Abstract
The maintenance of oxygen homeostasis in the gut is critical for the maintenance of a healthy gut microbiota. However, few studies have explored how the concentration of atmospheric oxygen affects the gut microbiota in natural populations. High altitude environments provide an opportunity to study the potential effects of atmospheric oxygen on the composition and function of the gut microbiota. Here, we characterized the cecal microbial communities of wild house mice (Mus musculus domesticus) in two independent altitudinal transects, one in Ecuador and one in Bolivia, from sea level to nearly 4000m. First, we found that differences in altitude were associated with differences in the gut microbial community after controlling for the effects of body mass, diet, reproductive status, and population of origin. Second, obligate anaerobes tended to show a positive correlation with altitude while all other microbes tended to show a negative correlation with altitude. These patterns were seen independently in both transects, consistent with the expected effects of atmospheric oxygen on gut microbes. Prevotella was the most-enriched genus at high elevations in both transects, consistent with observations in high-altitude populations of pikas, ruminants, and humans, and also consistent with observations of laboratory mice exposed to hypoxic conditions. Lastly, the renin-angiotensin system, a recently proposed microbiota-mediated pathway of blood pressure regulation, was the top predicted metagenomic pathway enriched in high altitudes in both transects. These results suggest that high altitude environments affect the composition and function of the gut microbiota in wild mammals.
Introduction
The gut microbiota can affect the health and fitness of animals (Knight et al., 2017; Suzuki, 2017). The maintenance of oxygen homeostasis in the gut is critical in maintaining a healthy gut microbiota (Zeitouni, Chotikatum, von Köckritz-Blickwede, & Naim, 2016) which is generally dominated by anaerobes and few aerobes. Alteration of the gut microbial community can lead to expansions of pathogenic aerobic bacteria (Byndloss et al., 2017; Rivera-Chávez et al., 2016). Thus, identifying the factors that affect the composition and function of the gut microbiota has broad implications for biology.
The oxygen concentration in the gut is one of the key factors that affects the assembly of the gut microbial community. The first colonizers of the infant gut are often aerobes that consume oxygen, and later colonizers tend to be obligate anaerobes (Matamoros, Gras-Leguen, Le Vacon, Potel, & De La Cochetiere, 2013; Palmer, Bik, DiGiulio, Relman, & Brown, 2007). Spatial variation of gut microbial communities also exists within the gastrointestinal (GI) tract. For example, consistent with the decrease in oxygen levels from the mouth to anus (He et al., 1999), aerobes tend to dominate the upper GI tract and anaerobes tend to dominate the lower GI tract in mice (Gu et al., 2013; Suzuki & Nachman, 2016) and humans (Hayashi, Takahashi, Nishi, Sakamoto, & Benno, 2005). Within the lower GI tract, anaerobes dominate the luminal content and aerobes are enriched in the oxygen-rich mucus layer where the oxygen diffuses from epithelial cells (Espey 2013; Albenberg et al. 2014; Yasuda et al. 2015). Hypoxic exposure in laboratory mice can induce changes in the gut microbial community, including an increase in obligate anaerobes, suggesting that an oxygen deficit can provide an advantage to anaerobes over aerobes (Moreno-Indias et al., 2015). Although other factors are well-known to influence the gut microbial composition in natural populations of mammals including diet (Wang et al., 2014; Wu et al., 2011), host genetics (Goodrich, Davenport, Waters, Clark, & Ley, 2016), body mass (Ley, Turnbaugh, Klein, & Gordon, 2006; Nishida & Ochman, 2017), and reproductive status (Nuriel-Ohayon, Neuman, & Koren, 2016), the influence of atmospheric oxygen on the gut microbial community has been less explored outside of laboratory settings.
High altitude environments provide an opportunity to study the effects of atmospheric oxygen on the composition and function of the gut microbiota. The reduced partial pressure of oxygen at higher elevations causes a variety of physiological changes related to hypoxic stress in animals (Grindlay & Regensteiner, 1983; Ivy & Scott, 2015; Simonson, 2015; Storz, Scott, & Cheviron, 2010). A few studies have characterized the gut microbiota from mammals living at different altitudes, including work on pikas (H. Li, Li, Beasley, et al., 2016; H. Li, Li, Yao, et al., 2016), ruminants (Z. Zhang et al., 2016), macaques (Sun et al., 2016), and humans (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015). In all of these studies, differences in altitude were associated with differences in the gut microbial composition. However, some of the observed patterns might also be explained by differences in diet, population structure, or culture, as these variables co-varied with altitude. Disentangling the effects of atmospheric oxygen and other co-variables on the gut microbiota remains a challenge.
Two major beneficial physiological influences of gut microbes in high-altitude environments have been proposed: gut microbiota-mediated energy harvest (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015; Z. Zhang et al., 2016; Zhao et al., 2018) and blood pressure regulation (Lan et al., 2017; L. Li & Zhao, 2015). First, high altitude environments may place greater energy demands on mammals due to thermoregulatory stress compared to lower altitude environments (Brooks et al., 1991; Cheviron, Bachman, Connaty, McClelland, & Storz, 2012; Hochachka, Buck, Doll, & Land, 1996; Schippers, Ramirez, Arana, Pinedo-Bernal, & McClelland, 2012). Many anaerobic gut bacteria produce short-chain fatty-acids (SCFAs) as end products of polysaccharide fermentation (Topping & Clifton, 2001). SCFAs are a major energy source for epithelial cells and provide about 10% of daily calories in humans (Bergman, 1990). A greater abundance of SCFA-producing obligate anaerobes has been reported for populations at high altitudes (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015; Z. Zhang et al., 2016; Zhao et al., 2018). Second, the regulation of blood pressure is one of the key physiological responses of animals that are exposed, acclimatized, or adapted to the low oxygen availability at high altitudes (Hainsworth & Drinkhill, 2007; Hanna, 1999; Ivy & Scott, 2015). Interestingly, SCFAs are not only an important energy source, but they also act as signaling molecules by traveling through the bloodstream and binding to SCFA-receptors in various tissues (Samuel et al., 2008). Pluznick et al. (2014) suggested that SCFAs produced by gut bacteria can mediate blood pressure in the host by binding to G-protein coupled receptors. For example, olfactory receptor 78 (Olfr78) in the kidney can increase blood pressure through SCFA-mediated renin release, an enzyme that plays a central role in regulating blood pressure through the renin-angiotensin system (Pluznick et al., 2013, 2009). These discoveries led some researchers to speculate that the gut microbiota may be involved in the regulation of blood pressure in high-altitude populations of humans (Lan et al., 2017; L. Li & Zhao, 2015).
House mice provide an opportunity to study the effects of a high-altitude environment on the composition and function of the gut microbiota. First, house mice (Mus musculus) are predominantly a lowland species, but they have successfully colonized high-altitude environments in the last few hundred years with human settlers including elevations over 4000m in Peru (Harland, 1958) and Bolivia (Storz et al., 2007). The ability to study multiple altitudinal transects within a single species provides an opportunity to look for parallel patterns in the gut microbiota in relation to altitude. Second, although the extent to which house mice have adapted to hypoxic environments remains unclear (Storz et al., 2007), there is evidence that lab mice kept at high altitudes for 30 generations show differences in physiology compared to mice from low altitudes (Jochmans-Lemoine, Shahare, Soliz, & Joseph, 2016; Jochmans-Lemoine et al., 2015). Lastly, house mice are a good mammalian model organism (Phifer-Rixey & Nachman, 2015). Genomic and physiological research related to hypoxia and the microbiota in laboratory mice provides useful information for interpreting patterns observed in natural populations.
Here we characterized the cecal microbial community of natural populations of house mice across two altitudinal transects in South America to test the effects of atmospheric oxygen on the gut microbiota. First, we tested whether altitude correlates with overall differences in the gut microbiota independently of covariates. Second, we tested whether obligate anaerobes show positive correlations with altitude and whether aerobes show negative correlations with altitude. Third, we looked for similar patterns in the gut microbiota in other high-altitude mammals and in laboratory mice under hypoxic exposure. Lastly, we identified predicted metagenomic pathways that correlate with altitude in both transects to generate hypotheses about the physiological influences of the gut microbiota in high-altitude environments. Overall, the results were consistent with the idea that atmospheric oxygen alters gut microbial composition. We also discuss potential beneficial effects of the gut microbiota in high-altitude environments.
Material and Methods
Sampling
Ten populations of house mice (Mus musculus domesticus) were collected across two altitudinal transects in South America (one in Ecuador and one in Bolivia) yielding a total of 92 individuals (Fig.1). The Ecuador transect included five populations: Portoviejo (n = 11, mean elev. = 32m), Santo Domingo (n = 10, mean elev. = 416m), Nanegalito (n= 9, mean elev. = 1643m), Tumbaco (n = 11, mean elev. = 2598m), and Latacunga (n = 8, mean elev. = 2918m) (Fig.1A). The Bolivia-Brazil transect also included five populations: Porto Velho (n = 9, mean elev. = 86m), Santa Cruz (n = 4, mean elev. = 320m), Cochabamba (n = 12, mean elev. 2615m), La Paz (n = 10, mean elev. 3435m), and Lake Titikaka (n = 8, mean elev. 3846m) (Fig. 1B). Detailed locality information for each animal is given in Table S1. Individuals were collected using Sherman live traps, and each individual was separated by at least 500m to avoid collecting close relatives, except at four sites in Ecuador where a few individuals were collected from the same site (Table S1). All of the Ecuador samples were collected in November and December, 2012. All of the Bolivia-Brazil samples were collected in August and September, 2014 except the Porto Velho population which was sampled in September, 2013. All mice were prepared as standard museum specimens (skins and skulls) and have been deposited in the collections of UC Berkeley Museum of Vertebrate Zoology (accession numbers given in Table S1).
