstrated, highlighting the presence of allelic imbalance[34,43,79,80,47,28]. In this review, we will begin by introducing allelic imbalance and haploinsufficiency, highlighting the broad role each plays within the spectrum of human disease. We will subsequently focus on the roles allelic imbalance and haploinsufficiency play within MYBPC3-linked HCM. Finally, we will explore the implications of these findings on future directions of HCM research. An improved understanding of allelic imbalance and haploinsufficiency may help us better understand genotype-phenotype relationships in HCM and develop novel targeted therapies, providing exciting future research opportunities.
Keywords: hypertrophic cardiomyopathy, myosin binding protein C, haploinsufficiency, allelic imbalance
Introduction
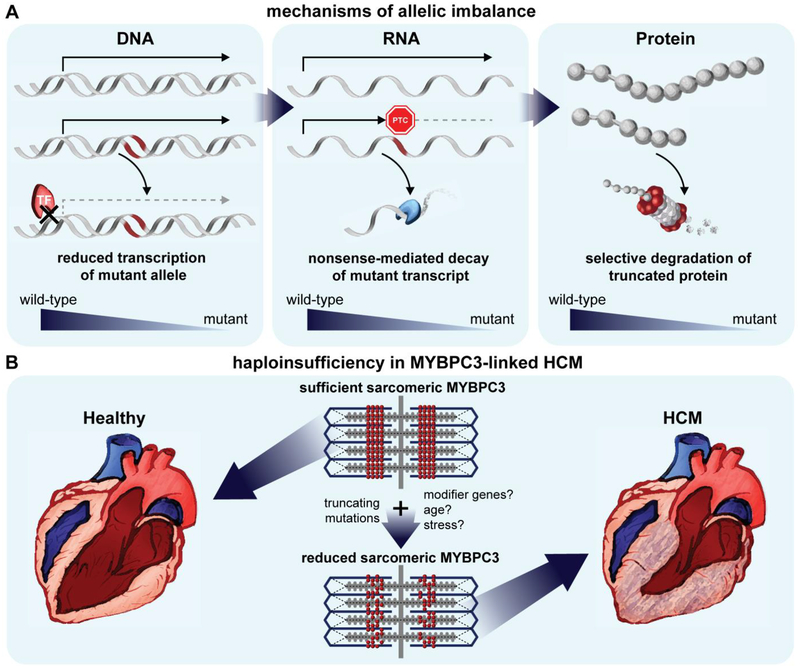
Allelic imbalance and haploinsufficiency represent disturbances in genetic and protein homeostasis, respectively. Allelic imbalance occurs when higher mRNA or protein expression is generated from one allele compared to the other (deviating from the expected 1:1 ratio of expression). Using allele-specific serial analysis of gene expression tags, it has been demonstrated that 25% of human genes display allelic imbalance[81]. Mechanisms leading to allelic imbalance can originate at the DNA, mRNA, and protein levels[20]. Epigenetic modifications of one allele such as gene imprinting or differential methylation/acetylation associated with cis-acting mutations can affect accessibility of a gene to transcription machinery. Meanwhile mutations in transacting regulatory machinery such as microRNA, transcription factors, and proteins can interact with wild-type and mutant alleles unequally. For example, on the mRNA level, mutations can lead to decreased transcript stability by affecting the 3’-untranslated region or microRNA binding. Further, mutant mRNA transcripts with premature termination codons are susceptible to nonsense-mediated mRNA decay (NMD)[68]. Protein allelic imbalance can occur if the mutation alters interaction with protein quality control machinery, protein-protein interactions, intrinsic protein folding, or cellular localization, typically leading to a decrease in mutant protein stability (Figure 1A).
Figure 1.
(A) Examples of mechanisms contributing to allelic imbalance at the DNA, RNA, and protein levels. In heterozygous individuals, the ratio of wild-type to mutant protein ultimately expressed under steady-state conditions could deviate from 1:1 as a result of mutations causing altered epigenetic or transcriptional regulation of the gene, premature termination of the transcript, or instability of the protein. TF: transcription factor; PTC: premature termination codon.
(B) The haploinsufficiency hypothesis as an underlying pathogenic mechanism in HCM. Reduced expression of wild-type MYBPC3 within the sarcomere may compromise contractile function and energy homeostasis, leading to hypertrophic remodeling. Other genetic and non-genetic factors may influence the magnitude of haploinsufficiency that results from the primary truncating mutation.
Haploinsufficiency occurs when a heterozygous mutation results in single functional copy of a gene that is insufficient to maintain normal function[17]. The expression of the functional protein does not necessarily have to be 50% of the normal total expression, but simply below the threshold of expression required for proper function. This can be caused by the same mechanisms which drive allelic imbalance including mechanisms of NMD and mutational-induced protein instability as discussed above. However, this may also occur when mutant protein is present but non-functional, as in the case of in-frame mutations resulting in loss of function. Allelic imbalance can theoretically exacerbate or compensate for haploinsufficiency by altering the relative expression of wild-type versus mutant protein.
Allelic imbalance and haploinsufficiency are the primary mechanisms underlying many Mendelian diseases. Of >220,000 disease-associated variants currently in the Human Gene Mutation Database (HGMD), ~50% of the mutations are nonsense, splice site, or frameshift mutations that would be predicted to result in loss-of-function. Interestingly, each human genome carries ~100 loss-of-function variants and ~20 completely inactivated genes[40,41]. Genes with loss-of-function alleles are less evolutionarily conserved and have more closely related gene paralogs, suggesting that at least partial redundancy may account for genetic robustness[40,32]. In addition, these genes tend to have lower connectivity in protein-protein interactions, and gene interaction networks, suggesting they also may be less central to essential cellular pathways[40]. Genes linked to hypertrophic, dilated and arrhythmogenic cardiomyopathies, harbor both truncating and non-truncating (mostly missense) mutations[4,26]. However, the majority of cardiomyopathy genes are enriched for missense mutations with only a small subset demonstrating strong evidence for dosage sensitivity[83]. Of ~50 unique cardiomyopathy genes, those with truncating mutations that exhibit a high ratio of cases compared to reference populations include DSP (desmoplakin), PKP2 (plakophilin 2), DSC2 (desmocollin 2), and DSG2 (desmoglein 2) for arrhythmogenic cardiomyopathy, LMNA (Lamin A/C), TTN (titin), and SCN5a (sodium voltage-gated channel) for dilated cardiomyopathy, and MYBPC3 (myosin binding protein C), TNNT2 (troponin T), and PLN (phospholamban) for hypertrophic cardiomyopathy[83]. MYBPC3 carries a particularly high odds ratio (118-fold) for the likelihood of hypertrophic cardiomyopathy for truncating variant carriers over noncarriers. Interestingly, mutations in TTN and MYBPC3 are the most common mutations in dilated cardiomyopathy and hypertrophic cardiomyopathy, respectively. Thus, while truncating mutations affect a minority of disease causing genes they account for a significant burden of disease.
Allelic imbalance in HCM
In hypertrophic cardiomyopathy, allelic imbalance for a select number of missense mutations in MYH7, TNNT2, MYL2, and MYBPC3 has been noted at the protein level[28,11,68]. In these studies, the proportion of mutant to wild-type protein varied according to the specific mutation, some samples with a higher and some with a lower percentage of mutant protein relative to the wild-type allele. This imbalance could be due to effects of the mutations on protein stability or avidness of binding within the sarcomere lattice[28]. It could also be due to allelic transcript imbalance. Our group previously demonstrated an equal abundance of wild-type and mutant transcripts for all 11 missense mutations studied[28], but another group observed differential abundance for wild-type and mutant transcripts for some of the mutations they studied[11,68]. Other than the difference in the techniques used (primer extension vs RT-qPCR with allele-specific digestion) and the mutations themselves, the reason for the discrepancy in these results is unclear.
