Abstract
Genetic engineering of probiotics, like bifidobacteria, may improve their microbial cell factory economy. This work designed a novel shuttle plasmid pBPES, which bears exogenous appA and is stable within Bifidobacterium longum JCM 1217. Cloning of three predicted promoters into pBPES proved that all of them drive appA expression in B. longum JCM 1217. Transformation of plasmids pBPES-tu and pBPES-groEL into B. longum JCM1217 resulted in much more phytase secretion suggests Ptu and PgroEL are strong promoters. Further in vitro and in vivo experiments suggested B. longum JCM 1217/pBPES-tu degrades phytate efficiently. In conclusion, the study screened two stronger promoters and constructed a recombinant live probiotic strain for effectively phytase secretion and phytate degradation in gut. The strategy used in the study provided a novel technique for improving the bioaccessibility of phytate and decreasing phosphorus excretion.
Keywords: Bifidobacterium longum JCM 1217, promoter, phytase, phytate, phosphorus
Introduction
Bifidobacteria are ideal hosts for food-grade delivery of useful enzymes. Strong promoters can regulate high-level gene expression thereby improving the efficiency of microbial cell factories. However, most of those finely defined promoters that had been used in other hosts cannot be readily applied in bifidobacteria (Sun et al., 2012). Therefore, researchers turn to isolate promoters from bifidobacteria themselves (Sun et al., 2014b). The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is widely expressed in both prokaryotic and eukaryotic cells. Therefore, its promoter Pgap was cloned and showed strong capacity for driving high-level expression of several proteins (Klijn et al., 2006; Khokhlova et al., 2010; Osswald et al., 2015a,b). Our previous study revealed that Pgap works in three species of Bifidobacterium (Sun, 2014). However, proteomic studies suggest that proteins of elongation factor Tu and groEL are more abundantly expressed than GAPDH in many bacteria, including B. longum NCC2705 and B. bifidum S17 (Liu et al., 2011; Wei et al., 2016). Hence, their promoters may be stronger than Pgap. B. longum JCM 1217 (also known as ATCC 15707) is a typical probiotic strain that protects host from enteropathogenic infection (Escherichia coli O157:H7) through production of acetate (Fukuda et al., 2011). The strain also reduced biofilm formation in pathogenic E. coli, reduced colitis in rats, and increased survival rate of Clostridium difficile infection in mice (Kim et al., 2012; Celiberto et al., 2017; Yun et al., 2017). If this strain was used as a host for foreign gene expression, its economy may improve as their synergistic effects.
Phytate is the main form of phosphorus storage in plant derived foods. However, due to the lack of endogenous phytase enzymes, monogastric animals cannot metabolize phytate (Lei et al., 2013). Therefore, inorganic phosphate is supplemented in diets of these animals to meet their nutritional requirement, while undigested phytate in excreta causes serious pollution. Meanwhile, undigested phytate significantly inhibits the absorption of zinc and iron from grains in both human and animals, leading to mineral deficiencies through the formation of insoluble chelates (Frontela et al., 2008). Further, tumor cells can employ extracellular phytate to improve their proliferation (Windhorst et al., 2013). Currently, there are two techniques for liberating phytate in grains. One is hydrolyzing it through food physicochemical processes, and the other is enzymatic degradation using phytases. Due to activation of the endogenous phytase that presented in plant foods or addition of exogenous phytase, phytate was degraded to various inositolphosphates, containing 1-5 phosphate groups per molecule of inositol. Commercial phytases are mainly produced by E. coli and Aspergillus niger (A. niger). Though these phytases were widely applied for reducing phytate in food and feed, they need purification, which significantly increases cost and causes activity loss during industrial processes (Lei et al., 2013). A few strains of wide-type phytase positive bifidobacteria had been applied in both feed and food for liberating phytate (García-Mantrana et al., 2014, 2015). However, the enzyme activity in these strains is much lower if detectable. Considering the important role of phytase and the increasing interest in its application, as well as available genetic tools in bifidobacteria, it is possible to engineer bifidobacteria for phytase over-expression (Parvaneh et al., 2014; Chen et al., 2016).
To overcome the shortage of genetic tools in B. longum JCM 1217 and support our hypothesis, we composed a vector and cloned three promoters for driving heterologous phytase secretion. One recombinant strain B. longum JCM 1217/pBPES-tu was tested for effective degradation of phytate in vitro and in vivo. The study aims at providing a recombinant probiotic strain that can effectively improve phytate accessibility in food or feed industry.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Type strain of E. coli DH5α obtained from Invitrogen was used for general cloning. The strain has standard genotypes and was grown in Luria Bertani (LB) broth at 37°C under agitation. Dry powder of strain B. longum JCM 1217 was purchased from Guangdong Institute of Microbiology Culture Center (GIMCC, China). It was propagated routinely in Lactobacilli MRS medium (BD DifcoTM, United States) supplemented with 0.5 g/L L-cysteine (Sigma-Aldrich, China) or adapted Reinforced Clostridium Medium (RCM, Qingdao Hopebiol Co., Ltd., China) without addition of 0.5 g/L agar (Solarbio Co., Ltd., China). Growth of bifidobacteria was carried out statically at 37°C in sealed jars under anaerobic condition using the MGC AnaeroPack (MitsubishiTM, Japan). Antibiotics and other chemical regents were purchased from Sigma-Aldrich. Chloramphenicol was added at a final concentration of 25 μg/mL for selection in both E. coli and B. longum. All strains and plasmids used in this study were listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant features or description | Sources |
|---|---|---|
| E. coli DH5a | Type strain | Invitrogen |
| B. longum JCM 1217 | Sequenced wide-type strain | GIMCC |
| B. longum JCM 1217/pBPES | JCM 1217 possessing plasmid pBPES | This study |
| B. longum JCM 1217/pBPES-tu | JCM 1217 possessing plasmid pBPES-tu | This study |
| B. longum JCM 1217/pBPES-groEL | JCM 1217 possessing plasmid pBPES-groEL | This study |
| B. longum JCM 1217/pBPES-gap | JCM 1217 possessing plasmid pBPES-gap | This study |
| pBPES | Escherichia coli-Bifidobacterium shuttle vector, no promoter, bearing signal peptide and appA gene | This study |
| pBPES-tu | pBPES containing 172 bp promoter region of gene elongation factor Tu (BLLJ_0515) | This study |
| pBPES-groEL | pBPES containing 241 bp promoter region of gene groEL (BLLJ_1448) | This study |
| pBPES-gap | pBPES containing 240 bp promoter region of gene GAPDH (BLLJ_1241) | This study |
GIMCC; Guangdong Institute of Microbiology Culture Center, Guangdong Province, China.
