Abstract
With few exceptions, the marked advances in knowledge about the genetic basis of schizophrenia have not converged on findings that can be confidently used for precise experimental modeling. Applying knowledge of the cellular taxonomy of the brain from single-cell RNA-sequencing, we evaluated whether the genomic loci implicated in schizophrenia map onto specific brain cell types. We found that the common variant genomic results consistently mapped to pyramidal cells, medium spiny neurons, and certain interneurons but far less consistently to embryonic, progenitor, or glial cells. These enrichments were due to sets of genes specifically expressed in each of these cell types. We also found that many of the diverse gene sets previously associated with schizophrenia (synaptic genes, FMRP interactors, antipsychotic targets, etc.) generally implicate the same brain cell types. Our results suggest a parsimonious explanation: the common-variant genetic results for schizophrenia point at a limited set of neurons, and the gene sets point to the same cells. The genetic risk associated with medium spiny neurons did not overlap with that of glutamatergic pyramidal cells and interneurons, suggesting that different cell types have biologically distinct roles in schizophrenia.
Introduction
Knowledge of the genetic basis of schizophrenia has markedly improved in the past five years1. We now know that much of the genetic basis and heritability of schizophrenia is due to common variation2,3. However, identifying “actionable” genes in sizable studies4,5 has proven difficult with a few exceptions6–8. For example, there is aggregated statistical evidence for diverse gene sets including genes expressed in brain or neurons3,5,9, genes highly intolerant of loss-of-function variation10, synaptic genes11, genes whose mRNA bind to FRMP12, and glial genes13 (Supplementary Table 1). Several gene sets have been implicated by both common and rare variant studies of schizophrenia, and this convergence strongly implicates these gene sets in the pathophysiology of schizophrenia. However, the gene sets in Supplementary Table 1 often contain hundreds of functionally distinctive genes that do not immediately suggest reductive targets for experimental modeling.
Connecting the genomic results to cellular studies is crucial since it would allow us to prioritize for cells fundamental to the genesis of schizophrenia. Enrichment of schizophrenia genomic findings in genes expressed in macroscopic samples of brain tissue has been reported3,14,15 but these results are insufficiently specific to guide subsequent experimentation.
A more precise approach has recently become feasible. Single-cell RNA-sequencing (scRNAseq) can be used to derive empirical taxonomies of brain cell types. We thus rigorously compared genomic results for schizophrenia to brain cell types defined by scRNAseq. Our goal was to connect human genomic findings to specific brain cell types defined by gene expression profiles: to what specific brain cell types do the common variant genetic findings for schizophrenia best “fit”? A schematic of our approach is shown in Figure 1.
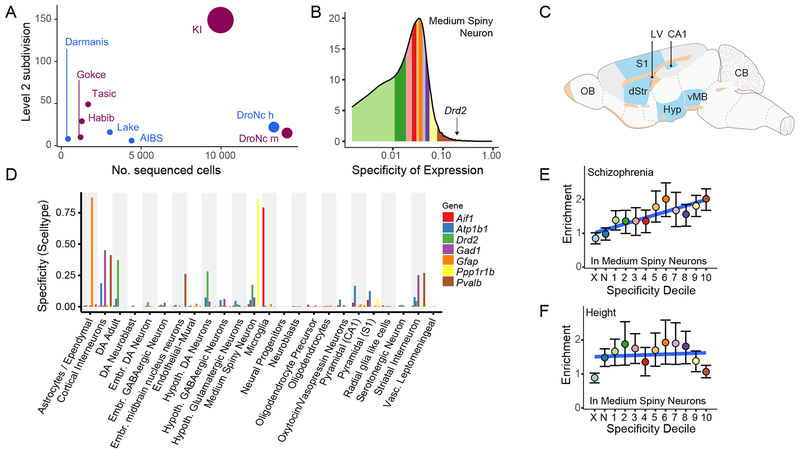
Figure 1. Specificity metric calculated from single cell transcriptome sequencing data can be used to test for increased burden of schizophrenia SNP-heritability in brain cell types.
(A) Comparison of Level 2 cell type categories and number of cells with snRNAseq or scRNAseq from adult brain tissue. Plum colored circles are mouse studies and blue are human studies. The number of different tissues is reflected in size of circle. See Supplementary Table 2 for citations. AIBS=Allen Institute for Brain Science. KI=Karolinska Institutet. (B) Histogram of specificity metric (SMSN,KI) for medium spiny neurons from the KI superset level 1. Colored regions indicate deciles (the brown region contains the genes most specific to MSNs). Specificity value for dopamine receptor D2 (Drd2, SMSN,KI,Drd2=0.17) is indicated by the arrow. (C) Schematic highlighting the brain regions sampled in the KI dataset in blue (D) Specificity values in the KI level 1 dataset for a range of known cell type markers. (E) Enrichment of schizophrenia SNP-heritability in each of the specificity deciles for medium spiny neurons (calculated using LDSC). Color of dots corresponds to regions of the specificity matrix in B. Error bars indicate the 95% confidence intervals. The light blue dot (marked ‘X’) represents all SNPs which map onto named transcripts which are not MGI annotated genes or which map onto a gene which does not have a 1:1 mouse:human homolog. The dark blue dot (marked ‘N’) represents all SNPs which map onto genes not expressed in MSNs. Blue line slows the linear regression slope fitted to the enrichment values. (F) Enrichment of height SNP-heritability in each of the specificity deciles for MSNs.
Results
Cell type specificity of gene expression
We assembled a superset of brain scRNAseq data from Karolinska Institutet (KI; Supplementary Tables 2–3). Brain regions in the KI superset include neocortex16, hippocampus16, hypothalamus17, striatum, and midbrain18 plus samples enriched for oligodendrocytes, dopaminergic neurons, and cortical parvalbuminergic interneurons (total of 9,970 cells, Figure 1c). These data were generated using identical methods from the same labs with unique molecular identifiers that allow for direct comparison of transcription across regions. Quality control and alignment are described elsewhere16. We did not identify important batch effects (Supplementary Figure 1). Based on the scRNAseq data and subsequent clustering each cell have been assigned to a Level 1 classification (e.g., pyramidal cell, microglia, or astrocyte). Level 2 classifications are subtypes of a Level 1 grouping (e.g., medium spiny neurons expressing Drd1 or Drd2). Clustering was based on patterns of correlations across hundreds of genes and not on single markers. After clustering, cell type identities were derived using known expression patterns, histology, and/or molecular studies16–18 (Supplementary Table 2). The KI mouse superset identified 24 Level 1 brain cell types (Supplementary Figure 2) and 149 Level 2 cell types (all sub-groupings of Level 1), far more than any other brain scRNAseq or single nuclei RNA-seq (snRNAseq) dataset presently available (Figure 1a).