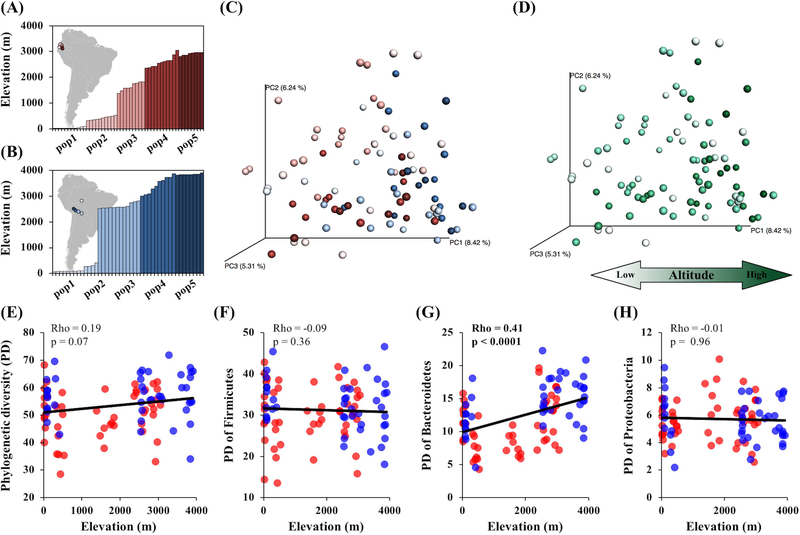
Figure 1.
Effects of altitude on the gut microbiota of wild mice. Wild house mice were collected across two altitudinal gradients from Ecuador transect (n=49) (A) and Bolivia-Brazil transect (n=43) (B). Each bar represents an individual collected from a given elevation and color coded by populations. PCoA plot of Bray-Curtis dissimilarity colored by populations (ADONIS r2 = 0.18, p<0.0001) (C) and altitude (ADONIS r2 = 0.04, p<0.0001) (D). The colors of Fig.1C correspond to Fig.1A and 1B. Darker colors in Fig.1D correspond to higher altitudes. A correlation between altitude and alpha-diversity measured by phylogenetic diversity (PD) (E). Correlation between altitude and PD of three dominant phyla; PD of Firmicutes (F), PD of Bacteroidetes (G), and PD of Proteobacteria (H). Only PD of Bacteroidetes significantly correlated with altitude among the dominant phyla (rho = 0.41, p < 0.0001). Red and blue colors correspond to individuals from Ecuador transect and Bolivia-Brazil transect, respectively.
Cecal samples and external measurements including body weight and body mass index [BMI = (body weight) / (body length)2] were collected within 24 hours after capture. The cecal samples were stored in RNAlater solution at 4°C overnight and then transferred to liquid nitrogen after 8–12 hours, except for the samples from Porto Velho where the cecal samples were placed directly in liquid nitrogen. After returning the samples to the laboratory, all samples were stored at −80°C until sequencing. While different sample preservation methods are known to affect the microbial community composition (Choo, Leong, & Rogers, 2015), individual effects tend to be larger than storage effects (Blekhman et al., 2016). Thus, we decided to include the Porto Velho population to increase the power of the analyses. Nonetheless, we accounted for the effect of sample preservation by including “sample storage method” in the linear mixed-effects models as a random effect (see below). We also conducted analyses after omitting the Proto Velho samples, and none of the major conclusions were altered (see Results). Carbon (δ13C) and nitrogen (δ15N) stable isotope ratios were analyzed from mouse hair to estimate diet following the protocol of Suzuki and Nachman (2016). Detailed information of individual measurements is summarized in Table S1. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Arizona (protocol 07–004) and at the University of California, Berkeley (protocol R361–0514). Animal collection permits were issued by the local governments where necessary.
DNA extraction and 16S rRNA gene sequencing
We removed ~200 mg of the distal portion of each cecal sample, containing both luminal and mucosal portions, under sterile conditions. We followed the DNA extraction protocol described in Suzuki and Nachman (2016). Briefly, we used a mechanical disruption step with sterile zirconia/silica beads (0.1mm, Research Products International Corp.) before step 4 (vortex and centrifugation) in the protocol from the QIAamp DNA stool Minikit (Qiagen). The samples were stored at −20°C before sequencing. The V4 region of the 16S rRNA gene was PCR-amplified, multiplexed, and sequenced using 150bp pair-end Illumina sequencing on a MiSeq machine at the Next Generation Sequencing Core Facility at Argonne National Laboratory. The PCR primers (515F and 806R) and the barcodes are described in Caporaso et al. (2012).
Data analyses
All of the 16S rRNA data were analyzed in QIIME version 1.9.0 (Caporaso et al., 2010). The forward reads were demultiplexed and quality-filtered with the default parameters using split_libraries_fastq.py. The subsampled open-reference OTU picking approach (pick_open_reference_otus.py) was used with default parameters. UCLUST was used to generate 97% OTUs (Edgar, 2010), and taxa were assigned based on the SILVA database (release 128) (Quast et al., 2013). Singletons were removed. A phylogenetic tree was created using FastTree (Price, Dehal, & Arkin, 2009). The OTU table was rarefied to an even depth of 5,000 reads.
Bray-Curtis dissimilarity was calculated and PCoA plots were generated including all individuals using jackknife_beta_diversity.py with default parameters. To identify variables that significantly affected the Bray-Curtis dissimilarity, we used ADONIS with 9999 permutations. We calculated correlations between altitude and other variables in the metadata using Spearman’s rho correlation. Population differences in stable isotope measurements (used to infer diet) were tested using ANOVA. P-values of ADONIS, Spearman’s rho correlation, and ANOVA were corrected for multiple testing using Bonferroni corrections. To test whether the correlation between altitude and Bray-Curtis dissimilarity was independent of other covariates, we conducted model comparisons using linear mixed-effects models with the “lmer” function in the package “lme4” in R (version 3.4.3). All of the variables were normalized by their standard deviation. The first three principle coordinates of Bray-Curtis dissimilarity, PC1 (8.4%), PC2 (6.2%), and PC3 (5.3%) were used as response variables. The full models included five fixed effects (i.e. altitude, body weight, BMI, carbon and nitrogen stable isotope measures) and three random effects (i.e. population, reproductive status, and sample storage methods). The full models were compared to models without altitude using Akaike information criterion with sample size correction (AICc) with the “AICc” function in the package “AICcmodavg”. We also used Likelihood ratio tests to compare between full models and models without altitude with the “lrtest” function in the package “lmtest”.
The overall alpha-diversity measurements (i.e. OTU counts, phylogenetic diversity, and Simpson’s index) were calculated based on the rarefied OTU table using alpha_diversity.py. In addition, to identify bacterial phyla that showed an increase in alpha-diversity in high-altitude environments, we calculated the phylogenetic diversity within each of the three dominant phyla (i.e. Firmicutes, Bacteroidetes, and Proteobacteria). Correlations between altitude and alpha-diversity measurements were based on Spearman’s rho correlation. To account for covariates, we conducted model comparisons using linear mixed-effects models with the same variables mentioned above and using phylogenetic diversity as the response variable.
The relative abundances of bacterial phyla and genera were calculated based on the rarefied OTU table using summarize_taxa.py. We focused on bacterial taxa that had an average relative abundance of 0.1% or greater across all samples and tested for correlations between altitude and relative abundance of bacterial taxa within each transect separately using Spearman’s rho correlation. When the slopes between altitude and relative abundances of bacterial taxa were in the same direction in both transects, Fisher’s method of combining p-values (Fisher, 1932) with Bonferroni correction was used to rank the bacterial taxa that correlated with altitude. Oxygen requirements of bacterial genera (i.e. obligate anaerobes, facultative anaerobes, aerotolerant anaerobes, microaerophiles, and obligate aerobes) were assigned based on Bergey’s Manual of Systematics of Archaea and Bacteria (Whitman et al., 2015) and recent literature (Hardham et al., 2008; Ouwerkerk et al., 2016; Reunanen et al., 2015; Robertson et al., 2005). Most genera were assigned to “obligate anaerobes” and only a few bacterial genera were assigned to the other four oxygen requirement categories. To gain statistical power, the oxygen requirements of bacterial genera were divided into two groups, “obligate anaerobes” and “non-anaerobes” (all other oxygen requirement types) as in (Albenberg et al., 2014). When the genera were unclassified, we assigned categories based on the oxygen requirements of the family. There were three instances where the genus was unclassified and the associated family included both obligate anaerobes and all other oxygen requirement types. In those cases, we searched for all the recognized genera within the family and assigned an oxygen requirement category based on a majority rule (i.e. two out of the three genera showed the same oxygen requirements). Omitting these three genera did not alter any conclusions (see Results).