For truncating mutations, the mechanism of allelic imbalance is more clearly defined. More than 90% of mutations in MYBPC3 associated with HCM contain premature termination codons and encode for truncated proteins.[68,11,4]. A reduction in the abundance of PTC-containing mutant transcripts relative to wild-type has been observed in human heart samples from multiple studies[28,82,57,23,59,79]. These observations are consistent with the known susceptibility of these transcripts to NMD which targets primarily nonsense mutant mRNA transcripts for degradation[82,68]. It is important to note, however, that NMD is not always 100% efficient and 5–25% of nonsense transcripts have been shown to escape degradation.[58] Our group has shown that for MYBPC3 mutations in human HCM, mutant transcripts constituted 0–44% of the total mRNA pool[28]. All of the transcripts were >55 nucleotides upstream from the terminal intron, but there was no association between the location of the PTC and the efficiency of NMD to degrade the PTC-containing transcripts. Despite mutant MYBPC3 mRNA levels being present in sufficient quantities to provide template for mutant protein synthesis[28], truncated mutant protein has not been detected[43,79,34]. This suggests that the protein is either not synthesized, or rapidly degraded leading to haploinsufficiency. The mechanisms driving this phenomenon will be discussed in more detail below.
Given the lack of detection of mutant proteins, the clinical significance of differential efficiency of NMD of mutant transcripts is unclear. Understanding additional mechanisms that drive allelic imbalance will be important as we strive to establish genotype-phenotype relationships. For example, allelic imbalance favoring the normal allele of MYBPC3 would represent a compensatory mechanism that could abrogate disease severity. Interestingly, level of allelic imbalance within MYH7 has been observed to be inherited within families, suggesting cis-acting variants contributing to allelic imbalance[55,76]. Emerging genome-wide allele-specific approaches will allow for high-throughput genome-wide evaluation of allelic imbalance[50,16,85]. Applying this technology to HCM patient samples could allow for a more direct evaluation of cis-regulatory variants and the relationship between allelic imbalance and disease severity for MYBPC3 mutation carriers[10,21,56,63].
Haploinsufficiency in HCM
Haploinsufficiency has been suspected as a potential pathogenic mechanism in HCM patients with truncating MYBPC3 mutations since the first truncating mutations were identified[87,88,5]. The possibility of truncated MYBPC3 acting as a poison peptide within the sarcomere, or a dominant negative (antimorph) mutation, has also been evaluated as some early murine disease models suggested truncated MYBPC3 was capable of aberrant sarcomeric incorporation[87]. However, over twenty years later, multiple independent studies have failed to detect truncated mutant proteins in myocardial tissue of HCM patients[43,79,34]. Therefore, haploinsufficiency is considered the most likely mechanism driving disease in these cases. In further support, the majority of studies using patient tissue also found reduced wild-type MYBPC3 protein expression. A summary of study results can be found in Table 1. As discussed above, allelic imbalance of mRNA favoring an increased ratio of wild-type to mutant transcript was also reported in some cases[43,79,28]. The combined results from studies of patient tissue indicate that on average, HCM patients with MYBPC3 truncating mutations express roughly 70% of wild-type MYBPC3 protein present in control individuals. This raises several important questions, such as: why is the reduction in MYBPC3 levels partially but not sufficiently compensated for by the wild-type allele, is there a threshold of MYBPC3 expression below which hypertrophic remodeling begins to develop, and do phenotype negative individuals carrying truncating mutations express higher levels of wild-type MYBPC3?
Table 1.
Studies in patient myocardium
| Study | Number of unique mutations | Allelic Imbalance % mutant MYBPC3 mRNA | Haploinsufficiency %Total MYBPC3 protein compared to donors | p-value for MYBPC3 protein level | Protein detection method |
|---|---|---|---|---|---|
| Jacques et al. (2008)[34] | 2 | Not reported | 76±4% | not reported | Immunoblot normalized to actin |
| Marston et al. (2009)[43] | 5 | ~40% | 76±3% | not reported | Immunoblot normalized to actin |
| Theis et al. (2009)[74] | 7 | Not reported | ~200% | p<0.001 | Immunoblot normalized to GAPDH |
| Van Dijk et al. (2009)[79] | 2 | ~20% | 67±5% | p<0.05 | Immunoblot normalized to α-actinin |
| Van Dijk et al. (2012)[80] | 4 | Not reported | 67±5% | p<0.0001 | SDS-PAGE and SYPRO stain normalized to α-actinin |
| Van Dijk et al. (2014)[78] | 3 | Not reported | 65±5% | not reported | SDS-PAGE and SYPRO stain normalized to α- actinin |
| Helms et al. (2014)[28] | 15 | 16±3% | 88±6% | not significant | Immunoblot normalized to GAPDH |
| McNamara et al. (2017)[47] | 8 | Not reported | ~68% | p<0.05 | Immunoblot normalized to α-actinin |
However, to truly differentiate haploinsufficiency from a dominant negative (antimorph) mutation it must be demonstrated that HCM phenotypes are exacerbated by decreases in MYBPC3 protein level and rescued by increasing MYBPC3 protein level. Both animal and in vitro studies have helped to begin the process of linking MYBPC3 protein level to disease phenotypes. As differentiation methods become more standardized, cardiomyocytes derived from human induced pluripotent stem cells (hiPS-CMs) and embryonic stem cells (hESCMs) are emerging as a powerful tool to study cardiomyopathies in a human model. A summary of MYBPC3 studies in hiPS-CMs and hES-CMs can be found in Table 2. These studies broadly suggest reduction of MYBPC3 is also present in both patient-derived and genetically engineered hiPS-CMs. Differentiated cardiomyocytes showed a mixture of phenotypes including cellular hypertrophy, myofibrillar disarray, reduced force generation, and Ca2+ dysregulation. In further support of the haploinsufficiency hypothesis, gene replacement therapy resulting in increased WT MYBPC3 expression was found to ameliorate hypertrophy and dysfunction in two independent studies within stem cell based systems[56,66].
Table 2.
Studies in hESC-CMs or hiPS-CMs
| Study | Model | Mutation | Allelic Imbalance % mutant MYBPC3 mRNA | Haploinsufficiency % MYBPC3 protein compared to control | p-value for protein level | Protein detection method | Phenotype |
|---|---|---|---|---|---|---|---|
| Tanaka et al. (2014)[73] | hiPS-CM embryoid bodies | p.Gly999_Gln1004del | Not reported | ~80% at day 60 | p<0.05 | Immunoblot normalized to GAPDH | Cellular hypertrophy, myofibrillar disarray |
| Birket et al. (2015)[9] | hiPS-CM | c.2373dupGp.Trp792Valfs*41 | Not reported | <50% at day 25 | p<0.05 | Immunoblot normalized to α-actinin | Reduced force generation No cellular hypertrophy |
| Monteiro da Rocha et al. (2016)[56] | hES-CM | c.2905+1 G>A | <10% | ~50% only at day 17 of differentiation. No difference at day 30. | p=0.04 | Immunoblot normalized to α-actinin | Cellular hypertrophy, sarcomere disarray, dysregulated Ca2+ homeostasis |
| Prondzyns ki et al. (2017)[66] | hiPS-CM | p.Val454Cysfs*21 | Not reported | ~50% | p=0.064 | Immunoblot normalized to α-actinin | Cellular hypertrophy |
| Ribeiro et al. (2017)[67] | hiPS-CM | TALEN-engineered KO via stop codon in exon 1 | ~50% | Reduced; not quantified | Not reported | Immunoblot normalized to α-actinin | Contractile defects, Reduced force generation |
Animal models of HCM provide an experimental platform to test whether ameliorating haploinsufficiency by increasing expression of MYBPC3 can rescue whole-organ disease phenotypes. A summary of relevant animal models can be found in Table 3. Several mouse models carrying null or truncating MYBPC3 mutations exist; however, there is considerable variability in severity and onset of phenotype between heterozygotes of different models, with some exhibiting very mild to no phenotype. It is possible that this variability is correlated to the amount of wild-type MYBPC3 each model expresses. In vivo experiments which have utilized approaches to correct or induce haploinsufficiency have helped mechanistically dissect whether reduction in MYBPC3 expression is a precursor or downstream consequence of cardiac hypertrophy. Heterozygous knock-in mice carrying a G>A transition in the last nucleotide of exon 6, which produces multiple products including a full-length missense transcript, a transcript terminating in exon 9, and a deletion of exon 6, express ~80% of control MYBPC3 levels and display diastolic dysfunction[82,19]. Exon skipping of exons 5 and 6 via antisense oligoribonucleotide injection increased expression of total MYBPC3 and temporarily rescued cardiomyopathy, though only in neonatal mice[22]. The protein product lacking the sequence encoded by exons 5 and 6 was stable and functional. In the same mouse model, wild-type MYBPC3 gene therapy delivered by adeno-associated virus to neonatal mice was able to partially prevent hypertrophic remodeling in a dose-dependent manner[49]. These studies support MYBPC3 haploinsufficiency as a key instigator of hypertrophic remodeling.