For each scRNAseq and snRNAseq dataset, we estimated the specificity of each gene and cell type. This measure represents the proportion of the total expression of a gene found in one cell type compared to all cell types (i.e., the mean expression in one cell type divided by the mean expression in all cell types). If the expression of a gene is shared between two or more cell types, it will get a lower specificity measure. For example, Drd2 is highly expressed in medium spiny neurons (MSNs), adult dopaminergic neurons, and hypothalamic interneurons, and its specificity measure in MSNs of 0.17, but this placed Drd2 in the top specificity decile for MSNs (Figure 1b). Figure 1c shows cell type specificity for seven genes with known expression patterns. Because expression is spread over several cell types, the pan-neuronal marker Atp1b1 has lower specificity than Ppp1r1b (DARPP-32, an MSN marker), Aif1 (a microglia marker), or Gfap (an astrocyte marker).
Cell type specificity of schizophrenia genetic associations
For each cell type, we ranked the expression specificity of each gene into groups (deciles or 40 quantiles). The underlying hypothesis is that if schizophrenia is associated with a particular cell type, then more of the genome-wide association (GWA) signal should be concentrated in genes with greater cell type specificity. For example, we plotted the enrichment of SNP-heritability for schizophrenia and human height in the cell-type specificity deciles of for MSNs and found a positive relationship for schizophrenia but no relationship with human height (Figures 1d–1e). To ensure rigor, we required that two different statistical methods (LDSC9 and MAGMA19) each give strong evidence for connecting schizophrenia GWA results to a cell type. These two methods are based on different assumptions and algorithms. LDSC assessed enrichment of the common SNP-heritability of schizophrenia in the most cell type-specific genes. MAGMA evaluated whether gene-level genetic association with schizophrenia linearly increased with cell type expression specificity. Both methods account in different ways for confounders like gene size and linkage disequilibrium. We required that both methods give similar results after correcting for multiple comparisons to minimize the chance of a spurious conclusion. As described in the Online Methods, we evaluated and excluded multiple potential threats to the validity of these analyses.
To identify brain cell types associated with schizophrenia, we used the largest available GWA study of schizophrenia: CLOZUK identified ~140 genome-wide significant loci in 40,675 cases and 64,643 controls20. We first compared the CLOZUK results to GTEx (RNA-seq of macroscopic samples from multiple human tissues)21 using MAGMA and confirmed3 that smaller schizophrenia GWA P-values were substantially enriched in brain and pituitary (Supplementary Figure 3).
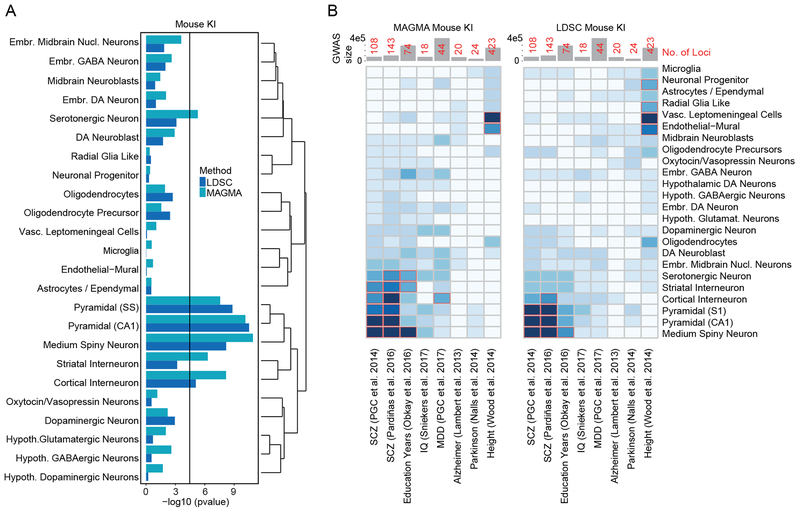
We evaluated the relation of the CLOZUK GWA schizophrenia results to the 24 KI Level 1 brain cell types. Both LDSC and MAGMA strongly highlighted only four cell types: hippocampal CA1 pyramidal cells, striatal medium spiny neurons, neocortical somatosensory pyramidal cells, and cortical interneurons (Figure 2a, Supplementary Figures 4–5). Each exceeded a Bonferroni significance level by several orders of magnitude. The results were not pan-neuronal as multiple other types of neurons did not show enrichment. Schizophrenia risk was greater in mature cells than in embryonic or progenitor cells. We extended the analysis to 149 KI Level 2 cell types (subtypes of Level 1 cells): for hippocampal CA1 pyramidal cells, both major subgroups were significant; for striatum, medium spiny neurons expressing Drd2, Drd1 and striatal Pvalb-expressing interneurons were consistently significant; and for neocortical somatosensory pyramidal cells, cortical layers 2/3, 4, 5, and 6 were significant (Supplementary Figure 6). The cortical Level 1 interneuron signal appeared to result from four interneuron subcategories all expressing Reln.
Figure 2. Evaluation of enrichment of common variant CLOZUK schizophrenia GWA results in the KI brain scRNAseq dataset from mouse.
(A) KI Level 1 brain cell types. Both LDSC and MAGMA show enrichment for pyramidal neurons (somatosensory cortex and hippocampus CA1), striatal medium spiny neurons, and cortical interneurons. The black line is the Bonferroni significance threshold (0.05/((24+149)*8). (B) Heat map of association pvalues of diverse human GWA with KI Level 1 mouse brain cell types using MAGMA (left panel) and LDSC (right panel). Bonferroni significant results are marked with red borders (0.05/((24+149)*8). Total number of cases and controls used in the GWAS are shown in the top bar plots, where numbers in red indicate the amount of genome-wide significant loci identified. The CLOZUK results do not generalize indiscriminately across human diseases/traits. In the more sensitive MAGMA analysis major depressive disorder (MDD) is primarily enriched in cortical interneurons and embryonic midbrain neurons, unlike schizophrenia. Similar but non-significant trends can be observed using LDSC.
Additional analyses showed that these results were not influenced by the total number of molecules detected per cell type or total number of cells per cell type (Supplementary Table 3). We conducted null simulations and confirmed that there was no Type 1 error inflation (Supplementary Figure 7). We also applied an alternative approach based on differential expression22, and replicated the association of MSNs, pyramidal CA1, and neocortical somatosensory pyramidal cells with schizophrenia using a third method (Supplementary Figure 8). These additional analyses suggest the robustness of our results.
We next evaluated whether these results were specific to schizophrenia or if they resulted from some feature common across human traits. Heat maps of KI Level 1 enrichment P-values for GWA results from eight studies of human complex traits are depicted in Figure 2b. Seven studies evaluated common variants associations for brain-related diseases or traits with ≥20,000 cases and ≥10 genome-wide significant associations. Human height was included as a non-brain comparator. The results from the earlier PGC GWA study of schizophrenia3 were similar to those from CLOZUK. Although we observed cell types being enriched in other sets, none had the specific signal observed in the two schizophrenia sets. For example, for major depressive disorder, we found that GABAergic interneurons, embryonic midbrain neurons, and dopaminergic interneurons were the most enriched cell types. For each cell type, we tested whether the enrichment observed in other GWA studies was significantly different from that in CLOZUK. We observed no significant difference for SCZ2 (a subset of CLOZUK) and years of education but all other studies contained significantly different cell type enrichments (Supplementary Figure 9).
