Abstract
Grain softness has been a major trait of interest in wheat because of its role in producing flour suitable for making high-quality biscuits, cookies, cakes and some other products. In the present study, marker-assisted backcross breeding scheme was deployed to develop advanced wheat lines with soft grains. The Australian soft-grained variety Barham was used as the donor parent to transfer the puroindoline grain softness gene Pina-D1a to the Indian variety, DBW14, which is hard grained and has PinaD1bPinbD1a genes. Foreground selection with allele-specific PCR-based primer for Pina-D1a (positive selection) was used to identify heterozygous BC1F1 plants. Background selection with 173 polymorphic SSR primers covering all the 21 chromosomes was also carried out, in the foreground-selected BC1F1 plants. BC1F2 plants were selected by ascertaining the presence of Pina-D1a (positive selection) and absence of Pina-D1b (negative selection). Using the approach of positive, negative and background selection with molecular markers, 15 BC1F2 and 31 BC2F1 plants were finally selected. The 15 BC1F2 plants were selfed and the 31 BC2F1 plants were further backcrossed and selfed to raise BC3F1 and BC2F2 progenies, respectively. A part of the BC2F2 seed of each of the 31 plants was analyzed for grain hardness index (GHI) with single-kernel characterization system. The GHI varied from 12.1 to 37.1 in the seeds borne on the 31 BC2F1 plants. The reasons for this variation and further course of action are discussed.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1717-5) contains supplementary material, which is available to authorized users.
Keywords: Grain softness, Marker-assisted backcross breeding, Puroindolines, Wheat grain quality
Introduction
Wheat is an important source of major nutrients required by the human body. The wheat kernel contains roughly 12% water, 70% carbohydrates, 12% protein, 2% fat, 1.8% minerals and 2.2% crude fibers (The Editors of Encyclopaedia Britannica 2018). Wheat is the second important food crop next to rice in India and contributes to nearly 25% of food grain production of the country (FAO in India 2018). Nearly 80% of wheat is consumed by Indian people in the form of ‘chapatti’: an unleavened flat bread (Misra 1998). Indian wheat is not segregated according to the end-product requirement though some amount of segregation does take place on the basis of grain color, hardness and luster which is sold at a higher price. The wheat breeding programs in India have been mainly focused on yield and disease resistance to develop verities to feed the ever-growing population. In the recent years, however, the milling and bakery industry in India has grown enormously (Indian Bakery Market 2018) and instant and ready-to-eat wheat-based foods have become popular. Several shopping malls, airports and similar centers have come up which has led to the establishment of bakeries serving ready-to-eat food. There is an increased production of breads, biscuits, cakes, buns, noodles, etc., in the country. India is now the world’s third largest country producing biscuits after USA and China (Kachave 2018). However, getting the desired quality of wheat flour for making specific kinds of bakery products is a challenging task for bakery industries. This results in use of external agents including potentially harmful chemicals for improving the texture and quality of end-products. The inconsistency in different wheat or flour lots also poses a challenge to get a consistent quality for baking. The need for development of product-specific varieties in the country has therefore been felt. The dearth of breeders, in the country, engaged in wheat quality enhancement makes this task difficult to achieve.
The transfer of quality traits during the development of elite genotypes or to the already released cultivars is time-consuming and inefficient by the conventional strategy (Hospital 2005). Moreover, most quality tests can be conducted only in the advanced generations, thereby increasing the time required to select the appropriate lines. Rapid advances in genomic research and molecular biology have led to the development of precise, rapid and efficient molecular markers for speedy development of new cultivars (Randhawa et al. 2009). Linked and gene-based markers have also become available for several quality traits in wheat (Ma et al. 2003; Wang et al. 2009, 2010). Therefore, combining marker-assisted selection with conventional phenotypic selection can enhance the efficiency of breeding and precise transfer of the target allele into the advanced progenies and sometimes in relatively shorter time (Hospital and Charcosset 1997).
A flour derived from soft grain endosperm with < 11% flour protein and producing weak and extensible gluten is considered ideal for biscuit quality and the alveo and farinographic indices, and the alleles of high molecular weight glutenin subunits (HMW-GS) and low molecular weight glutenin subunits (LMW-GS) have been suggested to achieve these criteria (Payne 1987; Rasheed et al. 2014). In our earlier study (Rai et al. 2018 communicated), 30 wheat varieties were screened for quality parameters and allele-specific PCR markers of PinaD1a, PinaD1b, PinbD1a, PinbD1b for grain texture and Glu-1 and Glu-3 genes for gluten strength and extensibility to identify candidate Indian wheat varieties for improving grain endosperm texture. Varieties HS490, HI1563 and DBW14 were found to possess desirable values of some of the related traits such as HMW-GS, LMW-GS, SDS-sedimentation value and farinograph quality number (FQN), and their values approached those in the Australian soft endosperm wheat lines Barham and Longreach Orion. Additionally, the variety DBW14 possessed HMW-GS (GluA1a, GluB1u, GluD1d) and LMW-GS (GluA3c, GluB3b, GluD3c) which were same as those of the Australian soft endosperm wheat varieties Barham and Longreach Orion and was therefore selected as the candidate variety to impart to it grain softness trait as it is a hard-grained variety. In this paper, we describe the process of marker-assisted backcross breeding (MABB) for grain softness trait with foreground and background markers in a DBW14/Barham cross.
Materials and methods
Plant materials
DBW14, an Indian awned wheat variety, was used as a recurrent parent (referred to as RP), and an Australian awnless variety, Barham, obtained in a collaborative program on marker-assisted breeding in wheat (Project no. 100157) was used as a donor parent (referred to as DP). DBW14 (RAJ 3765/PBW 343) is a semi-dwarf wheat variety released in the year 2002 for late sown irrigated conditions of North Eastern Plains Zone of India. These parents have been characterized in an earlier study by us (Rai et al. 2018, communicated). DBW 14 recorded a protein content of 10.19% in the crop season 2014–2015. It is a hard-grained variety and recorded a hardness index (HI) of 72.29 on single-kernel characterization system and has the allelic composition PinaD1b and PinbD1a. The donor parent Barham (Syn VO2697R) with the pedigree Bowie//Bersee/3*Bindawarrala 12697///Bowie is a wheat variety having moderate resistance to leaf and stem rust and possesses alien chromosome segment having Lr37/Yr17/SR38 genes. It is soft grained with HI of 26 on single-kernel characterization system (SKCS) and possesses the wild-type alleles (a) of the genes PinaD1 and PinbD1. Both DBW14 and Barham have similar alleles of HMW-GS and LMW-GS coding genes, and therefore no change in HMW or LMW alleles of DBW 14 would occur.
