Abstract
Many microbiome studies employ reference-based operational taxonomic unit (OTU)-picking methods, which in general, rely on databases cataloguing reference OTUs identified through clustering full-length 16S rRNA genes. Given that the rate of accumulation of mutations are not uniform throughout the length of a 16S rRNA gene across different taxonomic clades, results of OTU identification or taxonomic classification obtained using ‘short-read’ sequence queries (as generated by next-generation sequencing platforms) can be inconsistent and of suboptimal accuracy. De novo OTU clustering results too can significantly vary depending upon the hypervariable region (V-region) targeted for sequencing. As a consequence, comparison of microbiomes profiled in different scientific studies becomes difficult and often poses a challenge in analysing new findings in context of prior knowledge. The OTUX approach of reference-based OTU-picking proposes to overcome these limitations by using ‘customized’ OTU reference databases, which can cater to different sets of short-read sequences corresponding to different 16S V-regions. The results obtained with OTUX-approach (which are in terms of OTUX-OTU identifiers) can also be ‘mapped back’ or represented in terms of other OTU database identifiers/taxonomy, e.g. Greengenes, thus allowing for easy cross-study comparisons. Validation with simulated datasets indicates more efficient, accurate, and consistent taxonomic classifications obtained using OTUX-approach, as compared with conventional methods.
Keywords: database, metagenomics, sequence analysis
1. Introduction
Advances in high-throughput DNA sequencing technologies have enabled culture independent studies of diverse microbial communities (microbiome) inhabiting different environments. Targeted amplicon sequencing of phylogenetic markers genes (e.g. the bacterial 16S rRNA) forms the basis of most of these studies. The downstream steps of taxonomic classification involve a similarity search of the sequenced DNA fragments (reads) against a reference database of pre-classified sequences (with taxonomic annotations).1–4 Inspite of extensive sampling of microbiomes from different environments being performed over the past decade, it would be optimistic to claim the completeness of current generation reference databases, with respect to cataloguing (and annotating) the entire diversity of the microbial world. Given this, many of the sequenced reads in a microbiome study may not find close matches in the reference databases, and thereby remain as unclassified. OTU (operational taxonomic unit)-based methods constitute an alternate approach, wherein the sequenced reads are clustered based on some predefined similarity threshold (de novo OTU clustering). Sequences clustered into the same OTU may be inferred to have originated from the same taxonomic group. Given that all sequenced reads can be grouped into OTUs prior to analysis, the problem with unclassified sequences can be traversed using this method. Further, taxonomic affiliations of these OTUs (and sequences clustered therein) can still be ascertained using similarity searches against reference databases. In practice, ‘open-reference OTU-picking’ approaches have been the choice of a number of recent studies.5–10 Such an approach involves an initial similarity search step against a reference database (of pre-clustered OTUs) for identifying sequences corresponding to previously annotated OTUs, followed by a clustering step with the remaining sequenced reads for identifying de novo OTUs.
Conventional OTU-picking approaches also have certain limitations. Previous studies have indicated how DNA sequencing errors can lead to an increased number of detected OTUs,11–13 or how the use of different clustering approaches may result in formation of alternate OTU clusters.14–17 However, some other important limitations of the OTU-based approaches, especially in the context of high-throughput DNA-sequencing technologies, have received relatively lesser attention. Given the limitations pertaining to the length of contiguous stretch of DNA (read length) that can be sequenced by currently available technologies, the targeted amplicon often consists of a selected region from the phylogenetic marker gene, instead of the entire gene. For example, in case of the bacterial 16S rRNA gene, stretches consisting of one or more specific ‘variable’ regions (spanning around 400–600 bp) are targeted for sequencing. However, since the reference databases catalogue sequences of full length marker genes, querying the same with ‘short’ sequence reads for OTU identification or taxonomic classification can yield suboptimal results.18 Furthermore, it may be noted that rate of evolution (accumulation of mutations) is not always uniform across the length of a chosen marker gene (or in its variable regions) across different taxonomic clades.19,20 It is possible that while a short region may remain more or less identical during the course of evolution, the flanking regions may be more prone to mutations. Alternately, barring a small hyper-variable stretch, a major fraction of the marker gene may remain unchanged through evolution. Given this scenario, OTU clustering results can vary significantly based on the choice of the target region. Although OTUs identified or classified using reference-based methods vs de novo clustering methods can provide different results, any comparison between the results obtained from studies utilizing different variable regions of a given marker gene also loses relevance.
The question whether the above limitations can be addressed using reference OTU databases that are specific to the targeted regions of the marker gene has been explored in this study. We present the OTUX (meta)database, which consists of 19 distinct OTU databases corresponding to the different stretches of variable regions (V-regions) of the bacterial 16S rRNA gene, that are commonly targeted for amplicon sequencing in microbiome studies. Each of the V-region-specific databases consists of OTUs (referred to as OTUX-OTUs) identified by clustering sequence fragments from corresponding stretches of V-regions cropped out from full-length 16S rRNA gene sequences catalogued in reference databases. Also presented is a ‘mapping matrix’ (MAPMAT), which lists the probabilities of association of any of the OTUX-OTUs to the reference OTUs present in the widely used Greengenes OTU database (consisting full-length marker genes). An open-reference-based OTU-picking approach against an appropriately selected OTUX V-region database is expected to provide results closer to those obtained with de novo OTU clustering. Further, the OTU abundance profiles, obtained in terms of OTUX-OTUs, can be ‘mapped back’ and represented in terms of Greengenes OTUs, using the MAPMAT. Mapping back enables comparing OTU-picking or taxonomic annotation results from different microbiome studies, even if the choice of targeted V-regions had been different. The utility of the proposed database and OTU-picking approach has been extensively validated with multiple simulated sequence datasets mimicking microbiome samples collected from diverse environments. The results indicate the benefits of using V-region-specific databases for classifying 16S amplicon data generated using short-read sequencing technologies.