Construction of Shuttle Plasmid Backbone
The plasmid backbone of pBPES was constructed as presented (Figure 1). The chloramphenicol resistance gene Cmr is a selection marker in both E. coli and Bifidobacterium. A pMB1 replicon derived from the bifidobacterial plasmid pTB60 preserves its replication in B. longum JCM 1217. BBIF_1761 was a surface protein and its signal peptide (SP) directed efficient secretion of appA in both B. longum E18 and B. bifidum S17 (Osswald et al., 2015b). Fukuda et al. reported that a cell surface protein (BLLJ_1900) in the genome of B. longum JCM 1271 has the highest similarity to protein BBIF_1761 (Fukuda et al., 2011), shown by protein Blast analysis. Therefore, the predicted SP encoding sequence of gene BLJJ_1900 was cloned (see SP prediction and detail sequence in Supplementary Information). Briefly, the SP sequence of BLLJ_1900 was amplified by PCR using primers SPF/SPR with JCM 1217 genome as template. The parameters for PCR were pre-denaturated at 98°C for 3 min; then 35 cycles by denaturing at 95°C for 10 s, annealing at 55°C for 10 s and elongation at 72°C for 30 s. The mature form of phytase encoding gene appA was amplified by PCR using primers PhyF/PhyR with E. coli DH5α genome as a template according to a previous report (Sun, 2014). The 1009 bp replicon repB, including its promoter region of plasmid pTB6, was chemically synthesized according to its sequence as the shortage of DNA template (Tanaka et al., 2005). The 660 bp antibiotic resistance gene catA1 (Cmr) was chemically synthesized as well (Accession No.: NG_047582.1). All chemical DNA synthesis service was provided by a commercial supplier (Nanjing GenScript Co., Ltd., China). After all DNA elements were obtained, SOEing PCR was conducted using primers Soe1F/Soe1R with the mixture of SP and appA or using primers Soe2F/Soe2R with the mixture of Cmr and repB fragments in equal molar as templates. The SOEing PCR experiments were performed for five cycles without primers and continued 25 cycles with primers. Parameters were similar to traditional PCR except the elongation time extended to 2 min. The commercial plasmid pUC57 and the amplicon Cmr-repB obtained through SOEing PCR were digested by Ssp I and Hind III and then ligated to form an intermediate cloning plasmid pBS. The plasmid pBS and the amplicon SP-appA were further digested by EcoR I and Nde I. Digested plasmid and amplicon were ligated for framing Bifidobacterial Phytase Expressing Shuttle vector pBPES, which will be used as a backbone for promoter screen. All molecular enzymes, including that used for restriction digest and ligation were purchased from NEB (New England Biolabs Ltd., Beijing, China).
Figure 1.
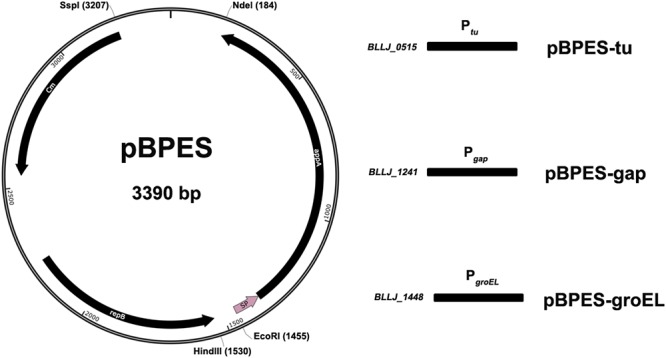
Schematic presentation of plasmid pBPES and its derivatives. Plasmid pBPES was the backbone used for screening promoters. The vector was an Escherichia coli – Bifidobacterium longum shuttle plasmid based on pUC57 using pMB1 cloned from pTB60 as a replicon, Cmr as a selection marker, SP as a signal peptide. Chloramphenicol was used for selection of the plasmid at a concentration of 25 μg/mL in both E. coli and B. longum. Ptu, Pgap, and PgroEL were predicted promoters of gene BLLJ_0515, BLLJ_1241, and BLLJ_1448, respectively. They were cloned into pBPES between restriction sites of Hind III and EcoR I. The resulted three derivative plasmids were named as pBPES-tu, pBPES-gap, and pBPES-groEL, respectively.
Prediction and Cloning of Promoters
Potential promoter regions of genes BLLJ_0515 (Elongation factor Tu), BLLJ_1241 (GAPDH), and BLLJ_1448 (groEL) were predicted by BPROM1 with their upstream intergenic sequences. Full intergenic sequences, including predicted core promoter regions spanning from 172 to 241 bp, were amplified with specific primers using B. longum JCM 1217 genomic DNA as a template (e g., primers tuF/tuR for Ptu, primers gapF/gapR for Pgap, and primers groF/groR for PgroEL). Then, they were digested and inserted into plasmid pBPES between restriction sites Hind III and EcoR I. Genomic DNA of B. longum JCM 1217 was extracted by a Rapid Bacterial Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., China). All DNA fragments were purified by the Universal DNA Purification Kit (Beijing Tiangen Biotech Co., Ltd., China). Transformation of E. coli DH5α and B. longum JCM 1217 was achieved through electroporation as described (Sun, 2014). Recombinant constructs were screened by colony PCR with primer PMF/PMR. Plasmid DNA was extracted with Plasmid Mini Extraction Kit (Tiangen Biotech. Co., Ltd., China) and sequenced by ABI3730 (Nanjing Genscript Biotech Co., Ltd., China). Sequence alignment was conducted with local BLAST program to make sure there are no mutations in promoter regions and open reading frames are correct. All primers used in this study were designed with Primer3 online and synthesized by a commercial supplier (Nanjing Genscript Biotech Co., Ltd., China) as listed (Table 2).