Grain characteristics
The parameters grain hardness index (GHI), thousand kernel weight (TKW) and diameter (DIA) were analyzed with single-kernel characterization system (SKCS) 4100 (Perten Instruments, Australia) using the procedure described in AACC (2000) method 55-31. Grain hardness was determined in each genotype on an approximately 300-kernel sample size. To determine TKW, 250 grains were taken at random, representing the whole sample, counted and weighed. The value so obtained was multiplied by four to get the 1000 kernel weight in ‘grams.’
Genotyping
Markers for the alleles of the grain texture genes Pina-D1a (Gautier et al. 1994), Pina-D1b (or null) (Feng et al. 2013) and PinbD1a (Giroux and Morris 1997) were validated in our earlier work (Rai et al. 2015). Two of the markers were used for the foreground selection (FG). The marker PinA (Gautier et al. 1994) was used for identifying the presence of grain softness gene Pina-D1a. Additionally, negative selection with a marker Pina-N1 (Feng et al. 2013) for Pina-D1b was carried out only in BC1F2 to identify homozygous individuals for PinaD1a. Primers were got synthesized by Sigma Pvt Ltd, India. PinA primer amplifies a ~ 450 bp fragment indicating the presence of Pina-D1a allele while Pina-N1 primer amplifies a 776 bp fragment indicating the presence of Pina-D1b allele. Supplementary Table 1 lists the primers, their respective sequence and fragment size obtained. A total of 1110 simple sequence repeat (SSR) markers covering all the 21 linkage groups of wheat were screened in the parents to find polymorphic SSR markers for use in background selection (BG) for the recovery of recurrent parent genome (RPG) in the segregating backcross progenies. The SSR primer sequences were obtained from http://wheat.pw.usda.gov/GG2/index.shtml (Grain Genes 2.0: a database for Triticeae and Avena) (Röder et al. 1998; Somers et al. 2004).
Marker-assisted backcross breeding
DNA isolation, PCR conditions and electrophoresis
DNA extraction of parental genotypes and their backcross or selfed progenies planted in the field was carried out from the leaves of 23–27-day-old plants using cetyltrimethylammonium bromide (CTAB) procedure (Doyle and Doyle 1987). PCR for PinA and Pina-N1 was performed using 0.3 µl Taq DNA polymerase in 15 µl reaction volumes containing approximately 50 ng of genomic DNA, 1 × PCR buffer with 1.5 MgCl2, 10 pmol of each PCR primer and 200 µM of each of dNTPs. PCR cycling conditions were 94 °C for 5 min following 35 cycles of 94 °C for 35 s, 60–65 °C for 35 s, 72 °C for 90 s and a final extension at 72 °C for 8 min. The PCR products were separated by electrophoresis in 1.5% agarose for foreground selection. For the background selection, the PCR products were run on 3% metaphor and visualized by ethidium bromide staining.
Breeding scheme
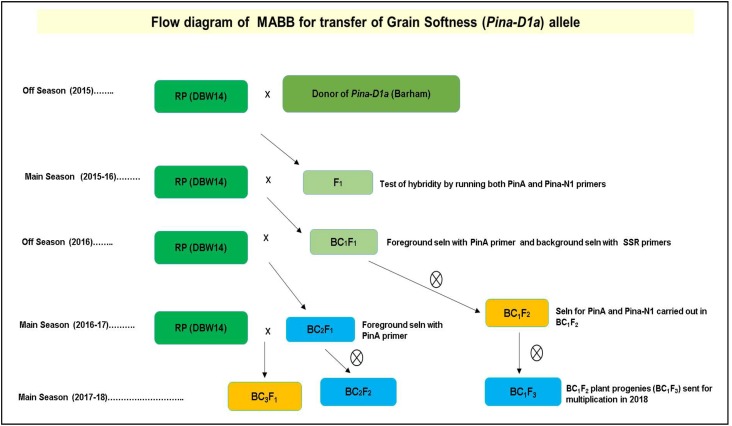
MABB scheme was followed to transfer Pina-D1a gene from Barham into the genetic background of DBW14 and is depicted in Fig. 1. Recurrent parent (RP) DBW14 with PinaD1b allele was used for crossing with Barham as donor of Pina-D1a allele. Although DBW14 was used as a female in every backcrosses made so as to get the cytoplasmic content of the recurrent parent to attain maximum genome recovery, in producing F1’s, DBW14 was used as pollen parent and Barham as female. Both foreground and background selections were carried out with rigorous molecular markers-based selection. The BC1F3 and BC2F2 seeds were generated till the submission of this article.
Fig. 1.
Flowchart of marker-assisted backcross breeding for transfer of Pina-D1a allele
Determination of recurrent parent genome recovery
The extent of RPG recovery in backcross generation was calculated using the following formula:
where R is the number of marker loci homozygous for recurrent parent allele; H is the number of marker loci remaining heterozygous and N is the total number of polymorphic markers used for background analysis (Miah et al. 2014). To represent RPG in the chromosome of the backcross progenies, Graphical Geno Type (GGT) software version 2 (Berloo 1999) was used.