2. Materials and methods
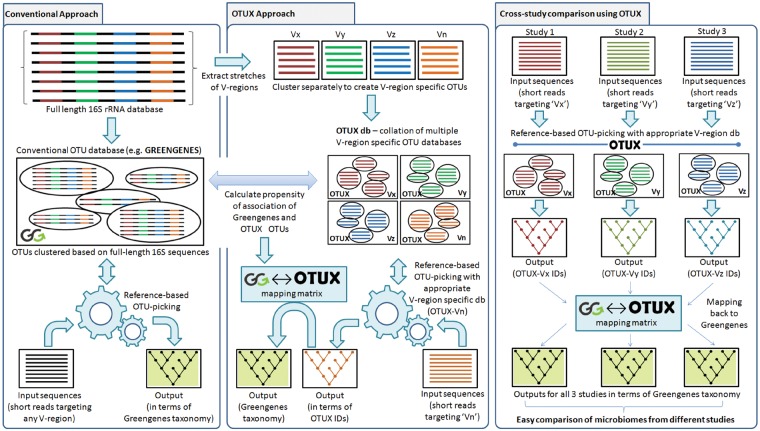
The OTUX workflow has two major components. This includes a one-time preprocessing step to generate customized V-region-specific reference databases (called OTUX databases) and a ‘MAPMAT’ for different stretches of V-regions. Subsequently, any amplicon sequencing dataset targeting 16S V-regions can be subjected to a reference-based OTU-picking cum taxonomic classification step using the OTUX reference database(s). Figure 1 gives a schematic representation of the OTUX method in context of conventional approach (CA) of OTU picking and taxonomic classification of 16S rRNA amplicon sequencing reads.
Figure 1.
A schematic representation of the OTUX method in context of CA of OTU picking and taxonomic classification of 16S rRNA amplicon sequencing reads. The utility of the ‘MAPMAT’ in enabling comparison of taxonomic profiles obtained using different V-regions is also depicted.
2.1. Building OTUX databases
The ‘prokMSA’ unaligned sequences from Greengenes database21 was downloaded through the following links: (i) http://greengenes.lbl.gov/Download/Sequence_Data/Fasta_data_files/current_prokMSA _unaligned.fasta.gz and (ii) ftp://greengenes.microbio.me/greengenes_release/gg_13_5/gg_13_5.fasta.gz. The downloaded files were decompressed to obtain the fasta formatted sequences, which were subsequently combined to create a final corpus of 1,079,252 unaligned sequences. The taxonomic classification of these sequences for different taxonomic hierarchical levels (including phylum, class, order, family, genus, species as well as Greengenes OTU IDs at 99% sequence identity) as annotated in Greengenes database version 13.8, were retrieved from ftp://greengenes.microbio.me/greengenes_release/gg_13_8_otus/taxonomy/99_otu_taxonomy.txt. Individual V-regions as well as stretches encompassing consecutive V-regions were extracted from each of the 16S rRNA gene sequences present in the sequence corpus using the software V-xtractor.22 The extracted sequences were then clustered based on sequence similarity using CD-HIT,23 wherein each resultant cluster constituted sequences sharing 99% sequence identity. Each of the clusters was assigned a V-region-specific OTU identifier (OTUXV ID) and was compiled to constitute an OTUXV reference database corresponding to a specific V-region.
2.2. Building MAPMAT
By definition, any OTU-picking approach, using one of the OTUX reference databases as reference, will result in taxonomic annotations in terms of OTUX OTU IDs corresponding to the specific OTUX database. To enable cross-comparisons between studies targeting different V-regions (thereby mandating use of different V-region-specific OTUX databases), as well as between studies using other CAs, a ‘MAPMAT’ between different OTUX OTU IDs and conventionally used Greengenes OTU IDs was created. The procedure of building mapping back matrices has been exemplified below using the V4 region-specific OTUX database. In this example, after compiling the OTUXV4 database, the propensity of association of a OTUXV4 ID (OTUXV4i) to a Greengenes OTU ID (GGj) is calculated using the following formula,
this is followed by populating the propensity matrix MAPMATV4, for the OTUXV4 database by computing all values for MAPMATV4ij wherein, i = 1 → NOTUX (i.e. total number of OTUXV4 OTUs); j = 1 → NGG (i.e. total number of Greengenes OTUs); and MAPMATV4 is a NGG × NOTUX matrix.
An example is shown in Supplementary Fig. S1, where an OTUX ID OTUXVi is mapped to corresponding Greengenes OTUX IDs. The percentages mentioned represent the propensities of OTUX ID OTUXVi to be associated to any of the hypothetical Greengenes OTU IDs GGx, GGy and GGz.
2.3. Creating validation datasets
Assessment of accuracy of taxonomic annotation of any OTU-picking or taxonomic classification method, on a real metagenomic dataset constituting short reads, poses a challenge since the origin (taxonomic affiliation) of the sequenced reads are ‘unknown’. The annotation results predicted by different available methods may vary from each other, with respect to the assigned lineage as well as depth of assignment, and cannot be used as a benchmark/standard for comparison. To overcome this problem, multiple validation sets of simulated short metagenomic reads were created and were classified into OTUs using our method. The simulated metagenomes pertaining to 4 different environments, viz. gut of healthy children (abbreviated as GUT),24 healthy human skin (abbreviated as SKIN),25 Mediterranean sea (abbreviated as SEA)26 and soil (SOIL),27 were generated using the following procedure. Publicly available datasets corresponding to metagenomic samples from the above mentioned environments were retrieved (Supplementary Table S1) and overall proportions of different genera present in each of the environments were obtained from the provided abundance tables (Supplementary Material S2). To create a simulated metagenome pertaining to an environment, full length 16S rRNA genes were randomly drawn from the RDP database (v10.3),28 ensuring that the proportions of the picked genera were similar to those present in the chosen environment (only top 50 genera in each environment considered). The sequences were drawn in the following manner. Let it be assumed that there are ‘N’ full-length sequences belonging to genus ‘X’ (which may belong to different species/strains of the same genus) in the RDP database. Further, let it be considered that in a simulated metagenome (of a pre-defined size or total number of sequences) pertaining to a specific environment, the genus ‘X’ needs to be represented with ‘i’ number of sequences. To achieve this, ‘i’ random sequences from the set of ‘N’ sequences (belonging to genus ‘X’) were picked with replacement. The process was repeated for every genus constituting the desired simulated metagenome. 100 simulated metagenomic datasets (each constituting 10,000 sequences) were created for each of the four environments (abbreviated as DGUT/F, DSKIN/F, DSEA/F and DSOIL/F). Specific V-regions or selected stretches of V-regions from each of the full-length 16S rRNA gene sequences (constituting these simulated metagenomes) were further cropped out to create corresponding simulated ‘short-read’ metagenomic datasets (abbreviated as DGUT/Vi, DSKIN/Vi, DSEA/Vi and DSOIL/Vi), mimicking targeted amplicon sequencing. The selected V-regions (and stretches of V-regions) considered for validation included V4, V1-V2, V1-V3, V2-V3, V3-V4, V3-V5, V3-V6, V4-V5, V4-V6, V5-V6 and V6-V8, which are typically employed in 16S rRNA-based microbiome studies.
