Abstract
BACKGROUND: Ependymomas (EPNs) are the third most common brain tumor in children. These tumors are resistant to available chemotherapeutic treatments, therefore new effective targeted therapeutics must be identified. Increasing evidence shows epigenetic alterations including histone posttranslational modifications (PTMs), are associated with malignancy, chemotherapeutic resistance and prognosis for pediatric EPNs. In this study we examined histone PTMs in EPNs and identified potential targets to improve chemotherapeutic efficacy. METHODS: Global histone H3 lysine 4 trimethylation (H3K4me3) levels were detected in pediatric EPN tumor samples with immunohistochemistry and immunoblots. Candidate genes conferring therapeutic resistance were profiled in pediatric EPN tumor samples with micro-array. Promoter H3K4me3 was examined for two candidate genes, CCND1 and ERBB2, with chromatin-immunoprecipitation coupled with real-time PCR (ChIP-PCR). These methods and MTS assay were used to verify a relationship between H3K4me3 levels and CCND1 and ERBB2, and to investigate cell viability in response to chemotherapeutic drugs in primary cultured pediatric EPN cells. RESULTS: H3K4me3 levels positively correlate with WHO grade malignancy in pediatric EPNs and are associated with progression free survival in patients with posterior fossa group A EPNs (PF-EPN-A). Reduction of H3K4me3 by silencing its methyltransferase SETD1A, in primary cultured EPN cells increased cell response to chemotherapy. CONCLUSIONS: Our results support the development of a novel treatment that targets H3K4me3 to increase chemotherapeutic efficacy in pediatric PF-EPN-A tumors.
Abbreviations: EPN, ependymoma; PTM, posttranslational modification; CNS, central nervous system; EMEM, Eagle's Minimum Essential Medium; CIMP+, CpG island methylator positive; TSS, transcription start site; PFS, progression free survival; VCR, vincristine; CPL, carboplatin; IRB, institutional review board; MTS, 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; FFPE, formalin-fixed paraffin-embedded; ChIP-PCR, chromatin-immunoprecipitation coupled with real-time PCR
Introduction
Ependymomas (EPNs) account for 3–6% of all central nervous system (CNS) tumors, with the highest frequency in children and young adults [1]. These malignancies represent the third most common pediatric CNS tumor [1], with infratentorial presentation being the most frequent [2]. Surgical resection with adjuvant radiation is used to treat nearly all EPN patients [3]. Numerous chemotherapeutic agents have been added and tested in clinical trials [4], [5]. With the exception of a single study by Strother et al. [6] which showed prolonged dose-intensive chemotherapy including cyclophosphamide, vincristine, cis-platinum and etoposide extending event-free survival in infants, there has been no clear benefit of adding chemotherapy.
Effective EPN management remains a challenge [7], in part because EPNs derived from different anatomical sites in the CNS are molecularly and clinically distinct [8], with nine molecular subgroups indicated via DNA methylation profiling. Among these subgroups, supratentorial EPNs with YAP (ST-EPN-YAP) and RELA (ST-EPN-RELA) fusions are most common, and posterior fossa group A (PF-EPN-A) are almost exclusively found in children [9]. Cells that fulfill stem cell criteria have been isolated from EPNs, including in a study by Meco et al. [10], the results of which showed impaired EPN growth in response to temozolomide through preferential depletion of a cell population with stem-like properties. To date, studies on pediatric EPN have not assessed the role of histone posttranslational modifications (PTMs) in stem cell populations or therapeutic response.
Histone PTMs occur primarily on histone amino-terminal tails protruding from the surface of nucleosomes [11]. These modifications impact gene expression by altering higher-order chromatin structure and recruiting transcriptional regulators. Histone modifying enzymes are potential therapeutic targets for treatment of EPNs. An example to support this, is the sensitivity of CpG island methylator-positive (CIMP+) hindbrain EPNs to drugs that target enzymes responsible for the methylation of histone H3 lysine 27 trimethylation (H3K27me3) [12]. Global reduction of H3K27me3 is exhibited in a subset of childhood posterior fossa EPNs and this reduction is a powerful outcome predictor [13]. H3K27me3 and H3K4me3 are bivalent markers in ~20% of the human genome [14]. To date, H3K4me3 has not been investigated in pediatric EPNs.
H3K4me3 is enriched near transcription start sites (TSS) of actively expressed genes. Levels of this mark have been linked to pathological changes in a variety of brain tumors including medulloblastoma [15], and glioma [16]. Such findings indicate that enzymes regulating H3K4me3 are potential therapeutic targets. In mammalian cells, six SET1-COMPASS complexes are responsible for H3K4 methylation, including human SETD1A (hSETD1A) [17]. Ablation of hSETD1A in breast cancer cells leads to a decrease in migration and invasiveness in vitro, and to a decrease in metastasis in nude mice through decreased H3K4me3 at the promoters of matrix metalloproteinase genes [18]. Furthermore, hSETD1A is recruited for H3K4me3 at the promoters of Wnt/β-catenin target genes, activation of which promotes growth in human colorectal cancer [19]. The relevance of hSETD1A as well as H3K4me3 and therapeutic response should be investigated in pediatric EPNs.
In this study, H3K4me3 and its role in regulating therapeutic resistance were investigated in pediatric EPNs. We found H3K4me3 positively correlates with WHO grade malignancy in pediatric EPNs, with lower levels associated with progression free survival (PFS) in patients with PF-EPN-A tumors. H3K4me3 and hSETD1A were upregulated in high-grade EPNs, with H3K4me3 enriched at promoters of genes, i.e. CCND1 and ERBB2, linked to therapeutic resistance and tumor malignancy. Reduction of H3K4me3 by downregulating hSETD1A decreased H3K4me3 promoter occupancy and gene expression and increased pediatric EPN cell response to chemotherapy. Our data suggest that H3K4me3 status is an important determinant of EPN therapeutic response and that hSETD1A, which regulates H3K4me3, is a promising target to increase therapeutic efficacy and improve prognosis for children with PF-EPN-A tumors.
Methods
Sample Information
Fifty formalin-fixed, paraffin-embedded (FFPE) primary pediatric EPN samples were used for immunohistochemical analysis. Samples were collected from patients diagnosed in the Department of Pathology, Ann & Robert H. Lurie Children's Hospital of Chicago (A&RHLCH, Chicago, IL) under IRB# 2005–12,252. Clinicopathological information is summarized in Table 1. Patients did not receive pre-treatment.
Table 1.