Results
Marker-assisted backcross breeding for PinaD1a (foreground selection)
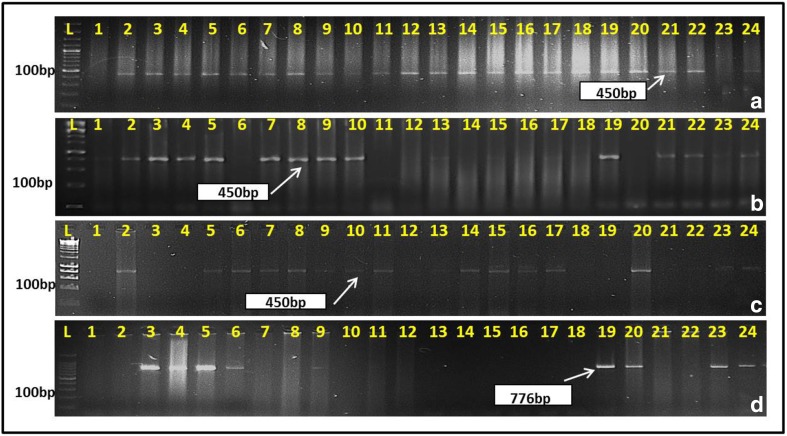
The cross was made between DBW14 which was used as pollen parent and Barham as female. Barham was used as female parent so that the selfed (PinaD1aPinaD1a) and the crossed seeds (PinaD1aPinaD1b) could be distinguished both morphologically and with markers for PinaD1a and PinaD1b. PINA primer was run in 105 F1 plants, of which 99 plants showed (Pina-D1a/Pina-D1b) allele while six plants showed (PinaD1a/PinaD1a) allele. These six plants were homozygous for PINA locus showing that these were selfed seeds of Barham and were soft in appearance and therefore were discarded and not taken forward. The F1 plants of the cross DBW14/Barham were screened with PinA (Fig. 2a). Further, DBW14 was used as a female in every backcrosses made so as to get the cytoplasmic content of the recurrent parent to attain maximum genome recovery. The 99 plants found positive for the allele (Pina-D1a) were backcrossed with RP, DBW14 to produce ~ 2000 BC1F1 seeds (Table 1). Out of these, 400 BC1F1 seeds were screened for foreground selection with PinA (Fig. 2b). Two hundred fifteen plants were positive for PinA. The 215 BC1F1 plants were strictly screened morphologically, and 112 plants were found to show close similarity with the recurrent DBW14. One hundred seventy-three polymorphic SSRs were run on these 112 BC1F1 plants, and only 19 BC1F1 plants were found to have high genome recovery of 80% and above. Now these 19 BC1F1 plants were bifurcated into two groups. In the first group, the 19 BC1F1 plants (4–5 spikes per plant) were backcrossed and in the second group same 19 BC1F1 plants (2 spikes per plant) were selfed to produce BC2F1 and BC1F2 generation, respectively. These 19 BC1F1 backcrossed plants produced 303 BC2F1 seeds. In the main season 2016–2017, 303 BC2F1 seeds were grown. All the plants were screened with PinA. One hundred seventeen plants were found to have Pina-D1a (Fig. 2c). These 117 BC2F1 were backcrossed to produce BC3F1 seeds, and simultaneously 2–3 spikes per plant were selfed to get BC2F2 seeds. Out of these 117 BC2F1 selfed plants, 31 BC2F1 selfed plants with high RPG % were harvested and its grain analysis was conducted. The seeds of BC2F2 borne on each of the plants were examined, each had both soft and hard grains, and therefore, the numbers were counted and are presented in Table 2. The 19 high RPG % BC1F2 plants seeds (selfed 19 BC1F1 plants) were also grown in the main season (2016–2017), and MAS was exercised with PinA and Pina-N1 markers in the 98 plants. DNA of 23 plants amplified PinA and did not amplify Pina-N1 marker, thus indicating that these were putative homozygotes for Pina-D1a (Fig. 2d). Further, these 23 plants were strictly screened for phenotypic similarity to the recurrent parent selected visually in the field and so 15 plants showing phenotypically similarity with recurrent parent were further taken forward. Rest eight plants showed less tillers, were shorter in height and therefore were discarded. The BC1F3 seeds from these 15 BC1F2 plants have been sown in off-season 2018 along with seeds from soft class of BC2F2.
Fig. 2.
PCR analysis for ascertaining the presence of target allele(s) (cropped gels displayed). a DNA amplification of Pina-D1a allele using the primer PinA in F1 population, b DNA amplification of Pina-D1a allele using the primer PinA in BC1F1, c DNA amplification of Pina-D1a allele using the primer PinA in BC2F1 population, d DNA amplification of Pina-N1 in BC1F2 population. L ladder 1–24 wells represent the plants of the respective generations
Table 1.
Generation-wise positive plants for puroindoline genes (Pina-D1a, Pina-D1b)
| Generation | Total plants analyzed with marker | No. of plants with amplification of markers | |||||
|---|---|---|---|---|---|---|---|
| PinA | Pina-N1 | PinA + Pina-N1 (Pina-D1a/Pina-D1b) | PinA alone; no amplicon of Pina-N1 seen (PinaD1a/PinaD1a) | No. of plants on which background selection was exercised | No. of plants taken forward | ||
| F1 | 105 | 105 | 99 | 99 | 6 | – | 99 |
| BC1F1 | 400 | 215 | nc | nc | – | 112 | 19 |
| BC1F2 | 98 | 52 | 75 | 29 | 23 | nc | 15 |
| BC2F1 | 303 | 117 | nc | nc | – | – | 117 |
nc not checked, – not expected, () indicates putative genotype
Table 2.