2.4. Classifying and annotating metagenomic sequences
Reference-based OTU-picking can be performed for a query set of metagenomic sequences using an OTUXVi reference database (wherein ‘i’ corresponds to the V-region or stretch of consecutive V-regions which were targeted during amplicon sequencing). For the purpose of validation, OTU-picking or taxonomic assignment exercise was performed with multiple simulated metagenomic datasets as well as using different OTUXVi reference databases (as appropriate for the individual datasets). The following section illustrates this process of annotation for a set of simulated query sequences corresponding to an amplicon sequencing experiment targeting the V4 region, using OTUXV4 as the reference database. The same methods can be extended for classifying sequences generated in sequencing experiments targeting a different V-region, by choosing an appropriate OTUXVi reference database. In this work, a naïve Bayesian classifier (Wang’s algorithm),4 as implemented in the software ‘mothur’,29 was used for classification, wherein a bootstrap confidence threshold of 80% was used for making assignments at the OTU level. Subsequently, an OTU abundance profile (TOTUX) was generated by cumulating the total number of sequenced reads from the given metagenomic sample that could be attributed to each of the OTUXV4 OTUs. It may be noted here that the choice of the naïve Bayesian classifier for the validation study was influenced by its wide use in microbiome studies and annotation tools (e.g. RDP classifier).4 Other reference-based OTU-picking and taxonomic classification methods can also be used while using OTUX as a reference database. The classification results (obtained in terms of OTUXV4 OTUs) were mapped back using the corresponding MAPMATV4 to represent the results in terms of Greengenes (v13.8 – OTUs obtained at 99% identity) OTU IDs. This ‘mapping back’ can be performed using either a ‘one-to-one mapping’ (one OTX OTU mapped back to a single Greengenes OTU) approach or a ‘one-to-many mapping’ (one OTX OTU mapped back to multiple Greengenes OTUs) approach. These mapping back approaches are described below.
2.4.1. One-to-one mapping
In this procedure, each query sequence is assigned to one particular Greengenes OTU ID. For example, if a particular query sequence ‘s’ has been assigned to the OTU ‘x’ corresponding to the reference database OTUXV4 (abbreviated as OTUXV4x), the MAPMATV4 elements {MAPMATV4xj} are retrieved (wherein ‘j’ = 1 → NGG i.e. the total no. of Greengenes OTUs). The maximum value of {MAPMATV4xj} is then computed. The sequence ‘s’ can subsequently be classified to the Greengenes OTU ‘y’ (abbreviated as GGy), wherein MAPMATV4xy = max{MAPMATV4xj}. In effect, the OTUX ID OTUXV4x is mapped to the Greengenes ID GGy since they have the highest propensity of association as recorded in the MAPMAT. The process, once repeated for all query sequences will subsequently lead to a new OTU abundance profile (TGG), represented in terms of Greengenes OTU IDs.
2.4.2. One-to-many mapping
Given that one OTUX ID may be associated with multiple Greengenes IDs (and vice versa), the mapping back matrix can also be utilized for one-to-many mapping to report abundance tables representing relative abundance of the OTUs (such as percentage normalized abundance). To begin with, the abundance profile TOTUX needs to be generated as described above, wherein the total number of sequences assigned to each of the OTUXV4 OTUs is represented.
For example, TOTUX can be represented in form of a column matrix (of size NOTUX × 1) as depicted above wherein ‘i’ varies from 1 to the total number of OTUXV4 OTUs (i.e. NOTUX), and wherein ‘a’ is the number of sequences assigned to the OTU OTUXV41, ‘b’ is the number of sequences assigned to OTUXV42, ‘c’ is the number of sequences assigned to OTUXV43 and so on. Next, an OTU abundance profile (TGGraw) is obtained for the set of query sequences, in terms of Greeengenes OTU IDs by multiplying the matrix MAPMATV4 with the matrix TOTUX. It may be noted that given the nature of the MAPMAT matrix, the abundance values for each of the Greengenes OTUs in TGGraw may be a fractional value.
wherein, TGGraw is a column matrix of size (NGG × 1), and NGG is the total number of Greengenes OTUs. Finally a percentage normalized OTU abundance profile (TGG%) is obtained by performing the following transformation on each element (Tj) of TGGraw
wherein, TGG% is a column matrix of size (NGG × 1), and NGG is the total number of Greengenes OTUs. The abundance of taxonomic groups present in the metagenomic sample, as obtained in the form of either of the three column matrices, viz. TOTUX, TGG and TGG%, can further be represented at any desired taxonomic level utilizing the taxonomic hierarchy information associated with the Greengenes OTUs.
As mentioned earlier, the current version of OTUX approach has been modelled around the Greengenes database and designed to pick OTUs at the most stringent threshold suggested by Greengenes (i.e. 99% identity). The choice of threshold was guided by the rationale that OTUs picked based on a more stringent identity threshold (e.g. 99%) can later be merged together to make larger OTU clusters corresponding to a lower threshold (e.g. 97%). Further, it has also been reported that stringent average nucleotide identity cut-off of >99% might be required to delineate a new species.30 Although clustering at 97% identity threshold would generate lesser number of clusters and make the data more amenable for analysis, a higher threshold of 99% is expected to be more suitable for accurate identification of strains and ecotypes. However, given that some studies may also prefer lower identity thresholds (e.g. 97%) for OTU clustering, the implementation of OTUX approach (available at https://web.rniapps.net/otux/) provides option to map back OTUX OTUs (originally picked at 99% identity threshold) to Greengenes OTU IDs corresponding to OTUs clustered at identity thresholds of 99% as well as 97%.
2.5. Classifying simulated metagenomic sequences (validation datasets)
The full length sequences belonging to each of the simulated metagenomic datasets (DGUT/F, DSKIN/F, DSEA/F and DSOIL/F) were first subjected to ‘OTU picking’ (taxonomic classification at the OTU level) against the Greengenes database (v13.8—ftp://greengenes.microbio.me/greengenes_release/gg_13_8_otus/rep_set/99_otus.fasta) using the naïve Bayesian classifier (Wang’s algorithm) as implemented in the software ‘mothur’ (with a bootstrap confidence threshold of 80%). Given that full-length 16S sequences were compared against a full-length 16S rRNA sequence database, the results obtained using the above procedure reflects the best achievable OTU-classification using 16S rRNA amplicon sequencing (using the given algorithm) and was considered as a ‘benchmark’ or the ‘gold standard’ (abbreviated as GS) of taxonomic classification for our validation study.