Lastly, to generate hypotheses about the physiological influences of the gut microbiota at high-altitudes, we identified predicted metagenomic functions that were significantly correlated with altitude using PICRUSt (Langille et al., 2013). A rarefied OTU table with OTUs that were present in the reference database (Greengenes 13_5) was used as an input for PICRUSt. Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog abundances were assigned and collapsed by KEGG pathway to hierarchy level 3. Correlations between relative abundances of KEGG pathway categories and altitude were calculated using Spearman’s rho correlation in both transects. Similar to the clinal tests of bacterial genera, when the slopes between altitude and relative abundances of KEGG pathway categories were in the same direction in both transects, Fisher’s method of combining p-values with Bonferroni correction was used to rank the bacterial taxa that correlated with altitude.
Results
Differences in altitude are associated with differences in the gut microbial composition
We characterized the microbial composition of cecal samples from 10 populations of house mice across two altitudinal transects, one in Ecuador (Fig.1A) and one in Bolivia and Brazil (Fig.1B). First, we identified factors that were significantly associated with variation in the gut microbial community using Bray-Curtis dissimilarity without controlling for covariates. Overall, we found that Bray-Curtis dissimilarity was significantly associated with differences in altitude and differences in populations of origin after correcting for multiple testing (Fig.1C&D, Table S2). BMI was significantly associated with Bray-Curtis dissimilarity in the Bolivia-Brazil transect, but not in the Ecuador transect (Table S2). Carbon and nitrogen stable isotope measurements, body weight, pregnancy, and sex showed no significant associations with Bray-Curtis dissimilarity (Table S2).
In principle, the observed association between altitude and Bray-Curtis dissimilarity could in part be explained by other variables that co-vary with altitude (Table S3). For example, animals living at higher altitude tend to be heavier (rho = 0.34, p = 0.0009) and have greater BMI (rho = 0.33, p = 0.0014) consistent with a trend reported in an altitudinal transect of house mice in Peru (Harland, 1958). Carbon and nitrogen stable isotope measurements also differed significantly between transects and among populations (Fig. S1). However, diet seems to be an unlikely explanation for the association between altitude and Bray-Curtis dissimilarity. Carbon and nitrogen stable isotope measurements showed a trend towards positive correlations with altitude in Ecuador and negative correlations with altitude in Bolivia-Brazil, although most of these correlations were not significant (Fig. S1, Table S3). In addition, differences in the method of sample storage do not account for the association between altitude and Bray Curtis dissimilarity (Table S4).
To test whether altitude was significantly associated with Bray-Curtis dissimilarity independently of the other variables we measured, we conducted model comparisons using linear mixed-effects models. We used Bray-Curtis dissimilarity PC1, PC2, and PC3 as response variables. Altitude, body weight, BMI, and carbon and nitrogen stable isotope measurements were used as fixed effects, and population, pregnancy, and sample storage method were used as random effects. We found that the models without altitude were worse than the full models based on AICc for Bray-Curtis dissimilarity PC1, PC2, and PC3 (Fig. S2). These differences were significant in likelihood ratio tests involving PC2 and PC3 (Fig. S2), indicating that altitude has a significant effect on Bray-Curtis dissimilarity independent of other covariates including diet, body mass, and population of origin.
We next asked whether alpha-diversity of the gut microbiota was correlated with altitude (Fig. 1E-H). Overall, phylogenetic diversity showed a weak (non-significant) positive trend of correlation with altitude (Fig. 1E, rho = 0.19, p = 0.07). Although the trend of greater species richness at higher altitude was also observed using other measures of alpha-diversity, none were significant after multiple testing (Table S5). To test whether alpha-diversity within individual phyla was correlated with altitude, phylogenetic diversity was calculated within each of the three dominant phyla (Fig. 1E-H). Only the phylogenetic diversity of Bacteroidetes showed a significant positive correlation with altitude (Fig. 1G). This correlation was driven by the Bolivia-Brazil transect (rho = 0.45, p = 0.003) and not by the Ecuador transect (rho = 0.18, p = 0.22). Using a linear mixed-effects model comparison, the correlation between phylogenetic diversity of Bacteroidetes and altitude remained significant after controlling for other covariates (likelihood ratio test p = 0.015).
Together, the results suggest that differences in altitude were associated with some differences in alpha- and beta-diversity of the gut microbial communities in natural populations of house mice.
Oxygen requirements of the bacteria predict the correlations between bacterial genera and altitude.
To identify bacterial taxa that show repeated clinal patterns across altitudinal gradients, we focused on 10 phyla and 38 genera that had an average relative abundance of 0.1% or greater across all samples. On average, Firmicutes, Bacteroidetes, and Proteobacteria comprised more than 94% of the total gut community (Fig. S3), which is typical for wild house mice (Linnenbrink et al., 2013; Suzuki & Nachman, 2016; Wang et al., 2014; Weldon et al., 2015). None of these three phyla showed a significant correlation between relative abundance and altitude after correcting for multiple testing (Table S6). In contrast, the relative abundance of Cyanobacteria was significantly correlated with altitude in Ecuador (rho = 0.29, p = 0.04) and in Bolivia-Brazil (rho = 0.31, p = 0.04) (Table S6). The specific taxon driving this relationship is the Order YS2, a group that was previously considered part of Cyanobacteria but has recently been placed in the candidate phylum Melainabacteria believed to be the sister lineage to Cyanobacteria (Di Rienzi et al., 2013).
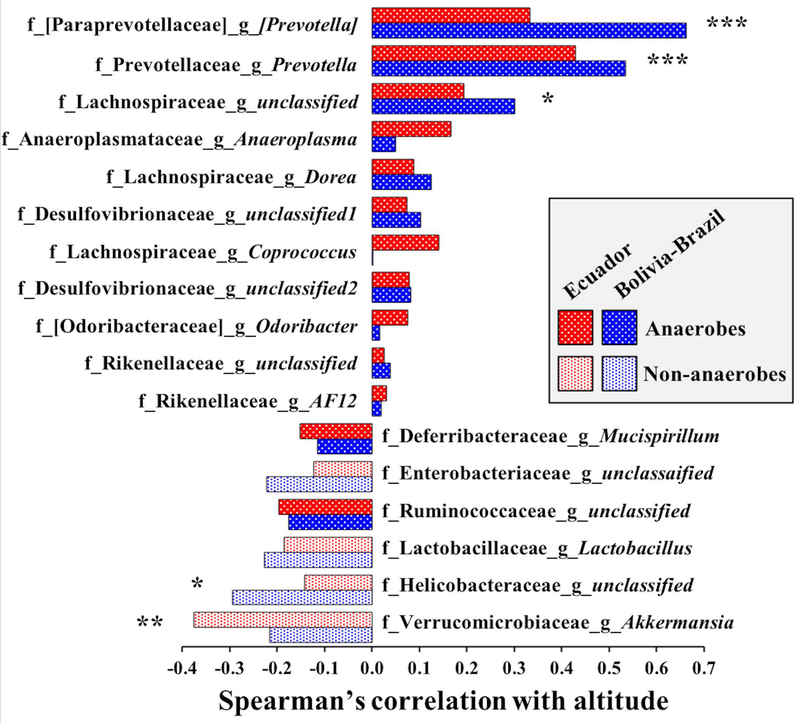
To test the hypothesis that obligate anaerobes have an advantage over other bacteria in hypoxic environments, we identified bacterial genera that show repeated patterns of clinal variation with altitude in both transects. We assigned oxygen requirements to bacterial genera from Bergey’s Manual of Systematics of Archaea and Bacteria as well as from recent literature (Fig.2, Table S7). Among the 38 bacterial genera, 23 showed correlations with altitude in the same direction in both transects (regardless of significance) (Table S7). We were able to assign oxygen requirements to 17 of these genera, and 15 of them showed correlations with altitude in the predicted direction; i.e. a positive correlation with altitude for obligate anaerobes, and a negative correlation with altitude for non-anaerobes (Fig.2, sign test, p = 0.002). Even when we excluded three unclassified genera for which we could not confidently assign oxygen requirements (see methods and Table S7), the pattern remained significant (sign test = 0.01). Moreover, when we limited the analysis to taxa that showed a Fisher’s combined p-value of less than 0.1, the correlations were all in the predicted direction without exceptions (Fig.2). For example, Akkermansia, an aerotolerant mucin-degrader that colonizes the mucus layer (Ouwerkerk et al., 2016; Reunanen et al., 2015), was more abundant at lower altitudes compared to higher altitudes. Differences in the method of sample storage did not account for the observed correlations between bacterial genera and altitude (Table S8). These results support the hypothesis that reduced atmospheric oxygen in high altitude environments provides an advantage to obligate anaerobes over other bacteria.
Figure 2.