Table 3.
In vivo studies in animal models.
| Study | Mutation type | Homozygous phenotype | Heterozygous phenotype | WT MYBPC3 protein levels compared to controls |
|---|---|---|---|---|
| Mouse Models | ||||
| McConnell et al. (1999)[45] McConnell et al. (2001)[44] Vignier et al. (2009)[82] Fraysse et al. (2012)[19] |
Truncation in exon 30 | At 8–12 weeks:
|
At >125 weeks:
|
+/−: 89.9±8.5% |
|
Harris et al. (2002)[27] Korte et al. (2003)[38] |
Truncation removing exon 3–10 | At 12 weeks:
|
|
+/−: no
reduction −/−: no WT protein |
| Carrier et al. (2004)[13] | Transcription start site knock out | At ~14 weeks:
|
At 10–11 months:
|
At 10–11 mo +/−: ~75% −/−: 0% |
| Vignier et al. (2009)[82] Fraysse et al. (2012)[19] Gedicke-Hornung et al. (2013)[22] Mearini et al. (2014)[49] |
Splice site (c.772G>A) multiple products. truncated and Δexon6 |
|
|
At 60 weeks +/−: 79% of WT −/−: 10% of WT |
| Chen et al. (2012)[14] | Conditional Nonsense (tamoxifen induces deletion of exons 3–5) | Knockout induced at 12wkBy 20wk
post tamoxifen (32wk):
|
Less than 10% of vehicle controls by 8 weeks after tamoxifen treatment | |
| Maine Coon Cat | ||||
| Meurs et al. (2005)[52] Sampedrano et al. (2009)[12] Van Dijk et al. (2016)[77] |
Ala31Pro |
|
|
Allelic imbalance in +/− (WT >A31P). Total MYBPC3 expression unchanged in both +/− and −/− |
| Zebrafish | ||||
| Chen et al. (2013)[15] | Knockdown using morpholinos against translation start site | At 72hr post fertilization,
dose dependent increase in:
|
Extent of knockdown not determined | |
Conversely, a tamoxifen-inducible homozygous MYBPC3 knockout mouse model was used to assess the consequences of loss of MYBPC3 expression in adult (12 week-old) mice. By 2 weeks after knockdown, MYBPC3 expression was reduced to ~40% of controls, and diastolic dysfunction was observed. At 20 weeks after knockdown, with MYBPC3 protein at ~10% of controls, left ventricular weight and wall thickness had significantly increased, suggesting that a marked reduction in MYBPC3 beginning in adulthood can induce significant functional and structural changes. An interesting unresolved question is whether MYBPC3 mutation carriers remain asymptomatic until a certain threshold of haploinsufficiency is reached. Some recent studies have raised the question of whether stress can trigger reduction in MYBPC3 levels and remodeling in asymptomatic individuals, which may explain the variable age of onset of MYBPC3-linked HCM[72]. Schlossarek et al. found that adrenergic challenge via one week of treatment with isoprenaline and phenylephrine induced ventricular hypertrophy and proteasome dysfunction in previously phenotype-negative heterozygous MYBPC3 mutant mice[71]. Additionally, Barefield et al. observed reduction in wild-type MYBPC3 expression in heterozygous, but not wild-type, mice following both transverse aortic constriction surgery and, surprisingly, sham surgery[7]. These findings tentatively suggest that stress may influence MYBPC3 expression particularly in truncating mutation carriers.
Despite substantial evidence for the haploinsufficiency hypothesis, the mechanism linking reduced MYBPC3 expression to hypertrophic remodeling has not been completely demonstrated. As with HCM caused by a subset of mutations in other sarcomere genes, Ca2+ sensitivity of the myofilament is increased in MYBPC3-linked HCM[79]. Increased Ca2+ sensitivity, potentially leading to hypercontractility, impaired relaxation, and alterations in Ca2+-dependent signaling pathways, has been suggested as a unifying pathogenic mechanism in HCM (Figure 1B). In support of this, partial extraction of MYBPC3 from rat cardiomyocytes has been shown to result in increased Ca2+ sensitivity, and total ablation of MYBPC3 is associated with increased Ca2+ sensitivity in some mouse models[31,19]. Another compelling hypothesis proposes HCM is the result of energetic deficits stemming from overuse of ATP by myosin due to altered contractility[6]. MYBPC3 acts as a “brake” on contraction, as evidenced in in vitro motility assays by slower actin sliding velocity in the C-zone compared to MYBPC3-free thick filament[65]. Haploinsufficiency of MYBPC3 would blunt this braking function, leading to a higher rate of cross-bridge cycling in the C-zone. Evidence in support of this includes recent findings in both mouse and human cardiomyocytes that MYBPC3 may stabilize the super-relaxed conformation of myosin, in which actin binding and ATP hydrolysis are severely restricted[48,47]. These reports demonstrated accelerated ATP turnover and a decrease in the proportion of super-relaxed myosin heads in patient cells and MYBPC3-knockout murine cells, respectively. Further studies are needed to fully illuminate the entire molecular pathway from haploinsufficiency to hypertrophy.
Further, the cellular mechanisms which drive haploinsufficiency are incompletely understood. As discussed above we know that there is allelic imbalance at the mRNA level favoring the wild-type allele. This finding and the presence of MYBPC3 at >50% of controls suggests a compensatory mechanism that incompletely overcomes the loss of mutant allele. This finding is consistent with the lack of evidence for widespread dosage compensation to maintain normal expression levels of genes affected by heterozygous PTC-containing variants.[68] However, the feedback mechanisms or transcription factors which would drive this compensatory response remain to be defined. Despite allelic imbalance, mutant mRNA remains present, suggesting mutant MYBPC3 protein is either not synthesized or rapidly degraded. In support of the latter mechanism, prior work has demonstrated that mutant MYBPC3 is degraded via the ubiquitin proteasome system (UPS)[82]. Further, in several studies, using cell models, animal models, and human samples, expression of truncated mutant MYBPC3 was associated with impairment of the UPS[69–71]. Deficits in the ubiquitin proteasome system (UPS) have been linked to human HCM particularly when caused by truncating mutations[64,75]. Thus, haploinsufficiency and UPS dysfunction may work in parallel to influence disease pathogenicity and severity. However, it remains unclear whether UPS dysfunction is a result of direct proteotoxicity of truncated MYBPC3 proteins, or a secondary consequence of myocardial remodeling. Our group found no proteotoxic effects or UPS dysfunction associated with expression of five MYBPC3 mutants in primary neonatal rat cardiomyocytes[24]. One approach to devising novel therapeutic strategies that aim to ameliorate both haploinsufficiency and UPS dysfunction would be to inhibit the mechanism by which MYBPC3 is targeted for degradation. Most recently our group has demonstrated that the HSC70 molecular chaperone regulates MYBPC3 protein homeostasis and turn-over rates, uncovering one such potential therapeutic target[24].