Clinicopathological information for pediatric primary EPNs IHC stained for H3K4me3.
| Variable | Category | N (%) | H3K4me3 IHC score |
P value |
||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||||
| Gender | Male | 27(54.0) | 1(25.0) | 3(60.0) | 6(42.9) | 11(61.1) | 6(66.7) | .5799 |
| Female | 23(46.0) | 3(75.0) | 2(40.0) | 8(57.1) | 7(38.9) | 3(33.3) | ||
| Age | <=3 y | 21(42.0) | 0 (0.0) | 4(80.0) | 5(35.7) | 7(38.9) | 5(55.6) | .1671 |
| >3 y | 29(58.0) | 4(100.0) | 1(20.0) | 9(64.3) | 11(61.1) | 4(44.4) | ||
| Tumor location | Supratentorial | 13(26.0) | 0(0.0) | 0(0.0) | 2(14.3) | 5(27.8) | 6(66.7) | .0022 |
| Infratentorial | 29(58.0) | 1(25.0) | 4(80.0) | 9(64.3) | 13(72.2) | 2(22.2) | ||
| Spine | 8(16.0) | 3(75.0) | 1(20.0) | 3(21.4) | 0(0.0) | 1(11.1) | ||
| Superatentorial | RELA Fusion+ | 6(46.2) | 0(0) | 0(0) | 1(33.3) | 2(40.0) | 3(60.0) | .6115 |
| RELA Fusion - | 7(53.8) | 0(0) | 0(0) | 2(66.7) | 3(60.0) | 2(40.0) | ||
| Posterior fossa | Group A | 24(82.8) | 1(100.0) | 3(100.0) | 7(87.5) | 10(76.9) | 3(75.0) | .2860 |
| Group B | 5(17.2) | 0(0) | 0(0) | 1(12.5) | 3(23.1) | 1(25.0) | ||
| WHO grade | I | 7(14.0) | 3(75.0) | 0(0.0) | 4(28.6) | 0(0.0) | 0(0.0) | <.0001 |
| II | 12(24.0) | 1(25.0) | 5(100.0) | 4(28.6) | 2(11.1) | 0(0.0) | ||
| III | 31(62.0) | 0(0.0) | 0(0.0) | 6(42.8) | 16(88.9) | 9(100.0) | ||
P value was calculated by Fisher's Exact test.
Twenty-two primary EPN tumor samples were used to obtain illumina microarray gene expression profiles. These samples were retrieved from the Falk Brain Tumor Tissue Bank at the Division of Pediatric Neurosurgery, A&RHLCH under IRB#s 2005–12252 and 2005–12,692. Control micro-dissected tissue was taken from areas surrounding the lateral and fourth ventricle, regions enriched in ependymal cells, from brain autopsies of 3 patients who died from non-brain-related diseases. Clinical pathology for these patients is summarized in Supplementary Table 1.
Ten fresh EPN samples (Supplementary Table 2) were obtained at the time of surgery and processed for primary cell culture within 30 min in accordance with protocols (IRB#s 2005–12252 and 2014–15,907 approved by institutional review boards at A&RLCH). These samples were used for primary cell culture, total and/or histone protein extraction and chromatin-immunoprecipitation.
Immunohistochemistry (IHC)
All tumors samples were reviewed by a senior pediatric pathologist (N.W), using World Health Organization 2007 criteria for tumor classification. IHC was performed on FFPE slides using antibodies against L1CAM (EMD Millipore, #ABT143, 1:100), to identify superatentorial (ST-EPN), H3K27me3 (Cell signaling Technology, #9733, 1:200), to identify posterior fossa (PF-EPN) EPNs, and H3K4me3 (Cell signaling Technology, #9727, 1:200) for all samples as per manufacturer's instructions. Images were captured on a Leica DMR-HC upright microscope (Leica Microsystem Inc., Buffalo Groove, IL, USA) and analyzed using OpenLab 5.0 software (PerkinElmer, Waltham, MA, USA). H3K4me3 positive staining was graded semi-quantitatively on a five-tier scale: 0 < 10%, 1 + =10–25%, 2 + =25–50%, 3 + =50–75%, 4 + = > 75% for positive tumor cell nuclei as described [20].
Primary Cell Culture
Primary cultured pediatric EPN cells, designated “LCH” were obtained during surgery under IRB# 2005–12,692 and cultured in vitro as described [21]. Briefly, single-cell suspensions were prepared by mincing tissue in HBSS, followed by dissociation with Accutase (Thermo Fisher Scientific, cat#A1110501) for ~10 min at 37 °C. The resulting suspension was added to fresh complete culture medium, filtered with a 40 μm cell strainer (BD Falcon, Cat#352340), and centrifuged at 500 g for 5 min. Supernatant was removed and the cell pellet was washed twice with HBSS prior to resuspension in Eagle's Minimum Essential Medium (EMEM) supplemented with 10% FBS, and incubated at 37 °C in humidified 5% CO2. Attached cells were passaged when confluent, with 0.05% trypsin in PBS containing 1 mM EDTA, and placed into culture medium, which included 10% conditioned medium from the previous passage. Using this method, pediatric EPN cell lines LCH-01-Ep, LCH-10-Ep and LCH-14-Ep were established in culture and for assay at early passages (<10 passages). Pediatric EPN primary cultured cell line 13–02-PBT was acquired from Children's Hospital of Los Angeles and cultured as described [22]. Pediatric EPN cell lines SF7183 and SF8070 were developed and contributed by Dr. Rintaro Hashizume. The EPN cells were obtained from surgical biopsies of tumors from patients admitted to the UCSF medical center, in accordance with an institution approved protocol. SF7183 was derived from a 1-year old female, diagnosed with posterior fossa anaplastic EPN (WHO grade III), and SF8070 was derived from a 4-year-old female diagnosed with posterior fossa EPN (WHO grade II).
Constructs and Transfections
Human His-tagged SETD1A expression plasmid, pET28-SETD1A-MHL, and its control pET28-MHL were gifts from Cheryl Arrowsmith (Addgene plasmid # 32868 and #26096, respectively). Transfection reagent TurboFectin 8.0 (Cat# TF81001) for cDNA cloning, short interference RNA (siRNA) against human SETD1A (Gene ID 9739, Cat# SR306505) and siTran 1.0 (Cat# TT300001) for siRNA transfection were purchased from Origene (Rockville, MD, USA). For gene knock-down experiments: primary cultured cells were transfected with 60 nM SETD1A and control siRNAs using siTran 1.0 as per manufacturer's instructions. Transfected cells were subjected to cell viability assays in response to chemotherapeutic reagents or lysed after 48 h for real-time PCR and western blot analysis.