Correlations between altitude and bacterial genera. Bacterial genera were included in the list when (1) the correlation between altitude and relative abundance of genera was in the same direction in both transects based on Spearman’s rho correlation, (2) average relative abundance >0.1% across all samples, and (3) at least named bacterial family was assigned to search for oxygen requirements. The brackets [ ] indicate recommended taxonomy. Red color indicates Ecuador transect and blue color indicate Bolivia-Brazil transect. Filled patterns show obligate anaerobes and open pattern show non-anaerobes (i.e. facultative anaerobes, aerotolerant anaerobes, microaerophiles, and obligate aerobes). Oxygen requirements were assigned to each genus based on Bergey’s Manual of Systematics of Archaea and Bacteria and recent literature. Fisher’s combined p-values of the Spearman’s rho raw p-values are indicated: * p < 0.1, ** p < 0.05, *** p < 0.0001. After Bonferroni correction (alpha = 0.05/17 = 0.003), only the two Prevotella genera were significant.
High altitude is associated with a greater relative abundance of Prevotella across multiple species of mammals
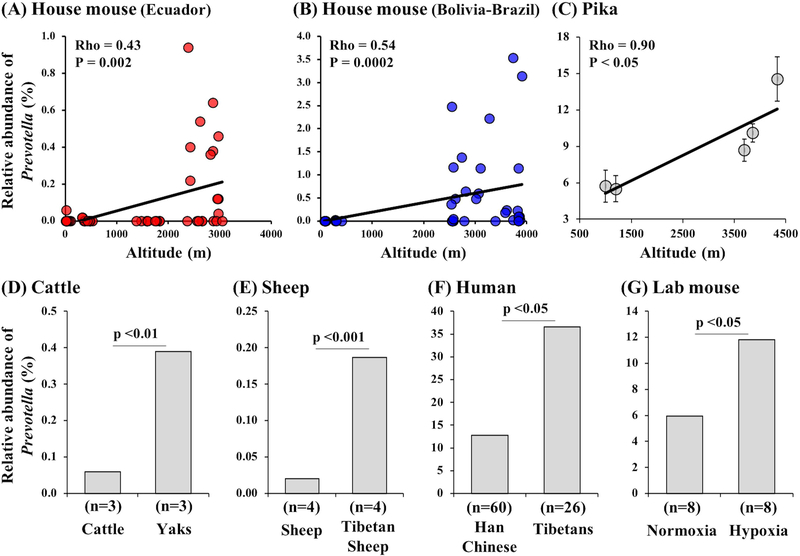
The strongest patterns of co-variation with altitude were seen for bacteria in the genus Prevotella (Fig.2, Table S6). In both transects, the relative abundance of Prevotella increased dramatically above 2000m in elevation (Fig. 3A&B). Interestingly, H. Li et al. (2016) observed a similar pattern in pika populations ranging from 1000m to 4331m (Fig. 3C). Using cecal samples, they found that the strongest associations with altitude were seen for Prevotella and, furthermore, that this pattern was independent of soil- and plant-associated microbial communities (H. Li, Li, Yao, et al., 2016). In addition, high-altitude Yaks and Tibetan sheep show a greater relative abundance of Prevotella compared to their lowland relatives (Fig.3D and E). Finally, a similar association between Prevotella and high-altitude has been observed in humans in some studies (Lan et al., 2017; K. Li et al., 2016), but not in others (L. Li & Zhao, 2015). For example, K. Li et al. (2016) found that Tibetan populations at high-altitudes had a higher relative abundance of Prevotella compared to Han populations at low-altitudes (Fig. 3D). Thus, the association between Prevotella and high altitude environments has been reported in house mice, pikas, cattle, sheep, and humans.
Figure 3.
Convergent associations between relative abundances of Prevotella and high-altitude environments in different species of mammals. Significant positive correlations were observed between altitude and the relative abundance of Prevotella in wild house mice from Ecuador transect (A) and Bolivia-Brazil transect (B). A similar correlation between altitude and relative abundance of Prevotella was found in Pika (n=102) (Figure generated from data in Li et al. 2016) (C). The relative abundance of Prevotella was higher in Yaks compared to cattle collected from the same farm (elev. 3000m) (D) and in Tibetan sheep (elev. 3000m) compared to sheep (elev. 2200m) (E) (Figure generated from data in Zhang et al. 2016). The relative abundance of Prevotella was higher in Tibetans (3600–4500m) living in high altitudes compared to Han (500–3600m) living in low altitudes (Figure generated from data in Li et al. 2016) (F). In controlled lab settings, intermittent hypoxic exposure in laboratory mice resulted in higher relative abundance of Prevotella compared to controls (Figure generated from data in Moreno-Indias et al. 2015) (G).
The observation that the relative abundance of Prevotella increases with altitude could be explained by differences in diet, climate, atmospheric oxygen, or any other variable that correlates with altitude. At least in house mice, differences in diet are unlikely to be driving this pattern because the correlations between altitude and stable isotope measurements (which reflect diet) tended to be in opposite directions in the two transects (Table S3). In contrast, we cannot exclude the possibility that climate is driving the observed patterns because climate and altitude are generally tightly correlated (Grindlay & Regensteiner, 1983). However, a recent study of sleep apnea in laboratory mice provides good evidence that reduced atmospheric oxygen alone can cause an increase in Prevotella (Moreno-Indias et al., 2015) (Fig.3E). Moreno-Indias et al. (2015) compared the gut microbiota of laboratory mice under intermittent hypoxia or normoxia treatments. They found that 6 out of 23 genera differed significantly in their relative abundances between the two treatments. In particular, Paraprevotella and Prevotella were enriched under intermittent hypoxia compared to controls (Moreno-Indias et al., 2015). These results provide experimental evidence that the increase in Prevotella at high altitudes may be driven by lower atmospheric oxygen levels.
Predicted metagenome functions suggest gut microbiota-mediated regulation of blood pressure
To explore the potential physiological influences of the gut microbiota in high altitude environments, we looked for predicted KEGG pathways that were correlated with altitude using PICRUSt (Langille et al., 2013). Among the 273 KEGG pathways (level 3) that were identified in both transects, 183 showed slopes in the same direction in both transects. Nineteen out of the 183 pathways showed significant Fisher’s combined p-values without correcting for multiple testing (raw p-value < 0.05) (Fig. S4). The top two KEGG pathways that showed positive correlations with altitude were “Renin-angiotensin system” (Ecuador; rho = 0.34, p = 0.02. Bolivia-Brazil; rho = 0.51, p = 0.0005. Fisher’s combined p-value = 0.0001) and “hypertrophic cardiomyopathy” (Ecuador; rho = 0.20, p = 0.17. Bolivia-Brazil; rho = 0.53, p = 0.0003, Fisher’s combined p-value = 0.0006) (Fig. S4). In contrast, the top KEGG pathway that negatively correlated with altitude was “Glycosphingolipid biosynthesis - lacto and neolacto series” (Ecuador; rho = −0.40, p = 0.004. Bolivia-Brazil; rho = −0.41, p = 0.006. Fisher’s combined p-value = 0.0003) (Fig. S4).
After correcting for multiple testing (Bonferroni corrected alpha = 0.05/183 = 0.0003), the only pathway that remained significant was the “renin-angiotensin system,” a pathway that plays a major role in blood pressure homeostasis (Sparks, Crowley, Gurley, Mirotsou, & Coffman, 2014). The significant positive correlation between altitude and “renin-angiotensin system” was driven by three predicted bacterial proteins that have homologs in diverse organisms including vertebrates; angiotensin I converting enzyme (K01283), prolyl endopeptidase (K01322), and thimet oligopeptidase 1 (K01392) (Fig. S5). Computational and biochemical studies suggest that these enzymes may have similar functions in bacteria and vertebrates (Kaushik & Sowdhamini, 2014; Rivière et al., 2007; Sugihara, Kawasaki, Tsujimoto, Matsui, & Watanabe, 2007). All three predicted KEGG genes showed positive correlations with altitude in both transects, and two of them had a significant Fisher’s combined p-value (< 0.05) based on Spearman’s rho correlations (Fig.S5).
Discussion
We tested whether altitudinal variation was associated with variation in the composition of the gut microbiota in wild house mice. We found that altitudinal differences were associated with differences in both alpha- and beta-diversity. Similar patterns have previously been reported for alpha diversity in other species (Lan et al., 2017; H. Li, Li, Beasley, et al., 2016; K. Li et al., 2016; L. Li & Zhao, 2015). In house mice, the trend towards higher alpha-diversity with increasing altitude was mainly driven by a significant positive correlation between the alpha-diversity of Bacteroidetes and altitude. Indeed, taxon-specific alpha-diversity measurements may shed light on the relationship between altitude and bacterial species richness overall. Beta-diversity was also significantly associated with altitude in house mice as previously reported in pikas (H. Li, Li, Beasley, et al., 2016; H. Li, Li, Yao, et al., 2016), ruminants (Z. Zhang et al., 2016), macaques (Sun et al., 2016), and humans (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015). In principal, these associations could be explained by other factors that co-vary with altitude. However, differences in diet appear to be unlikely as an explanation of the observed patterns in house mice since stable isotope measurements showed trends in opposite directions with altitude in the two transects (Table. S3, Fig.S1). Population history also seems to be an unlikely explanation for the observed patterns in house mice since the Andes are major biogeographic barriers for many taxa (Patterson & Costa, 2012) and the two transects occur on opposite slopes of the Andes. Thus, parallel patterns in the two transects are unlikely to reflect colonization history. This stands in contrast to patterns seen in humans, where correlations between altitude and the gut microbiota seem to be explained largely by population differences including differences in diet, culture, and genetic background (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015).