Grain hardness observations in BC2F2 grains of the cross DBW14*3/Barham
| Line number | No. of soft grains | No. of hard grains | Mottled | Hard + mottled | Total no. grains | GHI of soft class | GHI (SD) | TKW (g) | TKW (SD) | MOI (%) | MOI (SD) | DIA (mm) | DIA (SD) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16-92-7 | 317 | 275 | 33 | 308 | 625 | 28.8 | 8.1 | 42.1 | 7.3 | 11.4 | 0.1 | 2.8 | 0.2 |
| 16-86-6 | 328 | 182 | 24 | 206 | 534 | 25.7 | 10.9 | 33.7 | 4.6 | 11.6 | 0.2 | 2.7 | 0.3 |
| 16-75-2 | 341 | 364 | 60 | 424 | 765 | 32.3 | 9.0 | 39.9 | 4.5 | 11.3 | 0.1 | 2.7 | 0.2 |
| 16-126-6 | 207 | 110 | 68 | 178 | 385 | 35.5 | 14.8 | 36.4 | 8.3 | 11.3 | 0.2 | 2.8 | 0.4 |
| 16-128-5 | 374 | 291 | 0 | 291 | 665 | 20.7 | 10.1 | 39.6 | 7.8 | 11.3 | 0.1 | 2.8 | 0.4 |
| 16-86-7 | 100 | 165 | 52 | 217 | 317 | 22.0 | 17.0 | 35.7 | 5.8 | 10.9 | 0.2 | 2.7 | 0.3 |
| 16-148-1 | 242 | 353 | 5 | 358 | 600 | 28.8 | 11.2 | 28.6 | 5.0 | 11.4 | 0.3 | 2.5 | 0.2 |
| 16-70-4 | 125 | 125 | 25 | 150 | 275 | 12.1 | 9.3 | 30.9 | 5.7 | 11.6 | 0.6 | 2.5 | 0.4 |
| 16-125-8 | 520 | 180 | 0 | 180 | 700 | 18.2 | 10.2 | 40.4 | 6.8 | 11.4 | 0.2 | 2.9 | 0.2 |
| 16-125-4 | 224 | 189 | 1 | 190 | 414 | 34.1 | 9.2 | 40.7 | 4.7 | 11.7 | 0.1 | 3.0 | 0.2 |
| 16-125-10 | 137 | 181 | 7 | 188 | 325 | 35.0 | 8.6 | 29.7 | 6.1 | 11.5 | 0.1 | 2.6 | 0.2 |
| 16-69-4 | 108 | 250 | 92 | 342 | 450 | 31.8 | 7.9 | 37.1 | 5.5 | 11.2 | 0.2 | 2.8 | 0.2 |
| 16-142-6 | 186 | 250 | 14 | 264 | 450 | 25.8 | 12.2 | 39.5 | 8.2 | 11.2 | 0.2 | 2.8 | 0.3 |
| 16-149-10 | 72 | 129 | 96 | 225 | 297 | 25.1 | 10.1 | 38.8 | 5.0 | 11.1 | 0.2 | 3.0 | 0.3 |
| 16-138-4 | 310 | 343 | 77 | 420 | 730 | 32.8 | 12.2 | 34.9 | 5.7 | 11.5 | 0.2 | 2.8 | 0.3 |
| 16-113-1 | 850 | 300 | 0 | 300 | 1150 | 23.8 | 10.8 | 45.4 | 7.7 | 11.3 | 0.2 | 3.0 | 0.3 |
| 16-113-3 | 483 | 18 | 92 | 110 | 593 | 23.4 | 15.1 | 34.4 | 8.6 | 11.4 | 0.2 | 2.7 | 0.3 |
| 16-149-9 | 388 | 177 | 80 | 257 | 645 | 19.4 | 11.0 | 44.7 | 4.4 | 11.1 | 0.2 | 3.0 | 0.2 |
| 16-79-10 | 460 | 230 | 15 | 245 | 705 | 28.4 | 12.8 | 39.3 | 4.1 | 11.4 | 0.1 | 3.0 | 0.4 |
| 16-126-11 | 120 | 460 | 80 | 540 | 660 | 37.1 | 6.2 | 30.4 | 4.7 | 11.1 | 0.2 | 2.7 | 0.3 |
| 16-154-1 | 279 | 450 | 21 | 471 | 750 | 33.6 | 15.2 | 43.6 | 6.9 | 11.1 | 0.2 | 2.9 | 0.2 |
| 16-83-5 | 55 | 57 | 30 | 87 | 142 | 32.3 | 14.1 | 22.3 | 5.9 | 11.5 | 0.3 | 2.4 | 0.1 |
| 16-70-9 | 38 | 110 | 17 | 127 | 165 | 30.4 | 7.9 | 26.8 | 7.6 | 11.6 | 0.3 | 2.5 | 0.3 |
| 16-154-5 | 29 | 67 | 34 | 101 | 130 | 20.6 | 11.2 | 43.3 | 5.9 | 11.0 | 0.2 | 2.9 | 0.2 |
| 16-142-10 | 101 | 200 | 22 | 222 | 323 | 25.0 | 11.0 | 34.1 | 3.4 | 10.9 | 0.2 | 2.7 | 0.2 |
| 16-130-10 | 195 | 475 | 30 | 505 | 700 | 21.7 | 7.5 | 32.6 | 5.7 | 11.7 | 0.2 | 2.7 | 0.3 |
| 16-135-3 | 290 | 415 | 30 | 445 | 735 | 20.8 | 17.3 | 43.4 | 8.8 | 11.6 | 0.2 | 2.9 | 0.3 |
| 16-130-4 | 54 | 256 | 0 | 256 | 310 | 25.9 | 7.6 | 38.9 | 8.1 | 11.1 | 0.2 | 2.8 | 0.2 |
| 16-170-2 | 148 | 125 | 32 | 157 | 305 | 22.2 | 16.5 | 45.0 | 8.2 | 10.8 | 0.2 | 3.1 | 0.3 |
| 16-113-2 | 190 | 210 | 30 | 240 | 430 | 21.2 | 10.3 | 44.6 | 3.9 | 11.2 | 0.2 | 3.0 | 0.3 |
| 16-154-2 | 132 | 165 | 12 | 177 | 309 | 24.9 | 12.4 | 43.0 | 8.7 | 11.2 | 0.2 | 2.8 | 0.4 |
| DP | 26 | 21.3 | 26.3 | 8.3 | 14.3 | 0.5 | 2.3 | 0.2 | |||||
| RP | 72.3 | 14.8 | 36.2 | 9.1 | 11.2 | 0.3 | 2.7 | 0.4 |
HI hardness index, TKW thousand kernel weight, MOI moisture %, DIA diameter
Background selection
The parental polymorphism was carried out with 1110 SSR primers. Out of 1110 SSR primers used, only 173 SSRs were found to be polymorphic between DBW14 and Barham. The chromosome-wise distribution of the polymorphic markers is shown in Supplementary Table 3 and Supplementary Fig. S2. The individual SSR score for the respective polymorphic primers is given in Supplementary Table 4. These 173 primers were used for background selection. Some of them are depicted in Supplementary Fig. S1. The polymorphic markers were run on the BC1F1 plants, and RPG recovery was calculated. Nineteen BC1F1 plants with desirable foreground alleles and with RPG between 80 and 86.68% (Supplementary Table 2) were further backcrossed, and two spikes of each of these were selfed in off-season to obtain BC2F2 and BC1F2, respectively.