Table 2.
Primers used in this study.
| Primer | Nucleotide sequence | Amplicon | Size (bp) |
|---|---|---|---|
| SPR | ATCCGCTGCGTTGGCCGTG | SP | 99 |
| SPF | ATGAAATCACTGATGAAAAAGG | ||
| PhyR | TTACAAACTGCACGCCGGT | mature appA | 1233 |
| PhyF | CAGAGTGAGCCGGAGCTGA | ||
| Soe1R | GGAATTCCATATGTTACAAACTG CACGCCGGT | SP-appA | 1353 |
| Soe1F | CGGAATTCATGAAATCACTGAT GAAAAAGG | ||
| Soe2R | CCCAAGCTTTCAAACGCGCGA GAAACGGCATT | Cmr -pMB1 | 1669 |
| Soe2F | CGAATATTATGGAGAAAAAAATC ACTGGATATAC | ||
| tuR | CGGAATTCTACTTTTGTCCTCCT GGACGTCTC | Ptu | 172 |
| tuF | CCCAAGCTTCACGCGCCACTG CATGAAG | ||
| gapR | CGGAATTCGTAATTCTCCCTTGT AGGGTGG | Pgap | 240 |
| gapF | CCCAAGCTTCTTGCATGCCGC GCGCTTG | ||
| groR | CGGAATTCGAGTAACACGCAC GCAAGGATG | PgroEL | 241 |
| groF | CCCAAGCTTATGGTTGCGTATT CCTCCAAATGGCT | ||
| rPR | ATCCGCTGCGTTGGCCGTG | promoter- | variable |
| SP-appA | |||
| rPF | TCCGGAGACGTCAGCTGCT |
Underlined: digestion sites for restriction enzymes.
Enzymatic Assay of Recombinant Phytase Secretion in B. longum JCM 1217
All samples used for enzyme assays were prepared as described using ultra-pure water (Osswald et al., 2015b). Briefly, cell free medium (CFM) was prepared by collection of supernatants after centrifugation (5000 g, 5 min). Cell pellets were rinsed twice in 0.2 M sodium citrate buffer, pH5.5, and then disrupted using 0.1 mm glass beads (Sigma-Aldrich, China) by homogenization (Scientz-48, Ningbo, China). Then, total protein concentration in the cell extract was determined by the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Germany). Phytase assay was adapted from the Phytex method described elsewhere with minor changes (Kim and Lei, 2005). In brief, 100 μL pre-warmed supernatant of cell crude extract was mixed with equal volume 10.8 mM sodium phytate. Reaction lasted for 15 min at 37°C, and then stopped by 200 μL 15% tricholoroacetic acid. After centrifugation at 12000 rpm for 2 min, 20 μL supernatant was removed into 980 μL color solution (fresh mixture of 1 M sulfuric acid, 2.5% (w/v) ammonium molybdate in ddH2O, and 10% (w/v) ascorbic acid in ddH2O at a ratio of 3:1:1). Absorbance at 820 nm was read by a spectrophotometer (UV722N, Shanghai Yoke Instrument Ltd., China) in 1 cm plastic cuvettes. Phytase activity was calculated according to a standard curve using 9 mM potassium dihydrogen phosphate in water. One phytase unit (FTU) was defined as an enzyme that catalyzes the release of 1 μmol inorganic phosphate per minute in the above condition. Specific activities were expressed as FTU/mg in cell lysate and FTU/mL in CFM.
Phytate Degradation by B. longum JCM 1217/pBPES-tu in vitro
Phytate degradation in vitro was first confirmed as described by spotting bacteria on the agar plate of phytase screen medium, which supplemented insoluble Ca-phytate as a substrate (Sun, 2014). After 48 h incubation at 37°C, clean zone around colony was an obvious sign of phytate degradation. Second, the CFM was collected after growth of the recombinant strain B. longum JCM 1217/pBPES-tu in modified RCM broth, and was tested for degradation of 0.2% (w/v) maize powder (yellow, cultivar Huanong 101, provided by Da Bei Nong Group, Zhoukou, China), as Ptu is the strongest promoter in this study (Sanz-Penella et al., 2009). After fermentation of B. longum JCM 1217/pBPES-tu statically in the adapted RCM broth at 37°C for 48 h, spent CFM was prepared by centrifugation (10000RCF for 10 min) and stored at 4°C before assay. Degradation of phytate in maize powder was measured by test of released inorganic phosphate in CFM. Uncultured medium was used as a negative control, and commercial enzyme (Szym-PHY10G, SinoBios Imp. & Exp. Co., Ltd., Shanghai) was used as a positive control (PC). Third, B. longum JCM 1217/pBPES and B. longum JCM 1217/pBPES-tu were inoculated into adapted RCM supplemented 0.2% maize powder. Final bacterial counts and inorganic phosphorus in CFMs were measured after 48 h incubation. Bacterial counts were determined by plating of 100 μL culture fluids on the surface of MRS agar after 48 h incubation anaerobically. Soluble inorganic phosphorus was assayed as above enzymatic assay and calculated according to the same standard curve.