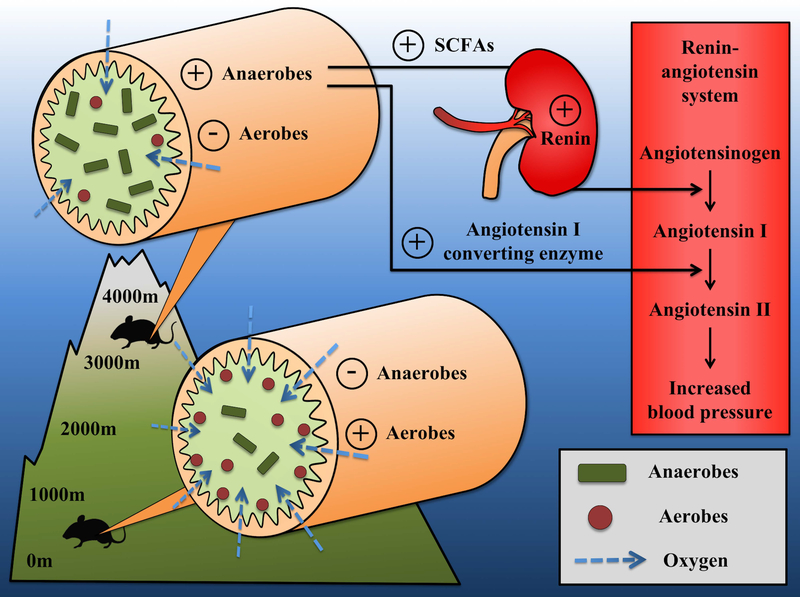
To tease apart the specific role of hypoxia in generating the observed correlations between microbial composition and altitude, we classified bacteria according to their oxygen requirements (i.e. obligate anaerobes, facultative anaerobes, aerotolerant anaerobes, microaerophiles, and obligate aerobes). Overall, we found that 15 out of 17 bacterial genera (for which we could assign oxygen requirements) showed correlations with altitude in the predicted direction (Fig. 2); strictly anaerobic bacteria were positively correlated with altitude, and facultative anaerobes, microaerophiles, and aerotolerant bacteria were all negatively correlated with altitude. For example, the most negatively correlated bacterial genus with altitude was Akkermansia, a bacterial genus that may benefit from oxygen in the mucus layer of the gut (Ouwerkerk et al., 2016; Reunanen et al., 2015). A similar negative association between Akkermansia and altitude has been seen in lizards (W. Zhang, Li, Tang, Liu, & Zhao, 2018). At the level of phyla, the only group that showed a positive correlation with altitude in both transects was the Cyanobacteria (Table S6). This pattern was driven by the Order YS2, a taxon that has recently been placed in the candidate phylum Melainabacteria, a group of anaerobic and obligately fermentative bacteria (Di Rienzi et al., 2013). Together, these results suggest that obligate anaerobes may have a competitive advantage under hypoxic conditions compared to bacteria that require or use oxygen (Moreno-Indias et al., 2015). The reduced partial pressure of oxygen at high altitudes lowers oxygen levels in the blood and may result in an insufficient supply of oxygen to epithelial cells (Zheng, Kelly, & Colgan, 2015) particularly during acute hypoxia exposure (Ivy & Scott, 2015). This, in turn, may reduce the niche for aerobes in the mucosal surfaces of the gut and concomitantly increase the niche for anaerobes in the luminal portion of the gut. In fact, the oxygen-rich mucus layer has a greater proportion of aerobes and a lower proportion of anaerobes compared to the luminal region of the gut in mice, macaques, and humans (Espey 2013; Albenberg et al. 2014; Yasuda et al. 2015). Several recent studies have shown that changes to the availability of oxygen in the gut can have profound effects on microbial communities; for example, antibiotic treatments can deplete some bacterial species and lead to an increase in epithelial oxygen, facilitating the spread of aerobic pathogens (Byndloss et al., 2017; Rivera-Chávez et al., 2016). Thus, atmospheric oxygen levels may affect anaerobes and aerobes differently by directly influencing their oxygen niche (Fig. 4). Future studies on other species would help test the generality of these results and assess whether different patterns exist among the four oxygen-requirement categories that we have lumped together as “non-anaerobes” in this study. Combining descriptions of the gut microbiota with physiological measurements of oxygen levels in epithelial tissue would also help to clarify the link between oxygen availability and specific bacterial niches in the gut.
Figure 4.
A proposed mechanism of the microbiota-mediated regulation of blood pressure in response to atmospheric oxygen at high altitudes. “Anaerobes” refer to obligate anaerobes and “Aerobes” refer to all other oxygen requirements (i.e. facultative anaerobes, aerotolerant anaerobes, microaerophiles, and obligate aerobes).
We found that Prevotella was the top bacterial genus correlated with altitude. This pattern was seen independently in both transects (Fig.3). Prevotella is a saccharolytic anaerobe whose major metabolic end products include SCFAs such as acetic acid (Shah & Collins, 1990). Interestingly, pikas (L. Li & Zhao, 2015), ruminants (Z. Zhang et al., 2016) and humans (Lan et al., 2017; K. Li et al., 2016) living in high altitudes also show an enrichment of Prevotella. However, two studies have reported negative correlations between Prevotella abundance and altitude, one in humans (L. Li & Zhao, 2015) and one in macaques (Zhao et al., 2018). The basis for these differences is unclear. However, the strongest evidence for the association between oxygen levels and Prevotella abundance comes from laboratory experiments with mice under hypoxic conditions (Moreno-Indias et al., 2015). In these experiments, Prevotella was one of the few genera that showed an increase under intermittent hypoxia exposure compared to normoxia controls (Moreno-Indias et al., 2015). The combination of laboratory experiments and field observations across multiple species of mammals suggests that atmospheric oxygen levels affect the abundance of some members of the gut microbiota such as Prevotella.
High-altitude environments impose thermoregulatory challenges and hypoxic stress on animals, and these stressors may affect both survival and reproduction (Cheviron et al., 2012; Grindlay & Regensteiner, 1983; Hochachka et al., 1996). Two major beneficial physiological influences of the gut microbiota at high-altitude have been proposed. One is the role of the gut microbiota in increased energy harvest, particularly at high altitude, by fermenting complex carbohydrates (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015; Z. Zhang et al., 2016; Zhao et al., 2018). A causal role of gut microbiota in increased energy harvest has been demonstrated in lab mice (Bäckhed et al., 2004; Turnbaugh et al., 2006). Many anaerobic gut bacteria produce SCFAs as end products of polysaccharide fermentation, and these SCFAs serve as an energy source for the host (den Besten et al., 2013). Previous studies involving high-altitude mammals have supported this hypothesis by reporting direct and indirect evidence of greater production of SCFAs by anaerobic bacteria at higher altitudes (Lan et al., 2017; K. Li et al., 2016; L. Li & Zhao, 2015; Z. Zhang et al., 2016; Zhao et al., 2018). Our results are also consistent with this hypothesis. We observed an increase in obligate anaerobes at higher elevations, including Prevotella and an unclassified genus of Lachnospiraceae. These taxa are known to produce SCFAs (Biddle, Stewart, Blanchard, & Leschine, 2013; Strobel, 1992). In particular, Prevotella-dominated gut communities are known to have an increased ability to ferment polysaccharides (Kovatcheva-Datchary et al., 2015) and an increased production of SCFAs (Chen et al., 2017). Predicted metagenomic functions also provide some support for this hypothesis, since starch and sucrose metabolism were positively, though non-significantly, correlated with altitude (Fig. S4). Greater reliance on anaerobic energy harvest by the gut microbiota at high-altitudes may be beneficial to the host by conserving oxygen.