Recurrent parent genome recovery
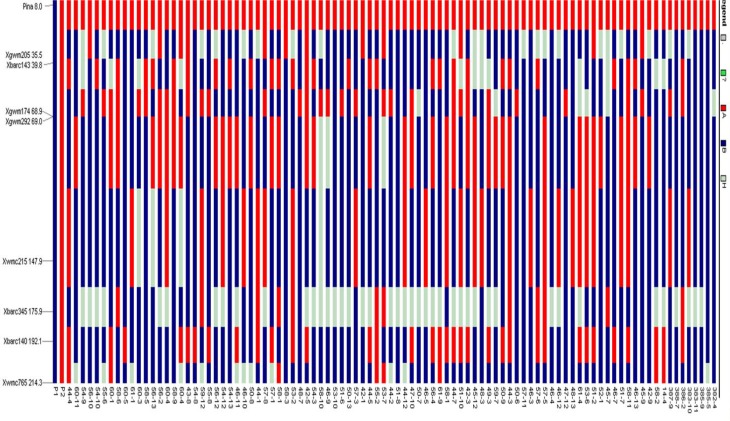
One hundred seventy-three markers were used to analyze RPG recovery in the 112 BC1F1 plants carrying PinaD1a allele (see Supplementary Table 4). Chromosome 5D (carrying the Pina-D1 gene locus) was found to have eight polymorphic SSRs between parents, viz., Xgwm205, Xbarc143, Xgwm174, Xgwm292, Xwmc215, Xbarc345, Xbarc140 and Xwmc765. Fourteen genotypes represented DBW14 allele of each of these primers. Similar exercise was conducted for other 20 other chromosomes. The % recovery based on these 173 SSRs ranged from 32.6 to 86.6%. The graphical genotyping (GGT) for chromosome 1A to 7D was carried out for the BC1F1 plants carrying Pina-D1a allele. The GGT for chromosome no. 5D is shown in Fig. 3. In Fig. 3, the red color bar segments represent the segments from donor parent Barham, while the blue color bar segment represents the genome segments from recurrent parent DBW14. More is the blue color, the more is the respective plant number close to its recurrent parent genome.
Fig. 3.
Graphical genotyping of BC1F1 plants for chromosome number 5D (P1 = DBW14, P2 = Barham, red segments from donor, blue segments from recurrent parent, gray segments showing heterozygosity and green segments are those not amplified by SSR)
Analysis of BC2F2 seeds for grain texture
The grain hardness data of the BC2F2 seeds are presented in Table 2. The grains from 31 plants and the donor parent and the recurrent parent were screened. Primary screening was based on the color and texture of the grain by the manual method. Grains were classified into hard and soft grain classes (Fig. 4). Further 30 grains from the putative soft category per segregating line were subjected to the SKCS (Table 2). The GHI in these grains ranged from 12.1 to 37.1. Line number 16-70-4 was observed to have the lowest GHI of 12.1, while line number 16-126-11 has the highest GHI of 37.1. TKW of soft grains ranged from 24.7 to 44.7 g and DIA from 2.8 to 3 mm.
Fig. 4.

Visual examination of the seeds of donor parent Barham, recurrent parent DBW14 and of their BC2F2 grains classified as hard, soft and mottled
Discussion
Marker-assisted selection (MAS) for transferring genes through plant breeding has been claimed to be cost- and time-effective. Foreground selection for a trait using molecular marker facilitates identification of positive plants for gene of interest at early plant stage, and thus, 50% population size is reduced in the breeding program enabling the breeders to decrease their efforts in handling of large populations (Collard and Mackill 2008). Molecular markers have also been acknowledged as a very powerful tool because of their reproducibility and precision (Gebremeskel et al. 2018; Gupta et al. 1999; Nadeem et al. 2018). One of the popular and successful schemes is the marker-assisted backcross breeding which is used to improve many available good varieties which need one or a few simple inherited traits such as rust resistance or a quality trait to be added to them (Kolchanov et al. 2017). Molecular markers for quality traits in wheat have been slow to develop but in the last few years, their numbers have increased rapidly (Xu et al. 2013). Molecular maker-assisted breeding for agronomic and quality traits is now being deployed in many countries’ national programs and private companies engaged in agriculture (Voosen 2009). De Bustos et al. (2001) carried out marker-assisted selection to improve HMW-GS in wheat. Similar, efforts were made by Vishwakarma et al. (2014) to introgress a high-quality protein Gpc-B1 in elite Indian variety. Reports on MAS for quality traits in wheat in India are relatively few (Gupta et al. 2008; Kumar et al. 2011; Tyagi et al. 2014). The knowledge on genes to improve end-use quality is diverse but its utilization in wheat breeding programs is way too less (Mondal et al. 2016).