Phytate Degradation by B. longum JCM 1217/pBPES-tu in vivo
Animal experiments were approved by the academic committee of Zhoukou Normal University. Forty-five 1-day-old Arbor Acres (AA) male broilers were randomly allocated into nine cages, with five in each cage, and were managed according to the Arbor Acres Management Guide (Manual 20092). All birds were fed a mixture of 1.9 kg commercial premix meal (LNBTM, Cargill, China) and 0.1 kg maize powder per kg of chicks’ body weight gain each day, unless indicated otherwise. The commercial premix meal is based on corn-soybean (55.3%: 38%) and contains 3% fish oil, 1.4% calcium hydrophosphate, 1% limestone, 0.3% salt, and 1% additives in extra (all in w/w). Cages 1–3 were assigned to non-treat group (NG), in which animals were always fed above the indicated meal. Cages 4–6 were assigned to test group (TG), in which animals were fed an adjusted diet that contains 109cfu B. longum JCM 1217/pBPES-tu per kg of meal for 2 weeks and then readapted to the diet used in NG group for two further weeks. Cages 7–9 were assigned to the control group (CG), who were fed an adjusted diet containing 109cfu B. longum JCM 1217/pBPES per kg of meal for 2 weeks and then readapted to the diet used in NG group for a further 2 weeks. Adjusted diets containing bacteria were prepared as previously described, using a granulating machine (GL-25A, Ronghua Machinery Manufactory Co., Ltd., China) (Li and Shi, 2016). Briefly, to produce enough bacterial cells, different recombinant B. longum were fermented in 1L MRS broth supplemented with antibiotics for 48 h, respectively. Bacterial cell pellets were collected by centrifugation and then double-washed by ddH2O. Then, bacterial cells suspended in 100 mL water were mixed with premix meal and maize powder at a certain ratio (2 × 109CFU bacteria: 1.9 kg premix meal: 0.1 kg maize powder). At last, adjusted diets were processed by a granulating machine before feeding. The weights of adjusted diets were 5% higher than that of un-supplemented diet due to the supplementation of bacterial suspension. Supplementation of bacteria has a negligible influence on the concentrations of total phosphorus in all diets and did not affect the body weight gain of birds after feeding (as shown in Supplementary Table S2). Plentiful distilled water was always supplied, and birds were raised under controlled conditions similar to that in commercial practiced during the study in a farm (Shandong Good Earth Company, China).
Assay of Total Phosphorus and Remnant Phytate Contents in Feces
During the experiment, live weights of all chicks in each cage were recorded daily and the body weight gain was calculated. Feces of chicks in each cage were collected at each weekend and dried in an oven at 60°C for 24 h before assay. Contents of total phosphorus (TP) in fecal samples were assayed according to a previous report (Bremer et al., 2008). Briefly, 1 g feces were ashed for 6 h and extracted with 10 mL of 3 N HCl. Then, water diluted extracts were filtered through filter paper (Whatman 42; 2.5 μm) for colorimetric analysis in duplicate with the molybdovanadate method by reading absorbance on a spectrophotometer (MultiScan FC, Thermo Fisher Scientific) and calibrated with standards developed in a similar procedure. Remnant phytate in feces was extracted and quantified by high performance liquid chromatography, according to a previous report with minor modification during extraction (Hotz and Gibson, 2001). Briefly, feces were dried in a 90°C oven until a constant weight was reached. Then, 5 g dried sample was extracted with 5 mL of 0.67 M HCl by vortexing in a metal bath at 37°C for 20 min. Following centrifugation at 5000RCF for 10 min, 2.5 mL of the supernatant was removed and mixed with 2.5 mL of ddH2O water. For parallel comparison, the relative contents of TP and remnant phytate were normalized by their contents in the feces of NG and expressed as percentages.
Isolation and Verification of Recombinant Bifidobacteria in Feces
To confirm the viability and possible colonization of engineered bacteria in vivo, strains of recombinant bifidobacteria were isolated from feces by culture-based method. Briefly, freshly collected feces at each weekend were suspended in sterile saline and then vortexed for 2 min. Supernatants obtained after centrifugation at 3000RCF for 2 min were spread on MRS agar supplemented with 25 μg/mL chloramphenicol. After anaerobical incubation at 37°C for 48 h, ten visible colonies were randomly picked and were amplified by colony PCR using primers rPF/rPR, which are specific sequences derived from plasmid pBPES (primers see Table 2). PCR positive colonies were further verified by enzymatic assays using a F6PPK ELISA Kit according to the user guide (Qingdao Hopebiol Co., Ltd., China).
Data Analysis
For all in vitro data, the mean of three independent experiments with triple replicates were calculated and expressed as mean ± standard deviations (SD). All in vivo data were obtained from three cages and expressed as relative mean ± SD. Statistical analysis for in vitro degradation of phytate was performed using Student’s t-test (significant with P < 0.01). Statistical analysis for in vivo study was performed using SPSS19.0 by one-way ANOVA with Bonferroni post-tests.
Results and Discussion
Stability of Shuttle Plasmid pBPES and Cloning of Predicted Promoter
The plasmid pBPES was constructed as designed in this study. Transformation into and successful isolation from both E. coli DH5α and B. longum JCM 1217 suggest that pBPES is indeed a shuttle vector that can be maintained in these hosts. The segregational stability of this plasmid in B. longum JCM 1217 was further evaluated by propagation in liquid MRS without antibiotics and the recovery rate under antibiotics reaches 100% after 100 generations and surpasses 95% after 500 generations (data not shown). High recovery rate indicates plasmid pBPES is very stable in JCM 1217 and can be used as a backbone for the construction of expression vectors. BPROM prediction of intergenic sequence yielded one potential promoter for each gene (Supplementary Table S1, Elongation factor Tu, GAPDH, and groEL, see detail results in Supplementary Information). Cloning of these predicted promoters into pBPES resulted in three expressional plasmids, namely pBPES-tu, pBPES-gap, and pBPES-groEL, respectively (Figure 1). Sequencing of the regions spanning promoter and appA confirmed that they were in correct open reading frames (ORFs).