The simulated ‘short-read’ metagenomic sequences in the validation datasets were subsequently subjected to taxonomic classification using the following three methods. For ease of explanation, examples corresponding to the simulated metagenomic datasets pertaining to V4 region (DGUT/V4, DSKIN/V4, DSEA/V4 and DSOIL/V4) have been used in the following sections. The same methods have been followed for validation using other simulated metagenomic datasets.
2.5.1. Conventional approach
Sequences present in each of the simulated metagenomes belonging to the sets DGUT/V4, DSKIN/V4, DSEA/V4 and DSOIL/V4 were classified using the naïve Bayesian classifier (Wang’s algorithm as implemented in the software ‘mothur’ with a bootstrap confidence threshold of 80%), considering the Greengenes OTU database as a reference (similar approach as GS). These results represent taxonomic classification that can be obtained using the CA of OTU-picking or taxonomic classification wherein short-read sequences (encompassing a certain region of a marker gene) are used as a query against a OTU database constituted of full length marker gene sequences. For ease of comparison, abundance profiles (as per Greengenes taxonomy) representing the proportion of OTUs (and other taxonomic hierarchical levels), both in terms of raw sequence counts as well as percentage normalized abundance, were generated.
2.5.2. De novo OTU-picking approach using CROP (CR)
Metagenomic sequences belonging to each of the validation sets (DGUT/V4, DSKIN/V4, DSEA/V4 and DSOIL/V4) were clustered de novo at 99% identity using the software CROP,31 wherein each cluster represents a OTU. Subsequently, in order to obtain taxonomic classification of the respective clusters/OTUs, the representative sequences from each of the clusters were classified using the Greengenes OTU database as a reference. The taxonomic profile TCR reports abundance of OTUs generated using CROP, wherein abundance of each OTU is equivalent to its cluster size.
2.5.3. OTUX approach (OTUX)
Metagenomic sequences belonging to each of the validation sets (DGUT/V4, DSKIN/V4, DSEA/V4 and DSOIL/V4) were also classified using the naïve Bayesian classifier (Wang’s algorithm as implemented in the software mother with a bootstrap confidence threshold of 80%), using the OTUXV4 database as a reference. These results represent the taxonomic classification that can be obtained using the novel OTUX approach of OTU-picking and taxonomic classification wherein short-read sequences (covering a certain region of a marker gene) are used as query against a pre-computed OTU database corresponding to a specific hypervariable region (V4 in this case). It may be noted that the obtained OTU abundance profile (TOTUX) reports the results in terms of OTUXV4 OTU IDs. For ease of comparison, these results are also mapped back in terms of Greengenes OTU IDs and are provided in form of OTU abundance profile TGG (using one-to-one mapping approach), wherein raw counts of sequences assigned to individual Greengenes OTUs are depicted. Furthermore, percentage normalized abundance profile(s) TGG% (using one-to-many mapping approach) are also generated, wherein abundance/proportion of OTUs (and/or other taxa) is represented in percent normalized terms.
2.6. Validation tests
The correctness of taxonomic assignments obtained with OTUX and CA approaches were assessed by comparing them against the benchmark/‘GS’, which corresponds to OTU assignments obtained using full-length 16S rRNA gene sequences searched against the Greengenes reference database. The comparison was based on the following parameters. First, the accuracy of taxonomic assignments by CA and OTUX at OTU, Genus, and Family level(s) were assessed in terms of correct number of assignments (with reference to GS results). During this assessment, one-to-one mapping of OTUX IDs to Greengenes IDs had been used. Subsequently, a t-test was performed to assess the performance of OTUX with respect to CA (based on results of 100 simulated metagenomic datasets). The approaches were also compared on the basis of computational time required for classification.
To evaluate the overall accuracy as well as coherence of taxonomic assignments obtained using different V-regions and methods, the widely used Unifrac distance metric was chosen.32 Unifrac is a dissimilarity measure which considers phylogenetic information of constituent microbes while comparing microbiome samples, thereby allowing assessment of overall accuracy considering all taxonomic hierarchical levels. Pairwise weighted Unifrac distances were calculated between percentage normalized abundance tables generated by GS, OTUX, CA and CR. In case of OTUX, CA and CR, taxonomic profiles obtained using all relevant V-regions or stretches of V-regions were considered. For this assessment, percentage normalized OTUX abundance profiles (TGG%) were generated using one-to-many mapping approach between OTUX IDs and Greengenes IDs. Dendograms were constructed from the pairwise Unifrac distances (averaged for 100 simulated metagenomic datasets) using UPGMA clustering, as implemented in the programme ‘Neighbor’ included in the Phylip package,33 considering GS as an outgroup. This process was repeated for the simulated metagenomes corresponding to all four selected environments (DGUT, DSKIN, DSEA and DSOIL). Different diversity measures for the taxonomic profiles obtained with GS, OTUX, CA and CR approaches, viz. Shannon diversity, Simpson index (evenness), Chao1 index (species richness) and number of observed species, were also computed for the DGUT environment, using the R Phyloseq package.34
In addition to checking for phylogenetic accuracy, it was also essential to validate whether OTUX results were closer to de novo OTU-picking results, when compared with the CA. For this purpose, the clusters generated using CROP (wherein each of these clusters represent an OTU) were compared against OTUs identified by OTUX and CA. 100 largest clusters/OTUs (for any selected V-region/stretches of V-regions) obtained from OTUX, CA and CROP were selected. Jaccard similarity was calculated between each cluster i obtained by OTUX against all the clusters obtained using CROP with the following formula,
where, j represents each cluster obtained using CROP and j = 1→ 100 (total number of selected clusters). The cluster pair with highest Jaccard similarity was considered. Similarly, a reciprocal search was also performed wherein the closest OTUX cluster to any of the CROP clusters were identified. Consequently, 200 pairs of closest clusters (in terms of Jaccard similarity) were obtained between OTUX and CROP, with a fraction of them being reciprocal best hits. An average Jaccard similarity score was calculated using the formula,
The similarity in OTU clustering between OTUX and CROP was subsequently evaluated in terms of average Jaccard similarity and the number of reciprocal best hits. For comparison, the similarity between CA and CROP approaches was also evaluated using the above mentioned steps. Additional analyses were also performed to evaluate the effect of different sequence identity thresholds for clustering as well as the effects of inclusion of relatively smaller clusters (in addition to the 100 largest clusters) during calculation of average Jaccard similarity. To this end, the above mentioned analyses were extended for the top 300 and 500 (most populous) clusters generated for the simulated gut microbiome (GUT) dataset with de novo clustering (CROP) thresholds set at 99%, 97% as well as 95%.