A second potential beneficial role of the gut microbiota at high altitude concerns the regulation of blood pressure (Pluznick, 2014; Yang & Zubcevic, 2017). Pluznick et al. (2013) have demonstrated microbiota-mediated controls on blood pressure in laboratory mouse models. They found that SCFAs produced by bacteria signal to two G-protein-coupled receptors (Gpr) that act in opposing directions to modulate blood pressure. In particular, propionate (a type of SCFA) acts as a signaling molecule on GPr41 to cause a dose-dependent drop in blood pressure in mice (Pluznick et al., 2013). Similar vasorelaxation effects have been observed in rats (Nutting, Islam, & Daugirdas, 1991) and humans (Mortensen, Nielsen, Mulvany, & Hessov, 1990). In contrast, olfactory receptor 78 (Olfr78) in the kidney can trigger an increase in blood pressure by SCFA-mediated renin release (Pluznick et al., 2013, 2009). Renin is an enzyme that converts angiotensinogen to angiotensin I in the renin-angiotensin system, leading to a downstream effect of increased blood pressure by vasoconstriction (Sparks et al., 2014). Our results suggest a hypothesis whereby the gut microbiota may influence blood pressure through two distinct mechanisms in high altitude environments (Fig. 4). The first mechanism, previously suggested for humans (Lan et al., 2017; L. Li & Zhao, 2015), involves increased production of SCFAs due to an increased abundance of anaerobic microbes at high elevations. These SCFAs then mediate blood pressure by acting on SCFA-receptors as described in laboratory experiments with mice (Pluznick, 2014). A second potential mechanism, not previously proposed, is that bacterial enzymes may act directly on the host renin-angiotensin system to affect blood pressure. The bacterial angiotensin I converting enzyme has been shown to convert mammalian angiotensin I to angiotensin II in vitro (Rivière et al., 2007). Here we observed a positive correlation between predicted genes in the renin-angiotensin pathway and altitude, suggesting that differences in the abundance of these gene products at different altitudes may have downstream effects on host blood pressure. Shotgun metagenomics will be useful to test whether bacterial angiotensin converting enzymes (ACEs) increase with altitude. Studying the physiological effects of bacterial ACEs in vivo will further test this hypothesis.
It is unclear whether the observed differences in microbial composition at different altitudes involve genetic adaptation of the host. For example, when high-altitude adapted populations have been compared with low-altitude populations at the same elevation, significant differences remained in the gut microbiota of the two groups, both in humans and in ruminants (Qiu et al., 2012; Wei et al., 2016). This suggests that the gut microbial composition is not simply responding to the availability of atmospheric oxygen, but that host genotype, vertical transmission, or dietary factors may contribute to observed differences. House mice are relatively recent colonizers in the Americas, but have been present for a sufficient number of generations for adaptive changes to have occurred (e.g. Lynch, 1992). Nonetheless, some of the “high altitude” populations sampled here came from elevations below 3000m, an altitude above which many of the challenges of hypobaric hypoxia are believed to occur in humans (Dempsey & Forster, 1982). Thus, it is possible that mice in these populations have not experienced strong selection associated with hypoxic stress. Further investigation of the physiological and genomic characteristics in high and low altitude populations of house mice, including at elevations well above 3000m, will help explain the role of the gut microbiome in the plastic and adaptive physiological responses to high altitude.
In summary, we found that altitude has a significant effect on the gut microbial composition of natural populations of house mice. Convergent patterns observed across two transects of house mice as well as in multiple species of mammals are consistent with laboratory experiments in suggesting that a reduced partial pressure of oxygen at high elevations provides an advantage for obligate anaerobes compared to other gut bacteria. The enrichment of obligate anaerobes which often produce SCFAs together with the predicted metagenomic functions related to the renin-angiotensin system at high elevations support potential roles of the gut microbiota in greater energy harvest and regulation of blood pressure in mice living in high altitudes.
Supplementary Material
Table S1. Sample information.
Table S2. Correlations between Bray-Curtis dissimilarity and predictor variables using ADONIS.
Table S3. Correlations between altitude and metadata.
Table S4. Correlations between Bray-Curtis dissimilarity and predictor variables using ADONIS with and without flash frozen samples (Porto Velho).
Table S5. Correlations between altitude and alpha-diversity measurements.
Table S6. Correlations between altitude and relative abundances of bacterial phyla.
Table S7. Correlations between altitude and the relative abundances of 23 bacterial genera with their oxygen requirements.
Table S8. Effects of flash frozen samples (Porto Velho) on correlations between altitude and relative abundances of bacterial genera.
Figure S1. Box plots of carbon and nitrogen stable isotope diet measurements. Carbon isotope measurements differed by population in Ecuador transect (ANOVA p = 0.04) (A) and Bolivia-Brazil transect (ANOVA p < 0.0001) (B). Nitrogen isotope measurements did not differ by population in Ecuador transect (ANOVA p = 0.4) (C), but did vary in Bolivia-Brazil transect (ANOVA p = 0.003) (D). The two transects significantly differ in their carbon (ANOVA p < 0.0001) and nitrogen (ANOVA p = 0.0009) stable isotope measurements.
Figure S2. Model comparisons using Linear Mixed-Effects models. The response variables were Bray-Curtis dissimilarity PC1 (8.4%), PC2 (6.2%), and PC3 (5.3%). The full model included five fixed effects (altitude, body weight, BMI, carbon, and nitrogen) and three random effects (population, reproductive status, and sample storage method). The full models were compared to models without altitude using Akaike information criterion with sample size correction (AICc). Significance is based on likelihood ratio test p-values; * p < 0.05, ns p > 0.05.
Figure S3. Average relative abundance of bacterial phyla per population. Pop1–5 are in the order of low to high altitude in Ecuador transect (A) and Bolivia-Brazil transect (B). The colors correspond to bacterial phyla that showed average relative abundance greater than 0.1% across all samples.
Figure S4. Correlations between altitude and predicted metagenomic functions. Spearman’s rho correlation between altitude and 19 KEGG pathways (level 3) that show Fisher’s combined p-value < 0.05 are shown. After correcting for multiple testing (Bonferroni correction; alpha = 0.05/183 = 0.0003) only Renin-angiotensin system remained significant (Ecuador; rho = 0.34, p = 0.0159. Bolivia-Brazil; rho = 0.51, p-value = 0.005. Fisher’s combined p-value = 0.0001). Spearman’s rho correlation for Ecuador transect is shown in red, and Bolivia-Brazil transect is shown in blue.
Figure S5. Correlations between altitude and predicted KEGG orthologs of renin-angiotensin system (ko04614). Spearman’s rho correlations between altitude and three KEGG orthologs are shown for both Ecuador transect and Bolivia-Brazil transect. Spearman’s rho correlations with Fisher’s combined p-value < 0.05 are indicated by *.
Acknowledgements
We thank Santiago Borneo, Alejandra Camacho, and Daniel Chavez at the Universidad Católica de Quito for facilitating work in Ecuador and Isabel Moya and staffs at Museo Nacional de Historia Natural Colección Boliviana de Fauna and Kawashima family for assisting the work in Bolivia. We also thank members of the Nachman lab and the reviewers for valuable comments and discussions. This work was supported by an NIH grant (RO1 GM074245) to MWN and an National Geographic Young Explorer Grant (152463) to TAS.
Footnotes
Data Accessibility
All museum specimens (skins and skulls) were prepared and have been deposited in the mammal collections of the Museum of Vertebrate Zoology at the University of California, Berkeley (MVZ specimen numbers are in Table S1) with ancillary data uploaded to ARCTOS. All 16S rRNA sequence data were uploaded to European Molecular Biology Laboratory, European Nucleotide Archive (ENA) database (accession number PRJEB28368).