Replication of results in additional single cell datasets
We replicated most findings in independent scRNAseq/snRNAseq mouse brain studies. We found significant enrichment for schizophrenia in hippocampal CA1 pyramidal cells, neocortical pyramidal cells, cortical interneurons (although not in all data sets), and medium spiny neurons23–26. We also saw enrichment in pyramidal neurons from CA3 and dentate gyrus granule cells. (Supplementary Figures 10a–d). Replication of our results in external other datasets again highlight the robustness of our cell type association results.
We identified an important technical issue for scRNAseq/snRNAseq studies of brain. scRNAseq is readily done in mouse brain but more difficult in larger and more fragile human brain neurons. Nearly all currently available human data have been generated using snRNAseq. The isolated nuclei used in snRNAseq lack the cytoplasmic compartment and proximal dendrites, and there are systematic differences between the types and amounts of mRNA in nucleus versus cell soma27. To evaluate the impact of this issue, we analyzed multiple mouse and human datasets. We confirmed that transcripts destined for export to synaptic neuropil28 were better captured by scRNAseq and specifically depleted in snRNAseq (Figure 3a). This is important for the purposes of this study because synaptic neuropil transcripts are enriched for genetic associations with schizophrenia (P=1.6×10−4). This places an important caveat on the use of snRNAseq to evaluate brain cell type associations with schizophrenia given that snRNAseq from human or mouse brain may not comprehensively capture the relevant transcriptome.
Figure 3. Comparison of single-cell and single-nuclei RNAseq, and evaluation of enrichment of common variant CLOZUK schizophrenia GWA results in brain single-nuclei RNAseq datasets from adult human.
(A) Each bar represents a comparison between two datasets (X vs Y), with the bootstrapped Z-scores representing the extent to which dendritically enriched transcripts28 have lower specificity for pyramidal neurons in dataset Y relative to X. Larger Z-scores indicate greater depletion of dendritically enriched transcripts, and red bars indicate a statistically significant depletion. Supplementary Table 2 describes the studies. (B) Human mid-temporal cortex brain cell type enrichment. Cortical pyramidal neurons and cortical interneurons show significant enrichment. Oligodendrocyte precursors also show enrichment that was not observed in the KI Level 1 data. The black line is the Bonferroni significance threshold (6×8 comparisons). (C) Human prefrontal cortex and hippocampus brain cell type enrichments from26. These data show enrichment in cortical and hippocampal glutamatergic (i.e., pyramidal and granule) cells. There is also an enrichment in cortical interneurons with the highest level in Reln/Vip cells. The black line is the Bonferroni significance threshold (15×8 comparisons). (D) Heat map of enrichment of diverse human GWA studies with human mid-temporal cortex Level 1 brain cell types using MAGMA and LDSC. The CLOZUK results do not generalize across human diseases. MDD again shows significant enrichments in cortical interneurons. Common variant genetic associations for Alzheimer’s disease were enriched in microglia. Bonferroni significant results are marked with red borders.
With these caveats in mind, we evaluated human snRNAseq datasets from mid-temporal cortex (Allen Institute for Brain Science, unpublished) and DroNc-seq in prefrontal cortex and hippocampus26. Using hierarchical clustering on specificity scores, we found that human and mouse cell types clustered together (Supplementary Figure 11); Level 1 cell types had greater similarity to the same cell type across species than to a different cell type in the same species. We confirmed enrichment of schizophrenia SNP-heritability in cortical pyramidal neurons (glutamatergic cells) and cortical interneurons (GABAergic cells) in two different human datasets (Figure 3b). In the DroNc-seq dataset26, we confirmed enrichment in hippocampal pyramidal neurons (glutamatergic cells) along with greater enrichment in Reln-expressing GABAergic interneurons compared to those expressing Pvalb. In both human studies, oligodendrocyte precursor cells (OPCs) were significant or close to significance but it is hard to judge if this is related to a loss of neuronal-specific signal in snRNAseq (note that OPCs showed stronger signal in OPCs in mouse snRNAseq vs scRNAseq; Figure 2 and Supplementary Figure 10d). In a small scRNAseq study29, human adult and fetal cortical neurons were significantly enriched for schizophrenia SNP-heritability. These are likely pyramidal cells but the small numbers of cells sequenced precluded further exploration. No significant enrichments were found in another snRNAseq study of a single human30, perhaps due to a lack of cellular diversity (data not shown). We are unaware of scRNAseq/snRNAseq data from human striatum. The specificity of the human cortical signal for schizophrenia was confirmed in relation to the same set of brain-specific GWA studies (Figure 2d). In summary, all major findings from the KI dataset were replicated in independent mouse or human studies.
Cell type enrichments of schizophrenia associated gene sets
A major question in the field regards interpretation of the large and diverse gene sets that have been compellingly related to schizophrenia (Supplementary Table 1). These gene sets are highly significant, replicate well, and are often implicated in both common and rare variant studies. However, their implications for an experimentalist are unclear: what do these large sets of genes really tell us? These gene sets are large, and could be expected to recapitulate the cell type enrichments found above. However, all neurons have synapses and NeuN (the protein product of Rbfox3) is a widely used neuronal marker, so another possibility is that the RBFOX, PSD95, and FMRP gene sets could simply be pan-neuronal.
We thus evaluated whether gene sets previously implicated in schizophrenia were specifically expressed in the KI level 1 brain cell types (using Expression Weighted Cell type Enrichment, EWCE)31. The inputs to EWCE are a list of genes (e.g., FRMP interacting genes or genes intolerant to loss-of-function variation) and the same scRNAseq cell type specificity matrix used in the MAGMA and LDSC analyses above. Association with schizophrenia is not a direct input although these data are incorporated indirectly (why a gene set was selected in the first place). However, these effects are subtle. For instance, there is a CLOZUK significant GWA hit in only 7.0% of genes that interact with FMRP versus 4.0% that do not interact with FMRP (using MAGMA gene-wise P-values), and there is a CLOZUK significant GWA hit in only 4.1% of genes with ExAC pLI > 0.9 versus 3.3% with low pLI. We also determined that overlap between gene sets was relatively low. For 10 key gene sets (antipsychotic targets, CELF4, FMRP, high or low dN/dS, high pLI, NMDAR, PSD, PSD95, and RBFOX), of 45 pairs of correlations (count of intersection/union), only two correlations exceeded 0.25 (RBFOX-CELF4 0.31 and RBFOX-high pLI 0.28), and most other correlations were near zero.