3. Results and discussion
3.1. OTUX approach provides higher proportion of correct taxonomic classifications
Table 1 provides a comparison of taxonomic classification accuracy of the proposed OTUX approach, with respect to CA, while classifying different simulated microbiome datasets. As mentioned earlier, CA involves reference-based OTU picking from a query set of short 16S rRNA gene reads using an OTU database consisting of full length 16S rRNA gene sequences. In this study, Greengenes version 13.8, containing OTUs clustered at 99% sequence identity, was used as the reference database for CA. In contrast, OTUX approach involves reference-based OTU picking of the same short reads using an appropriate OTUX V-region database (consisting sequences clustered with 99% identity). OTUs identified using OTUX approach, initially classified in terms of OTUX database identifiers (OTUX IDs), was ‘mapped back’ to be represented in terms of Greengenes IDs (see Materials and methods), so as to enable comparison against CA. Results obtained with both CA and OTUX approaches were evaluated with reference to a ‘GS’, which corresponded to the reference-based OTU-picking results (in terms of Greengenes IDs) that could be obtained using full length 16S rRNA gene amplicons instead of short-read sequences (see Materials and methods). In terms of number of sequences correctly assigned at the OTU level, OTUX approach significantly (P < 0.001) outperforms CA in almost all cases, irrespective of the type of microbiome sample or the targeted V-region (Table 1). Barring a couple of occasions, OTUX is observed to consistently assign over 50% of the query reads to appropriate OTUs, with accuracies reaching as high as 70% in some cases. More than often, OTUX could correctly assign almost double the number of reads compared with CA. CA provides better result than OTUX only in a single instance, wherein infant gut microbiome (GUT) samples were probed using V1–V3 region. In addition to the number of correct OTU assignments, the results of OTUX were also evaluated in terms of overall correctness of the obtained taxonomic profiles for the simulated microbiome samples considering different levels of taxonomic hierarchy (Table 1; Supplementary Table S2A and B; Supplementary Material S3). Weighted-UniFrac distances between the taxonomic abundance profiles obtained using OTUX (see Materials and methods) and those obtained using GS (using full-length 16S rRNA gene sequences) were computed for this purpose. For comparison, distances between taxonomic profiles obtained using CA and GS were also calculated. Echoing earlier results, taxonomic abundance profiles obtained with OTUX are found to be more similar to the GS taxonomic profiles as compared with the profiles obtained with CA, in almost all cases. The only exceptions pertain to the profiles generated for the infant gut (GUT) microbiome corresponding to experiments targeting the V1–V2 and V1–V3 regions. In summary, OTUX approach appears to be a more reliable method for reference-based OTU picking, providing greater accuracy in taxonomic classification than existing CAs utilizing full-length 16S rRNA sequence databases.
Table 1.
Taxonomic classification results obtained with the proposed OTUX approach and CA
| Simulated microbiomes | Classification accuracies |
Target V-region |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1V2 |
V1V3 |
V2V3 |
V3V4 |
V3V5 |
V3V6 |
V4 |
V4V5 |
V4V6 |
V5V6 |
V6V8 |
|||
| Comparison of taxonomic assignmentsa at OTU level (out of 10,000 reads; averaged for 100 randomly generated simulated datasets) | |||||||||||||
| GUT | OTUX | Correct assignments | 4386.8 | 4355.2 | 5461.2 | 6265.4 | 6600.5 | 6957.4 | 5795.3 | 6869.8 | 6849.7 | 5862.2 | 6771.3 |
| Wrong assignments | 47.2 | 84.3 | 102.5 | 151.9 | 229.2 | 97.4 | 175.9 | 57.4 | 78.5 | 110.1 | 40.3 | ||
| W-Unifracb from GS | 0.324 | 0.292 | 0.234 | 0.236 | 0.213 | 0.202 | 0.288 | 0.216 | 0.207 | 0.236 | 0.215 | ||
| CA | Correct assignments | 3618.6 | 4464.1 | 3808.6 | 3084.6 | 3759.3 | 4748.2 | 2318.5 | 2872.7 | 4162.0 | 3084.7 | 3429.6 | |
| Wrong assignments | 555.2 | 244.4 | 1020.9 | 1582.2 | 935.7 | 490.4 | 2631.3 | 814.5 | 531.8 | 1785.7 | 1154.7 | ||
| W-Unifracb from GS | 0.318 | 0.272 | 0.280 | 0.310 | 0.289 | 0.238 | 0.332 | 0.361 | 0.270 | 0.300 | 0.311 | ||
| SKIN | OTUX | Correct assignments | 4034.4 | 4257.3 | 5377.9 | 5431.8 | 5710.8 | 5759.2 | 3800.5 | 5027.3 | 5800.2 | 5472.8 | 5295.9 |
| Wrong assignments | 52.7 | 44.9 | 59.8 | 163.7 | 47.5 | 52.6 | 34.0 | 38.3 | 46.8 | 52.9 | 63.8 | ||
| W-Unifracb from GS | 0.218 | 0.193 | 0.101 | 0.123 | 0.129 | 0.115 | 0.237 | 0.118 | 0.125 | 0.104 | 0.104 | ||
| CA | Correct assignments | 1330.5 | 1789.3 | 1807.7 | 1478.2 | 2254.9 | 3238.6 | 320.4 | 1172.1 | 2164.7 | 1491.7 | 1816.6 | |
| Wrong assignments | 1508.7 | 625.9 | 1477.0 | 1965.6 | 1476.7 | 727.5 | 1652.0 | 1466.1 | 1460.9 | 2346.1 | 1692.2 | ||
| W-Unifracb from GS | 0.402 | 0.388 | 0.345 | 0.349 | 0.317 | 0.266 | 0.469 | 0.387 | 0.319 | 0.333 | 0.336 | ||
| SEA | OTUX | Correct assignments | 5017.8 | 6085.1 | 6348.9 | 6290.1 | 6297.6 | 7037.4 | 4932.7 | 6495.9 | 6652.1 | 6253.7 | 6695.5 |
| Wrong assignments | 18.1 | 11.0 | 24.0 | 77.8 | 9.2 | 16.7 | 1.7 | 7.6 | 15.9 | 13.3 | 32.6 | ||
| W-Unifracb from GS | 0.249 | 0.152 | 0.121 | 0.127 | 0.145 | 0.145 | 0.249 | 0.146 | 0.116 | 0.155 | 0.130 | ||
| CA | Correct assignments | 2579.8 | 3471.2 | 3180.7 | 3287.6 | 4562.6 | 4753.3 | 2183.1 | 3197.9 | 4240.4 | 2866.6 | 4047.9 | |
| Wrong assignments | 1450.0 | 1638.7 | 1415.3 | 1174.7 | 1108.1 | 1049.9 | 1531.6 | 925.8 | 1095.9 | 1644.5 | 1801.0 | ||
| W-Unifracb from GS | 0.385 | 0.316 | 0.333 | 0.319 | 0.224 | 0.218 | 0.397 | 0.328 | 0.256 | 0.335 | 0.256 | ||
| SOIL | OTUX | Correct assignments | 5109.9 | 5086.0 | 5540.1 | 5431.8 | 5948.4 | 5800.0 | 4155.5 | 5257.0 | 5613.2 | 5222.1 | 5814.4 |
| Wrong assignments | 46.5 | 45.4 | 54.6 | 225.5 | 69.0 | 51.4 | 68.3 | 71.5 | 47.7 | 45.5 | 85.7 | ||
| W-Unifracb from GS | 0.145 | 0.114 | 0.078 | 0.123 | 0.127 | 0.153 | 0.225 | 0.133 | 0.120 | 0.162 | 0.125 | ||
| CA | Correct assignments | 4089.1 | 4680.5 | 4703.2 | 1478.2 | 5176.7 | 5637.2 | 2287.0 | 3806.7 | 5357.6 | 3714.7 | 4388.2 | |
| Wrong assignments | 690.0 | 248.5 | 191.5 | 444.4 | 301.4 | 129.3 | 685.9 | 806.1 | 279.4 | 833.5 | 390.7 | ||