Implications for HCM Phenotypes
Phenotypic presentation in HCM is markedly heterogeneous, with severity and age of onset varying considerably. Family members of affected individuals who carry the same mutation often have different degrees of disease expression and complications, and some remain completely phenotype negative. This heterogeneity has made it difficult to connect genotypes to specific phenotypes and predict who is most at risk of developing disease. Conclusions from genotype-phenotype correlation studies have been limited by relatively small patient cohorts and possibly overrepresentation of specific mutations in areas with high frequencies of founder mutations. However, some trends have been observed. Patients with mutations in sarcomere genes have been reported to have worse clinical outcomes than patients for whom no mutation was identified (Ho et al, Circulation, 2018 in press)[18]. While multiple studies have aimed to differentiate disease severity, progression, and phenotype with specific sub-classes of mutations; there is currently no consensus regarding MYBPC3 mutations, truncating or otherwise, being predictive of any particular phenotype or prognosis[39]. Likewise, it has not yet been shown that the location of truncating mutations or missense mutations within MYBPC3 are predictive of clinical outcomes. The SHARE registry aims to address this gap in knowledge by capitalizing on a large multicenter patient cohort to understand the relationship of subtypes of mutations to clinical outcomes. Significant gaps in knowledge which will be important to consider in future genotype-phenotype correlation studies include the relationship between the magnitude of MYBPC3 haploinsufficiency and disease severity, and the role of modifier genes and environmental and behavioral factors in penetrance and phenotypic heterogeneity.
Future Directions and Potential Therapeutic Interventions
Recognition that allelic imbalance and haploinsufficiency play a central role in disease pathogenesis for MYBPC3 mutation-linked HCM raises interesting possibilities for disease-modifying therapies. Currently, medical management is supportive. Treatments include beta blockers, calcium channel blockers, surgical myectomy for symptomatic left ventricular outflow tract obstruction, implantable cardiac defibrillators for prevention of sudden cardiac death, and heart transplantation for a small subset of patients who progress to decompen sated heart failure. There is a great interest in exploring therapies that could prevent the emergence and/or progression of HCM. In a small randomized trial of diltiazem in sarcomere gene mutation carriers, LV chamber size increased toward normal in those taking diltiazem compared to placebo[29]. MYBPC3 carriers showed the greatest benefit with reductions in LV wall thickness and mass, improved diastolic filling, and lower cardiac troponin I levels in those taking diltiazem compared with controls. An ongoing clinical trial (VANISH) aims to evaluate if valsartan, an angiotensin-receptor blocker, can show similar or even greater benefits to halt disease progression across young patients carrying sarcomere gene mutations and exhibiting early signs of disease or overt HCM[30].
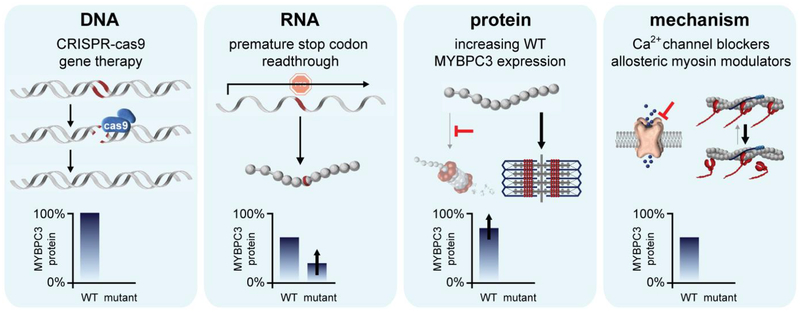
Correcting the fundamental defect that triggers pathological remodeling in HCM is the ultimate goal for disease prevention. For MYBPC3 mutation carriers, developing strategies to normalize MYBPC3 protein levels could hold tremendous promise. In cellular and mouse models, increasing WT MYBPC3 levels prevents the development of hypertrophy and other HCM phenotypic characteristics[51,49,56]. These studies raise the possibility of gene delivery of wild-type MYBPC3 via viral vectors into patients with HCM as a potential therapeutic option. Alternatively, with the advent of CRISPR technology, gene correction has been proposed as a treatment option (Figure 2). As a proof of concept CRISPR-mediated repair of a MYBPC3 mutation was recently successful performed in a human embryo[35]. This illustrates a proof of concept for mutation correction with reasonable efficiency in fertilized eggs. However, it is important to note that the first step in this process is preimplantation genetic diagnosis (PGD) to define the zygotes carrying the MBYPC3 mutation. PGD allows for the selection and implantation of mutation free zygotes, obviating the need to risk off-target mutations associated with gene editing[61]. The possibility of homology directed repair (HDR) directly in somatic cells, including cardiomyocytes, is also being explored. Although in theory HDR requires cellular replication, HDR in post-mitotic skeletal myofibers was achieved using a Cas9 expressed under the control of promoter active only in mature myofibers and inactive in stem cells[8]. Further, supporting the hypothesis that HDR can occur without cellular replication, recent studies have achieved HDR in mouse hearts[33,86]. Another method of gene editing is exon skipping. This strategy has been employed successfully for Duchenne muscular dystrophy[1,2]. In a mouse model carrying biallelic MYBPC3 mutations, AON-induced exon skipping produced an alternative transcript that restored partial protein expression and function[22]. However, this strategy is likely not applicable for heterozygous mutations given that the wild-type allele would also undergo exon skipping, leaving no full-length wild-type MYBPC3.
Figure 2.
Potential therapeutic approaches to correcting or ameliorating the effects of MYBPC3 haploinsufficiency in HCM at different mechanistic stages. Such approaches could include: correction of the mutant allele through gene editing technology, restoring full expression of wild-type MYBPC3; stop codon readthrough for truncating mutations, resulting in expression of a mutant but functional full-length protein; increasing expression of the wild-type protein by modulating its turnover; and targeting of downstream maladaptive mechanisms without affecting MYBPC3 protein expression.
Despite the obvious enthusiasm for applying gene corrective approaches to the treatment of Mendelian diseases like HCM, there are many technical obstacles and safety concerns that limit its applicability in the near future. HCM is characterized by incomplete penetrance of sarcomere gene mutations and highly variable expressivity and prognosis. In addition, disease progression occurs slowly, non-linearly, and over many decades, making the timing and duration of therapy needed potentially long and difficult to predict. Therefore, gene therapy approaches would require delivery of viral vectors to the heart capable of long term and robust expression and with an assurance of no off-target effects. Current techniques continue to carry risk of off-target effects and are largely untested in humans in regards to length and robustness of expression. For these reasons, a more attractive approach to targeted therapy for HCM is small molecule therapeutics. An allosteric myosin modulator (mavacamten) that inhibits myosin ATPase activity has been developed[25,36] and is currently in phase 3 (obstructive) and phase 2 (nonobstructive) clinical trials in HCM patients. However, it is not yet clear whether mavacamten will be effective for those who carry mutations in genes other than myosin. For MYBPC3 mutation carriers, the possibility of targeting haploinsufficiency therapeutically using small molecules could be very attractive for preventing or attenuating disease progression. One potential approach would be to employ read-through strategies. Small molecule drugs have been developed that enable ribosomal read-through of nonsense mutations in mRNA resulting in the production of full length protein. Altaluren, one such small molecule, is being tested in phase 3 clinic trials for the treatment of Duchene Muscular Dystrophy and has gained approval in Europe[46]. It is likewise being tested in cystic fibrosis patients[37]. This approach could be applicable to nonsense mutations within MYBPC3, but unfortunately these represent a minority of truncating mutations. The majority of MYBPC3 mutations are frameshift or splice site mutations and would not be amenable to this therapeutic approach. Thus, additional small molecule approaches are needed. As we uncover a more complete understanding surrounding how cis/trans-mutations, non-sense mediated mRNA decay, the ubiquitin proteasome system, and molecular chaperones regulate and control MYBPC3 protein levels, we may identify novel targets that can be regulated to restore wild-type MYBPC3 protein levels. Towards this goal, it has already been demonstrated that proteasome inhibition and inhibition of nonsense-mediated mRNA decay can increase MYBPC3 protein levels[82]. However, this approach could also increase levels of mutant protein which may not incorporate into the sarcomere or have deleterious effects. Thus, the phenotypic consequences of increased mutant protein levels must be defined. Further, these systems may change levels of other cellular proteins resulting in off target effects. A potentially more specific approach is to alter the activity of specific molecular chaperones. For example, our group has recently shown that inhibition of HSC70 prolongs the half-life of MYBPC3[24].