First, pharmacologically-defined molecular targets of antipsychotics (the mainstay of treatment for schizophrenia) have been associated with schizophrenia32, and we found that antipsychotic medication targets were associated with the same cell types as for the schizophrenia GWA results: neocortical S1 pyramidal cells, MSNs, and hippocampal CA1 pyramidal cells, while cortical interneurons were just above the significance threshold (Figure 4a). Expanding these analyses, we found that other gene sets associated with schizophrenia were specifically expressed in schizophrenia-relevant cell types (Figures 4b–d). The gene sets consistently associated with schizophrenia – intolerant to loss-of-function variation, NMDA receptor complex, post-synaptic density, PSD95 complex, RBFOX binding, CELF4 binding, and FMRP associated genes – all had more specific expression in neocortical S1 and hippocampal CA1 pyramidal cells, MSNs from the dorsal striatum, and cortical interneurons (with the exception of NMDA receptor complex genes). Because some of these gene sets are involved in diverse cellular functions, there were, as expected, associations with other Level 1 cell types. For example, genes intolerant to loss-of-function variation had significantly greater expression in progenitor cells (dopaminergic neuroblasts, neuroblasts, and embryonic GABAergic neurons). Notably, none of the gene sets previously associated with schizophrenia was pan-neuronal. A prior study33 reported that expert-curated glial gene sets were enriched for schizophrenia associations. We confirmed that those gene sets were significantly associated with glia (Supplementary Figure 12) but could not replicate the association of these gene sets with schizophrenia using MAGMA. Finally, we observed that gene sets previously associated with schizophrenia were substantially less associated with schizophrenia after controlling for the pyramidal neurons, MSNs, cortical interneurons (Supplementary Figure 13). Only loss of function intolerant, CELF4-binding and Rbfox-binding gene sets remained significant after controlling for the cell type enrichments. Our findings highlight that non-overlapping subsets of risk genes each point at the same cell types. Indeed, gene set analysis results can be further subdivided according to cell type-specific expression. Improved methods are thus needed for gene set analysis explicitly accounting for cell types – particularly given intensive efforts to conduct a census of the cellular complexity of the human body.
Figure 4. Cell type enrichment of gene sets associated with schizophrenia, neurological disorders, and the evolutionary divergence between human and mouse.
(A) Antipsychotic medication targets. (B-F) Gene sets previously shown to be enriched for schizophrenia SNP-heritability. (B) Genes intolerant to loss-of-function variation. (C) Synaptic gene sets. (D) Gene sets mediating DNA or RNA interactions. (E) Gene sets associated with neurological disorders. (F) The top 500 genes with lowest or highest dN/dS ratios between human and mouse (i.e., non-synonymous to synonymous exon changes). The Level 1 cell types associated with schizophrenia (MSNs, pyramidal CA1, pyramidal SS, and cortical interneurons) show enrichment in A-D but neurological diseases do not. Asterisks denote Benjamini–Hochberg corrected p-value <0.05 calculated using EWCE.
As neurological diseases are generally not genetically correlated with schizophrenia34, we evaluated the associations of Level 1 cell types with gene sets associated with neurological diseases. Genes associated with Alzheimer’s disease35,36 and multiple sclerosis37 were associated with microglia. Risk genes for leukodystrophy38 were associated with oligodendrocytes (Figure 4e). We analysed genes associated with neurological phenotypes from the Human Phenotype Ontology (HPO) and subcellular localization data from the Human Protein Atlas (Supplementary Figures 14–19). We found that these mostly targeted cell types distinct from those implicated in schizophrenia. For example, the HPO category “neural tube defect” was associated with neural progenitor cells (p=0.0002) and “abnormal myelination” was associated with oligodendrocytes (p<0.0001). We analysed genes with weak or strong conservation between human and mouse (low or high dN/dS scores), and found that highly conserved genes were specific to some types of neuron (e.g., serotonergic) while divergent genes were associated to other cell types (e.g., hypothalamic glutamatergic). None of the schizophrenia associated cell types showed unusually weak or strong evolutionary pressure on their coding sequences (Figure 4f).
Independence of genetic association between cell types
Finally, we assessed how much of cell type connections to schizophrenia was due to shared gene expression between cell types. For instance, the association of cortical interneurons with schizophrenia is weaker than for MSNs: are these independent connections to schizophrenia? Alternatively, given that both are GABAergic neurons, are both associations being driven by a common set of genes? We tested this using resampling without replacement: if the interneuron enrichment is driven solely by overlapping genes with MSNs, then an equivalent level of interneuron association should be found if the schizophrenia association scores of genes within each MSN specificity decile are randomized (Supplementary Figure 20). We performed 10,000 resamplings for each Level 1 cell type while controlling for all four of the significantly associated cell types (Figure 4a). We found that MSNs, cortical interneurons, and hippocampal CA1 pyramidal neurons were independently associated with schizophrenia. However, the association with somatosensory pyramidal neurons was largely due to shared expression with hippocampal CA1 pyramidal neurons. We confirmed this using conditional analysis (Supplementary Figure 21a). We then tested whether each cell type remained significant after conditioning on the three other significant cell types together. Strikingly, only MSNs remained significantly associated with schizophrenia (Supplementary Figure 21b), indicating that the association of MSNs with schizophrenia is independent from that of pyramidal neurons and cortical interneurons.
To evaluate whether the main sources of enrichment signal in different cell types were from overlapping genes, we used a qualitative measure. We plotted the overlap of the top 1,000 genes associated with schizophrenia (MAGMA gene-wise P-values) that also fell in the top decile of specificity scores for each of the four main cell types (Figure 5b). About half of the schizophrenia-associated genes enriched in pyramidal cells and MSNs were shared but those conferring risk-enrichment in interneurons were to a larger extent exclusive. We then evaluated enrichment of gene sets previously associated with schizophrenia (Rbfox, CELF4 or FMRP binding genes, loss of function intolerant gene, genes involved in synapse function, and dendritically transported genes) and genes involved in dopaminergic signalling (Online Methods) in the different areas of Figure 5b using a hypergeometric test. The most associated Rbfox genes were enriched in CA1 pyramidal cells, genes related to loss of function intolerant genes, and dopamine signalling were specifically enriched in medium spiny neurons (Figure 5c). A subset of synaptic genes associated with schizophrenia were shared by all cell types. These findings show that neuronal classes express a combination of overlapping and non-overlapping functional sets of risk genes.
Figure 5. CA1 pyramidal neurons, medium spiny neurons, and cortical Interneurons are independently associated with schizophrenia and distinct molecular pathways contribute to each cell type.
(A) Conditional enrichment analysis accounting for correlated gene expression between cell types. The left column shows baseline cell type enrichment probabilities values for schizophrenia calculated by fitting a linear model to specificity deciles against MAGMA gene enrichment Z-scores. The central four columns show the enrichment probabilities calculated using bootstrapping to control for correlated expression in other cell types; these probabilities approaching zero indicate that after accounting for expression of the other cell type, there is no enrichment remaining. The red box highlights that there is no longer enrichment in somatosensory pyramidal neurons after accounting for expression in CA1 pyramidal neurons; however, the converse is not true. The bar plot on the right shows the minimum value of the conditional probabilities (excluding self-self-comparisons). (B) Overlap of genes in the schizophrenia-associated cell types. Venn-diagram of the top 1,000 schizophrenia-associated genes from the highest enrichment-deciles in the four Level 1 cell types. (C) Benjamini-Hochberg corrected p-values for hypergeometric enrichment of genes in Figure 5b. We note enrichment for Rbfox in CA1 pyramidal cells, Mir137 targets and dopamine signaling in MSNs, along with shared synaptic genes between pyramidal cells but separate for GABAergic cells. Areas with striped shading indicates region with gene number <10.