| W-Unifracb from GS | 0.187 | 0.158 | 0.154 | 0.349 | 0.104 | 0.084 | 0.334 | 0.184 | 0.089 | 0.193 | 0.156 | ||
Performance of both the approaches have been evaluated with multiple simulated microbiome datasets mimicking different types of environmental samples, (viz., infant gut, skin, sea and soil), and amplicon sequencing experiments targeting different V-regions of the bacterial 16S rRNA gene. Cases wherein the number of correct assignments by OTUX are significantly higher (t-test; P < 0.001) have been highlighted in bold.
CA and OTUX approaches have been evaluated considering results obtained with full-length 16S rRNA genes to be ‘correct’ or the ‘GS’. Only number of average correct assignments and wrong assignments has been depicted. The remaining reads (out of 10,000) could not be classified at OTU level.
Weighted UNIFRAC distance of taxonomic abundance profile from actual taxonomic diversity (GS: gold-standard).
As mentioned earlier, one of the major reasons behind incorrect taxonomic assignment of short-read sequences to a different taxonomic lineage pertain to un-even evolutionary rate along the length of the 16S rRNA gene. The ‘mapping matrices’ introduced in the OTUX approach (see Building MAPMAT section) probably address this issue to some extent. Although in case of CA, finding high level of similarity of a very short stretch of sequence against a Greengenes (GG) OTU might result in a direct attribution of the short sequence to the lineage of the given GG OTU, the OTUX approach acknowledges that such a short semi-identical stretch could have alternate origins. The ‘mapping matrices’ presented in this work captures the propensities of association of such semi-identical short stretches of sequences (which have been clustered to form OTUX OTUs), to multiple GG OTUs/taxonomic lineages. It is based on these propensities that the final taxonomic abundance tables (according to GG lineages) are generated in the OTUX approach. Although this method (of propensity-based assignments) may also lead to incorrect assignments in some cases, overall the validation results indicate better performance of the OTUX approach compared with the direct assignments obtained with CA.
To evaluate whether the relatively better performance of OTUX spanned all the taxonomic lineages, or were limited to certain specific clades, the proportions of correct and incorrect assignments by OTUX and CA, cumulated at genus and family levels were analysed (Supplementary Material S4). Neither OTUX nor CA could provide unbiased performance for all the taxonomic lineages while using different V-regions/stretches of V-regions. However, it was observed that in most cases the proportions of correct assignments by OTUX (averaged over all families or genera) were higher compared with CA. Further, the standard deviations in proportions of correct assignments across families/genera were also observed to be mostly equivalent or slightly lower for OTUX, reiterating the utility of the newly proposed taxonomic classification approach.
3.2. OTU clusters identified by OTUX are similar to those obtained using de novo OTU-picking approach
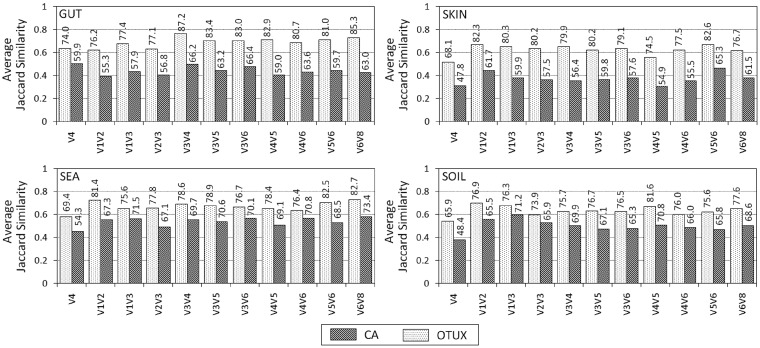
Given the observed improvements in reference-based OTU-picking and taxonomic classification achieved using OTUX, it was imperative to check how close are these results to those obtained using any sequence similarity based de novo OTU clustering method. Figure 2 depicts a comparison of OTU picking results obtained using CROP (CR), a popular de novo OTU clustering method, with those obtained using OTUX and CA. Since the three compared methods do not generate any common identifier(s) for the different OTU clusters obtained, ‘Jaccard similarity’ was computed to check the similarities between clusters generated by different methods (see Materials and methods). Similar clusters would share a greater proportion of query sequences, thereby resulting in a higher Jaccard similarity. Although comparing between OTUX and CR clusters, 100 largest clusters (in terms of number of query sequences in each cluster) generated by both the methods were selected, and pairwise Jaccard similarities were computed. For each of the 100 OTUX clusters, most similar CR clusters were identified based on Jaccard similarity. Similarly, for each of the 100 CR clusters, most similar OTUX clusters were also identified using reciprocal searches. Average Jaccard similarity values corresponding to these 2 × 100 closest pairs of clusters were considered as a measure of overall similarity in OTU clustering obtained using the two methods. Further, the number of reciprocal best hits between OTUX and CR clusters was also used as an indicator to evaluate the similarity between clustering results. Similarity between OTU clustering by CA and CR was also evaluated in a similar manner. As evident from Fig. 2, the results obtained using OTUX are relatively more similar to CR results when compared with results obtained with CA. The results of OTUX and CR are observed to be around 1.2–1.7 times more similar to each other in terms of average Jaccard similarity as compared with the similarity between CA and CR approaches. Even in terms of number of reciprocal best hits identified between the sets of clusters, OTUX and CR are more closely associated. Similar results were observed on extending the comparison for the top 300 and 500 (most populous) clusters generated for the simulated gut microbiome dataset with de novo clustering (CROP) thresholds set at 99, 97 and 95% (Supplementary Fig. S2). These results assume further significance when viewed in context of execution time. It is expected that both CA and OTUX, being reference-based OTU-picking approaches, would have quicker execution time when compared with de novo OTU clustering approaches.10 Comparison of execution times between OTUX and CA revealed an overall equivalent performance for both approaches, with minor variations depending on the targeted V-region (Supplementary Table S3A and B). A reasonable execution time coupled to a clustering solution closer to a de novo approach, as well as simultaneously improving on accuracy of taxonomic classification, certainly provides OTUX approach an edge over the conventional reference-based OTU picking.