In the present work, we have carried out MAS for transfer of grain softness allele to a hard-grained variety. In an earlier work (Rai et al. 2018), we have short-listed, based on an exhaustive quality and molecular analysis of genes for Puroindolines, HMW-GS and LMW-GS, three Indian wheat varieties out of the 30 analyzed, that could be improved for their texture and/or the gluten characteristics. One of them, DBW14 is grown in the region of northeastern plains zone (NEPZ). It is a hard-grained, awned variety. In the present work, MABB has been carried out in this variety to replace its Pina-D1b allele by Pina-D1a allele. Since it has Pinb-D1a allele, the resulting near-isogenic line (NIL) (PinaD1aPinbD1a) is expected to be a soft-grained genotype. The donor awnless variety Barham has the alleles PinaD1aPinbD1a. PinaD1a (amplified by PinA primer), Pina-D1b (amplified by Pina-N1 primer) were used for screening genotypes in F1, BC1F1, BC1F2 and BC2F1 plants (shown in Fig. 2a–d). The BC1F1 and BC2F1 plants were screened with PinA (Fig. 2b, c) while the F1 and BC1F2 plants were screened with Pina-N1 (for Pina-D1b) allele (Fig. 2d) in addition to the former marker. Along with the FG alleles, BG selection process was also practiced to transfer targeted genes to get NILs of DBW14. It was observed that even in individuals with high RPG recovery, the phenotypic similarity to the recurrent parent was less than optimum in several genotypes. Therefore, in BC1F1 and BC1F2 plants many of the genotypes that had positive target alleles by foreground selection were awnless and therefore, had to be rejected (Table 2). Similarly, many background-selected genotypes were lower tillering and much taller in height as compared to the RP and were therefore rejected despite having desired target alleles. Phenotypic selection could not be entirely substituted by background selection. A reason for this could be that only 173 polymorphic markers between parents were used for BG selection in BC1F1 generation. Given the size of wheat genome, these appear to be of limited value in selection of plants with a phenotype very similar to RP. Thus, combining phenotypic and genotypic selection in an MABB procedure is recommended. Similar experiment was designed by Zhou et al. (2003) which also conducted MAS combined with phenotypic screening in wheat. Another recent study by Kumar et al. (2018) demonstrated the synergistic effects of phenotyping along with genotyping done in the early generations on selection of better progenies. Plants with RPG ranging between 80 and 86.6% (Supplementary Table 2) were backcrossed or selfed further. The BC1F3 and BC2F2 seeds have been produced by shuttling the breeding materials between the main and the off-seasons per year for 2 years as shown in Fig. 1.
The seeds borne on BC2F1 (i.e., the BC2F2) were analyzed for their appearance and HI. Soft-looking, hard-looking and several mottled types of grains were observed. Selfing of a BC2F1 plant (PinaD1aPinbD1a/PinaD1bPinbD1a) should produce three types of zygotes: PinaD1aPinbD1a/PinaD1aPinbD1a, PinaD1aPinbD1a/PinaD1bPinbD1a and PinaD1bPinbD1a/PinaD1bPinbD1a in 1:2:1 ratio. Grain softness has been reported to be dominant over the hardness trait (Doekes and Belderok 1976; Guzmán et al. 2012); however, a mutation in any of the genes PinaD1 or PinbD1 produces a hard grain (Giroux and Morris 1997); therefore, the two classes in this case, viz., soft and hard grains in the ratio of 1:3, should be observed. However, it was found that the number of soft and hard grains did not fit into a 1:3 distribution; rather, no ratio fitted the soft and hard grains produced on different plants. When two laboratory personnel were asked one by one to classify these grains, they classified the grain differently as several grains were difficult to classify based on their appearance. There were also cases where the grains were mottled and appeared to have a yellow berry kind of appearance (Fig. 4). Yellow berry is a phenomenon commonly observed in durum grains and is said to be due to non-uniform deposition of protein in the grain (Headden 1915). The grains with such appearance were all hard as judged by crushing under teeth. Thus, on visual basis, it was found difficult to classify the seeds borne on a heterozygote.
Puroindoline proteins coded by the genes PinaD1a and PinbD1a have been shown to be exclusively produced in the starch cells of the endosperm (Wiley et al. 2007). It is stated that the Pin genes have tissue-specific promoters and are expressed in the endosperm cells during development of the grain. The endosperm is formed by double fertilization (primary endosperm nucleus fuses with one male gamete to give secondary nucleus while the other male gamete fuses with the egg cell) occurring in the megagametophyte. Therefore, each of the endosperm cells is 3n and has three alleles of each gene, two contributed by the female and one by the male gametophyte. Thus, the composition of endosperm upon the selfing of a plant heterozygous for PinaD1 gene, i.e., PinaD1aPinaD1b could be PinaD1aPinaD1aPinaD1a (referred to as type 1) or PinaD1aPinaD1aPinaD1b (referred to as type 2) or PinaD1aPinaD1bPinaD1b (referred to as type 3) or PinaD1bPinaD1bPinaD1b (referred to as type 4 here) depending upon the male and female gametes’ constitution. In an endosperm type 1, three doses of allele ‘a’ are present making threefold the protein (PINA) as compared to the other classes where twofold, onefold or no copies of protein may be present, respectively. Since PinaD1a-coded protein (PINA) physically binds along with the PinbD1a protein (PINB) to the lipids present on the starch cells of the endosperm (Kim et al. 2012; Pauly et al. 2013) giving rise to a soft endosperm texture, variation in the amount of PINA may influence the level of softness and thus, a reduction in softness of the level PinaD1aPinaD1aPinaD1a > PinaD1aPinaD1aPinaD1b > PinaD1aPinaD1bPinaD1b > PinaD1bPinaD1bPinaD1b is expected to appear. This may explain the reason for appearance of BC2F2 grains with varying degrees of hardness leading to confusion in their classification. There may be other reasons for varying degrees of hardness in the grains of different BC2F1 plants’ harvest. The fact that GH score of soft seeds borne on each of the 31 BC2F1 plants varied from 12.1 to 37.1 and also the fact that the cross produced soft grains in some BC1F1 with HI much less than that of soft-grained parent Barham imply that additional genes influencing grain texture are present in both the hard- and soft-grained parents.
Thus, in the present work, the allele Pina-D1a was successfully transferred using molecular marker-assisted selection in the hard-grained genotype DBW14. The presence of soft grains and hardness index varying between 12.1 and 37.1 among the BC2F2 grains provide proof of the presence of PinaD1a allele in the BC1F1 plants amplifying the PinA primer.