Discussion
A major issue in schizophrenia genomics is the meaning of the many GWA findings – how do we interpret the hundreds of common variant associations? Similarly, many sets of genes have been compellingly associated with schizophrenia: what are these diverse functional findings telling us? Thus, we attempted to connect human genomic findings for schizophrenia to specific brain cell types defined by their scRNAseq expression profiles: to what specific brain cell types do the common variant genetic findings for schizophrenia best “fit”? Other studies have addressed this question3,9,14, but using gene expression based on aggregates of millions of cells. As described more fully in the Online Methods (“Rationale”), we used scRNAseq data to answer this question. We set a high bar: we required that the connections to cell types be identified using two different methods and exceed an appropriately rigorous statistical threshold.
The results were not pan-neural, pan-neuronal, or in cell types prominent in early development. We found clear connections to just four of 24 main brain cell types: MSNs, pyramidal cells in hippocampal CA1, pyramidal cells in somatosensory cortex, and cortical interneurons. Most of the strong results found in the mouse data replicated in external mouse data and in the more limited human data sets. Intriguingly, many of the diverse gene sets (e.g., antipsychotic drug targets or genes that interact with FMRP or RBFOX proteins) robustly associated with schizophrenia connected to the same cell types. Our results suggest that these discrete cell types are central to the etiology of schizophrenia, and provide an empirical rationale for deeper investigation of these cell types in regard to the basis of schizophrenia. These results can be used to guide in vivo studies and in vitro modeling (e.g., patient-derived neurons from induced pluripotent stem cells) and provide a basis for analyzing how different risk genes interact to produce the symptoms of schizophrenia.
Our results also suggest that single-nuclei RNAseq of neurons leads to systematic underrepresentation of dendritically exported mRNA species. We hypothesize that this is due to destination-specific differences in rates of mRNA decay39. Our data on single-nuclei versus single-cell mRNA capture warrants caution when using single-nuclei data sets for the study of neuronal disorders or processes. This fact should be taken into consideration in the design or analysis of future large scale sequencing efforts.
There are several important caveats as described more fully in the Online Methods (“Limitations”, including discussion and analyses of gene conservation). Despite our use of multiple statistical methods and efforts to identify and resolve any spurious explanations for our findings, our work has to be considered in light of inevitable limitations. Although the KI scRNAseq data cover a broad range of brain regions thought to be relevant to the neurobiology of schizophrenia, extensive coverage of cortical and striatal development is lacking at present (gestation, early postnatal, or adolescence). The currently available functional genomic data in human brain are limited but improving rapidly via PsychENCODE40 and similar efforts, but precisely how schizophrenia GWAS signals impact cell-specific gene expression is not yet a solved problem. Finally, the genetic signals we captured were reflected in the expression levels of hundreds of genes. It is certainly possible for a gene to play an important role in schizophrenia and yet not be in one of the cell types we implicated. For example, genetic polymorphisms in C4a appear to be etiologically involved in schizophrenia7 but the expression of C4a is highest in astrocytes, vascular leptomeningeal cells, and microglia. We were thus careful with our conclusions: we can implicate a cell type (e.g., MSNs show positive evidence) but it is premature to exclude cell types for which we do not have data, or those with dissimilar function or under selection pressure between mouse and human.
In sum, our results support a parsimonious hypothesis: the common variant GWA results for schizophrenia point to a limited set of brain cells, and that subsets of these genes - the gene sets associated with schizophrenia (including antipsychotic medication targets) – each point at the same cell types.
Online Methods
Mouse-to-human gene mapping
We used the expert curated human-mouse homolog list (Mouse Genome Informatics, Jackson Laboratory, URLs, version of 11/22/2016). Only genes with a high-confidence, 1:1 mapping were retained. This is discussed further in the Supplementary Note.
Calculation of cell type expression specificity
A key metric used for our cell type analyses is the specificity (proportion of expression) for a given gene. This metric is calculated separately for each single cell dataset. This is a measure of cell type specificity scaled so that a value of 1 implies that the gene is completely specific to a cell type and a value of 0 implies the gene is not expressed in that cell type. It was calculated using the generate.celltype.data function of the EWCE package (URLs). See Supplementary Note for further details.
Thresholding of low expressed transcripts
Because sg,c is independent of the overall expression level of a gene, it is desirable to exclude genes with very low or sporadic gene expression levels, as a small number of reads in one cell can falsely make that gene appear to be a highly specific cell marker. Direct thresholding of low expressed genes is not ideal for performing this as thresholds need to be set individually for each dataset, and some individual cells can show exceptionally and anomalously high expression of the sporadically expressed gene. We reasoned that all the genes we want to include in the study should be differentially expressed in at least one Level 2 cell type included in the study. We thus excluded sporadically expressed genes via ANOVA with the Level 2 cell type annotations as groups, and excluding all genes with P > 0.00001. Gene filtering was performed separately for each single cell dataset; importantly though, the KI dataset was filtered as a merged superset. A consequence of this (and of differences in sample preparation and sequencing) is that different genes are used for example in the analysis of the KI superset than were used for the Habib et al (Mouse Hippocampus Div-Seq) dataset. For datasets where level 2 cell type annotations were not available (e.g. the Allan Brain Institute Human Cortex dataset) we used the same approach but with level 1 cell type annotations instead.
LD Score Regression (LDSC) and partitioning SNP-heritability
To partition SNP-heritability using LDSC (URLs)9, it is necessary to pass LDSC annotation files (one per chromosome) with a row per SNP and a column for each sub-annotation (1=a SNP is part of that sub-annotation). To map SNPs to genes, we used dbSNP SNPContigLocusId file (build 147 and hg19/NCBI Build 37 coordinates). All SNPs not annotated in this file were given a value of 0 in all sub-annotations. Template annotation files obtained from the LDSC Github repository were used as the basis for all cell type and gene set annotations (“cell_type_group.1*”). Only SNPs present in the template files were used. If an annotation had no SNPs, then 50 random SNPs from the same chromosome were selected as part of the annotation (if no SNPs are selected then the software fails to calculate SNP-heritability).
Annotation files were created for each cell type for which we applied partitioned LDSC. Twelve sub-annotations were created for each cell type. The first represented all SNPs which map onto named regions which are not MGI annotated genes or which map onto a gene which does not have a 1:1 mouse:human homolog. The second contained all SNPs which map onto genes not expressed in a cell type. The other 10 sub-annotations are associated with genes with increasing levels of expression specificity for that cell type. To assign these, the deciles of sg,c were calculated over all values of g (separately for each value of c) to give ten equal length sets of genes. These are then mapped to SNPs as described above. To partition SNP-heritability amongst the gene sets (not the cell types), a single set of annotation files was created with each of the gene sets used as a sub-annotation column.
LDSC was then run using associated data files from phase 3 of the 1000 Genomes Project41. We computed LD scores for cell type annotations using a 1 cM window (--ld-wind-cm 1). As recommended (LDSC Github Wiki, URLs), we restricted the analysis to using Hapmap3 SNPs, and, as in the original report9, excluded the major histocompatibility region due to its high gene density and exceptional LD. The LDSC “munge_sumstats.py” script was used to prepare the summary statistics files. The SNP-heritability is then partitioned to each sub-annotation. We used LD weights calculated for HapMap3 SNPs, excluding the MHC region, for the regression weights available from the Github page (files in the ‘weights_hm3_no_hla’ folder).