RNA Extraction
Freshly collected tissue samples, approximately 0.5 cm3, and primary cultured cells were used for RNA extraction. Total RNA was extracted from snap frozen tissue using a standard Trizol protocol (Invitrogen, Carlsbad, CA, 15596–026) and from cells with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) as per manufacturer's instructions. For FFPE samples, total RNA was isolated from five to twenty 10 μm-thick tissue sections using the Ambion RecoverAll kit (Ambion/Applied Biosystems, Foster City, CA) according to manufacturer's instructions. Total RNA quantity and quality were evaluated using Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, USA) and agarose gel electrophoresis prior to real-time PCR using primers shown in Supplementary Table 3.
Illumina Microarray Hybridization for Gene Expression Profiles and Data Analysis
See Supplementary Information. Pathway analysis was performed using the PANTHER pathway as described. [23]
Total Nuclear and Histone Protein Extraction and Immunoblotting
Total proteins were extracted with Tissue Extraction Buffer I (Life Technologies, cat# FNN0071) with proteinase (Cell Signaling Technology, Beverly, MA, USA), phosphatase (Sigma) inhibitor cocktails and phenylmethylsulfonyl fluoride (PMSF, Roche). Total histone was extracted using histone extraction kit (ab113476, Abcam) as per manufacturer's instructions. Protein concentrations were quantified with the BCA Protein Assay Kit (Thermo Fisher Scientific Inc.) with Nanodrop ND-1000 (Thermo Fisher Scientific Inc.). Equal amounts of cell lysate were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). Blocking was performed for 60 min with 5% nonfat dry milk in Tris-buffered saline and Tween 20, followed by blotting with primary antibodies overnight at 4 °C. Primary antibodies included: polyclonal anti-rabbit ERBB2 (Cat#2165, 1:1000), H3K4me3 (Cat#9727, 1:1000), H3 (Cat#9715, 1:2500), and Cyclin D1 (Cat#2922, 1:1000) Cell Signaling Technology; β-actin (ab8227,1:3000) from Abcam; rabbit polyclonal anti-GAPDH (sc-25,778, 1:2000) from Santa Cruz Biotechnology; and anti-human SETD1A (A300-289A, 1:1000) from Bethyl Laboratories. After washing with Tris-buffered saline and Tween 20, membranes were incubated for 1 h at room temperature with horseradish peroxidase conjugated donkey anti-rabbit antibody (sc-2305, 1:5000) or donkey anti-mouse antibody (sc-2306, 1:5000), and signal was detected with enhanced chemiluminescence substrate (Bio-Rad).
Chromatin-Immunoprecipitation (ChIP) and Real-Time PCR
ChIP using tissue was performed according to the manufacturer's instructions (Abcam). Briefly, ~20 mg frozen EPN tissue was minced into 1–3 mm3 pieces, thawed on ice, washed with ice cold PBS with proteinase inhibitor cocktail prior to fixation in 1% formaldehyde in PBS at room temperature for 10 min and quenched with 0.125 M glycine for 5 min. Tissue was collected and resuspended in 500 μl FA-lysis buffer (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL pepstatin) and disrupted while on ice, with a Wheaton Dounce Tissue Grinder (VWR International, 62,400–595). The tissue suspension was transferred into a 5 ml conical tube, sonicated and processed according to manufacturer's protocol using rabbit polyclonal H3K4me3 antibody (Cell Signaling Technology, Cat# 9727) and IgG from rabbit serum (Cell Signaling Technology, Cat# 2729) to collect immunoprecipitated DNA fragments. All DNA fragments were cleaned-up with GenElute PCR clean-Up Kit (Sigma Aldrich Co., St. Louis, MO, USA) prior to PCR analysis. ChIP using cells was performed with the SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic beads) (Cell Signaling Technology, Cat#9005) following manufacturer's instructions. Human ERBB2 and Cyclin D1 ChIP primers (Supplementary Table 3) were used following real-time PCR analysis.
MTS Assay for Assessing Chemosensitivity
Primary EPN cell lines, 13–02-PBT, LCH-14-pEP, SF8070 and LCH-01-pEP, LCH-10-pEP and SF7183, were used to determine if elevated or reduced levels of H3K4me3 affected cell viability in response to chemotherapeutic drugs, vincristine (VCR) and carboplatin (CPL). H3K4me3 was increased or globally reduced by increasing or decreasing its specific methyltransferase, hSETD1A, via transfection with SETD1A expression plasmid, pET28-SETD1A-MHL, or 60 nM small interfering SETD1A RNA (siSETD1A); control cells were transfected with pET28-MHL or siCtrl RNA. Cells were harvested 48 h after treatment. 2 × 104 cells/100 μl were plated in 96-well plates and incubated at 37 °C with 5% CO2 overnight. The cells were washed with serum free media, treated with culture media containing CPL (10−2 ~ 10−7 M) or VCR (10−5~10−10 M), and subjected to 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS, Promega) assay after 72 h. Cells not treated with CPL or VCR were used as normalization controls. Results were read on an ELISA Reader from TECAN Sunrise (TECAN, CA, USA). Each drug treatment was repeated three times in triplicate wells. Cell survival is presented as a percentage of viable cells compared to the viable cell number in the corresponding normalization control, with the control set as 100%. P values were calculated using Student's t test, with P < .05 considered significant. Statistical tests were 2-sided. Statistical analysis was carried out using GraphPad Prism 7 (GraphPad Software, Inc. La Jolla, CA, USA).