As we develop modulators of MBYPC3 protein homeostasis, it is also important to recognize that the dynamic relationship between MYBPC3 protein levels and HCM phenotypes remains undefined. More accurate techniques to measure and quantify allelic imbalance within mRNA and protein levels will facilitate a more refined understanding of this relationship. Some potential useful technologies include the use of human SNPs and high-density DNA arrays to detect allelic imbalance[50] and quantitative mass spectrometry-based analytical methods[62]. Single cell techniques hold promise in the deconvolution of cellular heterogeneity, potentially providing opportunities to correlate single cell phenotypes with single cell MYBPC3 protein levels[3,60,54,53,42]. Finally, the development of chemical probes that can dynamically control MYBPC3 protein levels or allow for the quantification of MYBPC3 protein levels within live cardiac tissue could significantly impact on our ability to evaluate how MYBPC3 protein level and disease phenotypes are linked.
Significant progress has been made in defining both allelic imbalance and haploinsufficiency as primary drivers of pathophysiology among patients carrying truncating MYBPC3 mutations. Future work should focus on developing improved methods to measure both protein levels and haploinsufficiency in disease models and patient samples to further determine the extent to which allelic imbalance and decreased MYBPC3 may drive disease severity. Novel tools which allow refined temporal and dose-dependent control of MYBPC3 protein levels could greatly improve our understanding of the dynamic relationship between HCM phenotypes and MYBPC3 protein level and clarify whether specific HCM phenotypes are able to be attenuated or reversed by modulation of MYBPC3 expression or stability. Finally, a more detailed understanding of the mechanisms which regulate MYBPC3 mRNA and protein homeostasis could potentially lead to a successful treatment strategy for HCM, where natural protein quality control mechanisms could be leveraged to selectively restore MYBPC3 protein homeostasis and overcome haploinsufficiency.
Acknowledgements:
This work was supported by: the National Heart, Lung and Blood Institute predoctoral fellowship grant HL131327–01 (to A.A. Glazier); The Children’s Cardiomyopathy Foundation (to S.M. Day); the Taubman Medical Institute (to S.M. Day); The Lefkofsky Foundation (to S.M. Day); and the University of Michigan Protein Folding Diseases Initiative (to S.M. Day).
References
- 1.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, den Dunnen JT (2009) Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Human mutation 30:293–299. doi: 10.1002/humu.20918 [DOI] [PubMed] [Google Scholar]
- 2.Aartsma-Rus A, Straub V, Hemmings R, Haas M, Schlosser-Weber G, Stoyanova-Beninska V, Mercuri E, Muntoni F, Sepodes B, Vroom E, Balabanov P (2017) Development of Exon Skipping Therapies for Duchenne Muscular Dystrophy: A Critical Review and a Perspective on the Outstanding Issues. Nucleic acid therapeutics 27:251–259. doi: 10.1089/nat.2017.0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albayrak C, Jordi CA, Zechner C, Lin J, Bichsel CA, Khammash M, Tay S (2016) Digital Quantification of Proteins and mRNA in Single Mammalian Cells. Molecular cell 61:914–924. doi: 10.1016/j.molcel.2016.02.030 [DOI] [PubMed] [Google Scholar]
- 4.Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL (2015) Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genetics in medicine: official journal of the American College of Medical Genetics. doi: 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- 5.Andersen PS, Havndrup O, Bundgaard H, Larsen LA, Vuust J, Pedersen AK, Kjeldsen K, Christiansen M (2004) Genetic and phenotypic characterization of mutations in myosin-binding protein C (MYBPC3) in 81 families with familial hypertrophic cardiomyopathy: total or partial haploinsufficiency. European journal of human genetics: EJHG 12:673–677. doi: 10.1038/sj.ejhg.5201190 [DOI] [PubMed] [Google Scholar]
- 6.Ashrafian H, Redwood C, Blair E, Watkins H (2003) Hypertrophic cardiomyopathy:a paradigm for myocardial energy depletion. Trends in genetics: TIG 19:263–268. doi: 10.1016/s0168-9525(03)00081-7 [DOI] [PubMed] [Google Scholar]
- 7.Barefield D, Kumar M, Gorham J, Seidman JG, Seidman CE, de Tombe PP, Sadayappan S (2015) Haploinsufficiency of MYBPC3 exacerbates the development of hypertrophic cardiomyopathy in heterozygous mice. Journal of molecular and cellular cardiology 79:234–243. doi: 10.1016/j.yjmcc.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS (2017) Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nature communications 8:14454. doi: 10.1038/ncomms14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, Atsma DE, Passier R, Mummery CL (2015) Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function. Cell reports 13:733–745. doi: 10.1016/j.celrep.2015.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjornsson HT, Albert TJ, Ladd-Acosta CM, Green RD, Rongione MA, Middle CM, Irizarry RA, Broman KW, Feinberg AP (2008) SNP-specific array-based allele-specific expression analysis. Genome Res 18:771–779. doi: 10.1101/gr.073254.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke MA, Cook SA, Seidman JG, Seidman CE (2016) Clinical and Mechanistic Insights Into the Genetics of Cardiomyopathy. Journal of the American College of Cardiology 68:2871–2886. doi: 10.1016/j.jacc.2016.08.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlos Sampedrano C, Chetboul V, Mary J, Tissier R, Abitbol M, Serres F, Gouni V, Thomas A, Pouchelon JL (2009) Prospective echocardiographic and tissue Doppler imaging screening of a population of Maine Coon cats tested for the A31P mutation in the myosin-binding protein C gene: a specific analysis of the heterozygous status. Journal of veterinary internal medicine 23:91–99. doi: 10.1111/j.1939-1676.2008.0218.x [DOI] [PubMed] [Google Scholar]
- 13.Carrier L, Knoll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J Jr., Schwartz K, Chien KR (2004) Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovascular research 63:293–304. doi: 10.1016/j.cardiores.2004.04.009 [DOI] [PubMed] [Google Scholar]
- 14.Chen PP, Patel JR, Powers PA, Fitzsimons DP, Moss RL (2012) Dissociation of structural and functional phenotypes in cardiac myosin-binding protein C conditional knockout mice. Circulation 126:1194–1205. doi: 10.1161/circulationaha.111.089219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YH, Pai CW, Huang SW, Chang SN, Lin LY, Chiang FT, Lin JL, Hwang JJ, Tsai CT (2013) Inactivation of Myosin binding protein C homolog in zebrafish as a model for human cardiac hypertrophy and diastolic dysfunction. Journal of the American Heart Association 2:e000231. doi: 10.1161/jaha.113.000231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH (1994) Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes, chromosomes & cancer 11:153–162 [DOI] [PubMed] [Google Scholar]
- 17.Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G (2005) Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169:1915–1925. doi: 10.1534/genetics.104.036871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erdmann J, Daehmlow S, Wischke S, Senyuva M, Werner U, Raible J, Tanis N, Dyachenko S, Hummel M, Hetzer R, Regitz-Zagrosek V (2003) Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clinical genetics 64:339–349 [DOI] [PubMed] [Google Scholar]