For the LD score files used as independent variables in LD Score regression we used the full baseline model9 and the annotations described above. We used the ‘--overlap-annot’ argument and the minor allele frequency files (‘1000G_Phase3_frq’ folder via the ‘--frqfile-chr’ argument).
Partitioned LDSC computes the proportion of SNP-heritability associated with each annotation column while taking into account all other annotations. Based on the proportion of total SNPs in an annotation, LDSC calculates an enrichment score and an associated enrichment P-value (one-tailed as we were only interested in annotations showing enrichments of SNP-heritability). All figures showing partitioned LDSC results show P-values associated with the enrichment of the most specific decile for each cell type.
Cell type identification using MAGMA
We used MAGMA (v1.04)19, a leading program for gene set analysis42, to evaluate the association of gene-level schizophrenia association statistics with cell-type specific expression under the hypothesis that, in relevant cell types, genes with greater cell type specificity should be more associated with schizophrenia. Gene level association statistics were obtained using MAGMA (window size 10 kb upstream and 1.5 kb downstream of each gene – see below for discussion window size) using an approach based on Brown’s method43 (model: snpwise-unweighted). This approach allows to combine P-values in the specified windows surrounding each gene into a gene-level pvalue while accounting for LD (computed using the European panel of 1000 Genomes Project Phase 341).
The tissue specific expression metric for each gene in each cell type was obtained by dividing the gene expression level in a particular cell type by the sum of the expression of the gene in all cell types (see sg,c, defined above). The distributions of sg,c were complex (point mass at zero expression, substantial right-skewing). For each cell type, we transformed S into 41 bins (0=not expressed, 1=below 2.5th percentile, 2=2.5–5th percentile, …, 40=above 97.5th percentile), so that each cell type would be comparable.
MAGMA was then used to test for a positive association (one-sided test) between the binned fractions in each cell type and the gene-level associations (option --gene-covar onesided). For a given mouse or human brain cell type, this tested whether increasing tissue specificity of gene expression is associated with increasing common-variant genetic findings for schizophrenia using information from all genes. By default, the linear regression performed by MAGMA is conditioned on the following covariates: gene size, log(gene size), gene density (representing the relative level of LD between SNPs in that gene) and log(gene density). The model also takes into account gene-gene correlations. For the conditional analysis, we used the condition modifier of the —gene-covar parameter to condition on each of the significant cell types
Random permutations of MAGMA
For the analysis in Supplementary Figure 7, we randomly permuted gene labels of gene-level association statistics of MAGMA and looked for cell type association with schizophrenia using 1,000 permutations. We observed a mean of 24.8 significant results across cell types at P<0.05 indicating that MAGMA is conservative using our approach (50 significant results expected by chance).
Schizophrenia association using alternative cell type specificity method
We tested another recent approach to associate cell types with traits using differentially expressed genes22. We computed a normalization factor for each single cell using the scran R package44 using the 50% of the genes with mean expression higher than the median. The normalization factors were computed after clustering cells using the scran quickcluster function to account for cell type heterogeneity. We then performed 24 differential expression analysis using BPSC45 testing each cell type against the 23 other cell types with the normalization factors as covariate. For each cell type, we then selected the 10% most upregulated genes and created bed files with the coordinate of these genes extended by 100kb upstream and 100kb downstream. SNPs of the baseline model from Finucane et al. located in the top 10% of the genes were used to create a cell type specific annotation that was added to the “baseline” model. We then used LDSC46 to test for association between the cell type specific annotations and schizophrenia using a one-sided P-value based on the coefficient Z-score from the output of LDSC.
Enrichment analyses of gene sets and antipsychotic drug targets
Expression Weighted Cell type Enrichment (EWCE, URLs)31 was used to test for cell types which show enriched expression of genes associated with particular schizophrenia-associated gene sets. These analyses used the same specificity (S) values for the KI Level 1 data that were used for the MAGMA and LDSC analyses. EWCE was run with 10,000 bootstrap samples. Enrichment P-values were corrected for multiple testing using the Bonferroni method calculated over all cell types and gene lists tested. EWCE returns a Z-score assessing standard deviations from the mean. Values < 0 (depletion of expression) were recoded to zero.
Schizophrenia common variant association results
The schizophrenia GWA results were from the CLOZUK and PGC studies3,20. CLOZUK is the largest currently obtainable GWA for schizophrenia (40,675 cases and 64,643 controls), and the authors identified ~150 genome-wide significant loci. It includes the schizophrenia samples from earlier PGC papers. For selected analyses, we also included the PGC schizophrenia results from the Nature 2014 report (URLs). This paper included 36,989 cases and 113,075 controls, and identified 108 loci associated with schizophrenia. Results from the published PGC and CLOZUK studies were qualitatively similar with the CLOZUK data generally showing increased significance owing to its larger sample size.
Comparison GWA results for other traits
We included comparisons for a selected set of brain related traits as well as height as a negative control. As power to identify cell types is directly proportional to the sample size of the GWA study, we only included traits with at least 20’000 samples that discovered at least 20 genome-wide significant loci. The GWA results were from the following sources: schizophrenia3 from the PGC; Alzheimer’s disease35; educational attainment47; IQ48; MDD from the PGC (unpublished); Parkinson’s disease49 and height50.
Test of cell-type association differences between traits
We tested whether the beta coefficient in MAGMA were significantly different between two traits for each cell type using the approach described in Paternoster et al, 199851. We first compute a Z-score for each cell type: . Where β1 and β2 are the SNP-heritability enrichments for trait 1 and 2 (or beta coefficients in MAGMA) and SE are the standard errors. A two-sided p-value is then computed based on the Z-score using the R pnorm function.
Gene sets associated with schizophrenia
The gene set results for schizophrenia are summarized in Supplementary Table 1. For CELF4 binding genes52, we used genes with iCLIP occupancy > 0.2 from Table S4. For FMRP binding genes, we used genes from Table S2A12. Genes intolerant to loss-of-function variation were from the Exome Aggregation Consortium (pLI > 0.9)10. Genes containing predicted miR-137 target sites were from microrna.org. NMDA receptor complex genes came from Genes-to-Cognition database entry L0000000753. The human post−synaptic density gene set was from Table S254. The PSD95 complex came from Table S1 using all genes marked with a cross in the ‘PSD-95 Core Complex’ column55. For RBFOX binding, we took all genes with RBFOX2 count > 4 or summed RBFOX1 and RBFOX3 > 12 from Table S156. For antipsychotic drug targets, we used a gene list provided by Drs Gerome Breen and Héléna Gaspar as reported in the biorXiv preprint32. The oligodendrocyte and astrocyte gene lists came from Supplementary Table 413. All EWCE P-values were corrected with the Benjamini–Hochberg method.