References
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin a, … Wu GD (2014). Correlation Between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota in Humans and Mice. Gastroenterology, 147(5), 1055–1063.e8. 10.1053/j.gastro.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, … Gordon JI (2004). The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America, 101(44), 15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman EN (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiological Reviews, 70(2), 567–590. [DOI] [PubMed] [Google Scholar]
- Biddle A, Stewart L, Blanchard J, & Leschine S (2013). Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity, 5(3), 627–640. 10.3390/d5030627 [DOI] [Google Scholar]
- Blekhman R, Tang K, Archie EA, Barreiro LB, Johnson ZP, Wilson ME, … Tung J (2016). Common methods for fecal sample storage in field studies yield consistent signatures of individual identity in microbiome sequencing data. Scientific Reports, 6(July), 1–5. 10.1038/srep31519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks G. a, Butterfield GE, Wolfe RR, Groves BM, Mazzeo RS, Sutton JR, … Reeves JT (1991). et al. dependence on blood glucose after acclimatization to 4,300 m. Journal of Applied Physiology (Bethesda, Md. : 1985), 70(2), 919–927. 10.1152/jappl.1991.70.2.919 [DOI] [PubMed] [Google Scholar]
- Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, … Bäumler AJ (2017). Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science, 357(6351), 570–575. 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Knight R (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5), 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters W.a, Berg-Lyons D, Huntley J, Fierer N, … Knight R (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal, 6(8), 1621–1624. 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Long W, Zhang C, Liu S, Zhao L, & Hamaker BR (2017). Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Scientific Reports, 7(1), 1–7. 10.1038/s41598-017-02995-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Bachman GC, Connaty AD, McClelland GB, & Storz JF (2012). Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proceedings of the National Academy of Sciences, 109(22), 8635–8640. 10.1073/pnas.1120523109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo JM, Leong LEX, & Rogers GB (2015). Sample storage conditions significantly influence faecal microbiome profiles. Scientific Reports, 5, 1–10. 10.1038/srep16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JA, & Forster HV (1982). Mediation of Ventilatory Adaptations. Physiological Reviews, 62(1), 262–346. 10.1152/physrev.1982.62.1.262 [DOI] [PubMed] [Google Scholar]
- den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, & Bakker BM (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res, 54(9), 2325–2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, … Ley RE (2013). The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. ELife, (2), 1–25. 10.7554/eLife.01102.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Espey MG (2013). Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radical Biology and Medicine, 55, 130–140. 10.1016/j.freeradbiomed.2012.10.554 [DOI] [PubMed] [Google Scholar]
- Fisher RA (1932). Statistical Methods for Research Workers. Edinburgh: Oliver and Boyd. [Google Scholar]
- Goodrich JK, Davenport ER, Waters JL, Clark AG, & Ley RE (2016). Cross-species comparisons of host genetic associations with the microbiome. Science, 352(6285), 29–32. 10.1126/science.aad9379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindlay L, & Regensteiner JG (1983). Adaptation to high altitude. Annual Review of Anthropology, 12, 285–304. [Google Scholar]
- Gu S, Chen D, Zhang J-N, Lv X, Wang K, Duan L-P, … Wu X-L (2013). Bacterial community mapping of the mouse gastrointestinal tract. PloS One, 8(10), e74957 10.1371/journal.pone.0074957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainsworth R, & Drinkhill MJ (2007). Cardiovascular adjustments for life at high altitude. Respiratory Physiology and Neurobiology, 158(2–3), 204–211. 10.1016/j.resp.2007.05.006 [DOI] [PubMed] [Google Scholar]
- Hanna JM (1999). Climate, altitude, and blood pressure. Human Biology, 71(4), 553–582. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10453102 [PubMed] [Google Scholar]
- Hardham JM, King KW, Dreier K, Wong J, Strietzel C, Eversole RR, … Evans RT (2008). Transfer of Bacteroides splanchnicus to Odoribacter gen. nov. as Odoribacter splanchnicus comb. nov., and description of Odoribacter denticanis sp. nov., isolated from the crevicular spaces of canine periodontitis patients. INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, 58(1), 103–109. 10.1099/ijs.0.63458-0 [DOI] [PubMed] [Google Scholar]
- Harland PSEG (1958). Skeletal variation in wild house mice from Peru. Annals and Magazine of Natural History, 1(3), 193–196. 10.1080/00222935808650937 [DOI] [Google Scholar]
- Hayashi H, Takahashi R, Nishi T, Sakamoto M, & Benno Y (2005). Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. Journal of Medical Microbiology, 54(Pt 11), 1093–1101. 10.1099/jmm.0.45935-0 [DOI] [PubMed] [Google Scholar]
- He G, Shankar R. a Chzhan M, Samouilov a, Kuppusamy P, & Zweier JL (1999). Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proceedings of the National Academy of Sciences of the United States of America, 96(8), 4586–4591. 10.1073/pnas.96.8.4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka PW, Buck LT, Doll CJ, & Land SC (1996). Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proceedings of the National Academy of Sciences, 93(18), 9493–9498. 10.1073/pnas.93.18.9493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy CM, & Scott GR (2015). Control of breathing and the circulation in high-altitude mammals and birds. Comparative Biochemistry and Physiology -Part A : Molecular and Integrative Physiology, 186, 66–74. 10.1016/j.cbpa.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Jochmans-Lemoine A, Shahare M, Soliz J, & Joseph V (2016). HIF1α and physiological responses to hypoxia are correlated in mice but not in rats. The Journal of Experimental Biology, 219(24), 3952–3961. 10.1242/jeb.142869 [DOI] [PubMed] [Google Scholar]
- Jochmans-Lemoine A, Villalpando G, Gonzales M, Valverde I, Soria R, & Joseph V (2015). Divergent physiological responses in laboratory rats and mice raised at high altitude. Journal of Experimental Biology, 218(7), 1035–1043. 10.1242/jeb.112862 [DOI] [PubMed] [Google Scholar]
- Kaushik S, & Sowdhamini R (2014). Distribution, classification, domain architectures and evolution of prolyl oligopeptidases in prokaryotic lineages. BMC Genomics, 15(1), 1–13. 10.1186/1471-2164-15-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R, Callewaert C, Marotz C, Hyde ER, Debelius JW, Mcdonald D, & Sogin ML (2017). The Microbiome and Human Biology. Annual Review of Genomics and Human Genetics, 183(March), 65–86. 10.1146/annurev-genom-083115 [DOI] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, … Bäckhed F (2015). Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metabolism, 22(6), 971–982. 10.1016/j.cmet.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Lan D, Ji W, Lin B, Chen Y, Huang C, Xiong X, … Li J (2017). Correlations between gut microbiota community structures of Tibetans and geography. Scientific Reports, 7(1), 1–9. 10.1038/s41598-017-17194-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, … Huttenhower C (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31(9), 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, & Gordon JI (2006). Microbial ecology: human gut microbes associated with obesity. Nature, 444(7122), 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Li H, Li T, Beasley DAE, Heděnec P, Xiao Z, Zhang S, … Li X (2016). Diet diversity is associated with beta but not alpha diversity of pika gut microbiota. Frontiers in Microbiology, 7(JUL), 1–9. 10.3389/fmicb.2016.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li T, Yao M, Li J, Zhang S, Wirth S, … Li X (2016). Pika gut may select for rare but diverse environmental bacteria. Frontiers in Microbiology, 7(AUG), 1–11. 10.3389/fmicb.2016.01269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Dan Z, Gesang L, Wang H, Zhou Y, Du Y, … Nie Y (2016). Comparative analysis of gut microbiota of native Tibetan and Han populations living at different altitudes. PLoS ONE, 11(5), 1–16. 10.1371/journal.pone.0155863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, & Zhao X (2015). Comparative analyses of fecal microbiota in Tibetan and Chinese Han living at low or high altitude by barcoded 454 pyrosequencing. Scientific Reports, 5(October), 1–10. 10.1038/srep14682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnenbrink M, Wang J, Hardouin E a, Künzel S, Metzler D, & Baines JF (2013). The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Molecular Ecology, 22, 1904–1916. 10.1111/mec.12206 [DOI] [PubMed] [Google Scholar]
- Lynch C (1992). Clinal variation in cold adaptation in Mus domesticus: verification of predictions from laboratory populations. American Naturalist, 139(6), 1219–1236. Retrieved from http://www.jstor.org/stable/2462338 [Google Scholar]
- Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, & De La Cochetiere MF (2013). Development of intestinal microbiota in infants and its impact on health. Trends in Microbiology, 21(4), 167–173. 10.1016/j.tim.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Moreno-Indias I, Torres M, Montserrat JM, Sanchez-Alcoholado L, Cardona F, Tinahones FJ, … Farré R (2015). Intermittent hypoxia alters gut microbiota diversity in a mouse model of sleep apnoea. European Respiratory Journal, 45(4), 1055–1065. 10.1183/09031936.00184314 [DOI] [PubMed] [Google Scholar]
- Mortensen FV, Nielsen H, Mulvany MJ, & Hessov I (1990). Short chain fatty acids dilate isolated human colonic resistance arteries. Gut, 31(12), 1391–1394. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/2265780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida AH, & Ochman H (2017). Rates of Gut Microbiome Divergence in Mammals. Molecular Ecology, 12(10), 3218–3221. 10.1111/mec.14473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuriel-Ohayon M, Neuman H, & Koren O (2016). Microbial changes during pregnancy, birth, and infancy. Frontiers in Microbiology, 7(JUL), 1–13. 10.3389/fmicb.2016.01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutting CW, Islam S, & Daugirdas JT (1991). Vasorelaxant effects of short chain fatty acid salts in rat caudal artery. The American Journal of Physiology, 261(2 Pt 2), H561–7. 10.1152/ajpheart.1991.261.2.H561 [DOI] [PubMed] [Google Scholar]
- Ouwerkerk JP, van der Ark KCH, Davids M, Claassens NJ, Finestra TR, de Vos WM, & Belzer C (2016). Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Applied and Environmental Microbiology, 82(23), 6983–6993. 10.1128/AEM.01641-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, & Brown PO (2007). Development of the human infant intestinal microbiota. PLoS Biology, 5(7), 1556–1573. 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson BD, & Costa LP (2012). Bones, Clones, and Biomes: The Role of the Andes in the Diversification and Biogeography of Neotropical Mammals. University of Chicago Press. 10.7208/chicago/9780226649214.001.0001 [DOI]