Ptu Is a Strong Promoter in B. longum JCM 1217
To compare the strength of these three promoters, they were transformed into B. longum JCM 1217, respectively. As expected, transformation of these plasmids into B. longum JCM 1217 does not influence growth in adapted RCM as all recombinant B. longum JCM 1217 strains have a similar biomass after incubation (Figure 2A). The strength of cloned promoters was tested by evaluating appA encoded phytase activity in B. longum JCM 1217. As shown, phytase activity was detected in the CFMs of all recombinant strains after 48h static incubation (Figure 2B). In fact, all promoters must constitutively drive appA expression in B. longum JCM 1217 as all constructs have high phytase activity in their CFMs. Among these three promoters, Ptu is significantly stronger than PgroEL and Pgap, though the last is the most extensively used promoter in bifidobacteria (Klijn et al., 2006; Shkoporov et al., 2008; Sun et al., 2014a; Osswald et al., 2015a,b). As shown, activities of phytase in the CFMs of JCM 1217/pBPES-tu and JCM 1217/pBPES-groEL are much higher than that in JCM 1217/pBPES-gap, which reaches 0.48 FTU/mL (Figure 2B). This value is comparable to a previous report in another bifidobacteria strain (Osswald et al., 2015b). Low phytase activity in B. lognum JCM/pBPES suggests wide-type bifidobacteria are either negative in phytases or inefficient in degrading phytate, agreeing with a previous report (Palacios et al., 2008). Further time course experiments suggest B. lognum JCM 1217/pBPES-tu has the highest level of relative activity at their exponential phases (Figure 2D). Therefore, the study showed promoters of other housekeeping genes, like groEL and elongation factor Tu, are stronger than Pgap To be precise, Ptu and PgroEL are 8- and 1-fold stronger than Pgap.
Figure 2.
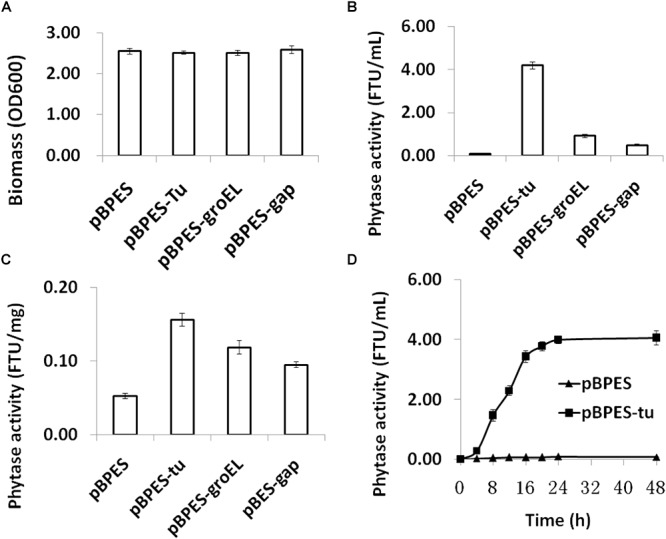
Biomass and phytase expression of recombinant B. longum JCM 1217 grown in modified reinforced clostridia medium. (A) biomass of different strains assayed by reading optical density at 600 nm after anaerobic incubation for 48 h; (B) phytase activity in the CFMs after 48 h incubation of different strains; (C) phytase activity in the cell lysate after 48 h incubation of different strains; (D) time course phytase activity in CFMs of recombinant B. longum JCM 1217 harboring plasmid pBPES or pBPES-tu. Samples were collected at different time points during fermentation in the modified RCM. CFM, cell free medium; RCM, reinforced clostridia medium.
Signal Peptide of BLLJ_1900 Directs Efficient Phytase Secretion in B. longum JCM 1217
The secretion of gene products is important for the functionality of many heterogonous expressed enzymes, especially when substrate bioavailability is low, like phytate. It was demonstrated that the SP of cell surface protein BBIF_1761 directs the efficient secretion of appA in B. bifidum S17 and B. longum E18 (Sun, 2014; Osswald et al., 2015b). Therefore, the predicted SP encoding sequence of its equivalent gene in B. longum JCM 1271 was cloned and fused ahead of the mature form of the appA gene. Phytase activity is much higher in CFMs and detectable but much lower in cell lysate, suggesting effective secretion of appA (Figure 2B,C). The results also indicated that the SP of BLLJ_1900 directs efficient secretion of appA in B. longum JCM 1271.
B. longum JCM 1217/pBPES-tu Degrades Phytate in vitro
Phytate is a main anti-nutritional ingredient in plant-derived diets for all monogastric animals and humans (Schlemmer et al., 2010). Currently, phytase degradation is one of the leading approaches for liberating phytate (Vohra and Satyanarayana, 2003; Jones et al., 2010). Although a few wide-type bifidobacteria strains are phytase positive, their activities are low (Haros et al., 2007, 2009; Oh and Lee, 2007; Sanz-Penella et al., 2012). In the present study, primary phytase degradation by engineered bifidobacteria was displayed phenotypically on the PSM agar containing insoluble Ca-phytate. Clear degradation zones were presented around colonies of B. longum JCM 1217/pBPES-tu but were presented much more weakly in B. longum JCM 1217/pBPES (Figure 3A). Secondly, the degradation potential of phytase-secreting B. longum JCM 1217/pBPES-tu on phytate was evaluated by adding 2% (w/v) maize powder into the spent CFM. The CFM of B. longum JCM 1217/pBPES resulted in a much faster degradation of phytate compared to the equal amount of commercial phytase did, though they both finally degraded phytate to similar levels (Figure 3B). Thirdly, strains of recombinant bifidobacteria were inoculated into the adapted medium (RCM) supplemented with 2% (w/v) maize powder. The medium had no influence on the growth as both recombinant strains have similar final bacteria counts (Figure 3C). Meanwhile, inoculation of B. longum JCM 1217/pBPES does not lead to obvious phosphate release. In contrast, inoculation of B. longum JCM 1217/pBPES-tu resulted in a higher concentration of phosphate in CFM, suggesting effective degradation of phytate in maize (Figure 3D). All these data confirmed that B. longum JCM 1217/pBPES-tu can degrade phytate in vitro.