Figure 2.
Similarity of OTU clusters obtained using OTUX and CA when compared with de novo OTU clusters obtained using CROP (CR) for different simulated microbiome datasets pertaining to (A) GUT, (B) SKIN, (C) SEA and (D) SOIL. The bars indicate average Jaccard similarity (in terms of query sequence reads) between 100 largest clusters identified by the compared methods (see Materials and methods). The number mentioned above each bar represents the percentage of reciprocal best hits identified between the 100 largest clusters identified by compared methods (see Materials and methods).
3.3. OTUX approach results obtained using different V-regions are coherent and better descriptors of taxonomic diversity
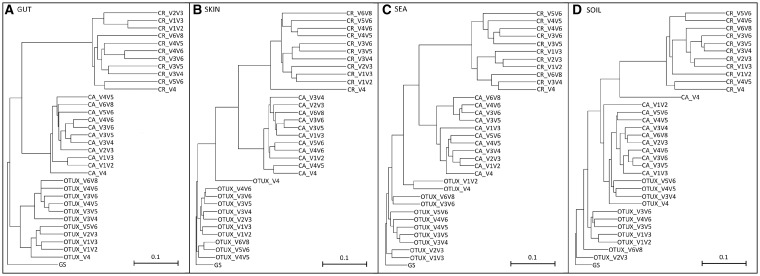
Despite the promising results mentioned above, utilizing OTUX for a cross-study comparison of microbiome datasets (targeting different V-regions) only finds relevance if OTUX results obtained with different V-regions are coherent. Figure 3 depicts four dendograms, wherein the taxonomic profiles corresponding to the simulated microbiome datasets obtained with OTUX, CA and CR, have been clustered based on pair wise weighted UniFrac distances (UPGMA method). Nodes in the tree correspond to the results obtained when different V-regions were targeted. Although OTUs identified using OTUX approach were mapped back to be represented in terms of Greengenes IDs and taxonomy, the cluster representatives of each of the OTUs generated using CR were also annotated in terms of Greengenes taxonomy to enable comparison with CA approach (see Materials and methods). The ‘GS’ result was used as an ‘outgroup’ during clustering. The clustering patterns in Fig. 3 indicate that results obtained using the same method (either of OTUX, CA or CR) group together irrespective of the targeted V-region. Although this observation assures of a certain level of coherence between results obtained using any given method, the proximity of the OTUX results cluster to the GS result is worth noting. Not only are the results obtained using OTUX approach coherent, but they are also consistently better representatives of the taxonomic diversity of the sampled environment, irrespective of the targeted V-region. In addition to the comparison based on weighted UniFrac distances, different diversity measures, like Shannon diversity, Simpson index (evenness), Chao1 index (species richness) and number of observed OTUs pertaining to the simulated gut microbiome, were computed based on the OTUs identified using CA, CR and OTUX approaches (Supplementary Table S4). These results were also compared in context of the diversity values corresponding to the GS taxonomic profiles. For most of the regions, diversity values obtained with OTUX were closer to the GS diversity values, when compared with CA and CROP methods.
Figure 3.
Dendograms depicting similarity between taxonomic profiles obtained with different OTU classification methods (viz. GS, CA, CR and OTUX) while using different V-regions. The trees have been constructed using the UPGMA method while using weighted UniFrac as the distance measure and considering the taxonomic profile generated with GS as an outgroup. Dendograms corresponding to simulated microbiomes pertaining to the environments (A) GUT, (B) SKIN, (C) SEA and (D) SOIL, have been plotted.
It is however important to note here that certain other biologically important differences and methodological biases (in addition to those resulting from the choice of V-regions) may be inherent to sequenced data from different studies and protocols. The possible effects of these potential biases, on the robustness of taxonomic assignments obtained with CA and OTUX was tested using real microbiome sampling data from an earlier study.35 Microbiomes pertaining to six different environments had been sampled in the earlier study, wherein the DNA sequencing for each of the samples were done on both Illumina (MiSeq 2 × 250 bp paired end protocol) and Roche 454 (GS FLX using Titanium) platforms. Taxonomic assignments for both the Illumina as well as Roche generated datasets from this study were performed with OTUX and CA, followed by a comparison of the similarity of the taxonomic profiles generated based on weighted Unifrac distances. Taxonomic profiles generated from Illumina and Roche datasets using OTUX method were observed to be more similar to each other than those generated by CA, in case of four (out of six) of the sampled environments (Supplementary Table S5). It may be noted here that the study specific biases pertaining to sequencing chemistry, sample storage/handling and other methodological differences, cannot be expected to be completely overcome by using in silico taxonomic classification methods such as OTUX or other CAs. However, given that the results indicate at OTUX’s ability to arrive at more coherent taxonomic profiles when compared with the CA, OTUX is expected to have better utility while performing cross-study taxonomic comparisons.
4. Conclusion
In summary, adopting OTUX approach of OTU picking can enable easy and accurate cross-study comparison of taxonomic profiles, even when targeted V-regions might vary across different studies. Although performing such comparisons at a large scale (with several microbiome datasets) using de novo OTU clustering approach might be computationally prohibitive, using CA like approaches might connote a compromise in accuracy of the comparison. In contrast, such comparisons using OTUX approach not only provides more accurate taxonomic annotations (even at the OTU level), but also attains the same with lesser computational expenses.