Results
H3K4me3 Levels Increase with Histopathological Malignancy in Pediatric EPNs
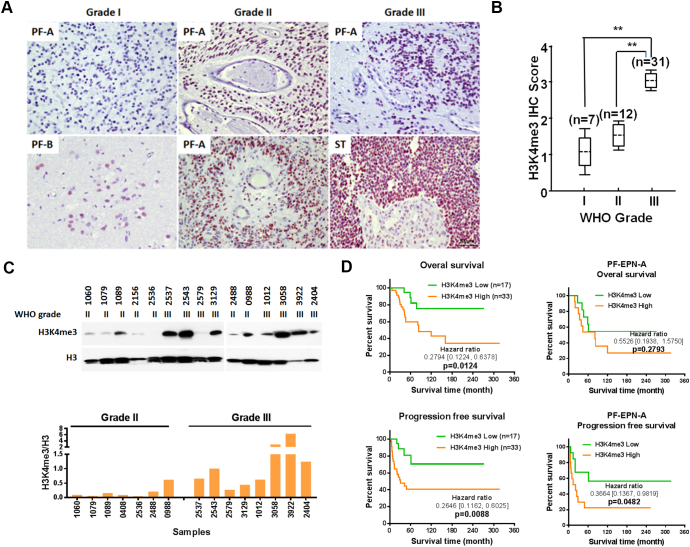
To investigate whether a correlation exists between clinicopathologic variables and H3K4me3, fifty primary FFPE specimens were IHC stained for H3K4me3 (Table 1). H3K4me3 was predominantly observed in cell nuclei with the frequency of immunopositive cells ranging from 0% to 100%. Patient age and gender were not associated with H3K4me3 levels. IHC results showed H3K4me3 significantly increased in WHO grade III EPNs (Figure 1, A and B, Supplementary Figure 1), relative to lower grade tumors, and a relation between IHC staining and tumor location was also indicated (P < .0001 and P = .0022, respectively). Western blots of protein extracts from tissue samples confirmed higher levels of H3K4me3 in WHO grade III versus grade II tumors (Figure 1C). Importantly, multivariate Cox proportional hazards analysis showed that overall survival (OS) for all patients and PFS in all (OS P = .0124; PFS P = .0088) PF-EPN-A (PFS P = .0482) patients were significantly shorter for patients with high levels of H3K4me3 (IHC scores ≥3) relative to patients with low levels of H3K4me3 (IHC scores ≤2) (Figure 1D, Supplementary Table 4). These results support H3K4me3 IHC staining as an accurate predictor of pediatric EPN malignancy especially in PF-EPN-A tumors.
Figure 1.
H3K4me3 is associated with malignancy in pediatric ependymomas. A, Representative images show H3K4me3 positive cells increase with WHO grade. B, Scores based on IHC for H3K4me3 positive cells. C, Western blots show H3K4me3 levels in WHO grade II and III ependymoma tissue, with total H3 as a control (top panel). The ratio of H3K4me3/H3 band intensity is indicated in the bottom panel. D, Overall survival and progression free survival for patients with low (IHC scores ≤2) and high (IHC scores ≥3) H3K4me3 IHC scores and for patients with posterior fossa group A ependymomas (PF-EPN-A).
Ependymoma Gene Expression Analysis
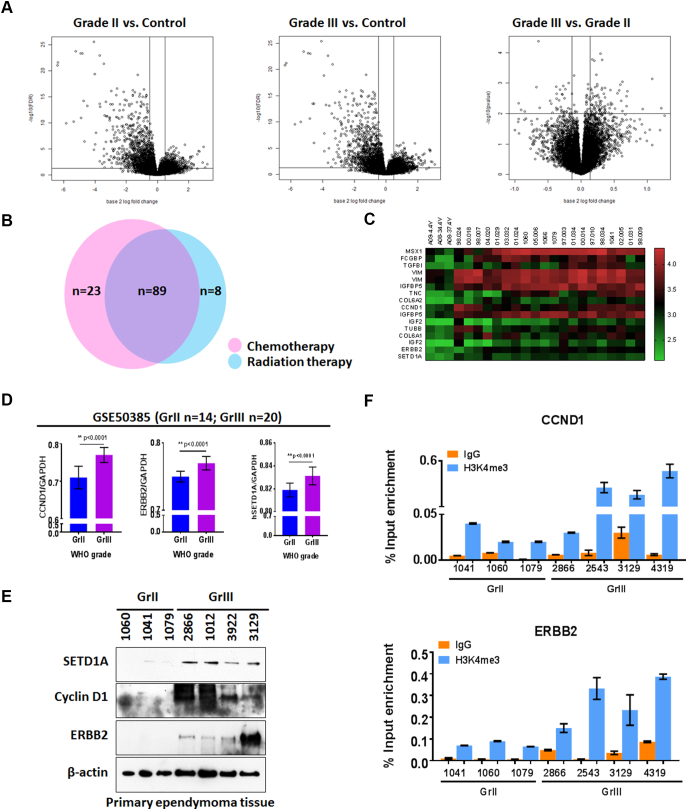
Comparison of 20 pediatric primary EPNs with 3 controls (Supplementary Table 1) showed 177 significant differences in gene expression between tumors with different WHO grades and control tissue (Figure 2A, Supplementary Figure 2, Fold >2, P < .05). To examine molecular interactions and reaction networks of these genes, we analyzed the data using the PANTHER database (Supplementary Figure 3). Pediatric EPNs showed significant enrichment of gene pathways that regulate developmental, cell differentiation and cell cycle processes. Detailed information on these signaling pathways is described in Supplementary file 1. We further noted that in human cancers, 23 of the differentially expressed genes are potentially associated with chemotherapeutic resistance, 8 with radiation resistance and 89 with both chemotherapeutic and radiation resistance (Figure 2B). Of these, 12 genes were further examined for a potential contribution to chemotherapeutic resistance (Figure 2C, Table 2). Although the expression differential for ERBB2 did not achieve our cutoff value of 4, for elevated gene expression in tumor tissues, we included it in the analysis which follows because of the frequency at which others have noted ERBB2 involvement with EPN high-grade malignancy [24], [25] and enrichment of H3K4me3 at its' promoter in human breast cancer tissue [26]. The H3K4me3 methyltransferase, SETD1A, was also examined (Table 2). Tumor samples from GSE50385 reanalyzed with GEO2R (Supplementary Table 5), were used to validate expression of these candidate genes in pediatric primary EPNs. Analysis results showed ERBB2, CCND1 and SETD1A highly overexpressed in grade III, compared to grade II primary pediatric EPNs (Figure 2D). The results from our in silico analysis are consistent with immunoblot results of protein extracts from patient tumor tissues (Figure 2E).
Figure 2.
Expression profiles of genes potentially involved in therapeutic resistance in pediatric primary EPN. A, Volcano plots showing differential gene expression profiles of 20 pediatric primary EPNs, 9 WHO grade II and 11 grade III tumors and 3 controls. B, Venn-diagram illustrating the number of genes associated with chemotherapeutic resistance (pink), radiation resistance (blue) or both chemotherapeutic and radiation resistance. C, Heat map highlighting 12 candidate overexpressed genes (fold change >4), associated with both chemotherapeutic and radiation resistance. D, Genes significantly associated with chemotherapeutic and radiation resistance, and identified through in silico analysis of dataset GSE50385. E, Immunoblots showing protein expression of cyclin D1 and ERBB2 in pediatric primary EPN tissue samples. F, CCND1 and ERBB2 expression shown with real-time PCR, after H3K4me3 chromatin immunoprecipitation of DNA from pediatric EPN tissues.