Figure 3.
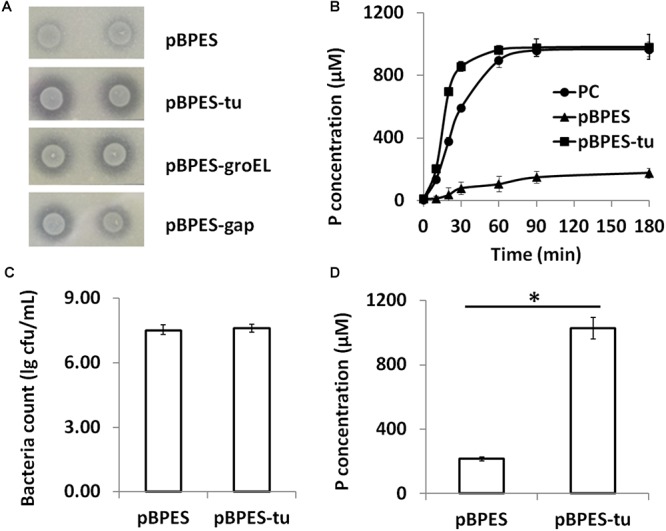
Phytate degradation by B. longum JCM 1217/pBPES-tu in vitro. (A) degradation of phytate-Ca on solid phytase screen medium by spotted bacterial colonies harboring plasmid pBPES or pBPES-tu; (B) degradation of phytate from maize powder in liquid phase by CFMs of different recombinant strains or commercial phytase; (C) bacteria counts of different recombinant strains grown in the modified RCM plus 2% maize powder; (D) degradation of phytate from maize powder by inoculation of different recombinant strains into the modified RCM. All data were mean of three independent experiments with triple replicates and expressed as mean ± SD. Statistical analysis for degradation of phytate was performed using Student’s t-test (∗significant with P < 0.01). CFM, cell free medium; RCM, reinforced clostridia medium; PC, 1 FTU/mL commercial phytase.
B. longum JCM 1217/pBPES-tu Degrades Phytate in vivo
Chicks were fed recombinant B. longum JCM 1217/pBPES-tu to evaluate its effectiveness on phytate degradation in vivo. Different diets had little influence on daily body weight gain of chicks during the survey (Supplementary Table S2). The result is in line with a former report that fed broilers with recombinant lactobacilli (Li and Shi, 2016). During treatment, the contents of TP and remnant phytate in feces were assayed at each weekend to reflect the extent of phytate degradation. The contents of TP didn’t differ between NG and CG groups, which suggested that B. longum JCM 1217/pBPES has little impact on the excretion of phosphorus (Supplementary Table S3). In contrast, the relative content of TP in the TG group is significantly lower than that in CG group from the second week to the end of the survey (Figure 4A, marked by asterisk, P < 0.01). The relative content of remnant phytate is also significantly lower in TG group than that in CG group during the whole treatment (Figure 4B, marked by asterisk, P < 0.01). These results indicated that B. longum JCM 1217/pBPES-tu degraded phytate effectively in vivo. Unexpectedly, at the weekend of the 4th week, the content of remnant phytate in CG group is also significantly lower than that in the NG group (Supplementary Table S4, marked by superscript a, P < 0.01). B. longum JCM 1217/pBPES-tu degraded phytate in vivo is in line with earlier studies using either purified phytases or other microbes producing phytases (Simons et al., 1990; Sanz-Penella et al., 2012; Li and Shi, 2016). However, conflicted results exist as B. longum JCM 1217/pBPES was also shown to reduce phytate excretion in vivo at the end of the survey, even though no obvious phytase activity can be detected in vitro. Although there is a 5% difference in weight between adjusted diets and un-supplemented diet, no difference in both TP and remnant phytate contents in NG and CG groups can be detected during the first 2 weeks of treatment with either diet. But F6PPK positive colonies were grown on MRS supplemented with antibiotics all over the survey even when the CG group was restored to the basal diet. Therefore, detection of recombinant bifidobacteria suggested either that B. longum JCM 1217/pBPES slowly colonized the gut or that the pBPES plasmid was horizontally transferred to endogenous bifidobacteria during the 2 week intake. Whatever the pathway, the slow decrease in phytate excretion cannot be related to the sole slow plasmid establishment in the gut as it is deprived in phytase code. It could, however, reflect the promotion of phytase-producing bacteria in the microbiota.
Figure 4.
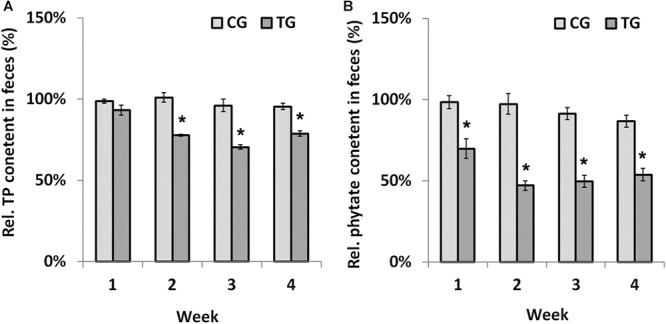
Phytate degradation by B. longum JCM 1217/pBPES-tu in chicks. (A) relative TP contents in the feces of chicks collected from different groups at each weekend; (B) relative remnant phytate in the feces of chicks collected from different groups at each weekend. Data are mean ± SD of three samples collected from three cages relative to the corresponding contents in NG group. Statistical analysis was performed using one-way ANOVA with Bonferroni post-test (∗significant with P < 0.01). NG, non-treat group; CG, control group; TG, test group.