Availability
Freely available for academic use on the web at https://web.rniapps.net/otux/
Supplementary Material
Acknowledgements
All three authors are employees of Tata Consultancy Services Ltd. (TCSL) and would like to acknowledge TCSL for its support in form of salaries.
Conflict of interest
None declared.
References
- 1. Caporaso J. G., Bittinger K., Bushman F. D., DeSantis T. Z., Andersen G. L., Knight R.. 2010, PyNAST: a flexible tool for aligning sequences to a template alignment, Bioinformatics, 26, 266–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edgar R.C. 2010, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 26, 2460–1. [DOI] [PubMed] [Google Scholar]
- 3. Ghosh T. S., Gajjalla P., Mohammed M. H., Mande S. S.. 2012, C16S - a Hidden Markov Model based algorithm for taxonomic classification of 16S rRNA gene sequences, Genomics, 99, 195–201. [DOI] [PubMed] [Google Scholar]
- 4. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R.. 2007, Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy, Appl. Environ. Microbiol., 73, 5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baron J. L., Vikram A., Duda S., Stout J. E., Bibby K.. 2014, Shift in the microbial ecology of a hospital hot water system following the introduction of an on-site monochloramine disinfection system, PLoS One, 9, e102679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harris J. K., Caporaso J. G., Walker J. J., et al. 2013, Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat, Isme J., 7, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jervis-Bardy J., Leong L. E. X., Marri S., et al. 2015, Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data, Microbiome, 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caporaso J. G., Paszkiewicz K., Field D., Knight R., Gilbert J. A.. 2012, The Western English Channel contains a persistent microbial seed bank, Isme J., 6, 1089–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pylro V. S., Roesch L. F. W., Morais D. K., Clark I. M., Hirsch P. R., Tótola M. R.. 2014, Data analysis for 16S microbial profiling from different benchtop sequencing platforms, J. Microbiol Methods, 107, 30–7. [DOI] [PubMed] [Google Scholar]
- 10. Rideout J. R., He Y., Navas-Molina J. A., et al. 2014, Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences, PeerJ., 2, e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dickie I. A. 2010, Insidious effects of sequencing errors on perceived diversity in molecular surveys, New Phytol., 188, 916–8. [DOI] [PubMed] [Google Scholar]
- 12. Poretsky R., Rodriguez-R L. M., Luo C., Tsementzi D., Konstantinidis K. T.. 2014, Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics, PLoS One, 9(4): e93827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quince C., Lanzen A., Davenport R. J., Turnbaugh P. J.. 2011, Removing noise from pyrosequenced amplicons, BMC Bioinformatics, 12, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barriuso J., Valverde J. R., Mellado R. P.. 2011, Estimation of bacterial diversity using next generation sequencing of 16S rDNA: a comparison of different workflows, BMC Bioinformatics, 12, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bonder M. J., Abeln S., Zaura E., Brandt B. W.. 2012, Comparing clustering and pre-processing in taxonomy analysis, Bioinformatics, 28, 2891–7. [DOI] [PubMed] [Google Scholar]
- 16. Chen W., Zhang C. K., Cheng Y., Zhang S., Zhao H.. 2013, A comparison of methods for clustering 16S rRNA sequences into OTUs, PLoS One, 8(8): e70837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schloss P. D., Westcott S. L.. 2011, Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis, Appl. Environ. Microbiol., 77, 3219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franzén O., Hu J., Bao X., Itzkowitz S. H., Peter I., Bashir A.. 2015, Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering, Microbiome, 3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim M., Morrison M., Yu Z.. 2011, Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes, J. Microbiol. Methods, 84, 81–7. [DOI] [PubMed] [Google Scholar]
- 20. Schloss P. D. 2010, The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies, PLoS Comput. Biol., 6, e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeSantis T. Z., Hugenholtz P., Larsen N., et al. 2006, Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB, Appl. Environ. Microbiol., 72, 5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hartmann M., Howes C. G., Abarenkov K., Mohn W. W., Nilsson R. H.. 2010, V-Xtractor: an open-source, high-throughput software tool to identify and extract hypervariable regions of small subunit (16S/18S) ribosomal RNA gene sequences, J. Microbiol. Methods, 83, 250–3. [DOI] [PubMed] [Google Scholar]
- 23. Li W., Godzik A.. 2006, Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences, Bioinformatics, 22, 1658–9. [DOI] [PubMed] [Google Scholar]
- 24. Ghosh T. S., Gupta S. S., Bhattacharya T., et al. 2014, Gut microbiomes of Indian children of varying nutritional status, PLoS One, 9, e95547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ganju P., Nagpal S., Mohammed M. H., et al. 2016, Microbial community profiling shows dysbiosis in the lesional skin of Vitiligo subjects, Sci. Rep., 6, 18761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sunagawa S., Coelho L. P., Chaffron S., et al. 2015, Ocean plankton. Structure and function of the global ocean microbiome, Science, 348, 1261359. [DOI] [PubMed] [Google Scholar]
- 27. Navarrete A. A., Tsai S. M., Mendes L. W., et al. 2015, Soil microbiome responses to the short-term effects of Amazonian deforestation, Mol. Ecol., 24, 2433–48. [DOI] [PubMed] [Google Scholar]
- 28. Cole J. R., Chai B., Farris R. J., et al. 2005, The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis, Nucleic Acids Res., 33, D294–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schloss P. D., Westcott S. L., Ryabin T., et al. 2009, Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 75, 7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Konstantinidis K. T., Tiedje J. M.. 2005, Genomic insights that advance the species definition for prokaryotes, Proc. Natl. Acad. Sci. U.S.A., 102, 2567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hao X., Jiang R., Chen T.. 2011, Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering, Bioinformatics, 27, 611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lozupone C., Lladser M. E., Knights D., Stombaugh J., Knight R.. 2011, UniFrac: an effective distance metric for microbial community comparison, Isme J., 5, 169–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Felsenstein J. 1981, Evolutionary trees from DNA sequences: a maximum likelihood approach, J. Mol. Evol., 17, 368–76. [DOI] [PubMed] [Google Scholar]
- 34. McMurdie P. J., Holmes S.. 2013, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, PLoS One, 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nelson M. C., Morrison H. G., Benjamino J., Grim S. L., Graf J.. 2014, Analysis, optimization and verification of illumina-generated 16S rRNA gene amplicon surveys, PLoS One, 9, e94249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.