- 19.Fraysse B, Weinberger F, Bardswell SC, Cuello F, Vignier N, Geertz B, Starbatty J, Kramer E, Coirault C, Eschenhagen T, Kentish JC, Avkiran M, Carrier L (2012) Increased myofilament Ca2+ sensitivity and diastolic dysfunction as early consequences of Mybpc3 mutation in heterozygous knock-in mice. J Mol Cell Cardiol 52:1299–1307. doi: 10.1016/j.yjmcc.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaur U, Li K, Mei S, Liu G (2013) Research progress in allele-specific expression and its regulatory mechanisms. Journal of applied genetics 54:271–283. doi: 10.1007/s13353-013-0148-y [DOI] [PubMed] [Google Scholar]
- 21.Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagne V, Dias J, Hoberman R, Montpetit A, Joly MM, Harvey EJ, Sinnett D, Beaulieu P, Hamon R, Graziani A, Dewar K, Harmsen E, Majewski J, Goring HH, Naumova AK, Blanchette M, Gunderson KL, Pastinen T (2009) Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet 41:1216–1222. doi: 10.1038/ng.473 [DOI] [PubMed] [Google Scholar]
- 22.Gedicke-Hornung C, Behrens-Gawlik V, Reischmann S, Geertz B, Stimpel D, Weinberger F, Schlossarek S, Precigout G, Braren I, Eschenhagen T, Mearini G, Lorain S, Voit T, Dreyfus PA, Garcia L, Carrier L (2013) Rescue of cardiomyopathy through U7snRNA-mediated exon skipping in Mybpc3-targeted knock-in mice. EMBO molecular medicine 5:1128–1145. doi: 10.1002/emmm.201202168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger SK, Bar H, Ehlermann P, Walde S, Rutschow D, Zeller R, Ivandic BT, Zentgraf H, Katus HA, Herrmann H, Weichenhan D (2008) Incomplete nonsense-mediated decay of mutant lamin A/C mRNA provokes dilated cardiomyopathy and ventricular tachycardia. Journal of molecular medicine (Berlin, Germany) 86:281–289. doi: 10.1007/s00109-007-0275-1 [DOI] [PubMed] [Google Scholar]
- 24.Glazier AA, Hafeez N, Mellacheruvu D, Basrur V, Nesvizhskii AI, Lee LM, Shao H, Tang V, Yob JM, Gestwicki JE, Helms AS, Day SM (2018) HSC70 is a chaperone for wild-type and mutant cardiac myosin binding protein C. JCI insight 3. doi: 10.1172/jci.insight.99319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, Song Y, Wan W, Leinwand LA, Spudich JA, McDowell RS, Seidman JG, Seidman CE (2016) A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science (New York, NY) 351:617–621. doi: 10.1126/science.aad3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Muller S, Kayvanpour E, Vogel B, Sedaghat-Hamedani F, Lim WK, Zhao X, Fradkin D, Kohler D, Fischer S, Franke J, Marquart S, Barb I, Li DT, Amr A, Ehlermann P, Mereles D, Weis T, Hassel S, Kremer A, King V, Wirsz E, Isnard R, Komajda M, Serio A, Grasso M, Syrris P, Wicks E, Plagnol V, Lopes L, Gadgaard T, Eiskjaer H, Jorgensen M, Garcia-Giustiniani D, Ortiz-Genga M, Crespo-Leiro MG, Deprez RH, Christiaans I, van Rijsingen IA, Wilde AA, Waldenstrom A, Bolognesi M, Bellazzi R, Morner S, Bermejo JL, Monserrat L, Villard E, Mogensen J, Pinto YM, Charron P, Elliott P, Arbustini E, Katus HA, Meder B (2015) Atlas of the clinical genetics of human dilated cardiomyopathy. European heart journal 36:1123–1135. doi: 10.1093/eurheartj/ehu301 [DOI] [PubMed] [Google Scholar]
- 27.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL (2002) Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res 90:594–601 [DOI] [PubMed] [Google Scholar]
- 28.Helms AS, Davis FM, Coleman D, Bartolone SN, Glazier AA, Pagani F, Yob JM, Sadayappan S, Pedersen E, Lyons R, Westfall MV, Jones R, Russell MW, Day SM (2014) Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circulation Cardiovascular genetics 7:434–443. doi: 10.1161/circgenetics.113.000448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho CY, Lakdawala NK, Cirino AL, Lipshultz SE, Sparks E, Abbasi SA, Kwong RY, Antman EM, Semsarian C, Gonzalez A, Lopez B, Diez J, Orav EJ, Colan SD, Seidman CE (2015) Diltiazem treatment for pre-clinical hypertrophic cardiomyopathy sarcomere mutation carriers: a pilot randomized trial to modify disease expression. JACC Heart failure 3:180–188. doi: 10.1016/j.jchf.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho CY, McMurray JJV, Cirino AL, Colan SD, Day SM, Desai AS, Lipshultz SE, MacRae CA, Shi L, Solomon SD, Orav EJ, Braunwald E (2017) The Design of the Valsartan for Attenuating Disease Evolution in Early Sarcomeric Hypertrophic Cardiomyopathy (VANISH) Trial. American heart journal 187:145–155. doi: 10.1016/j.ahj.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann PA, Hartzell HC, Moss RL (1991) Alterations in Ca2+ sensitive tension due to partial extraction of C-protein from rat skinned cardiac myocytes and rabbit skeletal muscle fibers. The Journal of general physiology 97:1141–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao TL, Vitkup D (2008) Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet 4:e1000014. doi: 10.1371/journal.pgen.1000014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishizu T, Higo S, Masumura Y, Kohama Y, Shiba M, Higo T, Shibamoto M, Nakagawa A, Morimoto S, Takashima S, Hikoso S, Sakata Y (2017) Targeted Genome Replacement via Homology-directed Repair in Non-dividing Cardiomyocytes. Scientific reports 7:9363. doi: 10.1038/s41598-017-09716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacques A, Hoskins AC, Kentish JC, Marston SB (2008) From genotype to phenotype: a longitudinal study of a patient with hypertrophic cardiomyopathy due to a mutation in the MYBPC3 gene. J Muscle Res Cell Motil 29:239–246. doi: 10.1007/s10974-009-9174-0 [DOI] [PubMed] [Google Scholar]
- 35.Kaul S, Heitner SB, Mitalipov S (2018) Sarcomere Gene Mutation correction. European heart journal 39:1506–1507. doi: 10.1093/eurheartj/ehy179 [DOI] [PubMed] [Google Scholar]
- 36.Kawas RF, Anderson RL, Ingle SRB, Song Y, Sran AS, Rodriguez HM (2017) A small-molecule modulator of cardiac myosin acts on multiple stages of the myosin chemomechanical cycle. The Journal of biological chemistry 292:16571–16577. doi: 10.1074/jbc.M117.776815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerem E, Konstan MW, De Boeck K, Accurso FJ, Sermet-Gaudelus I, Wilschanski M, Elborn JS, Melotti P, Bronsveld I, Fajac I, Malfroot A, Rosenbluth DB, Walker PA, McColley SA, Knoop C, Quattrucci S, Rietschel E, Zeitlin PL, Barth J, Elfring GL, Welch EM, Branstrom A, Spiegel RJ, Peltz SW, Ajayi T, Rowe SM (2014) Ataluren for the treatment of nonsense-mutation cystic fibrosis: a randomised, double-blind, placebo-controlled phase 3 trial. The Lancet Respiratory medicine 2:539–547. doi: 10.1016/s2213-2600(14)70100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korte FS, McDonald KS, Harris SP, Moss RL (2003) Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circulation research 93:752–758. doi: 10.1161/01.res.0000096363.85588.9a [DOI] [PubMed] [Google Scholar]
- 39.Lopes LR, Rahman MS, Elliott PM (2013) A systematic review and meta-analysis of genotype-phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart (British Cardiac Society) 99:1800–1811. doi: 10.1136/heartjnl-2013-303939 [DOI] [PubMed] [Google Scholar]