Conclusion
In conclusion, this study constructed a recombinant strain B. longum JCM 1217/pBPES-tu that constitutively secretes phytase appA and confirmed the effectiveness of this strain on phytate degradation both in vitro and in vivo. The study pioneered the potential of live genetically modified probiotics in the feedstuff industry, and also provided a novel strategy for solving the problems of phosphate utilization and pollution. However, application of this recombinant strain in the food industry is restrained due to legal issues and because further improvement is necessary.
Ethics Statement
This study was carried out in accordance with the recommendations of Arbor Acres Management Guide, and the protocol was approved by the Animal Ethic and Welfare Committee of Zhoukou Normal University. For this study, involving animals, all institutional and national guidelines for the care and use of laboratory animals were followed.
Author Contributions
ZS and CL designed the experiments and wrote the manuscript. ZS, ZY, XY, and CaC carried out the experiments. XH, MS, and CuC analyzed experimental results. LL provided technique support and discussed results.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the technician, B.Sc. Qiang Sun from Shandong Good Earth Company, for feeding of chicks and collection of samples.
Funding. This study was partially funded by a grant from the Education Department of Henan Province (No. 17A180017) and China Postdoctoral Science Foundation (No. 2018M642772) to ZS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00796/full#supplementary-material
References
- Bremer V. R., Buckner C. D., Erickson G. E., Klopfenstein T. J. (2008). Total and Water Soluble Phosphorus Content of Feedlot Cattle Feces and Manure. Nebraska Beef Cattle Reports. Available at: http://digitalcommons.unl.edu/animalscinbcr/27/ (accessed January 2008). [Google Scholar]
- Celiberto L. S., Bedani R., Dejani N. N., Ivo de Medeiros A., Sampaio Zuanon J. A., Spolidorio L. C., et al. (2017). Effect of a probiotic beverage consumption (Enterococcus faecium CRL 183 and Bifidobacterium longum ATCC 15707) in rats with chemically induced colitis. PLoS One 12:e0175935. 10.1371/journal.pone.0175935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Tian F., Sun Z. (2016). “Phosphorus nutrition and health: utilization of phytase-producing Bifidobacteria in food industry,” in Probiotics and Prebiotics in Human Nutrition and Health, eds Rao Venketeshwer, Rao G. Leticia. (London: IntechOpen; ). [Google Scholar]
- Frontela C., García-Alonso F. J., Ros G., Martinez C. (2008). Phytic acid and inositol phosphates in raw flours and infant cereals: The effect of processing. J. Food Compos. Anal. 21 343–350. 10.1016/j.jfca.2008.02.003 [DOI] [Google Scholar]
- Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469 543–547. 10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- García-Mantrana I., Monedero V., Haros M. (2014). Application of phytases from bifidobacteria in the development of cereal-based products with amaranth. Eur. Food Res. Technol. 238 853–862. 10.1007/s00217-014-2167-2 [DOI] [Google Scholar]
- García-Mantrana I., Monedero V., Haros M. (2015). Reduction of phytate in soy drink by fermentation with Lactobacillus casei, expressing phytases from Bifidobacteria. Plant Foods Hum. Nutr. 70 269–274. 10.1007/s11130-015-0489-2 [DOI] [PubMed] [Google Scholar]
- Haros M., Bielecka M., Honke J., Sanz Y. (2007). Myo-inositol hexakisphosphate degradation by Bifidobacterium infantis ATCC 15697. Int. J. Food Microbiol. 117 76–84. 10.1016/j.ijfoodmicro.2007.02.021 [DOI] [PubMed] [Google Scholar]
- Haros M., Carlsson N. G., Almgren A., Larsson-Alminger M., Sandberg A. S., Andlid T. (2009). Phytate degradation by human gut isolated Bifidobacterium pseudocatenulatum ATCC 27919 and its probiotic potential. Int. J. Food Microbiol. 135 7–14. 10.1016/j.ijfoodmicro.2009.07.015 [DOI] [PubMed] [Google Scholar]
- Hotz C., Gibson R. S. (2001). Assessment of home-based processing methods to reduce the phytate content and phytate/zinc molar ratio of white maize (Zea mays). J. Agric. Food Chem. 49:692. 10.1021/jf000462w [DOI] [PubMed] [Google Scholar]
- Jones C. K., Tokach M. D., Dritz S. S., Ratliff B. W., Horn N. L., Goodband R. D., et al. (2010). Efficacy of different commercial phytase enzymes and development of an available phosphorus release curve for Escherichia coli-derived phytases in nursery pigs. J. Anim. Sci. 88 3631–3644. 10.2527/jas.2010-2936 [DOI] [PubMed] [Google Scholar]
- Khokhlova E. V., Efimov B. A., Kafarskaia L. I., Shkoporov A. N. (2010). Heterologous expression of secreted biologically active human interleukin-10 in Bifidobacterium breve. Arch. Microbiol. 192 769–774. 10.1007/s00203-010-0606-4 [DOI] [PubMed] [Google Scholar]
- Kim T. W., Lei X. G. (2005). An improved method for a rapid determination of phytase activity in animal feed. J. Anim. Sci. 83 1062–1067. 10.2527/2005.8351062x [DOI] [PubMed] [Google Scholar]
- Kim Y., Lee J. W., Kang S. G., Oh S., Groffiths M. W. (2012). Bifidobacterium spp. influences the production of autoinducer-2 and biofilm formation by Escherichia coli O157:H7. Anaerobe 18 539–545. 10.1016/j.anaerobe.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Klijn A., Moine D., Delley M., Mercenier A., Arigoni F., Pridmore R. D. (2006). Construction of a reporter vector for the analysis of Bifidobacterium longum promoters. Appl. Environ. Microbiol. 72 7401–7405. 10.1128/AEM.01611-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X. G., Weaver J. D., Mullaney E., Ullah A. H., Azain M. J. (2013). Phytase, a new life for an “old” enzyme. Annu. Rev. Anim. Biosci. 1 283–309. 10.1146/annurev-animal-031412-103717 [DOI] [PubMed] [Google Scholar]