Table 2.
Candidate genes potentially associated with chemo- and radio-therapeutic resistance and histone H3K4me3 modification, identified by comparison of EPN samples with normal tissue from four-ventricular regions.
| ILMN probe ID | Gene symbol | Gene description | Fold change (Tumor vs Con) |
Diff P value (Tumor vs Con) | Chromosomal location |
|---|---|---|---|---|---|
| ILMN_1777397 | MSX1 | msh homeobox 1 | 8.40368379 | 0.001068928 | 4p16.2a |
| ILMN_2302757 | FCGBP | Fc fragment of IgG binding protein | 7.51850685 | 0.000212757 | 19q13.2b |
| ILMN_1663866 | TGFBI | transforming growth factor, beta-induced, 68 kDa | 7.18614540 | 0.000172186 | 5q31.1f-q31.2a |
| ILMN_1782538 | VIM | Vimentin | 6.90619675 | 3.86E-07 | 10p12.33c |
| ILMN_2058251 | VIM | Vimentin | 6.89964492 | 7.92E-07 | 10p12.33c |
| ILMN_1750324 | IGFBP5 | insulin-like growth factor binding protein 5 | 6.67755569 | 3.25E-07 | 2q35c |
| ILMN_1719759 | TNC | tenascin C | 6.56274410 | 0.008055601 | 9q33.1a |
| ILMN_1809928 | COL6A2 | collagen, type VI, alpha 2 | 5.40932324 | 2.59E-06 | 21q22.3f |
| ILMN_1688480 | CCND1 | cyclin D1 | 5.02124412 | 0.000523028 | 11q13.2c |
| ILMN_2132982 | IGFBP5 | insulin-like growth factor binding protein 5 | 4.96717083 | 5.37E-06 | 2q35c |
| ILMN_2413956 | IGF2 | insulin-like growth factor 2 (somatomedin A) | 4.90451748 | 0.001234166 | 11p15.5 |
| ILMN_1665583 | TUBB | tubulin, beta class I | 4.61522131 | 0.000125164 | 6p21.33b |
| ILMN_1732151 | COL6A1 | collagen, type VI, alpha 1 | 4.59061663 | 8.56E-05 | 21q22.3f |
| ILMN_1699867 | IGF2 | insulin-like growth factor 2 (somatomedin A) | 4.53155504 | 0.001128204 | 11p15.5 |
| ILMN_2352131 | ERBB2 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 3.18843308 | 0.000528173 | 17q12c |
| ILMN_1714327 | SETD1A | Homo sapiens SET domain containing 1A (SETD1A), mRNA. | 4.039779 | 0.0002234 | 16p11.2c |
H3K4me3 Linked to Expression of Genes that Confer Therapeutic Resistance
H3K4me3 is located at high levels at promoter regions near the TSS of virtually all active genes, and a strong positive correlation exists between this histone modification, transcription rates, and active polymerase II promoter occupancy [27]. To investigate levels of H3K4me3 at CCND1 and ERBB2 promoters, ChIP was conducted with rabbit H3K4me3 antibody prior to real-time PCR of immunoprecipitated DNA. The results showed that H3K4me3 at CCND1 and ERBB2 promoters', after normalization using input percentage, was higher in grade III compared to grade II EPNs (Figure 2F). This indicates H3K4me3 enrichment at key gene promoters and suggests a potential mechanism for increased expression of genes associated with therapeutic resistance.
H3K4me3 Promoter Occupancy at ERBB2 and CCND1 is Associated with Chemosensitivity
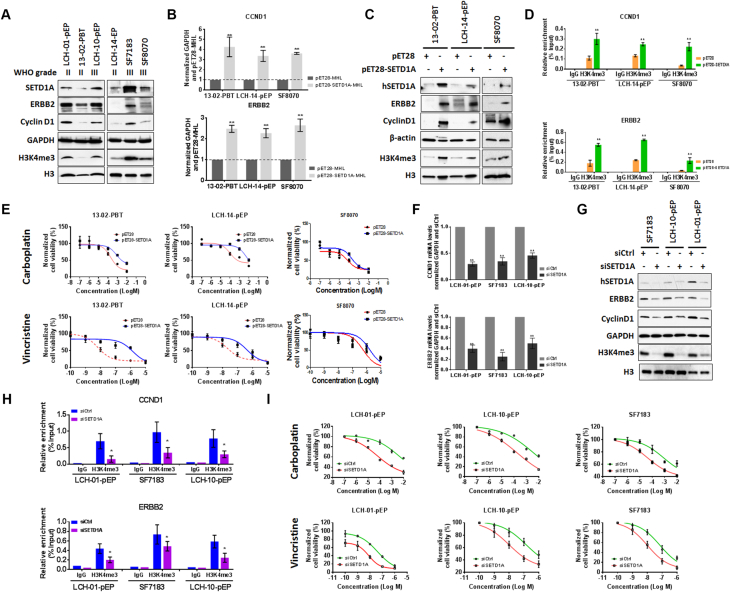
To validate the association between H3K4me3 and ERBB2 and CCND1 expression, transcript and H3K4me3 ChIP levels were examined in six primary pediatric EPN cell lines. Human SETD1A, a major methyltransferase for H3K4me3, was also investigated due to its elevated expression in WHO grade III EPNs, in comparison to grade II (Figure 2, D and E, Table 2). The results showed higher ERBB2, Cyclin D1, SETD1A and H3K4me3 in three of the six cell lines, LCH-01-pEP and LCH-10-pEP and SF7183, and lower levels in 13–02-PBT, LCH-14-pEP and SF8070 (Figure 3A). The relationship between SETD1A and ERBB2 and CCND1 expression was further investigated by treating 13–02-PBT, LCH-14-pEP and SF8070 cells with pET28-SETD1A-MHL; or LCH-01-pEP, LCH-10-pEP and SF7183 cells with small interfering SETD1A (siSETD1A) RNA. Real-time PCR and western blots showed that SETD1A elevation or knock-down increased or decreased ERBB2 and CCND1 transcript (Figure 3, B and F) and protein levels (Figure 3, C and G), relative to control pET28-MHL or siRNA, respectively. H3K4me3 also increased or decreased (Figure 3, C and G). Given that hSETD1A doesn't directly interact with promoters of target genes [28], the results indicate that SETD1A regulates expression of ERBB2 and CCND1 via adjusting H3K4me3 promoter occupancy. To confirm this, chromatin was immunoprecipitated with H3K4me3 from cells treated as above. PCR using primers for the promoters of these two genes, showed significantly increased or reduced H3K4me3 promoter occupancy in cells treated with pET28-SETd1A-MHL (Figure 3D) and siSETD1A RNA (Figure 3H), respectively. Altogether, these results demonstrate that ERBB2 and CCND1 expression is influenced by promoter H3K4me3 levels.