- 40.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner MM, Hunt T, Barnes IH, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Genomes Project C, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C (2012) A systematic survey of loss-of-function variants in human protein-coding genes. Science 335:823–828. doi: 10.1126/science.1215040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacArthur DG, Tyler-Smith C (2010) Loss-of-function variants in the genomes of healthy humans. Hum Mol Genet 19:R125–130. doi: 10.1093/hmg/ddq365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macaulay IC, Ponting CP, Voet T (2017) Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends in genetics: TIG 33:155–168. doi: 10.1016/j.tig.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C, Watkins H (2009) Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circulation research 105:219–222. doi: 10.1161/circresaha.109.202440 [DOI] [PubMed] [Google Scholar]
- 44.McConnell BK, Fatkin D, Semsarian C, Jones KA, Georgakopoulos D, Maguire CT, Healey MJ, Mudd JO, Moskowitz IP, Conner DA, Giewat M, Wakimoto H, Berul CI, Schoen FJ, Kass DA, Seidman CE, Seidman JG (2001) Comparison of two murine models of familial hypertrophic cardiomyopathy. Circulation research 88:383–389 [DOI] [PubMed] [Google Scholar]
- 45.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG (1999) Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. The Journal of clinical investigation 104:1235–1244. doi: 10.1172/jci7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald CM, Campbell C, Torricelli RE, Finkel RS, Flanigan KM, Goemans N, Heydemann P, Kaminska A, Kirschner J, Muntoni F, Osorio AN, Schara U, Sejersen T, Shieh PB, Sweeney HL, Topaloglu H, Tulinius M, Vilchez JJ, Voit T, Wong B, Elfring G, Kroger H, Luo X, McIntosh J, Ong T, Riebling P, Souza M, Spiegel RJ, Peltz SW, Mercuri E (2017) Ataluren in patients with nonsense mutation Duchenne muscular dystrophy (ACT DMD): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet (London, England) 390:1489–1498. doi: 10.1016/s0140-6736(17)31611-2 [DOI] [PubMed] [Google Scholar]
- 47.McNamara JW, Li A, Lal S, Bos JM, Harris SP, van der Velden J, Ackerman MJ, Cooke R, Dos Remedios CG (2017) MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One 12:e0180064. doi: 10.1371/journal.pone.0180064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNamara JW, Li A, Smith NJ, Lal S, Graham RM, Kooiker KB, van Dijk SJ, Remedios CGD, Harris SP, Cooke R (2016) Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. Journal of molecular and cellular cardiology 94:65–71. doi: 10.1016/j.yjmcc.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mearini G, Stimpel D, Geertz B, Weinberger F, Kramer E, Schlossarek S, Mourot-Filiatre J, Stoehr A, Dutsch A, Wijnker PJ, Braren I, Katus HA, Muller OJ, Voit T, Eschenhagen T, Carrier L (2014) Mybpc3 gene therapy for neonatal cardiomyopathy enables long-term disease prevention in mice. Nature communications 5:5515. doi: 10.1038/ncomms6515 [DOI] [PubMed] [Google Scholar]
- 50.Mei R, Galipeau PC, Prass C, Berno A, Ghandour G, Patil N, Wolff RK, Chee MS, Reid BJ, Lockhart DJ (2000) Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome research 10:1126–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merkulov S, Chen X, Chandler MP, Stelzer JE (2012) In vivo cardiac myosin binding protein C gene transfer rescues myofilament contractile dysfunction in cardiac myosin binding protein C null mice. Circ Heart Fail 5:635–644. doi: 10.1161/CIRCHEARTFAILURE.112.968941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, Kittleson JA, Munro MJ, Dryburgh K, Macdonald KA, Kittleson MD (2005) A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Human molecular genetics 14:3587–3593. doi: 10.1093/hmg/ddi386 [DOI] [PubMed] [Google Scholar]
- 53.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X (2016) High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proceedings of the National Academy of Sciences of the United States of America 113:11046–11051. doi: 10.1073/pnas.1612826113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montag J, Kowalski K, Makul M, Ernstberger P, Radocaj A, Beck J, Becker E, Tripathi S, Keyser B, Muhlfeld C, Wissel K, Pich A, van der Velden J, Dos Remedios CG, Perrot A, Francino A, Navarro-Lopez F, Brenner B, Kraft T (2018) Burst-Like Transcription of Mutant and Wildtype MYH7-Alleles as Possible Origin of Cell-to-Cell Contractile Imbalance in Hypertrophic Cardiomyopathy. Frontiers in physiology 9:359. doi: 10.3389/fphys.2018.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montag J, Syring M, Rose J, Weber AL, Ernstberger P, Mayer AK, Becker E, Keyser B, Dos Remedios C, Perrot A, van der Velden J, Francino A, Navarro-Lopez F, Ho CY, Brenner B, Kraft T (2017) Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy. Journal of muscle research and cell motility 38:291–302. doi: 10.1007/s10974-017-9486-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monteiro da Rocha A, Guerrero-Serna G, Helms A, Luzod C, Mironov S, Russell M, Jalife J, Day SM, Smith GD, Herron TJ (2016) Deficient cMyBP-C protein expression during cardiomyocyte differentiation underlies human hypertrophic cardiomyopathy cellular phenotypes in disease specific human ES cell derived cardiomyocytes. Journal of molecular and cellular cardiology 99:197–206. doi: 10.1016/j.yjmcc.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moolman JA, Reith S, Uhl K, Bailey S, Gautel M, Jeschke B, Fischer C, Ochs J, McKenna WJ, Klues H, Vosberg HP (2000) A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation 101:1396–1402 [DOI] [PubMed] [Google Scholar]
- 58.Mort M, Ivanov D, Cooper DN, Chuzhanova NA (2008) A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29:1037–1047. doi: 10.1002/humu.20763 [DOI] [PubMed] [Google Scholar]
- 59.Muchir A, Massart C, van Engelen BG, Lammens M, Bonne G, Worman HJ (2006) Proteasome-mediated degradation of integral inner nuclear membrane protein emerin in fibroblasts lacking A-type lamins. Biochemical and biophysical research communications 351:1011–1017. doi: 10.1016/j.bbrc.2006.10.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murgia M, Nagaraj N, Deshmukh AS, Zeiler M, Cancellara P, Moretti I, Reggiani C, Schiaffino S, Mann M (2015) Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO reports 16:387–395. doi: 10.15252/embr.201439757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohiri JC, McNally EM (2018) Gene Editing and Gene-Based Therapeutics for Cardiomyopathies. Heart failure clinics 14:179–188. doi: 10.1016/j.hfc.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panuwet P, Hunter RE Jr., D’Souza PE, Chen X, Radford SA, Cohen JR, Marder ME, Kartavenka K, Ryan PB, Barr DB (2016) Biological Matrix Effects in Quantitative Tandem Mass Spectrometry-Based Analytical Methods: Advancing Biomonitoring. Critical reviews in analytical chemistry 46:93–105. doi: 10.1080/10408347.2014.980775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pastinen T (2010) Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet 11:533–538. doi: 10.1038/nrg2815 [DOI] [PubMed] [Google Scholar]