- Phifer-Rixey M, & Nachman MW (2015). Insights into mammalian biology from the wild house mouse Mus musculus. ELife, 2015(4), 1–13. 10.7554/eLife.05959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluznick JL (2014). A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes, 5(2), 202–207. 10.4161/gmic.27492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, & Han J (2013). Olfactory receptor responding to gut microbiota- derived signals plays a role in renin secretion and blood pressure regulation. Proceedings of the National Academy of Sciences, 110(11), 4410–4415. 10.1073/pnas.1215927110/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1215927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluznick JL, Zou DJ, Zhang X, Yan Q, Rodriguez-Gil DJ, Eisner C, … Caplan MJ (2009). Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci U S A, 106(6), 2059–2064. 10.1073/pnas.0812859106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, & Arkin AP (2009). Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Molecular Biology and Evolution, 26(7), 1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Zhang G, Ma T, Qian W, Ye Z, Cao C, … Wang J (2012). The yak genome and adaptation to life at high altitude. Nature Genetics, 44(8), 946–949. 10.1038/ng.2343 [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, … Glöckner FO(2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research, 41(D1), 590–596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, … Satokaria R (2015). Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Applied and Environmental Microbiology, 81(11), 3655–3662. 10.1128/AEM.04050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, … Bäumler AJ. (2016). Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host and Microbe, 19(4), 443–454. 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière G, Michaud A, Corradi HR, Sturrock ED, Ravi Acharya K, Cogez V, … Corvol P (2007). Characterization of the first angiotensin-converting like enzyme in bacteria: Ancestor ACE is already active. Gene, 399(1), 81–90. 10.1016/j.gene.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SLW, Fox JG, & Lee A (2005). Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. International Journal of Systematic and Evolutionary Microbiology, 55(3), 1199–1204. 10.1099/ijs.0.63472-0 [DOI] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, … Gordon JI (2008). Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences, 105(43), 16767–16772. 10.1073/pnas.0808567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers MP, Ramirez O, Arana M, Pinedo-Bernal P, & McClelland GB (2012). Increase in carbohydrate utilization in high-altitude andean mice. Current Biology, 22(24), 2350–2354. 10.1016/j.cub.2012.10.043 [DOI] [PubMed] [Google Scholar]
- Shah HN, & Collins DM (1990). Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. International Journal of Systematic Bacteriology, 40(2), 205–208. 10.1099/00207713-40-2-205 [DOI] [PubMed] [Google Scholar]
- Simonson TS (2015). Altitude Adaptation: A Glimpse Through Various Lenses. High Altitude Medicine & Biology, 16(2), 125–137. 10.1089/ham.2015.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks MA, Crowley SD, Gurley SB, Mirotsou M, & Coffman TM (2014). Classical Renin-Angiotensin system in kidney physiology. Comprehensive Physiology, 4(3), 1201–1228. 10.1002/cphy.c130040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, & Hayes JP (2007). Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics, 177(1), 481–500. 10.1534/genetics.107.078550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Scott GR, & Cheviron ZA (2010). Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. Journal of Experimental Biology, 213(24), 4125–4136. 10.1242/jeb.048181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel HJ (1992). Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Applied and Environmental Microbiology, 58(7), 2331–2333. https://doi.org/0099-2240/92/072331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara Y, Kawasaki A, Tsujimoto Y, Matsui H, & Watanabe K (2007). Potencies of phosphine peptide inhibitors of mammalian thimet oligopeptidase and neurolysin on two bacterial pz peptidases. Bioscience, Biotechnology, and Biochemistry, 71(2), 594–597. 10.1271/bbb.60534 [DOI] [PubMed] [Google Scholar]
- Sun B, Wang X, Bernstein S, Huffman MA, Xia D-P, Gu Z, … Li J (2016). Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Scientific Reports, 6(April), 26035 10.1038/srep26035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki TA (2017). Links between Natural Variation in the Microbiome and Host Fitness in Wild Mammals. Integrative and Comparative Biology, 57(4), 756–769. 10.1093/icb/icx104 [DOI] [PubMed] [Google Scholar]
- Suzuki TA, & Nachman MW (2016). Spatial heterogeneity of gut microbial composition along the gastrointestinal tract in natural populations of house mice. PLoS ONE, 11(9), 1–15. 10.1371/journal.pone.0163720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping DL, & Clifton PM (2001). Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev, 81(3), 1031–1064. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald M. a, Magrini V, Mardis ER, & Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Wang J, Linnenbrink M, Künzel S, Fernandes R, Nadeau M-J, Rosenstiel P, & Baines JF (2014). Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proceedings of the National Academy of Sciences of the United States of America, 111(26), E2703–10. 10.1073/pnas.1402342111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Wang H, Liu G, Zhao F, Kijas JW, Ma Y, … Du L (2016). Genome-wide analysis reveals adaptation to high altitudes in Tibetan sheep. Scientific Reports, 6(May), 1–11. 10.1038/srep26770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, & Viney M (2015). The gut microbiota of wild mice. PLoS ONE, 10(8), 1–15. 10.1371/journal.pone.0134643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, … Dedysh S (Eds.). (2015). Bergey’s Manual of Systematics of Archaea and Bacteria. Chichester, UK: John Wiley & Sons, Ltd; 10.1002/9781118960608 [DOI] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Keilbaugh SA, … Lewis JD (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science (New York, N.Y.), 334(6052), 105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, & Zubcevic J (2017). Gut-brain axis in regulation of blood pressure. Frontiers in Physiology, 8(OCT), 1–12. 10.3389/fphys.2017.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, … Morgan XC (2015). Biogeography of the Intestinal Mucosal and Lumenal Microbiome in the Rhesus Macaque. Cell Host & Microbe, 17(3), 385–391. 10.1016/j.chom.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni NE, Chotikatum S, von Köckritz-Blickwede M, & Naim HY (2016). The impact of hypoxia on intestinal epithelial cell functions: consequences for invasion by bacterial pathogens. Molecular and Cellular Pediatrics, 3(1), 14 10.1186/s40348-016-0041-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li N, Tang X, Liu N, & Zhao W (2018). Changes in intestinal microbiota across an altitudinal gradient in the lizard Phrynocephalus vlangalii. Ecology and Evolution, 8(9), 4695–4703. 10.1002/ece3.4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu D, Wang L, Hao J, Wang J, Zhou X, … Shi P (2016). Convergent Evolution of Rumen Microbiomes in High-Altitude Mammals. Current Biology, 26(14), 1873–1879. 10.1016/j.cub.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Zhao J, Yao Y, Li D, Xu H, Wu J, Wen A, … Xu H (2018). Characterization of the Gut Microbiota in Six Geographical Populations of Chinese Rhesus Macaques (Macaca mulatta), Implying an Adaptation to High-Altitude Environment. Microbial Ecology. 10.1007/s00248-018-1146-8 [DOI] [PubMed] [Google Scholar]
- Zheng L, Kelly CJ, & Colgan SP (2015). Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. American Journal of Physiology - Cell Physiology, 309(6), C350–C360. 10.1152/ajpcell.00191.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sample information.
Table S2. Correlations between Bray-Curtis dissimilarity and predictor variables using ADONIS.
Table S3. Correlations between altitude and metadata.
Table S4. Correlations between Bray-Curtis dissimilarity and predictor variables using ADONIS with and without flash frozen samples (Porto Velho).
Table S5. Correlations between altitude and alpha-diversity measurements.
Table S6. Correlations between altitude and relative abundances of bacterial phyla.
Table S7. Correlations between altitude and the relative abundances of 23 bacterial genera with their oxygen requirements.
Table S8. Effects of flash frozen samples (Porto Velho) on correlations between altitude and relative abundances of bacterial genera.
Figure S1. Box plots of carbon and nitrogen stable isotope diet measurements. Carbon isotope measurements differed by population in Ecuador transect (ANOVA p = 0.04) (A) and Bolivia-Brazil transect (ANOVA p < 0.0001) (B). Nitrogen isotope measurements did not differ by population in Ecuador transect (ANOVA p = 0.4) (C), but did vary in Bolivia-Brazil transect (ANOVA p = 0.003) (D). The two transects significantly differ in their carbon (ANOVA p < 0.0001) and nitrogen (ANOVA p = 0.0009) stable isotope measurements.
Figure S2. Model comparisons using Linear Mixed-Effects models. The response variables were Bray-Curtis dissimilarity PC1 (8.4%), PC2 (6.2%), and PC3 (5.3%). The full model included five fixed effects (altitude, body weight, BMI, carbon, and nitrogen) and three random effects (population, reproductive status, and sample storage method). The full models were compared to models without altitude using Akaike information criterion with sample size correction (AICc). Significance is based on likelihood ratio test p-values; * p < 0.05, ns p > 0.05.
Figure S3. Average relative abundance of bacterial phyla per population. Pop1–5 are in the order of low to high altitude in Ecuador transect (A) and Bolivia-Brazil transect (B). The colors correspond to bacterial phyla that showed average relative abundance greater than 0.1% across all samples.
Figure S4. Correlations between altitude and predicted metagenomic functions. Spearman’s rho correlation between altitude and 19 KEGG pathways (level 3) that show Fisher’s combined p-value < 0.05 are shown. After correcting for multiple testing (Bonferroni correction; alpha = 0.05/183 = 0.0003) only Renin-angiotensin system remained significant (Ecuador; rho = 0.34, p = 0.0159. Bolivia-Brazil; rho = 0.51, p-value = 0.005. Fisher’s combined p-value = 0.0001). Spearman’s rho correlation for Ecuador transect is shown in red, and Bolivia-Brazil transect is shown in blue.
Figure S5. Correlations between altitude and predicted KEGG orthologs of renin-angiotensin system (ko04614). Spearman’s rho correlations between altitude and three KEGG orthologs are shown for both Ecuador transect and Bolivia-Brazil transect. Spearman’s rho correlations with Fisher’s combined p-value < 0.05 are indicated by *.