Conclusion
It has been shown in the above study that it is possible to utilize molecular markers for transfer of genes or their alleles governing major quality traits in wheat. Marker-assisted backcross breeding could be successfully utilized for developing wheat lines where the Pina-D1b allele could be replaced by the Pina-D1a allele using an allele-specific PCR-based primer. After selfing of the heterozygous plants, the homozygotes could be selected by combining the foreground selection with the marker for PinaD1a and negative selection with the marker for PinaD1b. Background selection with molecular markers has resulted in plants in early backcross generations with greater similarity to the recurrent parent. We are now multiplying the selected homozygotes (PinaD1aPinbD1a/PinaD1aPinbD1a) for carrying out yield and quality analysis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
AR acknowledges the senior research fellowship granted to her by the Indian Council of Agricultural Research for carrying out the work as part of her Ph.D. thesis. AMS acknowledges the funding support provided by the ICAR under the Indo-Australian Project on marker-assisted wheat breeding. Interactions with Drs Karen Cane and Howard Eagles during marker protocols exchange were helpful for this work and are acknowledged.
Author contributions
AMS was involved in planning and supervision of experiments, mentoring, financial facilitation and editing of manuscript. AR contributed to execution of experiments, molecular breeding and analysis of data and drafting of the manuscript. KR created F1’s and edited the manuscript. PS and TJK ran molecular markers (SSRs) in BC progenies. AKA contributed to grain hardness analysis. SKS, DG, MS and RBS were involved in field work in main and off-seasons. All the authors have read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
References
- AACC International . Approved methods of American Association of Cereal Chemists. 10. St. Paul: AACC International Press; 2000. pp. 55–131. [Google Scholar]
- Berloo VR. GGT: software for display of graphical genotypes. J Hered. 1999;90:328–329. doi: 10.1093/jhered/90.2.328. [DOI] [Google Scholar]
- Collard BCY, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bustos A, Rubio P, Soler C, Garcia P, Jouve N (2001) Marker assisted selection to improve HMW-glutenins in wheat. In: Wheat in a global environment, pp 171–176
- Doekes GJ, Belderok B. Kernel hardness and baking quality of wheat—a genetic analysis using chromosome substitution lines. Euphytica. 1976;25:565. doi: 10.1007/BF00041594. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- FAO in India (2018) India at a glance. http://www.fao.org/india/fao-in-india/india-at-a-glance/en/. Accessed 15 Dec 2018
- Feng C, Huanhuan L, Dangqun C. Discovery, distribution and diversity of Puroindoline-D1 genes in bread wheat from five countries (Triticum aestivum L.) BMC Plant Biol. 2013;13:125. doi: 10.1186/1471-2229-13-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier MF, Aleman ME, Guirao A, Marion D, Joudrier P. Triticum aestivum puroindolines, two basic cystine-rich seed proteins: cDNA sequence analysis and developmental gene expression. Plant Mol Biol. 1994;25:43–57. doi: 10.1007/BF00024197. [DOI] [PubMed] [Google Scholar]
- Gebremeskel S, Ana Luisa GO, Abebe M, Victor A, Melaku G. Effectiveness of predictive markers for marker assisted selection of pro-vitamin A carotenoids in medium-late maturing maize (Zea mays L.) inbred lines. J Cereal Sci. 2018;79:27–34. doi: 10.1016/j.jcs.2017.09.001. [DOI] [Google Scholar]
- Giroux MJ, Morris CF. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor Appl Genet. 1997;95:857–864. doi: 10.1007/s001220050636. [DOI] [Google Scholar]
- Gupta PK, Varshney RK, Sharma PC, Ramesh B. Molecular markers and their applications in wheat breeding. Plant Breed. 1999;118:369–390. doi: 10.1046/j.1439-0523.1999.00401.x. [DOI] [Google Scholar]
- Gupta PK, Balyan HS, Kumar J, Kulwal PK, Kumar N, Mir RR, Kumar A, Prabhu KV. QTL analysis and marker assisted selection for improvement in grain protein content and pre-harvest sprouting tolerance in bread wheat. Brisbane: 11th wheat genetics symposium (IWGS); 2008. pp. 1–3. [Google Scholar]
- Guzmán C, Caballero LM, Angela A, Juan B. Molecular characterization and diversity of the Pina and Pinb genes in cultivated and wild diploid wheat. Mol Breed. 2012;30:69–78. doi: 10.1007/s11032-011-9599-1. [DOI] [Google Scholar]
- Headden WP. Yellow-berry in wheat. Colo Agr Expt Sta Bui. 1915;205:38. [Google Scholar]
- Hospital F. Selection in backcross programmes. Phil Trans R Soc B. 2005;360:1503–1511. doi: 10.1098/rstb.2005.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Charcosset A. Marker-assisted introgression of quantitative trait loci. Genetics. 1997;147:1469–1485. doi: 10.1093/genetics/147.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indian Bakery Market (2018) Industry trends, share, size, growth, opportunity and forecast 2018–2023. https://www.imarcgroup.com/indian-bakery-market. Accessed 15 Dec 2018
- Kachave K (2018) Indian biscuit market projected to reach $7.25 billion in next four years. Bakery. http://www.fnbnews.com/Bakery-Biscuits/indian-biscuit-market-projected-to-reach-725-billionin-next-four-years-42107. Accessed 12 Dec 2018
- Kim KH, Feiz L, Martin JM, Giroux M. Puroindolines are associated with decreased polar lipid breakdown during wheat seed development. J Cereal Sci. 2012;56:142–146. doi: 10.1016/j.jcs.2012.02.011. [DOI] [Google Scholar]