Figure 3.
Reduction of H3K4me3 via down-regulation of hSETD1A decreases ERBB2 and CCND1 expression and increases chemosensitivity. A, Western blots showing hSETD1A, ERBB2, CCND1 and H3K4me3 in pediatric primary cultured cell lines. B and C, Real-time PCR (B) and western blots (C) show increased ERBB2 and CCND1 expression and H3K4me3 levels in pediatric primary cultured cells following treatment with pET28-SETD1A-MHL (pET-SETD1A), compared to pET28-MHL (pET28) control. D, Chromatin immunoprecipitation with H3K4me3 IgG antibodies coupled with real-time PCR (ChIP-PCR) using CCND1 and ERBB2 promoter primers in pediatric primary cultured EPN cells following treatment with pET-SETD1A, compared to pET28. E, MTS assays showing cell viability in response to CPL or VCR following treatment with pET-SETD1A or pET28 in pediatric primary EPN cells. F and G, Real-time PCR (F) and western blots (G) show decreased ERBB2 and CCND1 expression and H3K4me3 levels in pediatric primary cultured cells following treatment with siSETD1A (pET-SETD1A), compared to control siRNA (siCtrl). D, Chromatin immunoprecipitation with H3K4me3 IgG antibodies coupled with real-time PCR (ChIP-PCR) using CCND1 and ERBB2 promoter primers in pediatric primary cultured EPN cells following treatment with siSETD1A and siCtrl. I, MTS assays showing cell viability in response to CPL or VCR following treatment with siCtrl or siSETD1A in pediatric primary EPN cells. Error bars show the standard deviation of three independent experiments. * P < .05.
ERBB2 [29] and CCND1 [30] are associated with chemotherapeutic sensitivity in human cancers and H3K4me3 promoter occupancy positively correlates with their expression in human gastric and breast cancers [26], [31], [32], and as shown here with pediatric primary EPN cells. Given this relationship, reducing H3K4me3 should decrease ERBB2 and CCND1 expression and increase tumor cell chemosensitivity. To investigate this, 13–02-PBT, LCH-14-pEP and SF8070 cells were treated with pET28-SETD1A-MHL or pET28-MHL, and LCH-01-pEP, LCH-10-pEP and SF7183 cells with treated with siSETD1A or siCtrl RNA. Treated cells were then exposed to chemotherapeutic drugs CPL or VCR. Cell viability significantly increased in 13–02-PBT, LCH-14-pEP and SF8070 cells with SETD1A upregulation (Figure 3E), while viability decreased in LCH-01-pEP, LCH-10-pEP and SF7183 cells following knockdown with siSETD1A, in comparison to siCtrl (Figure 3I). These results demonstrate that H3K4me3 affects cytotoxic therapeutic efficacy in pediatric EPN cells.
Discussion
Higher order chromatin structure is an important factor in the regulation of gene expression [33], [34]. Histones are key components of chromatin, and are subject to a wide variety of dynamic PTMs [35]. Recent studies have shown differential histone modification in adult and pediatric brain tumors [36], [37], [38], including pediatric EPN [12], [13], [37], in relation to normal brain. In this study, global H3K4me3 levels directly correlate with pediatric EPN histopathologic malignancy, and inversely correlate with clinical prognosis. Furthermore, we show that global H3K4me3 levels influence therapeutic efficacy, in association with regulation of genes linked with therapeutic resistance, such as CCND1 and ERBB2, both validated EPN oncogenes [8], [24], [38], [39]. Therefore, a treatment regimen that reduces global H3K4me3 in combination with traditional chemotherapeutics may increase therapeutic efficacy and improve outcomes for children who present with EPN.
Over the last decade, the number of molecular drivers, identified in pediatric EPNs has significantly increased, with individual tumors in some cases exhibiting molecular heterogeneity in driver expression. Some of these drivers are associated with clinical outcome [40]. For instance, for pediatric posterior fossa EPN [9], [41], PF-EPN-A exhibit a heterogeneous picture of pathway alterations including several canonical cancer-associated signaling pathways such as HIF-1a, VEGF, EGFR, PDGF, TGF-β, tyrosine-receptor kinase, RAS etc., this heterogeneity has also been noted in our study with PANTHER database analysis.
Histone PTMs have also been associated with EPN prognosis. Sub-EPNs with higher H3K9Ac have a lower probability of recurrence and show less proliferation [37]. High H3K4me3, as shown here, correlates with WHO grade malignancy (Figure 1, A–C) and worse prognosis in PF-EPN-A tumors (Figure 1D) in which H3K27me3 is low or missing. PF-EPN-A tumors are more common in young children, and relatively aggressive [42], [43]. The current recommended treatment for these tumors is localized adjuvant radiation following total resection. Understanding the changes in histone PTMs in these tumors may lead to new more effective treatment options.
Future studies need to further examine the role of histone PTMs in tumorigenesis and cell proliferation in each subgroup of EPNs. For example, bivalent chromatin domains containing H3K4me3 and H3K27me3 silence developmental genes, while simultaneously keeping them poised for activation following differentiation [14]. These same domains may influence subsequent development of drug resistance and tumor progression in human cancers [45]. Chromatin domains containing these two marks, as well their relationship with other histone marks, such as H3K9Ac [37], must be examined in combination, rather than in isolation. Results for PTMs should also be related to isochromosome 1q status [44], [46], another prognostic EPN marker, and to molecular subgroups [47]. The majority of samples in this study were collected and archived over 20 years, limiting our ability to determine the relationship between H3K4me3, subgroups and 1q status. Future use of fresh tumor samples will overcome this problem.