Gene sets for neurological disorders & human phenotype ontology & dN/dS
For multiple sclerosis, we used results from the largest available GWAS (the Multiple Sclerosis Genomic Map); we used the genes listed in the Supplementary table37. For Alzheimer’s disease, we used the top results from the AlzGene database36 (URLs) as well as genome-wide significant genes35. For genes associated with leukodystrophy (HP:0002415,) we used the Human Phenotype Ontology38 (URLs). For amyotrophic lateral sclerosis we used the top results from the ALSGene database (URLs). For epilepsy, migraine, and stroke we used the EBI GWAS catalog. For the Human Phenotype Ontology (HPO) gene sets, the ‘ALL_SOURCES_ALL_FREQUENCIES_phenotype_to_genes.txt’ file was downloaded from build 133. To obtain the genes with the top 500 highest/lowest dN/dS between humans and mice we obtained the dN and dS values through BioMart.
Gene sets associated with subcellular localization
Subcellular localization data were downloaded from the Human Protein Atlas website (HPA, v17)57. Only gene lists with >100 genes were used. Lysosomal genes were downloaded from the Human Lysosome Gene Database58. Mitochondrial genes were obtained from Human MitoCarta2.059. Axonal (Adult) and Axonal (E17) were obtained from a study which used axon-TRAP-RiboTags to capture the mRNAs from retinal ganglion cell axons projecting to the superior colliculus60 (Supplementary Table 1). Presynaptic genes come from Supplementary Table 161. Synaptic vesicle genes came from Supplementary Table 162.
Depletion of dendritically enriched transcripts in nuclei datasets
Dendritically enriched transcripts were obtained from28 (Supplementary Table 10). This list was produced from pyramidal cells from rat hippocampus and human 1:1 homologs were obtained. We refer to this set of genes as Ldendritic. To enable direct comparisons between datasets, all datasets were reduced to contain a common set of six KI Level 1 cell types: pyramidal neurons, interneurons, astrocytes, interneurons, microglia, and oligodendrocyte precursors. For the KI dataset, we used S1 Pyramidal neurons. The specificity metric (denoted as sg,c) was recalculated for each dataset using this reduced set of cell types. Comparisons were then made between datasets (denoted in the graph with the format ‘X vs Y’). We denote the mean pyramidal neuron specificity scores for dendritically enriched genes in dataset X as . We then get the difference in pyramidal specificity of for list L between two datasets as . We then calculate values of DX,Y,L for 20,000 random gene lists, having the same length as the dendritically enriched gene list, with the genes randomly selected from the background gene set. We denote the nth random gene list as Rn. The mean and standard deviation of the bootstrapped DX,Y,L values are denoted and respectively. The depletion Z-score is then calculated as: . A large positive Z-score thus indicates that dendritically enriched transcripts are specifically depleted from pyramidal neurons from dataset Y relative to dataset X.
Conditional cell type enrichments
Gene association Z-scores for schizophrenia were calculated in MAGMA as described above. To enable randomization of the Z-scores and recalculation of the associations to be done programmatically, these were then loaded into R and associations with disease were calculated within this environment without external calls to MAGMA. All genes within the extended MHC region (chr6 25–34 mb) were removed due to its confounding effects. We controlled for gene size and gene density by regressing out the effect of NSNPS and NDENSITY parameters (and the log of each) on the Z-score. To ensure a meaningful number of genes were randomized within each group, associations were calculated over deciles rather than the smaller percentile bins used earlier with MAGMA. Probabilities of association are calculated using the lmFit and ebayes functions from the limma package to enable rapid computation. We denote the set of cells studied as C such that ci represents the ith celltype. The original Z-scores are denoted Z such that zi is the Z-score of the ith gene while the randomized Z-scores are denoted R. The set of genes in the ith specificity decile of the controlled cell type, cx and the jth specificity decile of target cell type, cy are denoted and thus contains all genes in the ith specificity decile of cell type, cx.
The basis of the approach (Supplementary Figure 23) is to randomise the Z-scores with respect to the specificity deciles of the target cell type, cy but not with respect to the specificity deciles of the controlled cell type, cx. Thus for each of the deciles indexed by i we randomly resampled without replacement the Z-scores such that and yet Rg ≠ Zg. In practical terms, this would mean that if we controlled for MSN’s and targeted cortical interneurons, the mean Z-score in the 10th MSN decile would remain the same but would be different in cortical interneurons; the question being tested is the degree to which this equates to total randomisation in terms of the schizophrenia association found in cortical interneurons.
The baseline association values shown in Figure 4a leftmost column (described as PcelltypeY,baseline) were calculated using Z. The values of PcelltypeY,celltypeX (probability of cell type y being associated with schizophrenia controlling for cell type x) are calculated using intermediate probabilities: 10,000 association p-values are calculated for resampled values of R. We selected the 500th lowest of these p-values (equivalent to the value which the baseline association probability would need to exceed to be declared independently associated with a probability of 95%) and denote this . The value of PcelltypeY,celltypeX is then calculated as exp(log(PcelltypeY,celltypeX)-log()). If the value of PcelltypeY,celltypeX exceeds 1 (indicating that the randomised samples were actually more significantly associated than was found to be the case) then it is set to 1. We were also able to evaluate whether the probability of schizophrenia association in cell type y is greater than would be expected based solely on the expression in cell type x by asking whether the actual association p-value was lower than 95% of the bootstrapped p-values. As expected, all self-self comparisons were found to be non-significant by this metric (i.e. after accounting for expression in CA1 pyramidal neurons, CA1 pyramidal neurons are no longer significant). In Figure 4a, a red box was placed around the CA1 Pyramidal vs Somatosensory Pyramidal square because this was the only comparison involving the four significantly associated cell types in which controlling for expression of a different cell type abolished the enrichment.
Venn diagram enrichments
The Venn diagram shown in Figure 5 was generated using by selecting the top 1000 genes most associated with schizophrenia based on the MAGMA gene specific Z-scores. All genes within the extended MHC region (chr6 25–34mb) were dropped from the analysis. We controlled for gene size and gene density by regressing out the effect of NSNPS and NDENSITY parameters (and the log of each) on the Z-score. We then took the intersection of the top 1000 genes with the top decile for each of the four significantly associated level 1 cell types and generated the Venn diagram using the R VennDiagram package. The dopamine gene set include all genes associated with any of the following GO terms: GO:0090494 (“dopamine uptake”), GO:0090493 (“catecholamine uptake”), GO:0051584 (“regulation of dopamine uptake involved in synaptic transmission”), GO:0032225 (“regulation of synaptic transmission, dopaminergic”), GO:0001963 (“synaptic transmission, dopaminergic”) and GO:0015872 (“dopamine transport”). The synaptic gene list comprised a combination of three published gene lists: the human post-synaptic density (referenced above); presynaptic active vesicle docking sites63 and synaptic vesicle genes62. For the presynaptic gene list, the data came from Supplementary table 1, the geneInfo numbers were converted from genInfo accessions to Refseq IDs using Entrez Batch then from Rat RefSeq to HGNC symbols keeping only 1:1 homologs. The synaptic vesicle gene list came from Supplementary table 1, and were converted from Rat RefSeq to HGNC symbols using only 1:1 homologs. Enrichment probabilities were calculated using a hypergeometric test against a background set of all MGI genes with 1:1 homologs in human (as described above).