- Kolchanov NA, Kochetov AV, Salina EA, Pershina LA, Khlestkina EK, Shumny VK. Status and prospects of marker-assisted and genomic plant breeding. Herald Russ Acad Sci. 2017;87:125–131. doi: 10.1134/S1019331617020113. [DOI] [Google Scholar]
- Kumar J, Jaiswal V, Kumar A, Kumar N, Mir RR, Kumar S, Dhariwal R, Tyagi S, Khandelwal M, Prabhu KV, Prasad R, Balyan HS, Gupta PK. Introgression of a major gene for high grain protein content in some Indian bread wheat cultivars. Field Crop Res. 2011;123:226–233. doi: 10.1016/j.fcr.2011.05.013. [DOI] [Google Scholar]
- Kumar A, Sandhu N, Dixit S, Yadav S, Swamy BPM, Shamsudin NAA. Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice. 2018;11:35. doi: 10.1186/s12284-018-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WG, Zhang W, Gale K. Multiplex-PCR typing of high molecular weight glutenin alleles in wheat. Euphytica. 2003;134:51–60. doi: 10.1023/a:1026191918704. [DOI] [Google Scholar]
- Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Latif MA. Recurrent parent genome recovery analysis in a marker-assisted backcrossing program of rice (Oryza sativa L.) C R Biol. 2014;338:83–94. doi: 10.1016/j.crvi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Misra BK. Quality needs for Indian traditional products. In: Nagarajan S, Singh G, Tyagi BS, editors. Wheat research needs beyond 2000AD. New Delhi: Narosa Publishing House; 1998. pp. 313–314. [Google Scholar]
- Mondal S, Jessica ER, Velu G, Singh PK, Crespo-Herrera LA, Guzman C, Bhavani S, Lan C, Xinyao H, Singh RP. Harnessing diversity in wheat to enhance grain yield, climate resilience, disease and insect pest resistance and nutrition through conventional and modern breeding approaches. Front Plant Sci. 2016;7:991. doi: 10.3389/fpls.2016.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H, Chung G, Baloch FS. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2018;32:261–285. doi: 10.1080/13102818.2017.1400401. [DOI] [Google Scholar]
- Pauly A, Pareyt B, Fierens E, Delcour JA. Wheat (Triticum aestivum L. and T. turgidum L. ssp. durum) kernel hardness: I. Current view on the role of puroindolines and polar lipids. Compr Rev Food Sci Food Saf. 2013;12:413–426. doi: 10.1111/1541-4337.12019. [DOI] [PubMed] [Google Scholar]
- Payne PI. Genetics of wheat storage proteins and the effect of allelic variation on bread making quality. Annu Rev Plant Physiol. 1987;8:141–153. doi: 10.1146/annurev.pp.38.060187.001041. [DOI] [Google Scholar]
- Rai A, Sharma P, Kumar S, Chaudhary S, Ahlawat AK, Singh AM. National symposium on germplasm to genes: harnessing biotechnology for food security and health. New Delhi: ICAR-Pusa Campus; 2015. p. 102. [Google Scholar]
- Rai A, Singh AM, Ganjewala D, Kumar RR, Ahlawat AK, Singh SK, Sharma P, Jain N (2018) Rheological evaluations and molecular marker analysis of cultivated bread wheat varieties of India (communicated) [DOI] [PMC free article] [PubMed]
- Randhawa HS, Mutti JS, Kidwell K, Morris CF, Chen X, Gill KS. Rapid and targeted introgression of genes into popular wheat cultivars using marker-assisted background selection. PLoS ONE. 2009;4:e5752. doi: 10.1371/journal.pone.0005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed A, Xianchun X, Yueming Y, Rudi A, Mahmood T, Zhonghu H. Wheat seed storage proteins: advances in molecular genetics, diversity and breeding applications. J Cereal Sci. 2014;60:11–24. doi: 10.1016/j.jcs.2014.01.020. [DOI] [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor Appl Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- The Editors of Encyclopaedia Britannica (2018) Wheat plant. Encyclopædia Britannica, Inc. https://www.britannica.com/plant/wheat
- Tyagi S, Mi RR, Chhuneja P, Ramesh B, Balyan HS, Gupta PK. Marker-assisted pyramiding of eight QTLs/genes for seven different traits in common wheat (Triticum aestivum L.) Mol Breed. 2014;34:167–175. doi: 10.1007/s11032-014-0027-1. [DOI] [Google Scholar]
- Vishwakarma MK, Mishra VK, Gupta PK, Yadav PS, Kumar H, Joshi AK. Introgression of the high grain protein gene Gpc-B1 in an elite wheat variety of Indo-Gangetic Plains through marker assisted backcross breeding. Curr Plant Biol. 2014;1:60–67. doi: 10.1016/j.cpb.2014.09.003. [DOI] [Google Scholar]
- Voosen P (2009) Molecular breeding makes crops hardier and more nutritious markers, knockouts and other technical advances improve breeding without modifying genes. Sci Am
- Wang LH, Zhao XL, He ZH, Ma W, Appels R, Peña RJ, Xia XC. Characterization of low-molecular-weight glutenin subunit Glu-B3 genes and development of STS markers in common wheat (Triticum aestivum L.) Theor Appl Genet. 2009;118:525–539. doi: 10.1007/s00122-008-0918-9. [DOI] [PubMed] [Google Scholar]
- Wang LH, Li GY, Pena RJ, Xia XC, He ZH. Development of STS markers and establishment of multiplex PCR for Glu-A3 alleles in common wheat (Triticum aestivum L.) J Cereal Sci. 2010;51:305–312. doi: 10.1016/j.jcs.2010.01.005. [DOI] [Google Scholar]
- Wiley PR, Tosi P, Evrard A, Lovegrove A, Jones HD, Shewry P. Promotor analysis and immunolocalisation show that puroindoline genes are exclusively expressed in starch endosperm cells of wheat grain. Plant Mol Biol. 2007;64:125–136. doi: 10.1007/s11103-007-9139-x. [DOI] [PubMed] [Google Scholar]
- Xu Y, Xie C, Wan J, He Z, Prasanna BM. Marker-assisted selection in cereals: platforms, strategies and examples. Cereal Genomics. 2013;II:375–411. doi: 10.1007/978-94-007-6401-9_14. [DOI] [Google Scholar]
- Zhou WC, Kolb FL, Bai GH, Domier LL, Boze LK, Smith NJ. Validation of a major QTL for scab resistance with SSR markers and use of marker-assisted selection in wheat. Plant Breed. 2003;122:40–46. doi: 10.1046/j.1439-0523.2003.00802.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.