- Li Q., Shi Y. (2016). Feeding of phytase over-expressed Lactobacillus decreases fecal phosphate in chicken. Res. J. Biotechnol. 11 9–16. [Google Scholar]
- Liu D., Wang S., Xu B., Guo Y., Zhao J., Liu W., et al. (2011). Proteomics analysis of Bifidobacterium longum NCC2705 growing on glucose, fructose, mannose, xylose, ribose, and galactose. Proteomics 11 2628–2638. 10.1002/pmic.201100035 [DOI] [PubMed] [Google Scholar]
- Oh N. S., Lee B. H. (2007). Phytase properties from Bifidobacterium animalis. Food Sci. Biotechnol. 16 580–583. 10.1016/j.foodpol.2006.11.001 17462768 [DOI] [Google Scholar]
- Osswald A., Sun Z., Grimm V., Ampem G., Riegel K., Westendorf A. M., et al. (2015a). Three-dimensional tumor spheroids for in vitro analysis of bacteria as gene delivery vectors in tumor therapy. Microb. Cell Fact. 14:199. 10.1186/s12934-015-0383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald A., Westermann C., Sun Z., Riedel C. U. (2015b). A phytase-based reporter system for identification of functional secretion signals in bifidobacteria. PLoS One 10:e0128802. 10.1371/journal.pone.0128802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios M. C., Haros M., Rosell C. M., Sanz Y. (2008). Selection of phytate-degrading human bifidobacteria and application in whole wheat dough fermentation. Food Microbiol. 25 169–176. 10.1016/j.fm.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Parvaneh K., Jamaluddin R., Karimi G., Erfani R. (2014). Effect of probiotics supplementation on bone mineral content and bone mass density. Sci. World J. 2014:595962. 10.1155/2014/595962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Penella J. M., Frontela C., Ros G., Martinez C., Monedero V., Haros M. (2012). Application of bifidobacterial phytases in infant cereals: effect on phytate contents and mineral dialyzability. J. Agric. Food Chem. 60 11787–11792. 10.1021/jf3034013 [DOI] [PubMed] [Google Scholar]
- Sanz-Penella J. M., Tamayo-Ramos J. A., Sanz Y., Haros M. (2009). Phytate reduction in bran-enriched bread by phytase-producing bifidobacteria. J. Agric. Food Chem. 57 10239–10244. 10.1021/jf9023678 [DOI] [PubMed] [Google Scholar]
- Schlemmer U., Frølich W., Prieto R. M., Grases F. (2010). Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 53 S330–S375. 10.1002/mnfr.200900099 [DOI] [PubMed] [Google Scholar]
- Shkoporov A. N., Efimov B. A., Khokhlova E. V., Kafarskaia L. I., Smeianov B. A. (2008). Production of human basic fibroblast growth factor (FGF-2) in Bifidobacterium breve using a series of novel expression/secretion vectors. Biotechnol. Lett. 30 1983–1988. 10.1021/pr800494a [DOI] [PubMed] [Google Scholar]
- Simons P., Versteegh H., Jongbloed A. W., Kemme P. A., Verschoor G. J. (1990). Improvement of phosphorus availability by microbial phytase in broilers and pigs. Br. J. Nutr. 64 525–540. 10.1079/BJN19900052 [DOI] [PubMed] [Google Scholar]
- Sun Z. (2014). Development of Gene Expression Systems in Bifidobacterium bifidum S17 and Their Application for Tumor Therapy. Ph.D. Thesis, Open Access Repositorium der Universität Ulm, Ulm. [Google Scholar]
- Sun Z., Baur A., Zhurina D., Yuan J., Riedel C. U. (2012). Accessing the inaccessible: molecular tools for bifidobacteria. Appl. Environ. Microbiol. 78 5035–5042. 10.1128/AEM.00551-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., He X., Brancaccio V. F., Yuan J., Riedel C. U. (2014a). Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PloS One 9:e88260. 10.1371/journal.pone.0088260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Westermann C., Yuan J., Riedel C. U. (2014b). Experimental determination and characterization of a gap promoter for high level gene expression in Bifidobacterium bifidum S17. Bioengineered 5 371–377. 10.4161/bioe.34423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Samura K., Kano Y. (2005). Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 69 422–425. 10.1271/bbb.69.422 [DOI] [PubMed] [Google Scholar]
- Vohra A., Satyanarayana T. (2003). Phytases: microbial sources, production, purification, and potential biotechnological applications. Crit. Rev. Biotechnol. 23 29–60. 10.1080/713609297 [DOI] [PubMed] [Google Scholar]
- Wei X., Wang S., Zhao X., Wang X., Li H., Lin W., et al. (2016). Proteomic profiling of Bifidobacterium bifidum S17 cultivated under in vitro conditions. Front. Microbiol. 7:97. 10.3389/fmicb.2016.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhorst S., Lin H., Blechner C., Fanick W., Brandt L., Brehm M., et al. (2013). Tumour cells can employ extracellular Ins(1,2,3,4,5,6)P(6) and multiple inositol-polyphosphate phosphatase 1 (MINPP1) dephosphorylation to improve their proliferation. Biochem. J. 450 115–125. 10.1042/BJ20121524 [DOI] [PubMed] [Google Scholar]
- Yun B., Song M., Park D. J., Oh S. (2017). Beneficial effect of Bifidobacterium longum ATCC 15707 on survival rate of Clostridium difficile infection in mice. Korean J. Food Sci. Anim. Resour. 37 368–375. 10.5851/kosfa.2017.37.3.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