Enzymes that regulate histone PTMs are also of interest as therapeutic targets and prognostic markers. Several enzymes regulate H3K4 methylation status. Six SET1-COMPASS methyltransferase complexes methylate H3K4, including human SETD1A/B, which specifically regulates trimethylation at the promoter region near the TSS. Jumonji (JmjC) domain-containing histone demethylases, in particular, the JARID1 family of histone demethylases (JARID1A − D) remove methyl groups from tri- and di- methylated H3K4 [14]. Methylases and demethylases are widely expressed and have unique functions in vivo that, when altered, can contribute to disease initiation and progression. WDR82, a subunit of SETD1A/B complexes, increases following chemotherapy [45], suggesting WDR82-SETD1A/B complex activity contributes to chemotherapeutic resistance. In this study human SETD1A was shown to be highly expressed in WHO grade III EPNs (Figure 2, D and E, Table 2). Elevation of SETD1A increased H3K4me3 promoter frequency at ERBB2 and CCND1 genes (Figure 3D), while down-regulation of SETD1A reduced H3K4me3 occupancy at ERBB2 and CCND1 promoters (Figure 3H), two genes that influence tumor cell response to chemotherapy and affect EPN cell response to chemotherapeutic drugs, CPL and VCR (Figure 3, E and I). Taken together, these results demonstrate that H3K4me3 alterations mediated by hSETD1A influence multiple cancer cell characteristics including chemosensitivity, and that SETD1A warrants consideration as a target for cancer treatment. SETD1A has been shown to act through several pathways. It modulates cell cycle progression through induction of a miRNA network to suppress P53 in human cancers independent of H3K4me3 [48], [49]. It also has a non-catalytic domain, “FLOS” (functional location on SETD1A), which interacts with CYCLIN K and regulates DNA damage response gene expression [50]. In the work presented in this study we hypothesize that the response is primarily through direct alteration of H3K4me3.
Conclusions
In conclusion our results added to the literature [12], [13] support a critical role for histone PTMs in influencing EPN chemotherapeutic sensitivity. Future studies should focus on investigating relationships among distinct EPN molecular variants and histone PTMs in defining their characteristic gene expression profiles.
Acknowledgments
Acknowledgements
We would like to extend our appreciation to Dr. Craig M. Horbinski for his critical reading of this manuscript and insightful comments. We would also like to thank Drs. Amanda M Saratisis, Tord D Alden, Robin M Bowman and Arthur J. DiPatri for collecting fresh tumor tissue and the histology laboratory at Ann & Robert H. Lurie Children’s Hospital for preparing the archived tissue and tissue slides.
Competing interests
The authors declare no competing interest.
Footnotes
Funding: This project was supported, in part, by the National Cancer Institute SPORE grant (P50CA221747-01A1) CAREER award (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health); and the Rory David Deutsch Foundation, the Surgical Neuro-Oncology Research Fund of Ann & Robert H. Lurie Children's Hospital (A&RLCH) of Chicago, and the Dr. Ralph and Marian C. Falk Medical Research Trust.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.03.012.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Ostrom QT, Gittleman H, de Blank PM, Finlay JL, Gurney JG, McKean-Cowdin R, Stearns DS, Wolff JE, Liu M, Wolinsky YL. American Brain Tumor Association Adolescent and Young Adult Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro-Oncology. 2016;18(Suppl. 1):i1–i50. doi: 10.1093/neuonc/nov297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilday JP, Rahman R, Dyer S, Ridley L, Lowe J, Coyle B, Grundy R. Pediatric ependymoma: biological perspectives. Mol Cancer Res. 2009;7(6):765–786. doi: 10.1158/1541-7786.MCR-08-0584. [DOI] [PubMed] [Google Scholar]
- 3.Massimino M, Miceli R, Giangaspero F, Boschetti L, Modena P, Antonelli M, Ferroli P, Bertin D, Pecori E, Valentini L. Final results of the second prospective AIEOP protocol for pediatric intracranial ependymoma. Neuro-Oncology. 2016;18(10):1451–1460. doi: 10.1093/neuonc/now108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouffet E, Foreman N. Chemotherapy for intracranial ependymomas. Childs Nerv Syst. 1999;15(10):563–570. doi: 10.1007/s003810050544. [DOI] [PubMed] [Google Scholar]
- 5.Bouffet E, Hawkins CE, Ballourah W, Taylor MD, Bartels UK, Schoenhoff N, Tsanqaris E, Huang A, Kulkarni A, Mabbot DJ. Survival benefit for pediatric patients with recurrent ependymoma treated with reirradiation. Int J Radiat Oncol Biol Phys. 2012;83(5):1541–1548. doi: 10.1016/j.ijrobp.2011.10.039. [DOI] [PubMed] [Google Scholar]
- 6.Strother DR, Lafay-Cousin L, Boyett JM, Burger P, Aronin P, Constine L, Duffner P, Kocak M, Kun LE, Horowitz ME. Benefit from prolonged dose-intensive chemotherapy for infants with malignant brain tumors is restricted to patients with ependymoma: a report of the Pediatric Oncology Group randomized controlled trial 9233/34. Neuro-Oncology. 2014;16(3):457–465. doi: 10.1093/neuonc/not163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merchant TE, Li C, Xiong X, Kun LE, Boop FA, Sanford RA. Conformal radiotherapy after surgery for paediatric ependymoma: a prospective study. Lancet Oncol. 2009;10(3):258–266. doi: 10.1016/S1470-2045(08)70342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, Magdaleno S, Dalton J, Calabrese C, Board J. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8(4):323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell. 2015;27(5):728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meco D, Servidei T, Lamorte G, Binda E, Arena V, Riccardi R. Ependymoma stem cells are highly sensitive to temozolomide in vitro and in orthotopic models. Neuro-Oncology. 2014;16(8):1067–1077. doi: 10.1093/neuonc/nou008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11(11):1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 12.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506(7489):445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panwalkar P, Clark J, Ramaswamy V, Hawes D, Yang F, Dunham C, Yip S, Hukin J, Sun Y, Schipper MJ. Immunohistochemical analysis of H3K27me3 demonstrates global reduction in group-A childhood posterior fossa ependymoma and is a powerful predictor of outcome. Acta Neuropathol. 2017;134(5):705–714. doi: 10.1007/s00401-017-1752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3(1):39. doi: 10.1186/2045-3701-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, Kool M, Jones DT, Unterberger A, Morrissy AS. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125(3):373–384. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandstrom RS, Foret MR, Grow DA, Haugen E, Rhodes CT, Cardona AE, Phelix CF, Wang Y, Berger MS, Lin CH. Epigenetic regulation by chromatin activation mark H3K4me3 in primate progenitor cells within adult neurogenic niche. Sci Rep. 2014;4:5371. doi: 10.1038/srep05371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282(18):13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 18.Salz T, Deng C, Pampo C, Siemann D, Qiu Y, Brown K, Huang S. Histone methyltransferase hSETD1A is a novel regulator of metastasis in breast cancer. Mol Cancer Res. 2015;13(3):461–469. doi: 10.1158/1541-7786.MCR-14-0389. [DOI] [PubMed] [Google Scholar]