Supplementary Material
Acknowledgements
JHL was funded by the Swedish Research Council (Vetenskapsrådet, award 2014–3863), StratNeuro, the Wellcome Trust (108726/Z/15/Z), and the Swedish Brain Foundation (Hjärnfonden). PFS gratefully acknowledges support from the Swedish Research Council (Vetenskapsrådet, award D0886501). NS was supported by the Wellcome Trust (108726/Z/15/Z). JB was supported by the Swiss National Science Foundation. The PGC has received major funding from the US National Institute of Mental Health (U01 MH109528 and U01 MH109532). HG was supported by a Psychiatric Genomics Consortium grant (1U01MH109514–01). Primary schizophrenia GWAS data was generated with support from Medical Research Council (MRC) Centre (MR/L010305/1), Program Grant (G0800509) and Project Grant (MR/L011794/1), and funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement n° 279227 (CRESTAR Consortium).
Footnotes
Conflicts of Interest
The authors reports the following potentially competing financial interests. PF Sullivan: Lundbeck (advisory committee), Pfizer (Scientific Advisory Board member), and Roche (grant recipient, speaker reimbursement). J Hjerling-Leffler: Cartana (Scientific Adviser) and Roche (grant recipient).
URLS
Expression Weighted Cell type Enrichment (EWCE), https://github.com/NathanSkene/EWCE
Linnarsson lab data, http://linnarssonlab.org/data
Mouse Genome Informatics, Jackson Laboratory, http://www.informatics.jax.org/homology.shtml
LDSC, https://github.com/bulik/ldsc and https://github.com/bulik/ldsc/wiki
PGC results, https://www.med.unc.edu/pgc/results-and-downloads
AlzGene database, http://www.alzgene.org/TopResult.asp
AlsGene database, http://www.alsgene.org/TopResult.asp
GREAT, http://great.stanford.edu/public/html
Hjerling-Leffler lab website, http://www.hjerling-leffler-lab.org/data/scz_singlecell
Human Phenotype Ontology, http://compbio.charite.de/hpoweb
MAGMA_Celltyping, https://github.com/NathanSkene/MAGMA_Celltyping
Data availability
The RNAseq data used in this report can be obtained from the Hjerling-Leffler lab website (URLs), and includes the KI single-cell RNAseq superset, processed versions of the human and mouse snRNAseq DroNc-seq data, and the Allan Brain Institute human snRNAseq data. The specificity values for the KI single-cell RNAseq superset are included in Supplementary Table 4. The dataset has also been made available in the ‘MAGMA_Celltyping’ R package (URLs).
Code availability
An R package which can be used for running the cell type association analysis can be obtained from https://github.com/NathanSkene/MAGMA_Celltyping
References
- 1.Sullivan PF, Daly MJ & O’Donovan M Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews Genetics 13, 537–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purcell SM et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fromer M et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genovese G et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nature Neuroscience 19, 1433–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh T et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci 19, 571–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekar A et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CNV Working Group of the Psychiatric Genomics Consortium & Schizophrenia Working Groups of the Psychiatric Genomics Consortium. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49, 27–35 (2016).27869829 [Google Scholar]
- 9.Finucane HK et al. Partitioning heritability by functional category using GWAS summary statistics. Nature Genetics 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Exome Aggregation Consortium et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lips ES et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Molecular psychiatry 17, 996–1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell JC et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goudriaan A et al. Specific glial functions contribute to schizophrenia susceptibility. Schizophrenia bulletin 40, 925–935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromer M et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nature Neuroscience 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pers TH et al. Comprehensive analysis of schizophrenia-associated loci highlights ion channel pathways and biologically plausible candidate causal genes. Hum Mol Genet 25, 1247–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisel A et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–42 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Romanov RA et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci 20, 176–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Manno G et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580 e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Leeuw CA, Mooij JM, Heskes T & Posthuma D MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardiñas AF et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and maintained by background selection. Nature Genetics (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finucane H et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nature Genetics (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gokce O et al. Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep 16, 1126–37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib N et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasic B et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habib N et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelmoez MN et al. Correlation of gene expressions between nucleus and cytoplasm reflects single-cell physiology. bioRxiv, 206672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cajigas IJ et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darmanis S et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lake BB et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skene NG & Grant SG Identification of Vulnerable Cell Types in Major Brain Disorders Using Single Cell Transcriptomes and Expression Weighted Cell Type Enrichment. Front Neurosci 10, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaspar HA & Breen G Pathway analyses of schizophrenia GWAS focusing on known and novel drug targets. bioRxiv (2017). [Google Scholar]
- 33.Goudriaan A et al. Specific glial functions contribute to schizophrenia susceptibility. Schizophr Bull 40, 925–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anttila V et al. Analysis of shared heritability in common disorders of the brain. Science (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert JC et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45, 1452–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertram L, McQueen MB, Mullin K, Blacker D & Tanzi RE Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nature genetics 39, 17–23 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Patsopoulos N et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Robinson PN & Wang K Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nature methods 12, 841–843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burow DA et al. Dynamic regulation of mRNA decay during neural development. Neural Dev 10, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Psych EC et al. The PsychENCODE project. Nat Neurosci 18, 1707–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genomes Project, C. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Leeuw CA, Neale BM, Heskes T & Posthuma D The statistical properties of gene-set analysis. Nat Rev Genet 17, 353–64 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Brown MB A Method for Combining Non-Independent, One-Sided Tests of Significance. Biometrics 31, 987–992 (1975). [Google Scholar]
- 44.Lun AT, McCarthy DJ & Marioni JC A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vu TN et al. Beta-Poisson model for single-cell RNA-seq data analyses. Bioinformatics 32, 2128–2135 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature genetics 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sniekers S et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat Genet 49, 1107–1112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nalls MA et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nature genetics 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood AR et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46, 1173–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paternoster R, Brame R, Mazerolle P & Piquero A Using the correct statistical test for the equality of regression coefficients. Criminology 36, 859–866 (1998). [Google Scholar]
- 52.Wagnon JL et al. CELF4 regulates translation and local abundance of a vast set of mRNAs, including genes associated with regulation of synaptic function. PLoS Genet 8, e1003067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins MO et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem 97 Suppl 1, 16–23 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Bayes A et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci 14, 19–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez E et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol 5, 269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weyn-Vanhentenryck SM et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 6, 1139–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thul PJ et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Brozzi A, Urbanelli L, Luc Germain P, Magini A & Emiliani C hLGDB: a database of human lysosomal genes and their regulation. Database 2013, bat024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calvo SE, Clauser KR & Mootha VK MitoCarta2. 0: an updated inventory of mammalian mitochondrial proteins. Nucleic acids research 44, D1251–D1257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shigeoka T et al. Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyken J et al. Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron 78, 285–297 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Takamori S et al. Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Boyken J et al. Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron 78, 285–97 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.