- 64.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, Pagani F, Powell SR, Day SM (2010) Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 121:997–1004. doi: 10.1161/circulationaha.109.904557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM (2012) Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science 337:1215–1218. doi:science.1223602 [pii] 10.1126/science.1223602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prondzynski M, Kramer E, Laufer SD, Shibamiya A, Pless O, Flenner F, Muller OJ, Munch J, Redwood C, Hansen A, Patten M, Eschenhagen T, Mearini G, Carrier L (2017) Evaluation of MYBPC3 trans-Splicing and Gene Replacement as Therapeutic Options in Human iPSC-Derived Cardiomyocytes. Molecular therapy Nucleic acids 7:475–486. doi: 10.1016/j.omtn.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribeiro AJS, Schwab O, Mandegar MA, Ang YS, Conklin BR, Srivastava D, Pruitt BL (2017) Multi-Imaging Method to Assay the Contractile Mechanical Output of Micropatterned Human iPSC-Derived Cardiac Myocytes. Circulation research 120:1572–1583. doi: 10.1161/circresaha.116.310363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, Ferreira PG, Smith KS, Zhang R, Zhao F, Banks E, Poplin R, Ruderfer DM, Purcell SM, Tukiainen T, Minikel EV, Stenson PD, Cooper DN, Huang KH, Sullivan TJ, Nedzel J, Consortium GT, Geuvadis C, Bustamante CD, Li JB, Daly MJ, Guigo R, Donnelly P, Ardlie K, Sammeth M, Dermitzakis ET, McCarthy MI, Montgomery SB, Lappalainen T, MacArthur DG (2015) Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348:666–669. doi: 10.1126/science.1261877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarikas A, Carrier L, Schenke C, Doll D, Flavigny J, Lindenberg KS, Eschenhagen T, Zolk O (2005) Impairment of the ubiquitin-proteasome system by truncated cardiac myosin binding protein C mutants. Cardiovascular research 66:33–44. doi: 10.1016/j.cardiores.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 70.Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, Carrier L (2012) Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic research in cardiology 107:235. doi: 10.1007/s00395-011-0235-3 [DOI] [PubMed] [Google Scholar]
- 71.Schlossarek S, Schuermann F, Geertz B, Mearini G, Eschenhagen T, Carrier L (2012) Adrenergic stress reveals septal hypertrophy and proteasome impairment in heterozygous Mybpc3-targeted knock-in mice. Journal of muscle research and cell motility 33:5–15. doi: 10.1007/s10974-011-9273-6 [DOI] [PubMed] [Google Scholar]
- 72.Strande JL (2015) Haploinsufficiency MYBPC3 mutations: another stress induced cardiomyopathy? Let’s take a look! Journal of molecular and cellular cardiology 79:284–286. doi: 10.1016/j.yjmcc.2014.12.008 [DOI] [PubMed] [Google Scholar]
- 73.Tanaka A, Yuasa S, Mearini G, Egashira T, Seki T, Kodaira M, Kusumoto D, Kuroda Y, Okata S, Suzuki T, Inohara T, Arimura T, Makino S, Kimura K, Kimura A, Furukawa T, Carrier L, Node K, Fukuda K (2014) Endothelin-1 induces myofibrillar disarray and contractile vector variability in hypertrophic cardiomyopathy-induced pluripotent stem cell-derived cardiomyocytes. Journal of the American Heart Association 3:e001263. doi: 10.1161/jaha.114.001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Theis JL, Bos JM, Theis JD, Miller DV, Dearani JA, Schaff HV, Gersh BJ, Ommen SR, Moss RL, Ackerman MJ (2009) Expression patterns of cardiac myofilament proteins: genomic and protein analysis of surgical myectomy tissue from patients with obstructive hypertrophic cardiomyopathy. Circulation Heart failure 2:325–333. doi: 10.1161/circheartfailure.108.789735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thottakara T, Friedrich FW, Reischmann S, Braumann S, Schlossarek S, Kramer E, Juhr D, Schluter H, van der Velden J, Munch J, Patten M, Eschenhagen T, Moog-Lutz C, Carrier L (2015) The E3 ubiquitin ligase Asb2beta is downregulated in a mouse model of hypertrophic cardiomyopathy and targets desmin for proteasomal degradation. Journal of molecular and cellular cardiology 87:214–224. doi: 10.1016/j.yjmcc.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 76.Tripathi S, Schultz I, Becker E, Montag J, Borchert B, Francino A, Navarro-Lopez F, Perrot A, Ozcelik C, Osterziel KJ, McKenna WJ, Brenner B, Kraft T (2011) Unequal allelic expression of wild-type and mutated beta-myosin in familial hypertrophic cardiomyopathy. Basic research in cardiology 106:1041–1055. doi: 10.1007/s00395-011-0205-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Dijk SJ, Bezold Kooiker K, Mazzalupo S, Yang Y, Kostyukova AS, Mustacich DJ, Hoye ER, Stern JA, Kittleson MD, Harris SP (2016) The A31P missense mutation in cardiac myosin binding protein C alters protein structure but does not cause haploinsufficiency. Archives of biochemistry and biophysics 601:133–140. doi: 10.1016/j.abb.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Dijk SJ, Boontje NM, Heymans MW, Ten Cate FJ, Michels M, Dos Remedios C, Dooijes D, van Slegtenhorst MA, van der Velden J, Stienen GJ (2014) Preserved cross-bridge kinetics in human hypertrophic cardiomyopathy patients with MYBPC3 mutations. Pflugers Archiv: European journal of physiology 466:1619–1633. doi: 10.1007/s00424-013-1391-0 [DOI] [PubMed] [Google Scholar]
- 79.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J (2009) Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation 119:1473–1483. doi:CIRCULATIONAHA.108.838672 [pii] 10.1161/CIRCULATIONAHA.108.838672 [DOI] [PubMed] [Google Scholar]
- 80.van Dijk SJ, Paalberends ER, Najafi A, Michels M, Sadayappan S, Carrier L, Boontje NM, Kuster DW, van Slegtenhorst M, Dooijes D, dos Remedios C, ten Cate FJ, Stienen GJ, van der Velden J (2012) Contractile dysfunction irrespective of the mutant protein in human hypertrophic cardiomyopathy with normal systolic function. Circulation Heart failure 5:36–46. doi: 10.1161/circheartfailure.111.963702 [DOI] [PubMed] [Google Scholar]
- 81.Vidal DO, de Souza JE, Pires LC, Masotti C, Salim AC, Costa MC, Galante PA, de Souza SJ, Camargo AA (2011) Analysis of allelic differential expression in the human genome using allele-specific serial analysis of gene expression tags. Genome 54:120–127. doi: 10.1139/g10-103 [DOI] [PubMed] [Google Scholar]
- 82.Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Pointu H, Mougenot N, Guiard J, Reimer R, Hohenberg H, Schwartz K, Vernet M, Eschenhagen T, Carrier L (2009) Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res 105:239–248. doi:CIRCRESAHA.109.201251 [pii] 10.1161/CIRCRESAHA.109.201251 [DOI] [PubMed] [Google Scholar]
- 83.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, Exome Aggregation C, MacArthur DG, Farrall M, Cook SA, Watkins H (2016) Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genetics in medicine: official journal of the American College of Medical Genetics. doi: 10.1038/gim.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EV, Exome Aggregation C, MacArthur DG, Farrall M, Cook SA, Watkins H (2017) Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 19:192–203. doi: 10.1038/gim.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong KK, Tsang YT, Shen J, Cheng RS, Chang YM, Man TK, Lau CC (2004) Allelic imbalance analysis by high-density single-nucleotide polymorphic allele (SNP) array with whole genome amplified DNA. Nucleic acids research 32:e69. doi: 10.1093/nar/gnh072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie C, Zhang YP, Song L, Luo J, Qi W, Hu J, Lu D, Yang Z, Zhang J, Xiao J, Zhou B, Du JL, Jing N, Liu Y, Wang Y, Li BL, Song BL, Yan Y (2016) Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell research 26:1099–1111. doi: 10.1038/cr.2016.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J (1998) A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. The Journal of clinical investigation 102:1292–1300. doi: 10.1172/JCI3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu B, French JA, Carrier L, Jeremy RW, McTaggart DR, Nicholson MR, Hambly B, Semsarian C, Richmond DR, Schwartz K, Trent RJ (1998) Molecular pathology of familial hypertrophic cardiomyopathy caused by mutations in the cardiac myosin binding protein C gene. Journal of medical genetics 35:205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]