- 19.Salz T, Li G, Kaye F, Zhou L, Qiu Y, Huang S. hSETD1A regulates Wnt target genes and controls tumor growth of colorectal cancer cells. Cancer Res. 2014;74(3):775–786. doi: 10.1158/0008-5472.CAN-13-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald JW, Pilgram TK. Nuclear expression of p53, p21 and cyclin D1 is increased in bronchioloalveolar carcinoma. Histopathology. 1999;34(5):439–446. doi: 10.1046/j.1365-2559.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 21.Kruse CA, Visonneau S, Kleinschmidt-DeMasters BK, Gup CJ, Gomez GG, Paul DB, Santoli D. The human leukemic T-cell line, TALL-104, is cytotoxic to human malignant brain tumors and traffics through brain tissue: implications for local adoptive immunotherapy. Cancer Res. 2000;60(20):5731–5739. [PubMed] [Google Scholar]
- 22.Gomez GG, Read SB, Gerschenson LE, Santoli D, Zweifach A, Kruse CA. Interactions of the allogeneic effector leukemic T cell line, TALL-104, with human malignant brain tumors. Neuro-Oncology. 2004;6(2):83–95. doi: 10.1215/S1152851703000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31(1):334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbertson RJ, Bentley L, Hernan R, Junttila TT, Frank AJ, Haapasalo H, Connelly M, Wetmore C, Curran T, Elenius K. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin Cancer Res. 2002;8(10):3054–3064. [PubMed] [Google Scholar]
- 25.de Bont JM, Packer RJ, Michiels EM, den Boer ML, Pieters R. Biological background of pediatric medulloblastoma and ependymoma: a review from a translational research perspective. Neuro-Oncology. 2008;10(6):1040–1060. doi: 10.1215/15228517-2008-059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mungamuri SK, Murk W, Grumolato L, Bernstein E, Aaronson SA. Chromatin modifications sequentially enhance ErbB2 expression in ErbB2-positive breast cancers. Cell Rep. 2013;5(2):302–313. doi: 10.1016/j.celrep.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 28.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28(24):7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollock NI, Wang L, Wallweber G, Gooding WE, Huang W, Chenna A, Winslow J, Sen M, DeGrave KA, Li H. Increased expression of HER2, HER3, and HER2:HER3 heterodimers in HPV-positive HNSCC using a novel proximity-based assay: implications for targeted therapies. Clin Cancer Res. 2015;21(20):4597–4606. doi: 10.1158/1078-0432.CCR-14-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest. 2012;122(10):3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun W, Guo F, Liu M. Up-regulated WDR5 promotes gastric cancer formation by induced cyclin D1 expression. J Cell Biochem. 2017;119(4):3304–3316. doi: 10.1002/jcb.26491. [DOI] [PubMed] [Google Scholar]
- 32.Simpson NE, Tryndyak VP, Pogribna M, Beland FA, Pogribny IP. Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics. 2012;7(12):1413–1420. doi: 10.4161/epi.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116(2):259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 36.Xi G, Mania-Farnell B, Lei T, Tomita T. Histone modification as a drug resistance driver in brain tumors. Oncol Transl Med. 2016;2:216–226. [Google Scholar]
- 37.Ebrahimi A, Schittenhelm J, Honegger J, Schluesener H. Prognostic relevance of global histone 3 lysine 9 acetylation in ependymal tumors. J Neurosurg. 2013;119(6):1424–1431. doi: 10.3171/2013.9.JNS13511. [DOI] [PubMed] [Google Scholar]
- 38.Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, Rand V, Leary SE, White E, Eden C. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466(7306):632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohankumar KM, Currle DS, White E, Bougos N, Dapper J, Eden C, Nimmervoll B, Thiruvenkatam R, Connellly M, Kranengurg TA. An in vivo screen identifies ependymoma oncogenes and tumor-suppressor genes. Nat Genet. 2015;47(8):878–887. doi: 10.1038/ng.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffman LM, Donson AM, Nakachi I, Griesinger AM, Birks DK, Amani V, Hemenway MS, Liu AK, Wang M, Hankinson TC. Molecular sub-group-specific immunophenotypic changes are associated with outcome in recurrent posterior fossa ependymoma. Acta Neuropathol. 2014;127(5):731–745. doi: 10.1007/s00401-013-1212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevosssian RG, Phoenix TN, Thiruvenkatam R. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506(7489):451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20(2):143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A, Hansford JR, von Hoff K, Wright KD, Hwang E. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2016;133(1):5–12. doi: 10.1007/s00401-016-1643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendrzyk F, Korshunov A, Benner A, Toedt G, Pfister S, Radlwimmer B, Lichter P. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12(7):2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. (PT 1) [DOI] [PubMed] [Google Scholar]
- 45.Chapman-Rothe N, Curry E, Zeller C, Liber D, Stronach E, Gabra H, Ghaem-Maghami S, Brown R. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene. 2013;32(38):4586–4592. doi: 10.1038/onc.2012.477. [DOI] [PubMed] [Google Scholar]
- 46.Kilday JP, Mitra B, Domerg C, Ward J, Andrejuolo F, Osteso-lbanez T, Mauquen A, Varlet P, le Deley MC, Lowe J. Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: a prospective European clinical trial cohort analysis on behalf of the Children's Cancer Leukaemia Group (CCLG), Societe Francaise d'Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP) Clin Cancer Res. 2012;18(7):2001–2011. doi: 10.1158/1078-0432.CCR-11-2489. [DOI] [PubMed] [Google Scholar]
- 47.Ramaswamy V, Hielscher T, Mack SC, Lassaletta A, Lin T, Pajtler KW, Jones DT, Luu B, Cavalli FM, Aldape K. Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: A retrospective multicohort analysis. J Clin Oncol. 2016;34(21):2468–2477. doi: 10.1200/JCO.2015.65.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yae T, Tajima K, Maheswaran S. SETD1A induced miRNA network suppresses the p53 gene expression module. Cell Cycle. 2016;15(4):487–488. doi: 10.1080/15384101.2015.1130572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tajima K, Yae T, Javaid S, Tam O, Comaills V, Morris R, Wittner BS, Liu M, Enqstrom A, Takahashi F. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat Commun. 2015;6:8257. doi: 10.1038/ncomms9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoshii T, Cifani P, Feng Z, Huang CH, Koche R, Chen CW, Delaney CD, Lowe SW, Kentsis A, Armstrong SA. A non-catalytic function of SETD1A regulates cyclin K and the DNA damage response. Cell. 2018;172(5):1007–1021. doi: 10.1016/j.cell.2018.01.032. (e1017) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2